Significance
Understanding early human development relies on studies of in vitro-generated tissues from human embryonic or induced pluripotent stem cells that recapitulate in vivo cell types. End-point analyses of specific cell types can be hindered by inappropriate cell-culture conditions that may drive cells to undesired fates. We address these issues by reconstructing the trajectory of gene expression dynamics of 90 marker genes during early human otic development at the single-cell level. To determine the efficacy of our guidance protocol we compare these cells to native cells from the developing mouse otocyst. We find that otic induction follows the predicted dynamics of gene expression and resembles otic cells from native mouse tissue up to 12 days of culture.
Keywords: posterior placode, ectoderm, gene expression analysis, inner ear, pluripotent stem cells
Abstract
Efficient pluripotent stem cell guidance protocols for the production of human posterior cranial placodes such as the otic placode that gives rise to the inner ear do not exist. Here we use a systematic approach including defined monolayer culture, signaling modulation, and single-cell gene expression analysis to delineate a developmental trajectory for human otic lineage specification in vitro. We found that modulation of bone morphogenetic protein (BMP) and WNT signaling combined with FGF and retinoic acid treatments over the course of 18 days generates cell populations that develop chronological expression of marker genes of non-neural ectoderm, preplacodal ectoderm, and early otic lineage. Gene expression along this differentiation path is distinct from other lineages such as endoderm, mesendoderm, and neural ectoderm. Single-cell analysis exposed the heterogeneity of differentiating cells and allowed discrimination of non-neural ectoderm and otic lineage cells from off-target populations. Pseudotemporal ordering of human embryonic stem cell and induced pluripotent stem cell-derived single-cell gene expression profiles revealed an initially synchronous guidance toward non-neural ectoderm, followed by comparatively asynchronous occurrences of preplacodal and otic marker genes. Positive correlation of marker gene expression between both cell lines and resemblance to mouse embryonic day 10.5 otocyst cells implied reasonable robustness of the guidance protocol. Single-cell trajectory analysis further revealed that otic progenitor cell types are induced in monolayer cultures, but further development appears impeded, likely because of lack of a lineage-stabilizing microenvironment. Our results provide a framework for future exploration of stabilizing microenvironments for efficient differentiation of stem cell-generated human otic cell types.
Vertebrate cranial placodes arise from a region of non-neural ectoderm (NNE) lateral to the rostral neural crest progenitor region and the neural plate (reviewed in refs. 1 and 2). In humans, the cranial placodes form during the first month of gestation, restricting our insight to experiments using pluripotent stem cell-based models. Generation of human NNE and derivation of anterior placodal cells (i.e., pituitary, lens, and trigeminal neurons) from human embryonic stem cells (hESCs) has previously been achieved (3, 4). Production of posterior human placodal fates such as the otic placode and epibranchial ganglia neurons remains elusive, despite indications that immature sensory hair cell-like cells might arise in hESC-derived aggregates via manipulation of TGFβ, WNT, and FGF signaling, albeit with low efficiency (5–7).
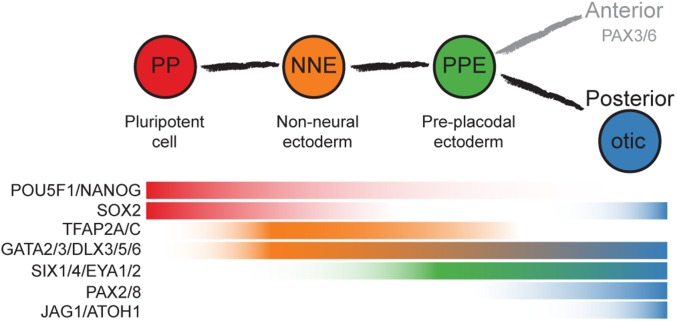
We used adherently grown hESCs and human induced pluripotent stem cells (iPSCs) to systematically test conditions for stepwise induction of NNE to posterior placode fates; specifically, the otic lineage. A challenge of in vitro guidance is the transient state of presumptive progenitor cells. In the case of early otic development, many marker genes are expressed in a fluctuating manner in which some early genes are transiently down-regulated and later reexpressed. Other genes are only transiently expressed or are persistently up-regulated (Fig. 1) (2). Moreover, it is possible that otic progenitor cells will change their fate or differentiate to alternative cell types if the culture environment is not suitable. It is therefore challenging to isolate presumptive progenitor cells for the analysis of cellular phenotypes. To address this confounding issue, we reconstructed the developmental trajectory of single cells based on their intrinsic temporal order orchestrated by the transcriptional dynamics as hESCs and iPSCs are guided toward NNE and, ultimately, in the direction of otic cells. Our analysis revealed robust generation of NNE followed by asynchronous occurrence of preplacodal marker genes. Retinoic acid treatment was essential for up-regulation of FGF-dependent early otic marker gene expression. Comparison with transcriptional profiles of native mouse otocyst cells revealed temporal boundaries of otic phenotypes along the differentiation timeline.
Fig. 1.
Otic induction from pluripotent cells. Shown is the expected sequence of marker gene expression based on studies of nonhuman vertebrate embryos.
Results
Induction of Non-neural Ectoderm from hESC Monolayer Cultures.
Differentiation of pluripotent stem cells into non-neural ectoderm requires suppression of mesodermal and endodermal lineages, which can be efficiently achieved by blocking TGFβ and WNT signaling (6, 8). We systematically tested various culture conditions for up-regulation of conserved NNE marker genes. TGFβ inhibitor SB431542 (SB) in combination with WNT inhibitor FH535 led to robust increase of transcription factors TFAP2A, GATA2/3, and DLX3 expression (Fig. 2 A and B and SI Appendix, Fig. S1 A and C).
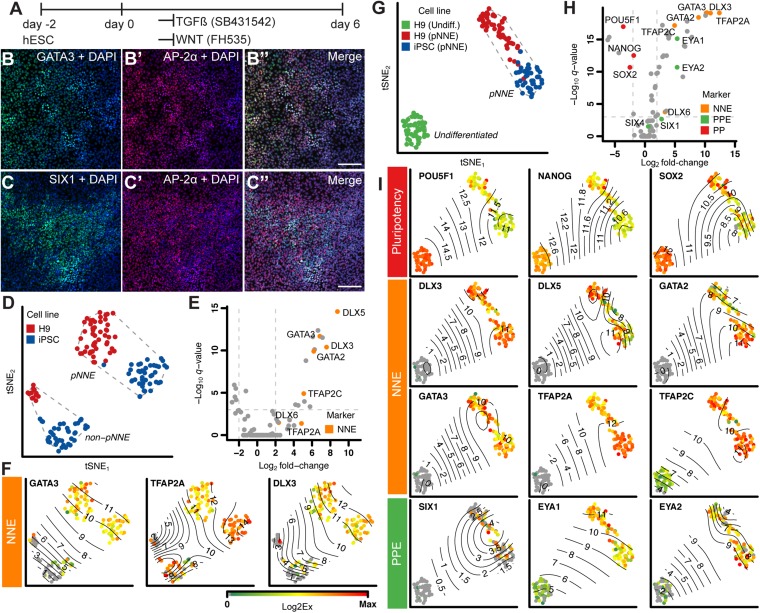
Fig. 2.
Generation of NNE. (A) Monolayer culture conditions. (B and C) Immunocytochemistry of cultures treated for 6 d with TGFβ inhibitor SB431452 and WNT inhibitor FH535 with antibodies to GATA3 [82.8 ± 16.3% (mean ± SD); n = 5, shown in green], AP-2α (69.7 ± 19.1%; n = 7; shown in red), and SIX1 (49.4 ± 13.8%; n = 3, shown in green). DAPI (blue) labels nuclei. (Scale bar, 100 μm.) (D) tSNE-based visualization of pNNE cultures from H9 hESCs (red) and iPSCs (blue) shows a distinct group of responding presumptive (p) NNE and nonresponding cells (non-pNNE). Dashed lines represent the convex hull of the classified cell populations. (E) Volcano plot of genes differentially expressed between pNNE and non-pNNE cells shows NNE marker genes up-regulated in pNNE cells. Dotted lines denote significance level of <10−3 values for multiple-testing corrected Mann-Whitney P value (q value) and fold change of 0.25 and 4. (F) Surface plots of single-cell expression values of selected NNE marker genes projected on the tSNE visualization map. Contour lines highlight the indicated Log2Ex values. (G) tSNE visualization of pNNE cells in relation to hESCs. H9 hESCs: green; H9 pNNE: red; iPSC pNNE: blue. (H) Volcano plot shows NNE marker genes are up-regulated in pNNE cells, and pluripotency markers are expressed in hESCs. (I) Surface plots for select pluripotency, NNE, and PPE marker genes. Color bar values represent dynamic range of Log2Ex values for each gene.
The majority of cells treated with SB/FH535 for 6 d coexpressed AP-2α (encoded by TFAP2A) and GATA3, whereas the preplacodal ectoderm (PPE) marker SIX1 was detectable in a subset of the AP-2α population (Fig. 2 B and C). SIX1 and EYA1 up-regulation (SI Appendix, Fig. S1D) suggests that a portion of cells express preplacodal genes, indicating that cell differentiation is not synchronous. We also noticed that prolonging the culture time from 6 to 8 or 10 d did not further increase expression of NNE markers and resulted in concurrent up-regulation of mesodermal and endodermal markers, which led us to conclude that 6 d was the most optimal culture time (SI Appendix, Fig. S1 C, F, and G).
To characterize the presumptive human NNE (pNNE) population in more detail, we conducted multiparallel quantitative (q) RT-PCR for 90 genes on single cells from 6-d NNE differentiation cultures of H9 hESCs and the iPSC line SUCI-0002. The qRT-PCR assay panel was designed to monitor marker genes for NNE, preplacodal ectoderm, and different placode fates, as well as controls for other tissue types (SI Appendix, Table S1). Relationships between single cells were visualized with t-distributed stochastic neighbor embedding (tSNE), a nonlinear dimension reduction technique (9). A population of nonresponding cells and cells that up-regulated NNE markers were distinguishable after 6 d of induction (Fig. 2 D–F). Of the 6-d culture cells, 64% (51 of 64 cells for hESCs and 43 of 82 cells for iPSCs) showed a NNE phenotype. The gap statistic indicated four clusters for the 6-d cells from both cell lines. Analyzing both cell lines together, using hierarchical clustering, intrinsically splits cells into two major clusters, defined as pNNE and non-pNNE, based on NNE marker expression (SI Appendix, Fig. S2 A and B). Comparison of the pNNE cells with undifferentiated hESCs (n = 44) confirmed that NNE markers are strongly up-regulated after SB/FH535 treatment (Fig. 2 G–I). In addition to NNE markers, PPE markers such as transcription factors EYA1/2 and SIX1 were also detectable, supporting our immunocytochemical observation that some cells are en route toward cranial placode (Fig. 2 H–I).
Presumptive Non-neural Ectoderm Is Distinct from Other Cell Lineages.
pNNE single-cell transcript profiles were compared with profiles from three different in vitro-generated germ layer lineages. Single-cell pNNE expression profiles were distinctively different from mesendoderm cells and resulted in two well-defined groups (Fig. 3 A–C). The top genes distinguishing the pNNE cells were TFAP2A, GATA3, DLX3, and EYA1, whereas early mesendoderm markers GATA4, T (Brachyury), and POU5F1 (OCT4) were among those that contributed most to the mesendoderm cell cluster (Fig. 3D and SI Appendix, Fig. S3A). pNNE cells were also different from intermediate mesoderm cells derived from hESCs, using a 9-d differentiation protocol developed for generating nephrogenic lineage cells (Fig. 3 E–H). Genes in the IRX and OTX gene families were the most differentially expressed genes between the pNNE cells and intermediate mesoderm (Fig. 3H and SI Appendix, Fig. S3B). The genes that defined the hESC-generated intermediate mesoderm the most were ISL1 and MSX1, which are expressed in developing mesodermal tissues (10, 11).
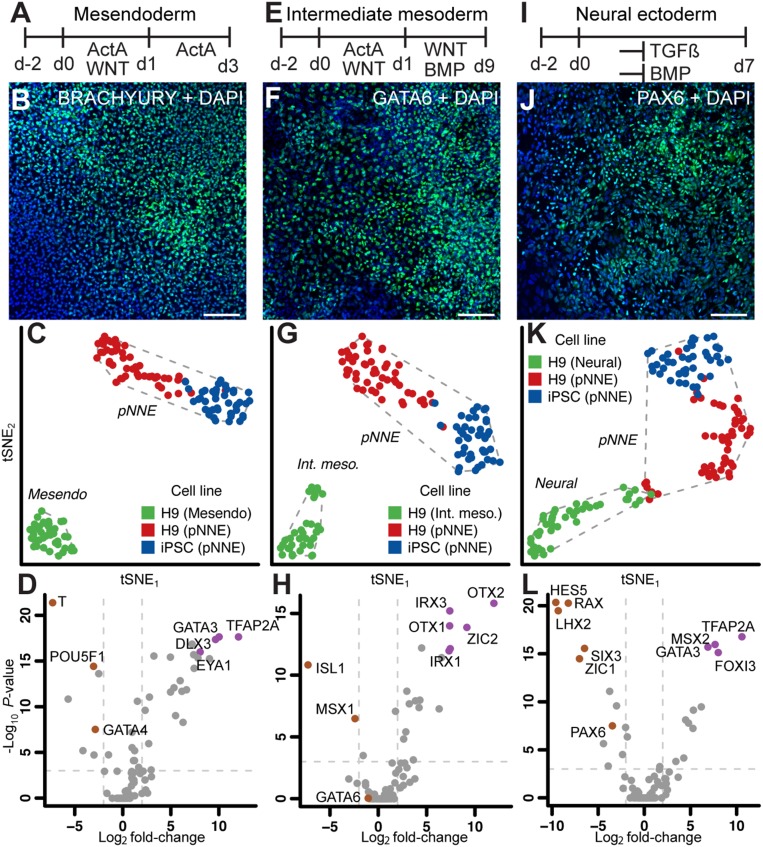
Fig. 3.
Single-cell analysis of pNNE and other in vitro-derived cell types. Single cells of hESC-derived cell types representing mesendoderm (A), intermediate mesoderm (E), and neural ectoderm (I) were compared with pNNE. (Scale bars, 100 μm.) Immunocytochemistry of cultures treated with factors to induce mesendoderm (B), intermediate mesoderm (F), and neural ectoderm (J). tSNE analysis of single cells from pNNE derived from H9 hESCs (blue) and iPSCs (red) and the control cell type (green) representing mesendoderm (C), intermediate mesoderm (G), and neural ectoderm (K). Dashed lines represent the convex hull of the classified cell populations. (D, H, and L) Volcano plot shows population-defining markers for each cell type. Dotted lines denote significance level of <10−3 values for multiple-testing corrected Mann-Whitney P value (q value) and fold change of 0.25 and 4.
Finally, we compared pNNE cells with neural ectoderm generated by TGFβ and bone morphogenetic protein (BMP) signaling inhibition for 7 d (Fig. 3 I–L) (12). Anterior neural plate (neural ectoderm) and NNE develop in close proximity during the transitional period of late gastrulation and early neurulation (2). NNE markers MSX2, TFAP2A, and GATA3 were among the genes that contributed most to separation of the pNNE cell population from the neural ectoderm group that was defined by expression of LHX2, RAX, and PAX6 (Fig. 3L and SI Appendix, Fig. S3C). A small cluster of cells in between neural and pNNE groups expressed higher levels of neural crest markers, indicating a potential overlap reflecting the natural relationship among neural, neural crest, and NNE (SI Appendix, Fig. S4). Across all comparisons, pNNE cells from H9 hESCs and iPSCs tended to cluster slightly away from each other, suggestive of small differences between the cell lines. However, the heterogeneity within the pNNE cell population was less than the observed variance between H9 pNNE and other H9-derived cell lineages (SI Appendix, Fig. S5).
Posterior Placode Cell Fates Require Combination of FGF, WNT, BMP, and Retinoic Acid Signaling.
After gastrulation, the NNE becomes restricted to PPE, and ultimately to different lineages, giving rise to the different cranial placodes (1). There is evidence that NNE partitions into anterior and posterior preplacodal regions driven by signals from surrounding tissue (13). Given that we detected PPE markers in pNNE cultures, we hypothesized that some cells in these cultures are competent to differentiate into cranial placode cell types (Fig. 2 C, H, and I and SI Appendix, Fig. S1D). In many vertebrates, FGF signaling is essential for restricting PPE to cranial placode fates (14). Different cranial placodes are further restricted through the action of other pathways such as sonic hedgehog (SHH), BMP, and WNT signaling (2, 13) (SI Appendix, Fig. S6A). pNNE cultures were readily competent to produce anterior placodal cells when we subjected them to factors known to promote anterior placode fates, such as adenohypophysial, lens, and trigeminal (SI Appendix, Fig. S6 A–F) (3).
We next determined the competency of the pNNE cells to differentiate into the posterior otic placode lineage. In other vertebrate models, otic-epibranchial placodes require FGF and WNT signaling (15, 16). We tested whether this induction mechanism in general can be applied to human cells and conducted a screen comparing 63 different culture conditions in which we cultured 6-d pNNE cells with different factors and small molecules for up to 12 additional days (SI Appendix, Fig. S7A). Combined treatment with FGF2 and the small molecule GSK3β inhibitor CHIR99021, an activator of WNT signaling, did not lead to detectable expression of posterior placode markers such as transcription factors PAX2 or PAX8 (Fig. 4 A, B, and D). Moreover, the anterior placode marker PAX6 was expressed in cells cultured in these conditions (Fig. 4B′), suggesting other signals are needed to steer the cultures toward posterior placodal fates.
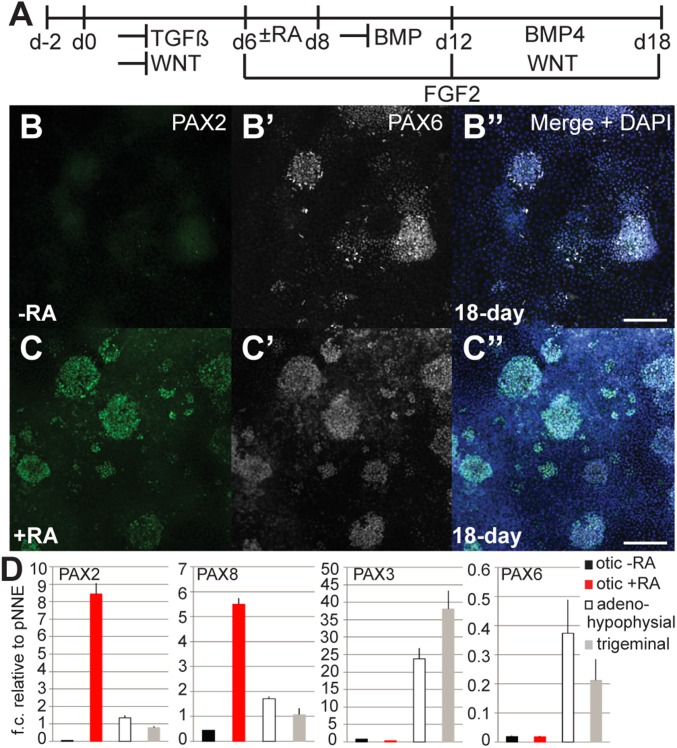
Fig. 4.
Posterior placode marker expression upon treatment with retinoic acid. (A) Monolayer culture conditions for otic induction. (B–B″) Immunocytochemistry showed no detectable expression of PAX2, but expression of PAX6 in cultures without retinoic acid (RA) treatment. (C–C″) Adding retinoic acid between days 6 and 8 resulted in expression of PAX2 detected at day 18 (14.1 ± 10.5%; n = 4). (D) qRT-PCR analysis of 18-d cultures for anterior and posterior placode markers. NNE cultures treated for otic induction with or without retinoic acid, adenohypophysial, and trigeminal placode induction. (Scale bars, 100 μm.)
We hypothesized that retinoic acid might be effective in promoting posterior placode gene expression in pNNE cultures. In zebrafish, retinoic acid treatment leads to anterior expansion of the posterior placode field (17). When we applied retinoic acid for 2 d, we detected PAX2 gene expression as well as PAX2 protein expression (Fig. 4 C and D and SI Appendix, Fig. S7 A and B). Interestingly, PAX6 expression did not cease in these cultures, suggesting that addition of retinoic acid does not restrict expression of this anterior placode marker (Fig. 4C′). Some cells appeared to coexpress PAX2 and PAX6 proteins, perhaps indicative of a mixed phenotype. PAX2 protein was found in islands of cells in the 18-d cultures; however, this accounted for only a small number of positively stained cells (2.6–28.9% of cells; n = 4). Quantitative gene expression analysis of cultures treated with retinoic acid revealed up-regulation of both posterior placode markers, PAX2 and PAX8 (Fig. 4D). Analyses of PAX2 and PAX8 expression in cultures treated with retinoic acid from day 6 onward for 1–12 d indicated that a maximum of 2 d retinoic acid treatment was optimal to induce the two posterior placode marker genes (SI Appendix, Fig. S7 B and C). Longer exposure reduced the level of posterior placode marker expression. Most robust expression of posterior markers occurred when pNNE cells were cultured for 12 d in the presence of FGF2, inhibition of BMP signaling for the first 6 d followed by 6-d treatment with BMP4 and simultaneous promotion of WNT signaling, and addition of retinoic acid for the first 2 d (Fig. 4A).
The robust tendency of the pNNE cells to express PAX6 suggests that differentiation into anterior fates is a much more predominant feature of these cells. We specifically tested previously established conditions that led to differentiation into trigeminal fates (3) and found robust up-regulation of PAX3 in these conditions (Fig. 4D). Promotion of the most anterior adenohypophysial lineage with SHH (3) increased expression of PAX6, indicating that the overall competence of pNNE cells to differentiate into this lineage is robust.
Temporal Trajectory Analysis of Early Otic Induction.
To delineate transcriptional dynamics of early human otic development, we analyzed single cells at distinct points of differentiation, using multiparallel qRT-PCR followed by computational trajectory reconstruction. We used cells collected at day 0 (H9 hESCs only), day 6 pNNE, day 12, and day 18 in conjunction with filtered genes exhibiting stage-dependent variance in the induction protocol (SI Appendix, Fig. S8A). Temporal cell–cell relations were weighted on the basis of their spatial proximity in the lower-dimensional space obtained by independent component analysis of the cellular expression profiles. All cells were connected, ensuring the minimal total weighting and simultaneously prohibiting links if cells were nonadjacent in terms of their stage. The longest shortest path through this minimum spanning tree denoted the backbone of the trajectory. We applied the Monocle algorithm to determine the optimal pseudotemporal ordering of the cells along and beside the backbone (18). Gene expression dynamics over pseudotime were fitted with a likelihood-based regression model.
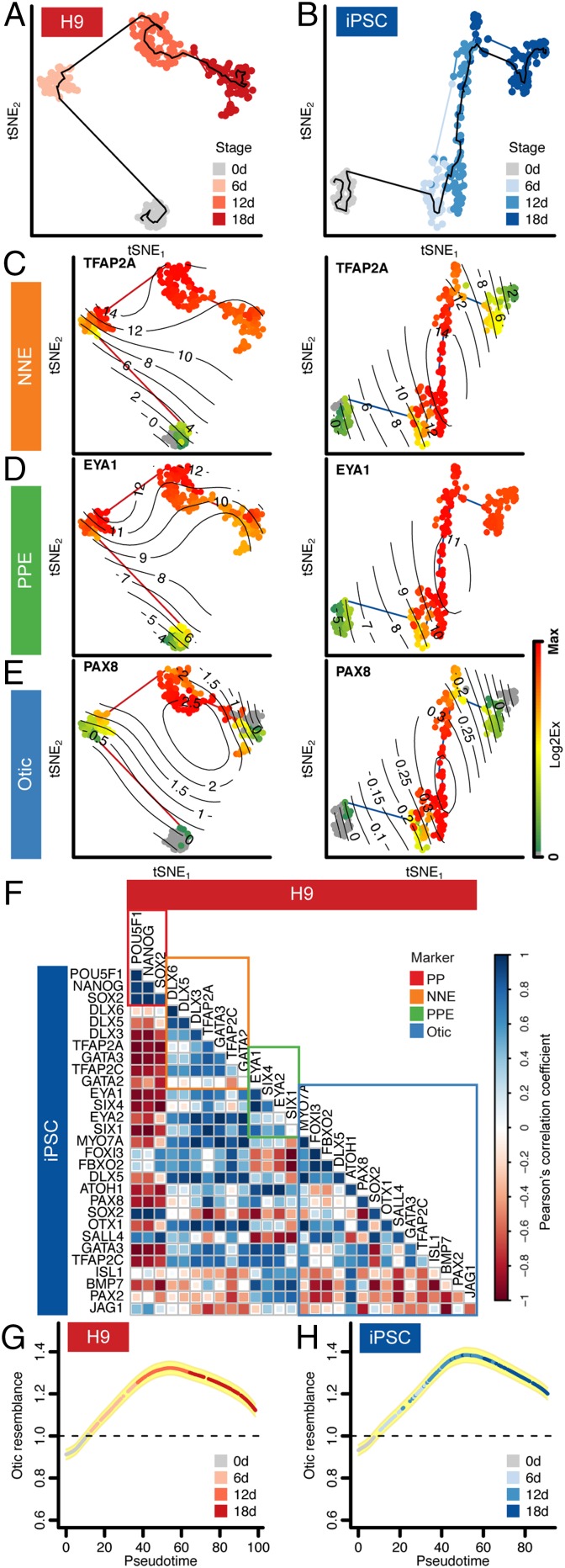
We expected that the temporal order of expression of known marker genes for lineage progression (Fig. 1) would be well reflected in the reconstructed trajectories of hESC and iPSC expression profiles (Fig. 5 A–E). Pluripotency markers were down-regulated over the course of otic induction (SI Appendix, Fig. S8B). NNE and PPE markers were up-regulated and maintained (Fig. 5 C and D and SI Appendix, Fig. S8B). Otic markers, including PAX8, in general were induced along the projected trajectory; they occurred more robustly in hESC-derived cultures than in iPSC cultures (Fig. 5E). A subset of the 18-d cells derived from hESCs expressed prosensory markers SOX2 and JAG1, as well as the otic marker ATOH1 (SI Appendix, Fig. S8B). iPSC cultures showed delayed up-regulation of PPE markers compared with hESC-derived cells, and up-regulation of otic markers was less coordinated in iPSC-derived cultures.
Fig. 5.
Computed single-cell trajectory of otic induction in monolayer cultures. (A and B) Pseudotemporal ordering of cells derived from H9 (A) and iPSC (B) cell lines visualized by tSNE. (C–E) Temporal transcriptional cascades of representative marker genes for both cell lines: (C) NNE, (D) PPE, and (E) otic. Surface plots of single-cell expression values of example marker genes are projected on the temporal trajectories. Color bar values represent dynamic range of Log2Ex values for each gene. (F) Expression dynamics of the otic induction in H9 and iPSCs were compared using Pearson’s correlation coefficient. Marker genes for each developmental point, pluripotency, NNE, PPE, and otic are highlighted. (G and H) Otic resemblance index of single cells along the temporal trajectory. Single-cell gene expression profiles for 23 genes were compared with native mouse otic cells. Yellow shading: ± SD.
Defining the temporal expression dynamics for otic guidance of two independent pluripotent stem cell lines provided the opportunity to assess general robustness of the approach. As such, we correlated temporal expression of all pluripotency, NNE, PPE, and otic marker genes between the computed trajectories. Overcoming the fact that each cell group was of different size, the cells were partitioned into 10 groups along the trajectory, and the average gene expression covariance was estimated using Pearson’s correlation coefficient (SI Appendix, Fig. S9). We found positive correlation for most marker genes including the majority of otic genes between the two cell lines (Fig. 5F). If the iPSC-derived cultures are indeed differentiating more asynchronously than the hESC-derived cells, one could conclude from our data that expression of the otic markers PAX2, JAG1, BMP7, and ISL1 would require additional time. We subjected 18-d differentiated cultures to prolonged cell differentiation, using previously established conditions (6), but did not observe any improvements with respect to otic differentiation, including generation of bona fide hair cell-like cells. This agrees with our hypothesis that further stabilization of otic progenitor cells likely requires additional factors that can potentially be provided in aggregate cultures in which heterogeneous cell types generate an otic lineage-promoting microenvironment (8, 19).
To further interpret the temporal trajectory for human otic guidance, we developed an otic resemblance index to assess which cells along the pseudotemporal trajectory were more closely related to native otic cells from embryonic day 10.5 mouse otocyst (20). Twenty-three genes present in both the human induction assay and previously reported mouse otocyst data were used for this analysis (SI Appendix, Fig. S10). We computed the similarity of each individual human otic induction cell to its nearest neighbors in the mouse otic reference population and compared it with that expected using a null reference cell population; a score greater than unity would represent a more otic phenotype than would occur by chance. Plotting this index for each cell in the temporal trajectory revealed that the cells closest to native mouse otocyst cells were those at day 12 of the induction process for both H9 and iPSCs (Fig. 5 G and H).
Discussion
Generation of human inner ear cell types from pluripotent stem cells is a challenging endeavor (5, 6). Most existing guidance protocols use cell aggregation to create indefinable microenvironments of multiple cell lineages (6, 8, 19, 21). Without a complex 3D microenvironment (19, 21), proper sensory hair cell differentiation only sporadically occurs in cocultures with primary embryonic otic cells (8). Whereas guidance protocols for mouse otic lineage cells are continuously evolving through novel tools such as cellular reprogramming (22) or highly specific novel otic lineage markers (23), generation of human hair cell-like cells remains perplexingly difficult. The lack of specific human otic lineage markers impedes identification and isolation of human otic lineage cells. Propensity of human preplacodal cells to adopt anterior placode cell fates exhibits another hurdle (3, 4).
Our goal was to advance the generation of human otic lineage cells from pluripotent stem cells by systematically exploring conditions that promote formation of posterior placode cells. We used a multiparallel qRT-PCR approach to monitor expression of 90 genes consisting of known markers for NNE, PPE, and the otic lineage, as well as other lineages. This minimized the limitation caused by the lack of individual specific human otic lineage markers.
We found that existing guidance strategies for suppression of mesoderm and endodermal lineages without distinct induction of neural ectoderm indeed promote efficient generation of NNE. Single-cell analyses revealed that in vitro guidance is a dynamic process. Once en route, cells continue along specific trajectories that may or may not represent naturally occurring developmental courses. For instance, some human pNNE cells appear to readily move along a trajectory that leads to up-regulation of PPE genes, which indicates that the monolayer culture conditions create a permissive or even inductive environment for the generation of different placodal lineages. Interestingly, retinoic acid was sufficient to reduce up-regulation of anterior placode genes PAX3 and PAX6 and to promote the expression of posterior placode genes PAX2 and PAX8. This requirement of retinoic acid for proper otic induction and the ability to expand posterior placodes has been previously shown in developing zebrafish (17). Nevertheless, we also noticed an occurrence of cells with a mixed gene expression phenotype, such as cells that appear to coexpress PAX2 and PAX6. We cannot be certain whether such a potential hybrid phenotype reflects a naturally occurring cell type or whether in vitro culture conditions inherently lead to situations that generate cells with novel phenotypes.
To reveal the dynamic progression of cell differentiation, we computed the pseudotemporal ordering of two pluripotent cell lines from a pluripotent state toward NNE through PPE to a presumptive early otic state. hESCs generally followed an expected pattern of successive up-regulation of NNE, PPE, and otic genes. The iPSC line followed a similar expected trajectory, but the order of gene expression was less organized and suggested a more heterogeneous process. Timing of specific factor treatments is important for efficient otic guidance of mouse ESCs (19, 24), and could therefore also account for differences between hESC and iPSC differentiation. Nevertheless, both differentiation trajectories independently revealed that guided cell differentiation in monolayers is able to produce presumptive early human otic cells that coexpress multiple otic markers. Further evidence for proper guidance is that the cells display proper lineage history marked by initial coexpression of NNE genes and successive up-regulation of PPE marker genes. In addition, they are distinct from intermediate mesodermal, mesendodermal, and neural ectodermal cell fates.
Quantification of otic resemblance to native murine otocyst cells revealed that the guided hESC- and iPSC-derived cells displayed the highest similarity at day 12 of culture. Although genes such as PAX2/8 are important for otic development, but not essential for otic induction (25), their expression pattern seems to be temporally and spatially restricted, and one would not expect these markers in all cells of the otic lineage (20). We assume that the lack of a proper microenvironment, such as presence of other lineages in aggregate cultures (19), eventually results in failure to stabilize the cells along a committed otic lineage path. These conclusions support the use of coculture or cell aggregation to provide yet-unidentified factors to stabilize the lineage trajectory of human otic progenitor cells generated from pluripotent stem cells.
In summary, our experimentation shows that single-cell gene expression analysis can be used for a refined dissection of stem cell guidance protocols. The method reveals intricate details of cellular heterogeneity and asynchrony, thereby allowing the identification of important points at which culture conditions need to be adapted or radically changed to further promote the desired lineage. Specifically, for cell lineages that are difficult to generate and foster, single-cell monitoring has the potential to aid in the development of novel guidance strategies.
Materials and Methods
hESC and iPSC guidance, immunocytochemical analyses, bulk and single-cell qRT-PCR, quality control, normalization of gene expression data, and additional methodological details about data analysis are described in detail in SI Appendix. Relationships between cells were visualized by projection to two dimensions, using tSNE (9). Data are represented as Log2 expression values (Log2Ex). Pseudotemporal orderings were computed by means of the Monocle algorithm (18). Cell counts are presented as mean ± SD; n is defined as number of independent experiments. Stem cell and human subject research were conducted with protocols approved by Stanford University’s Institutional Review Board.
Supplementary Material
Acknowledgments
We thank the S.H. laboratory members for critical discussion of data and the manuscript, Dr. G. Mostoslavsky for providing the EF1a-hSTEMCCA-loxp plasmid, and Dr. J. Waldhaus and Dr. M. Scheibinger for help with iPSC generation. This work was supported by NIH Grant DC012250 (to S.H.), by P30 core support (DC010363), by the Stanford Initiative to Cure Hearing Loss, and by FP7-Health-2013-Innovation, a cooperative grant by the European Commission. M.E. was partially supported by the Lucile Packard Foundation for Children’s Health, Stanford NIH-NCATS-CTSA UL TR001085, and Child Health Research Institute of Stanford University and NIH Grant F32DC014176. D.C.E. was partially supported by a Stanford School of Medicine Dean’s Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605537113/-/DCSupplemental.
References
- 1.Schlosser G. Making senses development of vertebrate cranial placodes. Int Rev Cell Mol Biol. 2010;283:129–234. doi: 10.1016/S1937-6448(10)83004-7. [DOI] [PubMed] [Google Scholar]
- 2.Lleras-Forero L, Streit A. Development of the sensory nervous system in the vertebrate head: The importance of being on time. Curr Opin Genet Dev. 2012;22(4):315–322. doi: 10.1016/j.gde.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 3.Dincer Z, et al. Specification of functional cranial placode derivatives from human pluripotent stem cells. Cell Reports. 2013;5(5):1387–1402. doi: 10.1016/j.celrep.2013.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung AW, Kent Morest D, Li JY. Differential BMP signaling controls formation and differentiation of multipotent preplacodal ectoderm progenitors from human embryonic stem cells. Dev Biol. 2013;379(2):208–220. doi: 10.1016/j.ydbio.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W, et al. Restoration of auditory evoked responses by human ES-cell-derived otic progenitors. Nature. 2012;490(7419):278–282. doi: 10.1038/nature11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ronaghi M, et al. Inner ear hair cell-like cells from human embryonic stem cells. Stem Cells Dev. 2014;23(11):1275–1284. doi: 10.1089/scd.2014.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohnishi H, et al. Limited hair cell induction from human induced pluripotent stem cells using a simple stepwise method. Neurosci Lett. 2015;599:49–54. doi: 10.1016/j.neulet.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 8.Oshima K, et al. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell. 2010;141(4):704–716. doi: 10.1016/j.cell.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Maaten LJP, Hinton GE. Visualizing high-dimensional data using t-SNE. J Mach Learn Res. 2008;9(Nov):2579–2605. [Google Scholar]
- 10.Kang J, Nathan E, Xu SM, Tzahor E, Black BL. Isl1 is a direct transcriptional target of Forkhead transcription factors in second-heart-field-derived mesoderm. Dev Biol. 2009;334(2):513–522. doi: 10.1016/j.ydbio.2009.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mackenzie A, Leeming GL, Jowett AK, Ferguson MW, Sharpe PT. The homeobox gene Hox 7.1 has specific regional and temporal expression patterns during early murine craniofacial embryogenesis, especially tooth development in vivo and in vitro. Development. 1991;111(2):269–285. doi: 10.1242/dev.111.2.269. [DOI] [PubMed] [Google Scholar]
- 12.Surmacz B, et al. Directing differentiation of human embryonic stem cells toward anterior neural ectoderm using small molecules. Stem Cells. 2012;30(9):1875–1884. doi: 10.1002/stem.1166. [DOI] [PubMed] [Google Scholar]
- 13.Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294(2):303–351. doi: 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Kwon HJ, Bhat N, Sweet EM, Cornell RA, Riley BB. Identification of early requirements for preplacodal ectoderm and sensory organ development. PLoS Genet. 2010;6(9):e1001133. doi: 10.1371/journal.pgen.1001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Streit A, Berliner AJ, Papanayotou C, Sirulnik A, Stern CD. Initiation of neural induction by FGF signalling before gastrulation. Nature. 2000;406(6791):74–78. doi: 10.1038/35017617. [DOI] [PubMed] [Google Scholar]
- 16.Wilson SI, Graziano E, Harland R, Jessell TM, Edlund T. An early requirement for FGF signalling in the acquisition of neural cell fate in the chick embryo. Curr Biol. 2000;10(8):421–429. doi: 10.1016/s0960-9822(00)00431-0. [DOI] [PubMed] [Google Scholar]
- 17.Hans S, Westerfield M. Changes in retinoic acid signaling alter otic patterning. Development. 2007;134(13):2449–2458. doi: 10.1242/dev.000448. [DOI] [PubMed] [Google Scholar]
- 18.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koehler KR, Mikosz AM, Molosh AI, Patel D, Hashino E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature. 2013;500(7461):217–221. doi: 10.1038/nature12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durruthy-Durruthy R, et al. Reconstruction of the mouse otocyst and early neuroblast lineage at single-cell resolution. Cell. 2014;157(4):964–978. doi: 10.1016/j.cell.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Roblin G, Liu H, Heller S. Generation of hair cells by stepwise differentiation of embryonic stem cells. Proc Natl Acad Sci USA. 2003;100(23):13495–13500. doi: 10.1073/pnas.2334503100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costa A, Henrique D. Transcriptome profiling of induced hair cells (iHCs) generated by combined expression of Gfi1, Pou4f3 and Atoh1 during embryonic stem cell differentiation. Genom Data. 2015;6:77–80. doi: 10.1016/j.gdata.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartman BH, Durruthy-Durruthy R, Laske RD, Losorelli S, Heller S. Identification and characterization of mouse otic sensory lineage genes. Front Cell Neurosci. 2015;9:79. doi: 10.3389/fncel.2015.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koehler KR, Hashino E. 3D mouse embryonic stem cell culture for generating inner ear organoids. Nat Protoc. 2014;9(6):1229–1244. doi: 10.1038/nprot.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Streit A. Induction of the inner ear: Stepwise specification of otic fate from multipotent progenitors. Hear Res. 2013;297:3–12. doi: 10.1016/j.heares.2012.11.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.