Significance
Mutations in the PI3K pathway are highly prevalent in cancers, and isoform-specific and pan-PI3K inhibitors have entered clinical trials in both solid and hematologic malignancies. The PI3K δ-specific inhibitor idelalisib (in combination with rituximab) was recently approved for the treatment of chronic lymphocytic leukemia. However, identifying tumor types and biological mechanisms that predict for response to PI3K inhibitors as single agents or in combination has been a challenge. Our data indicate that PI3K inhibitors induce DNA damage in tumors that have defects in DNA damage-repair pathways and that they do so by impairing the production of Rib phosphate and amino acids needed for deoxynucleotide synthesis.
Keywords: phosphoinositide 3-kinase, DNA damage, breast cancer, Parp-inhibition, tumor metabolism
Abstract
We previously reported that combining a phosphoinositide 3-kinase (PI3K) inhibitor with a poly-ADP Rib polymerase (PARP)-inhibitor enhanced DNA damage and cell death in breast cancers that have genetic aberrations in BRCA1 and TP53. Here, we show that enhanced DNA damage induced by PI3K inhibitors in this mutational background is a consequence of impaired production of nucleotides needed for DNA synthesis and DNA repair. Inhibition of PI3K causes a reduction in all four nucleotide triphosphates, whereas inhibition of the protein kinase AKT is less effective than inhibition of PI3K in suppressing nucleotide synthesis and inducing DNA damage. Carbon flux studies reveal that PI3K inhibition disproportionately affects the nonoxidative pentose phosphate pathway that delivers Rib-5-phosphate required for base ribosylation. In vivo in a mouse model of BRCA1-linked triple-negative breast cancer (K14-Cre BRCA1f/fp53f/f), the PI3K inhibitor BKM120 led to a precipitous drop in DNA synthesis within 8 h of drug treatment, whereas DNA synthesis in normal tissues was less affected. In this mouse model, combined PI3K and PARP inhibition was superior to either agent alone to induce durable remissions of established tumors.
Triple-negative breast cancers, including BRCA1-linked breast cancers, frequently show activation of the PI3K pathway as a result of overexpression of epidermal growth factor receptor (EGFR) (1, 2) or insulin-like growth factor 1 receptor (IGFR) (3), and inhibition of the EGFR (1, 2, 4) and/or the PI3K/Nrf2 axis (5) prevents cancers arising from BRCA1 mutant mammary epithelial cells (MECs). In addition, activating mutations of PIK3CA, or loss of the inhibitory lipid phosphatases PTEN (phosphatase and tensin homolog) and INPP4B (inositol polyphosphate 4-phosphatase type II) (6, 7), suggest that the PI3K pathway is contributing to tumor growth and survival. Aside from their role in regulating the homeostasis of phospho-inositides, PTEN and INPP4B may have independent roles in DNA damage repair. A role for PTEN in the maintenance of genomic stability was identified (8); more recently, INPP4B was found to directly interact with BRCA1 and the serine/threonine protein kinase ATR, and its loss destabilizes these DNA damage repair complexes, effectively sensitizing INPP4B-deficient cells to poly-ADP Rib polymerase (PARP) inhibition (9).
Despite the high incidence of predisposing lesions in the PI3K pathway, limited clinical activity has been observed with PI3K inhibitors as single-agent treatment in endocrine-resistant breast cancer, which may reflect bypass of PI3K-dependent mitogenic signaling by alternative signaling pathways such as the MAPK pathway. Therefore, concurrent inhibition of parallel and compensatory signaling networks to overcome resistance to PI3K inhibition is being investigated in clinical studies. This approach, however, carries the risk of overlapping toxicities of the targeted agents without sufficient efficacy because tumor cells may have greater plasticity for redundant signaling than normal tissues.
Multiple functional interdependencies between DNA damage induction/repair and signal transduction via PI3K have recently been discovered, providing the rationale for novel combination treatments. PI3K inhibitors PI-103 (10), GDC-0980 (11), and BKM120 (12, 13) have independently been found to induce DNA damage and to synergize with radiation, PARP-inhibitor or platinum-based treatments to augment deleterious DNA damage in breast cancer cells and xenograft models. However, the mechanisms underlying this interdependence are not well understood. Recently, we observed that Parp inhibition can augment the PI3K pathway, and that PI3K inhibition leads to a strong induction of PARP (12). Together, these observations provide the basis for a potential synthetic lethality of these two treatment modalities.
As a result of PI3K-mediated phosphorylation, the lipid-based second messengers PI(3,4,5)P3 and PI(4,5)P2 activate an array of downstream targets that lead to the activation of a complex signaling network to orchestrate cell division, and an acceleration of cellular metabolism, including glycolysis, reviewed in ref. 14. We have recently shown that PI3K directly coordinates glycolysis with cytoskeletal dynamics: PI3K-dependent activation of the GTPase Rac leads to an increased turnover of the actin cytoskeleton with release of the F-actin–bound glycolytic enzyme aldolase A into the cytoplasm where it is enzymatically active. Consistently, inhibitors of PI3K, but not inhibitors of the protein kinase AKT, SGK (serum/glucocorticoid regulated kinase), or mechanistic target of rapamycin (mTOR), cause a significant decrease in glycolysis at the step catalyzed by aldolase A, whereas activating PIK3CA mutations have the opposite effect (15). A product of the aldolase reaction is the triose glyceraldehyde 3-phosphate (Ga3P), which is a substrate for transketolase (TKT). Under conditions of enhanced glycolysis (16), Ga3P enters the nonoxidative pentose-phosphate pathway, is acted on by TKT, and used to generate the Rib-phosphate required for base ribosylation and ultimately the synthesis of DNA and RNA synthesis. Here, we examine the antimetabolic functions of PI3K inhibition for cancer treatment and show that PI3K inhibitors can lead to DNA damage through interference with base ribosylation, suggesting that PI3K inhibitors may augment the efficacy of antineoplastics that interfere with DNA synthesis or repair.
Results
PI3K Inhibitors Are More Effective Than AKT Inhibitors at Inducing DNA Damage in Breast Cancer Cells with BRCA1 and TP53 Defects.
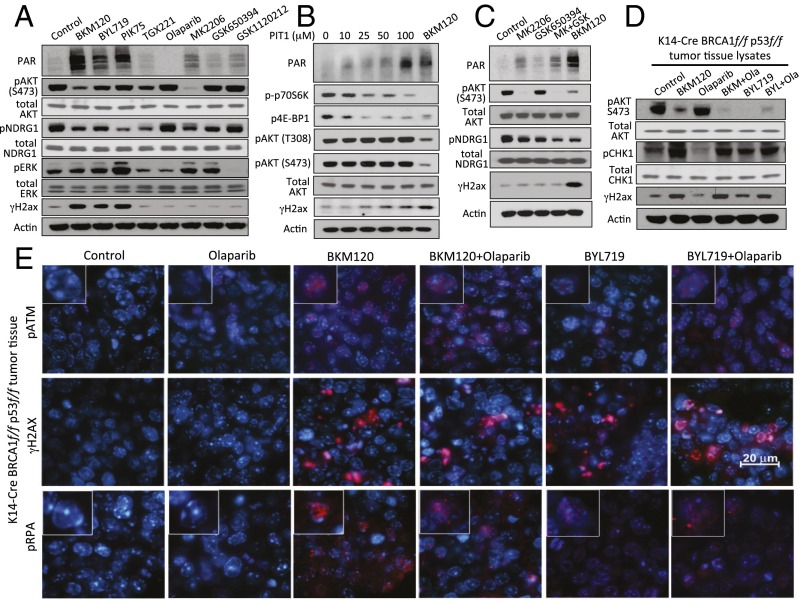
To determine the contributions of PI3K signaling to the DNA damage response, we examined the effects of specific signaling kinase inhibitors PI3K (BKM120), PI3Kα (BYL719, PIK75), PI3Kβ (TGX221), AKT (MK2206), SGK (GSK650394), or MAPKK (GSK1120212) on poly-(ADP)-ribosylation (PAR) and phosphorylation of histone H2AX (γH2AX) (Fig. 1A). HCC1937 breast cancer cells (BRCA1 5382insC/BRCA null, TP53-mutant, PTEN−/−, ER-, PR-, Her2neu-negative) were treated for 16 h. The three PI3K inhibitors BKM120, BYL719, and PIK75 were much more effective than the other inhibitors at inducing H2AX phosphorylation and increasing PAR (Fig. 1A). At the concentration used (Fig. 1A), the expected downstream targets were affected, i.e., the PIK3 and AKT inhibitors reduced AKT phosphorylation; the allosteric AKT inhibitor MK2206 was somewhat more effective than the pan-PI3K inhibitor in blocking AKT phosphorylation; and the isoform-specific PI3K inhibitors BYL719 and TGX221 were less effective than BKM120 at blocking AKT phosphorylation, suggesting that both PI3Kα and PI3Kβ contribute to AKT activation. Olaparib inhibited PAR of PARP, GSK1120212 impaired phosphorylation of extracellular signal-regulated kinases (ERK), and GSK650394 reduced phosphorylation of the SGK substrate N-Myc Downstream Regulated 1 (NDRG1), which was strong in these cells and responsive to BKM120 as noted (17). Response of pNDRG1 to AKT inhibition in breast cancer cell lines is variable. Some cell lines respond to AKT inhibition with a decrease in NDRG1 phosphorylation, whereas others do not (18). We found that NDRG1 phosphorylation was suppressed in cells treated with inhibitors of PI3K (BKM120, BYL719, PIK75, and TGX221) and AKT (MK2206 and GSK650394) but not inhibitors of PARP (Olaparib) or MAPK (GSK112021), i.e., we found that NDRG1 phosphorylation in HCC1937 cells was responsive to PI3K and AKT inhibition, confirming these previous reports of cross-talk between PI3K, AKT, and mTOR signaling (but excluding ERK signaling) with regard to SGK activity. These results suggest that the PI3K pathway protects HCC1937 cells from DNA damage by a pathway independent of AKT, SGK, and MAPKK signaling.
Fig. 1.
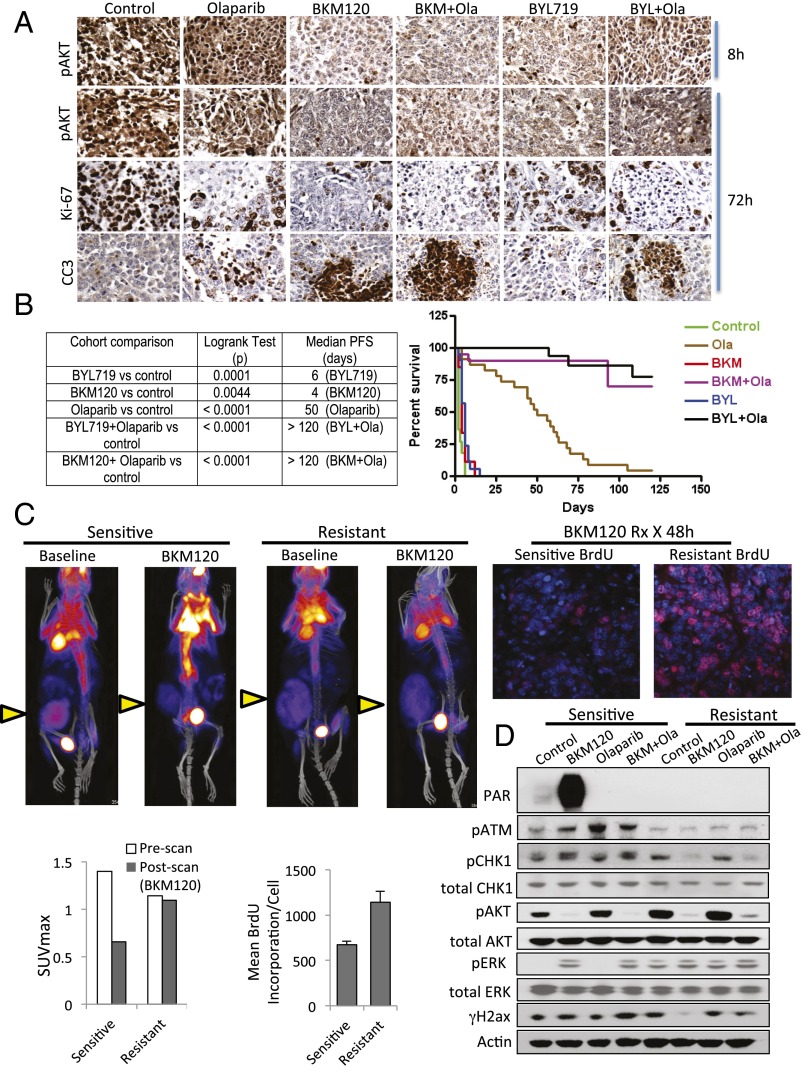
PI3K, but not AKT, MAPKK, or SGK inhibition induces markers of DNA damage in BRCA1-mutant breast cancers. For A–C, HCC1937 cells were treated for 16 h with inhibitors as indicated. Immunoblotting of total cell lysates was performed with antibodies as indicated. (A) Induction of PAR and H2ax phosphorylation (γH2ax) following treatment with inhibitors of pan-PI3K (BKM120, 1.5 μM), PI3Kα (BYL719, 3 μM; PIK75, 0.5 μM), PI3Kβ (TGX221, 30 μM), the PARP-inhibitor Olaparib (5 μM), and inhibitors of AKT (MK2206, 1 μM), SGK (GSK650394, 10 μM), or MAPKK (GSK1120212, 5 nM). (B) Induction of PAR and γH2ax by the PIP3-mimetic PIT1. (C) Neither AKT nor SGK, nor the combination of AKT and SGK inhibitors, induces PAR or γH2ax. (D and E) Induction of DNA damage indicators in vivo. K14-Cre BRCA1f/fp53f/f tumor-bearing mice were treated with two doses of drugs as indicated [BKM120 30 mg/kg by mouth (PO), BYL719 30 mg/kg PO, Olaparib 50 mg/kg i.p.] 8 and 2 h before killing. Tumors were immediately harvested and processed for immunoblotting of fresh tumor tissue lysates with antibodies as indicated (D) or fixed and stained for immunofluorescence with antibodies as indicated (E). Insets (400× magnification) show representative single cells stained for pATM (Upper) or pRPA (Lower).
PI3Ks mediate the phosphorylation of lipid phosphatidylinositol-4,5-bisphosphate (PIP2) to generate phosphatidylinositol-3,4,5-trisphosphate (PIP3), which specifically binds and activates pleckstrin-homology (PH) domain-containing target proteins. PIP3 signaling is terminated by the phosphatase PTEN, which dephosphorylates PIP3. Using a specific PIP3 mimetic, PIT1, that antagonizes the PIP3–PH domain interaction in a range of target proteins (19, 20), we could recapitulate the induction of PAR and γH2AX (Fig. 1B). Interestingly, and consistent with prior observations in PTEN-deficient cells (20), induction of the DNA damage response was observed at PIT1 concentrations lower than those required for AKT inhibition and coincided with abrogation of phosphorylation of ribosomal protein S6 and eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1). Direct inhibition of AKT with the competitive inhibitor MK2206 alone resulted in more modest induction of PAR and γH2AX and was not augmented by combination with the inhibitor of SGK (GSK650394) (Fig. 1C). These data confirm that the induction of DNA damage is specific to the inhibition of the lipid kinase activity of PI3K, and that inhibition of AKT is not sufficient and may not be required to induce DNA damage.
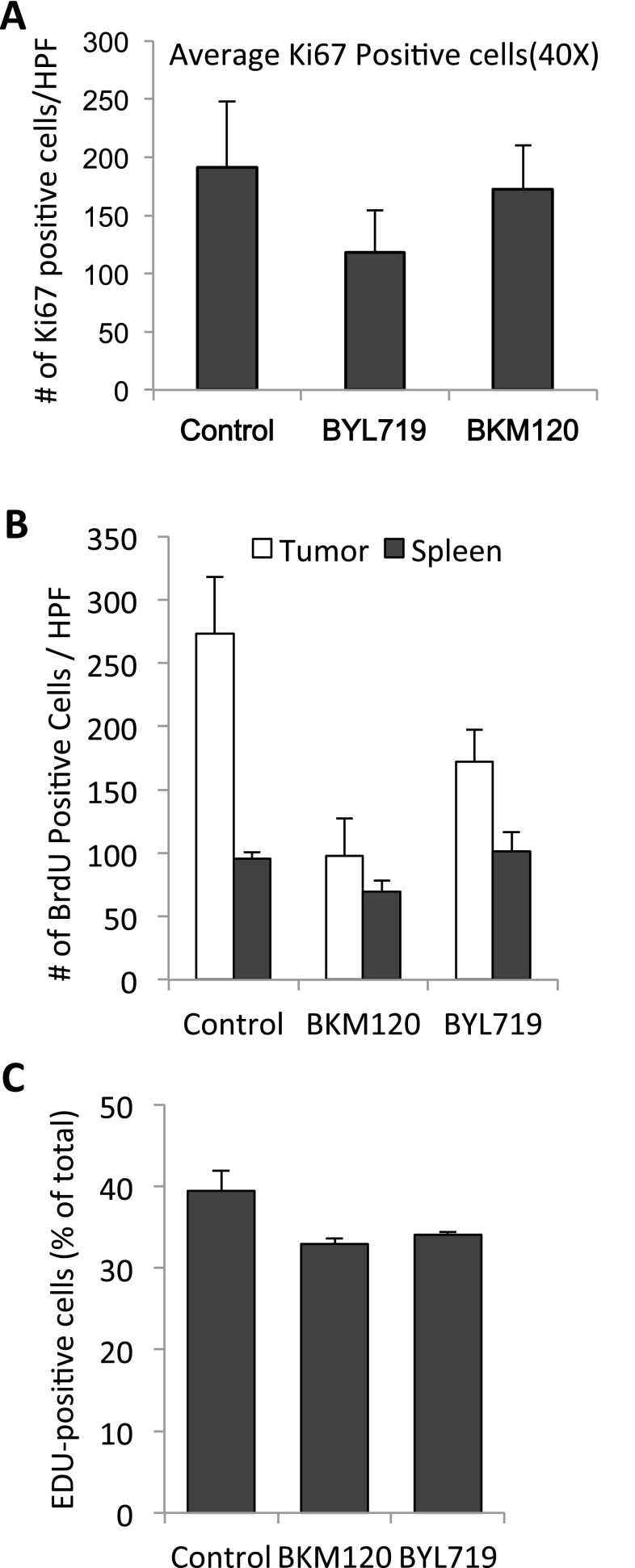
To examine whether this DNA damage response occurs in vivo, we used the K14-Cre BRCA1f/fp53f/f mouse model (21), in which loss of BRCA1 and p53 in the mammary gland is achieved by Lox P site-specific Cre recombinase driven by the cytokeratin 14 promoter. Tumors in this mouse model are typically high grade, triple negative, and respond with excellent regression, albeit not durable responses, to the PARP inhibitor Olaparib (21). Tumors generated in these mice were propagated in vivo through syngeneic transplantation into Cre− littermates as described (22), allowed to grow to 10 mm, and randomized to treatment groups. We found that effective inhibition of AKT phosphorylation (used here as a pharmacodynamics marker) occurred within 8 h of exposure to the pan-PI3K inhibitor BKM120 or the PI3Kα inhibitor BYL719 (Fig. 1D). Within this 8-h period, H2AX phosphorylation on serine 139 (γH2AX) could be observed in vivo in tumors treated with PI3K inhibitors BKM120 or BYL719, alone or in combination with PARP-inhibitor Olaparib (Fig. 1 D and E), as was autophosphorylation of the ATM kinase (Fig. 1E) and two further substrates of the ATR kinase, Checkpoint kinase 1 (pChk1 on serine 345) (Fig. 1D) and replication protein A (pRPA) (Fig. 1E). Activation of Chk1 by ATR occurs in response to replication stress (23) and is often associated with persistent accumulation of pRPA in nuclear speckles at sites of stalled replication (24). At this early timepoint, however, tumor cell proliferation as assessed by Ki67 stain, was not yet affected, i.e., indicators of DNA damage in response to PI3K inhibition preceded a decrease in proliferation (Fig. S1A). Taken together, the evidence of replication stress within 8 h of in vivo treatment suggests that PI3K inhibition might lead to impaired DNA synthesis.
Fig. S1.
(A) Proliferative activity as assessed by Ki67 stain in tumors after 8 h of treatment with BKM120 (30 mg/kg) or BYL719 (30 mg/kg). In each cohort, three mice were treated 8 and 2 h before killing, three mice per cohort, tumors fixed and stained with anti-Ki67 antibodies, and five high power fields (HPF, 400×) counted. Differences between the three cohorts were not significant by t test (two-tailed, unpaired). (B) Percentage of tumor cells entering S phase upon PI3K inhibition. K14-Cre BRCA1f/fp53f/f tumor-bearing mice were prepared, randomized, treated, and labeled with BrdU as described in Fig. 2A. Bar graphs display the number of BrdU+ cells in tumors (solid) or spleens (light) per 40× field, representing the mean of 3–5 different tumors. Significance was P < 0.05 for the bars marked with an asterisk. (C) HCC1927 cells were treated as described in Fig. 2B. Bar graphs display the percentage of EdU+ cells, means of experimental triplicates for each condition.
PI3K Inhibition Leads to Decreased DNA Aynthesis During S Phase in Tumor Cells in Vivo.
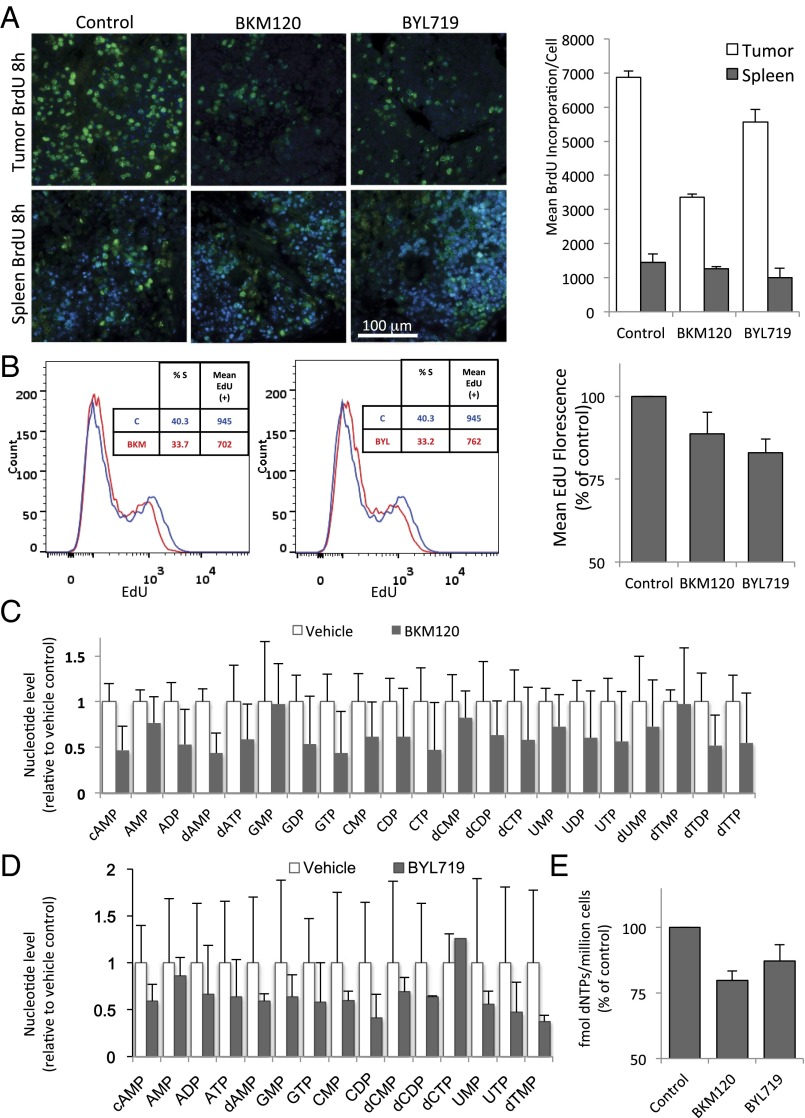
To examine S-phase progression, we examined the effects of the pan-PI3K inhibitor BKM120 (Buparlisib) and the PI3Kα-specific inhibitor BYL719 (Alpelisib) on the incorporation of the nucleotide analog BrdU in vivo. Tumor-bearing mice were treated with BKM120, BYL719, or vehicle control 8 h prior and again at 2 h before killing. BrdU was coinjected with the PI3K inhibitor or vehicle control during the second injection. This protocol was chosen to prevent labeling of cells that already entered S phase at the time of drug treatment. Actively dividing tumor and spleen cells showed a bright and robust BrdU signal in vehicle control-treated animals, indicative of unimpaired S phase entry (Fig. 2A). Within 8 h of PI3K inhibition, cells continued to enter S phase, albeit at a reduced rate (Fig. S1B). More striking, however, was that drug treatment led to a significant dimming of the BrdU signal within 8 h of treatment, suggestive of a decrease in DNA synthesis (Fig. 2A, columns 2 and 3). Dimming of the BrdU signal, measured as a decrease in mean fluorescence per tumor cell, at 8 h was more pronounced with BKM120 than with BYL719 treatments (Fig. 2A). In contrast, DNA synthesis in the nonmalignant spleen (Fig. 2A and Fig. S1B) was not significantly affected by PI3K inhibition.
Fig. 2.
PI3K inhibition leads to a decrease in DNA synthesis of cancer cells within 8 h. (A) K14-Cre BRCA1f/fp53f/f tumor-bearing mice were randomized to drug treatments as indicated and treated for 8 h (two doses, 8 and 2 h before killing). BrdU was given through intraperitoneal injection 2 h before killing. Upon necropsy, tumors and spleens (n = 3 for each group) were harvested, fixed, and stained with anti-BrdU antibodies for immunofluoresence (green). The mean fluorescence per cell of BrdU+ cells was measured by using volocity image analysis software. The bar graph represents mean fluorescence intensity of the BrdU+ cells ± SD. (B) HCC1937 cells were treated for 8 h with drugs as indicated, and EdU was added to the cultures before fixation. EdU was visualized by using the Click-IT technology and analyzed by using flow cytometry. The bar graph represents mean fluorescence intensity of the EdU+ population ± SD of experimental triplicates. (C and D) Determination of nucleotide levels in response to drug treatments in vivo. Tumor tissue from a MMTIV-Cre BRCA1f/fp53+/− (C) or a K14-Cre BRCA1f/f p53f/f tumor-bearing donor mouse (D) was transplanted into the mammary fat pad of NOD/Scid mice (C) or syngeneic K14-Cre− littermates (D) and allowed to grow to 10-mm diameter. Tumor-bearing recipients were then randomized to either control or drug treatment (BKM120 30 mg/kg by mouth or BYL719 30 mg/kg by mouth). Mice were treated 48, 24, and 2 h before killing in C and twice, 16 and 3 h, before killing in D. For raw data, see Datasets S1 and S2. (E) Determination of nucleotide levels in HCC1937 cells after 8 h of drug treatment with either BKM120 or BYL719 by competitive PCR (Materials and Methods).
To examine whether decreased DNA synthesis was a cell-autonomous effect of the PI3K inhibitor on the tumor cells, we conducted in vitro studies by using HCC1937 cells measuring EdU incorporation to quantify S-phase progression. Similar to the in vivo tumor model, in vitro in HCC1937 cells, EdU incorporation per cell was significantly reduced within 8 h of BKM120 and BYL719 treatments, seen as a decrease in the mean fluorescence of the EdU+ population (Fig. 2B and Fig. S1C). Taken together, these data suggest that tumor, but not normal, cells respond to PI3K inhibition with a cell-autonomous decrease in DNA synthesis within hours of drug exposure.
PI3K Inhibition Leads to the Depletion of Phosphorylated Nucleoside Pools.
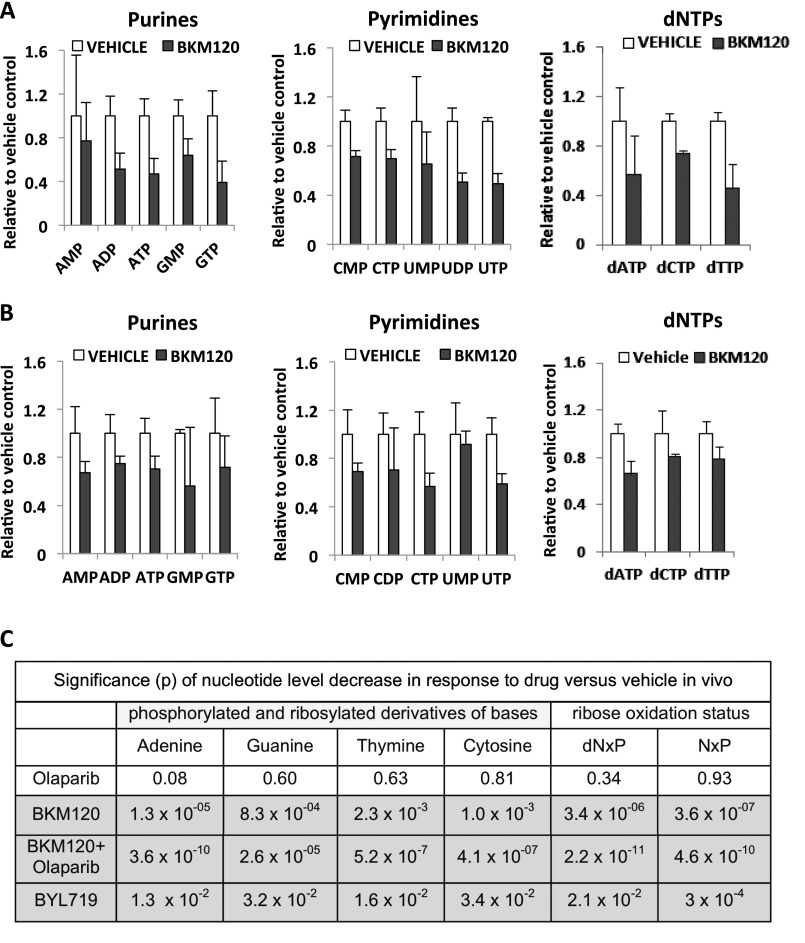
The synthesis of DNA in rapidly proliferating cultures and tissues requires an expansion of the nucleotide pool in early S phase. Nucleotide deficiency during S phase can lead to replication fork stalling in cancer cells and induce genomic instability with double-strand breaks (25). In addition, prolonged replication fork stalling exposes stretches of single-strand DNA that sequester RPA (26) (Fig. 1E). To determine whether the observed ineffective DNA synthesis in S phase was the result of nucleotide depletion, we measured nucleotide levels in vivo and in vitro. To test the effect of PI3K and combined PI3K/PARP inhibition on nucleotide levels in vivo, cohorts of breast tumor-bearing mice were prepared through allogeneic (Fig. 2C) or syngeneic (Fig. 2D) implantation of spontaneously developed tumor tissue collected from a MMTV-Cre BRCA1f/fp53+/− mouse (Fig. 2C) (27) or a K14-Cre BRCA1f/fp53f/f mouse (Fig. 2D) (21). For each condition, a cohort of four tumor-bearing mice was generated and tumors were allowed to grow to 10-mm diameter. Before killing, mice were given treatments with PI3K inhibitors BKM120 (30 mg/kg × 3 doses 40, 16, and 2 h prior) or BYL719 (30 mg/kg × 2 doses 16 and 3 h prior). Tumor tissue metabolites were analyzed by using mass spectrometry. Consistent in both mouse and tumor models, PI3K inhibition with either BKM120 (Fig. 2C) or BYL719 (Fig. 2D) led to a strong and highly significant drop in the levels of ribosylated and phosphorylated base derivatives (Fig. 2 C and D). This drop was not specific to the synthesis of purines or pyrimidines, but rather occurred in pools of all nucleoside derivatives (Fig. S2). In vitro in HCC1937 breast cancer cells (Fig. 2E), dNTP levels were measured using an HIV reverse transcriptase (RT)-based dNTP assay (28) after 3 h of exposure to BKM120 or BYL719 (Fig. 2E), and, in a second approach, a detailed analysis of nucleotides and their precursors was performed after exposure to BKM120 using mass spectrometry (Fig. S2 A and B and Datasets S3 and S4). We found that the abundance of nucleotides and of all their phosphorylated precursors were reduced after 3 h of PI3K inhibition (Fig. 2E and Fig. S2A), and this decrease was sustained after 16 h (Fig. S2B), similar to the in vivo observations (Fig. 2 A–D and Fig. S2C).
Fig. S2.
Decreased nucleotide levels in response to PI3K inhibition, determined by mass spectrometric measurement of nucleotide levels in HCC1937 cells in vitro (A and B) and in tumors in vivo (C). HCC1937 cells were cultured in the presence of the pan-PI3K inhibitor BKM120 (1 μM) for 3 (A) or 16 (B) h, and cell lysates were collected and processed for mass spectrometry in triplicates. For each metabolite, mean and SD are displayed, normalized to control. (C) Significance of nucleotide level decrease in vivo. Mass spectrometry data for a given metabolite were centered as a fraction of their median, and grouped according to treatment and base category as indicated. Displayed are the P values resulting from a two-tailed t test.
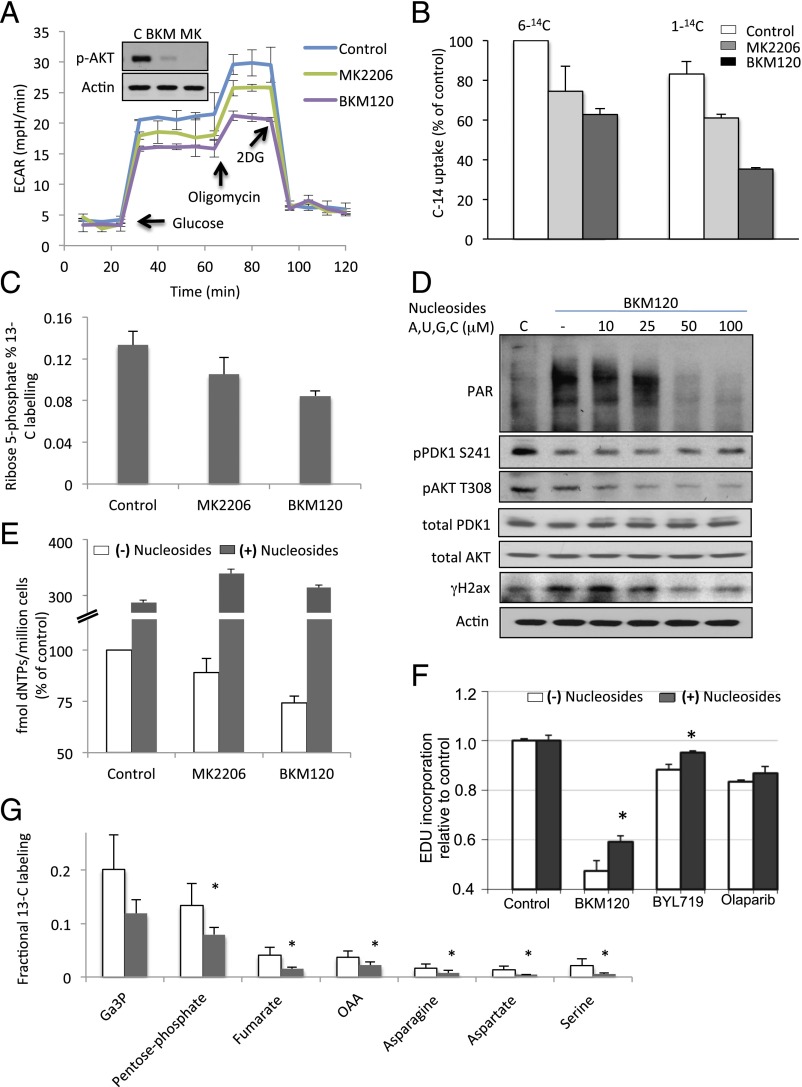
PI3K Inhibition Causes a Selective Decrease in Nonoxidative Pentose Phosphate Pathway Flux with Depletion of Rib Phosphate.
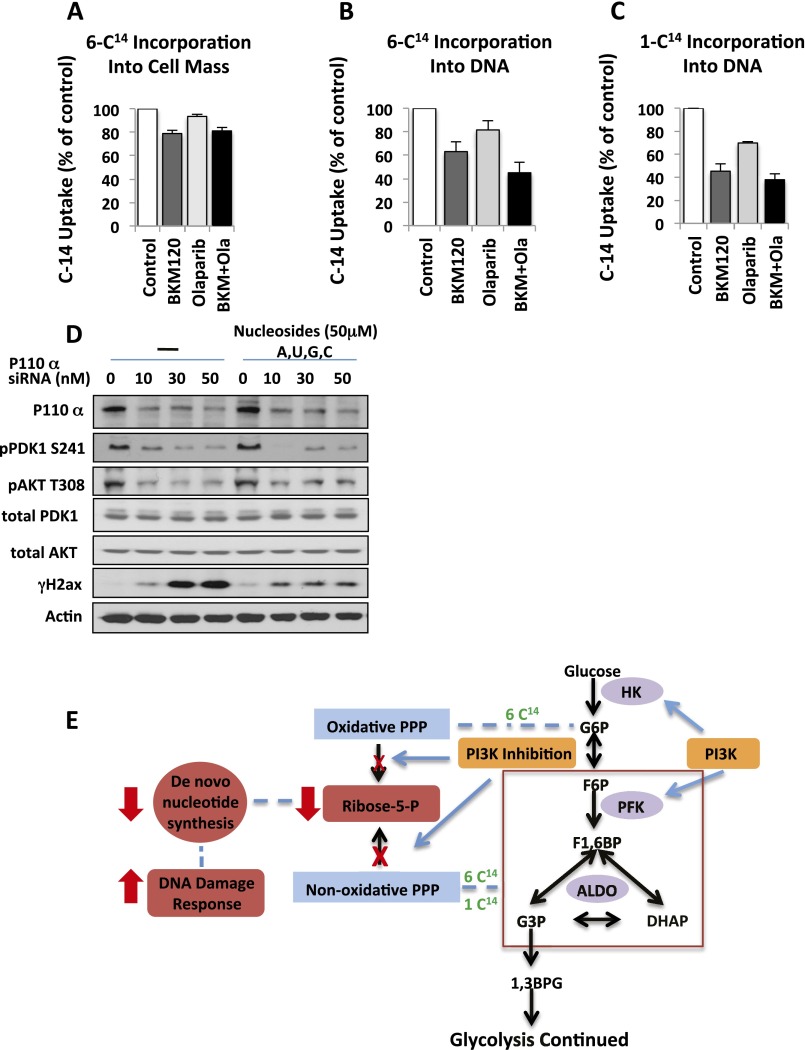
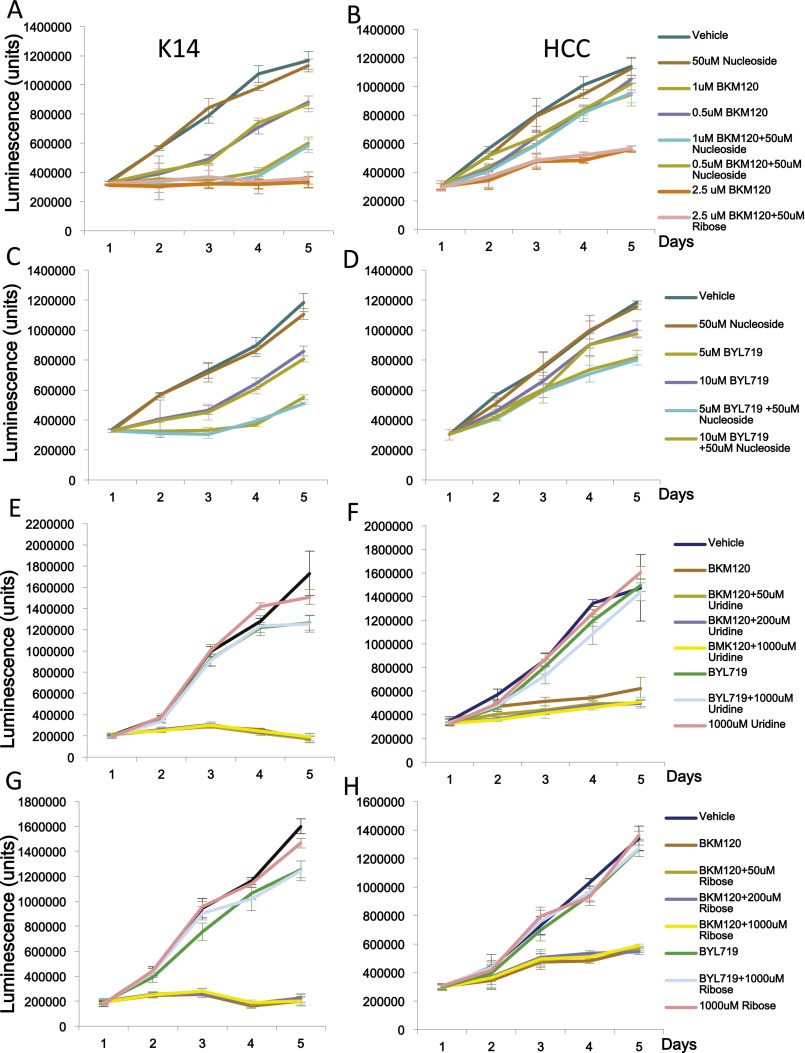
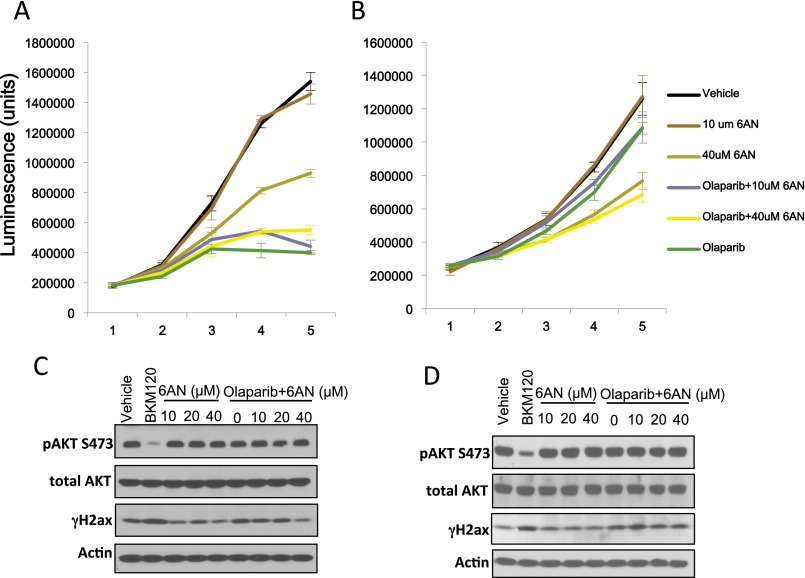
Cancer cells rely on de novo nucleotide synthesis for DNA synthesis (29). Because PI3K inhibition caused a reduction of all nucleotides regardless of base, phosphorylation, or Rib oxidation status (Fig. 2 and Fig. S2), we asked whether PI3K inhibition interfered with the production of Rib-5-phosphate (R5P). R5P is derived from glucose via the pentose-phosphate pathway, branching off glycolysis. Glycolytic flux in BRCA1-mutant HCC1937 cells was determined by measurement of the extracellular acidification rate (ECAR) using a Seahorse instrument after glucose challenge, mobilization of the glycolytic reserve after mitochondrial uncoupling with oligomycin, and blockade of glycolysis with 2-deoxy-glucose (2DG). As shown in Fig. 3A and reported earlier (15), glycolysis was only modestly reduced by AKT inhibitor MK2206 (Fig. 3A), whereas PI3K inhibitor BKM120 lowered the glycolytic rate strongly, particularly after mobilization of the glycolytic reserve with oligomycin (Fig. 3A). We used carbon tracing to follow the fate of radioactively labeled glucose into the deoxy-Rib–containing backbone of newly synthesized DNA (Fig. 3B and Fig. S3). When 6-14C-glucose is used, labeled Rib can be manufactured either through the oxidative or the nonoxidative arm of the pentose phosphate pathway (PPP) (Fig. 3B); however, a radioactive label of the 1-C atom in glucose is only preserved if Rib is produced via the nonoxidative PPP because the label will be lost as CO2 if C-1–labeled glucose is processed through the oxidative PPP (16). Therefore, radioactivity measured in the backbone of DNA in cells treated with 6-14C-glucose reflects Rib produced via the oxidative and the nonoxidative PPP, whereas radioactivity measured in cells treated with 1-14C-glucose stems from Rib derived only through the nonoxidative PPP (Fig. 3B). When cell cultures grown in parallel were offered equal concentrations of 1-14C- or 6-14C-glucose for 8 h, 1-14C-glucose uptake into DNA was >80% of 6-14C-glucose uptake, indicating that more than 80% of the Rib used to ribosylate nucleosides was produced through the nonoxidative PPP (Fig. 3B). A significant reduction in flow through the nonoxidative PPP occurred in response to both MK2206 and BKM120; however, the magnitude of Rib depletion (greater than 60% in the nonoxidative arm) was significantly greater with PI3K inhibition than with AKT inhibition (Fig. 3B). These results were corroborated with 13C-glucose tracing, which similarly illustrated that the PI3K inhibitor BKM120 was more effective than the AKT inhibitor MK2206 in reducing 13C-Rib-phosphate production (Fig. 3C). Because PARP inhibitors could potentially increase glucose metabolism (through preserving NAD levels), which might offset the decrease in Rib-phosphate, we examined whether the PARP-inhibitor Olaparib as a single agent or in combination with BKM120 affected Rib synthesis (Fig. S3) in this BRCA1-mutant breast cancer cell line. Within the 8-h timeframe examined, Olaparib did not have a significant impact on overall carbon metabolism (Fig. S3A). However, flux through the pentose-phosphate pathway (Fig. S3B) and, specifically, the nonoxidative arm of the PPP (Fig. S3B, Right) was independently and significantly reduced by PARP inhibition, and combined PI3K/PARP inhibition was more effective at reducing PPP flux than either single agent alone (Fig. S3 B and C). When we reconstituted the culture medium with ribosylated bases, we could prevent the induction of γH2AX and PAR induced by BKM120 (Fig. 3D, Upper) or by the RNAi-mediated ablation of PI3Kα (Fig. S3D). Importantly, ribosylated base treatment increased dNTP levels severalfold, in both control and PI3K inhibitor-treated cells (Fig. 3E). Consistently, nucleoside reconstitution at least partially rescued PI3K inhibitor-induced suppression of DNA synthesis as assessed by EdU incorporation (Fig. 3F and Fig. S3D). However, the only modest growth retardation of these cells by PI3K inhibitors in vitro was not reversed by nucleoside reconstitution (Fig. S4), suggesting that rescuing DNA damage caused by PI3K is essential, but not sufficient to rescue the growth inhibitory effect of PI3K inhibitors because they affect cell survival through mechanisms other than nucleoside synthesis such as TORC1 activation (30), FOXO transcriptional regulation (31), and suppression of GSK3 activity (32), which drives degradation of proteins such as MYC, SREBP1, and β-catenin. In addition, nucleoside reconstitution to supraphysiological levels (Fig. 3E) can lead to an imbalance that interferes with orderly cell division (33). Substitution with Rib or uridine was also not sufficient to rescue the growth retardation induced by PI3K inhibition in in vitro cell cultures (Fig. S4), suggesting that Rib cannot undergo phosphorylation directly in the cytoplasm and that uridine is not a significant substrate for ribosylation in these cells, different from HeLa cells (34). To summarize, our findings suggest that these cancer cells use the nonoxidative rather than the oxidative PPP to generate nucleosides, and consistently, inhibition of the oxidative pentose phosphate pathway with a competitive inhibitor of G6PD, 6-aminonicotinamide (6AN) did not induce a DNA damage response or inhibit cell growth, by itself or in combination with the Parp-inhibitor Olaparib (Fig. S5).
Fig. 3.
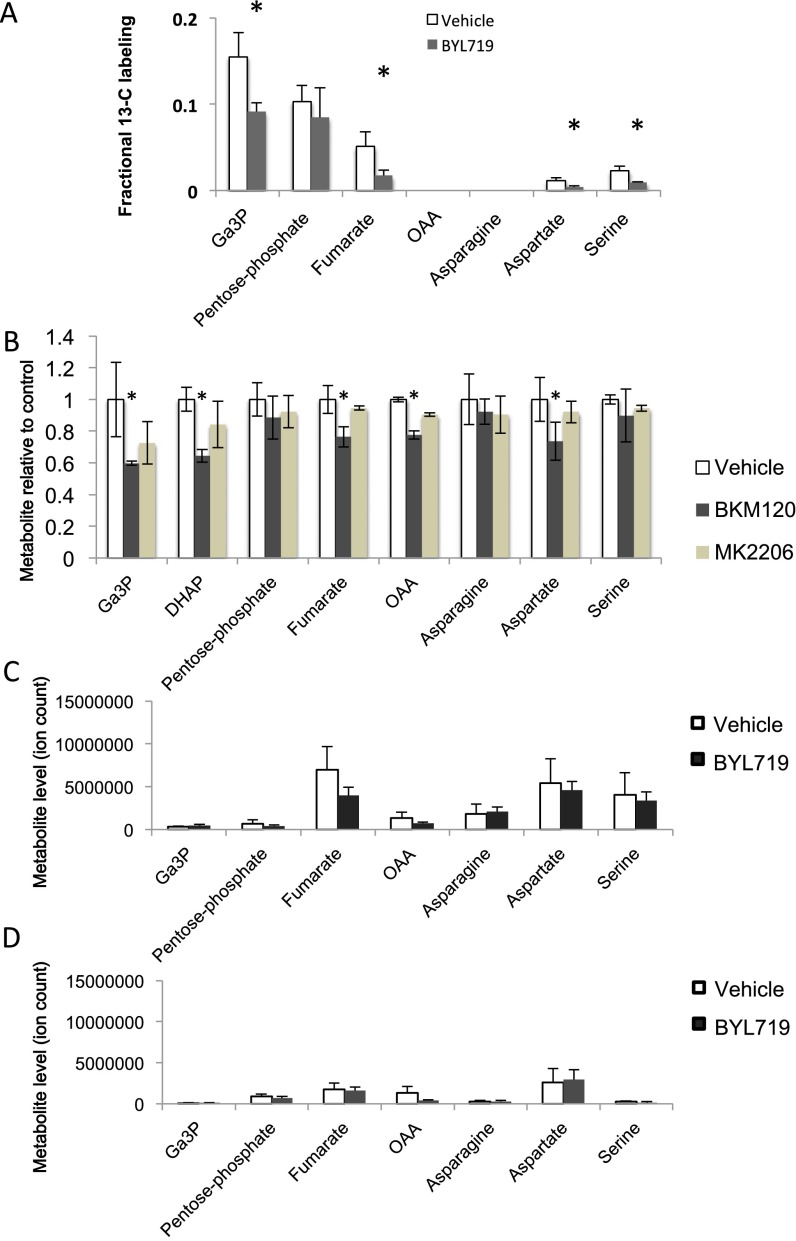
PI3K inhibition leads to decreased glycolytic flux through the nonoxidative pentose phosphate pathway. (A) Seahorse assay to determine overall glycolytic flux. Cells were seeded at 5,000 cells per well in a 24-well plate the night before the assay. Drugs were added at 3 h. ECAR was measured every minute for a total of 120 min in response to a glucose challenge, mitochondrial uncoupling to mobilize glycolytic reserve with oligomycin, and disruption of glycolysis with 2DG. Displayed is the ECAR over time of experimental quadruplicates ± SD. (B) Carbon flux from glucose to Rib as determined by 14C-glucose–derived carbon into DNA in response to PI3K and PARP inhibition. Cells were treated with 1-14C- or 6-14C-glucose and drugs as indicated 8 h before lysis. Scintillation count was done on purified genomic DNA. Displayed is the 14C-uptake of experimental triplicates ± SD, normalized to 14C-uptake in the 6-14C-glucose control. (C) Effect of PI3K and AKT inhibition on Rib-phosphate synthesis in HCC1937 cells. Cells were treated with vehicle control, BKM120 (1 μM), or MK2206 (1 μM) for 3 h, followed by labeling with [U-13C6]-glucose for 60 s and processed for mass spectrometry. (D) Nucleoside rescue of PI3K inhibitor-induced DNA damage indicators. Cells were treated for 16 h with drugs as indicated in the presence of a mixture of four nucleosides. (E) Determination of nucleotide levels in HCC1937 cells after 16 h of drug treatment with either BKM120 or MK2206 in the presence or absence of nucleosides. Determinations were done by competitive PCR, and displayed are the results of experimental triplicates. (F) Nucleoside rescue of PI3K inhibitor-induced decrease in DNA synthesis. Cells were treated for 8 h with drugs (BYL719, 2.5 μM; BKM120, 1 μM; or Olaparib, 5 μM) as indicated in the presence or absence of a mixture of four nucleosides. EdU was added in the last 2 h. Displayed are mean fluorescence of the EdU+ population relative to vehicle control in experimental triplicates. Significance was P < 0.05 for the bars indicated by the asterisk. (G) Effect of BYL719 on the nonox PPP and downstream metabolites in breast tumors in vivo. Tumor tissue from a K14-Cre BRCA1f/fp53f/f tumor-bearing donor mouse was transplanted into the mammary fat pad of syngeneic K14-Cre− littermates and allowed to grow to 10-mm diameter. Tumor-bearing recipients were then randomized to either control or BYL719 treatment. Mice were treated twice with BYL719 at 30 mg/kg per gavage, 16 h and again 3 h before killing. Mice were injected intraperitoneally with a [U-13C6]-glucose solution and metabolites extracted from the tumors. Data are presented as the relative fractional labeling of the pool, normalized to vehicle-treated tumors. Pentose phosphate includes R1P, R5P, X5P, and Ru5P because they cannot be distinguished by mass spec. Error bars represent the means ± SD of four tumors per treatment group. Significance was P < 0.05 for the bars marked with an asterisk. For raw data, see Dataset S5.
Fig. S3.
Carbon flux from glucose to Rib as determined by 14C-glucose–derived carbon into cell biomass (A) or DNA (B and C) in response to PI3K and PARP inhibition. HCC1937 cells were cultured in the presence of 14C6-glucose or 14C1-glucose, BKM120 (1 μM) or Olaparib (5 μM), or their combination for 3 h as indicated. Scintillation was counted for the entire cell lysate (A); genomic DNA was extracted and 14C measured (B and C). (D) Loss of PI3Kα leads to induction of H2AX phosphorylation (γH2AX) that can be rescued by nucleoside reconstitution. HCC1937 cells were depleted of PI3Kα by using siRNA for 48 h, and then treated with or without nucleosides for another 16 h. Cell lysates were subjected to immunoblotting with antibodies as indicated. (E) A radioactive label in the 6 position will measure flux through both oxidative and nonoxidative PPP, whereas a label in the 1 position will allow to measure flux through the nonoxidative PPP only.
Fig. S4.
Growth characteristics of K14-Cre BRCA1f/fp53f/f (A, C, E, and G) and HCC1937 (B, D, F, and H) cells in response to PI3K inhibitors BKM120 (A, B, G, and H) and BYL719 (C, D, G, and H) in the presence and absence of nucleosides (A–D) or uridine (E and F) or Rib (G and H). Cell growth, starting with 500 cells per well, was monitored in 96-well plates by using the CellTiter-Glo assay. Drug and supplement concentrations were as indicated.
Fig. S5.
Inhibition of the oxidative pentose-phosphate pathway with 6AN does not synergize with Olaparib treatments. The effects of 6AN alone or in combination with the Parp-inhibitor Olaparib were examined in K14-Cre BRCA1f/fp53f/f breast cancer cells (A and C) or HCC1937 human breast cancer cells (B and D). (A and B) Cell growth as determined by CellTiter-Glo assay over time (in days). (C and D) Cells were treated for 16 h with drugs as indicated and subjected to immunoblotting with antibodies as indicated.
To test whether PI3K inhibition had a similarly profound effect on nonoxidative PPP flux and de novo nucleotide synthesis in vivo, we used isotope tracing analyses with uniformly labeled ([U]-13C)-glucose in the mouse model of BRCA1-related breast cancer (Fig. 3G and Dataset S5), where we had previously observed a synergistic effect of PI3K and PARP inhibition (12). Cohorts of tumor-bearing mice were created through syngeneic transplantation (35). A bolus of [U]-13C-glucose was administered intraperitoneally 90 min before killing, followed by tumor harvest and mass spectrometry-based metabolomic analyses (36) (Fig. 3G). A sharp drop in the labeled fraction of 13C-glucose metabolites was seen in the aldolase products DHAP and Ga3P and in Rib-phosphate. Levels of glucose-derived serine, resulting from the oxidation and reductive amination of 3-phosphoglycerate, a product of lower glycolysis, were also decreased after BYL719 treatment. Interestingly, depletion of the aldolase products carried over to a reduction in 13C-flux into intermediates of the tricarboxylic acid (TCA) cycle with significant reduction of its intermediates fumarate and oxaloacetate. Consequently, 13C-incorporation into the oxaloacetate transamination products asparagine and aspartate was also reduced (Fig. 3G). Because aspartate donates its nitrogen atom to the synthesis of inosine, the basic building block for purines, and serine is required for both purine and pyrimidine synthesis, the decrease of these nonessential amino acids would further contribute to nucleotide depletion. In the nomal mammary glands from the tumor-bearing animals, fractional labeling of these intermediates was in general lower (Fig. S6A and Dataset S6), the effects of treatments with BYL719 on Rib-phosphate production were not significant, and TCA cycle intermediates were barely measurable (Fig. S6A) in this in vivo tracing analysis. Lowered levels of TCA cycle products, asparagine, and aspartic acid were also seen in vitro in cell lines treated with BKM120, but less so with the AKT inhibitor MK2206 (Fig. S6B and Datasets S7 and S8).
Fig. S6.
Metabolite changes in response to PI3K inhibition. (A) Effect of BYL719 on the nonox PPP in adjacent normal breast tissue in vivo (normal tissues from tumor-bearing mice described in Fig. 3F). Tumor tissue from a K14-Cre BRCA1f/fp53f/f tumor-bearing donor mouse was transplanted into the mammary fat pad of syngeneic K14-Cre− littermates and allowed to grow to 10-mm diameter. Tumor-bearing recipients were then randomized to either control or BYL719 treatment. Mice were treated twice with BYL719 at 30 mg/kg per gavage, 12 h and again 2 h before killing. Mice were injected intraperitoneally with a [U-13C6]-glucose solution and metabolites extracted from the normal mammary gland of the recipient mouse. Data are presented as the relative fractional labeling of the pool, normalized to vehicle-treated tumors. Error bars represent the means ± SD of four tumors per treatment group. (B) Effect of the pan-PI3K inhibitor BKM120 and the AKT-inhibitor MK2206 on aldolase products and TCA cycle products in HCC1937 cells. Cells were treated for 3 h, and metabolite abundance was determined by mass spectrometry. Experimental triplicates ± SD. Significance was P < 0.05 for the dark bars indicated by the asterisk: (A) comparison of BYL719 treatment with vehicle control; (B) comparison of BKM120 treatment with vehicle control. (C) Total metabolite levels of tumor tissue from K14-Cre BRCA1f/fp53f/f tumors. (D) Total metabolites levels of normal mammary tissue from K14-Cre BRCA1f/fp53f/f tumor-bearing mice.
Taken together, these data confirm the mass spectrometry data that PI3K inhibition did not affect nucleoside phosphorylation or oxidation (Fig. S2C). Instead, PI3K inhibition reduces overall glycolytic flux, resulting in a disproportionate reduction in flux through the PPP and, specifically, the nonoxidative arm. Decreased flux through the nonoxidative PPP leads to a depletion of R5P supplies that, in turn, are limiting for de novo nucleotide synthesis. The shortage of substrate for nucleotide biosynthesis is reinforced by a depletion of the nonessential amino acids necessary for purine or pyrimidine syntheses and generated from intermediates of lower glycolysis or the TCA cycle. AKT inhibition, while also reducing glycolytic flux to some extent, is less potent than the PI3K inhibitor in reducing PPP flux because it does not affect aldolase activity and, thus, the formation of the nonoxidative PPP substrate Ga3P.
The Antitumor Efficacy of the PI3K and PARP Inhibitor Combination Depends on Induction of the DNA Damage Phenotype.
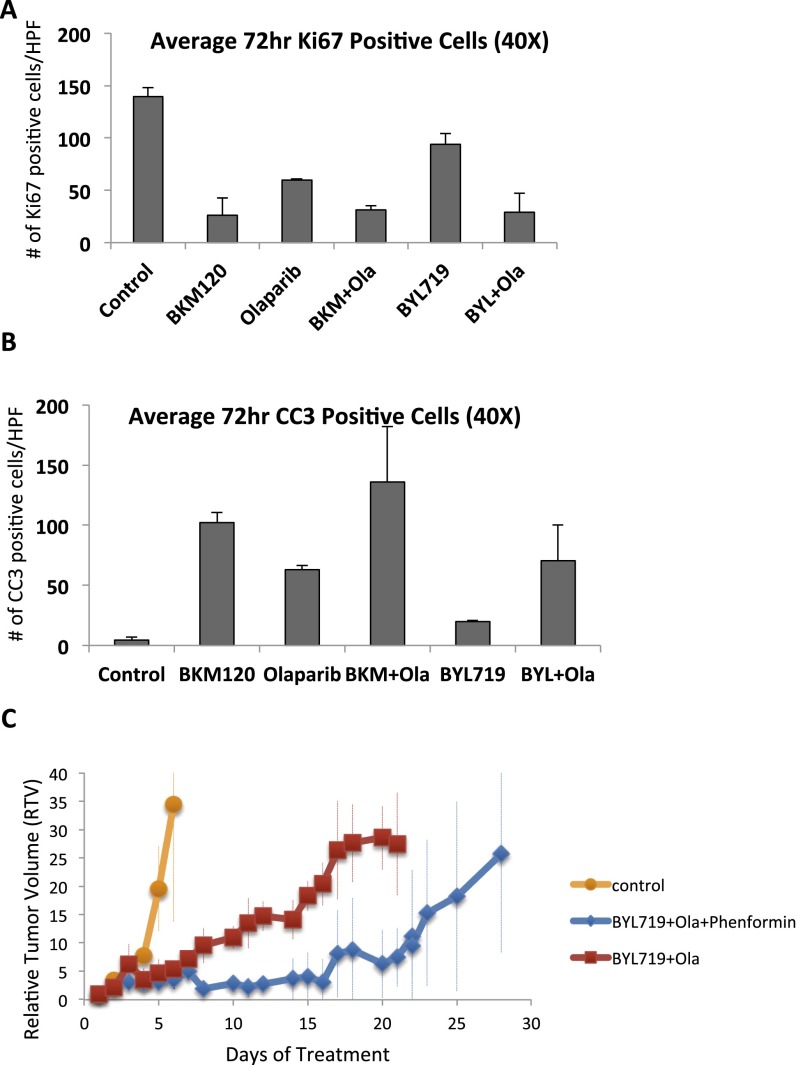
Given the augmentation of PARP inhibition by PI3K inhibition and building on our previous data in a mouse model with exon 11 loss of BRCA1 (12), we examined the efficacy of this combination in a mouse model with complete BRCA1 and p53 loss that is considered to be representative of human BRCA1-linked breast cancer (21). Synthetic lethality of the PI3K and the Parp inhibitor was expected because the PI3K inhibitor caused a strong induction of the product of Parp, PAR. Conversely, the Parp inhibitor was either neutral or enhanced AKT phosphorylation, indicative of an activated PI3K pathway (Figs. 1 A and C and 4D) (12). Both, the pan-PI3K inhibitor BKM120 and the PI3Kα-inhibitor BYL719 were dosed to suppress AKT phosphorylation, which was used here as a pharmacodynamic marker (Fig. 4A). This suppression was achieved within 8 h of drug exposure and sustained at 72 h. Tumor cell proliferation, as evidenced by a drop in Ki67 stain, was decreased in all treated tumors (Fig. S7A). A significant induction of tumor cell apoptosis was observed only in tumors treated with the combination of a PI3K and a PARP inhibitor, as evidenced by cleaved caspase 3 (CC3) stain (Fig. 4A and Fig. S7B).
Fig. 4.
Response to PI3K and PARP inhibition in vivo. Six different primary tumors from K14-Cre BRCA1f/fp53f/f females were propagated in vivo through syngeneic transplantation. Recipient females were randomized to treatments with BKM120 (30 mg⋅kg⋅−1d−1 by mouth), BYL719 (30 mg⋅kg⋅−1⋅d−1 by mouth), Olaparib (50 mg⋅kg⋅−1d−1 i.p.), or their combination, and tumor volume was recorded every 2 d. Treatment endpoint was time to progression as defined by tumor volume doubling. (A) Two mice per treatment condition were killed after 8 and 72 h of drug treatments to assess biomarkers of response, as assessed with Ki67 and CC3 by IHC (magnification: 200×). These mice were given a last dose of drugs 2 h before killing. (B) Survival statistics (log-rank test) and Kaplan–Meier analysis of 132 recipient females randomized to treatments as indicated. (C) Resistance to PI3K inhibition. 18FDG-PET-CT scan at baseline and after 48 h of administration of the PI3K inhibitor NVP-BKM120 (3 doses, 30 mg/kg PO) in sensitive and BKM120+Olaparib resistant K14-Cre BRCA1f/fp53f/f tumor-bearing mice. The maximum standardized uptake value (SUVmax) of tumors before and after PI3K inhibitor administration is displayed in the bar graph. Tumors are marked with a yellow arrow. Two hours before killing, mice were injected with BrdU and given an additional dose of BKM120. Tumor sections were stained with anti-BrdU antibodies for immunofluoresence (red) (magnification: 200×) and mean fluorescence per cell of BrdU+ cells was measured using volocity software. (D) A cell line was derived from the clinically resistant tumor and its PI3K/PARP-sensitive parental tumor. Cells were treated in vitro with drugs as indicated for 16 h, lysed, and blotted with antibodies as indicated.
Fig. S7.
Quantitative analysis of mitosis and apoptosis in response to PI3K and PARP inhibition after 3 d of treatment. Tumor-bearing mice were prepared and treated as described in Fig. 4A. For each treatment category, a minimum of two animals were killed at 72 h to determine biomarker responses. Immunohistochemistry was performed with antibodies against Ki67 (A) and CC3 (B), and three to five images were taken at 40x and analyzed with volocity software. (C) Effects of phenformin on treatment responses of a K14-Cre BRCA1f/fp53f/f tumor with acquired resistance to BKM120 and Olaparib. Tumors were implanted, allowed to grow to a size of 5 mm, and randomized to treatments with vehicle control (n = 2), BKM120 and Olaparib, or BKM120+Olaparib+Phenformin (n = 3). Tumors were assessed at the time points indicated. Tumor growth retardation with addition of Phenformin was significant between days 10 and 21 (P < 0.05).
In preparation for a randomized study, six different primary breast tumors generated in K14-Cre BRCA1f/fp53f/f genetically engineered mice were harvested and propagated through transplantation in the mammary fat pad of syngeneic female littermates. A total of 132 mice were randomized to treatments with either vehicle control, BKM120, BYL719, Olaparib, BKM120 plus Olaparib or BYL719 plus Olaparib (6-arm study). Treatments were started when tumor size reached 0.5–0.7 cm. Animals were followed every 48 h to time to progression (doubling of tumor size). Kaplan–Meier analysis revealed that these tumors progressed rapidly, and that both, BYL719 and BKM120, administered as single agents have only a minor growth-retarding effect. Olaparib, as seen previously (21), is an effective, although not curative, drug to treat these tumors, resulting in a median progression-free survival time of 50 d (Fig. 4B). Combined PI3K and PARP inhibition led to sustained long-term remissions in the majority of the tumor-bearing animals, resulting in a median progression-free survival of >120 d (Fig. 4B).
If the therapeutic activity of PI3K inhibitors depends on their ability to block DNA synthesis through inhibition of nucleoside synthesis, then PI3K inhibitors should fail to induce the DNA damage phenotype in tumors that are resistant to combined PI3K and Olaparib inhibition. To test this hypothesis, a resistant tumor was generated in vivo, by letting it grow to 1–1.5 cm in diameter, treating to induce a partial remission (down to 0.5 cm), allowing regrowth, and retreating until the tumor no longer responded. This tumor continued to grow, albeit more slowly than with vehicle control, when treated with BKM120 and Olaparib (Fig. S7C). The resulting treatment-resistant tumor was transplanted into a new recipient, where it was de novo treatment-resistant, confirming that treatment resistance was acquired by the tumor and not host-related. In this treatment-resistant tumor, fluorodeoxyglucose (FDG) uptake was lower than in its sensitive parental tumor, but importantly, FDG uptake was no longer lowered by PI3K inhibition, suggesting that glycolysis was no longer responsive to PI3K inhibition (Fig. 4C). Consistently, within 8 h of exposure to BKM120, the resistant tumor continued to incorporate BrdU, indicative of unimpaired DNA synthesis, whereas the sensitive tumor responded as in Fig. 2A, with a strong decrease in DNA synthesis. Treatment with the AMPK activator Phenformin slowed the progression of this PI3K and Parp inhibitor-resistant tumor, but did not prevent it (Fig. S7C), suggesting that neither mTOR activation nor mitochondrial complex I compensated for PI3K signaling in this tumor. In a cell line derived from the resistant tumor, BKM120 inhibited AKT phosphorylation as expected, but failed to induce PARP1 poly-(ADP)-ribosylation (Fig. 4D), γH2AX, or phosphorylation of ATM and chk1 (Fig. 4D). These data suggest that despite effective blockade of AKT signaling, nucleotide pools in these resistant tumor cells are less susceptible to depletion by PI3K inhibition, again pointing toward an AKT-independent mechanism for the antimetabolic function of PI3K inhibition. In summary, these data show that therapeutic activity of PI3K inhibitors depends to a significant extent on their ability to induce nucleotide depletion in cancer cells, triggering replication stress and DNA damage during S phase.
Discussion
PI3K has two distinct, yet related, functions for tumor cell growth: It serves as a nodal point for mitogenic signaling to coordinate cell division and mediates carbohydrate uptake and metabolism required for the generation of biomass. We recently found that activation of PI3K, and not AKT, enhances glycolytic flux at the step of aldolase (15). The reverse aldol condensation catalyzed by aldolase generates Ga3P, which is the triose acted on by TKT, the first step in the nonoxidative PPP that produces R5P required for base ribosylation. Here, we show that one aspect of the antimetabolic function of PI3K inhibition is the depletion of nucleotides as a consequence of reduced flux through glycolysis, and specifically the nonoxidative PPP. The consequence of decreased flux through the nonoxidative PPP is a decrease in R5P required for base ribosylation. In addition, the decrease of lower glycolysis affects the synthesis of amino acids required for the synthesis of bases themselves, thereby exacerbating the base shortage further (Fig. 3G). Consistent with PI3K’s role in aldolase activation, nucleotide depletion appears to be at least in part an AKT-independent effect of PI3K inhibition, because PI3K inhibition without AKT inhibition (Fig. 1C) induced the DNA damage phenotype whereas isolated AKT inhibition did not. In addition, AKT inhibition alone did not reduce flux through the nonoxidative PPP as effectively as PI3K inhibition (Fig. 3). Finally, in a PI3K and Parp inhibitor-resistant cell line, the DNA damage phenotype could not be elicited despite effective AKT inhibition (Fig. 4D).
Treatment with a PI3K inhibitor, and specifically a PI3Kα inhibitor, led to a depletion of nucleotides in rapidly proliferating cancer cells that could inflict replication stress within hours and interfered with the orderly progression of S phase. This effect appeared to be specific to tumor cells because it was not, or to a much lesser extent, seen in rapidly proliferating splenic lymphocytes. This metabolic effect may be more pronounced in tumor cells, because they rely more heavily on de novo nucleotide biosynthesis than normal tissues. The specificity for tumor cells may explain the favorable toxicity profile of PI3K inhibitors as toxicities typically associated with antineoplastic treatments such as myelosuppression or alopecia are not seen with PI3K inhibitors.
With this mechanism of action, to interfere with DNA synthesis in tumor cells through nucleotide depletion, PI3K inhibitors most closely resemble antineoplastics in the class of the antimetabolites: these include methotrexate, an inhibitor of dihydrofolate reductase that interferes with purine synthesis; Fluorouracil (5-FU), an inhibitor of thymidilate synthase that interferes with the synthesis of pyrimidine bases; and hydroxyurea, an inhibitor of ribonucleotide reductase. Two ongoing studies (NCT01300962 and NCT02000882; https://clinicaltrials.gov) will determine whether the addition of the PI3K inhibitor BKM120 improves the efficacy of a 5-FU–based drug, Capecitabine, in breast cancer.
PI3K inhibitor-induced nucleotide depletion resulted in DNA damage as a result of replication stress: Cells entered S phase, but DNA synthesis was decreased. Nucleotide shortage can induce replication stress and lead to error-prone replication. Bester et al. (25) showed that in keratinocytes driven by the E6/E7 oncogene nucleotide depletion led to replication fork stalling with ensuing DNA damage that was reversible upon nucleoside repletion. Consistent with those findings, we observed decreased DNA synthesis, leading to replication stress as evidenced by the appearance of cells with nuclear foci containing γH2AX, pRPA, and activation of Chk1 and ATM (Fig. 1) in response to PI3K inhibition. Through this mechanism of nucleotide depletion-mediated DNA damage, PI3K inhibition introduces a reliance on DNA damage repair mechanisms as evidenced also by a strong increase in PAR. This increased need for DNA damage repair becomes a vulnerability and creates an opportunity for PARP inhibition, particularly for tumor cells with a high degree of genomic instability. This explains why, in the case of BRCA1 mutant cancers examined here (Fig. 4B) and previously (12), addition of the PI3K inhibitor augments the synthetic lethality of the PARP inhibitor.
Cancer cells have developed a by-now well-understood redundancy in mitogenic kinase signaling that, at least in part, explains the frequently observed primary and often rapidly emerging secondary resistance to targeted inhibitors in solid tumors. Adaptive mechanisms that compensate for blocks in metabolic pathways are less well understood. We found that FDG uptake in tumors that were resistant to combined PI3K and PARP inhibition remained lower than in untreated tumors, suggesting a continued suppression of glucose uptake with continued PI3K-inhibitor treatment, whereas DNA synthesis had recovered (Fig. 4). Candidate mechanisms that could explain a metabolic bypass that allows for resurgent DNA synthesis while glucose uptake remains suppressed with ongoing PI3K inhibition potentially may include the following: (i) acquired myc amplification, which has been observed in PI3K inhibitor-resistant breast cancer cells (37, 38) and enhances the efficiency of nucleotide biosynthesis via increased glutaminolysis (38, 39); (ii) increased activity of phosphoribosyl-pyrophosphate synthetase 2 (PRPS2) (40); (iii) changes in the abundance or activity of rate-limiting enzymes that generate the trioses required for the nonoxidaitive PPP, such as the aldolase reaction, alone or in conjunction with transketolases; and (iv) a greater reliance on the recycling of preexisiting nucleotides.
Materials and Methods
The PI3K inhibitor NVP-BKM120 and NVP-BYL719 were obtained through a Material Transfer Agreement with Novartis Pharmaceuticals. Olaparib was purchased from LC Laboratories. SGK inhibitor GSK650394 and PIT-1 were purchased from Tocris Bioscience. The Akt inhibitor MK2206 was obtained through Haoyuan Chemexpress, China. All drugs were dissolved in DMSO. Nucleosides were obtained from Sigma Aldrich. BRCA1-mutant human breast cancer cell line HCC1937 was from American Type Culture Collection, CRL-2336, and maintained in DMEM supplemented with 10% (vol/vol) FBS. K14-Cre BRCA1f/fp53f/f cell line was generated from the parental tumors emerging in these mice and maintained as above.
Syngeneic Tumor Implants.
All animal experiments were conducted in accordance with Institutional Animal Care and Use Committee-approved protocols at Beth Israel Deaconess Medical Center. Tumors generated in K14-Cre BRCA1f/fp53f/f mice were syngeneically transplanted into the mammary pad of K14-Cre− mice to generate cohorts of mice. Tumors generated in MMTV-Cre BRCA1f/fp53+/− mice (12) were transplanted as allografts into NOD.CB17-Prkdc scid/J mice (Jackson Labs). D treatments were started once the tumors reached 5 mm and stopped once the tumors reached 20 mm in diameter.
Immunohistochemistry and Immunoflorescence.
For immunohistochemistry (IHC) and immunofluorescence (IF), we used anti–pAKT-S308 (ab38449; Abcam, rabbit polyclonal), anti-CC3 (9661S; Cell Signaling, rabbit polyclonal Asp175), anti-Ki67 (9106-S; Thermo Scientific, rabbit monoclonal SP6), anti-BrdU (5292, Cell Signaling, mouse monoclonal), anti-pRPA (A300-245A; Bethyl Laboratories, rabbit polyclonal), anti-ATM (phospho S1981, ab36810; Abcam) and anti-H2AX Ser139 (2577, Cell Signaling; rabbit polyclonal). All IHCs and IFs were done as described previously (12), including antigen retrieval with a citrate buffer.
Immunoblotting.
Cells were treated with either DMSO, NVP-BKM120, NVP-BYL719, Olaparib, MK2206, PIT-1, PIK75, TGX221, GSK1120212, or GSK650394 as single agents or combinations and lysed in cell lysis buffer (9803; Cell Signaling) as per the manufacturer's instructions. Immunoblots were conducted by using the Nupage System (Invitrogen). A total of 40 μg of protein was loaded. Primary antibodies used for Western blot analysis were phosphorylated anti-pAKT S473 (4058; Cell Signaling), anti-pAKT-S308 (ab38449; Abcam), anti-H2AX Ser139 (2577; Cell Signaling), p-PDK1 Ser241 (3061; Cell Signaling), p-P70 S6K T389 (9234; Cell Signaling), p-4E-BP1 S65 (9456; Cell Signaling), p-NDRG1 T346 (9485; Cell Signaling), p-CHK1 S345 (2348; Cell Signaling), p44/42 MAPK (4695; Cell Signaling), anti-Actin (ab6276; Abcam), and anti-pADPr (sc56198) from Santa Cruz Biotechnology.
siRNA Transfection.
The siRNAs were obtained from Dharmacon. HCC1937 cells were transfected in the presence or absence of 50 μM nucleosides with either a 0-, 10-, 30-, or 50-nM pool of four siRNA sequences targeting PIK3CA (catalog no. L-003018-00-0005). Transfections were performed by using HiPerFect Transfection Reagent (Qiagen) according to the manufacturer's protocol. Control cells were treated with control siRNA (sc-37007; Santa Cruz). Cells were grown and harvested 48 h after the transfection by using cell lysis buffer (9803; Cell Signaling) as per the manufacturer’s instructions and analyzed by immunoblotting.
14C-Labeling Experiment.
HCC1937 cells were grown to 60–70% confluence on a 10-cm tissue culture dish. Cells were treated with either DMSO, NVP-BKM120, Olaparib, NVP-BKM120+Olaparib, or MK2206 in DMEM media supplemented with 6-14C- or 1-14C-glucose for 8 h. DNA was extracted by using Qiagen DNeasy kit (catalog no. 69504) according to the manufacturer’s instructions. Equal volumes of DNA were added to scintillation vials, and radioactivity was measured by liquid scintillation counting and normalized to the DNA concentration. All experiments were done in triplicates.
dNTP Measurements.
HCC1937 cells were seeded at a density of 2 × 106 cells and treated with either DMSO, NVP-BKM120, Olaparib, NVP-BKM120+Olaparib, or MK2206 for 8 h. After treatment, the cells were washed with PBS and the cells were lysed in ice-cold 65% (vol/vol) methanol. The cells were scraped, and the pellet was vortexed vigorously and incubated at 95 °C for 3 min. The samples were shipped to the Kim Baek laboratory (Emory University, Atlanta), and the dNTPs were measured as described previously (28).
Seahorse Experiment.
HCC1937 cells were seeded overnight in 24-well culture plate at density 250,000 cells per well in Sigma (D5030) medium. The following day, the cell culture medium was removed and replaced with fresh medium, and cells were treated with either DMSO, NVP-BKM120, or MK2206 for 3 h and incubated at 37 °C. Per the manufacturer’s protocol, SeaHorse injection ports were loaded with either glucose; oligomycin, an inhibitor of ATP synthase (oxidative metabolism) that maximizes glycolytic metabolism (final concentration 1.0 μM); or 2-deoxy glucose an inhibitor of glycolysis. ECAR was measured by using the SeaHorse XF24 Extracellular Analyzer from SeaHorse Bioscience.
Metabolomics/13C Experiment.
HCC1937 cells were cultured to ∼50–60% confluence in growth media [DMEM, 10% (vol/vol) FBS] on a six-well plate in biological quadruplicate and treated as indicated for steady-state metabolomic analysis. The metabolites were extracted with 80% (vol/vol) ice-cold methanol and concentrated completely to dryness by using a SpeedVac. For the in vitro flux to Rib-phosphate, cells were treated with drugs as indicated for 3 h. The medium was replaced with medium of the same composition except for [U-13C6]-glucose instead of unlabeled glucose for 60 s, at which point metabolites were extracted with 80% (vol/vol) ice-cold methanol and processed as above.
Flow Cytometry.
HCC1937 cells (1 × 106) or K14cre breast cancer cells derived from K14-Cre BRCA1f/fp53f/f tumor were treated for 8 h with either DMSO, NVP-BKM120, NVP-BYL719, Olaparib, or the combination of NVP-BKM120+Olaparib in a six-well plate. EdU (10 μM) was added to the cell culture medium containing drugs as indicated above at 6 h timepoint, and the cells were allowed to be labeled for 2 h. For the negative staining control, HCC1937 cells were left untreated. The media was aspirated, and the cells were washed three times with 1% BSA in PBS before the cell pellet was fixed and permeabalized according to the manufacturer’s protocol (Click-iT EdU Flow Cytometry Assay Kits, Life Technologies, catalog no. C10418). Cell cycle analysis was performed by using a FACSAria II Cell Sorter 5-laser SORP instrument (BD Biosciences).
In Vivo Metabolomics.
To trace glucose metabolism in vivo, breast tumors were grown as described in Syngeneic Tumor Implants. The tumor bearing mice were treated orally with 30 mg/kg BYL719 16 and 3 h prior to killing. Before killing, mice were fasted overnight and intraperitoneally injected with a [U]-13C-glucose solution (2 g/kg in PBS) (Cambridge Isotope Labs). Blood glucose was monitored in 15- to 30-min intervals, and mice were killed when blood glucose levels began to decline. Tumors were harvested for metabolomic analysis by using 80% (vol/vol) aqueous methanol, and the metabolite equivalent of 10 mg of tumor tissue was dried. Metabolite fractions were resuspended in HPLC-grade water and analyzed by targeted liquid chromatography-tandem mass spectrometry (LC-MS/MS) using a 5500 QTRAP mass spectrometer (AB/SCIEX) coupled to a Prominence UFLC HPLC system (Shimadzu) with Amide HILIC chromatography (Waters). Data were acquired in SRM mode by using positive/negative ion polarity switching for steady-state polar profiling of more than 260 molecules. SRM transitions were also created for 13C-labeled precursor and fragment ions for monitoring 13C incorporation. Peak areas from the total ion current for each metabolite SRM transition were integrated by using MultiQuant v2.0 software (AB/SCIEX) (36).
Cell Viability Assay.
For cell viability assays, breast cancer cells were seeded at a density of 2,000 cells (HCC1937) or 1,000 cells (K14 cre) per well in 96-well plates in the absence or presence of drugs, and cell viability was determined by using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) according to the manufacturer’s instructions, using a Wallac 3 plate reader.
FDG-PET Scanning.
K14-Cre BRCA1f/fp53f/f mice were syngeneically transplanted into the mammary pad of K14-Cre− mice to generate cohorts of mice. The mice were treated with drugs as indicated, and 0.3–0.4 mCi of fluorine-18-deoxyglucose was injected intravenously through the retroorbital vein of the anesthetized mouse. After a “washout” period of 1 h, the mouse was imaged on a NanoPET/CT (Mediso) scanner. The NanoPET/CT is a high-resolution small-animal multimodality scanner consisting of 12 lutetium yttrium oxyorthosilicate (LYSO) detector blocks. The blocks comprise a total of 39,780 crystals each with a dimension of 1.2 × 1.2 × 13 mm3. Images were acquired in three dimensions. The mice remained supine and maintained their position throughout the procedure. First, a computed tomography scan was performed and second, a whole-body 18F-FDG PET emission scan was acquired covering the same area as the CT scan. Cpms were obtained, converted to becquerels (Bq), and values were normalized for ROI volume and injected dose. To correct for metabolic and injected activity variability between examinations and to determine tumor-specific uptake changes, FDG-uptake rates were calculated according to the following formula: (activity in tumor Bq)/[(injected activity in Bq)/(mouse weight)]. For studies involving repeat scanning, the change in tumor-specific FDG uptake was determined in percent [1 − (FDG-uptakepost/FDG-uptakepre) × 100]. Animals were housed in the Longwood SAIF satellite animal facility between scans.
Supplementary Material
Acknowledgments
L.C.C. and G.M.W. are supported by the Breast Cancer Research Foundation, the Mary Kay Ash Foundation, the Breast Cancer Alliance, the Men’s Initiative of the Dana–Farber Harvard Cancer Center, a Stand Up to Cancer Dream Team Translational Research Grant, and Program of the Entertainment Industry Foundation Grant SU2C-AACR-DT0209. The work was in part supported by NIH Grants NIHP50 CA168504 (to G.M.W.), R01 GM041890 (to L.C.C.), NIH 5P01CA120964-05 and 5P30CA006516-46 (to J.M.A.), and NIH GM104198 and AI049781 (to B.K.).
Footnotes
Conflict of interest statement: L.C.C. has consulted for Novartis Pharmaceuticals, which is developing NVP-BKM120 and NVP-BYL719 for cancer treatment; L.C.C. is a member of the board of directors and a consultant/advisory board member for Agios Pharmaceuticals, for which he also has ownership interests (including patents).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522223113/-/DCSupplemental.
References
- 1.Burga LN, et al. Altered proliferation and differentiation properties of primary mammary epithelial cells from BRCA1 mutation carriers. Cancer Res. 2009;69(4):1273–1278. doi: 10.1158/0008-5472.CAN-08-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burga LN, et al. Loss of BRCA1 leads to an increase in epidermal growth factor receptor expression in mammary epithelial cells, and epidermal growth factor receptor inhibition prevents estrogen receptor-negative cancers in BRCA1-mutant mice. Breast Cancer Res. 2011;13(2):R30. doi: 10.1186/bcr2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maor S, et al. Elevated insulin-like growth factor-I receptor (IGF-IR) levels in primary breast tumors associated with BRCA1 mutations. Cancer Lett. 2007;257(2):236–243. doi: 10.1016/j.canlet.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 4.Nowsheen S, Cooper T, Stanley JA, Yang ES. Synthetic lethal interactions between EGFR and PARP inhibition in human triple negative breast cancer cells. PLoS One. 2012;7(10):e46614. doi: 10.1371/journal.pone.0046614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorrini C, et al. Estrogen controls the survival of BRCA1-deficient cells via a PI3K-NRF2-regulated pathway. Proc Natl Acad Sci USA. 2014;111(12):4472–4477. doi: 10.1073/pnas.1324136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gewinner C, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16(2):115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fedele CG, et al. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proc Natl Acad Sci USA. 2010;107(51):22231–22236. doi: 10.1073/pnas.1015245107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen WH, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128(1):157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 9.Ip LR, et al. Loss of INPP4B causes a DNA repair defect through loss of BRCA1, ATM and ATR and can be targeted with PARP inhibitor treatment. Oncotarget. 2015;6(12):10548–10562. doi: 10.18632/oncotarget.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang NY, et al. Radiosensitization with combined use of olaparib and PI-103 in triple-negative breast cancer. BMC Cancer. 2015;15:89. doi: 10.1186/s12885-015-1090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De P, et al. Doubling down on the PI3K-AKT-mTOR pathway enhances the antitumor efficacy of PARP inhibitor in triple negative breast cancer model beyond BRCA-ness. Neoplasia. 2014;16(1):43–72. doi: 10.1593/neo.131694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juvekar A, et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2012;2(11):1048–1063. doi: 10.1158/2159-8290.CD-11-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibrahim YH, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2012;2(11):1036–1047. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7(8):606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 15.Hu H, et al. Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell. 2016;164(3):433–446. doi: 10.1016/j.cell.2015.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149(3):656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasser JA, et al. SGK3 mediates INPP4B-dependent PI3K signaling in breast cancer. Mol Cell. 2014;56(4):595–607. doi: 10.1016/j.molcel.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sommer EM, et al. Elevated SGK1 predicts resistance of breast cancer cells to Akt inhibitors. Biochem J. 2013;452(3):499–508. doi: 10.1042/BJ20130342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao B, Degterev A. Targeting phospshatidylinositol 3-kinase signaling with novel phosphatidylinositol 3,4,5-triphosphate antagonists. Autophagy. 2011;7(6):650–651. doi: 10.4161/auto.7.6.15248. [DOI] [PubMed] [Google Scholar]
- 20.Miao B, et al. Small molecule inhibition of phosphatidylinositol-3,4,5-triphosphate (PIP3) binding to pleckstrin homology domains. Proc Natl Acad Sci USA. 2010;107(46):20126–20131. doi: 10.1073/pnas.1004522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rottenberg S, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci USA. 2008;105(44):17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rottenberg S, et al. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc Natl Acad Sci USA. 2007;104(29):12117–12122. doi: 10.1073/pnas.0702955104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feijoo C, et al. Activation of mammalian Chk1 during DNA replication arrest: A role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154(5):913–923. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robison JG, Elliott J, Dixon K, Oakley GG. Replication protein A and the Mre11.Rad50.Nbs1 complex co-localize and interact at sites of stalled replication forks. J Biol Chem. 2004;279(33):34802–34810. doi: 10.1074/jbc.M404750200. [DOI] [PubMed] [Google Scholar]
- 25.Bester AC, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145(3):435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11(3):208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 27.Xu X, et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22(1):37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 28.Van Cor-Hosmer SK, Daddacha W, Kim B. Mechanistic interplay among the M184I HIV-1 reverse transcriptase mutant, the central polypurine tract, cellular dNTP concentrations and drug sensitivity. Virology. 2010;406(2):253–260. doi: 10.1016/j.virol.2010.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr Opin Genet Dev. 2009;19(1):32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menon S, et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell. 2014;156(4):771–785. doi: 10.1016/j.cell.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stitt TN, et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14(3):395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- 32.Denley A, Kang S, Karst U, Vogt PK. Oncogenic signaling of class I PI3K isoforms. Oncogene. 2008;27(18):2561–2574. doi: 10.1038/sj.onc.1210918. [DOI] [PubMed] [Google Scholar]
- 33.Kumar D, Viberg J, Nilsson AK, Chabes A. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010;38(12):3975–3983. doi: 10.1093/nar/gkq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wice BM, Reitzer LJ, Kennell D. The continuous growth of vertebrate cells in the absence of sugar. J Biol Chem. 1981;256(15):7812–7819. [PubMed] [Google Scholar]
- 35.Liu X, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA. 2007;104(29):12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc. 2012;7(5):872–881. doi: 10.1038/nprot.2012.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu P, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17(9):1116–1120. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang JB, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18(3):207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunningham JT, Moreno MV, Lodi A, Ronen SM, Ruggero D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell. 2014;157(5):1088–1103. doi: 10.1016/j.cell.2014.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.