ABSTRACT
Actin nucleation factors, which promote the formation of new actin filaments, have emerged in the last decade as key regulatory factors controlling asymmetric division in mammalian oocytes. Actin nucleators such as formin-2, spire, and the ARP2/3 complex have been found to be important regulators of actin remodeling during oocyte maturation. Another class of actin-binding proteins including cofilin, tropomyosin, myosin motors, capping proteins, tropomodulin, and Ezrin-Radixin-Moesin proteins are thought to control actin cytoskeleton dynamics at various steps of oocyte maturation. In addition, actin dynamics controlling asymmetric-symmetric transitions after fertilization is a new area of investigation. Taken together, defining the mechanisms by which actin-binding proteins regulate actin cytoskeletons is crucial for understanding the basic biology of mammalian gamete formation and pre-implantation development.
KEYWORDS: actin, actin-binding proteins, asymmetric division, oocyte maturation
Introduction
The most distinct difference between meiotic cell division in mammalian oocytes and somatic cell division in early embryogenesis is the asymmetric nature of cell division in meiosis.1 Because the cell division plane is determined by the position of the midbody in the spindle,2 meiotic spindle migration near the cortex is a crucial step for this asymmetric cell division process in mammalian gametes.3,4
Dynamic changes in actin filaments during mammalian oocyte maturation and the importance of actin filaments in asymmetric division in oocytes have been studied for over 30 y.5-9 One of the most dramatic changes in the actin cytoskeleton during mammalian oocyte maturation is the formation of the cortical actin cap near the migrating spindle.5 In addition to actin cap formation, cytoplasmic actin density10,11 and the thickness of cortical actin12,13 increases during oocyte maturation (Fig. 1). Although the actin cytoskeleton is the main driving force for asymmetric division in mammalian oocytes, including murine7 and porcine14,15 oocytes, the exact mechanism through which actin cytoskeleton remodeling contributes to asymmetric division in oocytes has not emerged until recently. The key molecules mediating actin filament remodeling are actin-binding proteins (ABPs),16 of which more than 100 exist in mammals, that can be sorted into several classes (Table 1). One of most extensively characterized families of ABPs, particularly regarding its role in oocyte maturation, is the actin nucleator.17 The generation of new actin filaments is a crucial regulatory step in actin filament remodeling.18 To overcome the thermodynamic barrier for monomeric actin to polymerize as filament actin, actin nucleation seed, which usually consists of 3 actin monomers, must be stabilized to accelerate actin filament formation (Fig. 2A).19 The ARP2/3 complex, a 7-subunit protein complex responsible for generating branched actin filaments from pre-existing filaments,20,21 is the most extensively studied actin nucleator and is implicated in oocyte maturation 22,23 and early embryogenesis.23-25 Formin family proteins are also involved in various aspects of actin filament remodeling.26 Among them, formin-2 (encoded by Fmn2 in mouse) is thought to be involved in oocyte maturation.27
Figure 1.
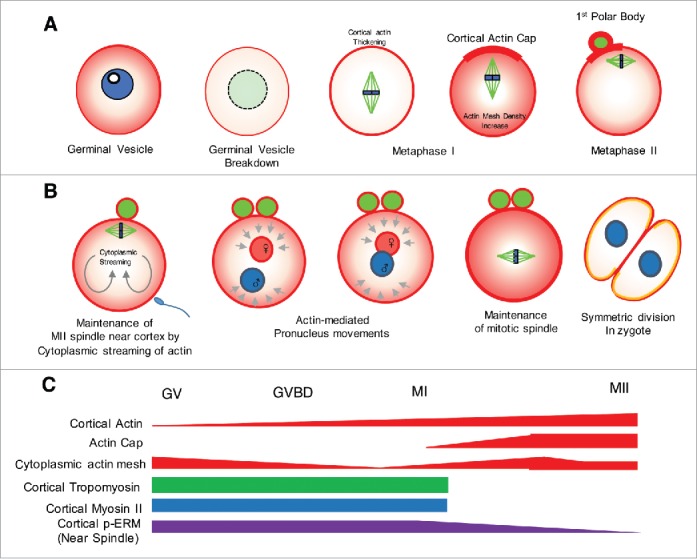
Dynamic changes in actin and actin-binding proteins during mammalian oocyte maturation. (A) During oocyte maturation, 3 types of actin reorganization processes occur. The first process is a change in cytoplasmic actin density. During the germinal vesicle (GV) stage, cytoplasmic actin mesh is formed at a higher density than germinal vesicle breakdown (GVBD) or that during the early MI stage,11 and is increased during spindle migration and the anaphase-telophase transition.10 The second process is the formation of the cortical actin cap,5 which appears at the vicinity of the approaching spindle. Formation of the cortical actin cap is mediated by the RANGTP signal associated with chromatin.76 Thickening of the cortex during the MI stage12 also occurs. Actin is indicated in red, microtubules in green, and chromatin in blue. (B) Stages of symmetric division in the mouse zygote and involvement of F-actin dynamics. In MII-stage oocytes, the spindle position is maintained near the cortex by cytoplasmic actin streaming generated by ARP2/3-mediated actin polymerization.23 The direction of actin-mediated cytoplasmic streaming is shown as an arrow. After fertilization, the second polar body is extruded after meiosis II. Pronuclei originating from male and female gametes (marked as blue and red, respectively) migrate to the center of zygotes, and their movement is dependent on actin dynamics and myosin Vb.143 The fine control of pronucleus centering involves high cortical tension mediated by myosin II recruitment.143 Maintenance of the mitotic spindle in the center of zygotes may depend on increased cytoplasm viscosity, which could be linked to an increased concentration of actin mesh.143 ARP2/3 (shown as yellow) is maintained in cell cortex regions but absent from the cleavage furrow. This polarization of ARP2/3 is maintained until the morula embryonic stage24 and thus results in apical polarization. Note that the SCMC,23,146 a protein complex essential for pre-implantation development and symmetric cell division, shows a similar localization during embryogenesis.142,146,147 The timing of each developmental stage of symmetric cell division is shown as the hour after fertilization. The directions of spindle or pronucleus movement mediated by actin polymerization are shown as arrows. (C) Temporal changes in actin and ABP levels during oocyte maturation. Temporal changes in actin or ABPs during each oocyte maturation stage are indicated by the height of the bars. Cortical actin thickness is increased after the MI stage12 and is associated with the exclusion of non-muscle myosin II from the cortex.12 ARP2/3-mediated actin polymerization is directly involved in this process.13 TPM3.1 is localized in the cortex during oocyte maturation but disappears in MII-stage oocytes.84 p-ERM is localized near the cortex but is excluded from the region near the spindle.77,123 The RANGTP signal is associated with exclusion of p-ERM near the spindle.77
Table 1.
Actin-binding proteins involved in oocyte maturation. Known biochemical functions, roles in oocyte maturation and embryogenesis are listed.
| Classes | Proteins | Known biochemical roles | Physiological roles in oocyte maturation |
|---|---|---|---|
| Actin nucleators and NPFs | ARP2/3 complex | Nucleate new actin filaments by branching from existing filaments | Cortical actin cap formation22; Subcortical actin thickening12,13; Generation of cytoplasmic flow to maintain position of spindle in MII oocytes23; Fast-phase migration of spindle65 |
| Formins | Nucleate actin filaments and facilitate filament elongation | Spindle migration27,36; Cytokinesis36; Cytoplasmic actin mesh formation (formin-2)10,11; Meiotic spindle formation (mDia family)45 | |
| Spire | Cooperate with formin-2 and form straight actin filaments | Cytoplasmic actin mesh formation in collaboration with formin-239 | |
| N-WASP | Activate the ARP2/3 complex via cdc42 | Cortical actin cap formation | |
| WAVE2 | Activate the ARP2/3 complex via Rac | Spindle formation and migration66; Subcortical actin formation12 | |
| JMY | Activate the ARP2/3 complex | Spindle formation and migration68 | |
| WHAMM | Activate the ARP2/3 complex | Spindle formation and migraton69 | |
| Filament severing | ADF/cofilin | Sever existing actin filaments and promote recycling of filaments | Recycling of cortical and cytoplasmic actin |
| Filament binding and protection | Tropomyosin(TPM3.1) | Bind actin filaments and protect from depolymerization/modulation of actin-myosin interactions | Maintenance of cortical actin integrity84 |
| Filament capping | Heterodimeric actin-capping protein (CP) | Bind to fast-growing ends of actin filaments and block elongation | Control of cytoplasmic actin mesh in competition with formin-299 |
| Tropomodulin(TMOD3) | Bind slow-growing ends of actin filaments and protect from depolymerization | Maintenance of cytoplasmic actin mesh level |
Figure 2.
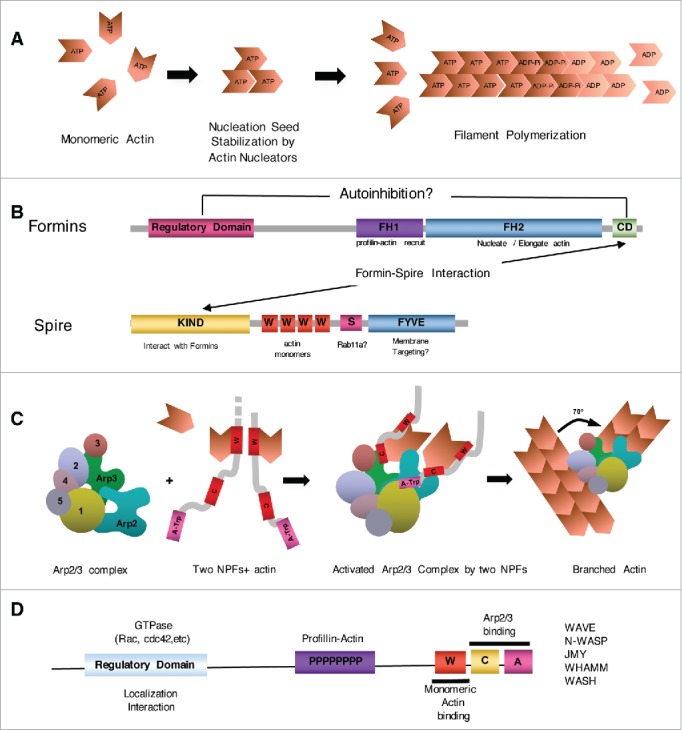
Actin nucleators and actin filament nucleation. (A) Actin polymerization is a kinetically unfavorable process and is accelerated by actin nucleators, which are families of proteins that stabilize unstable actin filament nucleation seeds. Whereas ATP-bound actin is mainly incorporated into fast-growing (also called barbed) ends of filaments, nucleotide hydrolysis occurs in the filaments, and ADP-bound actin is depolymerized at the slow-growing (also called pointed) ends of filaments. (B) Formin proteins and spire are important actin nucleators for the generation of actin filaments in oocytes. The domain architecture of canonical formin proteins (left). The proline-rich FH1 domain recruits profilin-actin complexes and directs them to the FH2 domain, which is responsible for the actin nucleation/elongation activity of the protein. Formins are usually regulated by an interaction between the C-terminal domain (CD) and the N-terminal regulatory domain. CD is essential for FMN2 catalytic activity and interaction with spire.154 The domain architecture of spire (right). Four tandem WH2 domains (marked as W) are responsible for binding with monomeric actin. Spire interacts with formin proteins via the N-terminal KIND.54 The putative Rab-binding domain56 (Spire-box, marked as S) and a modified FYVE domain,155 which usually target the protein to the membrane, are located in the CD. In mammalian oocytes, FMN2 interacts and cooperates with spire1/239 and is responsible for outer cortical actin and cytoplasmic actin mesh.12 (C) The ARP2/3 complex is a 7-protein complex responsible for the generation of branched actin.63 This complex contains 2 actin-related proteins (ARP2 and ARP3). Two NPFs recruit monomeric actin and bind to the ARP2/3 complex, activating ARP proteins.156,157 (D) Domain organization of typical NPFs. Regulatory domains can bind various GTPases, including Rac1 in the case of WAVEs158,159 and cdc42 in the case of n-WASP.160 Regulatory regions can also be involved in the localization of NPFs and actin polymerization, such as in the case of WHAMM161 or WASH.162 All known NPFs contain C-terminal ‘W-C-A’ motifs consisting of WH2 (marked as W), cofilin homology (C), and acidic (A) domains. The WH2 motif is responsible for monomeric actin recruitment, whereas ‘C-A’ motifs are required for ARP2/3 binding. Illustrations were adapted from Dominguez and Namgoong163 and modified to accommodate more recent findings.
In addition to actin nucleators, several other classes of ABPs exist, including monomeric actin (also called G-actin) binding proteins such as depolymerization/severing proteins,28 filament capping proteins,29,30 filament-binding/crosslinker protein,31 and myosins, a family of actin-based motor proteins.32 Considering that other cellular processes, including cell migration and cytokinesis, depend on classes of ABPs other than actin nucleators, ABPs may play crucial roles in actin cytoskeleton remodeling during oocyte maturation and early embryogenesis. However, the roles of these classes of ABPs in oocyte maturation have not been as well studied as those of actin nucleators.
Several previous reviews present general overviews of the roles of actin remodeling during oocyte maturation and asymmetric division.4,33-35 In this review, we mainly focus on the roles of ABPs in terms of their effect on actin dynamics during oocyte maturation. We also describe the roles of ABPs in oocyte maturation and embryogenesis in relation to their previously studied biochemical and cellular roles in other systems. In particular, we discuss ABPs that have not been extensively studied in oocyte maturation and suggest their potential roles in oocyte maturation and early embryogenesis.
Actin nucleator and nucleation-promoting factors
Formin-2 and other formin family proteins
In mammals, there are 15 formin proteins containing formin-homology domain 1 (FH1) and 2 (FH2)26; these domains are responsible for actin nucleation and elongation activity of the protein (Fig. 2B). Formin-2 (Fmn2) was the first actin nucleator shown to be associated with oocyte maturation.27 Knockout of mouse Fmn2 causes failure of spindle migration and fertilization and results in polyploidy due to arrest in metaphase I (MI) stage.27,36 During oocyte maturation, formin-2 is localized in the cortex10 and intracellular vesicles.37,38 Fmn2 is responsible for the generation of cytoplasmic actin mesh, which dynamically changes during oocyte maturation and is essential for spindle migration.10,38,39
Other formin proteins, including mouse homologs of Drosophila diaphanous (mDia) family formins, are regulated by autoinhibitory mechanisms and activated by the Rho-family GTPase RHOA,26 but the specific mechanisms regulating formin-2 are unclear. Biochemical and genetic studies using the Drosophila formin-2 homolog Cappuccino demonstrate the presence of an autoregulatory mechanism through an interaction between N-terminal and C-terminal FH2 domains,40,41 suggesting that formin-2 is regulated by similar autoregulatory mechanisms. However, the mechanisms through which upstream signaling molecules activate formin-2, particularly during oocyte maturation or embryogenesis, are unclear.
In addition to formin-2, at least 14 other formin family proteins have been identified in the mammalian genome, some of which may be involved in oocyte maturation. The biochemical properties of mDia family formins, including mDia1, mDia2, and mDia3, have been extensively studied,26,42,43 and mDia1 and mDia2 have been shown to be expressed in mouse oocytes.44,45 Knockdown of mDia1 or mDia2 decreases polar body extrusion and causes abnormal spindle morphology,45,46 indicating that mDia family formins are involved in proper meiotic spindle formation, in concord with previous studies showing that mDia formins are involved in maintaining the stability of microtubules in somatic cells.43,47 In addition to formin-2 and mDia family formins, mouse formin-like 1 (Fmnl1) was recently implicated in oocyte maturation.48 However, it is unclear how many formin family proteins are involved in oocyte maturation. The novel small molecule inhibitor SMIFH2,49 which binds to and inhibits the actin nucleation activity of the FH2 domain, inhibits several formin proteins in cells and has been used to treat immature oocytes, blocking various stages of oocyte maturation and resulting in phenotypes distinct from those of Fmn2(−/−) knockout oocytes.45 These results indicate that other formin proteins besides FMN2 are involved in oocyte maturation; however, their precise roles in oocyte maturation and early embryogenesis remain to be investigated.
Spire
Spire was first identified as a maternal effect locus that mediates Drosophila embryogenesis.50,51 Spire has 4 Wiskott-Aldrich syndrome homology 2 (WH2) domains,52 monomeric actin-binding motifs which are necessary for its actin nucleation activity.53 In addition to the WH2 domain, a kinase noncatalytic C-lobe domain (KIND), which interacts with the C-terminal of FMN2 or cappuccino (the Drosophila homolog of Fmn2), is located in the N-terminal region of spire,54,55 whereas the putative zinc-finger domain and modified FYVE domains are located in the C-terminal region.56
Knockdown of 2 genes encoding spire in the mouse, Spire1 and Spire2, impairs mouse oocyte maturation and ablation of actin mesh formation,39 similar to the phenotype of Fmn2 knockdown.10,11,27,36 Based on the evolutionarily conserved protein-protein interaction between formin-2 (cappuccino in Drosophila) and spire, and their colocalization in Drosophila55,57 and mouse oocytes,39 formin-2 and spire appear to function as a single unit for actin nucleation in oogenesis in both Drosophila and mammals. In vitro, the spire protein has relatively weak actin nucleation activity; however, its activity is significantly increased by interaction with cappuccino.55 Because formin family proteins exist as dimers, and artificial dimerization of the WH2 domain of spire significantly increases its in vitro actin nucleation activity58 and the binding of spire inhibits nucleation activity of formin-2/cappucinno,55,59 spire may function as an actin nucleator by associating with formin-2 only in vivo. Besides its interaction with formin-2, the regulatory mechanisms of spire are still elusive. Based on the presence of a putative rab-binding domain in spire and its colocalization with RAB11A and formin-2 at the vesicles and cortex in mouse oocytes,37,38 spire may localize at vesicles via its interaction with RAB11A. As the expression of dominant-negative RAB11A impairs spindle migration and actin mesh formation in mouse oocytes,38 localization of spire on vesicles could be a mechanism regulating spire activity and formation of cytoplasmic actin mesh. Mechanistic studies on the regulation of spire in concert with formin-2, particularly in terms of the signaling pathways involved in oocyte maturation such as the maturation promoting factor (MPF)60 or mitogen-activated protein kinase (MAPK) signaling pathways,61 would provide more detailed characterization of the roles of spire in oocyte maturation.
The ARP2/3 complex and nucleation-promoting factor
The ARP2/3 complex is a protein complex that initiates the formation of new actin branches on the side of pre-existing actin filaments (Fig. 2C).20,21 This complex was the first known and is the most extensively characterized actin nucleator,62 and its roles in various cellular processes, including cell migration, have been extensively studied.63 During oocyte maturation, chemical inhibition or RNAi-mediated knockdown of ARP2/3 complex components causes failure of asymmetric division in murine22,23 and porcine oocytes.64 Recent studies provide a more detailed understanding of the role of ARP2/3 in oocyte maturation. Spindle migration includes 2 phases; the first slow phase mainly depends on the action of formin-2, whereas the second rapid migration phase involves ARP2/3-mediated actin polymerization.65 In addition, ARP2/3-mediated actin polymerization generates cytoplasmic streaming inside oocyte cytoplasm and is the main driving force maintaining the localization of the meiotic spindle near the cortex in mature oocytes.23 Moreover, the ARP2/3 complex is responsible for the formation of a thicker cortex region called the 'subcortex'; the formation of the subcortex by ARP2/3 may be a factor mediating the exclusion of non-muscle myosin II and can cause changes in cortex softening during oocyte maturation.12,13
Nucleation-promoting factor (NPF) family proteins are necessary for the activation of the ARP2/3 complex, and several NPFs, including Wiskott-Aldrich syndrome protein family member 2 (WAVE2),66 neuronal Wiskott-Aldrich syndrome protein (N-WASP),67 junction-mediated regulatory protein (JMY),68 WASP homolog associated with actin, membranes, and microtubules (WHAMM),69 and WAS protein family homolog 1 (WASH),70 are implicated in oocyte maturation(Fig. 2D). Because activation of NPF is a pivotal regulatory step for ARP2/3-mediated actin polymerization, the upstream signal regulating NPF plays a crucial role in oocyte maturation. In the case of WAVE2, the small GTPase RAC1 activates WAVE2-mediated actin polymerization,71 and the expression of a dominant-negative mutant of RAC1 impairs actin cap formation during oocyte maturation,72 indicating that Rac1 is a pivotal signaling molecule upstream of ARP2/3-mediated actin polymerization during oocyte maturation. In addition, CDC42, which activates N-WASP, is involved in oocyte maturation and the protrusion of the polar body and formation of the actin cap.67,73 Compared with WAVE2 or N-WASP, regulatory mechanisms of other NPFs, including WHAMM, WASH, and JMY, are still elusive. Besides RAC1 or CDC42, the small GTPase RAN, which is localized as a gradient toward chromatin,74,75 is involved in ARP2/3-mediated actin polymerization, especially cortical cap formation.76 The RANGTP gradient is also involved in the polarization of the inactive Ezrin-Radixin-Moesin (ERM) protein in oocytes.77 However, the signaling pathway connecting RAN with ARP2/3-mediated actin polymerization is poorly understood. In addition to the small GTPase pathway, the actin polymerization pathway may be connected with well-known signaling pathways governing the cell cycle, including the MPF or Mos-MAP kinase pathways, and elucidation of these mechanisms could be critical for increasing our understanding of the regulation of ARP2/3-mediated actin polymerization based on cell cycle progression. Recent studies12,13 show that WAVE2 is phosphorylated by the extracellular signal-regulated kinase (ERK) pathway and activates ARP2/3-dependent actin polymerization as oocyte maturation progresses in mice, demonstrating the presence of crosstalk between the 2 signaling pathways. It is also possible that other NPFs and ABPs are regulated by MPF or MAPK pathways during oocyte maturation; however, the details of these mechanisms have not yet been elucidated.
Actin filament protection and depolymerization
Actin depolymerization factor/cofilin
Actin filaments initiated by actin nucleators must be depolymerized to recycle actin monomers. Therefore, active depolymerization or severing of existing actin filaments are crucial for dynamic actin filament reorganization.78 The main workhorse for actin depolymerization is the actin depolymerization factor/cofilin (ADF/cofilin) family of proteins.79 In mammals, there are 3 isoforms of cofilin (cofilin-1, cofilin-2, and destrin). Cofilins bind to actin filaments and monomers with a greater binding affinity for ADP-containing actin than for ATP-actin; therefore, cofilin binds to relatively old regions of the actin filament and severs them into shorter fragments(Fig. 3A).79 Negative regulation of cofilin is achieved by phosphorylation of the serine (Ser3) residue at the N-terminal end of cofilin(Fig. 3B).80,81 Phosphorylation by LIM kinase decreases the F-actin-binding affinity of cofilin.76 Additionally, LIM kinase activity is activated by Rho-associated kinase (ROCK) (Fig. 3B).82 Inhibition of ROCK by chemical inhibitors or siRNA decreases the phosphorylation of cofilin in oocytes, thereby increasing the activated portion of cofilin and impairing mouse oocyte maturation,83 indicating that the maintenance of proper levels of activated cofilin is important for oocyte maturation control. Inhibition of ROCK or expression of the dominant-active form of cofilin decreases cortical actin levels and impairs oocyte maturation,83,84 indicating that levels of activated cofilin should be regulated properly during oocyte maturation. Phosphorylated cofilin can be reactivated by slingshot phosphatases85 through dephosphorylation of phospho-Ser3 in cofilin, and functional studies in Xenopus oocytes show the importance of slingshot phosphatases in meiotic spindle assembly.86 However, the involvement of slingshot phosphatases in mammalian oocyte maturation has not yet been investigated.
Figure 3.
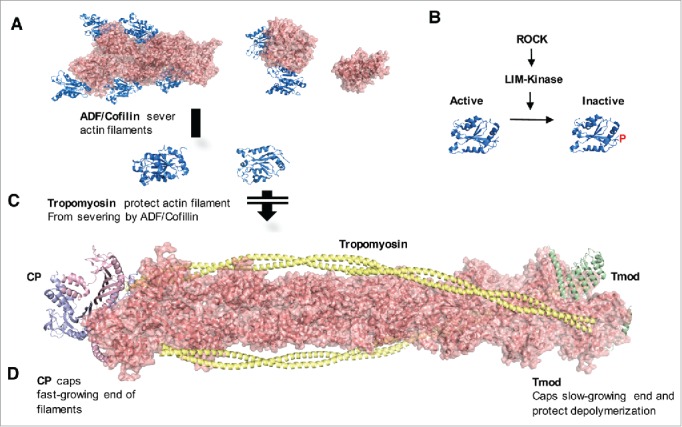
Roles of ABPs in the maintenance of actin filaments. (A) ADF/cofilin (shown as red) binds to and severs actin filaments, thereby promoting the depolymerization and recycling of actin filaments. The actin and cofilin binding model shown is based on the electron microscopy (EM) model of actin decorated by cofilin(PDB:3J0S).164 (B) ADF/cofilin activity is regulated by phosphorylation at Ser3 in the N-terminal80 and is mediated by the ROCK-LIM kinase signaling pathway.82 C. Tropomyosins (shown as yellow) are proteins that bind to the sides of filaments and protect actin filaments from ADF/cofilin.88,165 The actin-tropomyosin model shown is based on the EM-based actin-tropomyosin structure (PDB:3J8A).166 In oocytes, knockdown of tropomyosin decreases cortical actin levels and impairs cortical integrity during cytokinesis.84 (C) The 2 ends of actin filaments have different affinities for the addition of actin monomer.167 The fast-growing ends of actin filaments are capped by heterodimeric CP (shown as light blue and blue), which blocks the additional polymerization of actin filaments,168 whereas the slow-growing ends of actin filaments are protected from depolymerization by tropomodulin (shown as green), which also binds with tropomyosin.169 Both CP and Tmod3 are essential for maintaining cytoplasmic actin mesh levels in growing oocytes.99 Tmod3 and actin binding at the slow-growing end of actin filaments is shown based on the Tmod-actin complex structure (PDB:4PKI, 4PKH).170 CP and actin binding on the fast-growing ends of filaments was modeled based on the EM reconstitution of actin with CP171 and the structure of dynactin complex,172 which contains CP, Actin-related protein 1 (ARP1), and β actin (PDB:5ADX).
Tropomyosin
Whereas ADF/cofilin can sever and promote depolymerization of existing actin filaments, tropomyosin, a long coiled-coil protein that binds to the sides of actin,87 protects actin filaments from depolymerization.88 In mouse oocytes, knockdown of the non-muscle isoform of tropomyosin (TPM3.1) decreases cortical actin levels and promotes formation of the membrane bleb structure, which is caused by the collapse of cortical actin during cytokinesis,84 indicating that the maintenance of cortical integrity via the protection of actin filaments is crucial for proper asymmetric division. These phenotypes are similar to those induced by ROCK inhibition (i.e., increased amounts of active cofilin)83 or overexpression of dominant-active cofilin.84 Notably, decreasing cortical actin levels by the expression of dominant-active cofilin can be suppressed by co-expression of tropomyosin,84 demonstrating that non-muscle tropomyosin can protect cortical actin from depolymerization, presumably by protecting actin filaments from cofilin.
Cortical tropomyosin is excluded as oocytes mature to the MII stage.84 The moment of exclusion of tropomyosin from the cortex during oocyte maturation coincides with the exclusion of non-muscle myosin II,12 which is directly related to cortex softening, a driving force for asymmetric spindle migration.12,13,35,89 It is unclear whether exclusion of tropomyosin from the cortex is the consequence or cause of myosin II exclusion, and the relationship between these proteins needs to be clarified. However, based on the functional roles of tropomyosin in the regulation of myosin-actin interactions,90-93 the exclusion of tropomyosin could directly facilitate the exclusion of myosin II.
Filament end capping proteins and actin mesh control
Actin filaments have 2 ends; one is the fast-growing end (also called the barbed end), and the other is the slow-growing end (also called the pointed end).94 Because actin filaments can be depolymerized from both ends or can grow indefinitely, both ends should be capped by filament-binding proteins, called capping proteins(Fig. 3D).95,96
At the fast-growing barbed end of a filament, heterodimeric actin-capping protein (CP) binds and blocks further elongation.96,97 CP is a core factor involved in ARP2/3-mediated actin polymerization.98 When CP expression is reduced by RNAi or inhibited by overexpression of the CP antagonist CARMIL in mouse oocytes, asymmetric division of oocytes is compromised, and cytoplasmic actin mesh levels are significantly reduced.99 How can CP affect cytoplasmic actin mesh that has been nucleated by formin-2/spire10,39? Recent biochemical studies show that CP and formins can simultaneously bind to the barbed ends and form a ternary complex called a ‘decision-complex’ involved in elongation or filament end blocking,100,101 indicating that CPs regulate formin-mediated actin polymerization. Owing to the processivity of formin-mediated actin elongation in vitro,42 the length of actin filaments generated by formin should reach more than tens of micrometers before detachment,101 which is comparable to the diameter of mammalian oocytes. However, actin mesh lengths are much shorter than this,10,37 suggesting the presence of a regulatory mechanism preventing formin-mediated elongation and the start of a new cycle of actin polymerization. CP in the cytoplasm of oocytes may regulate actin filament length by competition with FMN2; however, the detailed mechanisms involved in these processes are still elusive. Whereas CP binds to and blocks elongation at the barbed ends of filaments, tropomodulin is responsible for capping the slow-growing pointed ends of actin filaments and protects actin filament from depolymerization.102,103 The physiological roles of tropomodulin have been investigated in various cells, including blood erythrocytes, skeletal muscle cells,104 and lens cells,105 and loss of tropomodulin impairs actin structure by promoting depolymerization from the pointed end. The ubiquitous isoform of tropomodulin, TMOD3, is expressed in mouse oocytes, and knockdown of tropomodulin causes a significant decrease in cytoplasmic actin mesh and impairs oocyte maturation (Jo et al., manuscript submitted).
Tropomodulins interact with tropomyosin and stabilize tropomyosin binding on actin filaments.106,107 In mouse oocytes, overexpression of the N-terminal part of TMOD3, which has a tropomyosin binding region, produces dominant-negative effects similar to the phenotype caused by TMOD3 knockdown (Jo et al., manuscript submitted). Simultaneous knockdown of TPM 3.1 and TMOD3 has an additive effect on decreasing cytoplasmic actin mesh levels, indicating that interaction and cooperation between TMOD3 and TPM 3.1 is essential for maintaining tropomyosin. Collectively, these results show that the capping of both ends of actin filaments by ABPs is essential for maintaining cytoplasmic actin mesh. However, it is not yet clear whether regulation of filament end-binding protein activities are responsible for the dynamic changes in cytoplasmic actin mesh levels during oocyte maturation. CPs are regulated by several factors, including phosphatidylinositol 4,5-bisphosphate108 and various ‘decapping’ proteins such as CARMIL109,110 and CD2AP.111 In the case of TMOD3, protein phosphorylation by AKT2 is implicated in the regulation of Tmod3 in mouse fibroblasts.112,113 Based on reports showing that chemical inhibition of AKT2 affects oocyte maturation,114-116 TMOD3 could be regulated by phosphorylation through AKT during oocyte maturation. However, the potential regulation of TMOD3 by AKT or other signaling pathways needs to be investigated.
Myosin motors in oocyte maturation
Non-muscle myosins play roles in various cellular processes including cell adhesion, cell migration, and cell division.117 In mouse oocytes, myosin IIA and IIB are expressed on the cortex in germinal vesicle (GV)-stage oocytes, whereas myosin IIA is only expressed on meiotic spindles in mature oocytes (Fig. 4A).118 Anti-myosin II antibody injection118 or chemical inhibition of myosin light chain kinase119 effectively blocks spindle migration, supporting the essential role of non-muscle myosin II in oocyte maturation. Recently, myosin II exclusion from mature oocytes was shown to mediate cortex softening in MII-stage oocytes and to be essential for spindle migration during meiosis.12,13 Interestingly, ARP2/3 complex-mediated actin polymerization in the cortex is implicated in myosin exclusion,13 although the underlying mechanisms are still not clear. Biochemical studies show that tropomyosin has antagonistic effects on ARP2/3-mediated actin polymerization,120 whereas ARP2/3 and cofilin, in turn, control the binding of tropomyosin to filamentous actin.121 A recent report showed that tropomyosin is excluded during the MII stage of mouse oocytes,84 suggesting that tropomyosin plays important regulatory roles in ARP2/3-mediated actin polymerization in the cortex and myosin exclusion in mature oocytes. However, the detailed roles of tropomyosin in ARP2/3-mediated actin polymerization and myosin recruitment/exclusion in the cortex need further investigation.
Figure 4.
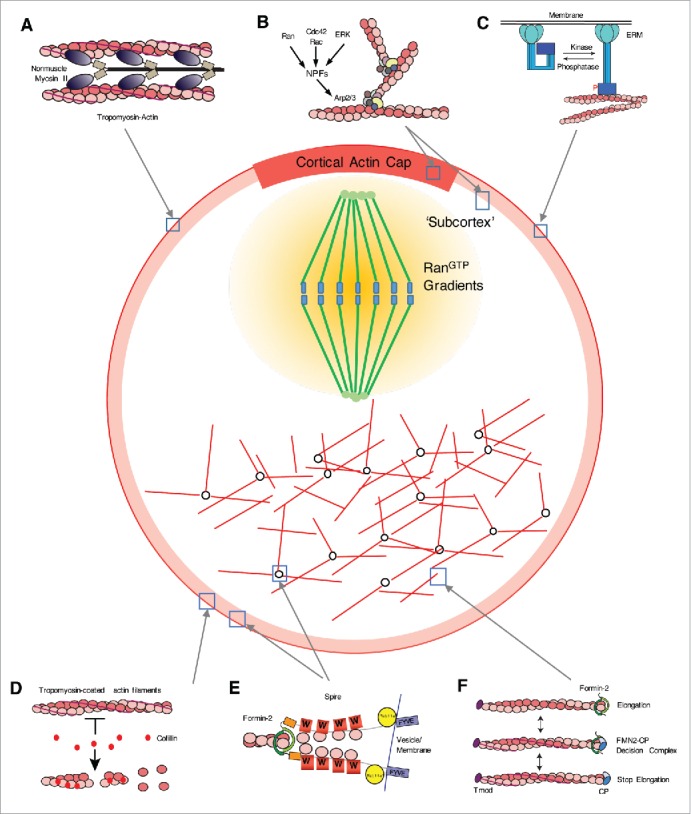
Various molecular events governed by ABPs in maturing oocytes. Localizations of ABPs in the oocyte are also indicated. Actin in oocytes were marked as red. (A) Actomyosin contraction between actin filaments and non-muscle myosin II is responsible for maintaining the cortical tension of the oocyte cortex.12,13,123 During oocyte maturation, delocalization of myosin II decreases cortical tension and is crucial for spindle migration.12 (B) ARP2/3-mediated actin polymerization is responsible for the formation of the cortical actin cap67,73,76 and cortex thickening during oocyte maturation.12,13 Small GTPases including RAC1,72 CDC42,67 and RANGTP/GDP76 are implicated in the activation of NPFs, which in turn activate the ARP2/3 complex. The MOS-MAPK pathway173 is also involved in triggering WAVE2-dependent ARP2/3-mediated actin polymerization.12 (C) ERM proteins localize in the oocyte cortex, act as link between the plasma membrane and cortical actin, and are involved in maintaining cortical tension.123 ERM protein is activated by phosphorylation on the C-terminal tail domain and deactivated by dephosphorylation.122 A recent study suggests that p-ERM is excluded at the cortex near the approaching spindle, which is mediated by RAN-based signaling.77 (D) In the oocyte cortex, Tpm3.1 is present with cortical actin and protects against depolymerization by cofilin.84 Cofilin activity in oocytes is regulated by phosphorylation by ROCK-LIM kinase cascades.83 (E) Formin-2 and spire are both responsible for the generation of cytoplasmic actin mesh or cortical actin10,11,38,39 in mammalian oocytes. Biochemical results55,59,154 suggest that both proteins synergistically promote actin nucleation. WH2 domain (marked as W) recruit monomeric actin. The presence of a potential Rab-binding domain and membrane-binding FYVE domain in the C-terminal part of spire56 suggests the possibility that spire mediates the recruitment of formin-2 in vesicles and the membrane in cooperation with Rab11a.38 (F) Formin-2 and heterodimeric CP compete against each other at the fast-growing filament end and regulate actin elogation100,101 Because the length of actin filaments generated by formins is much longer than that of cytoplasmic actin mesh10,37,101, some mechanism must regulate the elongation of actin filaments by formin-2. Impairment of CP in mouse oocytes decreases cytoplasmic actin density,99 suggesting that CP regulates actin mesh.
In addition to non-muscle myosin II, myosin Vb is also present in the cytoplasm in oocytes and plays roles in the movement of vesicles and the formation of cytoplasmic actin mesh.37,38 In contrast to the long-range movement observed in most somatic cells, vesicles in oocytes move along microfilaments, and myosin Vb carries vesicles as cargo along actin filaments formed by the actin nucleators FMN2 and spire localized at the vesicular surface.37,38 Expression of dominant-negative mutant myosin Vb impairs spindle migration,38 indicating the importance of myosin Vb in vesicle movement and actin mesh formation.
ERM family proteins and linking with the plasma membrane
ERM family proteins crosslink the plasma membrane and actin filaments, providing structural links to strengthen the cell cortex(Fig. 4C). These proteins also participate in cellular signaling cascades.122 In oocytes, ERM proteins are localized in the cellular cortex, and the activation of ERM by phosphorylation is important for maintaining cortex integrity and cortical tension.123 Interestingly, activated phospho-ERM (p-ERM) in the cortex is excluded when the meiotic spindle approaches the cortex,77,123 and polarization of p-ERM is induced by the RANGTP signal,77 which migrates along chromatin and is responsible for the formation of the cortical actin cap.76 These findings suggest that inactivation of ERM in the cortex near the actin cap and cortical granule-free domain is involved in polar body extrusion by modulation of cortical tension.123 How RANGTP/GDP induces cortical polarization of p-ERM and the actin cap is still not clear, but the polarization of p-ERM has been shown to be independent of RAC or CDC42 signaling.77
Recently, kinetochore-localized protein phosphatase 1 (PP1)-SDS22 holoenzymes were shown to dephosphorylate p-ERM in the cell cortex and break cortical symmetry.124 It is not clear whether PP1-SDS22 is responsible for p-ERM exclusion in the region of the cortical actin cap in oocytes, and further studies are needed to validate these results. However, because the PP1 targeting subunit SDS22 contains a predicted leucine-rich repeat domain, which is frequently found in many RAN-binding proteins including RANGAPs,125 this finding suggests the possibility that the protein phosphatase activity of SDS22-PP1 is regulated by the RANGTP gradient.
Actin monomer-binding proteins: Potential roles in oocyte maturation?
Because the cellular concentration of total actin is more than 0.1 mM, and actin tends to polymerize under physiological salt concentrations in vitro,126 most cellular actin should exist as filaments; however, approximately 40% of total actin in cells is present as monomers.127 One explanation for this disparity is that most monomeric actin exists as a complex with ABPs, including profilin and thymosin β4.128 Besides preventing spontaneous actin polymerization, profilin binds to actin monomers and facilitates rapid exchange of actin-bound ADP to ATP.129 The Drosophila profilin mutant(chickadee) is implicated in oogenesis defects,130 and ablation of profilin in bovine embryos impairs early embryonic development,130 indicating the essential roles of profilin in mammalian embryogenesis.
Although profilin-bound actin is a better substrate for formin-mediated elongation, it is a poor substrate for ARP2/3-mediated actin nucleation.131,132 Therefore, upregulation of the ratio of profilin-bound actin could promote actin polymerization by formin instead of ARP2/3. In budding yeast, the different actin structures generated by formin or the ARP2/3 complex can be regulated by manipulation of the fraction of profilin-bound actin in monomeric actin.133
Because ARP2/3- and formin-mediated actin polymerization processes are both important for oocyte maturation, profilin may play important roles in controlling the balance of the actin polymerization pathway between ARP2/3- and formin-mediated actin polymerization, thereby controlling oocyte maturation, as has been observed for Drosophila oogenesis.130 However, the specific roles of profilin in mammalian oocyte maturation are still unclear.
In addition to profilin, thymosin-β4, another monomeric actin-binding protein, is involved in maintaining monomeric actin pools.134 Apart from its role in monomeric actin sequestration, thymosin-β4 is involved in the regeneration and proliferation of cardiac cells135,136; therefore, thymosin-β4 may play a role in early embryogenesis (e.g., cell fate specification). However, its roles in oocyte maturation and embryogenesis have not yet been investigated. Collectively, actin-monomer—binding proteins, such as profilin and thymosin-β4, have the potential to function in oocyte maturation and embryogenesis, but further studies are needed to determine their specific roles in these processes.
Transition between asymmetric and symmetric division: Roles of F-actin
The most dramatic changes from meiotic cell division to the first mitotic division include the transition from asymmetric-to-symmetric cell division. Because the position of the midzone in the spindle determines the cleavage planes in cytokinesis,2 spindle movement near the cortex during oocyte maturation is the main reason for asymmetric division in oocytes and is mainly driven by actin cytoskeleton remodeling. We then wonder how the first mitotic spindle maintains its position in the zygote and mediates symmetric division. Although most animal cells depend on astral microtubules for the positioning of the spindle at the center of the cell,3 most mammalian zygotes lack astral microtubules137; therefore, the positioning of the spindle in zygotes may not rely on microtubule interactions.
As previously mentioned, both formin-2/spire- and ARP2/3-mediated actin polymerization processes are important for spindle migration in oocytes, and cytoplasmic streaming generated by ARP2/3-mediated actin filaments maintains the localization of meiotic spindles near cortex in MII oocytes. Studies have also examined changes that block the movement of the spindle in fertilized zygotes.
Early mitotic spindle formation until the 8-cell stage in oocytes is similar to the acentriolar microtubule-organizing center-mediated spindle formation that occurs during oocyte maturation138; therefore, transitions from asymmetric division in oocytes to symmetric division in zygotes may not be caused by different spindle assembly mechanisms. Moreover, because parthenogenetic activation of mammalian oocytes by chemical or electric stimulation results in symmetric division,139,140 asymmetric-to-symmetric division in mammalian oocytes may not be affected by sperm-derived factors.
Several studies141,142,143 show that the actin cytoskeleton is responsible for maintaining symmetric division in one-cell zygotes. For example, treatment with actin depolymerization agents (e.g., latrunculin-A) in one-cell zygotes impairs symmetric division in the first cleavage of mouse zygotes.141 A recent study shows that symmetric division in zygotes is achieved by modulation of F-actin dynamics in multiple steps.143 First, male and female pronuclei centering is mediated by F-actin/Myosin-Vb actin mesh. Second, cortical tension, which is implicated in spindle movement during meiosis,12 is responsible for the fine centering of the first mitotic spindle. Finally, maintenance of spindle position relies on a passive mechanism, presumably by increased cytoplasm viscosity, although the underlying mechanism is not clear. Previous work in oocytes38 showed that an increase of cytoplasmic actin mesh by overexpression of formin-2/spire or dominant Rab11a impairs spindles, therefore changes in cytoplasmic actin density may be responsible for restricting spindle movement.
Additional studies show that the subcortical maternal complex (SCMC) contains 4 proteins (MATER, FILIA, FLOPED, and TLE6)144-147 that are essential for maintaining early cell division in embryos and are involved in the control of F-actin dynamics, presumably by controlling ADF/cofilin.142 The potential interactions of other ABPs with SCMC are not well understood. In mouse embryos, ARP2/3 is localized in the cortex of embryos and not in the cell-to-cell junction,24 similar to the localization pattern of SCMC.142 Inhibition of ARP2/3 by chemical inhibitors results in the failure of embryogenesis in the 2- or 4-cell stage,142 similar to the phenotypes of Mater-, Filia-, or Floped-knockout embryos, suggesting that ARP2/3 interacts directly or indirectly with SCMC and that ARP2/3-mediated actin polymerization is involved in symmetric cell division in early embryos.
In addition to the ARP2/3 complex, FMN2/spire regulation after fertilization should be examined. Because knockout of FMN2 or RNAi-mediated downregulation of spire causes failure of spindle migration in oocytes,27,39 the activity of FMN2 and/or spire could be regulated after fertilization. However, our lack of knowledge of the mechanisms regulating FMN2 and spire prevents our understanding of how these proteins are regulated after fertilization; therefore, elucidation of the mechanisms regulating FMN2 and/or spire is crucial.
Concluding remarks
During the last 10 years, our understanding of the mechanism of asymmetric division in mammalian oocytes has increased substantially, particularly regarding the roles of actin nucleators and their regulators in spindle migration and asymmetric division, as summarized in the Figure 4. However, studies of the roles of other ABPs and their regulatory mechanisms during oocyte maturation and early embryogenesis are still in their infancy. Several profound questions remain. 1) How are FMN2/spire regulated during oocyte maturation and early embryogenesis? Because dynamic changes in cytoplasmic actin mesh are crucial during oocyte maturation and spindle migration, the regulation of FMN2/spire may be a critical step in these processes. 2) How are asymmetric division and spindle migration transformed into symmetric division during first cleavage at the zygote stage? Although the functional roles of several actin nucleators, including ARP2/3, FMN2, and spire, are involved in asymmetric division, their roles in symmetric division during embryogenesis are unclear. 3) Several NPFs, including n-WASP, WAVE2, JMY, WASH, and WHAMM, are implicated in oocyte maturation, but the mechanistic roles of each NPF are poorly understood. Some actin nucleators, especially WASH, WHAMM, and JMY, have been characterized in terms of their ability to activate ARP2/3-mediated actin polymerization at distinct cellular locations, including golgi, endocytic structure, and membrane ruffles.17 Further studies could clarify the exact functional roles of each NPF in terms of actin filament formation in different cellular contexts. 4) Are there other ABPs that have roles in oocyte maturation and embryogenesis, and if so, how is their activity regulated during these processes, particularly with regard to connections with signaling pathways governing the cell cycle? For example, filament crosslinker proteins like filamin,148 actin polymerases like Ena/VASP proteins,149 and filament-bundling proteins like α-actinin150 or fascin151 are actin binding-proteins studied extensively in other cellular process but not in oocyte maturation and early embryogenesis. In addition to cell division, recent studies suggest that controlling actin dynamics can also affect cell fate specification between trophoblasts and the inner cell mass during embryogenesis152 via the Hippo signaling pathway.153 The involvement of various ABPs in the regulation of the Hippo signaling pathway, and thus the control of cell fate specification during embryogenesis, should be elucidated in future studies.
Collectively, a mechanistic understanding of the functional roles of ABPs in biochemical or cellular studies in other model systems may provide insight into their roles in oocyte maturation and early embryogenesis. In turn, oocyte maturation and early embryogenesis could provide a simple and physiologically relevant model system for the functional study of ABPs.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Research performed by the authors described in this review was supported by grants from the Next Generation Biogreen 21 Program to S.N (PJ011206) and N.H.K. (PJ011126), Rural Development Administration, Republic of Korea.
References
- [1].Clift D, Schuh M. Restarting life: fertilization and the transition from meiosis to mitosis. Nat Rev Mol Cell Biol 2013; 14:549-62; PMID:23942453; http://dx.doi.org/ 10.1038/nrm3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rappaport R. Establishment of the mechanism of cytokinesis in animal cells. Int Rev Cytol 1986; 105:245-81; PMID:3539854; http://dx.doi.org/ 10.1016/S0074-7696(08)61065-7 [DOI] [PubMed] [Google Scholar]
- [3].McNally FJ. Mechanisms of spindle positioning. J Cell Biol 2013; 200:131-40; PMID:23337115; http://dx.doi.org/ 10.1083/jcb.201210007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sun SC, Kim NH. Molecular mechanisms of asymmetric division in oocytes. Microsc Microanal 2013; 19:883-97; PMID:23764118; http://dx.doi.org/ 10.1017/S1431927613001566 [DOI] [PubMed] [Google Scholar]
- [5].Maro B, Johnson MH, Pickering SJ, Flach G. Changes in actin distribution during fertilization of the mouse egg. J Embryol Exp Morphol 1984; 81:211-37; PMID:6540795 [PubMed] [Google Scholar]
- [6].Maro B, Johnson MH, Webb M, Flach G. Mechanism of polar body formation in the mouse oocyte: an interaction between the chromosomes, the cytoskeleton and the plasma membRANe. J Embryol Exp Morphol 1986; 92:11-32; PMID:3723057 [PubMed] [Google Scholar]
- [7].Longo FJ, Chen DY. Development of cortical polarity in mouse eggs: involvement of the meiotic apparatus. Dev Biol 1985; 107:382-94; PMID:4038667; http://dx.doi.org/ 10.1016/0012-1606(85)90320-3 [DOI] [PubMed] [Google Scholar]
- [8].Kim NH, Funahashi H, Prather RS, Schatten G, Day BN. Microtubule and microfilament dynamics in porcine oocytes during meiotic maturation. Mol Reprod Dev 1996; 43:248-55; PMID:8824923; http://dx.doi.org/ 10.1002/(SICI)1098-2795(199602)43:2%3c248::AID-MRD14%3e3.0.CO;2- [DOI] [PubMed] [Google Scholar]
- [9].Kim NH, Chung HM, Cha KY, Chung KS. Microtubule and microfilament organization in maturing human oocytes. Hum Reprod 1998; 13:2217-22; PMID:9756299; http://dx.doi.org/ 10.1093/humrep/13.8.2217 [DOI] [PubMed] [Google Scholar]
- [10].Azoury J, Lee KW, Georget V, Rassinier P, Leader B, Verlhac MH. Spindle positioning in mouse oocytes relies on a dynamic meshwork of actin filaments. Curr Biol 2008; 18:1514-9; PMID:18848445; http://dx.doi.org/ 10.1016/j.cub.2008.08.044 [DOI] [PubMed] [Google Scholar]
- [11].Azoury J, Lee KW, Georget V, Hikal P, Verlhac MH. Symmetry breaking in mouse oocytes requires transient F-actin meshwork destabilization. Development 2011; 138:2903-8; PMID:21653611; http://dx.doi.org/ 10.1242/dev.060269 [DOI] [PubMed] [Google Scholar]
- [12].Chaigne A, Campillo C, Gov NS, Voituriez R, Azoury J, Umana-Diaz C, Almonacid M, Queguiner I, Nassoy P, Sykes C, et al.. A soft cortex is essential for asymmetric spindle positioning in mouse oocytes. Nat Cell Biol 2013; 15:958-66; PMID:23851486; http://dx.doi.org/ 10.1038/ncb2799 [DOI] [PubMed] [Google Scholar]
- [13].Chaigne A, Campillo C, Gov NS, Voituriez R, Sykes C, Verlhac MH, Terret ME. A narrow window of cortical tension guides asymmetric spindle positioning in the mouse oocyte. Nat Commun 2015; 6:6027; PMID:25597399; http://dx.doi.org/ 10.1038/ncomms7027 [DOI] [PubMed] [Google Scholar]
- [14].Kim NH, Day BN, Lee HT, Chung KS. Microfilament assembly and cortical gRANule distribution during maturation, parthenogenetic activation and fertilisation in the porcine oocyte. Zygote 1996; 4:145-9; PMID:8913028; http://dx.doi.org/ 10.1017/S0967199400003026 [DOI] [PubMed] [Google Scholar]
- [15].Wang WH, Abeydeera LR, Prather RS, Day BN. Polymerization of nonfilamentous actin into microfilaments is an important process for porcine oocyte maturation and early embryo development. Biol Reprod 2000; 62:1177-83; PMID:10775164; http://dx.doi.org/ 10.1095/biolreprod62.5.1177 [DOI] [PubMed] [Google Scholar]
- [16].dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol Rev 2003; 83:433-73; PMID:12663865; http://dx.doi.org/ 10.1152/physrev.00026.2002 [DOI] [PubMed] [Google Scholar]
- [17].Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol 2010; 11:237-51; PMID:20237478; http://dx.doi.org/ 10.1038/nrm2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science 2009; 326:1208-12; PMID:19965462; http://dx.doi.org/ 10.1126/science.1175862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J 2001; 81:667-74; PMID:11463615; http://dx.doi.org/ 10.1016/S0006-3495(01)75731-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Amann KJ, Pollard TD. Direct real-time observation of actin filament bRANching mediated by ARP2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci U S A 2001; 98:15009-13; PMID:11742068; http://dx.doi.org/ 10.1073/pnas.211556398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Amann KJ, Pollard TD. The ARP2/3 complex nucleates actin filament bRANches from the sides of pre-existing filaments. Nat Cell Biol 2001; 3:306-10; PMID:11231582; http://dx.doi.org/ 10.1038/35060104 [DOI] [PubMed] [Google Scholar]
- [22].Sun SC, Wang ZB, Xu YN, Lee SE, Cui XS, Kim NH. ARP2/3 complex regulates asymmetric division and cytokinesis in mouse oocytes. PLoS One 2011; 6:e18392; PMID:21494665; http://dx.doi.org/ 10.1371/journal.pone.0018392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].IYi K, Unruh JR, Deng M, Slaughter BD, Rubinstein B, Li R. Dynamic maintenance of asymmetric meiotic spindle position through ARP2/3-complex-driven cytoplasmic streaming in mouse oocytes. Nat Cell Biol 2011; 13:1252-8; PMID:21874009; http://dx.doi.org/ 10.1038/ncb2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sun SC, Wang QL, Gao WW, Xu YN, Liu HL, Cui XS, Kim NH. Actin nucleator ARP2/3 complex is essential for mouse preimplantation embryo development. Reprod Fertil Dev 2013; 25:617-23; PMID:22951093; http://dx.doi.org/ 10.1071/RD12011 [DOI] [PubMed] [Google Scholar]
- [25].Wang QC, Liu J, Wang F, Duan X, Dai XX, Wang T, Liu HL, Cui XS, Sun SC, Kim NH. Role of nucleation-promoting factors in mouse early embryo development. Microsc Microanal 2013; 19:559-64; PMID:23552571; http://dx.doi.org/ 10.1017/S1431927613000032 [DOI] [PubMed] [Google Scholar]
- [26].Goode BL, Eck MJ. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem 2007; 76:593-627; PMID:17373907; http://dx.doi.org/ 10.1146/annurev.biochem.75.103004.142647 [DOI] [PubMed] [Google Scholar]
- [27].Leader B, Lim H, Carabatsos MJ, Harrington A, Ecsedy J, Pellman D, Maas R, Leder P. Formin-2, polyploidy, hypofertility and positioning of the meiotic spindle in mouse oocytes. Nat Cell Biol 2002; 4:921-8; PMID:12447394; http://dx.doi.org/ 10.1038/ncb880 [DOI] [PubMed] [Google Scholar]
- [28].Bamburg JR, Bernstein BW. Roles of ADF/cofilin in actin polymerization and beyond. F1000 Biol Rep 2010; 2:62; PMID:21173851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Cooper JA, Sept D. New insights into mechanism and regulation of actin capping protein. Int Rev Cell Mol Biol 2008; 267:183-206; PMID:18544499; http://dx.doi.org/ 10.1016/S1937-6448(08)00604-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fowler VM. Capping actin filament growth: tropomodulin in muscle and nonmuscle cells. Soc Gen Physiol Ser 1997; 52:79-89; PMID:9210222 [PubMed] [Google Scholar]
- [31].Uribe R, Jay D. A review of actin binding proteins: new perspectives. Mol Biol Rep 2009; 36:121-5; PMID:17939058; http://dx.doi.org/ 10.1007/s11033-007-9159-2 [DOI] [PubMed] [Google Scholar]
- [32].Hartman MA, Spudich JA. The myosin superfamily at a glance. J Cell Sci 2012; 125:1627-32; PMID:22566666; http://dx.doi.org/ 10.1242/jcs.094300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sun QY, Schatten H. Regulation of dynamic events by microfilaments during oocyte maturation and fertilization. Reproduction 2006; 131:193-205; PMID:16452714; http://dx.doi.org/ 10.1530/rep.1.00847 [DOI] [PubMed] [Google Scholar]
- [34].Yi K, Li R. Actin cytoskeleton in cell polarity and asymmetric division during mouse oocyte maturation. Cytoskeleton (Hoboken) 2012; 69:727-37; PMID:22753278; http://dx.doi.org/ 10.1002/cm.21048 [DOI] [PubMed] [Google Scholar]
- [35].Almonacid M, Terret ME, Verlhac MH. Actin-based spindle positioning: new insights from female gametes. J Cell Sci 2014; 127:477-83; PMID:24413163; http://dx.doi.org/ 10.1242/jcs.142711 [DOI] [PubMed] [Google Scholar]
- [36].Dumont J, Million K, Sunderland K, Rassinier P, Lim H, Leader B, Verlhac MH. Formin-2 is required for spindle migration and for the late steps of cytokinesis in mouse oocytes. Dev Biol 2007; 301:254-65; PMID:16989804; http://dx.doi.org/ 10.1016/j.ydbio.2006.08.044 [DOI] [PubMed] [Google Scholar]
- [37].Schuh M. An actin-dependent mechanism for long-RANge vesicle transport. Nat Cell Biol 2011; 13:1431-6; PMID:21983562; http://dx.doi.org/ 10.1038/ncb2353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Holubcova Z, Howard G, Schuh M. Vesicles modulate an actin network for asymmetric spindle positioning. Nat Cell Biol 2013; 15:937-47; PMID:23873150; http://dx.doi.org/ 10.1038/ncb2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pfender S, Kuznetsov V, Pleiser S, Kerkhoff E, Schuh M. Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr Biol 2011; 21:955-60; PMID:21620703; http://dx.doi.org/ 10.1016/j.cub.2011.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bor B, Bois JS, Quinlan ME. Regulation of the formin Cappuccino is critical for polarity of Drosophila oocytes. Cytoskeleton (Hoboken) 2015; 72:1-15; PMID:25557988; http://dx.doi.org/ 10.1002/cm.21205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Bor B, Vizcarra CL, Phillips ML, Quinlan ME. Autoinhibition of the formin Cappuccino in the absence of canonical autoinhibitory domains. Mol Biol Cell 2012; 23:3801-13; PMID:22875983; http://dx.doi.org/ 10.1091/mbc.E12-04-0288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 2006; 124:423-35; PMID:16439214; http://dx.doi.org/ 10.1016/j.cell.2005.11.038 [DOI] [PubMed] [Google Scholar]
- [43].Palazzo AF, Cook TA, Alberts AS, Gundersen GG. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat Cell Biol 2001; 3:723-9; PMID:11483957; http://dx.doi.org/ 10.1038/35087035 [DOI] [PubMed] [Google Scholar]
- [44].Kwon S, Shin H, Lim HJ. Dynamic interaction of formin proteins and cytoskeleton in mouse oocytes during meiotic maturation. Mol Hum Reprod 2011; 17:317-27; PMID:20971793; http://dx.doi.org/ 10.1093/molehr/gaq088 [DOI] [PubMed] [Google Scholar]
- [45].Kim HC, Jo YJ, Kim NH, Namgoong S. Small molecule inhibitor of formin homology 2 domains (SMIFH2) reveals the roles of the formin family of proteins in spindle assembly and asymmetric division in mouse oocytes. PLoS One 2015; 10:e0123438; PMID:25837661; http://dx.doi.org/ 10.1371/journal.pone.0123438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhang Y, Wang F, Niu YJ, Liu HL, Rui R, Cui XS, Kim NH, Sun SC. Formin mDia1, a downstream molecule of FMNL1, regulates Profilin1 for actin assembly and spindle organization during mouse oocyte meiosis. Biochim Biophys Acta 2015; 1853:317-27; PMID:25447542; http://dx.doi.org/ 10.1016/j.bbamcr.2014.11.005 [DOI] [PubMed] [Google Scholar]
- [47].Wen Y, Eng CH, SchmoRANzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol 2004; 6:820-30; PMID:15311282; http://dx.doi.org/ 10.1038/ncb1160 [DOI] [PubMed] [Google Scholar]
- [48].Wang F, Zhang L, Duan X, Zhang GL, Wang ZB, Wang Q, Xiong B, Sun SC. RhoA-mediated FMNL1 regulates GM130 for actin assembly and phosphorylates MAPK for spindle formation in mouse oocyte meiosis. Cell Cycle 2015; 14:2835-43; PMID:26083584; http://dx.doi.org/ 10.1080/15384101.2015.1031438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol 2009; 16:1158-68; PMID:19942139; http://dx.doi.org/ 10.1016/j.chembiol.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Manseau LJ, Schupbach T. cappuccino and spire: two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes Dev 1989; 3:1437-52; PMID:2514120; http://dx.doi.org/ 10.1101/gad.3.9.1437 [DOI] [PubMed] [Google Scholar]
- [51].Theurkauf WE. Premature microtubule-dependent cytoplasmic streaming in cappuccino and spire mutant oocytes. Science 1994; 265:2093-6; PMID:8091233; http://dx.doi.org/ 10.1126/science.8091233 [DOI] [PubMed] [Google Scholar]
- [52].Paunola E, Mattila PK, Lappalainen P. WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett 2002; 513:92-7; PMID:11911886; http://dx.doi.org/ 10.1016/S0014-5793(01)03242-2 [DOI] [PubMed] [Google Scholar]
- [53].Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature 2005; 433:382-8; PMID:15674283; http://dx.doi.org/ 10.1038/nature03241 [DOI] [PubMed] [Google Scholar]
- [54].Pechlivanis M, Samol A, Kerkhoff E. Identification of a short Spir interaction sequence at the C-terminal end of formin subgroup proteins. J Biol Chem 2009; 284:25324-33; PMID:19605360; http://dx.doi.org/ 10.1074/jbc.M109.030320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Quinlan ME, Hilgert S, Bedrossian A, Mullins RD, Kerkhoff E. Regulatory interactions between two actin nucleators, Spire and Cappuccino. J Cell Biol 2007; 179:117-28; PMID:17923532; http://dx.doi.org/ 10.1083/jcb.200706196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kerkhoff E, Simpson JC, Leberfinger CB, Otto IM, Doerks T, Bork P, Rapp UR, Raabe T, Pepperkok R. The Spir actin organizers are involved in vesicle transport processes. Curr Biol 2001; 11:1963-8; PMID:11747823; http://dx.doi.org/ 10.1016/S0960-9822(01)00602-9 [DOI] [PubMed] [Google Scholar]
- [57].Quinlan ME. Direct interaction between two actin nucleators is required in Drosophila oogenesis. Development 2013; 140:4417-25; PMID:24089467; http://dx.doi.org/ 10.1242/dev.097337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Namgoong S, Boczkowska M, Glista MJ, Winkelman JD, Rebowski G, Kovar DR, Dominguez R. Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat Struct Mol Biol 2011; 18:1060-7; PMID:21873985; http://dx.doi.org/ 10.1038/nsmb.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Montaville P, Jegou A, Pernier J, Compper C, Guichard B, Mogessie B, Schuh M, Romet-Lemonne G, Carlier MF. Spire and Formin 2 synergize and antagonize in regulating actin assembly in meiosis by a ping-pong mechanism. PLoS Biol 2014; 12:e1001795; PMID:24586110; http://dx.doi.org/ 10.1371/journal.pbio.1001795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Adhikari D, Liu K. The regulation of maturation promoting factor during prophase I arrest and meiotic entry in mammalian oocytes. Mol Cell Endocrinol 2014; 382:480-7; PMID:23916417; http://dx.doi.org/ 10.1016/j.mce.2013.07.027 [DOI] [PubMed] [Google Scholar]
- [61].Dupre A, Haccard O, Jessus C. Mos in the oocyte: how to use MAPK independently of growth factors and transcription to control meiotic divisions. J Signal TRANsduct 2011; 2011:350412; PMID:21637374; http://dx.doi.org/ 10.1155/2011/350412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Pollard TD. Regulation of actin filament assembly by ARP2/3 complex and formins. Annu Rev Biophys Biomol Struct 2007; 36:451-77; PMID:17477841; http://dx.doi.org/ 10.1146/annurev.biophys.35.040405.101936 [DOI] [PubMed] [Google Scholar]
- [63].Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol 2013; 14:7-12; PMID:23212475; http://dx.doi.org/ 10.1038/nrm3492 [DOI] [PubMed] [Google Scholar]
- [64].Wang F, An GY, Zhang Y, Liu HL, Cui XS, Kim NH, Sun SC. ARP2/3 complex inhibition prevents meiotic maturation in porcine oocytes. PLoS One 2014; 9:e87700; PMID:24498171; http://dx.doi.org/ 10.1371/journal.pone.0087700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yi K, Rubinstein B, Unruh JR, Guo F, Slaughter BD, Li R. Sequential actin-based pushing forces drive meiosis I chromosome migration and symmetry breaking in oocytes. J Cell Biol 2013; 200:567-76; PMID:23439682; http://dx.doi.org/ 10.1083/jcb.201211068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sun SC, Xu YN, Li YH, Lee SE, Jin YX, Cui XS, Kim NH. WAVE2 regulates meiotic spindle stability, peripheral positioning and polar body emission in mouse oocytes. Cell Cycle 2011; 10:1853-60; PMID:21543895; http://dx.doi.org/ 10.4161/cc.10.11.15796 [DOI] [PubMed] [Google Scholar]
- [67].Dehapiot B, Carriere V, Carroll J, Halet G. Polarized Cdc42 activation promotes polar body protrusion and asymmetric division in mouse oocytes. Dev Biol 2013; 377:202-12; PMID:23384564; http://dx.doi.org/ 10.1016/j.ydbio.2013.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sun SC, Sun QY, Kim NH. JMY is required for asymmetric division and cytokinesis in mouse oocytes. Mol Hum Reprod 2011; 17:296-304; PMID:21266449; http://dx.doi.org/ 10.1093/molehr/gar006 [DOI] [PubMed] [Google Scholar]
- [69].Huang X, Ding L, Pan R, Ma PF, Cheng PP, Zhang CH, Shen YT, Xu L, Liu Y, He XQ, et al.. WHAMM is required for meiotic spindle migration and asymmetric cytokinesis in mouse oocytes. Histochem Cell Biol 2013; 139:525-34; PMID:23160625; http://dx.doi.org/ 10.1007/s00418-012-1051-z [DOI] [PubMed] [Google Scholar]
- [70].Wang F, Zhang L, Zhang GL, Wang ZB, Cui XS, Kim NH, Sun SC. WASH complex regulates ARP2/3 complex for actin-based polar body extrusion in mouse oocytes. Sci Rep 2014; 4:5596; PMID:24998208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J 1998; 17:6932-41; PMID:9843499; http://dx.doi.org/ 10.1093/emboj/17.23.6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Halet G, Carroll J. Rac activity is polarized and regulates meiotic spindle stability and anchoring in mammalian oocytes. Dev Cell 2007; 12:309-17; PMID:17276347; http://dx.doi.org/ 10.1016/j.devcel.2006.12.010 [DOI] [PubMed] [Google Scholar]
- [73].Wang ZB, Jiang ZZ, Zhang QH, Hu MW, Huang L, Ou XH, Guo L, Ouyang YC, Hou Y, Brakebusch C, et al.. Specific deletion of Cdc42 does not affect meiotic spindle organization/migration and homologous chromosome segregation but disrupts polarity establishment and cytokinesis in mouse oocytes. Mol Biol Cell 2013; 24:3832-41; PMID:24131996; http://dx.doi.org/ 10.1091/mbc.E13-03-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Carazo-Salas RE, Gruss OJ, Mattaj IW, Karsenti E. RAN-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat Cell Biol 2001; 3:228-34; PMID:11231571; http://dx.doi.org/ 10.1038/35060009 [DOI] [PubMed] [Google Scholar]
- [75].Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. RAN induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 2001; 104:83-93; PMID:11163242; http://dx.doi.org/ 10.1016/S0092-8674(01)00193-3 [DOI] [PubMed] [Google Scholar]
- [76].Deng M, SuRANeni P, Schultz RM, Li R. The RAN GTPase mediates chromatin signaling to control cortical polarity during polar body extrusion in mouse oocytes. Dev Cell 2007; 12:301-8; PMID:17276346; http://dx.doi.org/ 10.1016/j.devcel.2006.11.008 [DOI] [PubMed] [Google Scholar]
- [77].Dehapiot B, Halet G. RAN GTPase promotes oocyte polarization by regulating ERM (Ezrin/Radixin/Moesin) inactivation. Cell Cycle 2013; 12:1672-8; PMID:23656777; http://dx.doi.org/ 10.4161/cc.24901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell 2003; 112:453-65; PMID:12600310; http://dx.doi.org/ 10.1016/S0092-8674(03)00120-X [DOI] [PubMed] [Google Scholar]
- [79].Bamburg JR, Bernstein BW. ADF/cofilin. Curr Biol 2008; 18:R273-5; PMID:18397729; http://dx.doi.org/ 10.1016/j.cub.2008.02.002 [DOI] [PubMed] [Google Scholar]
- [80].Yang N, Higuchi O, Ohashi K, Nagata K, Wada A, Kangawa K, Nishida E, Mizuno K. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature 1998; 393:809-12; PMID:9655398; http://dx.doi.org/ 10.1038/31735 [DOI] [PubMed] [Google Scholar]
- [81].Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature 1998; 393:805-9; PMID:9655397; http://dx.doi.org/ 10.1038/31729 [DOI] [PubMed] [Google Scholar]
- [82].Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S. Signaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinase. Science 1999; 285:895-8; PMID:10436159; http://dx.doi.org/ 10.1126/science.285.5429.895 [DOI] [PubMed] [Google Scholar]
- [83].Duan X, Liu J, Dai XX, Liu HL, Cui XS, Kim NH, Wang ZB, Wang Q, Sun SC. Rho-GTPase effector ROCK phosphorylates cofilin in actin-meditated cytokinesis during mouse oocyte meiosis. Biol Reprod 2014; 90:37; PMID:24429217; http://dx.doi.org/ 10.1095/biolreprod.113.113522 [DOI] [PubMed] [Google Scholar]
- [84].Jang WI, Jo YJ, Kim HC, Jia JL, Namgoong S, Kim NH. Non-muscle tropomyosin (Tpm3) is crucial for asymmetric cell division and maintenance of cortical integrity in mouse oocytes. Cell Cycle 2014; 13:2359-69; PMID:25483187; http://dx.doi.org/ 10.4161/cc.29333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell 2002; 108:233-46; PMID:11832213; http://dx.doi.org/ 10.1016/S0092-8674(01)00638-9 [DOI] [PubMed] [Google Scholar]
- [86].Iwase S, Sato R, De Bock PJ, Gevaert K, Fujiki S, Tawada T, Kuchitsu M, Yamagishi Y, Ono S, Abe H. Activation of ADF/cofilin by phosphorylation-regulated Slingshot phosphatase is required for the meiotic spindle assembly in Xenopus laevis oocytes. Mol Biol Cell 2013; 24:1933-46; PMID:23615437; http://dx.doi.org/ 10.1091/mbc.E12-12-0851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Gunning P, O'Neill G, Hardeman E. Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol Rev 2008; 88:1-35; PMID:18195081; http://dx.doi.org/ 10.1152/physrev.00001.2007 [DOI] [PubMed] [Google Scholar]
- [88].Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol 2002; 156:1065-76; PMID:11901171; http://dx.doi.org/ 10.1083/jcb.200110013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Chaigne A, Verlhac MH, Terret ME. [Cortex softening: a prerequisite for the asymmetry of oocyte first division]. Med Sci (Paris) 2014; 30:18-21; PMID:24472451; http://dx.doi.org/ 10.1051/medsci/20143001005 [DOI] [PubMed] [Google Scholar]
- [90].Eaton BL. Tropomyosin binding to F-actin induced by myosin heads. Science 1976; 192:1337-9; PMID:131972; http://dx.doi.org/ 10.1126/science.131972 [DOI] [PubMed] [Google Scholar]
- [91].Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev 2000; 80:853-924; PMID:10747208 [DOI] [PubMed] [Google Scholar]
- [92].Stark BC, Sladewski TE, Pollard LW, Lord M. Tropomyosin and myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Mol Biol Cell 2010; 21:989-1000; PMID:20110347; http://dx.doi.org/ 10.1091/mbc.E09-10-0852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Barua B, Nagy A, Sellers JR, Hitchcock-DeGregori SE. Regulation of nonmuscle myosin II by tropomyosin. Biochemistry 2014; 53:4015-24; PMID:24873380; http://dx.doi.org/ 10.1021/bi500162z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Woodrum DT, Rich SA, Pollard TD. Evidence for biased bidirectional polymerization of actin filaments using heavy meromyosin prepared by an improved method. J Cell Biol 1975; 67:231-7; PMID:240859; http://dx.doi.org/ 10.1083/jcb.67.1.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Cooper JA, Blum JD, Pollard TD. Acanthamoeba castellanii capping protein: properties, mechanism of action, immunologic cross-reactivity, and localization. J Cell Biol 1984; 99:217-25; PMID:6429155; http://dx.doi.org/ 10.1083/jcb.99.1.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Cooper JA, Pollard TD. Effect of capping protein on the kinetics of actin polymerization. Biochemistry 1985; 24:793-9; PMID:3994986; http://dx.doi.org/ 10.1021/bi00324a039 [DOI] [PubMed] [Google Scholar]
- [97].Caldwell JE, Heiss SG, Mermall V, Cooper JA. Effects of CapZ, an actin capping protein of muscle, on the polymerization of actin. Biochemistry 1989; 28:8506-14; PMID:2557904; http://dx.doi.org/ 10.1021/bi00447a036 [DOI] [PubMed] [Google Scholar]
- [98].Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 1999; 401:613-6; PMID:10524632; http://dx.doi.org/ 10.1038/44183 [DOI] [PubMed] [Google Scholar]
- [99].Jo YJ, Jang WI, Namgoong S, Kim NH. Actin-capping proteins play essential roles in the asymmetric division of maturing mouse oocytes. J Cell Sci 2015; 128:160-70; PMID:25395583; http://dx.doi.org/ 10.1242/jcs.163576 [DOI] [PubMed] [Google Scholar]
- [100].Bombardier JP, Eskin JA, Jaiswal R, Correa IR Jr., Xu MQ, Goode BL, Gelles J. Single-molecule visualization of a formin-capping protein 'decision complex' at the actin filament barbed end. Nat Commun 2015; 6:8707; PMID:26566078; http://dx.doi.org/ 10.1038/ncomms9707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Shekhar S, Kerleau M, Kuhn S, Pernier J, Romet-Lemonne G, Jegou A, Carlier MF. Formin and capping protein together embrace the actin filament in a menage a trois. Nat Commun 2015; 6:8730; PMID:26564775; http://dx.doi.org/ 10.1038/ncomms9730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Weber A, Pennise CR, Babcock GG, Fowler VM. Tropomodulin caps the pointed ends of actin filaments. J Cell Biol 1994; 127:1627-35; PMID:7798317; http://dx.doi.org/ 10.1083/jcb.127.6.1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Yamashiro S, Speicher KD, Speicher DW, Fowler VM. Mammalian tropomodulins nucleate actin polymerization via their actin monomer binding and filament pointed end-capping activities. J Biol Chem 2010; 285:33265-80; PMID:20650902; http://dx.doi.org/ 10.1074/jbc.M110.144873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Gregorio CC, Weber A, Bondad M, Pennise CR, Fowler VM. Requirement of pointed-end capping by tropomodulin to maintain actin filament length in embryonic chick cardiac myocytes. Nature 1995; 377:83-6; PMID:7544875; http://dx.doi.org/ 10.1038/377083a0 [DOI] [PubMed] [Google Scholar]
- [105].Woo MK, Lee A, Fischer RS, Moyer J, Fowler VM. The lens membRANe skeleton contains structures preferentially enriched in spectrin-actin or tropomodulin-actin complexes. Cell Motil Cytoskeleton 2000; 46:257-68; PMID:10962480; http://dx.doi.org/ 10.1002/1097-0169(200008)46:4%3c257::AID-CM3%3e3.0.CO;2-2 [DOI] [PubMed] [Google Scholar]
- [106].Kostyukova AS. Tropomodulin/tropomyosin interactions regulate actin pointed end dynamics. Adv Exp Med Biol 2008; 644:283-92; PMID:19209829; http://dx.doi.org/ 10.1007/978-0-387-85766-4_21 [DOI] [PubMed] [Google Scholar]
- [107].Kostyukova AS. Tropomodulins and tropomodulin/tropomyosin interactions. Cell Mol Life Sci 2008; 65:563-9; PMID:17965951; http://dx.doi.org/ 10.1007/s00018-007-7347-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Kim K, McCully ME, Bhattacharya N, Butler B, Sept D, Cooper JA. Structure/function analysis of the interaction of phosphatidylinositol 4,5-bisphosphate with actin-capping protein: implications for how capping protein binds the actin filament. J Biol Chem 2007; 282:5871-9; PMID:17182619; http://dx.doi.org/ 10.1074/jbc.M609850200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Jung G, Remmert K, Wu X, Volosky JM, Hammer JA 3rd. The Dictyostelium CARMIL protein links capping protein and the ARP2/3 complex to type I myosins through their SH3 domains. J Cell Biol 2001; 153:1479-97; PMID:11425877; http://dx.doi.org/ 10.1083/jcb.153.7.1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Yang C, Pring M, Wear MA, Huang M, Cooper JA, Svitkina TM, Zigmond SH. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev Cell 2005; 9:209-21; PMID:16054028; http://dx.doi.org/ 10.1016/j.devcel.2005.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Edwards M, Zwolak A, Schafer DA, Sept D, Dominguez R, Cooper JA. Capping protein regulators fine-tune actin assembly dynamics. Nat Rev Mol Cell Biol 2014; 15:677-89; PMID:25207437; http://dx.doi.org/ 10.1038/nrm3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Lim CY, Han W. Tropomodulin3 as the link between insulin-activated AKT2 and cortical actin remodeling in preparation of GLUT4 exocytosis. Bioarchitecture 2014; 4:210-4; PMID:26280982; http://dx.doi.org/ 10.1080/19490992.2015.1031949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Lim CY, Bi X, Wu D, Kim JB, Gunning PW, Hong W, Han W. Tropomodulin3 is a novel Akt2 effector regulating insulin-stimulated GLUT4 exocytosis through cortical actin remodeling. Nat Commun 2015; 6:5951; PMID:25575350; http://dx.doi.org/ 10.1038/ncomms6951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Tomek W, Smiljakovic T. Activation of Akt (protein kinase B) stimulates metaphase I to metaphase II transition in bovine oocytes. Reproduction 2005; 130:423-30; PMID:16183860; http://dx.doi.org/ 10.1530/rep.1.00754 [DOI] [PubMed] [Google Scholar]
- [115].Kalous J, Solc P, BaRAN V, Kubelka M, Schultz RM, Motlik J. PKB/AKT is involved in resumption of meiosis in mouse oocytes. Biol Cell 2006; 98:111-23; PMID:15842198; http://dx.doi.org/ 10.1042/BC20050020 [DOI] [PubMed] [Google Scholar]
- [116].Hoshino Y, Sato E. Protein kinase B (PKB/Akt) is required for the completion of meiosis in mouse oocytes. Dev Biol 2008; 314:215-23; PMID:18177853; http://dx.doi.org/ 10.1016/j.ydbio.2007.12.005 [DOI] [PubMed] [Google Scholar]
- [117].Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol 2009; 10:778-90; PMID:19851336; http://dx.doi.org/ 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Simerly C, Nowak G, de Lanerolle P, Schatten G. Differential expression and functions of cortical myosin IIA and IIB isotypes during meiotic maturation, fertilization, and mitosis in mouse oocytes and embryos. Mol Biol Cell 1998; 9:2509-25; PMID:9725909; http://dx.doi.org/ 10.1091/mbc.9.9.2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Schuh M, Ellenberg J. A new model for asymmetric spindle positioning in mouse oocytes. Curr Biol 2008; 18:1986-92; PMID:19062278; http://dx.doi.org/ 10.1016/j.cub.2008.11.022 [DOI] [PubMed] [Google Scholar]
- [120].Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. Inhibition of the ARP2/3 complex-nucleated actin polymerization and bRANch formation by tropomyosin. Curr Biol 2001; 11:1300-4; PMID:11525747; http://dx.doi.org/ 10.1016/S0960-9822(01)00395-5 [DOI] [PubMed] [Google Scholar]
- [121].Hsiao JY, Goins LM, Petek NA, Mullins RD. ARP2/3 complex and cofilin modulate binding of tropomyosin to bRANched actin networks. Curr Biol 2015; 25:1573-82; PMID:26028436; http://dx.doi.org/ 10.1016/j.cub.2015.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol 2010; 11:276-87; PMID:20308985; http://dx.doi.org/ 10.1038/nrm2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Larson SM, Lee HJ, Hung PH, Matthews LM, Robinson DN, Evans JP. Cortical mechanics and meiosis II completion in mammalian oocytes are mediated by myosin-II and Ezrin-Radixin-Moesin (ERM) proteins. Mol Biol Cell 2010; 21:3182-92; PMID:20660156; http://dx.doi.org/ 10.1091/mbc.E10-01-0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Rodrigues NT, Lekomtsev S, Jananji S, Kriston-Vizi J, Hickson GR, Baum B. Kinetochore-localized PP1-Sds22 couples chromosome segregation to polar relaxation. Nature 2015; 524:489-92; PMID:26168397; http://dx.doi.org/ 10.1038/nature14496 [DOI] [PubMed] [Google Scholar]
- [125].Hillig RC, Renault L, Vetter IR, Drell Tt, Wittinghofer A, Becker J. The crystal structure of rna1p: a new fold for a GTPase-activating protein. Mol Cell 1999; 3:781-91; PMID:10394366; http://dx.doi.org/ 10.1016/S1097-2765(01)80010-1 [DOI] [PubMed] [Google Scholar]
- [126].Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct 2000; 29:545-76; PMID:10940259; http://dx.doi.org/ 10.1146/annurev.biophys.29.1.545 [DOI] [PubMed] [Google Scholar]
- [127].Heacock CS, Bamburg JR. The quantitation of G- and F-actin in cultured cells. Anal Biochem 1983; 135:22-36; PMID:6670742; http://dx.doi.org/ 10.1016/0003-2697(83)90725-X [DOI] [PubMed] [Google Scholar]
- [128].Theriot JA. Actin filament dynamics in cell motility. Adv Exp Med Biol 1994; 358:133-45; PMID:7801800; http://dx.doi.org/ 10.1007/978-1-4615-2578-3_13 [DOI] [PubMed] [Google Scholar]
- [129].Selden LA, Kinosian HJ, Estes JE, Gershman LC. Impact of profilin on actin-bound nucleotide exchange and actin polymerization dynamics. Biochemistry 1999; 38:2769-78; PMID:10052948; http://dx.doi.org/ 10.1021/bi981543c [DOI] [PubMed] [Google Scholar]
- [130].Cooley L, Verheyen E, Ayers K. chickadee encodes a profilin required for intercellular cytoplasm transport during Drosophila oogenesis. Cell 1992; 69:173-84; PMID:1339308; http://dx.doi.org/ 10.1016/0092-8674(92)90128-Y [DOI] [PubMed] [Google Scholar]
- [131].Suarez C, Carroll RT, Burke TA, Christensen JR, Bestul AJ, Sees JA, James ML, Sirotkin V, Kovar DR. Profilin regulates F-actin network homeostasis by favoring formin over ARP2/3 complex. Dev Cell 2015; 32:43-53; PMID:25543282; http://dx.doi.org/ 10.1016/j.devcel.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Rotty JD, Wu C, Haynes EM, Suarez C, Winkelman JD, Johnson HE, Haugh JM, Kovar DR, Bear JE. Profilin-1 serves as a gatekeeper for actin assembly by ARP2/3-dependent and -independent pathways. Dev Cell 2015; 32:54-67; PMID:25543281; http://dx.doi.org/ 10.1016/j.devcel.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol 2014; 24:579-85; PMID:24560576; http://dx.doi.org/ 10.1016/j.cub.2014.01.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Mannherz HG, Hannappel E. The beta-thymosins: intracellular and extracellular activities of a versatile actin binding protein family. Cell Motil Cytoskeleton 2009; 66:839-51; PMID:19405116; http://dx.doi.org/ 10.1002/cm.20371 [DOI] [PubMed] [Google Scholar]
- [135].Bock-Marquette I, Saxena A, White MD, Dimaio JM, Srivastava D. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature 2004; 432:466-72; PMID:15565145; http://dx.doi.org/ 10.1038/nature03000 [DOI] [PubMed] [Google Scholar]
- [136].Shrivastava S, Srivastava D, Olson EN, DiMaio JM, Bock-Marquette I. Thymosin beta4 and cardiac repair. Ann N Y Acad Sci 2010; 1194:87-96; PMID:20536454; http://dx.doi.org/ 10.1111/j.1749-6632.2010.05468.x [DOI] [PubMed] [Google Scholar]
- [137].Wu GJ, Simerly C, ZoRAN SS, Funte LR, Schatten G. Microtubule and chromatin dynamics during fertilization and early development in rhesus monkeys, and regulation by intracellular calcium ions. Biol Reprod 1996; 55:260-70; PMID:8828828; http://dx.doi.org/ 10.1095/biolreprod55.2.260 [DOI] [PubMed] [Google Scholar]
- [138].Courtois A, Schuh M, Ellenberg J, Hiiragi T. The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J Cell Biol 2012; 198:357-70; PMID:22851319; http://dx.doi.org/ 10.1083/jcb.201202135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Fulton BP, Whittingham DG. Activation of mammalian oocytes by intracellular injection of calcium. Nature 1978; 273:149-51; PMID:565475; http://dx.doi.org/ 10.1038/273149a0 [DOI] [PubMed] [Google Scholar]
- [140].Onodera M, Tsunoda Y. Parthenogenetic activation of mouse and rabbit eggs by electric stimulation in vitro. Gamete Res 1989; 22:277-83; PMID:2707730; http://dx.doi.org/ 10.1002/mrd.1120220305 [DOI] [PubMed] [Google Scholar]
- [141].Chew TG, Lorthongpanich C, Ang WX, Knowles BB, Solter D. Symmetric cell division of the mouse zygote requires an actin network. Cytoskeleton (Hoboken) 2012; 69:1040-6; PMID:22887777; http://dx.doi.org/ 10.1002/cm.21062 [DOI] [PubMed] [Google Scholar]
- [142].Yu XJ, Yi Z, Gao Z, Qin D, Zhai Y, Chen X, Ou-Yang Y, Wang ZB, Zheng P, Zhu MS, et al.. The subcortical maternal complex controls symmetric division of mouse zygotes by regulating F-actin dynamics. Nat Commun 2014; 5:4887; PMID:25208553; http://dx.doi.org/ 10.1038/ncomms5887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Chaigne A, Campillo C, Voituriez R, Gov NS, Sykes C, Verlhac MH, Terret ME. F-actin mechanics control spindle centring in the mouse zygote. Nat Commun 2016; 7:10253; PMID:26727405; http://dx.doi.org/ 10.1038/ncomms10253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Tong ZB, Gold L, Pfeifer KE, Dorward H, Lee E, Bondy CA, Dean J, Nelson LM. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet 2000; 26:267-8; PMID:11062459; http://dx.doi.org/ 10.1038/81547 [DOI] [PubMed] [Google Scholar]
- [145].Tong ZB, Nelson LM, Dean J. Mater encodes a maternal protein in mice with a leucine-rich repeat domain homologous to porcine ribonuclease inhibitor. Mamm Genome 2000; 11:281-7; PMID:10754103; http://dx.doi.org/ 10.1007/s003350010053 [DOI] [PubMed] [Google Scholar]
- [146].Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell 2008; 15:416-25; PMID:18804437; http://dx.doi.org/ 10.1016/j.devcel.2008.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Ohsugi M, Zheng P, Baibakov B, Li L, Dean J. Maternally derived FILIA-MATER complex localizes asymmetrically in cleavage-stage mouse embryos. Development 2008; 135:259-69; PMID:18057100; http://dx.doi.org/ 10.1242/dev.011445 [DOI] [PubMed] [Google Scholar]
- [148].Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol 2004; 6:1034-8; PMID:15516996; http://dx.doi.org/ 10.1038/ncb1104-1034 [DOI] [PubMed] [Google Scholar]
- [149].Bear JE, Gertler FB. Ena/VASP: towards resolving a pointed controversy at the barbed end. J Cell Sci 2009; 122:1947-53; PMID:19494122; http://dx.doi.org/ 10.1242/jcs.038125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Sjoblom B, Salmazo A, Djinovic-Carugo K. Alpha-actinin structure and regulation. Cell Mol Life Sci 2008; 65:2688-701; PMID:18488141; http://dx.doi.org/ 10.1007/s00018-008-8080-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Machesky LM, Li A. Fascin: Invasive filopodia promoting metastasis. Commun Integr Biol 2010; 3:263-70; PMID:20714410; http://dx.doi.org/ 10.4161/cib.3.3.11556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Kono K, Tamashiro DA, Alarcon VB. Inhibition of RHO-ROCK signaling enhances ICM and suppresses TE characteristics through activation of Hippo signaling in the mouse blastocyst. Dev Biol 2014; 394:142-55; PMID:24997360; http://dx.doi.org/ 10.1016/j.ydbio.2014.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev 2013; 27:355-71; PMID:23431053; http://dx.doi.org/ 10.1101/gad.210773.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Vizcarra CL, Kreutz B, Rodal AA, Toms AV, Lu J, Zheng W, Quinlan ME, Eck MJ. Structure and function of the interacting domains of Spire and Fmn-family formins. Proc Natl Acad Sci U S A 2011; 108:11884-9; PMID:21730168; http://dx.doi.org/ 10.1073/pnas.1105703108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Stenmark H, Aasland R. FYVE-finger proteins–effectors of an inositol lipid. J Cell Sci 1999; 112(Pt 23):4175-83; PMID:10564636 [DOI] [PubMed] [Google Scholar]
- [156].Ti SC, Jurgenson CT, Nolen BJ, Pollard TD. Structural and biochemical characterization of two binding sites for nucleation-promoting factor WASp-VCA on ARP2/3 complex. Proc Natl Acad Sci U S A 2011; 108:E463-71; PMID:21676862; http://dx.doi.org/ 10.1073/pnas.1100125108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. ARP2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci U S A 2011; 108:E472-9; PMID:21676863; http://dx.doi.org/ 10.1073/pnas.1100236108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 2002; 418:790-3; PMID:12181570; http://dx.doi.org/ 10.1038/nature00859 [DOI] [PubMed] [Google Scholar]
- [159].Suetsugu S, Yamazaki D, Kurisu S, Takenawa T. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev Cell 2003; 5:595-609; PMID:14536061; http://dx.doi.org/ 10.1016/S1534-5807(03)00297-1 [DOI] [PubMed] [Google Scholar]
- [160].Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the ARP2/3 complex links Cdc42-dependent signals to actin assembly. Cell 1999; 97:221-31; PMID:10219243; http://dx.doi.org/ 10.1016/S0092-8674(00)80732-1 [DOI] [PubMed] [Google Scholar]
- [161].Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM is an ARP2/3 complex activator that binds microtubules and functions in ER to Golgi transport. Cell 2008; 134:148-61; PMID:18614018; http://dx.doi.org/ 10.1016/j.cell.2008.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The ARP2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell 2009; 17:712-23; PMID:19922875; http://dx.doi.org/ 10.1016/j.devcel.2009.09.010 [DOI] [PubMed] [Google Scholar]
- [163].Dominguez R, Namgoong S. Actin filament nucleation and elongation. In: Egelman EH, editor. Comprehensive Biophysics. Elsevier. 2012;31-47; http://dx.doi.org/ 10.1016/978-0-12-374920-8.00404-5 [DOI] [Google Scholar]
- [164].Galkin VE, Orlova A, Kudryashov DS, Solodukhin A, Reisler E, Schroder GF, Egelman EH. Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci U S A 2011; 108:20568-72; PMID:22158895; http://dx.doi.org/ 10.1073/pnas.1110109108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Kuhn TB, Bamburg JR. Tropomyosin and ADF/cofilin as collaborators and competitors. Adv Exp Med Biol 2008; 644:232-49; PMID:19209826; http://dx.doi.org/ 10.1007/978-0-387-85766-4_18 [DOI] [PubMed] [Google Scholar]
- [166].von der Ecken J, Muller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. Structure of the F-actin-tropomyosin complex. Nature 2015; 519:114-7; PMID:25470062; http://dx.doi.org/ 10.1038/nature14033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol 1986; 103:2747-54; PMID:3793756; http://dx.doi.org/ 10.1083/jcb.103.6.2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Wear MA, Cooper JA. Capping protein: new insights into mechanism and regulation. Trends Biochem Sci 2004; 29:418-28; PMID:15362226; http://dx.doi.org/ 10.1016/j.tibs.2004.06.003 [DOI] [PubMed] [Google Scholar]
- [169].Yamashiro S, Gokhin DS, Kimura S, Nowak RB, Fowler VM. Tropomodulins: pointed-end capping proteins that regulate actin filament architecture in diverse cell types. Cytoskeleton (Hoboken) 2012; 69:337-70; PMID:22488942; http://dx.doi.org/ 10.1002/cm.21031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Rao JN, Madasu Y, Dominguez R. Mechanism of actin filament pointed-end capping by tropomodulin. Science 2014; 345:463-7; PMID:25061212; http://dx.doi.org/ 10.1126/science.1256159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Narita A, Takeda S, Yamashita A, Maeda Y. Structural basis of actin filament capping at the barbed-end: a cryo-electron microscopy study. EMBO J 2006; 25:5626-33; PMID:17110933; http://dx.doi.org/ 10.1038/sj.emboj.7601395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, Yu M, Patel NA, Robinson CV, Carter AP. The structure of the dynactin complex and its interaction with dynein. Science 2015; 347:1441-6; PMID:25814576; http://dx.doi.org/ 10.1126/science.aaa4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Verlhac MH, Lefebvre C, Kubiak JZ, Umbhauer M, Rassinier P, Colledge W, Maro B. Mos activates MAP kinase in mouse oocytes through two opposite pathways. EMBO J 2000; 19:6065-74; PMID:11080153; http://dx.doi.org/ 10.1093/emboj/19.22.6065 [DOI] [PMC free article] [PubMed] [Google Scholar]


