Significance
The study of cell-to-cell genomic differences in complex multicellular systems such as cancer requires genome sequencing of large numbers of single cells. This in turn necessitates the uniform amplification of single-cell genomes with high reproducibility across large numbers of cells, which remains an outstanding challenge. Here, we introduce a method that uses commercially available liquid dispensing to perform inexpensive and high-throughput single-cell whole genome amplification (WGA) in nanoliter volumes. For the first time, to our knowledge, we demonstrate robust and highly uniform nanoliter-volume single-cell WGA across a large replicate set consisting of more than 100 single cells. Comparison with previous datasets shows that this method improves uniformity and achieves levels of genome coverage and genomic variant detection comparable or superior to existing methods.
Keywords: single-cell sequencing, whole genome amplification, multiple displacement amplification, microdroplet, nanoliter volume
Abstract
The genomes of large numbers of single cells must be sequenced to further understanding of the biological significance of genomic heterogeneity in complex systems. Whole genome amplification (WGA) of single cells is generally the first step in such studies, but is prone to nonuniformity that can compromise genomic measurement accuracy. Despite recent advances, robust performance in high-throughput single-cell WGA remains elusive. Here, we introduce droplet multiple displacement amplification (MDA), a method that uses commercially available liquid dispensing to perform high-throughput single-cell MDA in nanoliter volumes. The performance of droplet MDA is characterized using a large dataset of 129 normal diploid cells, and is shown to exceed previously reported single-cell WGA methods in amplification uniformity, genome coverage, and/or robustness. We achieve up to 80% coverage of a single-cell genome at 5× sequencing depth, and demonstrate excellent single-nucleotide variant (SNV) detection using targeted sequencing of droplet MDA product to achieve a median allelic dropout of 15%, and using whole genome sequencing to achieve false and true positive rates of 9.66 × 10−6 and 68.8%, respectively, in a G1-phase cell. We further show that droplet MDA allows for the detection of copy number variants (CNVs) as small as 30 kb in single cells of an ovarian cancer cell line and as small as 9 Mb in two high-grade serous ovarian cancer samples using only 0.02× depth. Droplet MDA provides an accessible and scalable method for performing robust and accurate CNV and SNV measurements on large numbers of single cells.
Genomic heterogeneity is now appreciated for its functional significance across oncology, development, neuroscience, and biotechnology. Rapid advances in cell handling, DNA amplification methods, and sequencing throughput have enabled genomic interrogation of human disease and physiology with single-cell resolution, including studies of heterogeneity in blood and solid tumors (1–4), circulating tumor cells (5), neurons (6), gametes (7, 8), and embryos (9). Despite the intense interest in single-cell genome sequencing, it remains widely inaccessible for two primary reasons: lack of robustness and high cost. In particular, whole genome amplification (WGA) is a critical step required to generate sufficient DNA mass for sequencing in most single-cell genomic studies, but it is prone to amplification bias, contamination, and poor coverage. Preferential amplification of some genomic regions results in distorted representation of the original template and compromises the accuracy of downstream measurements. High variation in single-cell genomic coverage and bias, both within experiments and between users, remains a fundamental problem.
In response to these challenges, there has been considerable effort and advancement in molecular strategies for amplifying low picogram amounts of genomic material. Of these, three WGA strategies have been most widely adopted, each with its relative strengths and weaknesses (10, 11): PCR-based protocols (1, 12), multiple displacement amplification (MDA) (2–4, 7), and combinations of displacement preamplification followed by PCR (8, 13–15). Of these strategies, MDA is most widely used when maximization of coverage breadth (fraction of reference genome covered at ≥1×) is the priority. Previous studies have shown that the implementation of MDA in nanoliter volumes can reduce amplification bias (7, 16, 17), minimize contamination and nonspecific products, and provide economies of scale that can significantly reduce reagent costs. However, to date, this approach has been demonstrated on only a limited number of single cells (<10). Although such datasets highlight the optimal performance of a method, these small sample sizes make it impossible to assess robustness, performance, or scalability, which are of utmost importance when evaluating a WGA method. Furthermore, nanoliter-volume MDA has been implemented exclusively in microfluidic devices that require specialized instrumentation and microfabrication, limiting accessibility.
To assess the robustness and performance of nanoliter-volume single-cell MDA, we adapted a commercially available noncontact liquid dispenser to systematically test amplification performance across large numbers of replicate reactions. For the first time, to our knowledge, we characterize the robustness and performance of a scalable nanoliter-volume single-cell MDA technology (referred to here as droplet MDA) across a large dataset composed of 129 normal diploid cells. Using both low-depth (0.02×) and higher-depth (6.5×) whole genome sequencing (WGS), we comprehensively compared the performance of droplet MDA to other recently reported state-of-the-art methods. This large single-cell dataset shows that our approach exceeds previously reported methods in one or more key performance metrics, including amplification bias, robustness, scalability, and coverage breadth. Our study further reveals that the observed variability in droplet MDA performance is driven by biological differences in cell state and cycle.
We then assessed the copy number variant (CNV) measurement capabilities enabled by droplet MDA using single cells from both a normal diploid cell line and an ovarian cancer cell line with CNVs on multiple length scales. We next evaluated single nucleotide variant (SNV) detection performance in single cells using both PCR-based targeted sequencing and deep WGS (48×) of droplet MDA product. Finally, to demonstrate applicability to primary samples, we used droplet MDA and low-depth WGS to perform CNV profiling of single nuclei from two high-grade serous ovarian cancer specimens. Taken together, this dataset demonstrates that droplet MDA provides a scalable and accessible method of performing robust and accurate CNV and SNV measurements on large numbers of single cells.
Results
Nanoliter-Volume Single-Cell MDA Reaction Formulation.
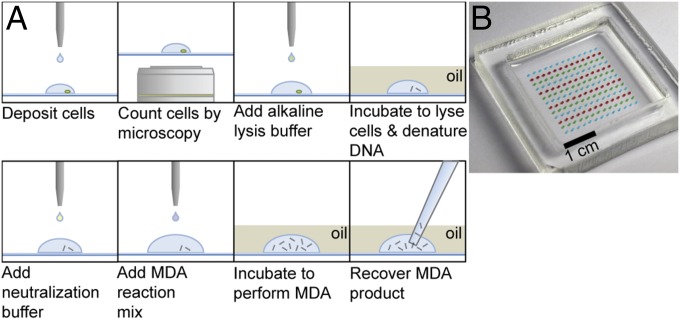
All cells and reagents were deposited onto a planar substrate using a commercially available piezoelectric noncontact liquid dispenser (Fig. 1 and SI Appendix, Fig. S1). Droplets containing single cells were obtained by dispensing cells at limiting dilution. Cells were stained with a fluorescent dye binding double-stranded DNA, and the number of cells in each droplet was then determined by fluorescence microscopy. We observed an average single-cell occupancy rate of 22% over all experiments described here. MDA reagents were then sequentially deposited onto each droplet. To minimize evaporation during heating of the substrate, the droplet array was covered with mineral oil before any heating steps (18). A total MDA reaction volume of 100 nL was used for this study. Additional details of methods and materials are available in SI Appendix, Materials and Methods.
Fig. 1.
Droplet MDA system. (A) Schematic of single-cell droplet MDA protocol. (B) Substrate with 100-nL droplets of food dye covered by light mineral oil.
Assessment of Amplification Bias from Low-Depth WGS.
To evaluate the amplification bias of droplet MDA, we analyzed cells from the 184-hTERT mammary epithelial cell line, which has been well characterized and has a chromosomally stable and nearly diploid karyotype (19). To make assessment of a large number of replicates practical, we initially performed low-depth (mean, 0.02×) WGS of single-cell MDA reaction products.
We developed an analysis pipeline to allow for the fair comparison of sample datasets having unequal average sequencing depths using the HMMcopy software package (20) (SI Appendix, Materials and Methods). We first evaluated the effect of amplification time and total MDA yield on bias and coverage. Amplification bias has been shown to scale with reaction gain (ratio of output DNA mass to input DNA mass) (10), and reduction of MDA time has been used to reduce bias (4, 17). In contrast to previous results, we observed a trend of reduced bias with increasing single-cell MDA reaction time and yield (SI Appendix, Table S1), as determined by the SD of reads per 1-Mb bin (SI Appendix, Fig. S2). We posit that this effect is due to the lower yield of nanoliter-volume reactions and losses in subsequent sequencing library preparation, because each genomic region must be amplified above a threshold to be represented in the library. Based on this result, we performed all subsequent MDA reactions for 18–20 h. The typical yield of our 100 nL-volume single-cell MDA reactions was ∼60 ng, representing roughly a 10,000-fold amplification of a normal diploid human genome.
Using this optimized protocol, we performed droplet MDA on 166 single 184-hTERT cells and nuclei in six separate experiments spanning an 8-mo period. Of these, 129 single-cell samples were selected at random for low depth (mean, 0.02×) WGS, of which FACS was used to obtain 10 nuclei in G1 phase, 11 nuclei in S phase, and 13 nuclei in G2 phase (SI Appendix, Fig. S3). The remaining 95 samples of the 129 samples sequenced were unsorted. Three reactions containing cell suspension fluid but no cells, which were processed on the same substrate directly adjacent to single-cell reactions, were also sequenced as stringent negative controls to detect potential cross-contamination between reactions or cell-free DNA in the suspension.
We assessed the bias of droplet MDA relative to the following published methods that have been shown to achieve high coverage breadth: multiple annealing and looping-based amplification cycles (MALBAC) on single cancer cell line cells (15), MDA of single sperm cells in custom microfluidic devices (referred to herein as “custom microfluidic MDA”) (7), microwell MDA of single neurons (MIDAS) (17), MDA of single cancer cell line cells in G2/M phase (nuc-seq) (4), WGA of genomic fragments partitioned into picoliter droplets performed on single endothelial cells (eWGA) (21), and MDA of single B-lymphoblast cell line cells in a commercially available microfluidic device (referred to herein as “commercial microfluidic MDA”) (22). We analyzed all publicly available single-cell WGS data in each study, using data from normal diploid cells for comparison wherever possible. We note that sperm cells are haploid and thus are expected to have reduced coverage and increased bias relative to samples with higher ploidy. Conversely, the nuc-seq method, which specifically selects single cells with at least four genome copies, is expected to benefit from increased ploidy.
With these caveats, we sought to perform a fair comparison of all methods using the aforementioned pipeline to reanalyze all raw datasets using the same analysis tools and parameters on equal quantities of aligned data. To reduce the confounding of bias measurements by true biological variation in single cell genomes, we performed CNV analysis on sequencing data from bulk DNA from nonnormal diploid cell lines where available, and omitted chromosomes with large-scale CNVs from our bias analysis (SI Appendix, Fig. S4 and Table S2). Although the omission of such chromosomes is necessary to perform as fair of a comparison as possible, we note that it often is not done in other comparisons in the literature. Chromosomes X and Y were omitted for the sperm cells.
We first computed basic alignment metrics using data from all samples randomly downsampled to the same number of total sequenced bases (SI Appendix, Table S3). Single-cell droplet MDA libraries (n = 129) had a mean duplicate rate of 0.11 ± 0.025%, a mean alignment rate of 97.99 ± 0.65%, and a mean coverage breadth of 0.71 ± 0.11%. This was the highest observed alignment rate and the lowest duplicate rate of all methods compared. The mean coverage breadth of droplet MDA samples was higher than that of all other datasets, with the exception of those reported from MIDAS. Libraries generated from droplet MDA negative control reactions containing cell suspension fluid but no cells had negligible coverage breadth (mean, 0.065 ± 0.025%; n = 3), showing that contamination, both from external sources and from other reactions on the same substrate, do not contribute appreciably to sequencing data.
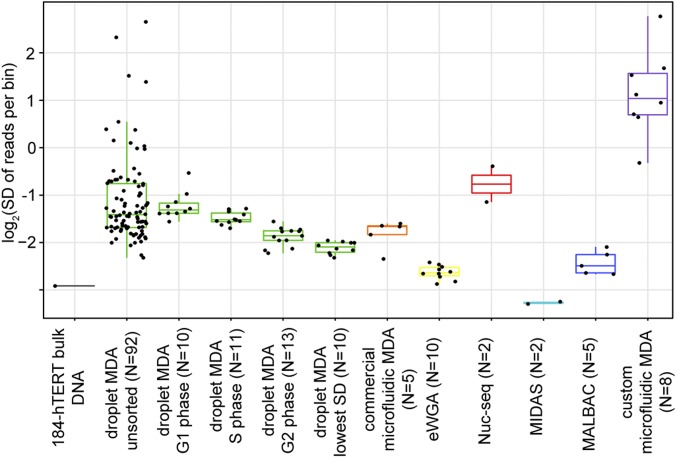
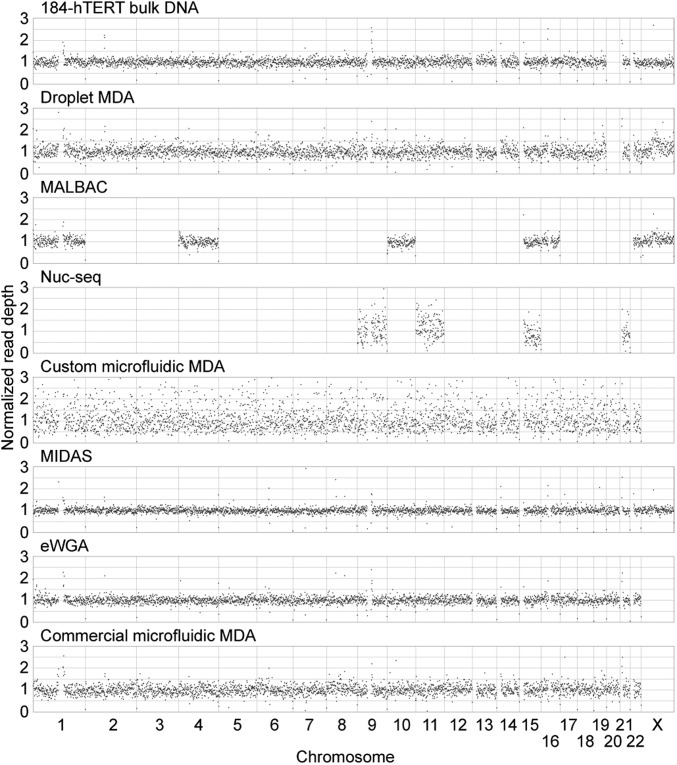
To accommodate the low WGS depth to which all included single-cell droplet MDA samples were sequenced, we performed a comparison using a bin size of 1 Mb (mean of 100 reads per bin). We then assessed the bias on this genomic length scale by comparing the SD in reads per bin (Fig. 2). When considering the 92 unsorted single-cell droplet MDA samples (excluding three extreme outliers), we observed a median SD lower than that of both nuc-seq and custom microfluidic MDA. When including only the 10 droplet MDA samples (the largest number of samples available from all other methods compared) with the lowest SD, the SD values of droplet MDA samples were significantly (P < 0.05) lower than those of commercial microfluidic MDA (P = 0.0376, one-sided Wilcoxon rank-sum test), nuc-seq (P = 0.0152), and custom microfluidic MDA (P = 2.29e-05). These results are qualitatively reflected in the read depth plots for the sample with the lowest SD from each method (Fig. 3). The MIDAS samples were found to exhibit SDs below that of the unamplified bulk 184-hTERT DNA sample (0.152 vs. 0.19). This result, based on reported data for the MIDAS method, is difficult to reconcile with the mechanism of MDA amplification, and we note that another group was unable to reproduce the MIDAS results (10). The SDs of all samples compared are listed in SI Appendix, Table S4.
Fig. 2.
Scatterplots and boxplots of SD of reads per 1-Mb bin comparing other published methods with all 184-hTERT single-cell droplet MDA samples sequenced to low depth. Unamplified bulk 184-hTERT gDNA is also included for comparison. Shown for the 184-hTERT single-cell droplet MDA samples are unsorted single cells (with three extreme outliers omitted), single cells FACS-sorted by cell phase (G1, S, and G2), and the 10 samples with the lowest SDs of all sorted and unsorted single cells.
Fig. 3.
Normalized read depth plots using 1-Mb bins for the sample from each method with the lowest SD in reads per bin.
Because visual inspection by fluorescence microscopy was used to identify cells in droplets, we hypothesize that many of the non–FACS-sorted samples included apoptotic cells or cellular debris, from which reduced amplification uniformity relative to intact and viable cells would be expected. This is supported by the observation that the SD of droplet MDA samples that were FACS-sorted before deposition into droplets, and gated to exclude such suboptimal samples, exhibited a much narrower distribution than unsorted samples (Fig. 2). The FACS-sorted samples also allowed us to test the effect of cell phase and ploidy on amplification bias. As noted elsewhere, amplification bias is dependent on cell phase (4). We observed that the median SD in reads per bin decreased from G1 phase to S phase to G2 phase (Fig. 2). Cells in G2 phase exhibited lower SD than the nuc-seq samples, which selects for cells in G2 phase (P = 0.0095, one-sided Wilcoxon rank-sum test). Within any given sorted cell population, the performance of the method displays low variability. These results strongly support the notion that the main contributor to MDA variability is biological, and that samples exhibiting the highest SD likely are cells with increased genomic variation owing to the biological state of the cell, and are not the result of variability in the performance of the method itself.
We also examined the GC content in each sample group (SI Appendix, Fig. S5) before correction for GC content bias. Interestingly, the curves for S phase 184-hTERT cells processed by droplet MDA exhibited distinct peaks at a GC content of ∼0.4, perhaps suggesting that DNA replication in S phase begins in regions with this level of GC content. Notably, the curves of samples from all other methods except nuc-seq exhibited very similar peaks and were distinct from those of the G1 phase 184-hTERT cells processed by droplet MDA, which decreased monotonically.
Assessment of Amplification Bias from High-Depth WGS.
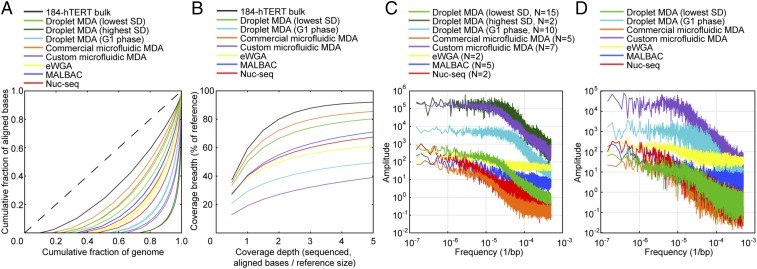
We next performed higher-depth (mean, 6.5×) WGS on 15 unsorted 184-hTERT single-cell droplet MDA samples among those with the lowest SD in reads per 1-Mb bin and the two samples with the highest SD (determined from low-depth WGS data as described above), and the 10 sorted G1 phase single-cell samples. We performed a comparison with data from other methods by downsampling all datasets to the same depth (3.73×), defined as the number of aligned bases divided by reference size (taking into consideration omitted genomic regions). From these downsampled data, we generated Lorenz curves for each sample to depict uniformity of coverage. This downsampling step, often omitted in other published comparisons, is crucial because higher depth can result in higher coverage breadth and thus a shift in the X-intercept of the Lorenz curve. Lorenz curves for the two samples with the lowest SD in each sample group are shown in Fig. 4A, and those of all samples analyzed are shown in SI Appendix, Fig. S6. MIDAS samples were not included in this analysis, because available data were at a depth of only 0.2×. The most uniform unsorted droplet MDA and commercial microfluidic MDA samples had the highest uniformity of all samples analyzed, whereas the G1-phase droplet MDA samples were less uniform than those of all other methods using diploid cells, again underscoring the important role of cell state.
Fig. 4.
Analysis of amplification bias from high-depth WGS. (A) Lorenz curves depicting uniformity of coverage for the two samples with the lowest SD in reads per 1-Mb bin from each amplification method. (B) Coverage breadth as a function of sequencing depth for the sample with the lowest SD in reads per 1-kb bin from each method. (C) Mean power spectra of 1-kb binned read depth for all samples analyzed from each amplification method. (D) Power spectra of 1-kb binned read depth for the sample with the lowest SD in reads per bin from each method.
We then used the analysis pipeline described above to bin reads into 1 kb-bins (mean of 30 reads per bin), computed the SD in reads per bin, and plotted coverage breadth as a function of sequencing depth (0.5–5×) for the sample with the lowest SD from each method (Fig. 4B). As before, the commercial microfluidic MDA and droplet MDA samples achieved the highest coverage breadth of all samples at any given sequencing depth, covering ∼84% and 80% of the reference genome, respectively, when sequenced to 5× depth.
Next, we used read depths from the 1-kb bins to analyze the characteristic length scale of coverage bias by computing the power spectra of read density variations. The mean power spectra of all samples from each sample group and the power spectrum of the sample with the lowest SD in reads per 1-kb bin from each sample group are shown in Fig. 4 C and D, respectively. As observed in other studies, the amplitudes of MDA samples generally showed a downward inflection point between 104 and 105 bp, corresponding to the mean fragment length of the MDA product (10, 15, 23). Whereas the SD in reads per 1-Mb bin of droplet MDA samples was higher compared with those of eWGA and MALBAC (Fig. 2), the mean power spectra of the droplet MDA samples dropped below that of eWGA and MALBAC at length scales smaller than 104–105 bp, explaining the superior performance of the droplet MDA samples in the Lorenz curve and coverage breadth vs. depth comparisons.
CNV Detection.
Because the 184-hTERT cell line is normal diploid and genomically stable, we first assessed CNV detection using droplet MDA by comparing the concordance of copy number calls between single-cell samples and unamplified bulk DNA as has been done elsewhere (14, 24). We again used the HMMcopy software package, which takes in normalized binned read depth, groups contiguous bins into segments predicted to have equal copy number, and assigns a copy number to bins in each segment using a hidden Markov model (20). We binned the high-depth WGS data from the 15 samples sequenced to high-depth and bulk DNA into bins of 1 Mb, 100 kb, and 10 kb (mean of 100 reads per bin for all bin sizes), assigned a copy number to each bin using HMMcopy, and computed the binwise concordance (fraction of genomic bins that have an identical copy number state for two samples) between each single-cell sample and bulk DNA (SI Appendix, Fig. S7). For 1-Mb, 100-kb, and 10-kb bin sizes, the median concordance rate was 100% (of 2,487 bins), 93.1% (of 24,162 bins), and 83.4% (of 227,172 bins), respectively. Using 1-Mb bins (mean of 100 reads per bin), the median concordance rate for all 129 184-hTERT single-cell samples sequenced to low depth is 100% (SI Appendix, Fig. S8).
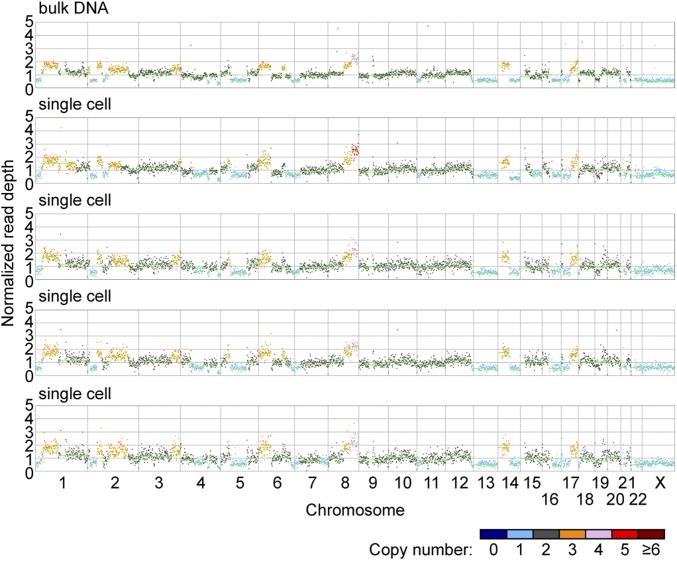
We next performed droplet MDA on 30 single cells from the TOV2295 cell line (25), derived from a high-grade serous ovarian cancer, which is genomically unstable and has CNVs on multiple length scales. Low-depth (mean, 0.02×) WGS was performed on all 30 samples, and higher-depth (mean, 6.5×) WGS was performed on the eight samples with the highest coverage breadth calculated from the low-depth WGS data. Using the high-depth data, reads were binned into 1-Mb, 100-kb, and 10-kb bins (mean of 100 reads per bin for all bin sizes). Read depth plots using 1-Mb bins from four representative single-cell samples qualitatively matched that of bulk DNA (Fig. 5), with many of the same large-scale variations discernible in both. We again used HMMcopy to find segments of contiguous bins with the same copy number. Because the TOV2295 cell line is genomically unstable, unlike the 184-hTERT cell line, there is likely to be true biological variation between the copy number profiles of individual single cells and bulk DNA. However, we were able to detect segments with identical genomic location, size, and copy number in two or more of the single-cell and bulk samples, as well as segments in two or more single-cell samples but not in the bulk sample (SI Appendix, Tables S5 and S6). We reason that these segments represent true CNV events and cell-to-cell heterogeneity, because it is highly unlikely that such identical events would be present by chance. Using a 10-kb bin size, the smallest such segment was 30 kb in length.
Fig. 5.
Normalized read depth plots using 1-Mb bins for TOV2295 bulk DNA and four single-cell samples. Horizontal green lines indicate segments of contiguous bins inferred to have the same copy number, where the read depth of each segment is equal to the median read depth of the bins in that segment. Inferred copy numbers are indicated by the color of the data point for each bin.
SNV Detection by Targeted Sequencing and WGS.
To evaluate the effectiveness of droplet MDA for nucleotide-level measurements, we then performed targeted sequencing on 29 randomly selected 184-hTERT single cell droplet MDA samples that underwent WGS. Identification of SNVs by targeted sequencing can be used to infer clonal structure and evolutionary history (2, 26) at relatively low cost. For each sample, a portion of the MDA product was used as template for a targeted sequencing library preparation pipeline targeting 39 known heterozygous loci identified in a previous study (27). For each sample, a binomial exact test was used to call the presence or absence of an allele at each locus covered by at least 50 reads, as described previously (26) (SI Appendix, Fig. S9). The median allele dropout (ADO) across all samples was calculated to be 15%, comparable with the best results that have been reported previously (2). Because we performed both targeted sequencing and WGS on these samples, we were also able to determine whether there was any correlation between targeted sequencing performance and bias measured by WGS. As expected, we observed that single-cell samples with lower SD in reads per bin generally also had lower ADO (SI Appendix, Fig. S10). When including only the quartile of samples with the lowest SD in reads per 1-Mb bin, the median ADO was reduced to 8%.
We next evaluated the performance of SNV discovery using WGS data. We performed deep WGS (48× depth) of a single G1 phase 184-hTERT single-cell droplet MDA sample, selected to have average uniformity as defined by the median of SD in reads per 1-Mb bin of the 10 G1 phase single-cell samples analyzed. SNV false-positive rate (FPR) and true-positive rate (TPR) were evaluated at sites from the dbSNP database using the mutationSeq software tool (28), applying various coverage depth and SNV-calling probability thresholds (SI Appendix, Table S7; details in SI Appendix, Materials and Methods). We found that a large fraction of false-positive SNVs occurred at the ends of single-cell sequencing “read islands” (SI Appendix, Figs. S11 and S12). Increasing the minimum single-cell coverage depth threshold removed many of these false positives at the expense of reducing the set of SNP sites considered. Because FPR and TPR have an inverse correlation (SI Appendix, Fig. S13), reporting of both metrics is crucial for the evaluation of SNV detection performance. For a single-cell coverage depth ≥15 and a mutationSeq SNV-calling probability threshold of 90%, the FPR and TPR were 9.66 × 10−6 and 68.8%, respectively. This FPR is comparable to that obtained by commercial microfluidic MDA using whole exome sequencing data from a cell in unknown cell phase (22).
Demonstration on High-Grade Serous Ovarian Cancers.
As a demonstration of applicability to primary samples, we applied droplet MDA to 39 single nuclei isolated from two high-grade serous ovarian cancer tumor specimens that were previously characterized in bulk using SNP genotyping arrays (29), performed low-depth (mean, 0.02×) WGS, and binned reads into 1-Mb bins. For each specimen, read depth plots of four single nuclei samples are shown in SI Appendix, Fig. S14. In both specimens, read depth profiles from three of the single nuclei shown closely matched that of bulk DNA, whereas a fourth nucleus appeared to have a normal diploid genome, likely from noncancerous tissue collected with the tumor. Using the binned read depths, we again found copy number segments with identical genomic location, length, and copy number common to two or more samples (single nuclei and bulk) for each specimen. We found 59 and 125 such segments ranging in length from 9 to 250 Mb and from 10 to 250 Mb for specimens 1 and 2 respectively (SI Appendix, Tables S8 and S9).
Discussion
We have implemented a new single-cell MDA method that exploits the improved reaction performance and reduced reagent consumption of nanoliter-scale volumes using a commercially available liquid dispensing system. This approach preserves the programmable and multistep nanoliter-volume processing of our previously reported microfluidic droplet-based device (13), but without the need for any specialized microfabrication. In contrast to closed-format microfluidic devices that require multiple sizes of trapping structures to isolate single cells of various diameters (22), the droplet MDA method is able to isolate cells of different sizes without any modification. Unlike similar systems that formulate nanoliter-volume single-cell reactions on an open array (17, 18), droplet MDA avoids the possibility of cross-contamination between reactions by using noncontact dispensing to place reactions in spatially distinct locations separated by oil. Depending on the speed of the dispenser used, this approach is also rapid and highly scalable. In this study, we formulated 154 100 nL-reactions per 3 cm × 3 cm substrate in ∼4 min.
Here, we have, for the first time to our knowledge, evaluated a single-cell nanoliter-volume WGA method across a large dataset comprising 129 normal diploid cells of known genotype. In contrast, previous studies have presented data from 10 or fewer replicates. In total, we analyzed 219 single-cell droplet MDA samples for this study. Through random selection of a large fraction of all samples that were processed, this dataset allows for evaluation of the method’s robustness and the variability in performance across samples. We believe that such efforts are essential to establish the reproducibility needed to drive such applications as single-cell tumor sequencing, where biological insight depends on a method that is consistent across large numbers of cells.
We have shown that the biological state of the processed cells has a major influence on the observed amplification performance. In particular, cells sorted to exclude apoptotic cells and cellular debris exhibit a much narrower distribution in amplification bias than unsorted cells. Furthermore, our experiments on cells sorted by cell phase demonstrate that ploidy is an important determinant of single-cell WGA performance, including both GC content bias and uniformity. Within each sorted cell population, the performance of droplet MDA was highly robust, displaying low variability.
Our analyses of low-depth and high-depth WGS data indicate that the performance of droplet MDA compares favorably to that of other methods considered, with our least-biased samples having comparably low bias on multiple length scales relative to other methods. Nonetheless, we emphasize that comparing our method with previous reports is complicated by differences in sample type, quality, and size. Our data demonstrate a clear effect of cell cycle on amplification uniformity, suggesting that direct comparison of methods should be done using datasets from normal diploid cells sorted for G1 phase. Apart from the nuc-seq protocol, which focuses on isolation of G2 phase cells, previous reports do not control for cell cycle. This factor makes it difficult to evaluate performance in cases where data are present from only a small number of cells, and where the rationale for selecting single cells for analysis is not made explicit. Similarly, the absence of large datasets makes it impossible to evaluate robustness and variability of any given method.
The uniform coverage enabled by droplet MDA allows for recovery of a high fraction of the genome with relatively little sequencing effort and also enables accurate CNV calling. We identified copy number segments as small as 30 kb using 6.5× WGS in a cancer cell line, which to our knowledge is the most sensitive CNV detection yet demonstrated from an MDA-amplified single-cell genome, and as small as 9 Mb in primary tumor specimens using only 0.02× WGS. In both tumor specimens, droplet MDA analysis was able to clearly distinguish populations of cells with diploid and low aberration content from those with multiple CNVs and chromosomal structure aberrations using very low-depth WGS. A significant advantage of MDA is that its high coverage breadth and large mean fragment length allow for application of both WGS and targeted sequencing to the same single cell. We demonstrated this by performing both on 29 single cells, achieving excellent SNV measurement performance, as quantified by low ADO. We also evaluated the performance of SNV discovery using deep WGS data from our median performing G1-phase cell and observed a false discovery rate that compares favorably with that reported in other methods from single cells in unknown cell phase.
These results demonstrate that droplet MDA provides an easily accessible way for researchers to exploit the benefits of nanoliter-volume processing for robust and cost-effective interrogation of single-cell genomic variation at both the copy number and single-nucleotide levels. We anticipate that this will be an increasingly useful tool as interest in single-cell studies continues to expand across numerous disciplines in the biological sciences.
Materials and Methods
More detailed information on the materials and methods used in this study is provided in SI Appendix, Materials and Methods. Ethical approval was obtained from the University of British Columbia Ethics Board. Women undergoing debulking surgery (primary or recurrent) for carcinoma of ovarian/peritoneal/fallopian tube origin were approached for informed consent for the banking of tumor tissue. Tissue was obtained from tumor sites in women histologically diagnosed with high-grade serous ovarian cancer and was collected before adjuvant therapy and frozen in cryovials. Consistent with the practice at University of British Columbia and the British Columbia Cancer Agency, all patients with high-grade serous cancer are referred to the hereditary cancer clinic and offered genetic testing for BRCA1 and BRCA2 mutations.
Supplementary Material
Acknowledgments
We thank Hans Zahn for assistance with operation of the piezoelectric dispenser and Ramunas Stepanauskas for valuable discussions regarding MDA. Funding support was provided by Genome British Columbia, Genome Canada, the National Science and Engineering Research Council of Canada, and the Canadian Institutes of Health Research.
Footnotes
Conflict of interest statement: K.L., A.K., S.A., S.P.S., and C.L.H. are coinventors on a patent application (PCT/CA2016/000031) that covers the methods and devices described in this paper, and have a potential financial interest in this work through the revenue-sharing policies of the University of British Columbia. Following submission of this manuscript, the aforementioned patent was exclusively licensed to AbCellera (www.abcellera.com), a University of British Columbia-based startup company in which K.L., A.K., and C.L.H. have an equity position.
This article is a PNAS Direct Submission. P.C.B. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the Sequence Read Archive database (accession no. SRP078069). Supporting datasets are available at www.msl.ubc.ca/sites/default/files/pdata/kaston-leung/Leung-et-al-supplementary-dataset-1.zip, www.msl.ubc.ca/sites/default/files/pdata/kaston-leung/Leung-et-al-supplementary-dataset-2.zip, www.msl.ubc.ca/sites/default/files/pdata/kaston-leung/Leung-et-al-supplementary-dataset-3.zip, and www.msl.ubc.ca/sites/default/files/pdata/kaston-leung/Leung-et-al-supplementary-dataset-4.zip as described in the SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520964113/-/DCSupplemental.
References
- 1.Navin N, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gawad C, Koh W, Quake SR. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc Natl Acad Sci USA. 2014;111(50):17947–17952. doi: 10.1073/pnas.1420822111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148(5):873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, et al. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512(7513):155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lohr JG, et al. Whole-exome sequencing of circulating tumor cells provides a window into metastatic prostate cancer. Nat Biotechnol. 2014;32(5):479–484. doi: 10.1038/nbt.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McConnell MJ, et al. Mosaic copy number variation in human neurons. Science. 2013;342(6158):632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150(2):402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu S, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338(6114):1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiorentino F, et al. Development and validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of embryos. Fertil Steril. 2014;101(5):1375–1382. doi: 10.1016/j.fertnstert.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 10.de Bourcy CFA, et al. A quantitative comparison of single-cell whole genome amplification methods. PLoS One. 2014;9(8):e105585. doi: 10.1371/journal.pone.0105585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou Y, et al. Comparison of variations detection between whole-genome amplification methods used in single-cell resequencing. Gigascience. 2015;4(1):37. doi: 10.1186/s13742-015-0068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baslan T, et al. Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Res. 2015;25(5):714–724. doi: 10.1101/gr.188060.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leung K, et al. A programmable droplet-based microfluidic device applied to multiparameter analysis of single microbes and microbial communities. Proc Natl Acad Sci USA. 2012;109(20):7665–7670. doi: 10.1073/pnas.1106752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voet T, et al. Single-cell paired-end genome sequencing reveals structural variation per cell cycle. Nucleic Acids Res. 2013;41(12):6119–6138. doi: 10.1093/nar/gkt345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338(6114):1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcy Y, et al. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 2007;3(9):1702–1708. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gole J, et al. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat Biotechnol. 2013;31(12):1126–1132. doi: 10.1038/nbt.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Y, et al. Printing 2-dimentional droplet array for single-cell reverse-transcription quantitative PCR assay with a microfluidic robot. Sci Rep. 2015;5:9551. doi: 10.1038/srep09551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raouf A, et al. Genomic instability of human mammary epithelial cells overexpressing a truncated form of EMSY. J Natl Cancer Inst. 2005;97(17):1302–1306. doi: 10.1093/jnci/dji254. [DOI] [PubMed] [Google Scholar]
- 20.Ha G, et al. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 2012;22(10):1995–2007. doi: 10.1101/gr.137570.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu Y, et al. Uniform and accurate single-cell sequencing based on emulsion whole-genome amplification. Proc Natl Acad Sci USA. 2015;112(38):11923–11928. doi: 10.1073/pnas.1513988112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szulwach KE, et al. Single-cell genetic analysis using automated microfluidics to resolve somatic mosaicism. PLoS One. 2015;10(8):e0135007. doi: 10.1371/journal.pone.0135007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C-Z, et al. Calibrating genomic and allelic coverage bias in single-cell sequencing. Nat Commun. 2015;6:6822. doi: 10.1038/ncomms7822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Macaulay IC, et al. G&T-seq: Parallel sequencing of single-cell genomes and transcriptomes. Nat Methods. 2015;12(6):519–522. doi: 10.1038/nmeth.3370. [DOI] [PubMed] [Google Scholar]
- 25.Létourneau IJ, et al. Derivation and characterization of matched cell lines from primary and recurrent serous ovarian cancer. BMC Cancer. 2012;12(1):379. doi: 10.1186/1471-2407-12-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eirew P, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518(7539):422–426. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roth A, et al. PyClone: Statistical inference of clonal population structure in cancer. Nat Methods. 2014;11(4):396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding J, et al. Feature-based classifiers for somatic mutation detection in tumour-normal paired sequencing data. Bioinformatics. 2012;28(2):167–175. doi: 10.1093/bioinformatics/btr629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bashashati A, et al. Distinct evolutionary trajectories of primary high-grade serous ovarian cancers revealed through spatial mutational profiling. J Pathol. 2013;231(1):21–34. doi: 10.1002/path.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.