Significance
DNA damage tolerance pathways like translesion synthesis and recombination facilitate the bypass of replication-blocking lesions. Such events are crucial for the survival of rapidly proliferating cells, including cancer and stem cells undergoing active duplication during tissue renewal. Herein, we characterize an unprecedented damage tolerance pathway that requires the combined function of a highly enigmatic translesion DNA polymerase ι (POLι) and the so-called guardian-of-the-genome, p53. We provide evidence demonstrating that p53 complexed with POLι triggers idling events that decelerate nascent DNA elongation at replication barriers, facilitating the resolution of stalled forks by specialized structure-specific enzymes. Our findings implicate p53 in the protection of quickly growing cancer and stem cells from endogenous and exogenous sources of replication stress.
Keywords: DNA damage bypass, DNA polymerase idling, nascent DNA elongation, polymerase ι, p53
Abstract
DNA damage tolerance facilitates the progression of replication forks that have encountered obstacles on the template strands. It involves either translesion DNA synthesis initiated by proliferating cell nuclear antigen monoubiquitination or less well-characterized fork reversal and template switch mechanisms. Herein, we characterize a novel tolerance pathway requiring the tumor suppressor p53, the translesion polymerase ι (POLι), the ubiquitin ligase Rad5-related helicase-like transcription factor (HLTF), and the SWI/SNF catalytic subunit (SNF2) translocase zinc finger ran-binding domain containing 3 (ZRANB3). This novel p53 activity is lost in the exonuclease-deficient but transcriptionally active p53(H115N) mutant. Wild-type p53, but not p53(H115N), associates with POLι in vivo. Strikingly, the concerted action of p53 and POLι decelerates nascent DNA elongation and promotes HLTF/ZRANB3-dependent recombination during unperturbed DNA replication. Particularly after cross-linker–induced replication stress, p53 and POLι also act together to promote meiotic recombination enzyme 11 (MRE11)-dependent accumulation of (phospho-)replication protein A (RPA)-coated ssDNA. These results implicate a direct role of p53 in the processing of replication forks encountering obstacles on the template strand. Our findings define an unprecedented function of p53 and POLι in the DNA damage response to endogenous or exogenous replication stress.
The tumor suppressor protein p53 has been called the guardian-of-the-genome due to its ability to transactivate downstream targets transcriptionally, which prevents S-phase entrance before facilitating DNA repair or eliminating cells with severe DNA damage via apoptosis (1). Interestingly, p53 also encodes an intrinsic 3′–5′ exonuclease activity located within its central DNA-binding domain (2–4). The contribution of the exonuclease proficiency to p53’s function has largely remained obscure. Exonucleases are involved in DNA replication, DNA repair, and recombination, increasing the fidelity or efficiency of these processes. The 3′–5′ exonuclease activity of DNA polymerases (POLs) catalyzes the correction of replication errors, thereby preventing genomic instability and cancer (5–7). The potential involvement of p53’s exonuclease in DNA repair has been ascribed to transcription-independent functions in nucleotide excision repair and base excision repair, in homologous recombination (HR), and in mitochondrial processes (8–10).
Regarding HR, in particular, reports indicate a dual role for p53. On the one hand, it has been reported that p53 down-regulates unscheduled and excessive HR in response to severe genotoxic stress, like formation of DNA double-strand breaks (DSBs) (8–10). This antirecombinogenic effect of p53 has been linked to the blockage of continued strand exchange by interactions with recombinase RAD51, RAD54, and nascent HR intermediates carrying specific mismatches (11, 12). On the other hand, p53 stimulates spontaneous HR during S-phase to overcome replication fork stalling and to prevent fork collapse (10, 13, 14). By this mechanism, p53 is believed to protect replicating DNA (14). However, the prorecombinogenic function of p53 during DNA synthesis has remained less well understood. p53 was found to associate with HR factors in S-phase cells after induced replication arrest (15–17) and was shown to interact with the replication factor RPA (replication protein A) and with POLα-primase (18, 19). Therefore, p53 seems to escort the replisome, at least after replication stress. Despite these pieces of evidence, the exact role of p53 in DNA replication remains unknown.
Recombination is one possible mechanism to resolve stalled and collapsed replication forks (20, 21). WT p53 exerts a prosurvival so-called healer effect on tumor cells in response to poly-(ADP ribose) polymerase (PARP) inhibition, which correlates with a stimulation of replication-associated recombination (14, 22). Because of the hypothesized role of p53 in HR and/or HR-driven replication events, we further examined the role of p53 in HR during unperturbed replication or after enforced replication fork stalling using DNA cross-linking, which is known to require HR for its resolution (23, 24). The recently identified p53 mutant with separated functions in transcription/cell cycle regulation and exonucleolytic DNA degradation enabled us to explore the specific contribution of p53’s exonuclease activity to the hypothesized role of p53 in HR-driven replication events, such as increasing the fidelity of these processes (2). Our study reveals that WT p53, in concert with POLι, protects the integrity of replication forks by mastering idling-like events, which either leads to successful DNA damage bypass or to pronounced meiotic recombination enzyme 11 (MRE11)-dependent resection of DNA. An epistasis-like functional and biochemical analysis unraveled the details of the DNA damage bypass mechanism, which involves a previously unknown complex between p53 and the specialized POLι, promoting fork reactivation via helicase-like transcription factor (HLTF) and zinc finger ran-binding domain containing 3 (ZRANB3).
Results
WT p53, but Not Its H115N Mutant, Stimulates Replication-Associated Recombination.
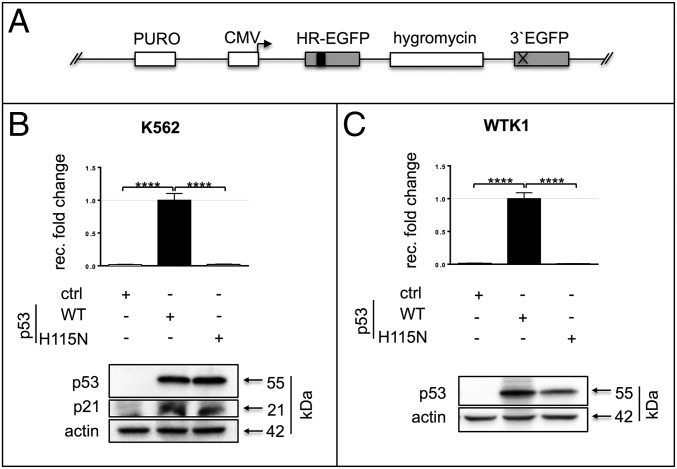
We have previously shown that spontaneous recombination events, which are independent of any targeted cleavage but strictly associated with DNA replication, can be detected in cells carrying a stably integrated EGFP-based substrate (13, 25) (Fig. 1A). Such recombination events are most likely triggered by the encounter of replication forks with endogenous DNA damage (23, 26). To determine the specific contribution of p53 to such events, we compared the spontaneous recombination frequencies of chromosomally integrated EGFP recombination substrates after expression of either p53(WT) or p53(H115N) (Fig. 1). In both p53-negative K562 leukemia cells and p53-mutated lymphoblastoid WTK1 cells, expression of p53(WT) led to a robust increase of the recombination frequency (Fig. 1 B and C). Intriguingly, an increased recombination frequency was not evident in K562 or WTK1 cells expressing the p53(H115N) mutant, although this mutant is transcriptionally active and modulates the cell cycle and apoptosis to a similar extent as the WT (Fig. 1 B and C and Fig. S1A). The augmentation of the global DNA damaging response by means of treatment with mitomycin C (MMC; 3 μM, 45 min) did not substantially increase the frequency of spontaneous recombination compared with the untreated controls in p53(WT)-expressing cells (Fig. S1B). Therefore, we conclude that the major trigger for spontaneous recombination by p53 is dependent on local signals at the replicating EGFP region.
Fig. 1.
p53 modulates DNA recombination in different cell types. (A) Schematic presentation of the recombination substrate (HR-EGFP/3′EGFP) chromosomally integrated in K562 cells [K562(HR-EGFP/3′EGFP)], which is used for the determination of recombination fold changes (25). Hygromycin, hygromycin resistance cassette; PURO, puromycin resistance cassette. The kinked arrow points to the promoter; the black square indicates a frame-shifting insertion in the EGFP chromophore coding region generating the inactive variant HR-EGFP; and the cross indicates replacement of the EGFP start codon by two stop codons, resulting in the inactive variant 3′EGFP. (B, Upper) Relative recombination frequencies in K562(HR-EGFP/3′EGFP) transfected with expression plasmids for p53(WT), p53(H115N), or empty vector (ctrl). Recombination (rec.) fold changes were analyzed by flow cytometry via quantification of EGFP+ cells 72 h after transfection. Measurements were individually corrected for transfection efficiencies. Mean values from untreated p53(WT)-expressing samples were set to 1 (absolute mean frequencies are provided in SI Materials and Methods). Data were obtained from 20 measurements. For graphic presentation, calculation of SEM and statistically significant differences via the two-tailed Mann–Whitney U test, we used GraphPad Prism 6.0f software. (B, Lower) p53 protein levels for samples used in recombination experiments. α-Actin served as a loading control. (C, Upper) Recombination fold changes in WTK1(HR-EGFP/3′EGFP) cells with chromosomally integrated recombination substrate. The experimental setup was the same as in B. (C, Lower) Western blot analysis shows p53 expression versus the loading control α-actin. ****P < 0.0001.
Fig. S1.
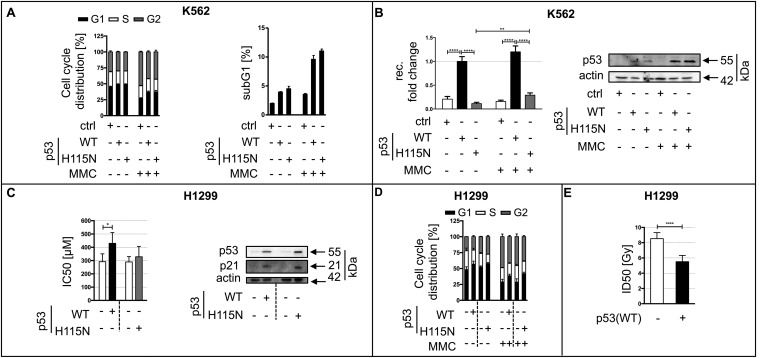
Effect of p53 on cell cycle distribution, recombination after MMC treatment, and cell survival following MMC or γ-ray treatment. (A) Cell cycle analysis in K562(HR-EGFP/3′EGFP) cells transfected with expression plasmids for p53(WT), p53(H115N), or empty vector (ctrl). Forty-eight hours after transfection, cells were mock-treated or MMC-treated (3 μM) for 45 min, washed, and reincubated in fresh media. The cell cycle distribution (Left) and subG1 fractions (Right) were determined by flow cytometry of propidium iodide-stained cells 72 h after transfection. Viable cells were divided into G1 (black), S (white), and G2 (gray) cell cycle phases (presented as percentages). Columns represent mean values and SEM from six measurements. (B, Left) Relative recombination frequencies in K562(HR-EGFP/3′EGFP) cells transfected and treated as described in A. Recombination (rec.) fold changes were analyzed by flow cytometry via quantification of EGFP+ cells 72 h after transfection. Measurements were individually corrected for transfection efficiencies. Mean values from untreated p53(WT)-expressing samples were set to 1 (absolute mean frequency: 2 × 10−5). Data were obtained from 18 to 20 measurements. For graphic presentation, calculation of SEM and statistically significant differences via the two-tailed Mann–Whitney U test (GraphPad Prism6.0f software) was used. (B, Right) p53 protein levels for samples used in recombination experiments. (C, Left) p53-negative H1299 cells, controlled with tetracycline, expressing p53(WT) or p53(H115N) were treated with increasing doses of MMC (1–1,000 μM) for 45 min, transferred to fresh medium, and subjected to MTT assay 48 h later. IC50 values and SEM were calculated from the results of eight measurements. Statistically significant differences between IC50 values of p53-negative controls and paired p53-expressing cells [(p53(WT)-cell and p53(H115N)-cell clones are separated by stippled lines] were defined using the extra-sum-of-square F test of log IC50 values. (C, Right) Immunoblotting revealing p53 and p21 levels in the presence/absence of tetracycline. (D) Cell cycle analysis in H1299 cell clones with tetracycline-regulated expression of p53(WT) and p53(H115N). Cell cycle distribution was determined by flow cytometry in H1299-cell clones without (−) and with (+) expression of p53(WT) or p53(H115N) [(p53(WT)-cell and p53(H115N)-cell clones separated by stippled lines] following mock treatment or MMC treatment (3 μM, 45 min, 3-h release). Columns represent mean values and SEM from four to six measurements. (E) Radiosensitivity after p53(WT) expression. Cell survival analysis was performed in an H1299-cell clone without or with expression of p53(WT). An MTT assay was performed 6 d after treatment with increasing doses of IR (0.5–16 Gy). Mean ID50 values and SEM were calculated from six measurements using GraphPad Prism6 software. Statistically significant differences between ID50 values of p53-expressing and p53-negative cells were determined using the F test. ****P < 0.0001; **P < 0.01; *P < 0.05.
Interestingly, in H1299 cells expressing tetracycline-regulated p53 variants (27), p53(WT) caused a statistically significant increase (1.5-fold; P = 0.0169) in the IC50 value following MMC treatment. In contrast, p53(H115N) expression did not alter the IC50 value (P = 0.5986), despite the increase in both p53 and p21 expression levels (Fig. S1C) and a similar effect on the cell cycle distribution as observed for p53(WT) expression (Fig. S1D). When interpreting these results, it is important to consider that although it is unlikely that MMC will trigger lesions within the EGFP coding region, the survival assay is monitoring the effect of MMC-induced interstrand cross-links (ICLs) in the whole genome. Given that ICLs, although representing only one MMC-DNA adduct out of many, are the major source of cytotoxicity (28–31), it is tempting to speculate that the survival assay is revealing the contribution of p53 to the resolution of ICLs. It is interesting that this scenario is different from the one observed after introduction of DSBs by ionizing radiation (IR). In such a setup, p53(WT) reduced the ID50 value from 8.5 to 5.5 Gy (Fig. S1E; P = 0.0001). Thus, although sensitization of cells to IR concurs with the well-described down-regulatory effect of p53(WT) on HR in response to DSBs (8–10), the desensitization to MMC is consistent with the reported p53(WT)-dependent stimulation of recombination during replication stress (13, 14). Taken together, our results suggest that p53 is involved in the recombinative bypass of replication blocks.
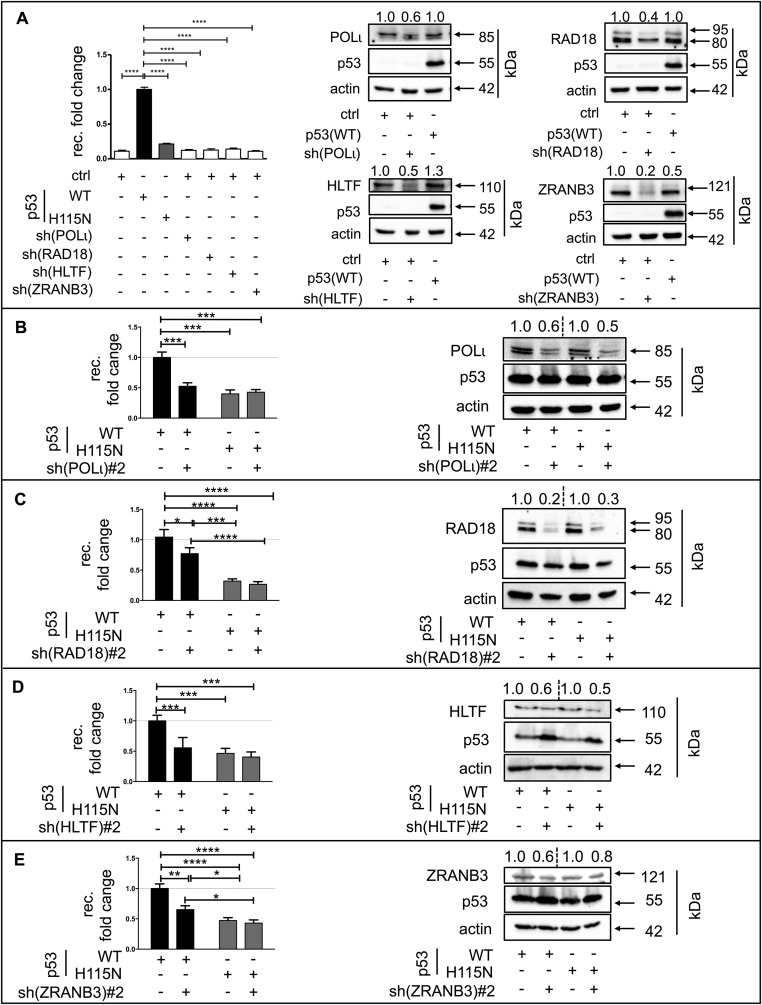
RAD18, HLTF, ZRANB3, and POLι cooperate with p53(WT), but Not with p53(H115N), to Stimulate Replication-Associated Recombination.
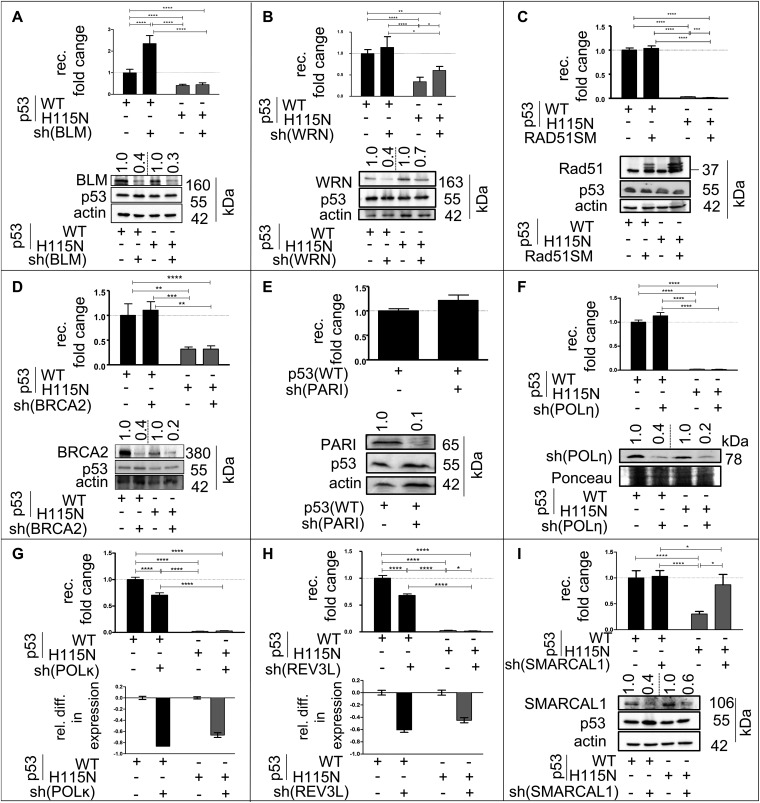
To investigate the molecular mechanism underlying p53(WT)-mediated recombination stimulation, we silenced factors implicated in the bypass of blocked replication forks. p53 inhibits the helicase and the branch-migrating activities of Bloom syndrome protein (BLM) and Werner syndrome protein (WRN) helicases, which are involved in the regulation of HR and in the bypass of replication barriers (32, 33), whereas RAD51 and breast cancer 2 (BRCA2) are involved in HR-dependent postreplication repair (34, 35). Proliferating cell nuclear antigen (PCNA)-associated recombination inhibitor (PARI) associates with DNA damage sites via SUMOylated PCNA and blocks recombination by inhibition of RAD51-DNA filament formation (36). Surprisingly, BLM, WRN, RAD51, BRCA2, and PARI were not required for the p53(WT)-mediated stimulation of recombination, hence suggesting an insignificant contribution of any RAD51-dependent pathway to this recombination event (Fig. S2 A–E). Consequently, RAD51-independent bypass mechanisms were explored by silencing different translesion synthesis (TLS)-POLs. Although silencing of POLη had no effect, silencing of POLκ and REV3L led to a 30% decrease of p53(WT)-induced recombination (Fig. S2 F–H). The most striking effect, however, was observed for POLι, with a 50% decrease in the recombination frequency in p53(WT)-expressing cells (Fig. 2A).
Fig. S2.
Recombination measurements after down-regulation of different helicases, recombination-related factors, and TLS-POLs. (Upper) Experimental setup and analysis of recombination fold changes were as described in the legend for Fig. 2, except that cells were cotransfected with shRNA (sh) plasmids for down-regulation of BLM (A), WRN (B), BRCA2 (D), PARI (E), POLη (F), POLκ (G), REV3L (H), or SMARCAL1 (I), or with expression plasmid for dominant-negative Rad51SM for inactivation of endogenous RAD51 (C). Bars indicate SEM. Data were obtained from 12 to 30 measurements of each. (Lower) To verify knockdown of respective targets or expression of RAD51/Rad51SM, either immunoblotting (A–F and I) or qRT-PCR (G and H) was performed, and results are shown. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05.
Fig. 2.
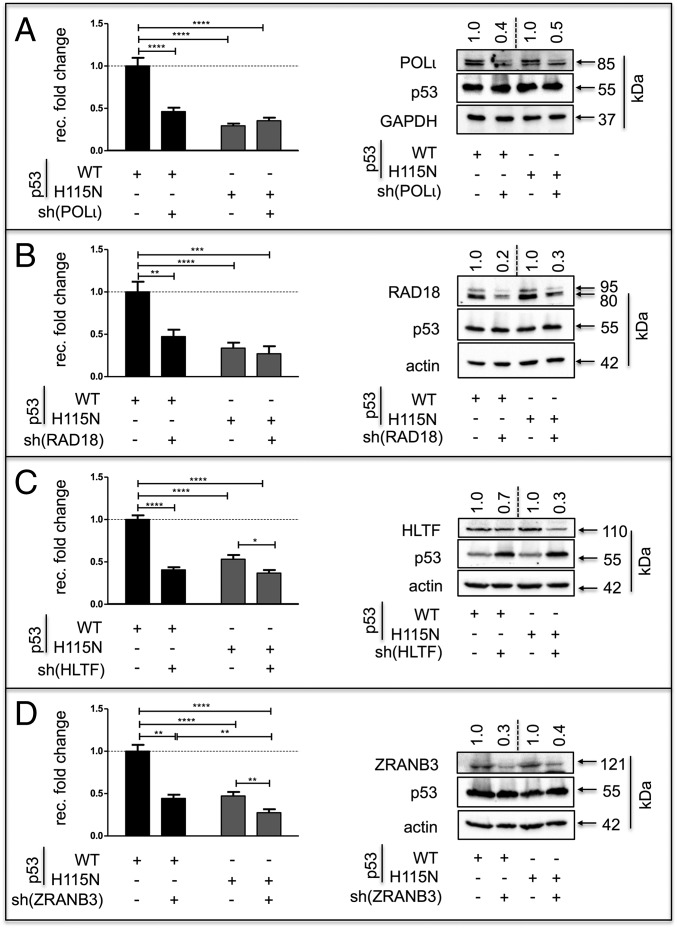
Stimulation of recombination by p53 requires POLι, RAD18, HLTF, and ZRANB3. (Left) K562(HR-EGFP/3′EGFP) cells were transfected with expression plasmids for either p53(WT) or p53(H115N), together with shRNA plasmid specific for POLι (A), RAD18 (B), HLTF (C), or ZRANB3 (D). Recombination fold changes were determined as described in Fig. 1. Data were obtained from 12 to 18 measurements. (Right) In all cases, immunoblotting was performed to verify knockdown of specific targets. Relative expression levels are indicated on the top of each panel. GAPDH (A) and α-actin (B–D) served as loading controls. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05.
PCNA monoubiquitination is a prerequisite for switching from replicative POLs to TLS-POLs at DNA damage sites (37–41). Silencing the E3-ubiquitin ligase RAD18, which mediates PCNA monoubiquitination (37, 38, 41) induced a 50–60% recombination decrease in p53(WT)-expressing cells (Fig. 2B). Similarly, silencing the E3-ubiquitin ligase and yeast Rad5 ortholog HLTF (also called SMARCA3 or RNF80 mediating PCNA polyubiquitination and fork reversal) (42–44) or silencing the structure-specific translocase ZRANB3 (45) induced a similar decrease in p53(WT)-expressing cells (Fig. 2 C and D). Importantly, knockdown of the annealing helicase SMARCAL1, which also functions downstream of HLTF (46, 47), displayed no effect (Fig. S2I). Also noteworthy, the silencing of POLι and RAD18 did not affect residual recombination activities in the presence of p53(H115N). The silencing of POLι, RAD18, HLTF, or ZRANB3 did not affect basal recombination activities in p53-negative cells (Fig. S3A). To exclude potential off-target effects, the genes of interest were also silenced with a second set of shRNA plasmids with a comparable effect (Fig. S3 B–E). Hence, p53 facilitates a PCNA ubiquitination-mediated bypass mechanism also involving RAD18, HLTF, ZRANB3, and POLι.
Fig. S3.
Verification of POLι, RAD18, HLTF, and ZRANB3 involvement in p53-mediated recombination. (A) K562(HR-EGFP/3′EGFP) cells were transfected with empty vector or expression plasmids for either p53(WT) or p53(H115N), together with the same shRNA plasmids specific for POLι, RAD18, HLTF, or ZRANB3 used in Fig. 2. Recombination fold changes were determined from 12 measurements. Knockdown of specific targets was verified via Western blot analysis. α-Actin staining served as a loading control. Relative expression levels are indicated on the top of each panel. K562(HR-EGFP/3′EGFP) cells were transfected with expression plasmids for either p53(WT) or p53(H115N), together with shRNA plasmids specific for POLι (B), RAD18 (C), HLTF (D), or ZRANB3 (E) targeting alternative sequences than shRNA plasmids used in Fig. 2 (#2). Recombination fold changes were determined from 12 to 18 measurements. (Right) In all cases, immunoblotting was performed to verify knockdown of specific targets. α-Actin staining served as a loading control. Expression levels relative to the corresponding empty shRNA vector control are indicated on the top of each panel. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05.
Interactions between PCNA, POLι, and p53.
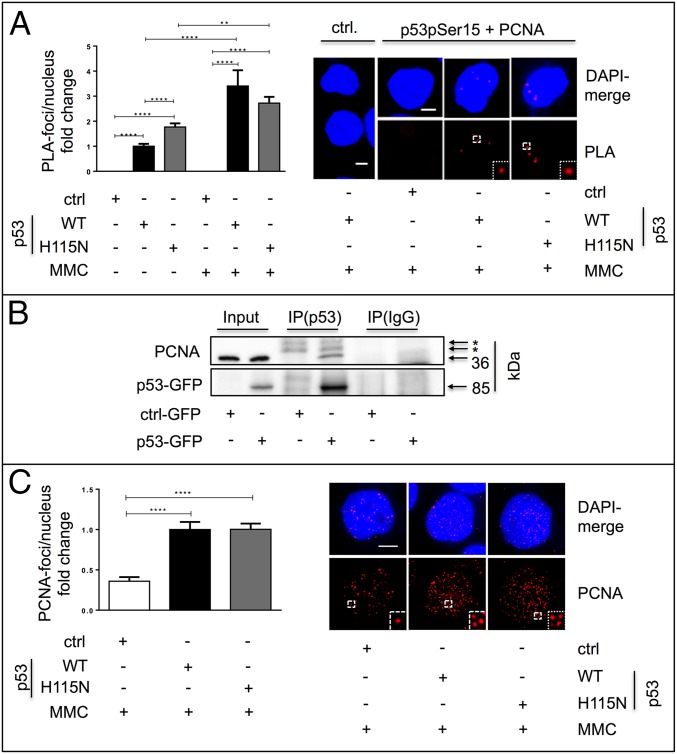
Having established a firm link between p53 and replication-associated recombination, we explored the potential interaction of p53 with key factors identified in our epistasis analysis. To this end, we performed an in situ proximity ligation assay (PLA) (Fig. 3A) for PCNA and the phosphorylated form of p53 (p53pSer15), because p53pSer15 was shown to associate with stalled replication forks (16, 17). The PLA indeed revealed an association between p53(WT) and PCNA, as well as between p53(H115N) and PCNA, which became more pronounced after MMC treatment. Moreover, PCNA was coimmunoprecipitated with GFP-tagged p53 (Fig. 3B), implying a physical interaction between p53 and PCNA. A functional link between p53 and PCNA was also suggested by a 2.8-fold increase in the number of PCNA foci per nucleus in p53-expressing cells after MMC treatment (Fig. 3C).
Fig. 3.
Analysis of the interaction between p53 and PCNA. (A) Association of p53pSer15 and PCNA by in situ PLA. After transfection of K562 cells with p53(WT) or p53(H115N) expression vectors or empty vector, the PLA assay was performed to detect interaction between p53pSer15 and PCNA as described in Materials and Methods. Forty-eight hours after transfection, K562 cells were mock-treated or MMC-treated (3 μM, 45 min), released by reincubation for an additional 3 h, and processed for PLA. Negative control (ctrl.) samples were processed accordingly, omitting primary antibodies against p53pSer15 and PCNA. Two hundred nuclei in two independent experiments were scored, whereby mean values from mock-treated p53(WT)-expressing cells were set to 1 (on average, one focus per nucleus). Bars indicate SEM. (Insets) Magnification (2.5×) of the highlighted region. (Scale bars: 5 μm.) (B) Coimmunoprecipitation analysis. Forty-eight hours after transfection with expression vectors for GFP-tagged p53 (p53-GFP) or GFP (ctrl-GFP), p53 was immunoprecipitated from K562 cells using the antibodies DO1 and Pab421, followed by immunoblotting for PCNA and p53. Asterisks indicate unspecific bands. IP, immunoprecipitation. (C) Immunofluorescence microscopy of PCNA signals as a function of the p53 status. Seventy-two hours after transfection with expression plasmids for p53(WT), p53(H115N), or empty vector, K562 cells were treated with MMC (3 μM, 45 min, 3-h release) and processed for immunofluorescence-based microscopy to visualize PCNA foci accumulation. The number of foci per nucleus was quantified using Keyence BZ-II Analyzer software. (Left) Average numbers were calculated from 68 nuclei in two independent experiments. Mean values in p53(WT)-expressing samples were set to 1 (on average, 27 foci per nucleus). Bars indicate SEM. (Right) Representative images with PCNA foci and merged images with a DAPI-stained nucleus are shown. (Insets) Magnification (2.5×) of the highlighted region. (Scale bar: 5 μm.) ****P < 0.0001; **P < 0.01.
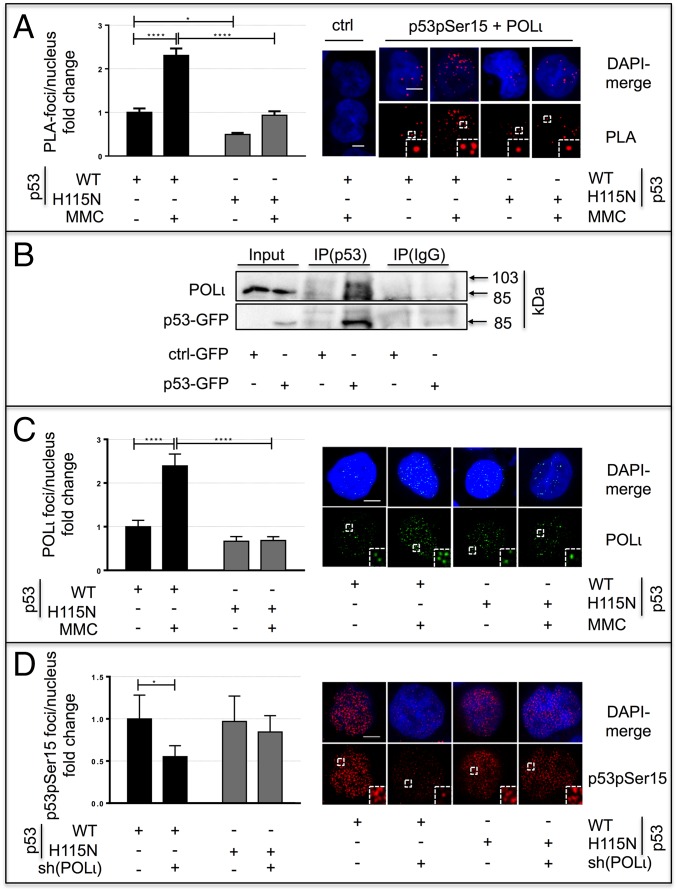
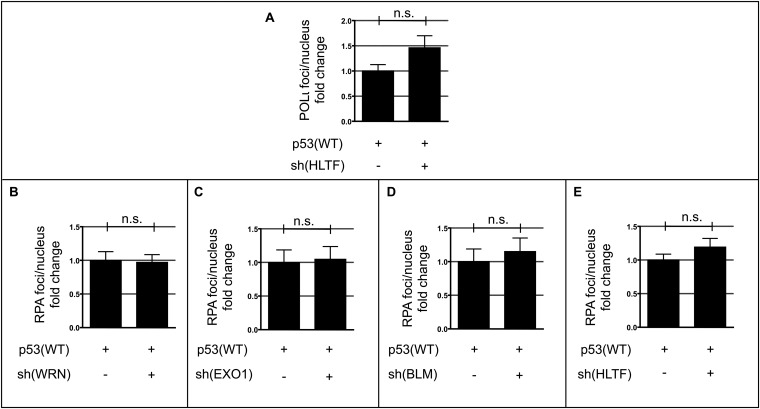
Because TLS-POLs are recruited to replication sites by PCNA ubiquitination (48), and because we observed an epistatic relationship between POLι and p53(WT) in our screening, the possibility of an interaction between p53(WT) and POLι was evaluated. PLA and POLι coimmunoprecipitation (Fig. 4 A and B) revealed an association between p53(WT) and POLι. Anti–POLι-antibodies did not support reciprocal p53 coimmunoprecipitation. However, the PLA also showed that association of p53(H115N) with POLι was significantly reduced compared with p53(WT), thus suggesting that the H115N mutation weakens the physical interaction between p53 and POLι (Fig. 4A). To elucidate the hierarchy of events downstream of PCNA, we quantified POLι-foci after p53(WT) and p53(H115N) expression and p53pSer15 foci with and without silencing of POLι. We detected a 2.4-fold increase in POLι-foci per nucleus upon expression of p53(WT) but not p53(H115N) (Fig. 4C). Interestingly, this POLι-foci accumulation was independent of HLTF (Fig. S4A). Because p53(WT), but not p53(H115N), enhanced POLι-foci formation, we wondered if POLι also affected p53 association with replication barriers. Silencing of POLι decreased p53pSer15 foci formation by 50% in p53(WT)-expressing cells (P = 0.0148), but not in cells expressing p53(H115N) (Fig. 4D). Altogether, our data indicate a complex interaction network between PCNA, p53(WT), and POLι. PCNA foci number is governed by p53(WT) and p53(H115N) in the same manner. However, because the recruitment of POLι required p53(WT) and was impaired upon p53(H115N) expression, we propose that p53(WT) favors the recruitment of POLι to replication barriers. Conversely, POLι is required to consolidate pSer15-modified p53(WT) foci but not p53(H115N) foci.
Fig. 4.
Analysis of the interaction between p53 and POLι. (A) Association of p53pSer15 and POLι by PLA. After transfection of K562 cells with p53(WT) or p53(H115N) expression vectors, the PLA assay was performed to detect interaction between p53pSer15 and POLι as described in Materials and Methods. Forty-eight hours after transfection with expression plasmids for p53(WT) or p53(H115N), K562-cells were mock-treated or MMC-treated (3 μM, 45 min, 3-h release). Negative control samples were processed accordingly, omitting primary antibodies against p53pSer15 and POLι. Two hundred nuclei in two independent experiments were scored, whereby mean values from mock-treated p53(WT)-expressing cells were set to 1 (on average, four foci per nucleus). (Insets) Magnification (2.5×) of the highlighted region. (Scale bars: 5 μm.) (B) POLι was detected in p53-GPF immunoprecipitations after MMC treatment (3 μM, 45 min, 3-h release) of K562 cells. Blots were first incubated with antibody against POLι and then with antibody against p53. (C) Subnuclear distribution of POLι is modulated by the p53 status. K562 cells were transfected and treated as in Fig. 4A, and samples were used for the immunofluorescence-based visualization of POLι-foci accumulation per nucleus. (Left) One hundred nuclei in two independent experiments were scored. (Right) Representative images are displayed. (Insets) Magnification (2.5×) of the highlighted region. Mean values of POLι-foci in p53(WT)-expressing cells after mock treatment were set to 1 (on average, four foci per nucleus). Bars indicate SEM. (Scale bar: 5 μm.) (D) Recruitment of exonuclease-proficient p53 into nuclear foci is affected by silencing of POLι. K562 cells transfected with expression plasmids for p53(WT) or p53(H115N) and with an shRNA plasmid specific for POLι [sh(POLι)] were treated with MMC (3 μM, 45 min, 3-h release). The number of p53pSer15 foci was scored in 100 nuclei and two independent experiments. Quantifications (Left) and representative images (Right) are shown. (Insets) Magnification (2.5×) of the highlighted region. Mean values of p53pSer15-foci in p53(WT)-expressing cells were set to 1. Bars indicate SEM. ****P < 0.0001; *P < 0.05.
Fig. S4.
Analysis of nuclear POLι-foci accumulation after HLTF silencing and RPA foci accumulation after knockdown of helicases and exonucleases related to HR. K562 cells were cotransfected with expression plasmid for p53(WT) and shRNA plasmids for HLTF [sh(HLTF), A and E], WRN [sh(WRN), B], EXO1 [sh(EXO1), C] or BLM [sh(BLM), D] to down-regulate expression of the respective targets. Forty-eight hours later, cells were treated with MMC (3 μM, 45 min) and processed for immunofluorescence-based microscopy to visualize POLɩ or RPA-foci accumulation 3 h later. Mean values for controls without knockdown were set to 1. n.s., not significant. Bars indicate SEM. (A) POLɩ-foci after HLTF knockdown. (B) RPA-foci after WRN knockdown. (C) RPA-foci after EXO1 knockdown. (D) RPA-foci after BLM knockdown. (E) RPA-foci after HLTF knockdown.
p53(WT) Promotes RPA Foci Accumulation in a Manner Dependent on POLι and MRE11.
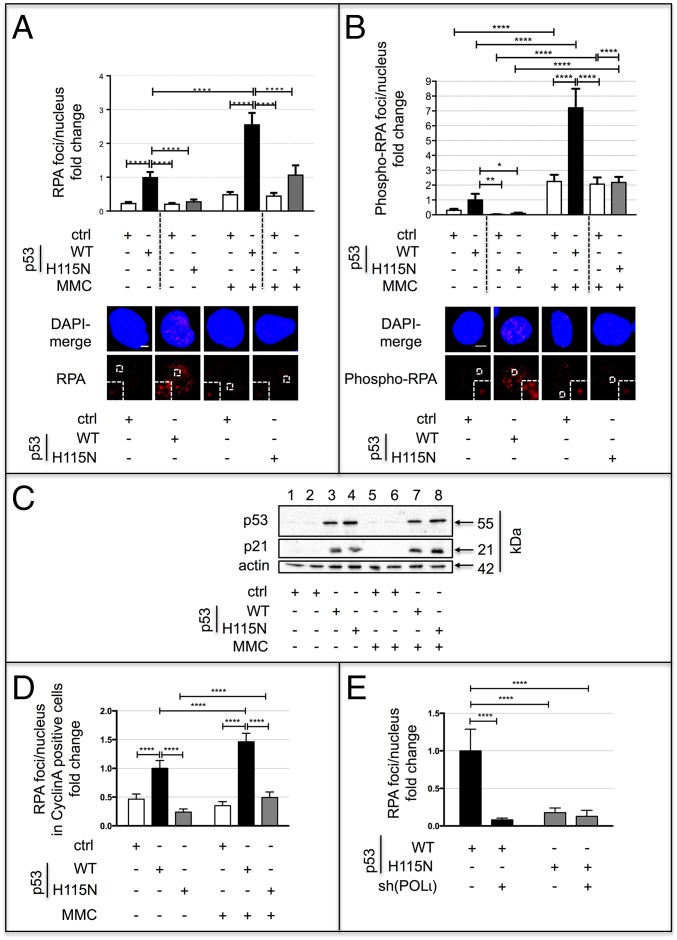
RPA foci reveal stretches of ssDNA exposed upon replication stress (23, 49). Using the tetracycline-controlled expression system in H1299 cells, we observed a fivefold increase of RPA foci per nucleus after expression of p53(WT) but not of p53(H115N) (Fig. 5A). Similarly, enforced replication blockage by MMC treatment was followed by a 2.7-fold increase in RPA foci for p53(WT), whereas there were no observable changes in p53-negative and p53(H115N)-expressing cells (Fig. 5A). These results were further strengthened by the analysis of phospho-S33-RPA foci accumulation, a marker of DNA replication lesions (50), which also supported a specific role of p53(WT) lost in p53(H115N)-expressing cells (Fig. 5B). Both p53 variants showed comparable p53 transcriptional activity revealed by their similar expression levels of p53 and p21 (Fig. 5C). Because K562 cells exhibit a p53(WT)-mediated recombination stimulation, we reexamined RPA foci formation in S- and/or G2-phase cells, defined by the expression of cyclin A (51). Consistently, we detected increased RPA foci numbers in p53(WT) compared with p53(H115N) or p53-negative cells before and after MMC treatment (Fig. 5D).
Fig. 5.
Effect of p53 and POLι on ssDNA accumulation. (A) Accumulation of RPA foci in H1299-cell clones. H1299 cells, controlled with tetracycline, expressing or not expressing either p53(WT) or p53(H115N) were mock-treated or MMC-treated (3 μM, 45 min, 3-h release) and processed for the detection of RPA foci accumulation. (Upper) Number of RPA foci per nucleus was quantified and expressed as fold changes. (Lower) Representative images with 2.5-fold enlarged magnifications (Insets) of highlighted regions for MMC-treatment are shown. Mean values for p53(WT)-expressing cells after mock treatment were defined as 1 (on average, eight foci per nucleus). Bars indicate SEM. Stippled lines separate individual cell clones with and without tetracycline treatment for suppression (−) and release (+) of p53 [p53(WT) and p53(H115N)] expression, respectively. (Scale bar: 5 μm.) (B) RPA-phospho-Ser33 focal accumulation. H1299-cell clones expressing or not expressing p53(WT) or p53(H115N) were subjected to mock or MMC treatment (3 μM, 45 min, 3-h release). Samples were inspected for phospho-RPA foci accumulation as in A. Mean values for p53(WT)-expressing cells after mock treatment were defined as 1 (on average, six foci per nucleus). Bars indicate SEM. Stippled lines separate cell clones with and without tetracycline treatment for suppression (−) and release (+) of p53 expression. (Scale bar: 5 μm.) (C) p53 protein levels in tetracycline-regulated H1299-cell clones. H1299-cell clones were treated with or without tetracycline for suppression of p53 expression [lanes 1 and 5, p53(WT) clone; lanes 2 and 6, p53(H115N) clone] and release of p53 expression [lanes 3 and 7, p53(WT) clone; lanes 4 and 8, p53(H115N) clone], respectively. After mock or MMC treatment (3 μM, 45 min, 3-h release), cells were lysed and subjected to immunoblotting to visualize p53 and p21 protein levels. α-Actin served as a loading control. (D) RPA foci analysis in K562 cells. K562 cells transfected with p53(WT) or p53(H115N) expression vector were mock-treated or MMC-treated (3 μM, 45 min, 3-h release) and processed for immunofluorescence-based microscopy to visualize RPA foci, which were quantified in cyclin A-costained cells. Mean values for p53(WT)-expressing cells after mock treatment were defined as 1 (on average, 14 foci per nucleus). (E) RPA foci formation after down-regulation of POLι. K562 cells were transfected with shRNA plasmid specific for POLι [sh(POLι)] and either p53(WT) or p53(H115N) expression plasmids, followed by MMC treatment (3 μM, 45 min, and 3-h release). Mean values for p53(WT)-expressing cells were defined as 1 (on average, nine foci per nucleus). ****P < 0.0001; **P < 0.01; *P < 0.05.
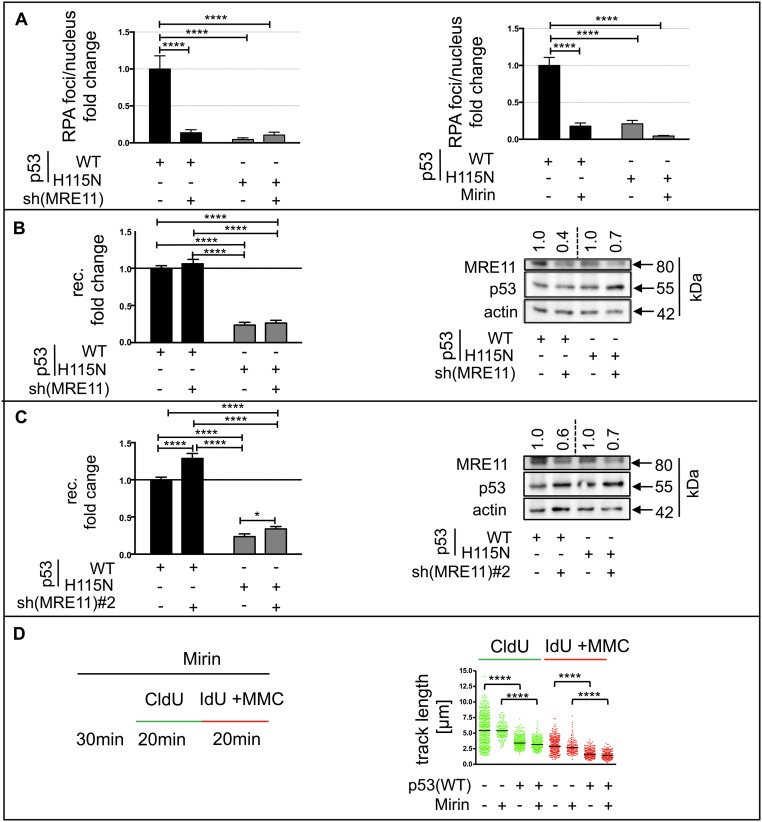
The increased p53–POLι interaction after DNA cross-linking (Fig. 4A) suggested a potential role of POLι in RPA accumulation after MMC treatment. Strikingly, silencing of POLι abrogated the p53(WT)-mediated RPA foci formation (Fig. 5E). Therefore, we investigated the involvement of other nucleases in ssDNA formation. Silencing the nucleases WRN and EXO1 did not affect RPA foci accumulation in p53(WT) cells (Fig. S4 B and C). Silencing BLM, the partner of the endonuclease DNA2 (52), or HLTF, which is involved in p53(WT)-mediated recombination, also did not alter RPA foci accumulation in p53(WT) cells (Fig. S4 D and E). However, silencing MRE11 or inhibiting its 3′–5′ exonuclease activity with the MRE11 exonuclease inhibitor Mirin (53) was sufficient to reduce RPA foci accumulation to a level similar to the level found in p53(H115N) cells (Fig. S5A). This result was intriguing, because MRE11 mediates DNA end resection at stalled replication forks (54). Notably, although p53(WT)-dependent RPA foci accumulation required the exonuclease activity of MRE11, stimulation of recombination in the reporter assay was not modulated by MRE11 down-regulation, as shown for two shRNAs (Fig. S5 B and C). These results revealed that MRE11 creates RPA-coated ssDNA stretches in concert with p53(WT) and POLι, although it does not contribute to p53(WT)- and POLι-dependent replication-associated recombination.
Fig. S5.
Effect of the MRE11 status on the p53-mediated RPA-foci accumulation, recombination efficiency, and DNA elongation rate. (A, Left) RPA-foci formation. K562 cells were transfected with p53(WT) or p53(H115N) expression plasmid and the shRNA plasmid specific for MRE11 [sh(MRE11)]. Forty-eight hours later, cells were treated with MMC (3 μM, 45 min) and processed for immunofluorescence microscopy to visualize RPA-foci accumulation 3 h later. (A, Right) For the catalytic inhibition of MRE11-exonuclease activity, cells were treated with Mirin (100 μM) starting 30 min before MMC treatment. Bars indicate SEM. (B and C) p53(WT)-mediated recombination. (Left) Experimental setup and analysis of recombination fold changes were as described in the legend for Fig. 2, except that cells were cotransfected with two different shRNA plasmids for down-regulation of MRE11 [sh(MRE11) in B; sh(MRE11)#2 in C]. Data were obtained from 18 measurements. (Right) To verify MRE11 down-regulation, immunoblotting was performed, and results are shown. Bars indicate SEM. (D) Track length distribution of CldU/IdU-labeled cells. DNA fiber-spreading assay was performed in H1299(p53WT) cells in the presence or absence of tetracycline for suppression (−) and release (+) of p53(WT) expression. For inhibition of MRE11-exonuclease activity, Mirin was added 30 min before and was kept throughout the whole length of the experiment. During IdU labeling, cells were treated with MMC (3 μM). Experimental setup and calculation of statistically significant differences were as described in the legend for Fig. 6. Mean values were calculated by measurement of ≥171 single DNA fibers. ****P < 0.0001; *P < 0.05.
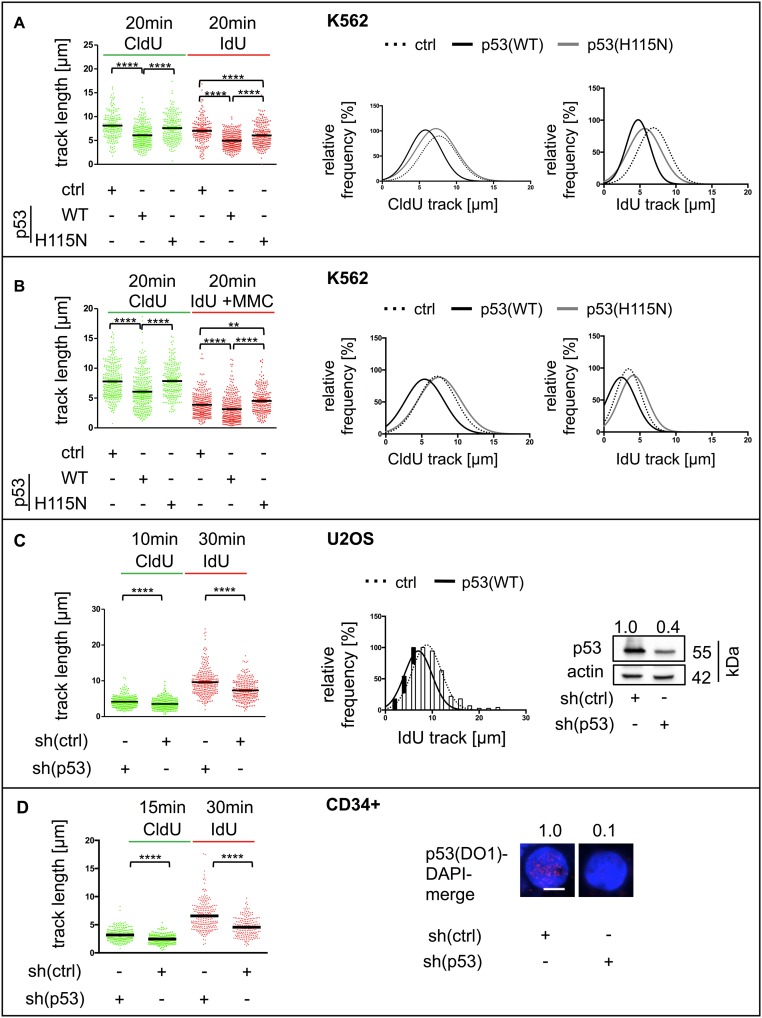
p53(WT), but Not p53(H115N), Restrains DNA Elongation in a POLι-Dependent Manner.
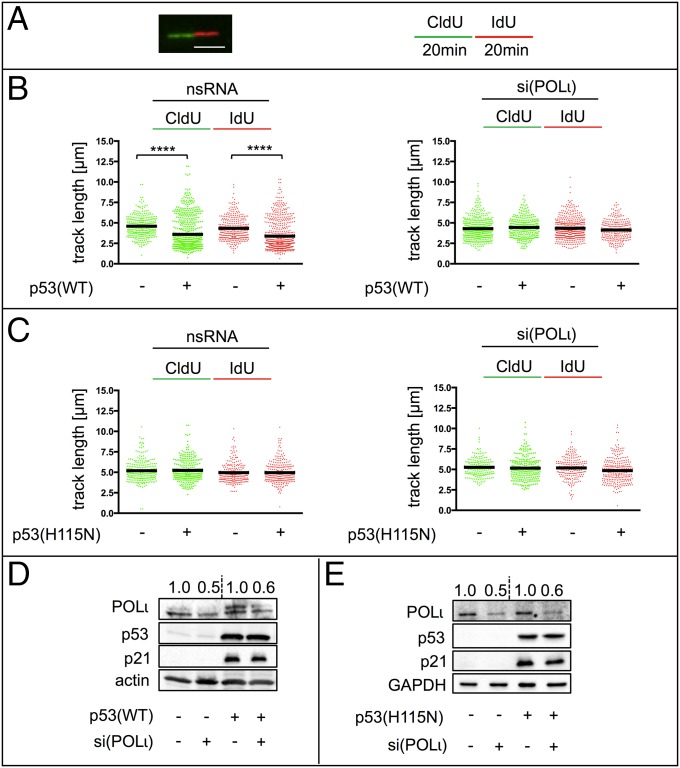
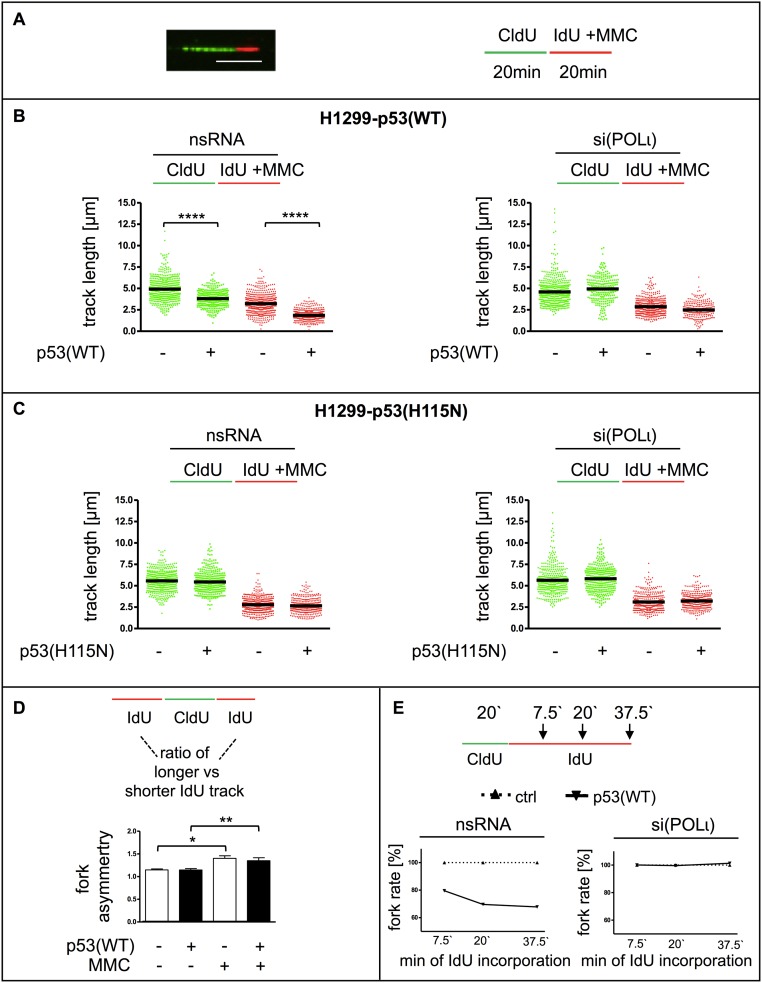
WT p53, together with POLι, promoted stimulation of replication-dependent recombination and RPA foci accumulation, suggesting a role for p53 and POLι in the replication process itself. To measure the elongation rate of replication directly, DNA fiber assays (55, 56) were applied to H1299 cells (Fig. 6 and Fig. S6) and K562 cells (Fig. S7 A and B). p53(WT) expression persistently led to a decrease in the length of the two DNA tracks resulting from subsequent incorporation of 5-chloro-2-deoxyuridine (CldU) and 5-iodo-2-deoxyuridine (IdU). This track shortening can be interpreted as exonuclease-mediated DNA degradation, increased replication stalling, and/or a continuous deceleration of the replication elongation speed. After MMC treatment of H1299 cells, the IdU track length was also shortened in p53(WT) cells, but not in p53(H115N) cells (Fig. S6 B and C). Mean replication fork rates calculated from track lengths in time-course experiments were 0.6 kb⋅min−1 or 0.5 kb⋅min−1 in control cells and 0.5 kb⋅min−1 or 0.4 kb⋅min−1 in p53(WT) cells after mock and MMC treatment, respectively. On average, this rate reduction suggests a p53(WT)-mediated track shortening of 120 nucleotides per minute (±27) and 143 nucleotides per minute (±13), respectively. Track length shortening was not detected in p53(H115N)-expressing cells (Fig. 6C and Fig. S7B). If the track shortening depends on fork stalling, the two tracks originating from the same point are differentially affected, leading to a difference in track length (57). Thus, stalling leads to an increase of the ratio of the two IdU track lengths called “fork asymmetry.” As expected, MMC treatment significantly increased fork asymmetry (Fig. S6D). Nevertheless, fork symmetry ratios in p53(WT) did not differ from fork symmetry ratios in control cells before or after MMC treatment. Hence, track length shortening after p53(WT) expression was not associated with stalling, and rather supports a continuous role for p53 (lost in the H115N mutant) on the synthesis of nascent DNA. These findings were cell type-independent, because track lengths expressing p53(WT) in K562 cells were also shorter compared with cells expressing p53(H115N) or p53-negative controls independent of MMC treatment (Fig. S7 A and B). Remarkably, we also observed similar results in U2OS cells (Fig. S7C) and in cycling primary human cord blood-derived hematopoietic stem and progenitor (CD34+) cells after silencing of endogenous p53 (Fig. S7D). In both of those cellular models, after 30 min of IdU incorporation, control samples elongated significantly less than p53-depleted samples. It is unclear to us whether p53 triggers a reduction in the synthesis of nascent DNA at random positions or if it performs a more continuous task. However, if we accept the second scenario, the impact of p53(WT) provides an average decrease of 171 nucleotides per minute (±85) in U2OS cells (Fig. S7C) and 165 nucleotides per minute (±41) in primary samples (Fig. S7D).
Fig. 6.
p53 modulates nascent DNA elongation. A DNA fiber-spreading assay was performed in H1299-cell clones inducibly expressing p53(WT) or p53(H115N), which had been transfected with nonspecific RNA [nsRNA; B (Left) and C (Left)] or siRNA specific for POLι [si(POLι); B (Right) and C (Right)] 48 h previously. Mean values were calculated by measuring fiber track lengths of ≥250 single fibers in two independent experiments [Left, 5-chloro-2-deoxyuridine (CldU); Right, IdU]. Statistically significant differences between p53-negative control cells and p53-expressing cells were calculated using Dunn’s test. (A) Representative fiber image and a schematic overview illustrate the technical procedure. (Scale bar: 5 μm.) (B) H1299-cell clone without and with p53(WT) expression. ****P < 0.0001. (C) H1299-cell clone without and with p53(H115N) expression. (D) POLι, p53, and p21 protein levels. Knockdown of POLι in H1299 cells without and with p53(WT) expression was examined by Western blot analysis. α-Actin served as a loading control. (E) POLι, p53, and p21 protein levels. Knockdown of POLι in H1299 cells without and with p53(H115N) expression was examined by Western blot analysis. GAPDH served as a loading control.
Fig. S6.
p53 modulates nascent DNA elongation after MMC treatment. A DNA fiber-spreading assay was performed in H1299 cells without or with tetracycline-regulated expression of p53(WT) or p53(H115N) and 48 h after transfection with nonspecific RNA (nsRNA; B, Left and C, Left) or siRNA specific for POLι [si(POLι), B, Right and C, Right]. Mean values and SEM were obtained by measuring fiber track lengths of ≥250 single fibers in two independent experiments (CldU, Left; IdU, Right). Statistically significant differences between p53-negative control cells and p53-expressing cells were calculated using Dunn’s test. (A) Representative fiber image and a schematic overview illustrate the technical procedure. (Scale bar: 5 μm.) (B) H1299-cell clone without and with p53(WT) expression. (C) H1299-cell clone without and with p53(H115N) expression. (D) Graphic presentation of fork asymmetry in H1299 cells. DNA fibers from the experiment in B and Fig. 6 were reanalyzed regarding fork asymmetry, comparing track lengths of IdU incorporation (red) departing from the same origin (green, CldU track). (Upper) Schematic overview. (Lower) Graph shows the ratio of longer track versus shorter track lengths. (E) Graphic presentation of relative FRs calculated from total track lengths of time-course experiments after MMC treatment in H1299 cells. The mean p53-negative control value for each time point was set to 100% [corresponding to 0.57 kb⋅min−1, 0.53 kb⋅min−1, and 0.47 kb⋅min−1 in nsRNA-transfected cells and 0.59 kb⋅min−1, 0.48 kb⋅min−1, and 0.49 kb⋅min−1 in si(POLɩ)-transfected cells for sample retrieval after 7.5 min, 20 min, and 37.5 min, respectively].
Fig. S7.
DNA fiber-spreading experiments in K562, U2OS, and CD34+ cells. (A) DNA fiber-spreading analysis in K562 cells after mock treatment. Cells were transfected with expression plasmids for p53(WT), p53(H115N), or empty vector. (Left) Graphic presentation shows track lengths obtained from ≥275 fibers (n = 3). Bars indicate SEM. (Right) Corresponding lengths of nascent DNA replication tracks plotted as the distribution of relative track length frequency, whereby the maximum value of the bin center of each dataset was defined as 100%. (B) DNA fiber-spreading analysis in K562 cells after MMC treatment. The experimental procedure and graphic presentation were the same as used in A, except that cells were MMC-treated during IdU labeling. (C) DNA fiber-spreading analysis as a function of endogenous p53 in U2OS cells. Cells transfected with p53 shRNA plasmid [sh(p53)] or empty vector [sh(ctrl)] were subjected to DNA fiber-spreading analysis. Mean values from ≥293 fibers (n = 3). Bars indicate SEM. (Center) Relative IdU track length frequency is shown, whereby the maximum value of the bin center of the dataset was set to 100%. (Right) p53 knockdown was verified by Western blot. (D, Left) DNA fiber-spreading analysis as a function of endogenous p53 in CD34+ cells. Cycling C34+ human hematopoietic stem and progenitor cells, nucleofected with p53 shRNA plasmid or empty vector were subjected to DNA fiber-spreading analysis. Mean values from ≥198 fibers obtained from three cord blood-derived samples. (Right) p53 knockdown was verified via immunofluorescence signal intensities per 50 nuclei each (i.e., mAb DO1-positivity in 50 DAPI-stained nuclei). Representative images are shown. (Scale bars: 5 μm.) ****P < 0.0001; **P < 0.01.
Strikingly, after down-regulating POLι with specific siRNAs, the differences in track lengths and fork rates between p53(WT)-expressing and p53-negative H1299 cells were lost (Fig. 6B and Fig. S6 B and E). Because the accumulation of RPA foci in p53(WT) cells after MMC treatment showed a dependency on the catalytic activity of MRE11, we also performed DNA fiber-spreading assays in the presence of Mirin in H1299 cells. Surprisingly, Mirin treatment did not cause any increase in track lengths in either controls or p53(WT) cells (Fig. S5D), thus matching the lack of effect in recombination measurements (Fig. S5 B and C). Altogether, these data demonstrate that exogenously and endogenously expressed, exonuclease-proficient p53(WT) decreased replication elongation through a mechanism other than fork stalling. Although TLS-POLι was necessary for p53(WT)-mediated recombination, the shortening of nascent DNA replication tracks, and RPA foci accumulation, the nuclease MRE11 was only required for the last function and dispensable for the first two functions of p53(WT).
Discussion
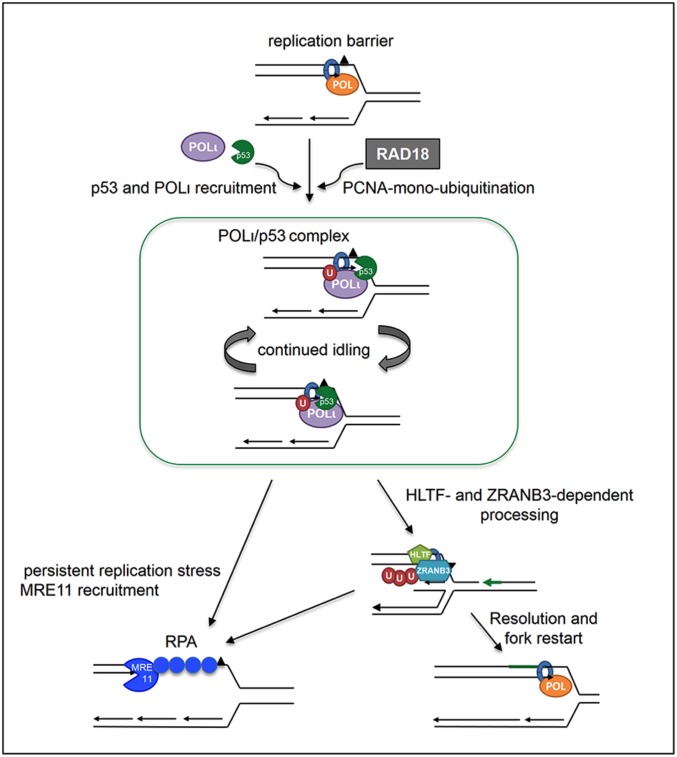
Our results suggest that the role of p53 in genome stabilization under both normal and stressed conditions may not only involve the control of cell cycle entrance and the apoptotic decision but might be a direct contribution to the maintenance of optimal rates of nascent DNA elongation and replication-associated recombination. Because p53(H115N) is not transcriptionally impaired, this function of p53 is clearly independent of its positive effect on its transcriptional targets. At this point, it is, however, important to emphasize that the p53(H115N) mutant was reported to have a slightly increased p53 transcriptional activity compared with p53(WT). Notably, in our experimental setting, p53(H115N) was not transcriptionally superior to p53(WT) in inducing p21 (Figs. 1B and 5C and Fig. S1C) Moreover, p53(H115N) was reported to have an enhanced capacity to bind DNA (27), a feature that may also contribute to the phenotype described in this report. As a prominent feature, the exonuclease activity of p53 is reduced by 85% in the p53(H115N) mutant, whereas the modifications in the transcriptional activity and DNA binding are much more modest. Therefore, we speculate that such an exonuclease activity, first described 20 y ago (2) and confirmed by several groups (3, 4, 18, 27, 58), may be implicated in the replication phenotype revealed in this study. In particular, we show that cells expressing the exonuclease-deficient but transcriptionally proficient mutant p53(H115N) do not exhibit the ability of p53(WT)-expressing cells to stimulate recombination in reporter assays and to modulate nascent DNA elongation in vivo. Collectively, our data suggest that a DNA damage tolerance against replication-blocking lesions can be achieved by the concerted action of RAD18, an exonuclease-proficient p53(WT)–POLι complex, and the fork-reactivating abilities of HLTF and ZRANB3 (Fig. 7). Notably, our work elucidates a new role of p53(WT), together with POLι, an extremely error-prone and highly enigmatic POL in humans (59) without paralogs in bacteria, yeast, or nematodes (48).
Fig. 7.
Model for p53-mediated resolution of replication barriers. When encountering replication barriers, the replication machinery stops, triggering PCNA monoubiquitination (U) and recruitment of p53, which is followed by POLι. The p53–POLι complexes favor continued idling, leading to polyubiquitination of PCNA (chain of U’s) via HLTF; subsequently, to error-free resolution/bypass via HLTF and ZRANB3; and, finally, to replication restart. Current models of the ZRANB3-mediated DNA damage tolerance pathway (63, 67) suggest that the structure-specific endonuclease of ZRANB3 introduces a nick into the unreplicated template strand ahead of the fork, which serves as a primer end for displacement DNA synthesis (green arrow). Concomitantly, ZRANB3, together with HLTF, promotes fork reversal. As a result, the replication-blocking lesion is replaced by a patch of newly synthesized DNA (green line), thus permitting unrestricted progression of the restarted fork. Persistent replication stress can alternatively lead to MRE11-dependent ssDNA formation, which is coated by RPA.
p53 has previously been reported to confer resistance to replication stress via PARP inhibition (14) and to stimulate recombination events during S-phase (13). Here, we demonstrated that p53(WT) cells were protected against replication-blocking MMC treatment, whereas p53(H115N) cells were not. Because only p53(WT) stimulated replication-associated recombination, we hypothesized that the exonuclease activities of p53 promoted resolution of replication lesions. Alternative explanations, such as cell cycle changes, are unlikely because p53(H115N) represents a true separation-of-function mutant with loss of exonuclease but not of cell cycle-regulatory activities (27) (Fig. S1 A and D).
p53-Mediated Recombination Engages the PCNA Switchboard.
Remarkably, screening targets for genetic interactions with p53(WT) in the recombination reporter assay excluded the involvement of RAD51, BRCA2, and the HR antagonist PARI, which all act downstream of PCNA SUMOylation (34). Alternative routes mediating a replicative lesion bypass are known to be triggered by PCNA ubiquitination, namely, TLS or template switching. The latter can further be subdivided into strand invasion or fork reversal mechanisms (36, 60). The stimulation of recombination by p53(WT) was fully epistatic with the E3-ubiquitin ligase RAD18, which monoubiquitinates PCNA in conjunction with the E2-conjugating enzyme RAD6 (60), and with POLι, which recognizes monoubiquitinated PCNA (48). Intriguingly, it has previously been speculated that p53 is required for efficient, UV-induced PCNA monoubiquitination (39, 61). The p53 effect on replication-associated recombination was partially epistatic [residual effect in cells with p53(H115N)] with the Rad5 functional homolog HLTF, which, in conjunction with UBC13/UEV1, polyubiquitinates PCNA (42), and with the translocase ZRANB3, which binds polyubiquitinated PCNA and stabilizes replication forks (45). Intriguingly, HLTF and ZRANB3 may also support a RAD51-independent mechanism of lesion bypass. Both enzymes have been reported to be able to create and resolve HR intermediates such as D-loops independent of RAD51, which may provide primers for the repair of gaps generated during replication of damaged DNA (62, 63). A major function of HLTF appears to be the promotion of fork reversal upon replication block (43, 64, 65). ZRANB3, a SWI/SNF catalytic subunit (SNF2) DNA translocase like HLTF, has been proposed to cooperate with HLTF in the remodeling of the blocked fork, additionally contributing a structure-specific endonuclease for the fork remodeling (45, 66, 67).
PCNA ubiquitination has been described to mediate a switch of POLs and to induce TLS (38). We noticed complete dependency of p53-mediated recombination with one specific TLS-POL, namely POLι. The observed moderate influences of TLS-POLκ and the TLS-POLζ catalytic subunit REV3L could be explained by functional overlap and cooperation with TLS-POLι, respectively (48, 68). Because p53(WT), but not p53(H115N), facilitated the accumulation of POLι-foci and, conversely, POLι silencing impaired accumulation of p53pSer15 foci in cross-linker–treated cells expressing exonuclease-proficient p53, we propose exonuclease-dependent stabilization of a p53–POLι complex at replication lesions. The identification of a feature in p53 that allows its physical and functional interaction with DNA POLs is not unprecedented (18, 69–71).
p53 and POLι Allow Damage Bypass via HLTF and ZRANB3.
An epistasis analysis thus indicates that p53(WT) and Polι represent one branch of the replication stress response pathway (Fig. 7). This pathway is initiated by Rad6/Rad18-dependent monoubiquitination of PCNA, commonly followed by recruitment of a suitable TLS-POL (72–74). Given that p53-induced deceleration of DNA replication was fully dependent on POLι and the exonuclease-proficient p53(WT), we propose exonuclease-dependent idling by the p53–POLι complex, leading to accumulation of POLι at replication lesions, which thus is dependent on p53’s exonuclease activity and, ultimately, stabilizing the complex (5, 75). In the literature, “idling” is described as a function achieved by some DNA POLs, where the exonuclease activity removes the same base that is preferentially incorporated (5). Idling may also act as a kinetic boundary to TLS, preventing stable incorporation of bases opposite DNA lesions (5). TLS-POLs do not possess an intrinsic exonuclease activity (48, 74, 76), and p53 might provide the missing exonuclease. The p53–POLι idling complex would transiently stabilize the fork at replication barriers and may lead to the observed replication slowdown. The persistent p53- and POLι-driven idling events might also prevent RAD51-dependent recombination (8–10), which is further blocked by HLTF-dependent PCNA polyubiquitination. Because POLι-foci accumulation was unaffected by HLTF silencing, POLι acts upstream of PCNA polyubiquitination in cells with p53(WT). PCNA polyubiquitination may ultimately lead to ZRANB3 recruitment for the successful bypass of replication barriers and fork restart (63, 66, 67). Notably, ZRANB3 possesses a unique, structure-specific endonuclease activity, which is able to incise the DNA 5′ of a blocking lesion on the leading strand template. In this way, an accessible 3′-OH group is generated that can serve as a primer to displace the lesion on the leading strand template (67) (Fig. 7). ZRANB3, together with HLTF, thus facilitates fork reversal, damage bypass, and replication restart (43, 62, 67). The same mechanism is also suitable to explain the recombination-dependent, but RAD51-independent, recombination events observed (Figs. 1 and 2 and Fig. S2).
Further clues to the hierarchy of events come from data on RPA foci. Accumulation of RPA foci required p53(WT) and POLι, and depended on the exonuclease activity of MRE11. Conversely, MRE11 was required for neither p53-induced replication slowdown nor increased recombination. Therefore, MRE11 is involved in neither p53-dependent idling nor the mechanism causing recombination. Therefore, exonucleolytic attack by MRE11 may be ultimately triggered by a persistent replication block that cannot be resolved by HLTF/ZRANB3 (77).
All in all, we propose that exonuclease-proficient p53(WT) resolves replication barriers via continued idling in complex with POLι, which allows PCNA polyubiquitination and damage bypass by HLTF and ZRANB3. The proposed mechanism shows how p53 stimulates spontaneous recombination events during S-phase or after DNA cross-linking, and may explain how it protects rapidly growing cells, such as cancer cells (14) or hematopoietic stem cells (78), directly against replicative stress.
Materials and Methods
Additional experimental details are provided in SI Materials and Methods.
Cell Survival Assay.
For assessment of cell viabilities, the colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used (14). The assay was performed 48 h after 45 min of MMC treatment (1–1,000 μM). Details are provided in SI Materials and Methods.
Recombination Measurements.
K562 or WTK1 cells with chromosomally integrated recombination substrate [i.e., K562(HR-EGFP/3′EGFP), WTK1(HR-EGFP/3′EGFP-SV40)] (14, 25) were cotransfected with p53 expression plasmids or shRNA plasmids as detailed in the figure legends. Recombination frequencies were measured as described (13, 14) and are detailed in SI Materials and Methods.
Coimmunoprecipitation and Expression Analysis.
K562 cells were transfected with expression plasmids, and immunoprecipitation was performed using a mixture of mAbs Pab421 and DO1 (Calbiochem) directed against p53 as described (13, 17) and detailed in SI Materials and Methods. Western blot analysis and quantitative real-time PCR were performed as described in SI Materials and Methods.
DNA Fiber-Spreading Assay.
The DNA fiber assay was performed as described by Speroni et al. (56) and is detailed in SI Materials and Methods. Human CD34+ hematopoietic stem and progenitor cells were obtained from cord blood samples [approval by the Ethics Committee of Ulm University (no.155/13)] and cultured as described. Informed consent was obtained from mothers before or after having delivered a child within 24 h after birth, informing them about the type of investigations planned with the cord blood as well as about the absence of any risk for the child.
Immunofluorescence Staining.
H1299 cells were grown on coverslips, whereas K562 and human hematopoietic stem and progenitor cells were spun onto cytospin glass slides. Cells were fixed at indicated time points after MMC treatment, followed by processing for immunofluorescence microscopy as detailed in SI Materials and Methods.
In Situ PLA.
The in situ PLA was carried according to the manufacturer’s instruction (DUO92102; Sigma). Details are provided in SI Materials and Methods.
Plasmids, siRNA, and Transfection.
Plasmids, siRNA, and transfection methods used in this study are described in SI Materials and Methods.
Statistics.
Graphic presentation of data was performed using GraphPad Prism 6.0f software. For calculation of statistically significant differences, the Kruskal–Wallis test (Dunn’s multiple comparison test), two-tailed Mann–Whitney U test, and/or extra sum-of-squares F test was used (****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05). Details are provided in SI Materials and Methods.
SI Materials and Methods
Cell Culture and Drug Treatment.
K562, K562(HR-EGFP/3′EGFP), and WTK1(HR-EGFP/3′EGFP-SV40) cells were cultivated in RPMI 1640 medium (Gibco) supplemented with 13% (vol/vol) FBS (Merck Millipore), and 1.3% (vol/vol) penicillin-streptomycin-glutamine (Gibco). Human CD34+ hematopoietic stem and progenitor cells were isolated from cord blood samples and cultured as described (79). Human non–small-cell lung cancer cell lines H1299(p53WT) and H1299(p53H115N) contain a chromosomally integrated Tet-off system for controlled expression of either p53(WT) or p53(H115N), and they were cultivated in DMEM (Gibco) and supplemented with 10% (vol/vol) FCS, puromycin (2 μg/mL), G418 (300 μg/mL), and tetracycline (4.5 μg/mL) (27). Before starting experiments, cells were washed two times with medium, split, and cultivated in medium without tetracycline for induction of p53 expression and in medium with tetracycline for suppression of p53 expression in control cells, respectively. Twenty-four hours later, cells were washed again and reincubated in medium without or with tetracycline for an additional 24 h. Experiments were then performed. For DNA cross-linker treatment, cells were washed, incubated with MMC (Sigma) containing medium at a final concentration of 3 μM or mock-treated with the solvent H2O for 45 min, washed, and reincubated with fresh medium for an additional 3 h. For the catalytic inhibition of MRE11-exonuclease activity, cells were treated with Mirin (Tocris) at a final concentration of 100 μM or mock-treated with the solvent DMSO starting 30 min before the experiment, and Mirin was kept throughout the whole length of the experiment.
Cell Survival Assay.
For assessment of cell viabilities, the colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay was used. The assay was performed 48 h after MMC treatment or 6 d after irradiation. Cells were incubated in 0.5 mg/mL MTT for 3.5 h and lysed in 0.04 M HCl diluted in isopropanol, and light absorbance was measured at 570 nM with an ELISA reader. Each dataset was corrected for mock treatment: MMC value/H2O value × 100. Cell viability curves were plotted, and IC50 values were calculated using GraphPad Prism 6.0f software. Statistically significant differences between IC50 values of p53-expressing cell lines and control cell lines were calculated using an extra sum-of-squares F test of log IC50 values. For graphic presentation, mean IC50 values and SEM from the independent experiments were shown as columns.
Cell Cycle Distribution.
For the analysis of the distribution in cell cycle phases, K562 cells were collected by centrifugation and H1299 cells were trypsinized; both cell types were washed once with PBS, resuspended with 0.5 mL of PBS, fixed drop-wise in 4.5 mL of fixing solution (1:1 mixture of acetone and 80% (vol/vol) ethanol, stored at −20 °C) while mixing gently, and kept on ice for 15 min. Fixed cells were washed twice with ice-cold PBS, resuspended in 200 mL of propidium iodide staining solution [freshly added 50 μg/mL RNase A, 50 μg/mL propidium iodide (Sigma–Aldrich) in PBS], and incubated for 30 min in the dark. After diluting the suspension with 100 mL of PBS with 0.2% EDTA, the stained cells were analyzed in a FACSCalibur flow cytometer (BD Biosciences).
Recombination Measurements.
To measure recombination frequencies, K562 or WTK1 cells with chromosomally integrated recombination substrate [K562(HR-EGFP/3′EGFP) and WTK1(HR-EGFP/3′EGFP-SV40, respectively] (14, 25) were cotransfected via electroporation with expression plasmids for p53(WT), p53(H115N), or empty vector (ctrl), and with shRNA plasmids for silencing various targets as detailed in the figure legends. Cells were transfected in a total of 20–40 μg of plasmid DNA [10 μg of pBS, 10 μg of empty vector or expression plasmid for p53(WT) or p53(H115N), and 10–20 μg of shRNA plasmid or empty vector in silencing experiments]. Each measurement was paralleled by the same cotransfection, including a WT EGFP control plasmid instead of pBS plasmid for determination of transfection efficiency. Recombination frequencies were measured 72 h after transfection by quantification of 1 million cells from EGFP+ cells within the life cell population [side scatter (SSC)/forward scatter (FSC) gate], and were individually corrected for transfection efficiencies. Mean values of recombination frequencies of mock-treated p53(WT)-expressing cells were set to 1 (absolute mean frequencies were 2 × 10−5 for K562 cells and 4 × 10−5 for WTK1 cells). For experiments with DNA cross-linker treatment, cells were treated for 45 min with MMC (3 μM) 48 h after electroporation, washed, and reincubated in fresh medium for an additional 24 h. Flow cytometric analysis of EGFP+ cells was then performed.
Coimmunoprecipitation.
K562 cells were transfected with control EGFP expression plasmid pEGFPN1 (ctrl-GFP) or expression plasmid for p53-EGFP fusion protein (p53-GFP). Forty-eight hours after transfection, immunoprecipitation was performed. Cells were lysed in 50 mM Tris (pH 8), 150 mM NaCl, 1% Nonidet P-40, and complete protease inhibitor (Roche). p53 and a mixture of murine mAb Pab421 (Calbiochem) and murine mAb DO1 (Calbiochem) were used for immunoprecipitation; as a control, mouse IgG fraction (Santa Cruz Biotechnology) was used. Protein extract and Protein G Sepharose (PGS) were rotated overnight at 4 °C to remove components unspecifically binding to PGS. In parallel, antibody/PGS mixtures were rotated at 4 °C. Afterward, protein extracts were separated from PGS by centrifugation and the supernatants were transferred to the antibody/PGS mixtures, followed by rotation at 4 °C for an additional 4 h. After spin-down, precipitated proteins were washed five times with lysis buffer and dissolved in SDS/PAGE sample buffer.
Western Blot Analysis.
For Western blot analysis, protein extracts were separated electrophoretically and transferred to membranes, and proteins were immunodetected via chemiluminescence as described (14). For immunostaining, the following antibodies were used: anti-BLM (rabbit, polyclonal; ab476, Abcam), anti-BRCA2 (mouse; mAb Ab-1, Calbiochem), anti-EXO1 (rabbit, polyclonal; GTX109891, GeneTex), anti–HA-tag (mouse; mAb peroxidase-conjugated 3F10, Roche), anti-HLTF (rabbit, polyclonal; A300-229A, Bethyl), anti-MRE11 (rabbit, polyclonal; NB100-142, Novus), anti-p21 (mouse; mAb SX118, BD Biosciences), anti-p53 (mouse; mAb DO1, Calbiochem), anti-PARI (rabbit, polyclonal; ab107308, Abcam), anti-PCNA (mouse; mAb PC10, Abcam), anti-POLη (rabbit, polyclonal; NB100-60424, Novus), anti-POLι (rabbit, polyclonal; ab1324, Abcam), anti-RAD18 (rabbit, polyclonal; A301-340A, Bethyl), anti-RAD51 (rabbit, polyclonal; H-92, Santa-Cruz Biotechnology), anti-SMARCAL1 (rabbit, polyclonal; A301-616A, Bethyl), anti-WRN (rabbit, polyclonal; H-300, Santa Cruz Biotechnology), and anti-ZRANB3 (rabbit, polyclonal; A303-033A, Bethyl). To verify equal amounts of proteins, the membranes were prestained with Ponceau Red (Sigma) or reincubated with anti–α-actin antibody (goat, polyclonal; I-19, Santa Cruz Biotechnology) or anti-GAPDH (mouse mAb; Abcam). Chemiluminescence detection and quantification of protein levels were carried out in the linear range using ImageLab software on a ChemiDocMP System (BioRad). Values for the protein of interest were corrected with values of the loading control.
Quantitative Real-Time PCR.
For knockdown verification of TLS POLs REV3L and POLκ, quantitative real-time PCR (qRT-PCR) was performed. Total mRNA was isolated using the RNeasy Mini Kit (Qiagen) and reverse-transcribed with the QuantiTect Reverse Transcription Kit (Qiagen), and qRT-PCR was performed using the QuantiTect SYBR Green PCR Kit (Qiagen) on a Viia7 RUO thermocycler (Applied Biosystems). Gene-specific mRNAs were detected using QuantiTect primer assays: Hs.232021 (REV3L) and Hs.715657 (POLκ) (both from Qiagen). Hs_GAPDH_2_SG (Qiagen) was used as an internal control. The differences in target mRNA expression were calculated with the comparative cycle treshold (ΔΔCt) method.
DNA Fiber-Spreading Assay.
Cells were labeled with 5-chloro-2-deoxyuridine (CldU; Sigma) for the indicated times and washed twice with prewarmed PBS before labeling cells with IdU (Sigma) in a 10-fold higher concentration for the indicated times. Here, cells were either treated with MMC (3 μM) or H2O as a control. Cells were washed, harvested, and resuspended in PBS. A total of 2,500 cells were transferred to a slide; lysed with 6 μL of 0.5% SDS, 200 mM Tris⋅HCl (pH 7.4), and 50 mM EDTA; and incubated at room temperature for 6 min. Slides were tilted about 20° to allow DNA to spread via gravity, covered with aluminum foil, air-dried for 7 min, fixed for 5 min with 3:1 methanol/acetic acid (prepared fresh), air-dried for 7 min, and stored in 70% (vol/vol) ethanol at 4 °C overnight or directly afterward processed for denaturation/deproteination in 2.5 M HCl for 1 h, followed by immunofluorescence staining.
Immunofluorescence Staining.
Before fixing cells in 3.7% (vol/vol) formaldehyde in PBS, K562/hematopoietic stem cells and progenitor cells were spun onto cytospin glass slides, and adherently growing H1299 cells were grown on coverslips. Blocking unspecific binding sites was performed using 5% (vol/vol) goat serum in PBS for 1 h. Immunostaining for 1 h at 37 °C was performed with the primary antibodies: anti-cyclin A (rabbit, polyclonal; Santa-Cruz Biotechnology), anti-p53pSer15 (rabbit, polyclonal; Cell Signaling), anti-p53pSer15 16G8 (mouse, monoclonal; Cell Signaling), anti-p53 (mouse, mAb DO1; Calbiochem), anti-PCNA PC10 (mouse, mAb; PC10, Abcam), anti-POLι (rabbit, polyclonal; Abcam), anti–P-RPA32 S33 (rabbit, polyclonal; Bethyl), and anti-RPA ab-2 (mouse, monoclonal; Calbiochem). As secondary antibodies (incubation for 45 min at 37 °C), we used Alexa Fluor 555 (anti-mouse or anti-rabbit; Invitrogen) or Alexa Fluor 488 (anti-rabbit; Invitrogen). Immunofluorescence microscopy of nuclear signals was performed on an Olympus fluorescence microscope BX51 with a 100× oil immersion objective equipped with a cooled charge-coupled device camera (Colorview 12; Olympus) or on a BZ-9000 microscope (Keyence). Automated quantification of foci was carried out either with Cell^F imaging software, version 2.5 (Olympus Soft Imaging Solutions) or BZ-II Analyzer software. Intensity threshold and minimal focus size were maintained throughout one set of simultaneously treated and processed samples, when detecting single green or red foci.
Immunofluorescence staining after DNA fiber-spreading assays was performed by incubation with primary antibodies (1 h, room temperature): anti-BrdU (mouse, mAb; clone B44, 347580, BD BioSciences) for detection of IdU and anti-BrdU (rat, mAb; clone BU1/75, [ICR1] OBT0030, BioRad) for detection of CldU after blocking with 5% (wt/vol) BSA (45 min). As a secondary antibody, Alexa Fluor 555 (anti-mouse; Invitrogen) or Alexa Fluor 488 (anti-rat; Invitrogen) was used. DNA fibers were imaged with a BZ-9000 microscope or with a Zeiss Axioplan confocal microscope. Measurements of DNA fiber track lengths were carried out with BZ-II Analyzer software or using Zeiss LSM Image Browser software. Fork rate (FR) was calculated based on the length of the measured DNA fiber tracks using the following formula: FR (kb⋅min−1) = [(2.59 (kb⋅μm−1) × track length (μm)/pulse time (min)].
In Situ PLA.
K562 cells were transfected with empty vector or expression plasmids for p53(WT) or p53(H115N). The cells were spun onto cytospin glass slides and fixed with 3.7% (vol/vol) formaldehyde in PBS. Blocking was performed using 5% (vol/vol) goat serum in PBS for 1 h at room temperature. Cells were then double-stained with the primary antibodies anti-p53pSer15 (16G8, mouse, monoclonal; 9286S, Cell Signaling), anti-DNA POLι (rabbit, polyclonal; ab1324, Abcam) or anti-p53pSer15 (rabbit, polyclonal; Cell Signaling), and anti-PCNA (mouse, mAb; PC10, Abcam). PLA staining was then performed using a Duolink In Situ Orange Starter Kit Mouse/Rabbit (DUO92102; Sigma). Therefore, glass slides were stained with Duolink In Situ PLA Probe Anti-Rabbit PLUS [affinity-purified donkey anti-rabbit IgG (H+L); DUO92002, Sigma] and Duolink In Situ PLA Probe Anti-Mouse MINUS [affinity-purified donkey anti-mouse IgG (H+L); DUO92004, Sigma] for 1 h at 37 °C. After washing, the samples were incubated with the ligation-ligase solution for 30 min at 37 °C to hybridize oligonucleotides tagged on probes. After two short washing steps, the glass slides were incubated with the amplification POL solution for 100 min at 37 °C to amplify hybridized oligonucleotides and fluorescently label the amplification products. Then, glass slides were covered with coverslips using Duolink In Situ Mounting Medium with DAPI (DUO82040; Sigma). Imaging was performed using a BZ-9000 microscope. Automated quantification of PLA foci was carried out with BZ-II Analyzer software.
Plasmids and siRNA.
The following plasmids have been described previously: pSUPER-p53 for p53 silencing (14), pcDNA3.1-Rad51SM for expression of the RAD51SM chimera, pCMV-Wtp53 for expression of WT p53, pSUPER-WRN for silencing WRN (13), and pcDNA3-p53H115N for expression plasmid of p53(H115N) (27). For silencing other targets, the following gene-specific RNAi sequences were used for the design and cloning of the corresponding pSUPER-based shRNA-expressing plasmids:
shRNA(BRCA2): CAGTGGTATGTGGGAGTT
shRNA(BLM): catgagcgtttccaaagtc.
Alternatively, shRNA plasmids from Origene with the following shRNA sequences were used:
shRNA(RAD18): TTGGAACCTGACAGAGAAGAGGATTCTTC
shRNA(RAD18) no. 2: TGCGATGCTTTGCATCC TAAATCAGCTGC
shRNA(HLTF): ATGGAACCAGCTGAGGCTATTGAAACACC
shRNA(HLTF) no. 2: GTTTATTATGGTCCTGATCGTATTAGAGA
shRNA(ZRANB3): GTGTGAGACTCCTCAAGGCAGTGCTGTTA
shRNA(ZRANB3) no. 2: CAGCCAGGTAATGAACATGTGAAGAGTTC
shRNA(POLι): CAACTACTTCACGCTCTGGCAAGCACAGT
shRNA(POLι) no. 2: ATCCGTCGGTATTCCTCTGAGAAGCACTA
shRNA(POLη): GATGATGCTAAGAAGTTATGTCCAGATCT
shRNA(POLκ): AGCCATGCCAGGATTTATTGCTAAGAGGC
shRNA(REV3L): CAGTATCGACAGAGCACTTAATGTGGCTT
shRNA(SMARCAL1): TGACACCAAGACGTGGAACTTCAGCATGA, GGACTTCGCTACTGTGATGCCAAACGGAT
shRNA(c12orf48 = PARI): CCTAAGAGAACGCATCTGRGRGRCAATGC
shRNA(EXO1): CCAACTAACTCCAGAAGCGGAAGAGGATA, AGGTTGTGACTACCTGTCATCACTGCTGCTGT, TAAAGAGCGAGGAGTCCAGTCCAGTGACGATGAG;
shRNA(MRE11): GCCAGAGAAGCCTCTTGTACGACTGCGAG.
FlexiTube Gene-Solutions GS11201 siRNAs with pools of four target gene-specific siRNA duplexes were used to silence POLι (Qiagen). The control (nonsilencing) siRNA 1022076 (Qiagen) was used as a negative control.
Transfection.
HiPerFect (Qiagen) was used for siRNA transfection for silencing of POLι in H1299 cells. Plasmids were transiently introduced in K562, K562[HR-EGFP/3′EGFP], and WTK1(HR-EGFP/3′EGFP-SV40) via electroporation (GenePulser Xcell; BioRad) into U2OS cells via lipofection (jetPRIME; PolyPlus) and into human hematopoietic stem and progenitor cells via nucleofection according to the Amaxa protocol (Lonza) (13, 79).
Statistics.
Graphic presentation of data was performed using GraphPad Prism 6.0f software. Mean values ± SEM were calculated. For calculation of statistically significant differences in recombination measurements and foci accumulation experiments, the Mann–Whitney two-tailed test was used. For calculation of statistically significant differences in DNA fiber-spreading analysis, either the nonparametric Mann–Whitney two-tailed test or Kruskal–Wallis test (Dunn’s multiple comparison test) was used. Calculation of IC50 values in cell survival studies was based on the cell viability curves determined using GraphPad Prism 6.0f software. Statistically significant differences were calculated using an extra sum-of-squares F test comparing IC50 values using Graph PadPrism 6.0f software.
Acknowledgments
We thank Frank Grosse for extremely helpful discussions and expert advice regarding p53 and Veronika Winkelmann for experimental help. This work was supported by German Research Foundation (DFG) Grants Project A3 (PA3) in Research Training Group 1789 “Cellular and Molecular Mechanisms in Aging” (to L.W.) and Proyecto de investigación científica y/o tecnológica (PICT) 2013-1049 (to V.G.); a DFG (Graduate School of Molecular Medicine, Ulm University) PhD fellowship (to S.H.), a PhD fellowship from the State of Baden-Wurttemberg (to S.H.), and a Dr.med scholarship for Experimental Medicine (Ulm University) (to K.B.).
Footnotes
Conflict of interest statement: L.W. is an inventor of a patent on a test system for determining genotoxicities, which is owned by L.W.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605828113/-/DCSupplemental.
References
- 1.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358(6381):15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 2.Mummenbrauer T, et al. p53 protein exhibits 3′-to-5′ exonuclease activity. Cell. 1996;85(7):1089–1099. doi: 10.1016/s0092-8674(00)81309-4. [DOI] [PubMed] [Google Scholar]
- 3.Skalski V, Lin ZY, Choi BY, Brown KR. Substrate specificity of the p53-associated 3′-5′ exonuclease. Oncogene. 2000;19(29):3321–3329. doi: 10.1038/sj.onc.1203649. [DOI] [PubMed] [Google Scholar]
- 4.Bakhanashvili M. Exonucleolytic proofreading by p53 protein. Eur J Biochem. 2001;268(7):2047–2054. doi: 10.1046/j.1432-1327.2001.02075.x. [DOI] [PubMed] [Google Scholar]
- 5.Khare V, Eckert KA. The proofreading 3′-->5′ exonuclease activity of DNA polymerases: A kinetic barrier to translesion DNA synthesis. Mutat Res. 2002;510(1-2):45–54. doi: 10.1016/s0027-5107(02)00251-8. [DOI] [PubMed] [Google Scholar]
- 6.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11(3):208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 7.Heitzer E, Tomlinson I. Replicative DNA polymerase mutations in cancer. Curr Opin Genet Dev. 2014;24:107–113. doi: 10.1016/j.gde.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertrand P, Saintigny Y, Lopez BS. p53’s double life: Transactivation-independent repression of homologous recombination. Trends Genet. 2004;20(6):235–243. doi: 10.1016/j.tig.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Sengupta S, Harris CC. p53: Traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6(1):44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 10.Gatz SA, Wiesmüller L. p53 in recombination and repair. Cell Death Differ. 2006;13(6):1003–1016. doi: 10.1038/sj.cdd.4401903. [DOI] [PubMed] [Google Scholar]
- 11.Janz C, Süsse S, Wiesmüller L. p53 and recombination intermediates: Role of tetramerization at DNA junctions in complex formation and exonucleolytic degradation. Oncogene. 2002;21(14):2130–2140. doi: 10.1038/sj.onc.1205292. [DOI] [PubMed] [Google Scholar]
- 12.Linke SP, et al. p53 interacts with hRAD51 and hRAD54, and directly modulates homologous recombination. Cancer Res. 2003;63(10):2596–2605. [PubMed] [Google Scholar]
- 13.Restle A, et al. Dissecting the role of p53 phosphorylation in homologous recombination provides new clues for gain-of-function mutants. Nucleic Acids Res. 2008;36(16):5362–5375. doi: 10.1093/nar/gkn503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ireno IC, et al. Modulation of the poly (ADP-ribose) polymerase inhibitor response and DNA recombination in breast cancer cells by drugs affecting endogenous wild-type p53. Carcinogenesis. 2014;35(10):2273–2282. doi: 10.1093/carcin/bgu160. [DOI] [PubMed] [Google Scholar]
- 15.Zink D, Mayr C, Janz C, Wiesmüller L. Association of p53 and MSH2 with recombinative repair complexes during S phase. Oncogene. 2002;21(31):4788–4800. doi: 10.1038/sj.onc.1205614. [DOI] [PubMed] [Google Scholar]
- 16.Sengupta S, et al. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 2003;22(5):1210–1222. doi: 10.1093/emboj/cdg114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Restle A, Janz C, Wiesmüller L. Differences in the association of p53 phosphorylated on serine 15 and key enzymes of homologous recombination. Oncogene. 2005;24(27):4380–4387. doi: 10.1038/sj.onc.1208639. [DOI] [PubMed] [Google Scholar]
- 18.Melle C, Nasheuer HP. Physical and functional interactions of the tumor suppressor protein p53 and DNA polymerase α-primase. Nucleic Acids Res. 2002;30(7):1493–1499. doi: 10.1093/nar/30.7.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romanova LY, Willers H, Blagosklonny MV, Powell SN. The interaction of p53 with replication protein A mediates suppression of homologous recombination. Oncogene. 2004;23(56):9025–9033. doi: 10.1038/sj.onc.1207982. [DOI] [PubMed] [Google Scholar]
- 20.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8(3):193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 21.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11(10):683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 22.Gudkov AV. Converting p53 from a killer into a healer. Nat Med. 2002;8(11):1196–1198. doi: 10.1038/nm1102-1196. [DOI] [PubMed] [Google Scholar]
- 23.Thompson LH, Hinz JM. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: mechanistic insights. Mutat Res. 2009;668(1-2):54–72. doi: 10.1016/j.mrfmmm.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho TV, Schärer OD. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ Mol Mutagen. 2010;51(6):552–566. doi: 10.1002/em.20573. [DOI] [PubMed] [Google Scholar]
- 25.Akyüz N, et al. DNA substrate dependence of p53-mediated regulation of double-strand break repair. Mol Cell Biol. 2002;22(17):6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J Mol Biol. 2001;307(5):1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 27.Ahn J, et al. Dissection of the sequence-specific DNA binding and exonuclease activities reveals a superactive yet apoptotically impaired mutant p53 protein. Cell Cycle. 2009;8(10):1603–1615. doi: 10.4161/cc.8.10.8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer VN, Szybalski W. Mitomycins and porfiromycin: Chemical mechanism of activation and cross-linking of DNA. Science. 1964;145(3627):55–58. doi: 10.1126/science.145.3627.55. [DOI] [PubMed] [Google Scholar]
- 29.Bizanek R, McGuinness BF, Nakanishi K, Tomasz M. Isolation and structure of an intrastrand cross-link adduct of mitomycin C and DNA. Biochemistry. 1992;31(12):3084–3091. doi: 10.1021/bi00127a008. [DOI] [PubMed] [Google Scholar]
- 30.Bizanek R, et al. Adducts of mitomycin C and DNA in EMT6 mouse mammary tumor cells: Effects of hypoxia and dicumarol on adduct patterns. Cancer Res. 1993;53(21):5127–5134. [PubMed] [Google Scholar]
- 31.Warren AJ, Maccubbin AE, Hamilton JW. Detection of mitomycin C-DNA adducts in vivo by 32P-postlabeling: Time course for formation and removal of adducts and biochemical modulation. Cancer Res. 1998;58(3):453–461. [PubMed] [Google Scholar]
- 32.Yang Q, et al. The processing of Holliday junctions by BLM and WRN helicases is regulated by p53. J Biol Chem. 2002;277(35):31980–31987. doi: 10.1074/jbc.M204111200. [DOI] [PubMed] [Google Scholar]
- 33.Sommers JA, et al. p53 modulates RPA-dependent and RPA-independent WRN helicase activity. Cancer Res. 2005;65(4):1223–1233. doi: 10.1158/0008-5472.CAN-03-0231. [DOI] [PubMed] [Google Scholar]
- 34.Yang H, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297(5588):1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 35.Carreira A, et al. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136(6):1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moldovan GL, et al. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell. 2012;45(1):75–86. doi: 10.1016/j.molcel.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14(4):491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 38.Kannouche PL, Lehmann AR. Ubiquitination of PCNA and the polymerase switch in human cells. Cell Cycle. 2004;3(8):1011–1013. [PubMed] [Google Scholar]
- 39.Avkin S, et al. p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol Cell. 2006;22(3):407–413. doi: 10.1016/j.molcel.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 40.Soria G, Podhajcer O, Prives C, Gottifredi V. P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene. 2006;25(20):2829–2838. doi: 10.1038/sj.onc.1209315. [DOI] [PubMed] [Google Scholar]
- 41.Lehmann AR, et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6(7):891–899. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 42.Unk I, Hajdú I, Blastyák A, Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair (Amst) 2010;9(3):257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 43.Blastyák A, Hajdú I, Unk I, Haracska L. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol Cell Biol. 2010;30(3):684–693. doi: 10.1128/MCB.00863-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saugar I, Ortiz-Bazán MA, Tercero JA. Tolerating DNA damage during eukaryotic chromosome replication. Exp Cell Res. 2014;329(1):170–177. doi: 10.1016/j.yexcr.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 45.Ciccia A, et al. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell. 2012;47(3):396–409. doi: 10.1016/j.molcel.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bétous R, et al. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012;26(2):151–162. doi: 10.1101/gad.178459.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Couch FB, et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013;27(14):1610–1623. doi: 10.1101/gad.214080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo C, Kosarek-Stancel JN, Tang TS, Friedberg EC. Y-family DNA polymerases in mammalian cells. Cell Mol Life Sci. 2009;66(14):2363–2381. doi: 10.1007/s00018-009-0024-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 50.Vassin VM, Anantha RW, Sokolova E, Kanner S, Borowiec JA. Human RPA phosphorylation by ATR stimulates DNA synthesis and prevents ssDNA accumulation during DNA-replication stress. J Cell Sci. 2009;122(Pt 22):4070–4080. doi: 10.1242/jcs.053702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldstone S, Pavey S, Forrest A, Sinnamon J, Gabrielli B. Cdc25-dependent activation of cyclin A/cdk2 is blocked in G2 phase arrested cells independently of ATM/ATR. Oncogene. 2001;20(8):921–932. doi: 10.1038/sj.onc.1204177. [DOI] [PubMed] [Google Scholar]
- 52.Nimonkar AV, et al. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25(4):350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dupré A, et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol. 2008;4(2):119–125. doi: 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Costanzo V. Brca2, Rad51 and Mre11: Performing balancing acts on replication forks. DNA Repair (Amst) 2011;10(10):1060–1065. doi: 10.1016/j.dnarep.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 55.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol. 1998;140(6):1285–1295. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Speroni J, Federico MB, Mansilla SF, Soria G, Gottifredi V. Kinase-independent function of checkpoint kinase 1 (Chk1) in the replication of damaged DNA. Proc Natl Acad Sci USA. 2012;109(19):7344–7349. doi: 10.1073/pnas.1116345109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Técher H, et al. Replication dynamics: Biases and robustness of DNA fiber analysis. J Mol Biol. 2013;425(23):4845–4855. doi: 10.1016/j.jmb.2013.03.040. [DOI] [PubMed] [Google Scholar]
- 58.Shakked Z, et al. DNA binding and 3′-5′ exonuclease activity in the murine alternatively-spliced p53 protein. Oncogene. 2002;21(33):5117–5126. doi: 10.1038/sj.onc.1205667. [DOI] [PubMed] [Google Scholar]
- 59.Vidal AE, Woodgate R. Insights into the cellular role of enigmatic DNA polymerase iota. DNA Repair (Amst) 2009;8(3):420–423. doi: 10.1016/j.dnarep.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 60.Ulrich HD. Timing and spacing of ubiquitin-dependent DNA damage bypass. FEBS Lett. 2011;585(18):2861–2867. doi: 10.1016/j.febslet.2011.05.028. [DOI] [PubMed] [Google Scholar]
- 61.Livneh Z. Keeping mammalian mutation load in check: Regulation of the activity of error-prone DNA polymerases by p53 and p21. Cell Cycle. 2006;5(17):1918–1922. doi: 10.4161/cc.5.17.3193. [DOI] [PubMed] [Google Scholar]
- 62.Burkovics P, Sebesta M, Balogh D, Haracska L, Krejci L. Strand invasion by HLTF as a mechanism for template switch in fork rescue. Nucleic Acids Res. 2014;42(3):1711–1720. doi: 10.1093/nar/gkt1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeman MK, Cimprich KA. Finally, polyubiquitinated PCNA gets recognized. Mol Cell. 2012;47(3):333–334. doi: 10.1016/j.molcel.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 64.Achar YJ, Balogh D, Haracska L. Coordinated protein and DNA remodeling by human HLTF on stalled replication fork. Proc Natl Acad Sci USA. 2011;108(34):14073–14078. doi: 10.1073/pnas.1101951108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kile AC, et al. HLTF’s ancient HIRAN domain binds 3′ DNA ends to drive replication fork reversal. Mol Cell. 2015;58(6):1090–1100. doi: 10.1016/j.molcel.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuan J, Ghosal G, Chen J. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol Cell. 2012;47(3):410–421. doi: 10.1016/j.molcel.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weston R, Peeters H, Ahel D. ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Genes Dev. 2012;26(14):1558–1572. doi: 10.1101/gad.193516.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hübscher U, Maga G. DNA replication and repair bypass machines. Curr Opin Chem Biol. 2011;15(5):627–635. doi: 10.1016/j.cbpa.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 69.Zhou J, Ahn J, Wilson SH, Prives C. A role for p53 in base excision repair. EMBO J. 2001;20(4):914–923. doi: 10.1093/emboj/20.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bakhanashvili M, et al. p53 in mitochondria enhances the accuracy of DNA synthesis. Cell Death Differ. 2008;15(12):1865–1874. doi: 10.1038/cdd.2008.122. [DOI] [PubMed] [Google Scholar]
- 71.Bakhanashvili M, Hizi A, Rahav G. The interaction of p53 with 3′-terminal mismatched DNA. Cell Cycle. 2010;9(7):1380–1389. doi: 10.4161/cc.9.7.11201. [DOI] [PubMed] [Google Scholar]
- 72.Livneh Z, Ziv O, Shachar S. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle. 2010;9(4):729–735. doi: 10.4161/cc.9.4.10727. [DOI] [PubMed] [Google Scholar]
- 73.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11(2):96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Makarova AV, Kulbachinskiy AV. Structure of human DNA polymerase iota and the mechanism of DNA synthesis. Biochemistry (Mosc) 2012;77(6):547–561. doi: 10.1134/S0006297912060016. [DOI] [PubMed] [Google Scholar]
- 75.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase δ maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18(22):2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tissier A, McDonald JP, Frank EG, Woodgate R. poliota, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14(13):1642–1650. [PMC free article] [PubMed] [Google Scholar]
- 77.Rein K, Stracker TH. The MRE11 complex: An important source of stress relief. Exp Cell Res. 2014;329(1):162–169. doi: 10.1016/j.yexcr.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 78.Milyavsky M, et al. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell. 2010;7(2):186–197. doi: 10.1016/j.stem.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 79.Kraft D, et al. NF-κB-dependent DNA damage-signaling differentially regulates DNA double-strand break repair mechanisms in immature and mature human hematopoietic cells. Leukemia. 2015;29(7):1543–1554. doi: 10.1038/leu.2015.28. [DOI] [PubMed] [Google Scholar]