In the glycolysis pathway, pyruvate kinase (PK) and phosphoglycerate kinase 1 (PGK1) are the only 2 ATP-generating enzymes. PK is a rate-limiting glycolytic enzyme that catalyzes the conversion of phosphoenolpyruvate (PEP) and ADP to pyruvate and ATP. Splicing inclusion of exon 9 and exon 10 from PKM pre-mRNA results in expression of the PKM1 and PKM2 isoforms,1 respectively. Our previous studies demonstrated that PKM2 but not PKM1 has dual enzymatic activities as both a glycolytic enzyme and protein kinase. PKM2 uses PEP as a phosphate donor to phosphorylate its protein substrates, which include histone H3 T11, STAT3 Y705, Bub3 Y207, and MLC2 Y118.2 In addition, more than 100 protein substrates of PKM2 were identified via protein array screening analysis.3 Of note, a recent report debated that PKM2 lacks evidence of being a protein kinase.4 However, these results may owe to the drawbacks of using one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis, which cannot distinguish differences in the phosphorylation spectrum between cells wild-type for PKM2 and PKM2-null cells. Importantly, a more recent publication confirmed that the yeast PKM2 homolog, acting as a protein kinase, directly phosphorylated histone H3 T11 in the presence of PEP but not ATP.5 Additionally, it was demonstrated that AKT1 substrate 1 is a new protein substrate of PKM2.6 Taken together, these reports support that PKM2 possesses protein kinase activity (Fig. 1).
Figure 1.
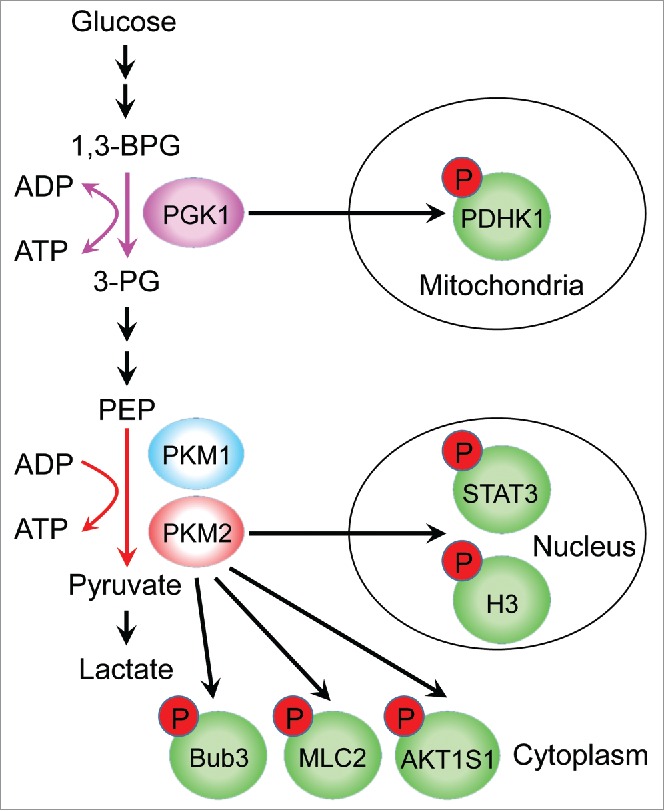
PGK1 and PKM2 possess dual glycolytic enzyme and protein kinase functions. PGK1 and PKM2 catalyze the 2 ATP-generating reactions in the glycolysis pathway. Mitochondrial PGK1 phosphorylates PDHK1 using ATP as a phosphate donor, and PKM2, but not PKM1, phosphorylates its protein substrates using PEP as a phosphate donor.
PGK1 is the first ATP-generating enzyme in the glycolysis pathway. It catalyzes the reversible conversion of 1,3-diphosphoglycerate and ADP to 3-phosphoglycerate and ATP, respectively. Our recent studies demonstrated that PGK1 translocates into mitochondria under hypoxic stress, epidermal growth factor receptor (EGFR) activation, or expression of oncogenic K-Ras G12V or the B-Raf V600E mutation. Mechanistically, mitochondrial translocation of PGK1 is mediated by extracellular signal-regulated kinase (ERK) 1/2-dependent S203 phosphorylation, which results in PGK1 isomerization by Peptidyl-prolyl cis/trans isomerase NIMA-interacting 1 (PIN1), and subsequent exposure of the presequence of PGK1 (38-QRIKAA-43) to recognition by the mitochondrial translocase of the outer membrane (TOM) complex. In mitochondria, PGK1 directly phosphorylated pyruvate dehydrogenase kinase isozyme 1 (PDHK1) at T338 using ATP as a phosphate donor (Fig. 1). PGK1-dependent phosphorylation of PDHK1 activated PDHK1 and enhanced PDHK1-mediated pyruvate dehydrogenase E1α S293 phosphorylation, which inactivated pyruvate dehydrogenase complex, an enzyme complex that converts pyruvate and coenzyme A to acetyl-coenzyme A and CO2. Thus, mitochondrial translocation of PGK1 resulted in inhibition of pyruvate oxidation in mitochondria and enhancement of lactate production from pyruvate in cytosol. Deficiency in mitochondrial translocation of PGK1 via CRISPR/Cas9-mediated knock-in of PGK1 S203A or reconstituted expression of a kinase-dead PGK1 T378P mutant in PGK1-depleted cells blocked hypoxia- and EGFR activation-induced lactate production and attenuation of mitochondrial pyruvate oxidation. These results indicated that the protein kinase activity of mitochondrial PGK1 is a gatekeeper for the tricarboxylic acid (TCA) cycle by shunting pyruvate from the mitochondria into the cytosol for lactate production. In addition, we revealed that hypoxia-enhanced expression of PDHK1 had a limited effect on pyruvate dehydrogenase E1α S293 phosphorylation in PGK1 mitochondrial translocation-deficient cells, which indicated that PDHK1 relied on mitochondrial PGK1-dependent phosphorylation of PDHK1 more than PDHK1 protein expression for its activity. Functional studies demonstrated that replacement of endogenous PGK1 with mitochondrial translocation-deficient mutant PGK1 S203A dramatically reduced the growth of tumors orthotopically transplanted in the brains of mice, which resulted from a low proliferation rate and enhanced apoptosis of tumor cells. Immunohistochemical staining of human glioblastoma tissues demonstrated that PGK1 pS203 and PDHK1 pT338 are positively correlated with each other, and the levels of PGK1 pS203 and PDHK1 pT338 staining are inversely correlated with survival duration in glioblastoma patients, supporting a pivotal role for PGK1-dependent PDHK1 phosphorylation in glioblastoma progression.7
In summary, we expanded the protein kinome via characterization and demonstration of the protein kinase activities of the glycolytic enzymes PKM2 and PGK1. Mitochondrial PGK1, although accounting for only about 12% of total intracellular PGK1, rewires the metabolic pathway to support the rapid growth of cancer cells by shunting mitochondrial pyruvate for lactate production, which promotes glycolysis by anaplerosis of NAD+ under hypoxic stress and activation of oncogenes, such as EGFR, K-Ras G12V, and B-Raf V600E.7 Deciphering the PGK1/PDHK1/pyruvate dehydrogenase complex axis, which coordinates the regulation of mitochondrial metabolism and glycolysis, provides a molecular basis for developing new therapeutic interventions for human cancer.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Lu Z. Cell Cycle 2012; 11:4101-2; PMID:23070542; http://dx.doi.org/ 10.4161/cc.22325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yang W, Lu Z. J Cell Sci 2015; 128:1655-60; PMID:25770102; http://dx.doi.org/ 10.1242/jcs.166629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Keller KE, et al. Mol Cell 2014; 53:700-9; PMID:24606918; http://dx.doi.org/ 10.1016/j.molcel.2014.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hosios AM, et al. Mol Cell 2015; 59:850-7; PMID:26300261; http://dx.doi.org/ 10.1016/j.molcel.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li S, et al.. Mol Cell 2015; 60:408-21.; PMID:26527276; http://dx.doi.org/ 10.1016/j.molcel.2015.09.024 [DOI] [PubMed] [Google Scholar]
- [6].He CL, et al.. Sci Rep 2016; 6:21524.; PMID:26876154; http://dx.doi.org/ 10.1038/srep21524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li X, et al.. Mol Cell 2016; 61:705-19.; PMID:26942675; http://dx.doi.org/ 10.1016/j.molcel.2016.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]


