Significance
The architecture of neuronal dendritic trees is crucial for brain connectivity, whereas dendritic abnormalities are associated with psychiatric disorders. Rare copy number variations (CNVs) are a category of mutations often associated with schizophrenia, autism, and other mental disorders. However, the genetic complexity of CNV disorders poses a serious challenge for the identification of experimentally approachable mechanisms. Here we used network analysis to identify the most topologically important hub in protein–protein interaction networks within such a CNV, and in broader gene networks of psychiatric CNVs. We then show that pharmacological targeting of this node reverses dendritic alterations in a neuronal model of this CNV, outlining a strategy for the analysis and reversal of cellular phenotypes in CNV-related psychiatric disorders.
Keywords: schizophrenia, autism, ERK, CNV, MAPK3
Abstract
The architecture of dendritic arbors contributes to neuronal connectivity in the brain. Conversely, abnormalities in dendrites have been reported in multiple mental disorders and are thought to contribute to pathogenesis. Rare copy number variations (CNVs) are genetic alterations that are associated with a wide range of mental disorders and are highly penetrant. The 16p11.2 microduplication is one of the CNVs most strongly associated with schizophrenia and autism, spanning multiple genes possibly involved in synaptic neurotransmission. However, disease-relevant cellular phenotypes of 16p11.2 microduplication and the driver gene(s) remain to be identified. We found increased dendritic arborization in isolated cortical pyramidal neurons from a mouse model of 16p11.2 duplication (dp/+). Network analysis identified MAPK3, which encodes ERK1 MAP kinase, as the most topologically important hub in protein–protein interaction networks within the 16p11.2 region and broader gene networks of schizophrenia-associated CNVs. Pharmacological targeting of ERK reversed dendritic alterations associated with dp/+ neurons, outlining a strategy for the analysis and reversal of cellular phenotypes in CNV-related psychiatric disorders.
The architecture of the dendritic arbors defines a pyramidal neuron's dendritic receptive field (1). Moreover, patterns of dendritic arborization are essential to the computational ability of the neuron (1). Generating and maintaining proper dendritic receptive fields is therefore crucial for neural circuit function that underlies complex behaviors. Conversely, alterations in dendrites occur in psychiatric disorders, including schizophrenia and autism spectrum disorder (ASD) (2).
Psychiatric disorders have complex genetic architecture that is partly explained by rare variants with high penetrance (3, 4). Though incidences of these rare mutations are low (4–7), the accrued burden of rare variants on disease risk may account for a significant proportion of cases in linked disorders (8). The most well-characterized forms of rare variations are large genomic regional duplications or deletions known as copy number variations (CNVs). CNVs represent large genomic alterations, often encompassing multiple genes, which confer significant risk (odds ratio = 3–30) (9). The increased rare CNV burden is associated with numerous neurodevelopmental psychiatric conditions, including schizophrenia, ASD, and intellectual disability (5, 7, 10, 11). However, due to the large number of genes within CNVs, understanding the relationship between genotype and phenotypes and the rational identification of potential targets for reversing pathologically relevant phenotypes in CNV disorders has been challenging.
Recently, much attention has been paid to recurrent microduplication or deletion at the 16p11.2 locus. The ∼600-kb 16p11.2 CNV region encompasses 29 known protein-coding genes and is a hot spot for chromosomal rearrangement (3); 16p11.2 CNVs have been linked to multiple disorders and phenotypes; CNVs of this region are associated with schizophrenia (12–14), bipolar disorder (13), mental retardation, seizure disorders (15), ASD (13, 16–18), psychosis in Alzheimer's disease (19), and obesity (20, 21). Mouse models of 16p11.2 deletion and duplication have been generated, and heterozygotes of both the duplication and the deletion exhibit dosage-dependent alterations in gene expression, behavioral phenotypes, and brain morphology (22). Mice with deletion of the 16p11.2 syntenic region have been studied much more extensively than mice with duplication of this region. Mice with the 16p11.2 syntenic region deletion had cortical and striatal anatomical and cellular abnormalities (23), including elevated numbers of striatal medium spiny neurons expressing the dopamine D2 receptor, fewer dopamine-sensitive neurons in the cortex and synaptic defects in striatal medium spiny neurons. These mice also displayed behavioral deficits, including hyperactivity, circling, deficits in movement control as well as complete lack of habituation, suggesting abnormal basal ganglia circuitry function.
However, because multiple organ systems and brain regions are affected by the CNV (22, 23), it is difficult to distinguish cell type-autonomous from systemic effects. To address this confound, here we examined cortical pyramidal neurons from primary cultures from mice heterozygous for the 16p11.2-orthologous duplication (dp/+) and their wild-type littermates (+/+).
The complexity of the genetic architecture of psychiatric disorders poses a serious challenge for the identification of experimentally approachable mechanisms. Though determining causation may be complex and experimentally difficult to approach, even within the same CNV, we reasoned that phenotype reversal may be more accessible experimentally. It has been hypothesized that disease phenotypes are a consequence of altered gene pathways or networks, and the optimal therapeutic targets should be nodes that are highly influential to the function of the entire network (24). Because use of network-based approaches have been speculated to be able to help to identify major drivers of pathogenesis or candidate therapeutic targets (25), we further reasoned that use of network analysis might provide an approach to reduce complexity and identify salient mechanisms that can be harnessed therapeutically.
Here we used network analysis to identify potential candidate drivers of phenotype reversal in dp/+ mice, and followed up by experimental validation of the hypothesized mechanism. We found that dp/+ neurons showed significantly increased dendritic complexity compared with +/+ neurons. Network analysis identified MAPK3, which encodes ERK1 MAP kinase, as the most topologically important node in the protein–protein interaction (PPI) network related to schizophrenia-associated CNV genes, including 16p11.2. Pharmacological inhibition of ERK signaling resulted in a reversal of dendritic alterations, suggesting potential approaches for treatment.
Results
Increased Dendritic Arborization in dp/+ Neurons.
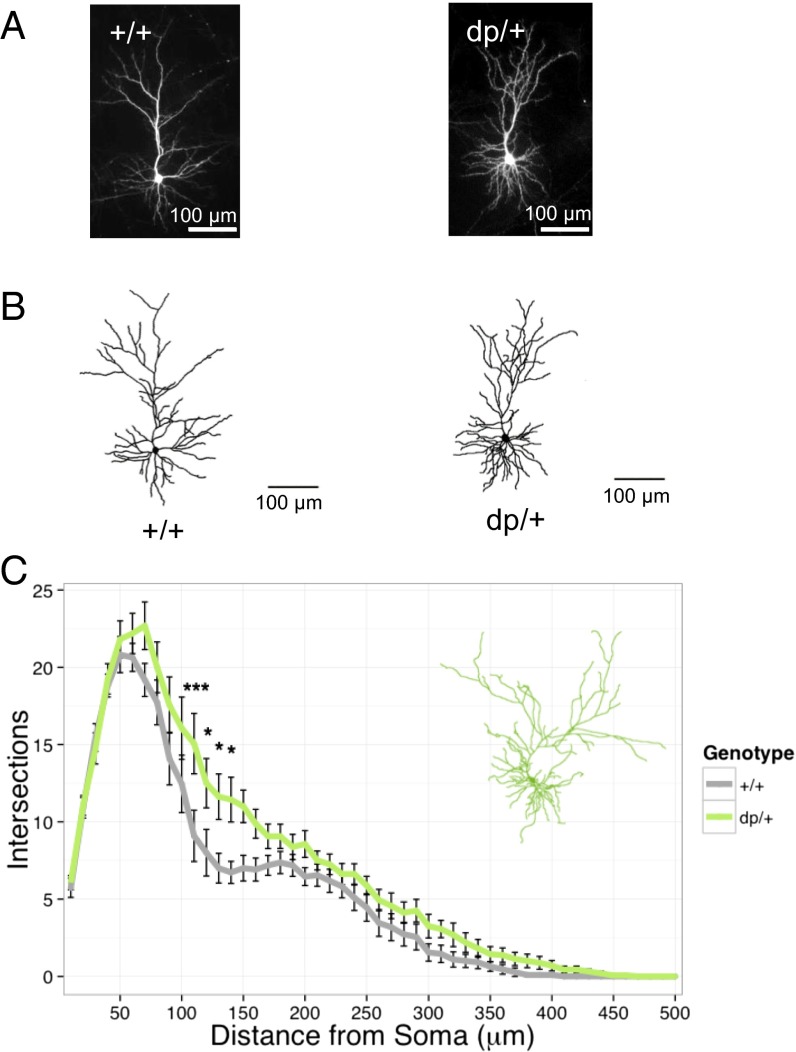
Dendrites constitute the receptive field of neurons, and alterations in dendritic morphology could contribute to abnormalities in cortical connectivity (26). We therefore examined dendrite morphology in cortical pyramidal neurons from mice heterozygous for the 16p11.2-orthologous duplication (dp/+) and their wild-type littermates (+/+). To enhance interspecies phenotype conservation, and to avoid confounds caused by the effects of CNV on multiple organ systems, we examined dendrites in cultured primary neurons. At 21–24 d in vitro, we transfected cultured neurons with GFP-expressing plasmid DNA to allow us to visualize cell morphology (Fig. 1 A and B). Single-plane images were taken using a 10× objective and traced and used for Sholl analysis. We found that dp/+ pyramidal neurons exhibited a significant increase in dendritic complexity compared with +/+ neurons [genotype: F(1, 25) = 6.21, P = 0.02; genotype by distance: F(49, 1225) = 1.63, P = 0.005] (Fig. 1C).
Fig. 1.
Increased dendritic arborization in dp/+ neurons. (A) Representative low-magnification images of wild-type (+/+) and duplication (dp/+) pyramidal neurons used in analyses. (B) Representative traces of +/+ and dp/+ pyramidal neurons. (C) Sholl analysis of total dendrites in +/+ (n = 11) and dp/+ (n = 16); significant effect of genotype (P = 0.02) and genotype–distance interaction (P = 0.005). Values are means ± SEM. Individual data points that were significant after Bonferroni correction are indicated: *P < 0.05; **P < 0.01; ***P < 0.001.
Network Analyses Identifies MAPK3 in the 16p11.2 CNV as a Central Hub in Schizophrenia-Associated CNV Gene Networks.
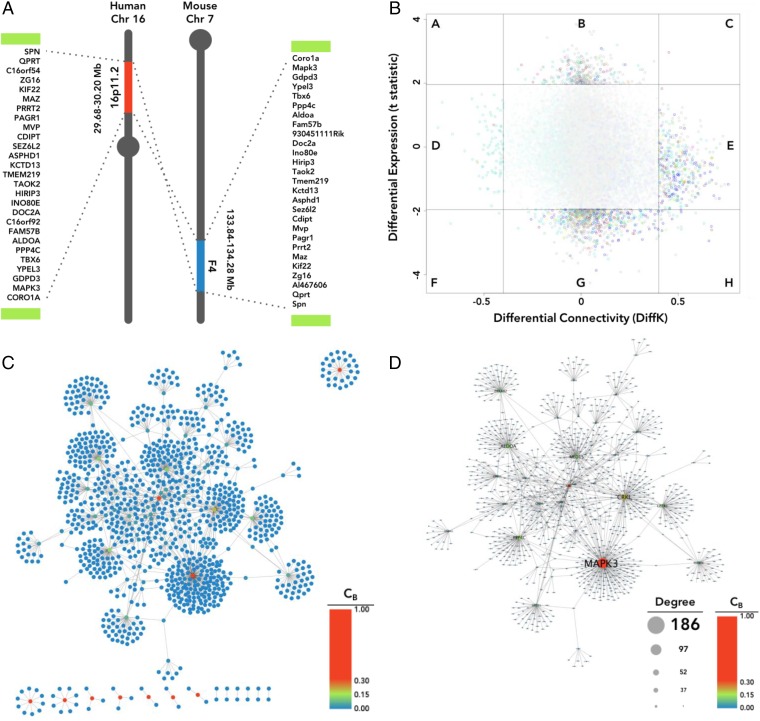
Past research has consistently related psychiatric disorders, including schizophrenia and ASD, to aberrations in neurodevelopment and synaptic function (27–29). A number of genes from within the 16p11.2 microduplication region have been shown to play pertinent roles in neurodevelopmental and synaptic processes (Fig. 2A); however, establishing their relative importance is challenging. We reasoned that an alternative strategy could be to identify genes that may “drive” phenotype reversal. Such “reversal drivers” could be highly connected nodes in PPI networks within and outside of this region. We first investigated changes in 16p11.2 gene expression and connectivity in schizophrenia. Using microarray-based transcriptomic data of frontocortical tissues of schizophrenia subjects and controls (from the Stanley Medical Research Institute), we found relatively little case-control differential expression for 16p11.2 CNV genes (with false discovery rate correction; Fig. 2B, y axis). Likewise, differential connectivity network analysis (DCNA) (30) did not show significant changes in 16p11.2 CNV gene connectivity (Fig. 2B, x axis). We reasoned that these expression-based analyses may be underpowered due to small sample size. We next analyzed PPI networks to identify highly connected network hubs within the 16p11.2 CNV. Because of the relative high penetrance of rare CNVs in schizophrenia, we attempted to identify the gene networks connected by five major schizophrenia-associated CNVs (1q21.1, NRXN1, 15q13.2, 16p11.2, and 22q11.2) (14). We used PINA2 (31) to construct a PPI network. The 107 Refseq genes within these CNV regions formed 1,261 PPIs, involving 1,110 nodes (Fig. 2C). Of all genes from the five included CNVs, ERK1 showed the highest betweenness, closeness, and degree centrality (CB = 0.30; CC = 0.17; CD = 0.168) and degree of connectivity (D = 186) of any CNV gene (Fig. 2D). Our analysis indicates that MAPK3/ERK1 is the most topologically important node in the PPI network related to the 16p11.2 network and broader schizophrenia-associated CNV gene networks.
Fig. 2.
MAPK3 is a central hub in schizophrenia-associated CNV networks. (A) Schematic of the 16p11.2 CNV region and the synonymous region in mouse chromosome 7. Green bars represent the low-copy repeat sequences flanking the 16p11.2 and 7F4 regions. A number of genes from within the 16p11.2 microduplication region have been shown to play pertinent roles in neurodevelopmental and synaptic processes. (B) DCNA. Shaded region in the center (containing the majority of genes) is comprised of genes that are neither significantly different in expression or connectivity between schizophrenia and control networks; all 16p11.2 CNV genes are located within this shaded region. (C) PPI network for genes from five schizophrenia-associated CNVs. Nodes are color-scaled by betweenness centrality score (CB). (D) The primary CNV PPI network from C; nodes are size-scaled by degree (D) and color-scaled by CB.
Dosage-Dependent Increased ERK1 Expression and Phosphorylation in dp/+ Neurons.
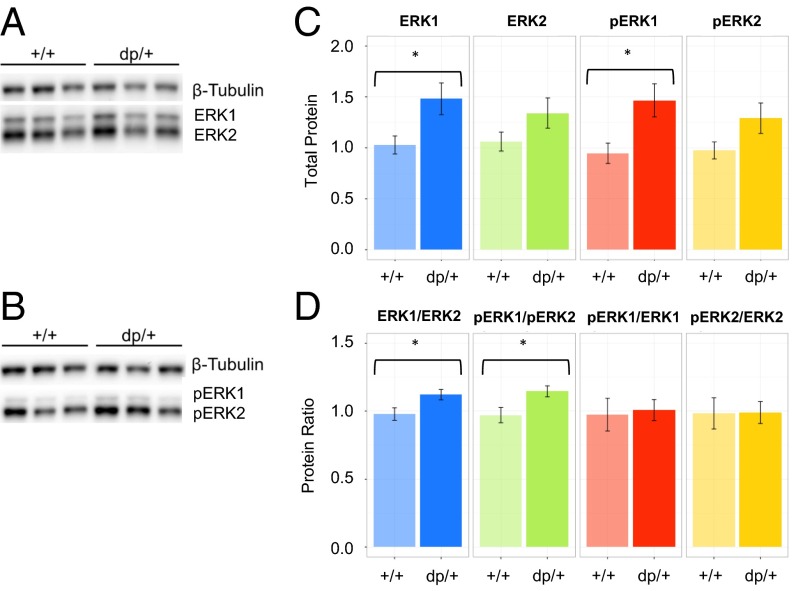
Because MAPK3 emerged as a central hub in the 16p11.2 network and broader gene networks of schizophrenia-associated CNVs, we examined the levels of ERK1 expression and phosphorylation, as well as their relation to the expression and activation of ERK2. Phosphorylation of ERK1 and ERK2 on the T202 and Y204 residues is reflective of their activation by upstream MEK. Homogenates were generated from individual coverslips of +/+ and dp/+ cultured neurons and analyzed for expression of ERK1, ERK2, phosphorylated ERK1 (pERK1), and phosphorylated ERK2 (pERK2) by Western blotting (Fig. 3 A and B). There was a significant effect of genotype on total ERK expression [F(1,150) = 12.53, P < 0.001], as well as an interaction between genotype and type of ERK protein [ERK1 vs. ERK2; F(2, 150) = 5.16, P = 0.007]. This effect was primarily driven by 44% and 55% increases in ERK1 [t(12.7) = 2.73, P = 0.03] (Fig. 3C) and pERK1 [t(13.5) = 2.73, P = 0.02] (Fig. 3C), respectively, in dp/+ samples. These shifts also reversed ERK1-to-ERK2 and pERK1-to-pERK2 ratios to a significant extent [t(17.9) = 2.39, P = 0.03; and t(17.3) = 2.54, P = 0.02, respectively] (Fig. 3D). Though reduced, pERK-to-ERK ratios were not significantly altered in the dp/+ samples, suggesting that increases in phosphorylated ERK1 was proportionate to increases in total ERK1 expression (Fig. 3D).
Fig. 3.
ERK1 protein levels and phosphorylation parallel gene dosage. (A) Western blot of ERK1 and ERK2 in +/+ (n = 11) and dp/+ (n = 9) neuronal homogenates. (B) Western blot of pERK1 and pERK2. (C) Quantification of blots in A and B. (D) Quantification of ERK1/ERK2 and pERK1/pERK2 ratios. Values are means ± SEM. *P < 0.05.
Reversal of Cellular Phenotypes by Pharmacological Inhibition of ERK Signaling.
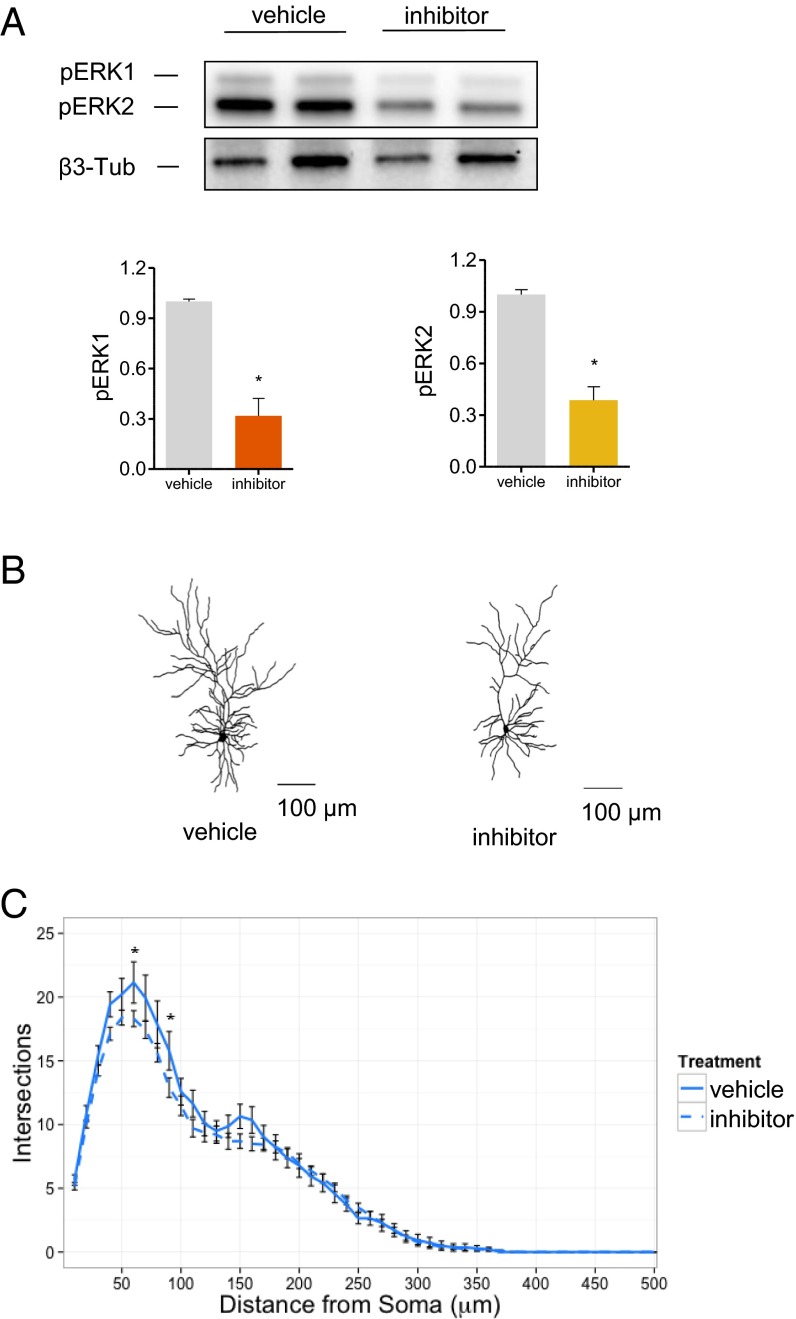
ERK signaling has been implicated in dendrite growth and maintenance. Because our network analysis suggested MAPK3 as the most highly connected hub in the PPI network related to genes within schizophrenia-associated CNVs, including the 16p11.2 CNV, we reasoned that inhibiting ERK function might reverse some of the cellular phenotypes observed in dup/+ neurons. We tested the efficacy of a known ERK signaling inhibitor (U0126) in reducing ERK activation in cultured neurons. Treatment with U0126 did not affect ERK1 expression but, as expected, strongly reduced ERK phosphorylation (Fig. 4A). We therefore incubated dup/+ neurons with the ERK signaling inhibitor and examined dendritic arborization, as described previously (Fig. 4B). Treatment with ERK inhibitor reduced dendritic complexity over whole dendritic content, compared with DMSO-treated dp/+ neurons [treatment–distance interaction: F(49, 2989) = 1.44, P = 0.0238] (Fig. 4C).
Fig. 4.
Reversal of dendritic phenotypes in dp/+ neurons. (A) Effect of MEK inhibitor (U0126) on ERK phosphorylation in dp/+ neurons. Western blotting of two representative samples are presented for each condition (n = 3 for quantification) (B) Representative neuron traces. (C) Sholl analysis of total dendrites in vehicle-treated (DMSO, n = 14) or inhibitor-treated (U0126, n = 49) dp/+ neurons. Significant effect of treatment–distance interaction (P = 0.02). Values are means ± SEM. Individual data points that were significant after Bonferroni correction are indicated: *P < 0.05.
Discussion
Here we used network analysis to identify a candidate driver gene within a CNV, capable of cell-autonomous phenotype reversal. We followed up our predictions by experimental validation using targeted pharmacology. Though network analysis has been commonly used in large-scale psychiatric genomics studies, such as the genome-wide association study (GWAS), to implicate specific biological pathways, it has rarely been used to generate and experimentally validate specific hypotheses.
We chose a highly simplified system where individual neurons can be directly manipulated and examined without confounding effects of the mutation on other brain regions or organ systems; this is particularly important, because the 16p11.2 CNV is associated with abnormalities in other brain regions or organ systems, such as striatal defects, head size abnormalities, and obesity, which could have indirect effects on the phenotypes of cortical neurons. In addition, pharmacological treatments of mice with MAPK inhibitor will also have effects on multiple organ systems, making it difficult to discern direct effects on cortical neuronal phenotypes. Future studies will therefore be needed to extend our cellular findings to whole animals.
Our network analysis shows that MAPK3 is the most topologically central hub in a highly connected network consisting of genes and encoded proteins within schizophrenia-associated CNVs. Expression of ERK1 in its active (pERK1) and inactive (ERK1) states were increased in homogenates of dp/+ neurons, as would be expected by the gene dosage effect of the microduplication. Because ERK1 is encoded by the MAPK3 gene located within the 16p11.2 region, and all genes in this region are duplicated in the dup/+ mouse, it is predicted that the protein expression level of ERK1 will be increased relative to ERK2, encoded by a gene on a different chromosome. This increased level of ERK1 will, when phosphorylated, result in an increased level of pERK1, relative to pERK2; indeed, this is what we found in our Western blots of dp/+ mouse brains. Increased ERK1 expression is consistent with increased dendrite complexity, because ERK1 promotes dendritic growth (32, 33). Furthermore, a possible role of ERK1 in psychiatric-related phenotypes is also supported by the most recent schizophrenia GWAS (34), where common variants spanning MAPK3 (and other adjacent genes) showed genome-wide significant association to schizophrenia. Interestingly, MAPK3 and MAP kinase signaling in general is also a central hub in gene networks implicated by ASD-associated CNVs and SNVs (35). Based on the network centrality of MAPK3 and the physiological functions of ERK1 in regulating dendrites, we reasoned that targeting this highly connected and central hub could alter the output of the entire network and thus modify specific phenotypes. A similar theoretical approach has been proposed to target MAP kinase signaling in cancer (24). Interestingly, chronic treatment with a negative allosteric modulator of mGluR5 reversed the cognitive deficits in 16p11.2 deletion model mice (36). ERK and mGluR5 signaling converge on disease phenotypes in the ASD-related Fmr1 knockout mouse model, suggesting these pathways may be important cellular targets for reversing neuropsychiatric phenotypes (37).
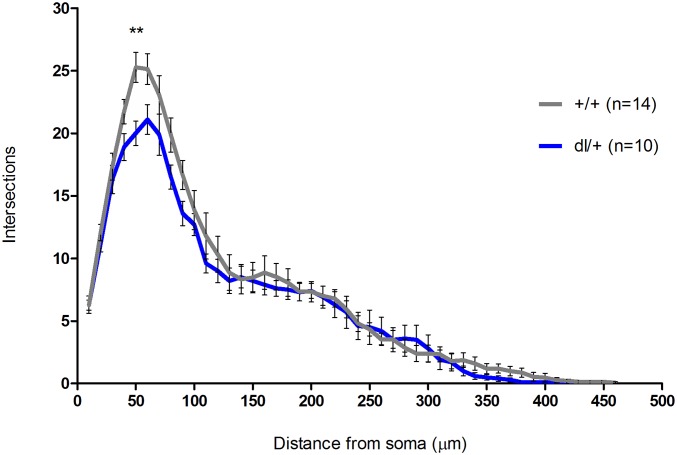
Our findings show, to our knowledge for the first time, that duplication of the mouse chromosomal region syntenic with human 16p11.2 results in cortical pyramidal neuronal dysregulation, manifested at the level of dendrites. We found that dp/+ neurons showed increased dendritic arborization over whole dendritic content in comparison with +/+ neurons. However, we did not detect dendritic alterations in pyramidal neurons from 16p11.2 deletion model mice, similar to recent reports of lack of dendritic alterations in MSN in 16p11.2 deletion model mice (Fig. S1) (23). This finding is unexpected, because deletions have been thought of as having more severe consequences than the duplication, and the phenotypes have been thought as being opposite, or mirrored, in duplication and deletion models (22). For example, head circumference is reduced in duplication carriers, and increased in deletion carriers (38). Body mass index is reduced in duplication and increased in deletion (20). However, further analysis of phenotypes in human subjects and mouse models revealed several phenotypes that do not conform to this pattern. For example, verbal memory encoding and verbal memory delayed recall are increased in duplication carriers, but are not altered in deletion carriers (39). In mice, hippocampal long-term potentiation is reduced in dp/+ but is unaltered in del/+ (40). Moreover, some other phenotypes, such as full-scale intelligence quotient or frequency of ASD, are altered in the same direction in duplication and deletion carriers (38). We have not detected dendritic abnormalities in del/+ neurons, but found increased arborization in dp/+ neurons. Our data are consistent with no changes in dendrite morphology in medium spiny neurons reported by Portmann et al. (23), which suggests that genes in this region may not be required for the maintenance of dendritic arborization, and their reduced expression does not affect dendrites; however, their abnormally increased expression may promote dendrite overgrowth.
Fig. S1.
Dendritic arborization in dl/+ neurons. Sholl analysis of total dendrites in +/+ (n = 14) and dl/+ (n = 10); no significant effect of genotype (P = 0.13) and no significant effect of genotype–distance interaction (P = 0.21). Values are means ± SEM. Individual data points that were significant after Bonferroni correction are indicated: *P < 0.05; **P < 0.01.
Abnormalities in dendrite arborization have been reported in several neuropsychiatric disorders. Surprisingly, our findings of increased dendrite arborization did not parallel previous postmortem findings in schizophrenia (41); despite this, the increased dendrite arborization associated with 16p11.2 duplication during development may lead to premature maturation of synapses, as has been shown when schizophrenia-associated MIR137 expression is reduced (42, 43). However, 16p11.2 duplications are also associated with ASD and seizure disorder, and the cellular phenotypes we identified may be more reflective of these disorders (16, 44, 45), such as the hypothesized increased brain connectivity in ASD. Alternatively, 16p11.2 microduplication carriers may have specific cellular phenotypes that are unique manifestations of this microduplication syndrome.
It is important to note that here we did not address mechanisms that contribute to the causation of 16p11.2 CNV; rather, we focused on “drivers” of the cellular phenotype reversal. Our network analysis and pharmacological phenotype reversal suggests MAPK3 could be such a driver gene. Causal etiological mechanisms may be overlapping or distinct from our findings. For example, a network analysis study that found that dysregulation of the KCTD13-Cul3-RhoA pathway in layer 4 of the inner cortical plate may be a potential determinant of 16p11.2 CNV brain size and connectivity phenotypes (46). Another important point to make is that networks that control dendrites may be similar, overlapping, or different from networks controlling other phenotypes, such as head size. For example, KCTD13 has been shown to be a driver of head size phenotypes in zebrafish (47).
Taken together, our data provide novel insight into the cellular phenotypes of CNV-associated psychiatric disorders, and suggest modalities to reverse such cellular phenotypes by targeting central network nodes.
Materials and Methods
Expression Plasmids and Antibodies.
Empty pEGFP-N2 expression plasmids were purchased from Clontech and overexpressed in cultured neurons to outline the cells for morphometric analysis. Primary antibodies against ERK1 (Santa Cruz) and pERK1/2 (Cell Signaling) were used in these studies.
Mouse Model.
Male dp/+ (C57-Black 6/129S7 mixed background) mice, bearing an ∼0.39-Mb duplication of the mouse chromosome 7 region between Giyd2 and Sept1 genes (22), were purchased from Jackson Laboratories and bred to wild-type C57-Black 6/129S7 (+/+) females. Offspring were housed according to gender, with mixed populations of dp/+ and +/+ animals. The colony was maintained via +/+ female–dp/+ male crosses to obviate the need to control for maternal strategy in subsequent generations.
Primary Neuronal Cultures and Transfection.
Dissociated cultures of primary cortical neurons from dp/+ and +/+ animals were procured according to previously described procedures (48). Each individual experiment was derived from one litter of P1 mouse pups. Briefly, brains were extracted from P1 pups in ice-cold HBSS (Corning), and cortical tissues were isolated and kept separate from one another. Following digestion in papain (Sigma-Aldrich) [diluted in Neurobasal (Thermo Fisher Scientific) with 0.5 mM EDTA (Sigma-Aldrich), DNaseI (Sigma-Aldrich) at 2 units/mL, and activated with 1 mM l-cysteine (Sigma-Aldrich) at 37 °C], cortical cells were mechanically dissociated in neuronal feeding media [Neurobasal + B27 supplement (Thermo Fisher Scientific) + 0.5 mM glutamine (Thermo Fisher Scientific) + penicillin/streptomycin (Thermo Fisher Scientific)] and plated on poly-d-lysine-coated (Sigma-Aldrich) coverslips. After 1 h, media was replaced with fresh media to remove dead or nonadherent cells. Neuronal cultures were maintained at 37 °C in 5% (vol/vol) CO2. After 4 d, neuronal media was supplemented with 200 μM dl-2-amino-5-phosphonopentanoic acid (APV; Abcam Biochemicals), and 100 μM APV supplemented media was used for feeding every 3 d hence.
Plasmids (1–10 μg total DNA) and Lipofectamine 2000 (Thermo Fisher Scientific) were diluted in DMEM (Corning) and Hepes (10 mM; Corning), mixed thoroughly, and incubated for 20–30 min at 37 °C, after which the mixture was added to cultured cells at 21–24 d in vitro in antibiotic-free medium. Following transfection, coverslips were replaced into feeding media containing half-conditioned, half-fresh media with antibiotics, and cells were allowed to express constructs for 3 d.
Dendrite Analysis.
To examine dendritic morphology, neurons expressing GFP were fixed, visualized with an antibody against GFP, and imaged as previously described (48). Confocal images of healthy immunostained neurons with intact secondary and tertiary dendrites were obtained with a Zeiss LSM5 Pascal confocal microscope. Single-plane images were taken using the 10× objective (N.A. 0.17) with 1,024 × 1,024 pixel resolution as previously described (49) and imported into Fiji (NIH) for analysis. Twenty-seven neurons (11 +/+ and 16 dp/+) from three separate cultures of P1 mouse litters were used in analyses of dendritic complexity. Binary images were created in Fiji from manual tracings of the entire dendritic arbor for each neuron and analyzed using the Fiji Sholl analysis plug-in. Sholl analysis was performed on total dendritic content using 10-μm incremental increases in concentric circular diameter. Distributions of these measures were assessed for normality, and group differences were assessed via repeated measures two-way ANOVA, followed by Bonferroni correction for multiple comparisons. For phenotype reversal experiments, 63 dp/+ neurons (14 treated with DMSO and 49 treated U0126) with were used for Sholl analysis. Two days after GFP transfection, neurons were treated with either DMSO or 0.5 μM U0126 (dissolved in DMSO) for 24 h and subsequently fixed for image analysis.
Western Blotting.
Quantification of total protein expression was measured using Western blotting methods as previously described (48). Homogenates were generated from individual coverslips of +/+ and dp/+ cultured neurons and analyzed for expression of ERK1, ERK2, pERK1, and pERK2 by Western blotting (Figs. 3 A and B and 4A). Phosphorylation of ERK1 and ERK2 on the T202 and Y204 residues is reflective of their activation by upstream MEK. Western blotting experiments were carried out on 20 samples (11 +/+ and 9 dp/+) from five separate cultured litters. For ERK inhibition experiments, dp/+ neurons were treated with DMSO (n = 3) or 0.5 μM U0126 (dissolved in DMSO, n = 3) for 24 h. A one-tailed Mann–Whitney t test was used to determine statistical significance of inhibition.
Protein–Protein Interaction Network Analysis.
Lists of the proteins encoded by genes within five rare schizophrenia-linked CNVs (1q21.1, NRXN1, 15q13.2, 16p11.2, and 22q11.2) (14) were assembled from gene lists adapted to protein accession numbers using DAVID (50, 51). Protein lists were imported into PINA2 for analysis (31, 52). Protein–protein interactions were uploaded from PINA2 into Cytoscape (53, 54) to visualize network topology. Values of degree (D) and centrality (betweenness centrality, CB; closeness centrality, CC; and degree centrality, CD) were extracted using Cytoscape’s integrated network analysis module.
Differential Connectivity Network Analysis.
SI Materials and Methods
Differential Connectivity Network Analysis.
Human dorsolateral prefrontal cortex gene expression data were downloaded from the Stanley Medical Research Institute (https://www.stanleygenomics.org; courtesy of Tadafumi Kato, Riken Brain Science Institute, Wako, Saitama, Japan). Probe intensity data from Human U133A Affymetrix GeneChip arrays were read into R (www.R-project.org) directly from CEL files using the affy package (55), and processed according to previously described methods (56). DCNA was performed on the expression data using the methods of Fuller et al. (30). Briefly, adjacency matrices were constructed and read into the R programming environment and analyzed with the WGCNA package (57). Adjacency matrices were normalized and transformed to resemble weighted scale-free network topology according to Zhang and Horvath (58). After transformation, topological overlap matrices (TOMs) were generated for each subject group (all subjects, schizophrenia subjects only, and control subjects only), which define network distance for each gene.
To identify network modules, TOMs were converted to dissimilarity matrices. Subsequently, hierarchical clustering and module identification was performed with the dynamicTreeCut algorithm (59), which is based on an adaptive process of cluster decomposition and combination; the process was iterated until the number of clusters became stable.
Schizophrenia and control networks were compared by calculating differential connectivity values, DiffKs, for each gene between the schizophrenia and the control conditions. DiffK of gene i is given by the formula
where
Here, k(i) represents the connectivity measure of gene i, and max(k) denotes the maximum network connectivity.
Acknowledgments
We thank P.V. Gejman for conceptual advice. This work was supported by NIH National Institute of Mental Health (NIMH) Grants MH071316 and MH097216 (to P.P.), MH071616 and MH084803 (to L.W. and J.G.C.), and R21MH102685 (to J.D.) and a Swiss National Science Foundation (SNSF) Early Postdoc Mobility Fellowship (to M.P.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607014113/-/DCSupplemental.
References
- 1.Spruston N. Pyramidal neurons: Dendritic structure and synaptic integration. Nat Rev Neurosci. 2008;9(3):206–221. doi: 10.1038/nrn2286. [DOI] [PubMed] [Google Scholar]
- 2.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14(3):285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duan J, Sanders AR, Gejman PV. Genome-wide approaches to schizophrenia. Brain Res Bull. 2010;83(3-4):93–102. doi: 10.1016/j.brainresbull.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Schizophrenia Consortium Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455(7210):237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirov G, et al. International Schizophrenia Consortium; Wellcome Trust Case Control Consortium Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum Mol Genet. 2009;18(8):1497–1503. doi: 10.1093/hmg/ddp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Den Bossche MJ, et al. Rare copy number variants in neuropsychiatric disorders: Specific phenotype or not? Am J Med Genet B Neuropsychiatr Genet. 2012;159B(7):812–822. doi: 10.1002/ajmg.b.32088. [DOI] [PubMed] [Google Scholar]
- 7.Kirov G, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17(2):142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Akil H, et al. Medicine. The future of psychiatric research: Genomes and neural circuits. Science. 2010;327(5973):1580–1581. doi: 10.1126/science.1188654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirov G. The role of copy number variation in schizophrenia. Expert Rev Neurother. 2010;10(1):25–32. doi: 10.1586/ern.09.133. [DOI] [PubMed] [Google Scholar]
- 11.Guilmatre A, et al. Recurrent rearrangements in synaptic and neurodevelopmental genes and shared biologic pathways in schizophrenia, autism, and mental retardation. Arch Gen Psychiatry. 2009;66(9):947–956. doi: 10.1001/archgenpsychiatry.2009.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szatkiewicz JP, et al. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19(7):762–773. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy SE, et al. Wellcome Trust Case Control Consortium Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet. 2009;41(11):1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levinson DF, et al. Copy number variants in schizophrenia: Confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168(3):302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghebranious N, Giampietro PF, Wesbrook FP, Rezkalla SH. A novel microdeletion at 16p11.2 harbors candidate genes for aortic valve development, seizure disorder, and mild mental retardation. Am J Med Genet A. 2007;143A(13):1462–1471. doi: 10.1002/ajmg.a.31837. [DOI] [PubMed] [Google Scholar]
- 16.Weiss LA, et al. Autism Consortium Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358(7):667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 17.Levy D, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70(5):886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 18.Sanders SJ, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70(5):863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng X, et al. A rare duplication on chromosome 16p11.2 is identified in patients with psychosis in Alzheimer’s disease. PLoS One. 2014;9(11):e111462. doi: 10.1371/journal.pone.0111462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacquemont S, et al. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478(7367):97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walters RG, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463(7281):671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horev G, et al. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc Natl Acad Sci USA. 2011;108(41):17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Portmann T, et al. Behavioral abnormalities and circuit defects in the basal ganglia of a mouse model of 16p11.2 deletion syndrome. Cell Reports. 2014;7(4):1077–1092. doi: 10.1016/j.celrep.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng Q, Schork NJ. Utility of network integrity methods in therapeutic target identification. Front Genet. 2014;5:12. doi: 10.3389/fgene.2014.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barabási AL, Gulbahce N, Loscalzo J. Network medicine: A network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jan YN, Jan LY. Branching out: Mechanisms of dendritic arborization. Nat Rev Neurosci. 2010;11(5):316–328. doi: 10.1038/nrn2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: On the matter of their convergence. Mol Psychiatry. 2005;10(1):40–68; image 45. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 28.Le-Niculescu H, et al. Towards understanding the schizophrenia code: An expanded convergent functional genomics approach. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(2):129–158. doi: 10.1002/ajmg.b.30481. [DOI] [PubMed] [Google Scholar]
- 29.Chu TT, Liu Y. An integrated genomic analysis of gene-function correlation on schizophrenia susceptibility genes. J Hum Genet. 2010;55(5):285–292. doi: 10.1038/jhg.2010.24. [DOI] [PubMed] [Google Scholar]
- 30.Fuller TF, et al. Weighted gene coexpression network analysis strategies applied to mouse weight. Mamm Genome. 2007;18(6-7):463–472. doi: 10.1007/s00335-007-9043-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cowley MJ, et al. PINA v2.0: Mining interactome modules. Nucleic Acids Res. 2012;40(Database issue):D862–D865. doi: 10.1093/nar/gkr967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaillant AR, et al. Signaling mechanisms underlying reversible, activity-dependent dendrite formation. Neuron. 2002;34(6):985–998. doi: 10.1016/s0896-6273(02)00717-1. [DOI] [PubMed] [Google Scholar]
- 33.Ha S, Redmond L. ERK mediates activity dependent neuronal complexity via sustained activity and CREB-mediated signaling. Dev Neurobiol. 2008;68(14):1565–1579. doi: 10.1002/dneu.20682. [DOI] [PubMed] [Google Scholar]
- 34.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511(7510):421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinto D, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Hum Genet. 2014;94(5):677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian D, et al. Contribution of mGluR5 to pathophysiology in a mouse model of human chromosome 16p11.2 microdeletion. Nat Neurosci. 2015;18(2):182–184. doi: 10.1038/nn.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osterweil EK, Krueger DD, Reinhold K, Bear MF. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J Neurosci. 2010;30(46):15616–15627. doi: 10.1523/JNEUROSCI.3888-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D’Angelo D, et al. Cardiff University Experiences of Children With Copy Number Variants (ECHO) Study; 16p11.2 European Consortium; Simons Variation in Individuals Project (VIP) Consortium Defining the effect of the 16p11.2 duplication on cognition, behavior, and medical comorbidities. JAMA Psychiatry. 2016;73(1):20–30. doi: 10.1001/jamapsychiatry.2015.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hippolyte L, et al. The number of genomic copies at the 16p11.2 locus modulates language, verbal memory, and inhibition. Biol Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 40.Arbogast T, et al. Reciprocal effects on neurocognitive and metabolic phenotypes in mouse models of 16p11.2 deletion and duplication syndromes. PLoS Genet. 2016;12(2):e1005709. doi: 10.1371/journal.pgen.1005709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis DA, Sweet RA. Schizophrenia from a neural circuitry perspective: Advancing toward rational pharmacological therapies. J Clin Invest. 2009;119(4):706–716. doi: 10.1172/JCI37335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olde Loohuis NF, et al. MicroRNA-137 controls AMPA-receptor-mediated transmission and mGluR-dependent LTD. Cell Reports. 2015;11(12):1876–1884. doi: 10.1016/j.celrep.2015.05.040. [DOI] [PubMed] [Google Scholar]
- 43.Duan J, et al. Molecular Genetics of Schizophrenia collaboration; Genomic Psychiatric Cohort consortium A rare functional noncoding variant at the GWAS-implicated MIR137/MIR2682 locus might confer risk to schizophrenia and bipolar disorder. Am J Hum Genet. 2014;95(6):744–753. doi: 10.1016/j.ajhg.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reinthaler EM, et al. 16p11.2 European Consortium; EPICURE Consortium; EuroEPINOMICS Consortium 16p11.2 600-kb duplications confer risk for typical and atypical Rolandic epilepsy. Hum Mol Genet. 2014;23(22):6069–6080. doi: 10.1093/hmg/ddu306. [DOI] [PubMed] [Google Scholar]
- 45.Shinawi M, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet. 2010;47(5):332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin GN, et al. Spatiotemporal 16p11.2 protein network implicates cortical late mid-fetal brain development and KCTD13-Cul3-RhoA pathway in psychiatric diseases. Neuron. 2015;85(4):742–754. doi: 10.1016/j.neuron.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golzio C, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485(7398):363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woolfrey KM, et al. Epac2 induces synapse remodeling and depression and its disease-associated forms alter spines. Nat Neurosci. 2009;12(10):1275–1284. doi: 10.1038/nn.2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones KA, et al. Rapid modulation of spine morphology by the 5-HT2A serotonin receptor through kalirin-7 signaling. Proc Natl Acad Sci USA. 2009;106(46):19575–19580. doi: 10.1073/pnas.0905884106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 51.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu J, et al. Integrated network analysis platform for protein-protein interactions. Nat Methods. 2009;6(1):75–77. doi: 10.1038/nmeth.1282. [DOI] [PubMed] [Google Scholar]
- 53.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yeung N, Cline MS, Kuchinsky A, Smoot ME, Bader GD. Exploring biological networks with Cytoscape software. Curr Protoc Bioinformatics. 2008 doi: 10.1002/0471250953.bi0813s23. [DOI] [PubMed] [Google Scholar]
- 55.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 56.Andrus BM, et al. Gene expression patterns in the hippocampus and amygdala of endogenous depression and chronic stress models. Mol Psychiatry. 2012;17(1):49–61. doi: 10.1038/mp.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4(1):Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 59.Langfelder P, Zhang B, Horvath S. Defining clusters from a hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinformatics. 2008;24(5):719–720. doi: 10.1093/bioinformatics/btm563. [DOI] [PubMed] [Google Scholar]