Abstract
In postmitotic neurons, nucleosomal turnover was long considered to be a static process that is inconsequential to transcription. However, our recent studies in human and rodent brain indicate that replication-independent (RI) nucleosomal turnover, which requires the histone variant H3.3, is dynamic throughout life and is necessary for activity-dependent gene expression, synaptic connectivity, and cognition. H3.3 turnover also facilitates cellular lineage specification and plays a role in suppressing the expression of heterochromatic repetitive elements, including mutagenic transposable sequences, in mouse embryonic stem cells. In this essay, we review mechanisms and functions for RI nucleosomal turnover in brain and present the hypothesis that defects in histone dynamics may represent a common mechanism underlying neurological aging and disease.
Keywords: aging, H3.3, histone variant, neurological disease, nucleosomal turnover, replication-independent
Introduction
Diseases of the central nervous system (CNS) are often challenging to understand on molecular and mechanistic levels due to the diversity of neuronal and glial sub-types expressed, the partitioning of functions between brain regions, and the complicated neural circuits that traverse the brain. Nevertheless, common features of most neurodevelopmental, age- and experience-related neurological diseases include abnormal synaptic development and/or plasticity [1]. To date, the majority of mechanistic studies of neurological disease have focused on roles of cytoplasmic proteins in the regulation of synaptic architecture and function [2]. However, recent advances in whole exome sequencing have revealed that prevalent neuropathologies, such as autism [3, 4] adult onset dementia [5], schizophrenia [6], and intellectual disability [4, 7] can derive from mutations in genes that regulate chromatin structure (Fig. 1). Such mutations often lead to aberrant patterns of neuronal transcription that are believed to play causative roles in the development and progression of these illnesses.
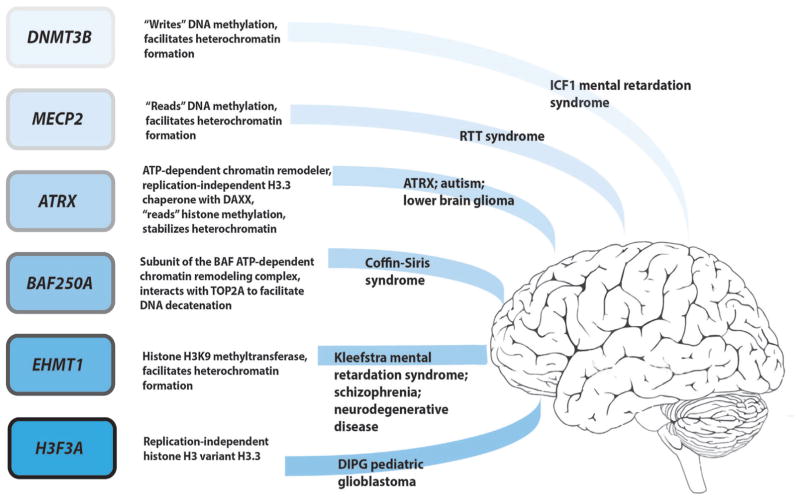
Figure 1.
Certain monogenic neurological diseases, particularly intellectual disability syndromes, are associated with mutations in chromatin modifying proteins. Shown at the left, examples include enzymes that “read” or “write” DNA and histone methylation, ATP-dependent chromatin remodelers, chaperone machinery for the RI histone variant H3.3, and H3.3 itself. A brief description of the functions for each gene product is listed to the right of each gene. Further to the right, the associated neurological diseases are given.
Chromatin exists as the protein-packaged form of our DNA. The basic unit of chromatin, the nucleosome, consists of an octamer of core histone proteins, H2A, H2B, H3, and H4 (or variants thereof), around which ~147 bp of DNA is wrapped 1.7 times [8]. Electrostatic interactions between basic (i.e. lysine and arginine rich) histone proteins and the negatively charged phosphate backbone of DNA lend energetic stability to the nucleosome [9]. These interactions are so strong, in fact, that nucleosomal DNA remains inaccessible to most DNA binding proteins, the transcriptional machinery, and DNA damaging agents [10]. Nucleosomes serve as the building blocks for higher-order chromatin structures that act to stabilize transcriptional programs. In the CNS, precise patterns of chromatin regulation are required to engender the diversity of neuronal and glial subtypes found throughout the brain and facilitate activity-dependent changes in gene expression that are necessary for neuronal plasticity.
Studies of chromatin regulation in the CNS, which fall within the newly minted field of “neuroepigenetics,” have centered on chemical modifications to DNA (e.g. 5-methyl cytosine [11], 5-hydroxymethyl cytosine [12], etc.) and histone proteins (e.g. acetylation, methylation) [13–17]. Such modifications can influence nucleosome stability and serve as binding sites for “reader” domain-containing proteins (e.g. methyl CpG binding protein 2-MeCP2; or the H3K9me3 reader, HP1, etc.) that recruit additional chromatin modifying complexes, assemble higher-order chromatin structures, and regulate transcription within a given cell type [18]. Chemical modifications on chromatin can profoundly impact neuronal functions, as evidenced by phenotypes arising from mutations in enzymes that “write,” “erase,” or “read” such marks (Fig. 1). Neurological syndromes associated with aberrant environmental stimuli, such as chronic illicit drug use [14, 16, 17, 19] and/or exposure to social stress [14], have also been linked to alterations in histone modifications. Such phenotypes reveal that chromatin is, at least in part, capable of influencing neurodevelopmental, age- and experience-related gene expression changes in brain.
In addition to chemical modifications on chromatin, we and others have recently uncovered a crucial role for nucleosomal dynamics (i.e. histone incorporation, sliding, and nucleosomal disassembly) during activity-dependent transcriptional responses in brain [20–23]. In particular, nucleosomal turnover, which involves incorporation of the replication-independent (RI) H3 variant, H3.3, is required for neuronal and glial specific gene expression patterns, synaptic development, dendritic spinogenesis, and cognition (Fig. 2) [20]. These findings suggest that nucleosomal turnover may serve as a fundamental regulator of transcriptional identity and plasticity within the CNS; however, more work is needed to fully understand the contribution of histone turnover to neurological function and disease.
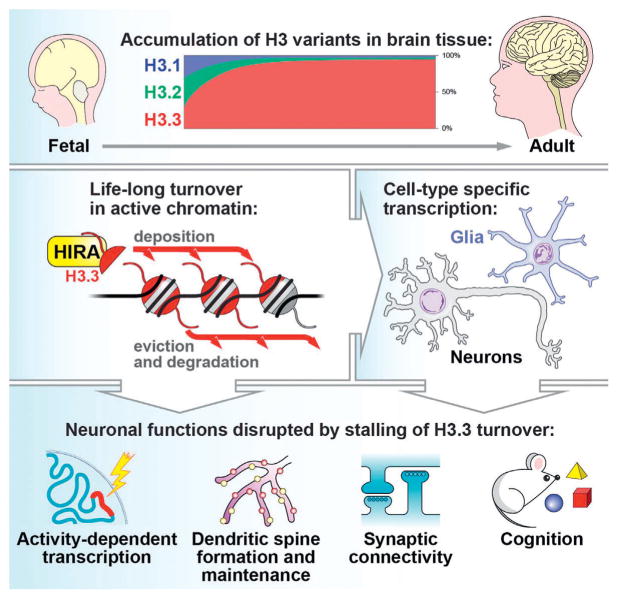
Figure 2.
Roles for H3.3 turnover in brain. Top: H3.3 accumulates with age in brain tissue, with levels reaching >90% of the total H3 pool in neuronal chromatin during mid-adolescence. Middle: H3.3 containing nucleosomes are deposited by HIRA and removed in a proteasomal-dependent manner to promote cell-type specific transcriptional responses in brain. Bottom: Reducing H3.3 turnover in neurons disrupts transcription of activity-dependent “late response” synaptic genes, decreases dendritic spines, reduces both excitatory (glutamatergic) and inhibitory (GABAergic) synapses, and impairs cognition.
In this essay, we review potential mechanisms and functions of nucleosomal turnover in brain. In addition, we present the hypothesis that defects in nucleosomal turnover may serve as a common driver of neurological aging and disease.
Neuronal histone turnover is dynamic throughout life and involves incorporation of RI histone variants
Seminal studies of histone synthesis during cell cycle progression indicate that the majority of histones are assembled onto chromatin during DNA replication [24, 25]. However, “non-canonical” histone variants, such as H2A.X, H2A.Z, and H3.3, as well as a small fraction of canonical H2B and H4, can be synthesized outside of cell division [24, 25]. Consistent with such findings, the levels of RI histone variants H2A.X, H2A.2, and H3.3 have been found to increase in rodent brain with postnatal age, while canonical variants H2A.1, H3.1, and H3.2 display decreased expression [26]. Interestingly, H2A.Z expression remained constant over time [26]. Although the functional significance of these events remained unclear for many decades, this work provided foundational evidence for the existence of RI nucleosomal exchange in brain and suggested that the neuronal chromatin landscape may eventually saturate with certain RI histone variants with age. This stands in stark contrast to the chromatin landscape in dividing cells, where RI histone variants, particularly H3.3, display punctate patterns of enrichment. Thus, it is important to consider as a general principle that the mechanisms of chromatin regulation in neurons may be distinct from those characterized in dividing eukaryotic cells.
Building on these earlier studies, we recently utilized “bomb-pulse” 14carbon dating to assess H3.3 turnover as a function of age in human brain. In doing so, we found that the rate of H3.3 turnover is extraordinarily high during early periods of neurodevelopment and into adolescence, after which point nucleosomal dynamics begin to slow while remaining constitutive through the remainder of life. This switch in the rate of H3.3 turnover coincides with saturation of the neuronal genome by H3.3, where H3.3 accumulates to constitute >90% of the total H3 pool (i.e. H3.1, H3.2, and H3.3) in neuronal chromatin [20]. These findings have been further confirmed using stable isotope labeling of amino acids in mice (SILAM) coupled to mass spectrometry [20]. Moreover, the rate of H3.3 turnover in adult mouse hippocampus has been demonstrated to increase in response to environmental enrichment – a common model of environmentally induced neural and behavioral plasticity – demonstrating that even mild environmental stimuli can have a profound impact on the rate of nucleosomal exchange in brain [20].
Nucleosomal dynamics: Basic mechanisms
From yeast to humans, high levels of nucleosomal “fluidity” – including nucleosomal sliding, partial disassembly/re-assembly, and complete nucleosomal exchange – have been observed during periods of active transcription. Such nucleosomal dynamics can unmask sequence motifs for DNA binding proteins and expose DNA for recruitment of the transcriptional machinery [10]. Equally important, nucleosomes must be constantly replaced to protect DNA from damage [27] and prevent cryptic transcription [28].
RI nucleosomal assembly
Nucleosomal assembly in neurons begins with the synthesis of RI histones. Of the two H3.3 genes expressed in brain (H3f3a and H3f3b), we recently found that H3f3b transcripts specifically are highly responsive to a variety of neuronal stimuli (Fig. 3) including GABA receptor inhibition, neurotrophic signaling (e.g. BDNF), and optogenetic stimulations [20]. Similarly, a recent study of double stranded break (DSBs)-induced gene expression in neurons revealed that the H3f3b gene, along with other activity-dependent loci (e.g. Npas4), is marked by γH2A.X (a marker of DNA DSBs) following membrane depolarization [29]. These data implicate H3f3b as a bona fide stimulus-responsive gene in brain. Thus, histone synthesis likely serves as an important mediating step in the process of activity-dependent nucleosomal turnover in neurons.
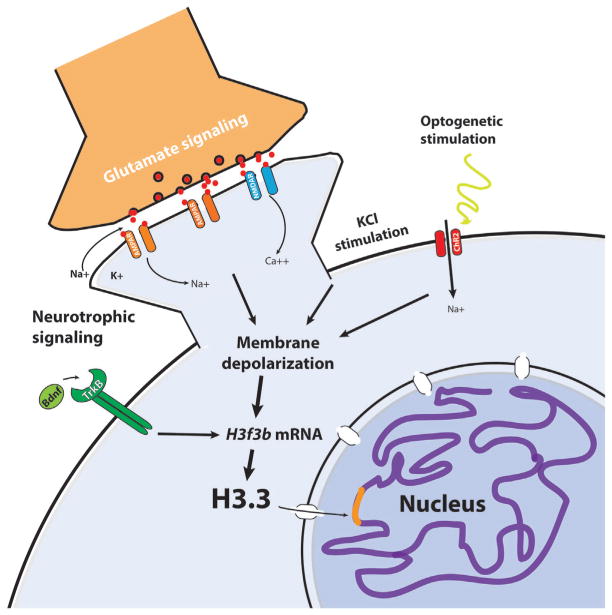
Figure 3.
H3.3 transcripts and protein levels in the soluble nuclear pool are induced by a variety of neuronal stimuli: neurotrophin signaling (e.g. brain-derived growth factor, BDNF), glutamate receptor activation, and membrane depolarization. Sufficient soluble H3.3 levels are required for nucleosomal turnover and activity-dependent neuronal transcription.
Studies of nucleosomal assembly in non-neuronal systems indicate that newly synthesized H3.3-H4 heterodimers are transported into the nucleus by the H3-H4 chaperone, ASF1 [30], and potentially with the aid of nuclear receptor Importin 4 [31]. In primary human mesenchymal stem cells, for example, ASF1a hands over H3.3-H4 heterodimers to the H3.3-specific chaperone DAXX [32], which then transports H3.3-H4 to nuclear promyelocytic leukemia (PML) bodies [30]. The ATP-dependent chromatin remodeler, ATRX, which is heavily mutated in X-linked alpha-thalessemia mental retardation syndrome (ATR-X), is also associated with PML bodies [30, 33] and has been shown to interact directly with DAXX during H3.3 deposition at pericentromeric and telomeric repeats [34–36]. In neurons, DAXX has recently been shown to deposit H3.3 at a subset of activity-dependent gene promoters including Bdnf [22]. While it remains unclear whether DAXX-ATRX assembles H3.3-H4 heterotetramers in neurons, this complex can assemble H3.3-H4 tetramers on DNA in vitro [34] and likely plays a critical role in neuronal genomic stability [37–40].
A second, distinct [36], nucleosomal predeposition complex contains histone cell cycle regulator (HIRA), the H3.3-specific chaperone Ubinuclein 1 and Calcineurin-binding protein 1 (CABIN1) [41]. HIRA, and not DAXX, appears to facilitate the majority of activity dependent H3.3 turnover in neuronal chromatin, primarily within in the promoters and gene bodies of “late-response” stimulus-dependent synaptic genes [20]. HIRA has similarly been shown to promote H3.3 deposition at promoters and gene bodies in yeast and fruit flies, suggesting a conserved role for HIRA in the regulation of H3.3 deposition throughout actively coding genic regions [20, 35, 42]. Structural studies have further revealed that H3.3 and ASF1a [43] bind to HIRA on opposite “faces” [44] indicating that HIRA and ASF1a may work together to assemble H3.3-H4 tetramers in the soluble nuclear pool (i.e. the non-chromatin bound fraction). In flies, HIRA has also been shown to interact with the ATP-dependent chromatin remodeler, CHD1 [45], a protein that suppresses neuronal differentiation from mESCs [46]. It will be vitally important in future studies to determine whether HIRA itself interacts with additional chromatin modifying proteins in brain to fine-tune patterns of neuronal transcription.
Finally, nucleosome assembly is completed upon the addition of H2A-H2B heterodimers onto the H3-H4 tetramer [47] by chaperones such as FACT [48, 49] and NAP1 [50]. This final step of nucleosomal assembly remains to be fully explored in brain and likely represents another facet of the increasingly complex regulatory mechanisms known to mediate gene expression in neurons and glia.
Nucleosomal disassembly
Nucleosomes can be partially or fully disassembled to facilitate transcription factor binding and to allow for engagement by the transcriptional machinery. During partial disassembly, H2A-H2B heterodimers are ejected sequentially from the nucleosome, leaving behind intact nucleosomal H3-H4 tetramers and the chemical modifications that they possess [51]. In mammalian brain, H2A.2 (or macroH2A), H2A.X, and H2A.Z are the only H2A variants known to be synthesized [26, 52]. Therefore, partial nucleosomal disassembly and exchange likely involves the incorporation of one, or combinations, of these variants. Evidence exists to suggest that H2A-H2B dimers may also be displaced to facilitate transcriptional elongation in the absence of complete nucleosomal turnover [51]. Specifically, H2A-H2B histone chaperones, such as FACT [53] and NAP1 [54], have been found to exchange H2A-H2B dimers while simultaneously stabilizing hexameric and tetrameric nucleosomes resulting in transient, partial nucleosomal disassembly during elongation. Reducing FACT expression in yeast has been found to increase spurious transcription [55], underscoring the potential importance of stabilizing nucleosomes in gene bodies during active transcription. Thus, partial nucleosomal disassembly by H2A-H2B ejection preserves nucleosomal density and maintains epigenetic memory on H3-H4 tetramers.
Disassembly of the +1 nucleosome relative to the transcriptional start site (TSS) also appears to be a crucial step during transcriptional elongation [23, 56]. In Drosophila cells, a large fraction of elongating RNA polymerase II has been observed to backtrack and pause at the +1 nucleosome, suggesting that this nucleosome acts a potent barrier to transcription [56]. Interestingly, the +1 nucleosome is often homotypic for the variant H2A.Z [57], which is associated with deeper transcription into the nucleosome, reduced RNA polymerase pausing, and decreased H3-H4 turnover [56]. H2A.Z may, therefore, reduce the +1 nucleosomal barrier by permitting transcription through partially disassembled nucleosomes. In murine CA1 hippocampal neurons, H2A.Z eviction from the +1 nucleosome of specific activity-induced genes is associated with transcriptional activity and enhanced cognition [23]. This suggests that H2A.Z eviction from the +1 nucleosome may be required to facilitate its role in decreasing nucleosomal barriers. Given its putative role in regulating transcriptional elongation, it is not surprising that H2A.Z localization within the genome is tightly regulated by the ATP-dependent chromatin remodelers INO80 [58] and SRCAP [59] and histone chaperones FACT [55] and ANP32E [60]. This may also explain why H2A.Z turnover remains dynamic throughout life but H2A.Z does not accumulate in brain [20, 26].
Complete nucleosomal disassembly, on the other hand, involves the total unwrapping of nucleosomal DNA and H3-H4 tetramer release [9]. Binding of multiple RNA polymerases at strongly induced genes is one mechanism through which H3-H4 tetramers may be ejected from chromatin [51, 61]. However, our recent studies in neurons implicate a prominent role for proteasomal-dependent degradation of H3.3 during complete nucleosomal disassembly, such that proteasome inhibition significantly reduces H3.3 eviction from neuronal chromatin [20]. The presence of neuronal activity-induced polyubiquitylation on chromatin-bound H3.3 further supports the notion that proteasomal-dependent degradation may directly regulate this process [20]. These data are in accordance with previous observations of H2A, H2B, and H3 polyubiquitylation in nuclei of elongating rat spermatids [62]. Importantly, these findings shed new light on the proteasome as a mediator of neural plasticity, whereby the proteasome coordinates activity-dependent synaptic proteostasis [63] with nucleosomal eviction [20].
Nucleosomal stability is modulated by ATP-dependent chromatin remodelers
ATP-dependent chromatin remodelers serve critical roles during CNS development [4] and have been implicated in the regulation of nucleosomal stability. While the impact of ATP-dependent chromatin remodeling complexes on nucleosomal turnover has yet to be explored in brain, studies in non-neuronal systems suggest that chromatin remodelers are intricately involved in the stabilization and positioning of nucleosomes at promoters and gene bodies, as well as at developmental enhancers. Specifically, chromatin remodelers of the SWI/SNF family (e.g. BRG1-associated factor/BAF and Remodel the structure of chromatin/RSC) have previously been implicated in the regulation of nucleosomal spacing near TSSs [64, 65] and are thought to contribute to transcriptional initiation and RNA polymerase pausing. The BAF complex additionally influences transcriptional initiation by increasing nucleosomal spacing at enhancer elements, a phenomenon that has recently been demonstrated during the process of hematopoiesis [66]. Similarly, ISW1 and CHD1 chromatin remodelers have been shown to stabilize gene body nucleosomes during transcriptional elongation [67]. It will, therefore, be insightful to learn whether nucleosomal positioning and/or turnover are indeed essential functions of chromatin remodelers during CNS development.
RI nucleosome turnover is sensitive to soluble H3.3 levels
Sufficient levels of non-chromatin bound H3.3 are required for both H3.3-dependent nucleosomal assembly and disassembly in neurons and mESCs [20, 42, 68]. Reducing the soluble nuclear pool of H3.3 by genetic knockdown in both embryonic and adult neurons significantly stalls nucleosomal turnover and leads to transcriptional, synaptic, and cognitive deficits [20]. Such findings indicate that mammalian cells have evolved an uncharacterized sensing mechanism to prevent nucleosomal depletion during times in which histones are unavailable for re-assembly. Conversely, excess histones have been shown to disrupt nucleosomal spacing and chromatin structure [69] and can increase sensitivity to DNA damage [70]. Therefore, soluble histone proteostasis may be crucial for both transcriptional responses and genomic stability in brain.
We have found that neurons modulate the level of soluble H3.3, at least in part, though activity-dependent transcription of H3f3b [20]. However, two additional mechanisms have been identified through which human cells maintain soluble histone levels: (i) DAXX-dependent H3.3-H4 storage at PML bodies [30] and (ii) protection from chaperone (Hsc70 and Hsp90)-mediated autophagy by the histone chaperone NASP1 [71]. While these mechanisms have yet to be explored in brain, the targeting of ATRX to PML bodies was found to be reduced by ATR-X patient mutations [72], suggesting that defects in the maintenance and/or mobilization of soluble H3.3 may contribute to intellectual disability.
A putative role for nucleosomal turnover in neuronal lineage specification and differentiation
RI nucleosomal turnover has been shown to facilitate targeting of the polycomb repressive complex 2 (PRC2) to developmental genes in flies and mESCs [42, 73], a process that may similarly play role in neurogenesis. The promoters of many important developmental genes are considered to be “bivalent,” as they bear both transcriptionally permissive (H3K4me3) and repressive (H3K27me3) methylation signatures [42]. While transcriptionally silent, these genes are “poised” for rapid induction upon cellular differentiation. Recently, it was demonstrated that knockdown of either H3.3 or its chaperone, HIRA, reduces RI nucleosomal turnover and dramatically diminishes the signal of PRC2 and its enzymatic mark, H3K27me3, at bivalent promoters in mESCs [42]. Such loss of PRC2 recruitment results in the aberrant activation of developmental genes and impairs restricted patterns of ESC differentiation toward the three embryonic germ layers. Importantly, HIRA was found to interact with PRC2 in an H3.3-dependent manner indicating that PRC2 recruitment is coupled to HIRA-dependent H3.3 nucleosomal deposition [42].
While it remains unclear whether H3.3 deposition-coupled PRC2 targeting is employed as a general mechanism of lineage specification in multipotent cells, including neural progenitor cells (NPCs), nucleosomal turnover is known to facilitate PRC2 targeting to cis-regulatory elements in Drosophila [73], suggesting that this process may be conserved across cell types and species. Additionally, H3.3 depletion studies have corroborated a role for nucleosomal turnover during cellular differentiation such that H3.3 is necessary for a diverse array of developmental processes including myogenesis [74], neural crest differentiation [75], gametogenesis [76, 77], and zygotic genomic activation [77, 78].
Heterochromatic H3.3 turnover: A potentially conserved role in repetitive element silencing
Repetitive elements constitute approximately two-thirds of the human genome [79] and are often buried in heterochromatin to prevent mutagenic recombination and retrotransposition events [80], as well as transcription-coupled repeat expansion [81]. Repetitive element unsilencing in brain has been associated with drug abuse [17], stress [82], psychiatric [80], and mood disorders [14]; neurodevelopmental disorders [83]; neurodegeneration [84]; and human brain cancers [85]. It is plausible that genomic instability resulting from the unsilencing of heterochromatic repetitive elements contributes to the onset and/or progression of these neurological diseases.
RI nucleosomal turnover occurs slowly in heterochromatin [86], however, H3.3 has been found to enrich at subsets of transposable elements in Drosophila [87] and mESCs [68], and at pericentromeric and telomeric satellite repeats in cycling mammalian cells [35, 86, 88]. Knockout studies have revealed that H3.3 is necessary for heterochromatic methylation on H3K9, H3K27, and H4K20 in mESCs [42, 89] and that H3.3 itself is a crucial substrate for H3K9 and H3K27 methyltransferase activity in early embryos [89, 90]. In mammalian brain, components of the DAXX/ATRX H3.3 deposition pathway have been implicated in silencing imprinted loci, ribosomal- [37], pericentromeric-, centromeric- [39], and simple intronic repeats [91]. These findings suggest a significant and potentially conserved role for heterochromatic H3.3 turnover in silencing repetitive elements.
A role for H3.3 in the suppression of repetitive element expansion in brain?
Repetitive element expansions are associated with over 40 neurodevelopmental, neuromuscular, and neurodegenerative disorders, including Huntington’s, Alzheimer’s, and Parkinson’s disease [92]. Unlike static mutations, repeat expansions are dynamic throughout life and produce genetic mosaicism within tissues [93]. The length of repeat expansion is correlated with increased disease severity and inversely correlates with the average age of onset of disease, a phenomenon referred to as genetic anticipation [92]. Trinucleotide repeat expansions, the most common form in human brain disease, involve the expansion of microsatellite sequences. These sequences are enriched near telomeres, long- or short interspersed nuclear element (LINE and SINE) retrotransposons, and in 3′ repeats of human Alu retrotransposons [94]. Neurological disease-causing microsatellite expansions can arise from replication fork slippage during DNA synthesis or from homologous repair of DNA:RNA hybrid structures (R-loops) formed during the transcription of repeats [81, 92]. Indeed, R-loop repair has been demonstrated to cause a trinucleotide repeat expansion found in Fragile X mental retardation syndrome 1 [81].
To our knowledge, the functional relationship between nucleosomal turnover and repeat expansion has not yet been studied in brain. If DAXX/ATRX-dependent H3.3 turnover promotes the transcriptional silencing of repetitive elements in brain as in mESCs, then heterochromatic H3.3 turnover may protect against neurological disease-causing repeat expansions. In line with this hypothesis, we have found that ATRX is necessary to repress transcription at pericentromeric and centromeric satellite repeats in mouse embryonic neurons [39]. Moreover, loss of function mutations in ATRX are found in approximately 40% of human glioma [95], brain tumors that exhibit microsatellite instability [96]. Thus, heterochromatic H3.3 turnover mediated by DAXX-ATRX may protect against deleterious repetitive element expansions in the CNS. Much more work is needed, however, to fully understand the role of histone turnover, or aberrations thereof, in the development of these devastating illnesses.
H3.3 nucleosomal deposition may silence transposable elements in brain
Retrotransposons are repetitive elements that can cause mutations by “copying-and-pasting” themselves into new genomic locations via an RNA intermediate. Several studies indicate that retrotransposon expression and/or insertion in mammalian brain is responsive to environmental stimuli including drug use [17], stress [14, 82], exercise [97], and membrane depolarization [39]. Excessive retrotransposon expression may be pathological, as patients with schizophrenia, bipolar disorder, and major depressive disorder exhibit elevated retrotransposition in frontal cortex [80]. In addition, high retrotransposon expression has been observed in human glioma, glioblastoma, and medullablastoma [85]. These findings suggest that the retrotransposon expression is actively regulated in brain and may become misregulated in disease.
Negative regulators of retrotransposition in brain known thus far include the transcriptional co-repressor KRAB-associated protein-1 (KAP1) [98] and methyl CpG-binding protein (MeCP2) [99]. Interestingly, a mechanism for H3.3 turnover-dependent, KAP1-mediated transcriptional silencing of endogenous retroviral elements (ERVs) was recently uncovered in mESCs [68]. Similar to the mechanism of PRC2 targeting described above, KAP1 was found to interact directly with H3.3-DAXX-ATRX pre-deposition complexes and co-recruit the H3K9 methyltransferase ESET to class I and II ERVs including early transposon (ETn)/MuSD and intracisternal A-type particles (IAPs) [68]. H3K9 trimethylation by ESET is known to transcriptionally silence ERVs [100] and indeed, H3.3 deposition-coupled targeting of KAP1 and ESET to ERVs extinguished their expression [68]. Consistent with these findings, KAP1 conditional knockout in NPCs leads to an increase the expression of IAP-1 and MERVK10C ERVs, concomitant with a decrease in H3K9 methylation at these sites [98]. These results indicate that KAP1 similarly targets ERVs for H3K9 methylation-dependent transcriptional silencing in both mESCs and NPCs. However, the complete mechanism for KAP1-dependent ERV silencing, including a role for H3.3 deposition, remains to be elucidated in NPCs.
MeCP2 knockout mice exhibit an elevated level of LINE1 retrotransposition in midbrain and cerebellar neurons [99]; however, the mechanistic basis for this phenomenon is not fully understood. MeCP2 has been identified in ATRX/DAXX protein complexes in brain and co-localizes with DAXX and ATRX at heterochromatic loci in neurons [22, 37]. Intriguingly, mutations in MECP2 that cause Rett syndrome and mutations in ATRX that cause ATR-X disrupt their interaction with one another [7], suggesting that the association of MeCP2 with ATRX may be crucial for neural development. These findings also provide circumstantial evidence that H3.3 nucleosomal dynamics may be involved in MeCP2-mediated LINE1 repression in brain.
In addition to the above mechanisms, we have shown that ATRX is a repressor of pericentromeric satellite expression in mouse embryonic neurons [39]. Pericentromeres are enriched with LINE and SINE retrotransposons and their expression has been linked to pericentromeric satellite transcription in other cell types [101, 102]. Upon membrane depolarization, we found that ATRX exhibits increased enrichment at pericentromeres, coincident with displacement of heterochromatin protein 1 (HP1) by activity-induced H3S10 phosphorylation/H3K9me3 [39]. However, RNAi-mediated knockdown of ATRX induces pericentromeric satellite expression in neurons [39], suggesting that the presence of ATRX at pericentromeres stabilizes heterochromatin. Although indirect, our findings shed light on a possible mechanism for ATRX-mediated H3.3 turnover in facilitating the heterochromatic silencing of neuronal retrotransposons.
Nucleosome dynamics in neurological aging and disease
In neurons, activity-dependent expression of late-response genes requires complete nucleosomal turnover [20]. The majority of such turnover depends upon nucleosomal deposition by the H3.3 chaperone complex HIRA, although Ca2+-dependent dephosphorylation of DAXX by calcineurin has also been shown to facilitate H3.3 incorporation at a subset of immediate early genes (IEGs) [22]. Blocking nucleosomal turnover by microRNA-mediated knockdown of H3.3 in embryonic and adult hippocampal neurons – a process that greatly depletes soluble nuclear H3.3 pools without reducing levels of chromatin bound H3.3 – results in a substantial loss of dendritic spines, decreased excitatory and inhibitory synapses, and deficits in memory that cannot be attributed to cell death [20]. As mentioned previously, the eviction of H2A.Z from the +1 nucleosome of subsets of activity-dependent neuronal genes, a process that would be predicted to increase H3.3 turnover [56], has also recently been implicated in memory consolidation [23]. Thus, complete nucleosomal turnover, as well as mechanisms that modulate such events, appear to play a fundamental role in synaptic plasticity and cognition.
Both during neurodevelopment and in adulthood, neuronal circuits are continuously established and remodeled through the careful maintenance and pruning of dendritic spines, which exist as the primary sites of excitatory glutamatergic input in brain [1]. Dendritic spine loss and synaptic dysregulation are common features of many neurological disorders, including autism [103], schizophrenia [104], drug addiction [16], stress [105], mood disorders [106], and later-life neurodegenerative diseases [107]. The significance of spine loss has been directly examined in the context of Alzheimer’s disease, where synaptic and dendritic spine reductions have been found to exhibit stronger correlations with cognitive decline than the presence of neurofibrillary tangles or neuronal loss itself [108]. Gradual loss of excitatory synaptic inputs is also associated with natural aging processes in primates and is thought to contribute to age-related cognitive decline [109].
Our recent studies of H3.3 dynamics in brain reveal that impaired nucleosomal turnover results in the loss of dendritic spines, reductions in excitatory synapses, and impaired cognitive capacity [20]. Given the shared phenotypes between genetically mediated loss of H3.3 in neurons and common neurological “spinopathies,” one could hypothesize that aberrant H3.3 turnover may represent a mechanistic point of convergence for multiple pathways involved in the development of neurological and psychiatric disease. Such assumptions, however, will require significant future study.
Nucleosomal turnover as a potential regulator of adaptive evolution
Mutations in cis-regulatory elements are becoming increasingly appreciated as drivers of evolution and may be caused by transposition events that are regulated by H3.3 turnover. Recent studies of adaptive evolution in fish and primates reveal that ~80% of the mutations linked with phenotypic change occur in regulatory elements, while only ~20% occur within coding regions [110, 111]. Examination of regulatory regions identified by DNase hypersensitivity from multiple cell types and species (ENCODE) reveal that ~44% of all regulatory elements lie within transposable elements [112]. Strikingly, this number increases to ~63% when considering primate-specific regions of open chromatin [112]. In line with this analysis, a recent study of neural crest enhancer divergence between chimpanzee and human revealed that species-biased enhancers tend to enrich for ERVs, particularly ERV1, ERVL-MaLR, ERVK, and LINE1 elements [113]. Thus, transposition events appear to have meaningfully contributed to adaptive evolution among primates.
Given that H3.3 nucleosomal turnover is necessary for the repression of certain ERVs in mESCs [68], it is plausible that H3.3 turnover in embryos of ancestral primates may have played a role in mediating transposition events that have shaped human evolution. While a relationship between H3.3 turnover and organismal adaptive evolution is difficult to assess experimentally, it may be possible to examine this relationship in cultured cells and/or tissues. The CNS may be particularly useful for such studies, as transposition in NPCs has recently been shown to produce genetic mosaicsm in brain and has been proposed as a mechanism through which cells of the CNS achieve subtype diversity [114]. Moreover, LINE1 and IAP element expression are inducible in neurons, and like the rate of RI nucleosomal turnover, can be directly manipulated by alterations in neural activity [20, 39]. Combining lineage tracing in brain with cause-effect studies probing the relationship between H3.3 turnover and transposition may reveal new insights into the molecular mechanisms underlying cellular, and possibly even organismal, adaptive evolution.
Conclusion and outlook
Histone turnover represents a newly identified and critical mediator of neuronal plasticity in mammalian brain [20, 22, 23]. Here, we propose that deficits in RI nucleosomal dynamics likely contribute to both age-related patterns of cognitive decline, as well as to the vast array of neurological diseases that plague our society. To test this hypothesis, neurological diseases with altered rates of histone turnover must first be identified. This can be accomplished by measuring histone turnover in healthy versus diseased human brain tissues using bomb-pulse labeling, or by metabolically labeling histones in cultures of patient iPSC-derived neurons or in rodent models of neurological disease. The functional significance of aberrant nucleosomal dynamics in disease can be determined from restoring histone turnover to a “healthy” rate (e.g. by modulating transcript levels of H3.3) and assessing the extent of rescue for the disease phenotype. Complementarily, adjusting histone turnover to mimic a disease-associated rate in healthy cells may further help to establish causal links between nucleosomal dynamics and a given neurological disease phenotype. However, such manipulations of histone turnover will require intimate knowledge of the mechanisms and regulation of nucleosomal dynamics in brain, which remain incompletely understood. Genetic and pharmacological screens for regulators of histone turnover in brain, as well as hypothesis-driven research based on the regulation of histone turnover in other cell types, may fill in these gaps in knowledge and inform future strategies for therapeutic intervention in neurological disease.
Acknowledgments
We express sincere apologies to colleagues whose work we could not include in this article. We thank members of the Maze laboratory for critical readings of this work, as well as Dr. Alexey Soshnev for assistance with illustrations. I.M. is supported by: (i) a 2014 NARSAD Young Investigator Award; (ii) an MQ Research Fellowship (MQ15FIP100011); (iii) a 2016 Sloan Research Fellowship in Neuroscience; (iv) the Rosen Family Research Scholar Award; and (v) the National Institute of Mental Health (NIMH): P50 MH096890.
Abbreviations
- CNS
central nervous system
- ERV
endogenous retroviral element
- IAP
intracisternal A-type particle
- LINE
long interspersed nuclear element
- NPC
neural progenitor cell
- PRC2
polycomb repressive complex 2
- RI
replication-independent
- SINE
short interspersed nuclear element
- TSS
transcriptional start site
Footnotes
The authors have declared no conflicts of interest.
References
- 1.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, et al. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14:285–93. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannerman DM, Sprengel R, Sanderson DJ, McHugh SB, et al. Hippocampal synaptic plasticity, spatial memory and anxiety. Nat Rev Neurosci. 2014;15:181–92. doi: 10.1038/nrn3677. [DOI] [PubMed] [Google Scholar]
- 3.Adegbola A, Gao H, Sommer S, Browning M. A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD) Am J Med Genet A. 2008;146A:505–11. doi: 10.1002/ajmg.a.32142. [DOI] [PubMed] [Google Scholar]
- 4.Lopez AJ, Wood MA. Role of nucleosome remodeling in neurodevelopmental and intellectual disability disorders. Front Behav Neurosci. 2015;9:100. doi: 10.3389/fnbeh.2015.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein CJ, Botuyan MV, Wu Y, Ward CJ, et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43:595–600. doi: 10.1038/ng.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirov G, Pocklington AJ, Holmans P, Ivanov D, et al. De novo CNV analysis implicates specific abnormalities of post-synaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–53. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nan X, Hou J, Maclean A, Nasir J, et al. Interaction between chromatin proteins MECP2 and ATRX is disrupted by mutations that cause inherited mental retardation. Proc Natl Acad Sci USA. 2007;104:2709–14. doi: 10.1073/pnas.0608056104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luger K, Mader A, Sargent DF, Richmond TJ. The atomic structure of the nucleosome core particle. J Biomol Struct Dyn. 2000;17:185–8. doi: 10.1080/07391102.2000.10506619. [DOI] [PubMed] [Google Scholar]
- 9.Weber CM, Henikoff S. Histone variants: dynamic punctuation in transcription. Genes Dev. 2014;28:672–82. doi: 10.1101/gad.238873.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–8. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 11.Guo JU, Ma DK, Mo H, Ball MP, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–51. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo JU, Su Y, Zhong C, Ming GL, et al. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–34. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar A, Choi KH, Renthal W, Tsankova NM, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–14. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 14.Covington HE, 3rd, Maze I, Sun H, Bomze HM, et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71:656–70. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maze I, Chaudhury D, Dietz DM, Von Schimmelmann M, et al. G9a influences neuronal subtype specification in striatum. Nat Neurosci. 2014;17:533–9. doi: 10.1038/nn.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–6. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maze I, Feng J, Wilkinson MB, Sun H, et al. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci USA. 2011;108:3035–40. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20:259–66. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 19.Feng J, Wilkinson M, Liu X, Purushothaman I, et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014;15:R65. doi: 10.1186/gb-2014-15-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maze I, Wenderski W, Noh KM, Bagot RC, et al. Critical role of histone turnover in neuronal transcription and plasticity. Neuron. 2015;87:77–94. doi: 10.1016/j.neuron.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun H, Damez-Werno DM, Scobie KN, Shao NY, et al. ACF chromatin-remodeling complex mediates stress-induced depressive-like behavior. Nat Med. 2015;21:1146–53. doi: 10.1038/nm.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michod D, Bartesaghi S, Khelifi A, Bellodi C, et al. Calcium-dependent dephosphorylation of the histone chaperone DAXX regulates H3.3 loading and transcription upon neuronal activation. Neuron. 2012;74:122–35. doi: 10.1016/j.neuron.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zovkic IB, Paulukaitis BS, Day JJ, Etikala DM, et al. Histone H2A.Z. subunit exchange controls consolidation of recent and remote memory. Nature. 2014;515:582–6. doi: 10.1038/nature13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu RS, Tsai S, Bonner WM. Patterns of histone variant synthesis can distinguish G0 from G1 cells. Cell. 1982;31:367–74. doi: 10.1016/0092-8674(82)90130-1. [DOI] [PubMed] [Google Scholar]
- 25.Wu RS, Bonner WM. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981;27:321–30. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]
- 26.Pina B, Suau P. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev Biol. 1987;123:51–8. doi: 10.1016/0012-1606(87)90426-x. [DOI] [PubMed] [Google Scholar]
- 27.Enright HU, Miller WJ, Hebbel RP. Nucleosomal histone protein protects DNA from iron-mediated damage. Nucleic Acids Res. 1992;20:3341–6. doi: 10.1093/nar/20.13.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hennig BP, Bendrin K, Zhou Y, Fischer T. Chd1 chromatin remodelers maintain nucleosome organization and repress cryptic transcription. EMBO Rep. 2012;13:997–1003. doi: 10.1038/embor.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madabhushi R, Gao F, Pfenning AR, Pan L, et al. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell. 2015;161:1592–605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delbarre E, Ivanauskiene K, Kuntziger T, Collas P. DAXX-dependent supply of soluble (H3.3-H4) dimers to PML bodies pending deposition into chromatin. Genome Res. 2013;23:440–51. doi: 10.1101/gr.142703.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jasencakova Z, Scharf AN, Ask K, Corpet A, et al. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell. 2010;37:736–43. doi: 10.1016/j.molcel.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 32.Elsasser SJ, Huang H, Lewis PW, Chin JW, et al. DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature. 2012;491:560–5. doi: 10.1038/nature11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xue Y, Gibbons R, Yan Z, Yang D, et al. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci USA. 2003;100:10635–40. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, et al. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA. 2010;107:14075–80. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goldberg AD, Banaszynski LA, Noh KM, Lewis PW, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–91. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elsaesser SJ, Allis CD. HIRA and Daxx constitute two independent histone H3.3-containing predeposition complexes. Cold Spring Harb Symp Quant Biol. 2010;75:27–34. doi: 10.1101/sqb.2010.75.008. [DOI] [PubMed] [Google Scholar]
- 37.Kernohan KD, Jiang Y, Tremblay DC, Bonvissuto AC, et al. ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev Cell. 2010;18:191–202. doi: 10.1016/j.devcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 38.Kernohan KD, Vernimmen D, Gloor GB, Berube NG. Analysis of neonatal brain lacking ATRX or MeCP2 reveals changes in nucleosome density, CTCF binding and chromatin looping. Nucleic Acids Res. 2014;42:8356–68. doi: 10.1093/nar/gku564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noh KM, Maze I, Zhao D, Xiang B, et al. ATRX tolerates activity-dependent histone H3 methyl/phos switching to maintain repetitive element silencing in neurons. Proc Natl Acad Sci USA. 2015;112:6820–7. doi: 10.1073/pnas.1411258112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–31. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 41.Daniel Ricketts M, Frederick B, Hoff H, Tang Y, et al. Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat Commun. 2015;6:7711. doi: 10.1038/ncomms8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banaszynski LA, Wen D, Dewell S, Whit-comb SJ, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–20. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang Y, Poustovoitov MV, Zhao K, Garfinkel M, et al. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat Struct Mol Biol. 2006;13:921–9. doi: 10.1038/nsmb1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natsume R, Eitoku M, Akai Y, Sano N, et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–41. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 45.Konev AY, Tribus M, Park SY, Podhraski V, et al. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science. 2007;317:1087–90. doi: 10.1126/science.1145339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460:863–8. doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan HF, Liu ZN, Chow SY, Lu YH, et al. Histone chaperone-mediated nucleosome assembly process. PloS ONE. 2015;10:e0115007. doi: 10.1371/journal.pone.0115007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hondele M, Stuwe T, Hassler M, Halbach F, et al. Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature. 2013;499:111–4. doi: 10.1038/nature12242. [DOI] [PubMed] [Google Scholar]
- 49.Winkler DD, Muthurajan UM, Hieb AR, Luger K. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events. J Biol Chem. 2011;286:41883–92. doi: 10.1074/jbc.M111.301465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrews AJ, Chen X, Zevin A, Stargell LA, et al. The histone chaperone Nap1 promotes nucleosome assembly by eliminating nonnucleosomal histone DNA interactions. Mol Cell. 2010;37:834–42. doi: 10.1016/j.molcel.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kulaeva OI, Hsieh FK, Chang HW, Luse DS, et al. Mechanism of transcription through a nucleosome by RNA polymerase II. Biochim Biophys Acta. 2013;1829:76–83. doi: 10.1016/j.bbagrm.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Magri L, Swiss VA, Jablonska B, Lei L, et al. E2F1 coregulates cell cycle genes and chromatin components during the transition of oligodendrocyte progenitors from proliferation to differentiation. J Neurosci. 2014;34:1481–93. doi: 10.1523/JNEUROSCI.2840-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, et al. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–3. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 54.Kuryan BG, Kim J, Tran NN, Lombardo SR, et al. Histone density is maintained during transcription mediated by the chromatin remodeler RSC and histone chaperone NAP1 in vitro. Proc Natl Acad Sci USA. 2012;109:1931–6. doi: 10.1073/pnas.1109994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jeronimo C, Watanabe S, Kaplan CD, Peterson CL, et al. The histone chaperones FACT and Spt6 restrict H2A.Z. from intragenic locations. Mol Cell. 2015;58:1113–23. doi: 10.1016/j.molcel.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weber CM, Ramachandran S, Henikoff S. Nucleosomes are context-specific, H2A.Z-modulated barriers to RNA polymerase. Mol Cell. 2014;53:819–30. doi: 10.1016/j.molcel.2014.02.014. [DOI] [PubMed] [Google Scholar]
- 57.Weber CM, Henikoff JG, Henikoff S. H2A.Z. nucleosomes enriched over active genes are homotypic. Nat Struct Mol Biol. 2010;17:1500–7. doi: 10.1038/nsmb.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z. localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–13. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ruhl DD, Jin J, Cai Y, Swanson S, et al. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z. into nucleosomes. Biochemistry. 2006;45:5671–7. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- 60.Obri A, Ouararhni K, Papin C, Diebold ML, et al. ANP32E is a histone chaperone that removes H2A.Z. from chromatin. Nature. 2014;505:648–53. doi: 10.1038/nature12922. [DOI] [PubMed] [Google Scholar]
- 61.Ray-Gallet D, Woolfe A, Vassias I, Pellentz C, et al. Dynamics of histone H3 deposition in vivo reveal a nucleosome gap-filling mechanism for H3.3 to maintain chromatin integrity. Mol Cell. 2011;44:928–41. doi: 10.1016/j.molcel.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 62.Chen HY, Sun JM, Zhang Y, Davie JR, et al. Ubiquitination of histone H3 in elongating spermatids of rat testes. J Biol Chem. 1998;273:13165–9. doi: 10.1074/jbc.273.21.13165. [DOI] [PubMed] [Google Scholar]
- 63.Hamilton AM, Oh WC, Vega-Ramirez H, Stein IS, et al. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron. 2012;74:1023–30. doi: 10.1016/j.neuron.2012.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yen K, Vinayachandran V, Batta K, Koerber RT, et al. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–73. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tolstorukov MY, Sansam CG, Lu P, Koellhoffer EC, et al. Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc Natl Acad Sci USA. 2013;110:10165–70. doi: 10.1073/pnas.1302209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu G, Schones DE, Cui K, Ybarra R, et al. Regulation of nucleosome landscape and transcription factor targeting at tissue-specific enhancers by BRG1. Genome Res. 2011;21:1650–8. doi: 10.1101/gr.121145.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smolle M, Venkatesh S, Gogol MM, Li H, et al. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol. 2012;19:884–92. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Elsasser SJ, Noh KM, Diaz N, Allis CD, et al. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522:240–4. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh RK, Liang D, Gajjalaiahvari UR, Kabbaj MH, et al. Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell Cycle. 2010;9:4236–44. doi: 10.4161/cc.9.20.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–49. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 71.Cook AJ, Gurard-Levin ZA, Vassias I, Almouzni G. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol Cell. 2011;44:918–27. doi: 10.1016/j.molcel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 72.Berube NG, Healy J, Medina CF, Wu S, et al. Patient mutations alter ATRX targeting to PML nuclear bodies. Eur J Hum Genet. 2008;16:192–201. doi: 10.1038/sj.ejhg.5201943. [DOI] [PubMed] [Google Scholar]
- 73.Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–11. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 74.Yang JH, Song Y, Seol JH, Park JY, et al. Myogenic transcriptional activation of MyoD mediated by replication-independent histone deposition. Proc Natl Acad Sci USA. 2011;108:85–90. doi: 10.1073/pnas.1009830108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cox SG, Kim H, Garnett AT, Medeiros DM, et al. An essential role of variant histone H3.3 for ectomesenchyme potential of the cranial neural crest. PLoS Genet. 2012;8:e1002938. doi: 10.1371/journal.pgen.1002938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yuen BT, Bush KM, Barrilleaux BL, Cotterman R, et al. Histone H3.3 regulates dynamic chromatin states during spermatogenesis. Development. 2014;141:3483–94. doi: 10.1242/dev.106450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tang MC, Jacobs SA, Mattiske DM, Soh YM, et al. Contribution of the two genes encoding histone variant h3.3 to viability and fertility in mice. PLoS Genet. 2015;11:e1004964. doi: 10.1371/journal.pgen.1004964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lin CJ, Koh FM, Wong P, Conti M, et al. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell. 2014;30:268–79. doi: 10.1016/j.devcel.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Koning AP, Gu W, Castoe TA, Batzer MA, et al. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bundo M, Toyoshima M, Okada Y, Akamatsu W, et al. Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron. 2014;81:306–13. doi: 10.1016/j.neuron.2013.10.053. [DOI] [PubMed] [Google Scholar]
- 81.Loomis EW, Sanz LA, Chedin F, Hagerman PJ. Transcription-associated R-loop formation across the human FMR1 CGG-repeat region. PLoS Genet. 2014;10:e1004294. doi: 10.1371/journal.pgen.1004294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hunter RG, Murakami G, Dewell S, Seligsohn M, et al. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc Natl Acad Sci USA. 2012;109:17657–62. doi: 10.1073/pnas.1215810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xu GL, Bestor TH, Bourc’his D, Hsieh CL, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–91. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 84.Li W, Prazak L, Chatterjee N, Gruninger S, et al. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat Neurosci. 2013;16:529–31. doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Balaj L, Lessard R, Dai L, Cho YJ, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kraushaar DC, Jin W, Maunakea A, Abraham B, et al. Genome-wide incorporation dynamics reveal distinct categories of turnover for the histone variant H3.3. Genome Biol. 2013;14:R121. doi: 10.1186/gb-2013-14-10-r121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–7. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 88.Wong LH, McGhie JD, Sim M, Anderson MA, et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–60. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Udugama M, Chang FT, Chan FL, Tang MC, et al. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res. 2015;43:10227–37. doi: 10.1093/nar/gkv847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Santenard A, Ziegler-Birling C, Koch M, Tora L, et al. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat Cell Biol. 2010;12:853–62. doi: 10.1038/ncb2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Levy MA, Kernohan KD, Jiang Y, Berube NG. ATRX promotes gene expression by facilitating transcriptional elongation through guanine-rich coding regions. Hum Mol Genet. 2015;24:1824–35. doi: 10.1093/hmg/ddu596. [DOI] [PubMed] [Google Scholar]
- 92.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–42. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 93.Hashida H, Goto J, Kurisaki H, Mizusawa H, et al. Brain regional differences in the expansion of a CAG repeat in the spinocerebellar ataxias: dentatorubral-pallidoluysian atrophy, Machado-Joseph disease, and spinocerebellar ataxia type 1. Ann Neurol. 1997;41:505–11. doi: 10.1002/ana.410410414. [DOI] [PubMed] [Google Scholar]
- 94.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat Rev Genet. 2004;5:435–45. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 95.Johnson BE, Mazor T, Hong C, Barnes M, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–93. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Viana-Pereira M, Lee A, Popov S, Bax DA, et al. Microsatellite instability in pediatric high grade glioma is associated with genomic profile and differential target gene inactivation. PLoS ONE. 2011;6:e20588. doi: 10.1371/journal.pone.0020588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Muotri AR, Zhao C, Marchetto MC, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19:1002–7. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fasching L, Kapopoulou A, Sachdeva R, Petri R, et al. TRIM28 represses transcription of endogenous retroviruses in neural progenitor cells. Cell Rep. 2015;10:20–8. doi: 10.1016/j.celrep.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Muotri AR, Marchetto MC, Coufal NG, Oefner R, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–6. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Matsui T, Leung D, Miyashita H, Maksakova IA, et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–31. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 101.Ting DT, Lipson D, Paul S, Brannigan BW, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331:593–6. doi: 10.1126/science.1200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rizzi N, Denegri M, Chiodi I, Corioni M, et al. Transcriptional activation of a constitutive heterochromatic domain of the human genome in response to heat shock. Mol Biol Cell. 2004;15:543–51. doi: 10.1091/mbc.E03-07-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010;1309:83–94. doi: 10.1016/j.brainres.2009.09.120. [DOI] [PubMed] [Google Scholar]
- 104.Sekar A, Bialas AR, de Rivera H, Davis A, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–83. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Christoffel DJ, Golden SA, Dumitriu D, Robison AJ, et al. IkappaB kinase regulates social defeat stress-induced synaptic and behavioral plasticity. J Neurosci. 2011;31:314–21. doi: 10.1523/JNEUROSCI.4763-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kang HJ, Voleti B, Hajszan T, Rajkowska G, et al. Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nat Med. 2012;18:1413–7. doi: 10.1038/nm.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Herms J, Dorostkar MM. Dendritic spine pathology in neurodegenerative diseases. Annu Rev Pathol. 2016 doi: 10.1146/annurev-pathol-012615-044216. in press. [DOI] [PubMed] [Google Scholar]
- 108.DeKosky ST, Scheff SW. Synapse loss in frontal cortex biopsies in Alzheimer’s disease: correlation with cognitive severity. Ann Neurol. 1990;27:457–64. doi: 10.1002/ana.410270502. [DOI] [PubMed] [Google Scholar]
- 109.Dumitriu D, Hao J, Hara Y, Kaufmann J, et al. Selective changes in thin spine density and morphology in monkey pre-frontal cortex correlate with aging-related cognitive impairment. J Neurosci. 2010;30:7507–15. doi: 10.1523/JNEUROSCI.6410-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jones FC, Grabherr MG, Chan YF, Russell P, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McLean CY, Reno PL, Pollen AA, Bassan AI, et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature. 2011;471:216–9. doi: 10.1038/nature09774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jacques PE, Jeyakani J, Bourque G. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 2013;9:e1003504. doi: 10.1371/journal.pgen.1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Prescott SL, Srinivasan R, Marchetto MC, Grishina I, et al. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell. 2015;163:68–83. doi: 10.1016/j.cell.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–31. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]