Abstract
Foodborne pathogens, including Plesiomonas shigelloides and Shigella flexneri B, are the major cause of diarrheal endemics worldwide. Antibiotic drug resistance is increasing. Therefore, bioactive compounds with antibacterial activity, such as gallic acid (GA), are needed. Gold nanoparticles (AuNPs) are used as drug delivery agents. This study aimed to conjugate and characterize AuNP–GA and to evaluate the antibacterial activity. AuNP was conjugated with GA, and the core–shell structures were characterized by small-angle X-ray scattering and transmission electron microscopy. Antibacterial activity of AuNP–GA against P. shigelloides and S. flexneri B was evaluated by well diffusion method. AuNP–GA bactericidal mechanism was elucidated by Fourier transform infrared microspectroscopic analysis. The results of small-angle X-ray scattering showed that AuNP–GA conjugation was successful. Antibacterial activity of GA against both bacteria was improved by conjugation with AuNP because the minimum inhibitory concentration value of AuNP–GA was significantly decreased (P<0.0001) compared to that of GA. Fourier transform infrared analysis revealed that AuNP–GA resulted in alterations of lipids, proteins, and nucleic acids at the bacterial cell membrane. Our findings show that AuNP–GA has potential for further application in biomedical sciences.
Keywords: gold nanoparticles, gallic acid, antibacterial activity, foodborne bacteria, small-angle X-ray scattering (SAXS)
Introduction
Bacterial foodborne illness is one of the most important causes of morbidity and mortality in developing countries. Plesiomonas shigelloides and Shigella flexneri are Gram-negative bacilli of the family Enterobacteriaceae, which have been responsible for producing diarrheal epidemics worldwide.1 Most of the P. shigelloides-infected patients had a history of seafood consumption or drinking untreated water.2,3 P. shigelloides infection is highly severe and has caused over 80% mortality.4 Antibiotic treatment is required to reduce the severity of diarrhea or its duration.5,6 P. shigelloides is sensitive to quinolones and cephalosporins. Owing to beta-lactam production, partial resistance to ampicillin, tetracycline, cotrimoxazole, and chloramphenicol has been reported (72%, 67%, 12%, and 5%, respectively).7 At present, increasing antibiotic resistance of P. shigelloides implies that fewer antibiotic drugs can produce bactericidal minimum inhibitory concentration (MIC).6,8,9 S. flexneri is highly infectious at low counts.10 First-line antimicrobial drugs, including sulfonamides, tetracycline, ampicillin, and trimethoprim–sulfamethoxazole, have become ineffective due to plasmid- encoded resistance to S. flexneri, which causes more than a million deaths worldwide.11,12 Recently, the emergence of resistance to fluoroquinolones and cephalosporins in S. flexneri was found in the People’s Republic of China.11 Multidrug resistance in Shigella spp is also found at high rate.12 Therefore, investigation of new drugs and materials to face the emerging resistance is needed.
Currently, the study of plants as a source of bioactive compounds with known antimicrobial activities is having a great impact on the search for new antimicrobial drugs.13 Gallic acid (GA) is a natural phenolic compound found in many fruits and plants. It exhibits promising therapeutic effects such as antioxidant,14 anti-inflammatory,15,16 anticancer,17,18 and antimicrobial19–21 activities. Its metabolites and derivatives have no observed toxic effects in rats or human beings.22–25 GA affects irreversible changes in the bacterial membrane properties, including charge and intra- and extracellular permeabilities. It also affects bacterial cell membrane physicochemical properties by changing the hydrophobicity, decreasing the negative surface charge, and disrupting pore formation, with a consequent leakage of essential intracellular constituents.26
Recent studies reveal that gold nanoparticles (AuNP) have gained much attention in drug delivery due to their unique dimensions, tunable functionalities on the surface, and controllable drug release. AuNP conjugated with bio-active compounds is easy to prepare, stable, and can be incorporated into secondary tags without toxicity to cells.27,28 AuNP–GA has been evaluated for its bactericidal activity against Streptococcus mutans.29 Considering the potential use of AuNP–GA in medical applications, the investigation of AuNP–GA antibacterial activity is essential. In the present study, we report the preparation and characterization of AuNP–GA. Antibacterial activity of the prepared AuNP–GA against P. shigelloides and S. flexneri B and the mechanism of action were evaluated.
Materials and methods
Conjugation of AuNP with GA
Bare spherical AuNP (Nanopartz Inc., Loveland, CO, USA), 17 nm in diameter, was conjugated with GA (Sigma-Aldrich Co., St Louis, MO, USA) as follows. GA was added to the AuNP solution to make a final concentration of 10 mM, 30 mM, and 50 mM GA at pH 7.51 and stirred in the dark for 4 hours at 50°C. AuNP–GA was kept at room temperature for further analysis. This study was approved by the Khon Kaen institutional review board.
Characterization of dispersion of AuNP–GA by ultraviolet-visible spectrometer
Dispersion of AuNP and AuNP–GA were characterized by measuring the absorbance of the sample with ultraviolet-visible (UV-Vis) spectrophotometer (UV mini-1240; Kyoto, Shimadzu, Japan) at room temperature. The spectra of AuNP, AuNP–10 mM GA, AuNP–30 mM GA, and AuNP–50 mM GA were plotted.
Microstructural investigation of AuNP–GA by transmission electron microscope
A core–shell nanostructure of AuNP–GA was studied by using transmission electron microscope (TEM). AuNP–GA sample was dropped on a carbon film copper grid (Electron Microscopy Sciences, Hatfield, PA, USA) and dried in an oven before analysis by TEM (Tecnai G2 20 model).
Characterization of AuNP–GA by synchrotron small-angle X-ray scattering
Small-angle X-ray scattering (SAXS) was also used to characterize the prepared AuNP–GA. It was performed at BL1.3W. The synchrotron X-ray was monochromatized with a double multilayer monochromator for an X-ray energy of 8 keV. Each sample was in a liquid cell with Kapton windows. The measurement with two sample-detector distance setups of 1,002.92 mm and 3,033.95 mm, calibrated with silver behenate and Styrene-Ethylene-Butadiene-Styrene block copolymer, respectively, were conducted to cover the scattering vector range (q-range) of ~0.1–3.0 nm−1. A Mar SX165 CCD was used to record the scattering patterns. Liquid cells containing deionized water were measured as background. The background was subtracted from the measured sample patterns, which were circularly averaged to obtain the SAXS profiles as functions of the scattering vector q =4πsinθ/λ, where θ denotes half of the scattering angle and λ denotes the X-ray wavelength.
Evaluation for antibacterial activity of AuNP–GA
The prepared AuNP–GA was evaluated for antibacterial activity against two human foodborne bacterial strains, P. shigelloides and S. flexneri B, which were obtained from the Department of Clinical Microbiology at the Faculty of Associated Medical Sciences, Khon Kaen University.
The MIC of AuNP–GA and GA was investigated by a well diffusion method.30 For each bacterial species, 0.5 McFarland inoculum was swabbed onto Mueller-Hinton agar plate. The 6-mm-diameter wells were drilled into the agar plate with a sterilized cork borer; then 100 µL of various concentrations of GA (10 mM, 30 mM, 50 mM, 70 mM, 90 mM, and 110 mM) and AuNP–GA (at 50 mM, 30 mM, and 10 mM) were added to the agar wells. After incubation at 37°C for 16–24 hours, the diameters of the clear zones were measured. The lowest concentration of the tested compound that showed no visible growth was reported as the MIC. All experiments were conducted in triplicate, and AuNP was used as a control.
Detection of bacterial morphology by scanning electron microscope
The bacterial membrane was investigated after treatment with AuNP–GA by scanning electron microscope (SEM), which was performed according to the method described in Uawonggul et al,31 with slight modification. P. shigelloides and S. flexneri B were cultured in nutrient broth and harvested at the logarithmic phase of growth by centrifugation at 3,000× g for 5 minutes. The bacterial cells were then washed twice with phosphate-buffered saline, pH 7.0, and resuspended to yield a final concentration of 106 CFU/mL. Aliquots of the bacterial suspension (100 µL) were individually incubated with GA at the MIC concentration that was previously determined (110 mM for P. shigelloides and 50 mM for S. flexneri B at 37°C for 1 hour and 2 hours, respectively). The incubated bacterium (150 µL) was fixed with equal volume of 2.5% glutaraldehyde (Sigma-Aldrich Co.) in a 0.1 M phosphate buffer, pH 7.2, for 2 hours. The fixed cells were pipetted and settled onto 0.2 µM polycarbonate Whatman membrane filter (GE Healthcare, Chicago, IL, USA) for 5 minutes. The materials were then dehydrated in graded ethanol concentrations (30%, 50%, 70%, and 90%) for 15 minutes each. The dehydrated materials in the absolute ethanol were dried in a critical point drier (CPD7510; Thermo Fisher Scientific, Waltham, MA, USA), with carbon dioxide used as the drying agent. Dry materials were coated with gold palladium by a sputter coater (SC7620; Polaron, Hertfordshire, England) and examined with SEM (LEO1450VP; LEO Electron Microscopy Ltd., Cambridgeshire, England) operated at 12–20 kV. For the negative control, all procedures were performed in a similar manner, except that the bacterial cells were incubated with a phosphate-buffered saline instead of GA.
Elucidation of AuNP–GA bactericidal mechanism by Fourier transform infrared microspectroscopic method
All bacterial cultures were grown in nutrient broth and incubated at 37°C for 24 hours. The concentration of the cultures was then adjusted to 0.5 McFarland standards. Bacterial cultures were individually incubated with the previously determined MIC of AuNP–GA (50 mM for P. shigelloides and 30 mM for S. flexneri B) at 37°C for 1 hour and 2 hours. The treated cells were recovered from prepared 1 mL broth cultures by centrifugation at 12,000 rpm for 15 minutes. The supernatant was discarded, and the pellet was washed twice in 0.85% NaCl. For sample preparation, 5 µL of each bacterial sample was deposited onto infrared (IR) low-e slides (Kevley Technologies, Chesterland, OH, USA) to form a homogeneous dried film of bacterial cells. The slides were air dried and stored in a desiccator.
Fourier transform infrared (FTIR) microspectroscopic system consisted of a Vertex 70 FTIR spectrometer (Bruker Optik GmbH, Ettlingen, Germany) coupled to a Hyperion 2000 IR microscope with a liquid nitrogen cooled mercury cadmium telluride detector. The bacterial samples coated on the low-e slides were placed under the microscope objective, and the IR spectra were recorded in the reflection mode of 4,000–400 cm−1 at a spectral resolution of 4 cm−1. One hundred spectra were obtained for each sample. Spectra from all samples were extracted using a macro converter of OPUS7.0 software (Bruker Optik GmbH, Ettlingen, Germany) prior to multivariate data analysis. Approximately 50 representative spectra from each sample were pretreated by performing a second derivative carried out by applying the Savitzky–Golay algorithm with 13 smoothed data points and then normalized with extended multiplicative signal correction. Pretreated spectra were then analyzed by principal component analysis (PCA) using the Unscrambler® 9.7 software (Camo Inc., Oslo, Norway) to distinguish between the control and AuNP–GA-treated cells.
Results and discussion
Preparation of AuNP–GA
In the present study, the GA solution was colorless, whereas the 17-nm-diameter AuNP solution was pale red. When GA was mixed with AuNP, the pale purple solution was observed, which indicated that GA may conjugate with AuNP. UV-Vis is a spectral technique used to confirm the presence of AuNP–GA. The absorption spectra (400–800 nm) obtained from the prepared AuNP and AuNP–GA (Figure 1) showed that the absorption maximum (λmax) of AuNP was 520 nm and there was a redshift of λmax of AuNP–GA. This confirms that AuNP was conjugated with GA because the redshift can occur when the particle size increases. Our results agree with a previous study of Moreno-Alvarez et al,29 which showed that the UV-Vis absorption spectrum of GA-stabilized AuNP shifted to a broader band.
Figure 1.
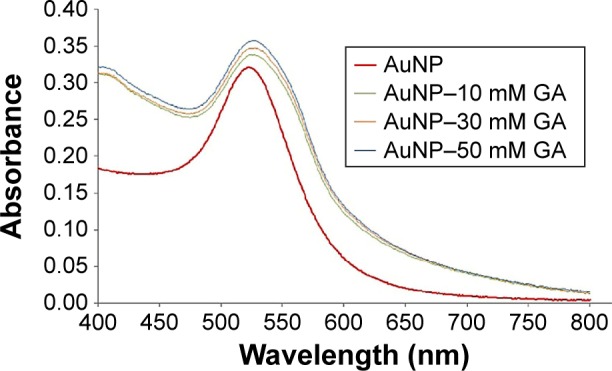
Absorption spectra of AuNP (red line), AuNP–10 mM GA (green line), AuNP–30 mM GA (orange line), and AuNP–50 mM GA (blue line).
Abbreviations: AuNP, gold nanoparticles; GA, gallic acid.
Characterization of AuNP–GA
To verify the particle size of the studied nanoparticles, TEM images of AuNP and AuNP–10 mM GA were taken (data not shown). The TEM results did not show a significant increase in the particle size of AuNP–GA compared to a 17-mm-diameter AuNP. However, the AuNP and GA could form a core–shell structure with a thin GA shell, but this was not clearly observed in the TEM image due to low contrast between GA and the sample substrate.
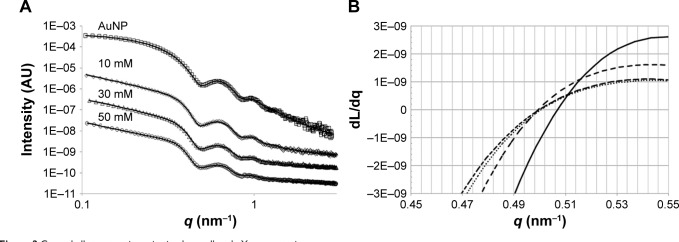
SAXS was then conducted for investigation of the AuNP–GA core–shell structure because X-ray scattering is less affected by sample conditions. The SAXS profile (Figure 2A) clearly shows characteristics of spherical nanoparticles with a narrow size distribution. The SAXS data were fitted using a hard sphere form factor with a lognormal size distribution. The theoretical SAXS intensity is given by:
| (1) |
Figure 2.
Core–shell structure investigation by small angle X-ray scattering.
Notes: (A) Measured SAXS intensity profiles of AuNP (squares) and AuNP–GA with GA concentrations of 10 mM (diamonds), 30 mM (triangles), and 50 mM (circles). The solid lines indicate the hard sphere model fit (for AuNP) and core–shell sticky hard sphere model (for AuNP–GA). (B) Derivatives of the measured SAXS intensity profiles for the AuNP (solid line) and AuNP–GA with GA concentrations of 10 mM (long dashed line), 30 mM (dashed line), and 50 mM (dotted line).
Abbreviations: SAXS, small-angle X-ray scattering; AuNP, gold nanoparticles; GA, gallic acid; AU, arbitrary unit.
In Equation 1, the hard sphere form factor as given by Glatter and Kratky is:32
| (2) |
where C is a constant, taken as a scaling factor in the fit, and j1(qr) is the first-order spherical Bessel function. In Equation 1,G (r, m, σ) denotes a lognormal distribution:
| (3) |
where m and σ are the median and shape parameters, respectively, of the lognormal distribution. The fitting was carried out through a chi-square minimization procedure, using the SASfit software program (Paul Scherrer Institut, Villigen, Switzerland).33 The fitted SAXS profile, using Equation 1 with a constant background, is indicated by a solid line in Figure 2B. The fit result produces values of m =8.95 nm and σ =0.102 nm for the lognormal distribution, values that are equivalent to a mean particle radius of 8.99 nm and a size distribution with a standard deviation of 0.92 nm.34
The SAXS profiles of the AuNP–GA (Figure 2A) reveal that the mixed AuNP–GA solutions continue to show characteristics of a scattering profile of nanoparticles with a narrow size distribution. However, the shapes of the scattering profiles in the regions of the small and large scattering vectors imply some structural change. To confirm the core–shell structure, we first investigated the particle size by considering the first minima of the scattering profiles. For the scattering of the nanoparticles, the radius of the particles can be approximated by the first minimum in the scattering profile. From the SAXS intensity of a hard sphere, Equation 1, it can be observed that the first minimum in intensity, located at the q value where the derivative is zero, is at the first zero of the spherical Bessel function, j1 (qr). The particle radius R may therefore be estimated from j1 (q0R) =0, which yields q0R =4.49,35 where q0 is the scattering vector at the location of the zero derivative. In the case of core–shell structures, the form factor is given by:
| (4) |
where Rcs and R denote the radii of the core + shell and the core, respectively, and µ is the scattering length density (SLD) ratio between the core and the shell. The constant Ccs is again treated as a scaling factor. In this case, it would be difficult to estimate the particle size from the first minimum in scattering intensity due to complex interferences from the core and shell contributions. One may nevertheless expect the core–shell scattering profile to have a first minimum at a smaller q than that of the core particle because the core–shell particle is larger. The formation of the core–shell particles, in this case, may therefore be determined by comparing the first minimum locations in the scattering profiles of the core–shell and the core particles.
The measured SAXS profiles of the dispersed AuNP and the mixed AuNP–GA solutions were then smoothed and numerically differentiated with respect to the scattering vector q. The derivatives obtained are shown in Figure 2B. These data show that the first minima in the three mixed solution samples had indeed shifted to smaller q values. Moreover, the shifted first minima appeared to be at similar locations, which may further suggest that the AuNP–GA core–shell particles formed by the mixed solutions have similar shell thicknesses for all three GA concentrations.
To quantitatively characterize the core–shell structure, the measured SAXS profiles of the mixed solutions were fitted with a theoretical model. The theoretical SAXS intensity model is given by:
| (5) |
The first term is the contribution of the core–shell particles, where the form-factor term is:
| (6) |
and where the core–shell form factor is given by Equation 4, with the lognormal size distribution given by Equation 3. S (q, RHS, τ, f) in Equation 5 is the structure factor responsible for the interparticle interaction. This interaction is expected to arise from the interaction between the polymer shells surrounding the core particles and can be modeled by the Baxter’s sticky hard sphere model,36 which is parameterized by the hard sphere radius RHS, the stickiness parameter τ, and the volume fraction f. The Lorentzian profile appearing in the second term in Equation 5 is the Ornstein–Zernike term, parameterized by the correlation length ξ, with the scaling factor indicated by I0. This term has been used to account for the fluctuation of the polymer chains in solutions.37 For the fittings, the radius and size distribution of the core particles were fixed by the parameters obtained from the fitting of the AuNP data mentioned earlier. The SLD ratio µ in the core–shell form factor was calculated from the ratio of the SLD values of Au and GA, where the SLD of a molecule is given by:
| (7) |
where n is the number of atoms in the molecule, Zi is the atomic number of the ith atom, re is the classical electron radius, and V is the effective atomic volume (in the case of Au) or molecular volume (in the case of GA). The SLD ratio used in the fitting was calculated as µ =8.85. For the sticky hard sphere structure factor, the hard sphere radius RHS is considered an imaginary barrier preventing the particles from getting closer. This parameter was therefore treated as a fit parameter, together with the stickiness parameter and the volume fraction. The fitting results are shown in Figure 2B. In the AuNP–GA results, the structure factor was responsible for the modified shape, compared to that of the AuNP, of the SAXS intensity at low q. However, the Ornstein–Zernike term also has an effect on a large q region. The oscillations in the SAXS intensity originated solely from the core–shell sphere form factor. The fitted core–shell parameters are summarized in Table 1. The SAXS results suggest that the core–shell structure was formed by a thin GA shell around the AuNP core. In addition, the results indicate that the GA shell thickness does not depend on the GA concentration.
Table 1.
Fitted parameters for the mean radius and SD of the Au core particles (〈Rcore〉 and SDcore), the GA shell thickness (Rshell = Rcs − 〈Rcore〉), the hard sphere radius (RHS), the stickiness parameter (τ) and the volume fraction (f) of the sticky hard sphere structure factor, and the correlation length (ξ) of the Ornstein–Zernike term
| Sample | 〈Rcore〉 (nm) | SDcore (nm) | Rshell (nm) | RHS (nm) | τ | f | ξ (nm) |
|---|---|---|---|---|---|---|---|
| AuNP | 8.99 | 0.92 | – | – | – | – | – |
| AuNP–10 mM GA | 8.99 | 0.92 | 1.36 | 9.42 | 0.09 | 0.07 | 15.94 |
| AuNP–30 mM GA | 8.99 | 0.92 | 1.25 | 9.41 | 0.09 | 0.06 | 2.88 |
| AuNP–50 mM GA | 8.99 | 0.92 | 1.67 | 9.84 | 0.10 | 0.12 | 4.13 |
Abbreviations: SD, standard deviation; Au, gold; GA, gallic acid; AuNP, gold nanoparticles.
Antibacterial activity of AuNP–GA
The antibacterial activities of GA and AuNP–GA against P. shigelloides and S. flexneri B are shown in Figure 3. The MIC of GA (range 50–110 mM) was significantly higher than that of AuNP–GA against these bacteria (range 30–50 mM) (P<0.0001). Only AuNP had no antibacterial effect on the studied bacteria. These results indicated that the antibacterial activity of GA could be improved by conjugation with AuNP, which is in close agreement with a previous study.29
Figure 3.
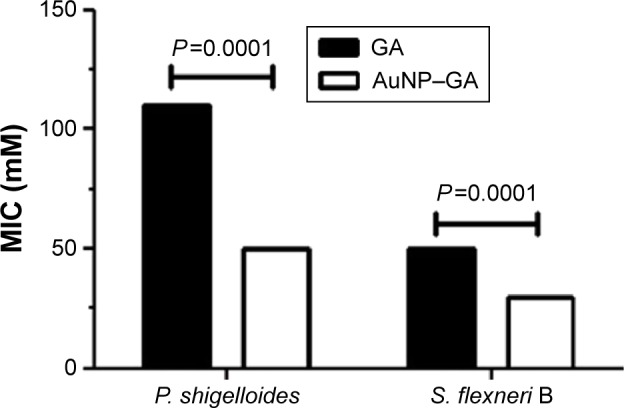
The MIC of GA and AuNP–GA on P. shigelloides and S. flexneri B.
Abbreviations: MIC, minimum inhibitory concentration; GA, gallic acid; AuNP, gold nanoparticles; P. shigelloides, Plesiomonas shigelloides; S. flexneri B, Shigella flexneri B.
Bactericidal mechanism of AuNP–GA
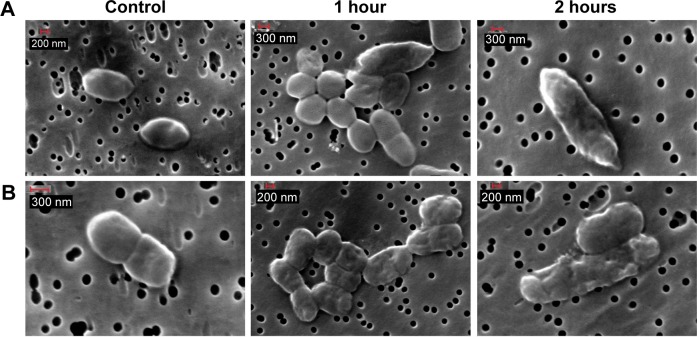
SEM images show that the bacterial membrane became wrinkled after adding 110 mM GA to P. shigelloides (Figure 4A) and 50 mM GA to S. flexneri B (Figure 4B) for 1 hour and 2 hours, respectively. There are several studies that reported that phenolic compounds can penetrate a cell due to alterations in cell membrane permeability.38 Zhou et al39 showed that poly(allylamine hydrochloride) helped accelerating cell wall breakdown and cytoplasm release. It has also been reported that cationic-coated AuNP had more toxicity than anionic-coated AuNP,40 which supports the hypothesis that poly(allylamine hydrochloride)–AuNP cause immediate cell lysis, while citrate AuNP does not. These findings support our results. We assumed that AuNP–GA affected the bacterial cell membrane in the same manner as GA.
Figure 4.
Bacterial membrane disruption after treatment with GA at MIC concentration of (A) P. shigelloides and (B) S. flexneri B.
Abbreviations: GA, gallic acid; MIC, minimum inhibitory concentration; P. shigelloides, Plesiomonas shigelloides; S. flexneri B, Shigella flexneri B.
FTIR microspectroscopic analysis was used for studying the AuNP–GA bactericidal mechanism of action because of its sensitivity, rapidity, low cost, and simplicity. This technique has been also used to detect and identify different types of microorganisms and evaluate the activity of antibacterial agents.41,42 PCA was applied to the data set, in which two regions are of interest: i) the biochemical fingerprint region from 1,800 cm−1 to 1,000 cm−1 influenced by stretching vibrations of ester functional groups in lipids (1,740 cm−1), amide I and amide II groups in proteins and peptides (1,655–1,637 cm−1), P=O stretching in nucleic acids (1,250–1,220 cm−1, 1084–1088 cm−1), and carbohydrate and polysaccharide bands (1,200–900 cm−1) and ii) the region from 3,000 cm−1 to 2850 cm−1 dominated by the functional groups of membrane fatty acids and some amino acid side chains.43
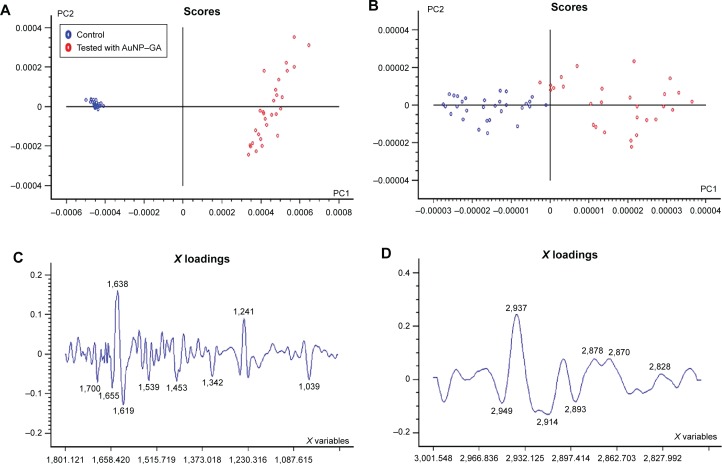
PCA score plots showed distinct group separation along PC1 when comparing the spectra from P. shigelloides (control cells; blue dots) and P. shigelloides (tested with AuNP–GA; red dots) as shown in Figure 5A and B, respectively. In loading plots corresponding to this comparison, PC1 and PC2 separately controlled and tested spectra for changes in the lipid signal (2,949 cm−1, 2,893 cm−1), protein signal (1,655 cm−1, 1,619 cm−1, 1,539 cm−1, 1,342 cm−1), and cellular phosphorylation (1,039 cm−1), as shown in Figure 5C and D.
Figure 5.
FTIR spectral analysis at wave number region of (A and C) 1,800–1,000 cm−1 and (B and D) 3,000–2,827 cm−1 by PCA using control and tested cells with AuNP–GA in P. shigelloides.
Abbreviations: FTIR, Fourier transform infrared; PCA, principal component analysis; AuNP, gold nanoparticles; GA, gallic acid; P. shigelloides, Plesiomonas shigelloides; PC1, principal component 1; PC2, principal component 2.
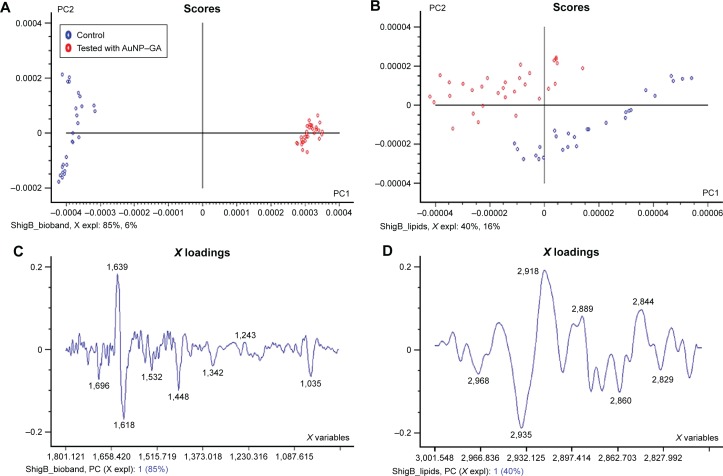
For the spectra of S. flexneri B control cells (blue dots) and S. flexneri B tested with AuNP–GA (red dots), PC1 and PC2 explained the spectral differences between the groups (Figure 6A and B). The corresponding loading plots similarly elucidated in the spectral regions of P. shigelloides contained changes primarily in the lipid signal (2,918 cm−1, 2,889 cm−1, 2,844 cm−1), protein signal (1,618 cm−1, 1,532 cm−1, 1,342 cm−1), and cellular phosphorylation (1,035 cm−1), as shown in Figure 6C and D. PCA of spectral data revealed segregation of control and AuNP–GA-treated cells. The spectra obtained for both bacteria were similar to those previously described for other bacteria.44,45
Figure 6.
FTIR spectral analysis at a wave number region of (A and C) 1,800–1,000 cm−1 and (B and D) 3,000–2,827 cm−1 by PCA using control and tested cells with AuNP–GA in S. flexneri B.
Abbreviations: FTIR, Fourier transform infrared; PCA, principal component analysis; AuNP, gold nanoparticles; GA, gallic acid; S. flexneri B, Shigella flexneri B; PC1, principal component 1; PC2, principal component 2.
The spectral profiles imply biochemical alterations in the bacterial cells, especially in lipids (2,918 cm−1, 2,889 cm−1, 2,844 cm−1) and proteins (1,618 cm−1, 1,532 cm−1, 1,342 cm−1). The outer cell membranes are composed of lipids and proteins, and the spectral changes in these regions suggest disruption of the cell membrane structure and integrity. The SEM images (Figure 4) show the disruption of bacterial membranes after treatment with the MIC of GA. Moreover, alteration in the DNA region at 1,035 cm−1 was observed, which may indicate that AuNP acted as a vehicle to deliver GA through the cell membrane and interact directly with DNA. The analysis of control and AuNP–GA-treated bacterial FTIR spectra revealed the molecular mechanisms of action that lead to AuNP–GA-induced cell death.
Conclusion
GA was successfully conjugated with AuNP, and the core–shell structure was investigated by TEM and SAXS. SAXS showed that AuNP–GA had similar shell thicknesses regardless of the GA concentration. The antibacterial activity of GA, against foodborne bacteria P. shigelloides and S. flexneri B, was improved by conjugation with AuNP. AuNP–GA decrease bacterial pathogenicity by altering bacterial membrane compositions. This was supported by FTIR analysis that the AuNP–GA significantly altered lipids or fatty acids in cell membranes, proteins, and nucleic acids. With these antibacterial properties, AuNP–GA is a promising bioactive compound that is potentially suitable for biomedical applications.
Acknowledgments
This work was supported by the Higher Education Research Promotion and National Research University Project of Thailand; Office of the Higher Education Commission, through the Health Cluster (SHeP-GMS), Khon Kaen University; and the Centre for Research and Development of Medical Diagnostic Laboratories, Faculty of Associated Medical Sciences, Khon Kaen University. The authors would like to thank Doctor Alisa Sandee, Doctor Sakda Daduang, Doctor Ratree Tavichakorntrakool, and Mr Seksit Sungkeeree for their assistance. This work was supported by a research grant from Khon Kaen University (grant number PhD 54208).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Kotloff KL, Winickoff JP, Ivanoff B, et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull World Health Organ. 1999;77(8):651–666. [PMC free article] [PubMed] [Google Scholar]
- 2.Brenden RA, Miller MA, Janda JM. Clinical disease spectrum and pathogenic factors associated with Plesiomonas shigelloides infections in humans. Rev Infect Dis. 1988;10(2):303–316. doi: 10.1093/clinids/10.2.303. [DOI] [PubMed] [Google Scholar]
- 3.Das SK, Ahmed S, Ferdous F, et al. Changing emergence of Shigella sero-groups in Bangladesh: observation from four different diarrheal disease hospitals. PLoS One. 2013;8(4):e62029. doi: 10.1371/journal.pone.0062029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider F, Lang N, Reibke R, Michaely HJ, Hiddemann W, Ostermann H. Plesiomonas shigelloides pneumonia. Med Mal Infect. 2009;39(6):397–400. doi: 10.1016/j.medmal.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Sur D, Ramamurthy T, Deen J, et al. Shigellosis: challenges and management issues. Indian J Med Res. 2004;120(5):454–462. [PubMed] [Google Scholar]
- 6.Pazhani GP, Niyogi SK, Singh AK, et al. Molecular characterization of multidrug-resistant Shigella species isolated from epidemic and endemic cases of shigellosis in India. J Med Microbiol. 2008;57(pt 7):856–863. doi: 10.1099/jmm.0.2008/000521-0. [DOI] [PubMed] [Google Scholar]
- 7.Kain KC, Kelly MT. Antimicrobial susceptibility of Plesiomonas shigelloides from patients with diarrhea. Antimicrob Agents Chemother. 1989;33(9):1609–1610. doi: 10.1128/aac.33.9.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Med. 2006;119(6 suppl 1):S3–S10. doi: 10.1016/j.amjmed.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Maluping RP, Lavilla-Pitogo CR, DePaola A, et al. Antimicrobial susceptibility of Aeromonas spp., Vibrio spp. and Plesiomonas shigelloides isolated in the Philippines and Thailand. Int J Antimicrob Agents. 2005;25(4):348–350. doi: 10.1016/j.ijantimicag.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. Acid and base resistance in Escherichia coli and Shigella flexneri role of rpoS and growth pH. J Bacteriol. 1994;176(6):1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu S, Xu X, Wang V, et al. Emergence of resistance to fluoroquinolones and third-generation cephalosporins in Shigella flexneri subserotype 1c isolates from China. Clin Microbiol Infect. 2012;18(4):E95–E98. doi: 10.1111/j.1469-0691.2012.03768.x. [DOI] [PubMed] [Google Scholar]
- 12.Taneja N, Mewara A, Kumar A, et al. Cephalosporin-resistant Shigella flexneri over 9 years (2001–2009) in India. J Antimicrob Chemother. 2012;67(6):1347–1353. doi: 10.1093/jac/dks061. [DOI] [PubMed] [Google Scholar]
- 13.Li D, Liu Z, Yuan Y, et al. Green synthesis of gallic acid-coated silver nanoparticles with high antimicrobial activity and low cytotoxicity to normal cells. Process Biochem. 2015;50:357–366. [Google Scholar]
- 14.Yilmaz Y, Toledo RT. Major flavonoids in grape seeds and skins: antioxidant capacity of catechin, epicatechin, and gallic acid. J Agric Food Chem. 2004;52:255–260. doi: 10.1021/jf030117h. [DOI] [PubMed] [Google Scholar]
- 15.Kroes BH, Van den Berg AJJ, Quarles van Ufford HC, van Dijk H, Labadie RP. Anti-inflammatory of gallic acid. Planta Med. 1992;58(6):499–504. doi: 10.1055/s-2006-961535. [DOI] [PubMed] [Google Scholar]
- 16.Kim SH, Jun CD, Suk K, et al. Gallic acid inhibits histamine release and pro-inflammatory cytokine production in mast cells. Toxicol Sci. 2006;91(1):123–131. doi: 10.1093/toxsci/kfj063. [DOI] [PubMed] [Google Scholar]
- 17.You BR, Moon HJ, Han YH, Park WH. Gallic acid inhibits the growth of HeLa cervical cancer cells via apoptosis and/or necrosis. Food Chem Toxicol. 2010;48(5):1334–1340. doi: 10.1016/j.fct.2010.02.034. [DOI] [PubMed] [Google Scholar]
- 18.Daduang J, Palasap A, Daduang S, Boonsiri P, Suwannalert P, Limpaiboon T. Gallic acid enhancement of gold nanoparticle anticancer activity in cervical cancer cells. Asian Pac J Cancer Prev. 2015;16(1):169–174. doi: 10.7314/apjcp.2015.16.1.169. [DOI] [PubMed] [Google Scholar]
- 19.Chanwitheesuk A, Teerawutgulrag A, Kilburn JD, et al. Antimicrobial gallic acid from Caesalpinia mimosoides Lamk. Food Chem. 2007;100:1044–1048. [Google Scholar]
- 20.Li A, Chen J, Zhu W, et al. Antibacterial activity of gallic acid from the flowers of Rosa chinensis Jacq against fish pathogens. Aquac Res. 2007;38:1110–1112. [Google Scholar]
- 21.Nakamura K, Yamada Y, Ikai H, et al. Bactericidal action of photo-irradiated gallic acid via reactive oxygen species formation. J Agric Food Chem. 2012;60(40):10048–10054. doi: 10.1021/jf303177p. [DOI] [PubMed] [Google Scholar]
- 22.Shahrzad S, Bitsch I. Determination of gallic acid and its metabolites in human plasma and urine by high-performance liquid chromatography. J Chromatogr B Biomed Sci Appl. 1998;705(1):87–95. doi: 10.1016/s0378-4347(97)00487-8. [DOI] [PubMed] [Google Scholar]
- 23.Zong L, Inoue M, Nose M, et al. Metabolic fate of gallic acid orally administered to rats. Biol Pharm Bull. 1999;22(3):326–329. doi: 10.1248/bpb.22.326. [DOI] [PubMed] [Google Scholar]
- 24.Hodgson JM, Morton LW, Puddey IB, Beilin LJ, Croft KD. Gallic acid metabolites are markers of black tea intake in humans. J Agric Food Chem. 2000;48(6):2276–2280. doi: 10.1021/jf000089s. [DOI] [PubMed] [Google Scholar]
- 25.Shahrzad S, Aoyagi K, Winter A, Koyama A, Bitsch I. Pharmacokinetics of gallic acid and its relative bioavailability from tea in healthy humans. J Nutr. 2001;131(4):1207–1210. doi: 10.1093/jn/131.4.1207. [DOI] [PubMed] [Google Scholar]
- 26.Borges A, Ferreira C, Saavedra MJ, Simões M. Antibacterial activity and mode of action of ferulic and gallic acids against pathogenic bacteria. Microb Drug Resist. 2013;19(4):256–265. doi: 10.1089/mdr.2012.0244. [DOI] [PubMed] [Google Scholar]
- 27.Murphy CJ, Gole AM, Stone JW, et al. Gold nanoparticles in biology: beyond toxicity to cellular imaging. Acc Chem Res. 2008;41(12):1721–1730. doi: 10.1021/ar800035u. [DOI] [PubMed] [Google Scholar]
- 28.Shukla R, Bansal V, Chaudhary M, Basu A, Bhonde RR, Sastry M. Biocompatibility of gold nanoparticles and their endocytotic fate inside the cellular compartment: a microscopic overview. Langmuir. 2005;21(23):10644–10654. doi: 10.1021/la0513712. [DOI] [PubMed] [Google Scholar]
- 29.Moreno-Alvarez SA, Martinez-Castanon GA, Nino-Martinez N, et al. Preparation and bactericide activity of gallic acid stabilized gold nanoparticles. J Nanopart Res. 2010;12:2741–2746. [Google Scholar]
- 30.Kande JA, Hayashi Y. Potency of extract contents from selected tropical chewing sticks against Staphylococcus aureus and Staphylococcus auricularis. World J Microbiol. 1998;14:235–238. [Google Scholar]
- 31.Uawonggul N, Thammasirirak S, Chaveerach A, et al. Purification and characterization of heteroscorpine-1 (HS-1) toxin from Heterometrus laoticus scorpion venom. Toxicon. 2007;49(1):19–29. doi: 10.1016/j.toxicon.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Glatter O, Kratky O. Small Angle X-ray Scattering. London: Academic Press; 1982. [Google Scholar]
- 33.Kohlbrecher J. A Program for Fitting Elementary Structure Models of Small Angle Scattering Data. [Accessed January 29, 2015]. webpage on the Internet. Available from: http://kur.web.psi.ch/sans1/SANSSoft/sasfit.html.
- 34.Forbes C, Evans M, Hastings N, et al. Statistical Distributions. New York, NY: John Wiley and Sons Inc; 2011. [Google Scholar]
- 35.Abramowitz M, Stegun IA. Handbook of Mathematical Functions. New York, NY: Dover Publications Inc; 1972. [Google Scholar]
- 36.Baxter RJ. Percus-Yevick equation for hard spheres with surface adhesion. J Chem Phys. 1968;49:2770–2774. [Google Scholar]
- 37.Shibayama M, Tanaka T, Han CC. Small angle neutron scattering study on poly (n-isopropylacrylamide) gels near their volume-phase transition. J Chem Phys. 1992;97:6829–6841. [Google Scholar]
- 38.Moreno S, Scheyer T, Romano CS, Vojnov AA. Antioxidant and antimicrobial activities of rosemary extracts linked to their polyphenol composition. Free Radic Res. 2006;40(2):223–231. doi: 10.1080/10715760500473834. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Y, Kong Y, Kundu S, Cirillo JD, Liang H. Antibacterial activities of gold and silver nanoparticles against Escherichia coli and bacillus Calmette-Guérin. J Nanobiotechnology. 2012;10:19. doi: 10.1186/1477-3155-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodman CM, McCusker CD, Yilmaz T, Rotello VM. Toxicity of gold nanoparticles functionalized with cationic and anionic side chains. Bioconjug Chem. 2004;15(4):897–900. doi: 10.1021/bc049951i. [DOI] [PubMed] [Google Scholar]
- 41.Burgula Y, Khali D, Kim S, et al. Review of Mid-infrared Fourier transform infrared spectroscopy applications for bacterial detection. J Rapid Methods Autom Microbiol. 2007;15:146–175. [Google Scholar]
- 42.Riding MJ, Martin FL, Trevisan J, et al. Concentration-dependent effects of carbon nanoparticles in gram-negative bacteria determined by infrared spectroscopy with multivariate analysis. Environ Pollut. 2012;163:226–234. doi: 10.1016/j.envpol.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 43.Alvarez-Ordóñez A, Mouwen DJ, López M, Prieto M. Fourier transform infrared spectroscopy as a tool to characterize molecular composition and stress response in foodborne pathogenic bacteria. J Microbiol Methods. 2011;84(3):369–378. doi: 10.1016/j.mimet.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Zoumpopoulou G, Papadimitriou K, Polissiou MG, Tarantilis PA, Tsakalidou E. Detection of changes in the cellular composition of Salmonella enterica serovar Typhimurium in the presence of antimicrobial compound(s) of Lactobacillus strains using Fourier transform infrared spectroscopy. Int J Food Microbiol. 2010;144(1):202–207. doi: 10.1016/j.ijfoodmicro.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 45.Booyens J, Thantsha MS. Fourier transform infra-red spectroscopy and flow cytometric assessment of the antibacterial mechanism of action of aqueous extract of garlic (Allium sativum) against selected probiotic Bifidobacterium strains. BMC Complement Altern Med. 2014;14:289. doi: 10.1186/1472-6882-14-289. [DOI] [PMC free article] [PubMed] [Google Scholar]