Significance
Plant tissues are resistant to the potentially damaging UV-B radiation intrinsic to sunlight. UV-B photoreception by a UV RESISTANCE LOCUS 8 (UVR8) photoreceptor regulates gene expression in plants associated with UV-B acclimation and stress tolerance and with morphological changes. Mechanistically, UV-B photon reception by specific tryptophan residues of UVR8 homodimers results in monomerization and enhanced nuclear accumulation of UVR8. Active UVR8 monomers interact with the key signaling protein CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1). This UV-B–dependent interaction is a crucial step in signal propagation, but the link between this mechanism and UVR8 nuclear accumulation and gene expression remains ill defined. Our results emphasize the importance of nuclear-localized UVR8 and highlight a previously unknown activity of COP1 in mediating UVR8 nuclear accumulation in response to UV-B.
Keywords: nuclear accumulation, UV-B photoreceptor, COP1, glucocorticoid receptor, UVR8
Abstract
The UV-B photoreceptor UV RESISTANCE LOCUS 8 (UVR8) promotes UV-B acclimation and tolerance in Arabidopsis thaliana. UVR8 localizes to both cytosol and nucleus, but its main activity is assumed to be nuclear. UV-B photoreception stimulates nuclear accumulation of UVR8 in a presently unknown manner. Here, we show that CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) is required for UV-B–induced nuclear accumulation of UVR8, but bypassing the COP1 requirement for UVR8 nuclear accumulation did not rescue the cop1 mutant UV-B phenotype. Using a glucocorticoid receptor (GR)-based fusion protein system to conditionally localize GR-UVR8 to the nucleus, we have demonstrated that both photoactivation and nuclear localization of UVR8 are required for UV-B–induced photomorphogenic responses. In contrast, there was no UV-B response when UV-B–activated UVR8 was artificially retained in the cytosol. In agreement with a predominantly nuclear activity, constitutively active UVR8W285A accumulated in the nucleus also in the absence of UV-B. Furthermore, GR-COP1 expression lines suggested that UV-B–activated UVR8 can be coimported into the nucleus by COP1. Our data strongly support localization of UVR8 signaling in the nucleus and a dual role for COP1 in the regulation of UV-B–induced UVR8 nuclear accumulation and in UVR8-mediated UV-B signaling.
The UV-B radiation intrinsic to sunlight is potentially damaging to living tissues. However, a biochemical pathway exists in plants by which UV-B radiation induces UV-B stress tolerance through the activation of acclimation responses (1–4). The UV-B radiation inducing these responses is perceived by the UV RESISTANCE LOCUS 8 (UVR8) sensory photoreceptor that converts from a biologically inactive homodimeric to an active monomeric conformer (5). In contrast to visible light photoreceptors, UVR8 has no external chromophore but includes specific intrinsic tryptophan residues whose standard aromatic side chains act as a chromophore (5–7). Trp-285 is of major importance for UV-B responsiveness; mutation to Phe results in a “UV-B blind” constitutively homodimeric UVR8W285F, whereas mutation to Ala leads to a constitutively partially active UVR8W285A (5, 8). By inactivation, UVR8 reverts to the dimeric ground state in association with REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 (RUP1) and RUP2 (9, 10).
Activated monomeric UVR8 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) (1), an E3 ubiquitin ligase that is not only a key factor in UV-B signaling but also acts as a repressor of photomorphogenesis in the dark and in visible light (11–13). COP1 forms stable complexes with the four partially redundant SUPPRESSOR OF PHYA-105 (SPA) protein family members SPA1–SPA4 that are crucial for the majority of COP1 activities (14–16). As an exception, the SPA proteins are not required for COP1 activity in early seedling development or for UV-B signaling (11, 17).
The COP1–SPA complex mediates ubiquitination of several positive regulators of photomorphogenesis in the dark, including the bZIP transcription factor ELONGATED HYPOCOTYL 5 (HY5) (18). In visible light, COP1–SPA is inactivated by the phytochrome red/far-red and the cryptochrome blue light photoreceptors, especially through their light-dependent interaction with the SPA proteins (19–23). In addition to direct inhibition through the photoreceptors, COP1 is influenced by light-regulated nucleocytosolic partitioning, with nuclear accumulation in the dark and nuclear exclusion in the light (12, 24, 25). In agreement, COP1 includes both a nuclear localization signal (NLS) and a nuclear export signal (NES) (12). However, UV-B counteracts nuclear exclusion of COP1 in white light, resulting in its nuclear accumulation under supplemental UV-B (11). This response is associated with an increase in COP1 level under supplemental UV-B due to transcriptional activation and posttranslational stabilization (1, 11, 26). Similarly, HY5 accumulates in response to UV-B in a UVR8-dependent manner, also mediated by transcriptional activation and posttranslational stabilization (1, 4, 11, 27, 28). HY5 associates with the promoters of its target genes and is required for activation of a large fraction of UV-B–responsive genes (4, 27, 29).
Photoactivation of primarily cytosolic UVR8 triggers its rapid nuclear accumulation in an unknown manner, except that it depends on the N-terminal 23 amino acids of UVR8 (30). Here, we show that COP1 is required for nuclear accumulation of UV-B–activated UVR8 photoreceptor and can potentially coimport UVR8 in response to UV-B. The nuclear localization of UVR8 is essential to its activity and COP1 plays a dual role in UV-B signaling and UVR8 nuclear accumulation.
Results
Nuclear Accumulation of UV-B–Activated UVR8 Is Impaired in cop1-4 Mutants.
To investigate the nucleocytosolic partitioning of UVR8 in response to UV-B, Arabidopsis wild-type seedlings were grown under continuous white light or white light supplemented with UV-B. Total protein extracts were separated into nuclear and cytosolic fractions, and their purity was verified by immunoblot analysis of nuclear histone H3 and a cytosolic UGPase. Consistent with published data (30), UVR8 accumulation was detected in nuclear fractions within a few hours of photomorphogenic narrowband UV-B treatment (Fig. 1A). The nuclear accumulation of UV-B–activated UVR8 was accompanied by a slight reduction in the cytosolic fraction (Fig. 1A).
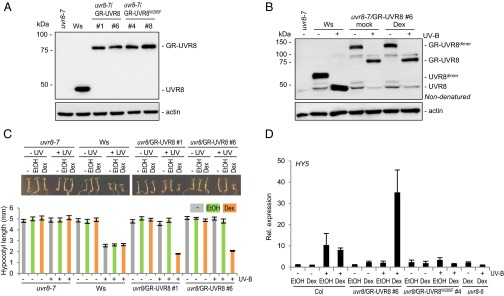
Fig. 1.
Nuclear accumulation of UVR8 requires COP1. (A-C) Immunoblot analyses: (A) UVR8, histone H3 (nuclear control), and UGPase (cytosolic control) in cytosolic and nuclear proteins of 7-d-old wild-type plants (Col) grown in white light without (0 h) or with UV-B for 4 h or 24 h. (B) UVR8, histone H3, and UGPase in nuclear (Left) and cytosolic proteins (Right) of uvr8-6, wild-type (Col), cop1-4, and cop1-20 plants grown in white light without (−UV-B) or with UV-B for 6 h (+UV-B). (C) Total UVR8 protein levels of uvr8-6, wild-type (Col), cop1-4, and cop1-20 plants grown in white light without (−UV-B) or with UV-B for 6 h (+UV-B). (D) UVR8, YFP-UVR8, and actin (loading control) proteins in 4-d-old uvr8-6, wild-type (Col), uvr8-6/Pro35S:YFP-UVR8 (uvr8-6/YFP-UVR8 3), and cop1-4/Pro35S:YFP-UVR8 (cop1-4/YFP-UVR8 6 and 11) lines. (E) YFP and DAPI fluorescence in cotyledon adaxial epidermis of 4-d-old uvr8-6/Pro35S:YFP-UVR8 line 3 and cop1-4/Pro35S:YFP-UVR8 line 6. (Scale bars: 10 µm.) (F) Yeast two-hybrid growth assays of COP1 interactions with wild-type UVR8, UVR8ΔN23 truncation, and constitutively active variant UVR8W285A on selective –His medium (SD/-Trp/-Leu/-His) in the presence or absence of UV-B. Growth on +His medium (SD/-Trp/-Leu) as a transformation control. AD, activation domain; BD, binding domain. (G) Coimmunoprecipitation of COP1 using anti-GFP antibodies in extracts from uvr8-7 (negative control), uvr8-7/Pro35S:YFP-NLS-UVR8, and uvr8-7/Pro35S:YFP-NLS-UVR8ΔN23 lines. Seven-day-old seedlings were treated with broadband UV-B for 15 min (+UV-B) or not (−UV-B). IB, immunoblotting; IP, immunoprecipitation (H and I) Immunoblot analyses: (H) UVR8, histone H3, and UGPase nuclear (Upper) and cytosolic proteins (lower) of 7-d-old uvr8-7, wild-type (Ws), uvr8-7/Pro35S:UVR8W285A line 4 (uvr8-7/UVR8W285A), and cop1-4 uvr8-7/Pro35S:UVR8W285A (cop1-4 uvr8-7/UVR8W285A) treated with 9-h narrowband UV-B or not. (I) UVR8, histone H3, and UGPase in nuclear (Upper) and cytosolic proteins (lower) of 7-d-old wild-type (Col) and uvr8-6, and rup1 rup2 treated with 6-h narrowband UV-B or not.
Examining whether COP1 affects nuclear accumulation of UV-B–activated UVR8, we found no UV-B–mediated UVR8 nuclear accumulation in cop1-4 and cop1-20 mutant seedlings (Fig. 1B and SI Appendix, Fig. S1). Total UVR8 protein levels were not affected in the cop1 mutants compared with the wild type (Fig. 1C). To verify a requirement for COP1 in UVR8 nuclear accumulation, we generated transgenic Arabidopsis lines expressing yellow fluorescent protein (YFP)-UVR8 under the control of the CaMV 35S promoter in cop1-4 mutant backgrounds. As a control, we used a transgenic complementation line with comparable YFP-UVR8 protein levels in a uvr8-6 mutant background (Fig. 1D and SI Appendix, Fig. S2). An enhanced nuclear YFP-UVR8 signal was detected in cotyledons of the uvr8/YFP-UVR8 control line in response to UV-B (Fig. 1E). In sharp contrast, UV-B–activated YFP-UVR8 did not accumulate in nuclei in the absence of functional COP1 in the cop1-4 mutant backgrounds (Fig. 1E). It is of note, however, that UVR8 monomerization is similar in cop1-4 and the wild type (5, 10). Thus, we conclude that COP1 is required for UVR8 nuclear accumulation in response to UV-B.
cop1eid6 mutant seedlings show normal etiolated growth in the dark but are hypersensitive to visible light similar to cop1-4, including dwarf growth, early flowering, and elevated pigment levels (31). In COP1EID6, the conserved His-69 residue of the RING finger motif is changed to a Tyr (31). Interestingly, COP1EID6 is impaired in visible light signaling but not in UV-B signaling, including the UV-B–dependent interaction with UVR8 (1, 11). Therefore, we tested UVR8 nuclear accumulation in response to UV-B in cop1eid6 mutants. In agreement with its ability to respond to UV-B, UVR8 nuclear accumulation in cop1eid6 was comparable to the wild type (SI Appendix, Fig. S3). This finding suggests that the UVR8 nuclear accumulation defect in cop1-4 mutants is independent of the enhanced photomorphogenic phenotype characteristic of both cop1-4 and cop1eid6, and further highlights the different structural requirements and activities of COP1 in the visible and UV-B pathways (1, 11).
Interestingly, nuclear accumulation of UVR8 was shown previously to depend on its N-terminal 23 amino acids (30). In contrast to the GFP-UVR8 wild-type control, the GFP-tagged N-terminal deletion variant GFP-ΔNUVR8 (here written GFP-UVR8ΔN23 for consistency) showed no UV-B–dependent nuclear accumulation (30). Thus, we hypothesized that the nuclear accumulation defect of UVR8ΔN23 is due to impaired UV-B–dependent interaction with COP1. We tested this hypothesis in a yeast two-hybrid assay in which wild-type UVR8 interacts with COP1 specifically in the presence of UV-B (Fig. 1F), as reported (5). Indeed, in agreement with the importance of COP1 for UV-B–dependent UVR8 nuclear accumulation in Arabidopsis and the defect in this process in UVR8ΔN23, UVR8ΔN23 in yeast was impaired in UV-B–dependent interaction with COP1 (Fig. 1F). Moreover, we generated transgenic lines expressing YFP-NLS-UVR8 or YFP-NLS-UVR8ΔN23 in a uvr8 mutant background. In contrast to YFP-NLS-UVR8, YFP-NLS-UVR8ΔN23 did not complement the hypocotyl growth inhibition phenotype (SI Appendix, Fig. S4) and did not coimmunoprecipitate endogenous COP1 in response to UV-B (Fig. 1G), in agreement with the yeast two-hybrid data.
UVR8 Triggers UV-B Photomorphogenic Responses in the Nucleus.
UVR8W285A has a constitutive signaling activity that involves interaction with COP1 and UV-B acclimation (5). In agreement with its activity, UVR8W285A showed constitutive nuclear protein levels higher than wild-type UVR8 (Fig. 1H and SI Appendix, Fig. S5), supporting the notion of predominant nuclear activity of UVR8. The nuclear accumulation of UVR8W285A was impaired in the cop1-4 mutant and, thus, depends on wild-type COP1 (Fig. 1H). In sharp contrast to UVR8, UVR8W285F did not accumulate in the nucleus in response to UV-B (SI Appendix, Fig. S5), in agreement with the absence of a response to UV-B by monomerization and of COP1 interaction (5, 8, 32). Moreover, rup1 rup2 double mutants are hypersensitive to UV-B because of impairment of UVR8 inactivation by redimerization (9, 10). Therefore, we tested whether UVR8 nuclear accumulation in response to UV-B is higher in rup1 rup2 than in the wild type and found this to be the case (Fig. 1I). This finding is a further indication that active UVR8 accumulates in the nucleus in response to UV-B.
To investigate whether UVR8 regulates photomorphogenic responses primarily in the nucleus or in the cytosol, we generated transgenic Arabidopsis lines expressing UVR8 fused to mammalian glucocorticoid receptor (GR-UVR8) and driven by the constitutive CaMV 35S promoter in a uvr8-7 null mutant background. The GR-based fusion protein system has been widely used to chemically control nuclear transport of plant proteins (33–35), including the photoreceptors phytochrome B (phyB) and cryptochrome 2 (cry2) (36, 37). In addition to GR-UVR8, we generated a GR fusion with “UV-B blind” and constitutively homodimeric UVR8W285F (5) as a negative control (GR-UVR8W285F). We selected several independent transgenic lines with expression levels comparable to endogenous UVR8 in the wild type (Fig. 2A). Whereas GR-UVR8 fusion proteins were retained in the cytosol of these lines, dexamethasone treatment resulted in partial translocation into the nucleus (SI Appendix, Fig. S6), providing a conditional nuclear accumulation system for UVR8. We further tested whether the GR fusion affects UVR8 activation by UV-B. Similar to wild-type UVR8, GR-UVR8 is a homodimer in the absence of UV-B and monomerizes in response to UV-B, both with dexamethasone (partially nuclear) and mock treatments (cytosolic) (Fig. 2B). It should be noted, that UVR8 homodimers are only detectable by SDS/PAGE of nonheat-denatured protein samples and that they migrate aberrantly at an apparent molecular mass of approximately 70 kDa, which is significantly smaller than predicted for the combination of two fully denatured UVR8 (47.1 + 47.1 = 94.2 kDa; see ref. 5). Most important, comparable aberrant migration in SDS/PAGE is seen with nonheat-denatured highly purified homodimeric recombinant UVR8 (6, 38). Similar to endogenous UVR8 homodimers, GR-UVR8 homodimers also migrated aberrantly at an apparent molecular mass of approximately 140 kDa (the monomer runs at approximately 80 kDa). Thus, we conclude that GR-UVR8 can be UV-B–activated independent of its subcellular localization and independent of dexamethasone treatment. However, UV-B activation of GR-UVR8 did not result in nuclear accumulation in the absence of dexamethasone (SI Appendix, Fig. S6), indicating that GR-mediated cytosolic retention is tight.
Fig. 2.
Nuclear UVR8 regulates UV-B responses. (A and B) Immunoblot analyses: (A) UVR8, GR-UVR8, and actin (control) proteins in 4-d-old uvr8-7, wild-type (Ws), uvr8-7/Pro35S:GR-UVR8 (uvr8-7/GR-UVR8 1 and 6), and uvr8-7/Pro35S:GR-UVR8W285F (uvr8-7/GR-UVR8W285F 4 and 8) lines. (B) UVR8 and GR-UVR8 homodimers and monomers in nonheat-denatured protein extracts of 4-d-old uvr8-7, wild-type (Ws), and uvr8-7/Pro35S:GR-UVR8 line 6 seedlings. The latter seedlings were treated with 10 µM dexamethasone (Dex) or ethanol (mock). Seedlings were irradiated for 15 min with (+) or without (−) broadband UV-B. Actin was the loading control. (C) Hypocotyl growth in weak white light with or without narrowband UV-B and treated with 10 µM dexamethasone (Dex) or ethanol (EtOH) or no treatment (−). Images of representative individuals (Upper) and hypocotyl lengths (Lower) of 4-d-old seedlings; means with SE; n = 15. (D) Quantitative RT-PCR analysis of HY5 mRNA in 7-d-old seedlings incubated for 3 h in one-half-strength MS (Ctrl.) with 10 µM dexamethasone (Dex) or ethanol (EtOH) before irradiation for 2 h with narrowband UV-B (+) or without (−). Data for Col with ethanol treatment were set to 1; means with SE; n = 3.
We further tested whether GR-UVR8 can complement uvr8 mutant phenotypes and, if so, whether nuclear localization is required for GR-UVR8 activity. For this experiment uvr8/Pro35S:GR-UVR8 transgenic lines were grown in the absence or presence of dexamethasone in white light or white light supplemented with UV-B. Hypocotyl growth was strongly inhibited only in the combined presence of dexamethasone and UV-B (Fig. 2C). This result demonstrates (i) that GR-UVR8 can functionally complement uvr8 mutants and (ii) that GR-UVR8 regulates hypocotyl growth inhibition specifically in the nucleus; cytosolic monomerization had no effect. Importantly, dexamethasone treatment did not affect wild-type, uvr8-7, or uvr8-7/Pro35S:GR-UVR8W285F control seedlings (Fig. 2C and SI Appendix, Fig. S7).
UV-B–mediated activation of UVR8 is followed by a transcriptional response. Expression of the HY5, MYB12, and ELIP2 marker genes was induced by UV-B only after concomitant treatment of GR-UVR8 transgenic lines with dexamethasone (Fig. 2D and SI Appendix, Fig. S8). In sharp contrast to GR-UVR8, treatment of GR-UVR8W285F lines with UV-B and dexamethasone did not induce transcription of those marker genes (Fig. 2D and SI Appendix, Fig. S8). Taken together, we conclude from the experiments with GR-UVR8 that nuclear UVR8 is required to regulate UV-B–induced hypocotyl growth inhibition and transcriptional induction of the examined marker genes; cytosolic UVR8 has no respective biological activity.
COP1 Is Required for UVR8 Signaling, Not only for UVR8 Nuclear Accumulation in Response to UV-B.
We tested whether COP1 is required solely for UV-B–dependent nuclear accumulation of UVR8, which could explain the previously described UV-B–insensitive phenotype of cop1 mutants (11). To test this hypothesis, we introduced GR-UVR8 into cop1-4 mutants, thus allowing the chemical induction of GR-UVR8 nuclear accumulation in the absence of functional COP1 (i.e., circumventing the COP1 requirement for nuclear accumulation) (Fig. 3A). However, in contrast to the functional complementation of uvr8 mutants, expression of GR-UVR8 in cop1-4 mutants together with dexamethasone treatment did not restore UV-B–dependent transcriptional induction of marker genes (Fig. 3B and SI Appendix, Fig. S9). Similarly, expression of NLS-UVR8, which provides a signal promoting nuclear accumulation (Fig. 3 C and D), complemented the uvr8-6 mutant but not the cop1-4 uvr8-6 double mutant (Fig. 3E). We thus conclude that COP1 is active in UVR8 nuclear accumulation but also in signaling.
Fig. 3.
Nuclear-localized NLS-UVR8 and GR-UVR8 do not rescue the cop1-4 UV-B phenotype. (A) Immunoblot analysis of UVR8, GR-UVR8, and actin (control) proteins in 4-d-old wild-type (Col), uvr8-6, uvr8-6/Pro35S:GR-UVR8 (uvr8-6/GR-UVR8 6), and cop1-4/Pro35S:GR-UVR8 (cop1-4/GR-UVR8 7 and 8) lines. (B) Quantitative RT-PCR analysis of HY5 mRNA in 7-d-old seedlings of uvr8-6/Pro35S:GR-UVR8 line 6 and cop1-4/Pro35S:GR-UVR8 lines 7 and 8 incubated for 3 h in one-half-strength MS with 10 µM dexamethasone (Dex) or ethanol (EtOH) before irradiation for 2 h with narrowband UV-B (+) or without (−); means with SE; n = 3. (C) Immunoblot analysis of UVR8, NLS-UVR8, and histone H3 proteins in nuclear fractions of uvr8-6, wild-type (Col), uvr8-6/Pro35S:NLS-UVR8 (uvr8-6/NLS-UVR8 5), and uvr8-6 cop1-4/Pro35S:NLS-UVR8 (uvr8-6 cop1-4/NLS-UVR8 7) lines grown in white light with (only wild-type Col) or without UV-B for 6 h. (D) Immunoblot analysis of UVR8 and actin (control) in total protein from 4-d-old uvr8-6, wild-type (Col), uvr8-6/Pro35S:NLS-UVR8 (uvr8-6/NLS-UVR8 5 and 9), and uvr8-6 cop1-4/Pro35S:NLS-UVR8 (uvr8-6 cop1-4/NLS-UVR8 7 and 8) lines. (E) Quantitative RT-PCR analysis of HY5 mRNA of 7-d-old seedlings irradiated for 2 h with (+) or without (−) narrowband UV-B, relative to wild-type Col minus UV-B; means with SE, n = 3.
Chemically Regulated Nuclear Import of GR-COP1 Coimports UVR8 Specifically in Response to UV-B.
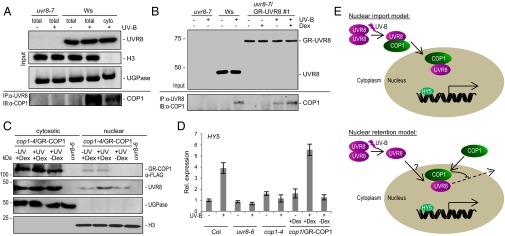
Our data indicate that COP1 may coimport UVR8 in a UV-B–dependent manner. We first tested whether cytosolic UVR8 can interact with COP1 in a UV-B–dependent manner, which would be a requirement for coimport. UVR8–COP1 interaction can be detected within minutes of UV-B irradiation of Arabidopsis seedlings (1). We performed coimmunoprecipitation assays by using a cytosolic protein fraction from UV-B–treated wild-type seedlings compared with total protein extracts as a positive control. Indeed, COP1 coimmunoprecipitated with endogenous UVR8 in the cytosolic protein fraction of UV-B–treated seedlings, albeit to a lower extent than in the total protein control (Fig. 4A). We further found that cytosolically retained, UV-B–activated GR-UVR8 coimmunoprecipitated COP1 with or without dexamethasone treatment (Fig. 4B). Altogether, we conclude that UVR8 can interact with COP1 in the cytosol, where both proteins are primarily localized in visible light in the absence of UV-B.
Fig. 4.
GR-COP1 mediation of nuclear coimport and accumulation of UVR8. (A) Coimmunoprecipitation of COP1 with UVR8 from total protein extracts of 7-d-old wild-type (Ws) and uvr8-7 seedlings (total) or Ws cytosolic fraction (cyto.). Seedlings were irradiated for 15 min with broadband UV-B or not; IB, immunoblotting; IP, immunoprecipitation. (B) Coimmunoprecipitation of COP1 with UVR8 and GR-UVR8 from total protein extracts of 7-d-old uvr8-7, wild-type (Ws), and uvr8-7/Pro35S:GR-UVR8 (line 1) seedlings irradiated for 15 min with broadband UV-B or not in the absence (−Dex) or presence of 10 µM dexamethasone (+ Dex). (C) Immunoblot analysis of GR-COP1 (FLAG-tagged), UVR8, UGPase, and histone H3 in cytosolic and nuclear fractions of 7-d-old cop1-4/Pro35S:GR-COP1 treated for 30 min with broadband UV-B or not, followed by 100 µM dexamethasone (+Dex) for 60 min or an ethanol mock treatment (−Dex); uvr8-6 seedlings +UV and +Dex as a control. (D) Quantitative RT-PCR analysis of HY5 mRNA in 7-d-old seedlings irradiated for 30 min with broadband UV-B or not. Expression levels were normalized against a Col -UV-B control; means with SE, n = 3. (E) Nuclear coimport model (Upper): UVR8 monomerization and binding to COP1 in the cytosol in response to UV-B, followed by nuclear coimport of UVR8 mediated by the COP1 NLS. Nuclear retention model (Lower): UV-B promotes UVR8 monomerization followed by translocation into the nucleus by an unknown mechanism (cryptic intrinsic NLS, coimport with a presently unknown protein); nuclear COP1 retains UVR8 in the nucleus by inhibiting its immediate nuclear export.
To test whether COP1 can mediate nuclear coimport of UV-B–activated UVR8, we established transgenic Arabidopsis lines expressing GR-COP1 in the cop1-4 mutant background. We first tested whether GR-COP1 is functional and can complement the constitutively photomorphogenic (cop) phenotype of cop1-4 in the dark. In the absence of dexamethasone, cop1-4/GR-COP1 seedlings showed a cop phenotype comparable to cop1-4 (SI Appendix, Fig. S10). However, dexamethasone treatment resulted in functional complementation, as shown by the longer hypocotyls of cop1-4/GR-COP1 compared with cop1-4 or the absence of dexamethasone (SI Appendix, Fig. S10). It should be noted, however, that GR-COP1 complemented the hypocotyl growth phenotype of cop1-4 in the presence of dexamethasone, whereas it did not complement the open cotyledon phenotype of cop1-4 (SI Appendix, Fig. S10). To test whether chemically induced nuclear translocation of GR-COP1 can coimport endogenous UVR8, we treated seedlings expressing GR-COP1 with UV-B for 30 min, followed by 60 min of dexamethasone or mock treatment. Indeed, the nuclear level of UVR8 was higher in the GR-COP1 line when UV-B activation was followed by dexamethasone treatment (Fig. 4C). Moreover, the increase was clearly detectable, despite the fact that dexamethasone treatment resulted in GR-COP1 accumulation in the nucleus only to a minor extent and, in particular, that there was no significant reduction in cytosolic GR-COP1 under these conditions (Fig. 4C). Nevertheless, dexamethasone alone in the absence of UV-B did not increase the nuclear level of UVR8. Thus, we conclude that the UV-B–dependent interaction of UVR8 with COP1 can lead to nuclear coimport of activated UVR8. Importantly, coimport of activated UVR8 with GR-COP1 functionally complemented the cop1-4 UV-B phenotype by restoring HY5 marker gene activation, specifically after UV-B and dexamethasone treatment (Fig. 4D).
Discussion
UV-B triggers UVR8 monomerization and UVR8–COP1 interaction (1, 5) and also stimulates UVR8 migration to the nucleus (30). However, a large fraction of UVR8 remains cytosolic, even under conditions most favorable for nuclear accumulation (30). The questions remained of how UVR8 nucleocytosolic partitioning is regulated and whether nuclear accumulation of active UVR8 is crucial to the observed physiological responses. The results of this study demonstrate that COP1 is required for UV-B–induced nuclear accumulation of UVR8 and that nuclear localization of UVR8 is essential for the UV-B responses tested.
Using the dexamethasone-inducible mammalian GR-based fusion protein system to chemically control nuclear transport, we have shown that GR-UVR8 is retained in the cytosol and is UV-B responsive. However, the signaling activity of UV-B–activated GR-UVR8 requires dexamethasone treatment. This finding clearly indicates that UVR8 functions primarily in the nucleus.
Although we have shown using cop1-4 mutants that COP1 is required for the nuclear accumulation of UVR8 in response to UV-B, neither NLS-UVR8 nor dexamethasone-treated GR-UVR8 rescued the cop1 mutant UV-B phenotype. Thus, COP1 appears to be active not only in UVR8 nuclear accumulation but also in UVR8-mediated signaling. Consequently, the cop1 mutant could not be used to address whether nuclear accumulation of UVR8 is required, as against simply its nuclear localization, for the induction of UV-B–induced photomorphogenic responses. It was also shown that NES-GFP-UVR8 accumulates in the nucleus in response to UV-B (30). However, nuclear levels of the N-terminal deletion variant UVR8ΔN23 (30) linked to GFP were similar to GFP-UVR8 in the absence of UV-B but did not further accumulate in the nucleus in response to UV-B. Interestingly, expression of GFP-UVR8ΔN23 did not complement the uvr8 null mutant phenotype although it appeared to interact with chromatin in the HY5 promoter similar to GFP-UVR8 (30). However, it should be noted that chromatin association of UVR8 has recently been challenged (39). Independent of this controversy, the GFP-UVR8ΔN23 responses indicate that simple nuclear localization is not sufficient and that UV-B–induced nuclear accumulation of UVR8 is required for UV-B signaling. However, UVR8ΔN23 was impaired in UV-B–dependent interaction with COP1 in yeast two-hybrid and coimmunoprecipitation assays, suggesting that this defect may be why it failed to complement the uvr8 mutant phenotype. It is likely that the UVR8 β-propeller structure is affected by the deletion of the N-terminal 23 amino acids (6, 7), which may be a reason for the impaired interaction with COP1 (32) and failure to complement uvr8 mutant phenotypes (ref. 30 and this work). Altogether, we conclude that UVR8 rapidly and strongly accumulates in the nucleus, that UVR8 nuclear localization is required for signaling, and that COP1 is required both for UVR8 nuclear accumulation in response to UV-B and for UV-B signaling.
In contrast to UVR8, COP1 contains NLS and NES sequences (12, 40). Thus, COP1 may shuttle back and forth between the cytosol and nucleus. Indeed, COP1 shows nucleocytosolic partitioning influenced by light: COP1 resides mainly in the nucleus in the dark but is excluded in white light (24, 25). Furthermore, supplemental UV-B counteracts the nuclear exclusion of COP1 in white light, resulting in its nuclear accumulation (11). The UV-B–sensitive nuclear accumulation of COP1 is associated with UVR8-dependent COP1 protein stabilization and accumulation (1). It is tempting to speculate that cytosolic UVR8-COP1 interaction leads to their combined COP1-NLS–mediated nuclear import. The GR-COP1 mediated coimport of UVR8 after UV-B treatment and in the presence of dexamethasone indicates that such a “piggy-back” coimport mechanism is possible. Interestingly, similar mechanisms have been postulated for the light-responsive nuclear import of phyA and phyB (41–43). FAR-RED ELONGATED HYPOCOTYL 1 (FHY1) and FHY1 LIKE (FHL) play important roles in phyA nuclear import, which requires only their NLS and binding domains specific for active phyA (Pfr form) (41, 42). As well as coimport by interaction with the PHYTOCHROME-INTERACTING FACTOR 3 (PIF3) transcription factor (43), the light-dependent unmasking of a cryptic NLS was postulated as a nuclear import mechanism for phyB (44). Presently, we cannot exclude a similar cryptic NLS mechanism for UVR8 nuclear accumulation. However, if such a mechanism exists, nuclear COP1 would retain imported UVR8 in the nucleus. Given the COP1 interaction that occurs in the cytosol and the presence of a COP1 NLS, we favor a model in which UVR8 and COP1 are coimported into the nucleus in response to UV-B (Fig. 4E, Upper). In bimolecular fluorescence complementation experiments, UVR8–COP1 heterodimers were detected mainly in nuclei of plant cells after exposure to UV-B (1). However, these experiments did not differentiate between nuclear coimport and nuclear retention through UVR8–COP1 interaction, or a combination of the two mechanisms. A third possibility of differential stabilization of UVR8 through interaction with COP1 specifically in the nucleus is doubtful, given that UVR8 accumulates in the nucleus without change in total protein level and that nuclear accumulation is associated with reduced cytosolic levels (ref. 30 and this work). Although we cannot exclude a nuclear retention mechanism (Fig. 4E, Lower), our data favor COP1-enhanced UVR8 nuclear accumulation due to nuclear coimport based on the COP1 NLS (Fig. 4E).
It is not clear whether cytosolic active UVR8 induces physiological responses. In previous work in which NES was fused to UVR8, UV-B treatment led to nuclear accumulation of NES-UVR8, which precluded the investigation of possible cytosolic activity of NES-UVR8 (30). In this present study, we retained GR-UVR8 in the cytosol in the absence of dexamethasone and showed that GR-UVR8 remained in the cytosol after UV-B exposure. In the absence of dexamethasone, cytosolic GR-UVR8 monomerized and interacted with COP1 in response to UV-B. In contrast, hypocotyl shortening and changes in gene expression depended on dexamethasone treatment and, thus, nuclear translocation of GR-UVR8. However, we cannot exclude that some untested (e.g., stomatal closure; ref. 45) or as yet unknown physiological response to UV-B is activated by cytosolic UVR8. The uvr8/Pro35S:GR-UVR8 described here will allow further investigation of possible cytosolic UVR8 activity.
Materials and Methods
Plant Material, Growth Conditions, and UV-B Irradiation.
The uvr8-6 and rup1-1 rup2-1 mutants are in the Columbia (Col), uvr8-7 in the Wassilewskija (Ws), and cop1eid6 in the Landsberg erecta (Ler) backgrounds (1, 9). The cop1-4 mutant allele used in this work is in the Col background (46) except for the results in SI Appendix, Fig. S1, where a cop1-4 allele in the Ws background was used (11). The SALK_137391 line (47) is a T-DNA–tagged cop1 mutant allele, designated herein as cop1-20 (SI Appendix, Fig. S11). The uvr8-7/Pro35S:UVR8W285A and uvr8-7/Pro35S:UVR8W285F transgenic lines were as described (8). The uvr8-7/Pro35S:UVR8W285A line 4 (8) was crossed with cop1-4 (Ws) (11) to generate cop1-4 uvr8-7/ Pro35S:UVR8W285A.
Arabidopsis seeds were surface-sterilized and sown on one-half-strength Murashige and Skoog basal salt medium (MS; Duchefa Biochemie) containing 1% (wt/vol) sucrose and 1% (wt/vol) phytagel (Sigma-Aldrich). Seeds were stratified for 2 d in the dark at 4 °C, and seedlings were grown at 22 °C under continuous irradiation in a white-light field with Osram L18W/30 tubes (1). For dexamethasone treatment, seedlings were grown as described above in medium supplemented with dexamethasone (Sigma-Aldrich) dissolved in ethanol, with an equivalent volume of ethanol as control. UV-B treatments were performed by using established conditions with broadband (Philips TL40W/12RS; 21 μmol⋅m−2⋅s−1) (27) or narrowband UV-B lamps (Philips TL20W/01RS; 1.5 μmol⋅m−2⋅s−1) (11) as indicated.
Generation of Transgenic Arabidopsis Lines.
The coding sequence of UVR8 (At5g63860) was cloned into pDONR207 (Thermo Fisher Scientific). The correct sequence was confirmed by sequencing and further inserted into the Gateway-compatible vectors pJAN33-FLAG-GR (34), pB7WGY2, and pB2GW7 (48).
To clone NLS-UVR8Δ23, forward primer (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGCTGCAGCCTAAGAAGAAGAGAAAGGTTGGAGGAGCTAGCCACTCCGTCGCTCTTCTC-3′) and reverse primer (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTCAAATTCGTACACGCTTGAC-3′) were used. To clone NLS-UVR8, forward primer (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGCTGCAGCCTAAGAAGAAGAGAAAGGTTGGAGGAATGGCGGAGGATATGGCTGCCGAC-3′) and reverse primer (5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTCAAATTCGTACACGCTTGAC-3′) were used. The PCR products were cloned into pDONR207. The NLS-UVR8Δ23 and NLS-UVR8 sequences were further inserted into the Gateway-compatible vector pB7WGY2 (48).
The coding sequence of COP1 (At2g32950) was first cloned into pENTR1A (Thermo Fisher Scientific) and further inserted into pJAN33-FLAG-GR (34). The binary vectors were used for Agrobacterium-mediated transformation of plants (49). The resulting transgenic lines were shown to have the transgene integrated at a single locus. Homozygous transgenic lines were used for experiments.
Cell Fractionation.
Seven-day-old seedlings were collected for total protein isolation in extraction buffer [20 mM Tris⋅HCl, pH 7.4, 25% (vol/vol) glycerol, 20 mM KCl, 2 mM EDTA, 2.5 mM MgCl2, 250 mM sucrose, 1 mM DTT, and 1 mM PMSF] at 4 °C. Total protein extracts were filtered through three layers of Miracloth. After centrifugation at 1,500 × g for 10 min at 4 °C, the clear supernatant was taken as the cytosolic fraction. The pellet was washed twice with nuclei resuspension Triton buffer [20 mM Tris⋅HCl, pH 7.4, 25% (vol/vol) glycerol, 2.5 mM MgCl2, 0.2% Triton X-100] and once with nuclei resuspension buffer [20 mM Tris⋅HCl, pH 7.4, 25% (vol/vol) glycerol, 2.5 mM MgCl2]. For protein gel blots, 30 µg of the cytosolic and 10 µg of the nuclear fraction were separated by SDS/PAGE and transferred to polyvinylidene difluoride (PVDF) membranes.
Protein Extraction, Immunoprecipitation, and Protein Gel Blots.
For immunoprecipitation, total or cytosolic protein extracts were incubated with anti-UVR8(426–440) antibodies (10). Immunoprecipitates were captured with protein A-agarose (Roche Applied Science) for 1 h. For protein gel blot analysis, proteins were separated by SDS/PAGE and transferred to PVDF membranes according to the manufacturer’s instructions (Bio-Rad). Polyclonal anti-UVR8(426–440) (1), anti-COP1(13–26) (10), anti-histone H3 (Abcam), anti-UGPase (Agrisera), and anti-actin (Sigma-Aldrich) were used as primary antibodies, with horseradish peroxidase (HRP)-conjugated protein A (Pierce), anti-rabbit immunoglobulins or anti-mouse immunoglobulins (Dako A/S) as secondary antibodies. Chemiluminescent signals were generated by using the ECL Western Detection Kit and detected with an ImageQuant LAS 4000 mini CCD camera system (GE Healthcare).
Quantitative Real-Time PCR.
Arabidopsis total RNA was isolated with Plant RNeasy kit (Qiagen) and treated with DNaseI according to the manufacturer’s instructions. cDNA was synthesized with a 1:1 mix of poly-A primer and hexamers with a TaqMan Reverse Transcription Reagents Kit (Applied Biosystems). PCRs were performed by using the ABsolute QPCR Rox Mix Kit according to the manufacturer’s instructions (ABgene). The primers used were as follows: for HY5 (At5g11260), HY5-Fd (5′-GCTCTTTTCCTCTTTATCCTTTTCAC-3′) and HY5-Rv (5′-TGTTCCTGCATTTTTCTTACTCTTTG-3′) (50); for ELIP2 (At4g14690), ELIP2-Fd (5′-GTGAGTACGAAGTTTGGAGATTTGC-3′) and ELIP2-Rv (5′-TTGCTAGTCTCCCGTTGATCCT-3′) (8); for MYB12 (At2g47460), MYB12-Fd (5′-AAAAACTCGTAAAACGAAGAAAACG-3′) and MYB12-Rv (5′-TCTTTATCAGCCCCAGCTACATC-3′) (51). cDNA concentrations were normalized to the 18S rRNA transcript levels as standard by using the Eukaryotic 18S rRNA Kit (Applied Biosystems). Quantitative real-time PCR was carried out by using a 7900HT Real-Time PCR System (Applied Biosystems). Expression was determined in biological triplicates.
Subcellular Localization Analysis.
Transgenic seedlings were grown on one-half-strength MS phytagel plates supplemented with 1% sucrose for 4 d. Seedlings were grown for 3 d under weak white light followed by 24 h with (+UV-B) or without (−UV-B) supplementary narrowband UV-B. To stain nuclei, individual seedlings were mounted in PBS with 5 μg/mL 4'-6-diamidino-2-phenylindole (DAPI) (SERVA Electrophoresis) under a coverslip and incubated for 15 min before imaging. Imaging of YFP-UVR8 and DAPI was performed with an LSM 780 confocal laser-scanning microscope (Zeiss) (Bioimaging Center, University of Geneva) by using a water 40× N.A. 1.2 C-Apochromat lens. DAPI and YFP were excited at 405 nm and 514 nm, respectively, and emission was determined between 436 and 482 nm with a PMT (DAPI) and between 525 and 561 nm with a GaAsP detector (YFP). Acquisition settings and image processing were the same for all treatments and genotypes. Z stacks of all images were processed in Fiji by using the maximum projection function (52).
Supplementary Material
Acknowledgments
We thank Stephan Wenkel (University of Copenhagen) for providing GR binary vectors, and Joël Nicolet, Christophe Buser, and Marie-Charlotte Benagli for excellent technical assistance. This study was supported by the Canton Geneva, Swiss National Science Foundation Grant 31003A_153475, and European Research Council Grant 310539 (UV-B Perception) under the European Union’s Seventh Framework Programme.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607074113/-/DCSupplemental.
References
- 1.Favory JJ, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28(5):591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.González Besteiro MA, Bartels S, Albert A, Ulm R. Arabidopsis MAP kinase phosphatase 1 and its target MAP kinases 3 and 6 antagonistically determine UV-B stress tolerance, independent of the UVR8 photoreceptor pathway. Plant J. 2011;68(4):727–737. doi: 10.1111/j.1365-313X.2011.04725.x. [DOI] [PubMed] [Google Scholar]
- 3.Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 2002;130(1):234–243. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown BA, et al. A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci USA. 2005;102(50):18225–18230. doi: 10.1073/pnas.0507187102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rizzini L, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332(6025):103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 6.Christie JM, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335(6075):1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu D, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484(7393):214–219. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- 8.Heijde M, et al. Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc Natl Acad Sci USA. 2013;110(50):20326–20331. doi: 10.1073/pnas.1314336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruber H, et al. Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci USA. 2010;107(46):20132–20137. doi: 10.1073/pnas.0914532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heijde M, Ulm R. Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci USA. 2013;110(3):1113–1118. doi: 10.1073/pnas.1214237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oravecz A, et al. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 2006;18(8):1975–1990. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17(10):584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Deng XW, et al. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992;71(5):791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- 14.Zhu D, et al. Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell. 2008;20(9):2307–2323. doi: 10.1105/tpc.107.056580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laubinger S, Fittinghoff K, Hoecker U. The SPA quartet: A family of WD-repeat proteins with a central role in suppression of photomorphogenesis in arabidopsis. Plant Cell. 2004;16(9):2293–2306. doi: 10.1105/tpc.104.024216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoecker U, Tepperman JM, Quail PH. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science. 1999;284(5413):496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- 17.Ordoñez-Herrera N, et al. A cop1 spa mutant deficient in COP1 and SPA proteins reveals partial co-action of COP1 and SPA during Arabidopsis post-embryonic development and photomorphogenesis. Mol Plant. 2015;8(3):479–481. doi: 10.1016/j.molp.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 18.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405(6785):462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 19.Lian HL, et al. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011;25(10):1023–1028. doi: 10.1101/gad.2025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, Zuo Z, Liu H, Liu X, Lin C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011;25(10):1029–1034. doi: 10.1101/gad.2025011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuo Z, Liu H, Liu B, Liu X, Lin C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr Biol. 2011;21(10):841–847. doi: 10.1016/j.cub.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu XD, et al. Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant. 2015;8(3):467–478. doi: 10.1016/j.molp.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 23.Sheerin DJ, et al. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell. 2015;27(1):189–201. doi: 10.1105/tpc.114.134775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Arnim AG, Deng XW. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell. 1994;79(6):1035–1045. doi: 10.1016/0092-8674(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 25.Pacín M, Legris M, Casal JJ. Rapid decline in nuclear costitutive photomorphogenesis1 abundance anticipates the stabilization of its target elongated hypocotyl5 in the light. Plant Physiol. 2014;164(3):1134–1138. doi: 10.1104/pp.113.234245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, et al. Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell. 2012;24(11):4590–4606. doi: 10.1105/tpc.112.103994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulm R, et al. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA. 2004;101(5):1397–1402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang X, et al. Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc Natl Acad Sci USA. 2013;110(41):16669–16674. doi: 10.1073/pnas.1316622110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Binkert M, et al. UV-B-responsive association of the Arabidopsis bZIP transcription factor ELONGATED HYPOCOTYL5 with target genes, including its own promoter. Plant Cell. 2014;26(10):4200–4213. doi: 10.1105/tpc.114.130716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaiserli E, Jenkins GI. UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell. 2007;19(8):2662–2673. doi: 10.1105/tpc.107.053330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dieterle M, Buche C, Schafer E, Kretsch T. Characterization of a novel non-constitutive photomorphogenic cop1 allele. Plant Physiol. 2003;133(4):1557–1564. doi: 10.1104/pp.103.028654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin R, Arongaus AB, Binkert M, Ulm R. Two distinct domains of the UVR8 photoreceptor interact with COP1 to initiate UV-B signaling in Arabidopsis. Plant Cell. 2015;27(1):202–213. doi: 10.1105/tpc.114.133868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schena M, Lloyd AM, Davis RW. A steroid-inducible gene expression system for plant cells. Proc Natl Acad Sci USA. 1991;88(23):10421–10425. doi: 10.1073/pnas.88.23.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brandt R, et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 2012;72(1):31–42. doi: 10.1111/j.1365-313X.2012.05049.x. [DOI] [PubMed] [Google Scholar]
- 35.Samach A, et al. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288(5471):1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- 36.Huq E, Al-Sady B, Quail PH. Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 2003;35(5):660–664. doi: 10.1046/j.1365-313x.2003.01836.x. [DOI] [PubMed] [Google Scholar]
- 37.Yu X, et al. Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell. 2007;19(10):3146–3156. doi: 10.1105/tpc.107.053017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heilmann M, Jenkins GI. Rapid reversion from monomer to dimer regenerates the ultraviolet-B photoreceptor UV RESISTANCE LOCUS8 in intact Arabidopsis plants. Plant Physiol. 2013;161(1):547–555. doi: 10.1104/pp.112.206805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Binkert M, et al. Revisiting chromatin binding of the Arabidopsis UV-B photoreceptor UVR8. BMC Plant Biol. 2016;16(1):42. doi: 10.1186/s12870-016-0732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian C, et al. The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: Mutational analysis by bioluminescence resonance energy transfer. Proc Natl Acad Sci USA. 2004;101(17):6798–6802. doi: 10.1073/pnas.0307964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Genoud T, et al. FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 2008;4(8):e1000143. doi: 10.1371/journal.pgen.1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rausenberger J, et al. Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell. 2011;146(5):813–825. doi: 10.1016/j.cell.2011.07.023. [DOI] [PubMed] [Google Scholar]
- 43.Pfeiffer A, et al. Interaction with plant transcription factors can mediate nuclear import of phytochrome B. Proc Natl Acad Sci USA. 2012;109(15):5892–5897. doi: 10.1073/pnas.1120764109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen M, Tao Y, Lim J, Shaw A, Chory J. Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr Biol. 2005;15(7):637–642. doi: 10.1016/j.cub.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 45.Tossi V, Lamattina L, Jenkins GI, Cassia RO. Ultraviolet-B-induced stomatal closure in Arabidopsis is regulated by the UV RESISTANCE LOCUS8 photoreceptor in a nitric oxide-dependent mechanism. Plant Physiol. 2014;164(4):2220–2230. doi: 10.1104/pp.113.231753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McNellis TW, et al. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell. 1994;6(4):487–500. doi: 10.1105/tpc.6.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301(5633):653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 48.Karimi M, Inzé D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7(5):193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 49.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 50.Tilbrook K, et al. UV-B perception and acclimation in Chlamydomonas reinhardtii. Plant Cell. 2016;28(4):966–983. doi: 10.1105/tpc.15.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stracke R, et al. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2010;33(1):88–103. doi: 10.1111/j.1365-3040.2009.02061.x. [DOI] [PubMed] [Google Scholar]
- 52.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.