ABSTRACT
Although the role of cyclins in controlling nuclear division is well established, their function in ciliate meiosis remains unknown. In ciliates, the cyclin family has undergone massive expansion which suggests that diverse cell cycle systems exist, and this warrants further investigation. A screen for cyclins in the model ciliate Tetrahymena thermophila showed that there are 34 cyclins in this organism. Only 1 cyclin, Cyc17, contains the complete cyclin core and is specifically expressed during meiosis. Deletion of CYC17 led to meiotic arrest at the diakinesis-like metaphase I stage. Expression of genes involved in DNA metabolism and chromosome organization (chromatin remodeling and basic chromosomal structure) was repressed in cyc17 knockout matings. Further investigation suggested that Cyc17 is involved in regulating spindle pole attachment, and is thus essential for chromosome segregation at meiosis. These findings suggest a simple model in which chromosome segregation is influenced by Cyc17.
KEYWORDS: anaphase initiation, cyclin, chromosome segregation, meiosis, Tetrahymena thermophila
Introduction
Meiosis is a specialized form of nuclear division in which chromosomes are duplicated only once before 2 cell divisions to generate cells containing half the normal complement of chromosomes. Meiotic progression occurs through a series of orderly events, such as DNA replication, homologous chromosome recombination and pairing, and chromosome condensation and segregation. Of these processes, homologous chromosome segregation at anaphase I is important for the correct distribution of genetic material to future generations. However, several steps must be completed before anaphase initiation.1 First, DNA replication must be completed and chromosomes organized correctly.1,2 Second, microtubule–chromosome attachment is necessary to allow chromosome segregation during anaphase. The major site of microtubule attachment is the kinetochore, comprising Cenp-A, Cenp-C, Cenp-H, Ndc80, Dam1, and many other components.1,3-9 Microtubule attachment is facilitated by Aurora B and some components of the spindle assembly checkpoint (SAC).10-12 Third, completion of microtubule–chromosome attachment leads to anaphase promoting complex (APCCdc20) activation, which release sister–chromatid cohesion to allow separation.1,13,14
Cyclins, cyclin-dependent kinases (CDKs) and their regulators are key components of the cell cycle control system. Cyclins are a diverse family of proteins that bind to CDKs and target genes, thus enabling the former to phosphorylate the latter. These target genes are involved in numerous cellular processes.1 In budding yeast, 4 cyclins (Clb1, Clb2, Clb3, and Clb4) have been well studied; Clb2 is the main mitotic cyclin that promote G2 to M phase transition.15,16 In mammals, a group of at least 16 cyclins has been reported.17 Most cyclins possess a complete cyclin core including N- and C-terminal cyclin domains, while some do not contain a C-terminal cyclin domain. The cyclin domain enables the cyclin to bind to CDK.17,18 However, our understanding of the function of cyclins is limited to those of several well-studied organisms. Compared with those in yeasts and animals, cyclin family genes in ciliates and angiosperms have undergone massive expansion.19-22 Genome annotation has revealed at least 50 cyclins in Arabidopsis, 49 cyclins in rice, 59 cyclins in maize,20-22 and a staggering 140 cyclin homologs in Paramecium tetraurelia.19 The extensive expansion of cyclin family suggests that diverse cell cycle systems exist, and this warrants further investigation.
Tetrahymena thermophila is an excellent model unicellular eukaryote. Like other ciliates, it possesses 2 nuclei, a diploid germline micronucleus (MIC) and a polyploid somatic macronucleus (MAC).23 Cells of complementary mating types can conjugate under starvation conditions and initiate synchronous meioses of their MICs.24 Meiosis has been well studied in T. thermophila over last few decades.4,25-30 In T. thermophila, 26 cyclins have been found based on previous gene predictions,19,31,32 thus providing good starting material to study the function of cyclins in ciliates.
In this study, we screened for cyclin domain containing proteins in T. thermophila based on the latest gene predictions and RNA-Seq data. Only 1 protein, Cyc17, contained a complete cyclin core (both N- and C-terminal cyclin domains) was specifically expressed during conjugation (or meiosis). A functional investigation showed that Cyc17 is essential for anaphase onset; it may promote microtubule–chromosome attachment and cohesion resolution rather than spindle formation for chromosome segregation. These findings have improved our understanding of the ciliate cyclins.
Results and discussion
Cyc17 is the only meiosis-specific cyclin with a complete cyclin core in T. thermophila
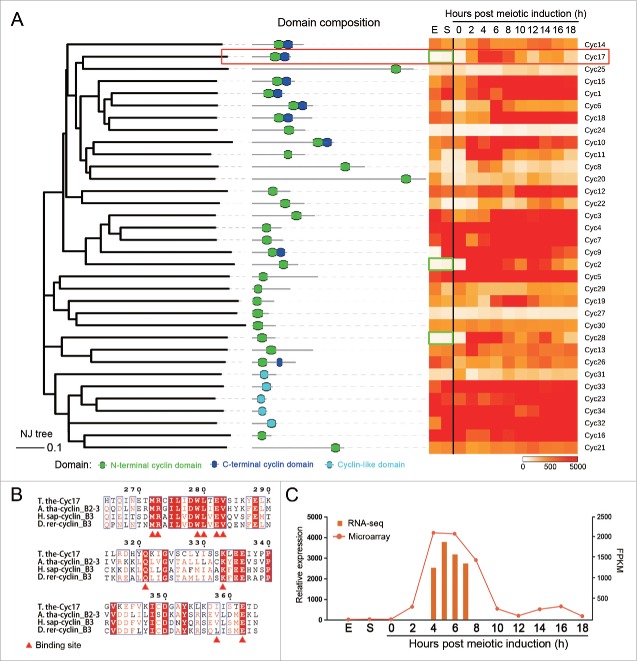
Previous studies suggested that there are 26 cyclins in T. thermophila based on previous gene predictions (version 2008).19,31,32 Each cyclin has only one distinct expression peak during conjugation (i.e. when meiosis occurs); they are therefore likely to have unique roles in advancing the meiotic cell cycle.19,31,32 Since an RNA-Seq study has shown many mispredictions in a previously T. thermophila gene annotation,33 we rescreened for cyclin domain containing proteins in a new T. thermophila gene prediction (version 2014) and in RNA-Seq data. We identified a total of 34 potential cyclins (Table S1). These included all 26 previously identified cyclins (Cyc1 – Cyc26), except for the misprediction of TTHERM_00698650 (Table S1). We therefore retained the naming system used in the previous study (Table S1), and named the 8 novel cyclin domain containing proteins sequentially from Cyc27 to Cyc34 (Table S1).
Analysis of domain composition suggested that these cyclins could be classified into 3 categories (Fig. 1A): (1) 9 cyclins contained both N- and C-terminal cyclin domains (i.e., a complete cyclin core); (2) 20 cyclins contained only an N-terminal cyclin domain; and (3) 5 cyclins contained a cyclin-like domain. The latter group are probably related to the Pho80 protein previously identified in budding yeast, where it regulates both the cell cycle and other processes.34 Next, we determined their expression profiles during vegetative propagation, starvation and conjugation.32 We found that 3 (Cyc2, Cyc17, and Cyc28) were specifically expressed during conjugation, whereas the others were also highly expressed during vegetative propagation and/or starvation conditions (Fig. 1A). Therefore, the 3 cyclins specifically expressed during conjugation are more likely to regulate conjugation processes, such as meiosis.
Figure 1.
Cyc17 is a meiosis-specific cyclin in T. thermophila. (A) Phylogeny, domain composition and expression profile of 34 cyclins in T. thermophila. Green box, genes not expressed during vegetative propagation and starvation; red box, Cyc17. (B) CYC17 Expression profile based on microarray and RNA-Seq data. T.the, T. thermophila; A.tha, Arabidopsis thaliana; H.sap, Homo sapiens; D.rer, Danio rerio. (C) Multiple sequence alignment showing a conserved cyclin domain in Cyc17. E, exponentially growing cells; S, starved cells. Microarray data were retrieved from TetraFGD.
Since most well-studied cyclins in other organisms contain both N- and C-terminal domains, this study focuses on Cyc17 (TTHERM_00693080) that is the only meiosis-specific cyclin containing a complete cyclin core (both the N- and C-terminal cyclin domain). A BLASTP search of non-redundant protein sequences showed that Cyc17 may be a Cyclin B3 homolog. Sequence alignment of Cyc17 and its homologs in humans, zebrafish, and Arabidopsis showed that this protein contains the conserved CDK-binding sites (Fig. 1B),22,35,36 suggesting that it may regulate the cell cycle via CDK binding. Although the exact role of Cyclin B3 in meiosis is yet not clear, it has been identified as a meiotic cyclin in mammals35 and as a marker for spermatogonia and spermatocytes in zebrafish.36 A recent study in mice showed that Cyclin B3 controls anaphase onset in meiotic oocytes.37 These reports suggest that Cyc17 might also be involved in meiosis in T. thermophila.
Our RNA-Seq analysis found that CYC17 expression corresponds to that of a previous microarray study (Fig. 1C),31,32 with the highest expression level at 5 h post meiotic induction (Fig. 1C). This result indicates that Cyc17 may play an important role at 5 h after meiosis is initiated.
Meiosis is arrested at diakinesis-like metaphase I in cyc17Δ cells
In wild-type (WT) T. thermophila, meiosis is initiated by the mating of 2 starved cells. The MIC elongates during meiotic prophase (3 – 4 h post meiotic induction) in response to double-strand break (DSB) formation (∼2 h post meiotic induction). Homologous chromosome pairing occurs at maximal MIC elongation, after which DSBs are repaired by homologous recombination. Both MIC elongation and DSB repair are essential for metaphase progression.29 Thereafter, the MIC shortens and 5 condensed bivalents appear at diakinesis-like stage to metaphase I (diakinesis-like metaphase I; ∼5 h post meiotic induction). Homologous chromosomes then segregate at anaphase I and sister–chromatids segregate at anaphase II (5 – 7 h post meiotic induction).28
To investigate Cyc17 function during meiosis, Cyc17 knockout strains (cyc17Δ) of 2 mating types were constructed. RNA-Seq analysis showed that CYC17 expression was totally abolished during meiosis (Fig. S1). Comparison of the doubling time of cyc17Δ and WT strains, confirmed that Cyc17 deficiency does not affect vegetative propagation (i.e. mitotic cell division) (Fig. S2). We subsequently compared MIC development in WT and cyc17Δ strains during meiosis. Similar to WT cells, meiosis in cyc17Δ cells progressed normally through early prophase I (leptonema-like, zygonema-like, pachynema-like, and diplonema-like stages; 1 – 4 h post meiotic induction) (Fig. 2A). As DSB repair via homologous recombination requires DNA synthesis, meiotic BrdU incorporation was used to examine DSB repair in MICs. As shown in Figure 2B, BrdU staining foci were observed in both MICs of WT and cyc17Δ cells, indicating that DSB repair occurs in cyc17Δ cells. We next assessed homologous chromosome pairing by fluorescence in situ hybridization (FISH) which usually shows a single merged or 2 adjacent foci at diakinesis-like metaphase I (Fig. 2C).27 Like in WT cells, all homologous chromosomes underwent normal pairing in cyc17Δ cells (about 44% had single merged foci and 56% had 2 adjacent foci; n = 100) (Fig. 2C). These results strongly suggested that meiotic events in cyc17Δ cells are normal before diakinesis-like metaphase I.
Figure 2.
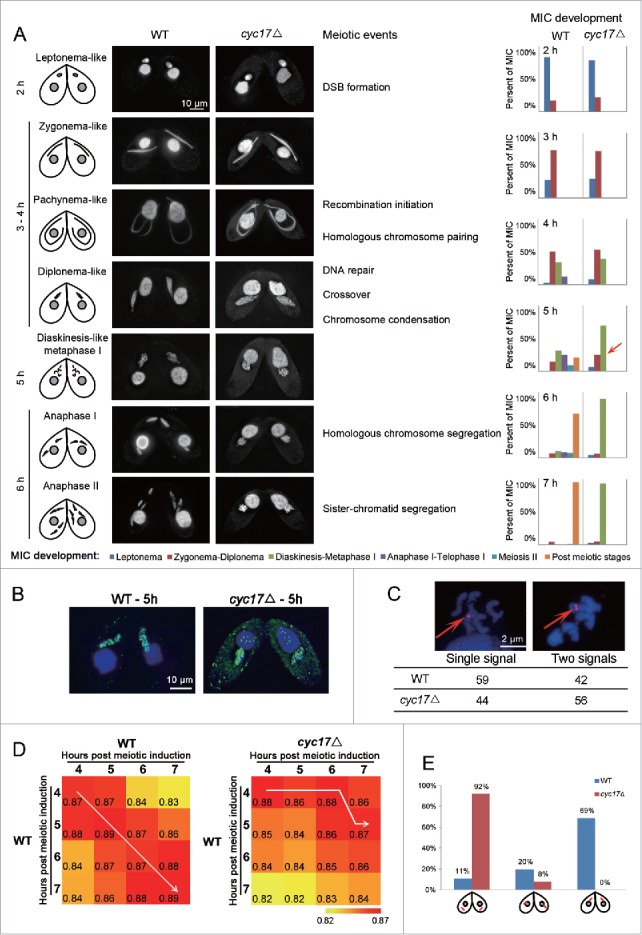
Meiosis is arrested at 5 h post meiotic induction in cyc17Δ cells. (A) Meiotic progression of WT and cyc17Δ cells from 2 h to 7 h post meiotic induction.25,29,57 Red arrow, cyc17Δ cells arrested at diakinesis-like metaphase I stage (B) BrdU staining. (C) FISH shows normal homologous chromosome pairing in cyc17Δ cells. Red arrow, FISH signal. (D) Correlation of genes expression between different samples. (E) Quantification of MIC positions in WT and cyc17Δ cells.
However, meiosis is arrested at diakinesis-like metaphase I in cyc17Δ cells, and there is a failure to initiate anaphase I (∼5 h post meiotic induction) (Fig. 2A). In comparison, WT cells completed homologous and sister chromosome separation at the first and second meiotic divisions (Fig. 2A). We next compared gene expression in WT and cyc17Δ cells at time points from 4 h to 7 h post meiotic induction. There was a diagonal distribution of correlation coefficients (r) for gene expression value between different paired time points in WT cells (Fig. 2D, left panel, white arrow). In contrast, gene expression in cyc17Δ cells at 6 h and 7 h post meiotic induction showed the highest degree of correlation with those at 4 h and 5 h post meiotic induction in WT cells (Fig. 2D, right panel, white arrow), respectively. This result suggests that MIC development was arrested at 5 h post meiotic induction in the transcriptional level.
We also noticed that the MIC and MAC had exchanged position in cyc17Δ cells (Fig. 2E). From diakinesis-like to metaphase I of meiosis in WT cells, MICs tend to occupy the region next to the conjugational junction, where the 2 mating partners are connected, and the MAC relocates to the posterior part of the cell (Fig. 2E). In the mutant, MAC and MIC adopt the reverse localization. In Paramaecium and Tetrahymena, the cellular position of a nucleus is known to determine its development.38,39 Therefore, it is possible that failure of the MIC to progress to metaphase and anaphase I is a consequence of its mislocalization.
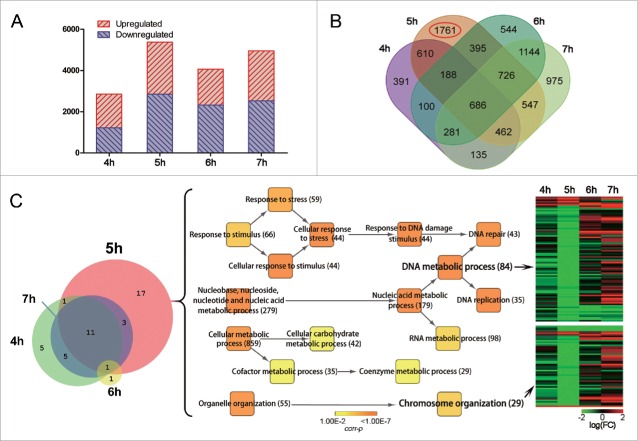
Next, differentially expressed genes (DEGs) were screened using a two-fold change cutoff of cyc17Δ versus WT at each time points (see Material and Methods for details). DEG numbers were greatly increased at 5 h compared with 4 h post meiotic induction (Fig. 3A). Moreover, overlapping analysis of DEGs at 4, 5, 6, and 7 h time points showed that most unique DEGs (1761 genes; Fig. 3B, red circle) were detected at 5 h post meiotic induction. This result also shows that most differences in gene expression resulting from CYC17 deletion occur at 5 h post meiotic induction, corresponding to the period of diakinesis-like metaphase I meiotic arrest. As changes in gene transcription usually occur before changes in related cellular events, changes in the expression of some genes responsible for the cyc17Δ phenotype might be abnormal before 5 h.
Figure 3.
Differential gene expression in WT and cyc17Δ cells. (A) Distribution of DEGs at 4, 5, 6, and 7 h post meiotic induction. (B) Venn diagram showing overlapping DEGs at 4 time points. Red circle, unique DEGs at 5 h post meiotic induction. (C) Functional enrichment of DEGs at 5 h post meiotic induction. Heatmap shows genes related to DNA metabolism and chromosome organization. Heatmap color scheme depicts the log2(FC) in expression between cyc17Δ and WT at a given time point. FC, fold change.
Both cytological and gene expression analyses showed meiotic cell cycle arrest at diakinesis-like metaphase I (∼5 h post meiotic induction) in cyc17Δ cells. We therefore investigated the function of DEGs at 5 h post meiotic induction compared with those at other times points. 17 unique gene ontology (GO) categories for biological processes were found in 5 h post meiotic induction by enrichment analysis (Fig. 3C and Table S2). These were mainly involved in DNA metabolism (such as the DNA repair and DNA replication processes) and chromosome organization (chromatin remodeling and basic structure of chromosome). Almost all of these genes are downregulated in cyc17Δ cells, suggesting that these processes are repressed in the absence of Cyc17. As DNA metabolism and chromosome organization are important processes in cell division, DEGs involved in these processes at diakinesis-like metaphase I might be responsible for the cyc17Δ phenotype.
Cyc17 is essential for chromosome segregation
Our findings indicated that cell cycle arrest occurs at diakinesis-like metaphase I in cyc17Δ cells, thus blocking anaphase I progression. DEG analysis showed that genes related to DNA metabolism and chromosome organization are downregulated at 5 h post meiotic induction (diakinesis-like metaphase I) in cyc17Δ cells. Thus, we further investigated anaphase initiation and chromosome segregation in cyc17Δ cells. First, spindle behavior was assessed. α-Tubulin staining of the meiotic spindle suggested spindle formation was normal (Fig. 4A). Thus, we speculated that microtubule-chromosome association might be abnormal in cyc17Δ cells. Giemsa staining provides a good image of the bivalent structure at the point of arrest. It revealed that kinetochores were stretched in opposing directions (i.e., tension was applied to the kinetochores) in only 4% of pairs (n = 101) in cyc17Δ cells, compared with 67% of pairs (n = 100) in WT (Fig. 4B). This result suggests that cyc17Δ cells have a defect in spindle pole attachment to centromeres. We therefore investigated the expression of genes related to spindle pole attachment (Table S3), including: (1) CENP-A, CENP-C, NDC80, and MCAK (related to the kinetochore); and (2): SAC components (MAD1, BUD1B, BUB3, and CENP-E) and AURORA B, which assist in microtubule-chromosome attachment.
Figure 4.

Cyc17 may be involved in microtubule-chromosome attachment and cohesion resolution, but not spindle formation. (A) β-tubulin staining of the meiotic spindle. (B) Giemsa staining at 5 h shows the kinetochore is stretched by opposing forces in WT but nearly not in cyc17Δ cells. (C) Genes encoding kinetochore components and SAC factors are downregulated in cyc17Δ cells. Heatmap color scheme depicts the log2(FC) in expression between cyc17Δ and WT at the indicated time points. (D) Diagram showing genes downregulated in cyc17Δ cells in a predicted chromosome segregation network in T. thermophila. Blue, genes downregulated in cyc17Δcells. FC, fold change.
Previous studies established that kinetochores are responsible for chromosome segregation.1,3-5 Although the kinetochore structure in T. thermophila is unknown, it has been well studied in many other species and several components are conserved among different organisms. In the eukaryotes studied so far, the kinetochore is assembled onto a histone H3 variant called Cenp-A (Cna1 in T. thermophila), which is directly associated with the conserved protein Cenp-C. Cenp-A is known to be essential for chromosome segregation in T. thermophila.4 These 2 proteins, along with a large group of other components that may not be so well conserved, form the inner kinetochore.1,3-7 The outer kinetochore comprises of the Ndc80 complex and another 10-subunit complex known as the Dam1. These 2 complexes are connected to both the inner kinetochore and the chromosome, together with a variety of associated microtubule-regulating and motor proteins such as Mcak (a microtubule-regulator).1,6,8,9,40 In cyc17Δ cells, all of the genes encoding these proteins were downregulated (Fig. 4C and 4D).Thus, the lack of these structural proteins could inhibit microtubule attachment to the chromosome, and the absence of Mcak could decrease the pulling force from the microtubule.
Although the precise function of SAC during meiosis remains unknown, there is no doubt that it helps to control the onset of anaphase I. Unlike in mitosis, the function of SAC in meiosis is more likely to assist microtubule-chromosome attachment along with Aurora B rather than to inhibit cohesion resolution.10-12 According to this model, downregulation of Aurora B and genes involved in SAC could lead to a defect in microtubule-chromosome attachment in cyc17Δ cells (Fig. 4C and 4D).
Besides the kinetochore and SAC, the cohesin complex also has an important function in chromosome segregation. Cohesin complex components form a ring-shaped structure around chromosomes to prevent their separation.13,14 Separase (Esp1 in T. thermophila) can open the ring when securin is removed by activated APCCdc20. In T. thermophila, most Esp1 knockdown cells become arrested in anaphase I during meiosis.14 Our results show that inhibition of anaphase initiation accompanies the downregulation of homologous genes encoding APCCdc20-Esp1 and SAC protein in cyc17Δ cells. This finding supports previous reports that cohesion resolution is regulated by a SAC-independent pathway during meiosis,17,37,41 and suggests that Esp1 downregulation may be responsible for the cyc17Δ phenotype (Fig. 4C and 4D).
Our results show that all genes related to spindle pole attachment were downregulated in cyc17Δ cells. This strongly suggests that Cyc17 is involved in preparing for chromosome segregation and may promote the expression of genes related to this process. Downregulation of genes involved in microtubule-chromosome attachment could explain the defect in homologous chromosome segregation and the meiotic arrest at diakinesis-like metaphase I in cyc17Δ cells (Fig. 4C). Based on our findings, a hypothetical model of the chromosome segregation process influenced by Cyc17 after DNA metabolism and chromosome organization is suggested (Fig. 4D) by highlighting the downregulated genes in cyc17Δ (Fig. 4D, shown in blue). It seems that when Cyc17 is deleted, many genes related to structural proteins of kinetochore, microtubule-chromosome attachment and cohesion resolution are down regulated.
Materials and methods
Cell culture and conjugation induction
Both CU427 (VI) and CU428 (VII) T. thermophila WT strains were obtained from the Tetrahymena Stock Center, Cornell University (http://tetrahymena.vet.cornell.edu/). Cells were cultured in Super Proteose Peptone (SPP) medium (1% Proteose Peptone, 0.2% glucose, 0.1% yeast extract, 0.003% Sequestrene) at 135 rpm and 30°C.30 For conjugation, cells of 2 different mating types (∼2 × 105 cells/ml) were first starved in 10 mM Tris-Cl (pH 7.4) for 12–16 h, and then mixed in equal amounts.
Generation of CYC17 knockout strains
To construct CYC17 knockout strains (cyc17Δ), a gene knockout sequence was cloned. For this, ∼1 kb upstream and downstream DNA sequences of the CYC17 ORF were amplified using the following primers: upstream, CYC17-5f652-NotI 5′-AGTTCTAGAGCGGCCGCTAGATATTTTGGGTAGATAAGACAC-3′ and CYC17-5r1589-N4 5′-GTCAGGTGCCTGGTACCCTGCAATATTAAAATAAATAAAAATG-3′; downstream, CYC17-3f3368-N4 5′-CTGACGTCGCACCATCCCTTTTTTACTAAGACCTTAATTTATC-3′ and CYC17-3r4316-NotI 5′-ACCGCGGTGGCGGCCGCTCTATCATATTCAGTTTGAGGAG-3′. The 2 sequences were then assembled into a NEO4 cassette containing a Tetrahymena codon optimized neomycin resistance gene under the control of a Cd2+-inducible metallothionein (MTT1) promoter using fusion PCR, and cloned into the pBlueScript SK (+) vector. The knockout fragment was released by NotI digestion and transformed into starved CU427 (VI) and CU428 (VII) WT cells using a biolistic transformation. The transformants were selected using SPP medium containing CdCl2 (1 μg/ml at the start of selection and 0.05 μg/ml at the end) and increasing concentrations of paromomycin (from 120 μg/ml to 40 mg/ml) until all WT chromosomes in the MAC were replaced by knockout chromosomes.42
BrdU and α-tubulin immunostaining
For BrdU staining, a final concentration of 200 μM BrdU reagent (in DMSO) was added to conjugating cells at 3.5 h post meiotic induction. At 4 h and 5 h post meiotic induction, 5 ml cells were harvested, 250 µl 10 % Triton X-100 and 500 μl 37% formaldehyde were added, and cells were incubated at room temperature (RT) for 30 min. The cell suspension was centrifuged and resuspended in 500 µl fixative with a solution of 4% paraformaldehyde and 3.4% sucrose in water. Cells were labeled with anti-BrdU primary and fluorochrome-labeled secondary antibodies, and then mounted in Vectashield antifading agent (Vector Laboratories; Burlingame, CA, USA) supplemented with 0.05 mg/ml 4,6-diamidino-2-phenylindole (DAPI).29
For α-tubulin immunostaining, 5 ml conjugating cell suspension was centrifuged and resuspended in 5 ml PEM buffer for 1 – 2 min at RT; 243 µl formaldehyde was then added gently and incubated for 30 min at RT. Cells were sedimented and re-suspended in 0.5 – 1 ml PEM buffer with 0.1 M glycine. A total of 150 µl cell suspension was spread onto coated poly-lysine slides that had have been heated to 65°C for 3 h, and then air-dried. Finally, slides were washed twice with PBS and once with PBS+0.05 % Triton (for 5 min each), and then incubated with anti-α-tubulin antibody (1:150 dilution in PEM buffer; mouse monoclonal) overnight at 4°C. Cells were then washed and incubated with fluorescence-labeled secondary antibody for 1.5 h at RT, washed again, and then mounted in Vectashield antifading agent supplemented with DAPI.
Fluorescence in situ hybridization
Cells were fixed in Schaudinn solution (containing 2 parts of saturated HgCl2, 1 part of absolute ethanol, and 1% of acetic acid) and incubated for 1 h at RT. The cell suspension was then centrifuged and resuspended in 300 µl of M/A solution (containing 2 parts of metanol, 1 part of glacial acid). The compound probe was made by PCR amplification of a 22.1 kb intercalary chromosomal locus.25 PCR products were purified and Cy3 labeled by nick translation. Both the probe and chromosomal DNA were denatured by adding hot formamide and then hybridized for 36 – 48 h at 37°C.28 Slides were then washed in PBS and mounted in Vectashield antifading agent supplemented with DAPI.
Giemsa staining
Cells fixed with Schaudinn solution were treated with 5 N HCl solution (100 µl under a coverslip) for 2 min at RT, rinsed with distilled water, and air-dried. Slides were then stained in 4% Giemsa solution in 10 mM sodium phosphate buffer (pH 6.8) for 10 min, rinsed with distilled water, air-dried, and mounted in Euparal.
For all cytological experiments, images were taken under fluorescence microscopy or bright field microscopy with the appropriate filter and then deconvolved, projected, and assigned with false colors; multicolor images were merged.
Gene identification and phylogenetic and heatmap analyses
As almost all cyclins possess an N-terminal cyclin domain or a cyclin-like domain, the domain composition was examined by InterproScan,43 and we screened proteins with N-terminal cyclin domain or cyclin-like domain as cyclins for further study. Cyclin identification was based on the latest gene predictions (version 2014: http://ciliate.org/index.php/home/downloads).
For identifying homologous genes that function in chromosome segregation, BLASTP analysis44 was used to compare the amino acid sequence of T. thermophila with those of other model eukaryotes. To confirm that candidate genes are expressed during conjugation, gene expression profiles were determined using both TetraFGD31,32 and RNA-Seq data.
For phylogenetic analysis, a multiple sequence alignment was generated by clustalW.45 A phylogenetic tree was constructed using MEGA6 with the NJ method and 1500 bootstrap replicates.46 iTOL was used to visualize the phylogenetic tree, the domain composition and the heatmap.47,48 Sequence alignment was visualized with ESPript 3,49 and MeV software (version 4.9.0)50 was used for heatmap construction.
RNA-Seq analysis
Total RNA was extracted from WT and cyc17Δ cells at 4, 5, 6 and 7 h post meiotic induction using the RNeasy Protect Cell Mini Kit (Qiagen), as previously described (TetraFGD: http://tfgd.ihb.ac.cn/index/smphelp).33 Poly-A tailed mRNA was enriched using Sera‐Mag magnetic oligo (dT) beads (GE). Illumina sequencing libraries were constructed according to the manufacturer's protocols. Paired-end (2 × 150 bp) sequencing was performed for all samples using an Illumina Hiseq4000 sequencer. All sequence data have been submitted to GenBank databases under accession number GSE77585. Adaptors of raw reads were trimmed with Trim-Galore (version 0.4.0)51 and then mapped to the T. thermophila Macronuclear genome assembly (version 2014: http://ciliate.org/index.php/home/downloads) using TopHat (version 2.0.9).52 Expression values were calculated and normalized to FPKM (fragments per kilobase of exon per million fragments mapped) using the Cuffdiff program (version 2.1.1).53
Pearson correlation coefficients (r) between samples were calculated using R software based on the FPKM values.54 For DEG analysis, genes were excluded if its expression levels are very low (FPKM < 10), and DEGs between matched WT and cyc17Δ samples were identified using a two-fold change cutoff. GO enrichment analysis55 was carried out with BiNGO (version 3.0.2).56 FWER was used for multiple testing correction of false positives, and significantly enriched GO terms were defined by a corrected P-value of ≤ 0.01.
Supplementary Material
Abbreviations
- BrdU
5-Bromo-2-deoxyUridine
- CDKs
cyclin-dependent kinases
- cyc17Δ
CYC17 knockout
- DAPI
4,6-diamidino-2-phenylindole
- DEGs
differentially expressed genes
- DSBs
double-strand breaks
- FISH
fluorescence in situ hybridization
- FPKM
fragments per kilobase of exon per million fragments mapped
- GO
gene ontology
- MAC
macronucleus
- MIC
micronucleus
- r
Pearson correlation coefficient
- RNA-Seq
RNA sequencing
- SAC
spindle assembly checkpoint
- WT
wild type
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Prof. Josef Loidl, Center for Molecular Biology, University of Vienna, Austria and Prof. Alan Warren, Department of Life Sciences, Natural History Museum, United Kingdom, for their critical comments and for English improvement.
References
- [1].Morgan DO. The cell cycle: Principles of control. London: New Science Press Ltd, 2007 [Google Scholar]
- [2].Hirano T. Condensins: Organizing and segregating the genome. Curr Biol 2005; 15:R265-75; PMID:15823530; http://dx.doi.org/ 10.1016/j.cub.2005.03.037 [DOI] [PubMed] [Google Scholar]
- [3].Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol 2001; 155:1147-57; PMID:11756469; http://dx.doi.org/ 10.1083/jcb.200108125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cervantes MD, Xi X, Vermaak D, Yao MC, Malik HS. The CNA1 histone of the ciliate Tetrahymena thermophila is essential for chromosome segregation in the germline micronucleus. Mol Biol Cell 2006; 17:485-97; PMID:16251352; http://dx.doi.org/ 10.1091/mbc.E05-07-0698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Moore LL, Roth MB. HCP-4, a CENP-C-like protein in Caenorhabditis elegans, is required for resolution of sister centromeres. J Cell Biol 2001; 153:1199-208; PMID:11402064; http://dx.doi.org/ 10.1083/jcb.153.6.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Goldstein LSB. Kinetochore structure and its role in chromosome orientation during the first meiotic division in male D. melanogaster. Cell 1981; 25:591-602; PMID:6793236; http://dx.doi.org/ 10.1016/0092-8674(81)90167-7 [DOI] [PubMed] [Google Scholar]
- [7].Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 2008; 9:33-46; PMID:18097444; http://dx.doi.org/ 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- [8].Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci U S A 2005; 102:5363-7; PMID:15809444; http://dx.doi.org/ 10.1073/pnas.0501168102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wigge PA, Kilmartin JV. The Ndc80p complex from Saccharomyces cerevisiae contains Conserved centromere components and has a function in chromosome segregation. J Cell Biol 2001; 152:349-60; PMID:11266451; http://dx.doi.org/ 10.1083/jcb.152.2.349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yao X, Abrieu A, Zheng Y, Sullivan K, Cleveland D. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol 2000; 2:484-91; PMID:10934468; http://dx.doi.org/ 10.1038/35019518 [DOI] [PubMed] [Google Scholar]
- [11].Shonn MA, Mccarroll R, Murray AW. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science 2000; 289:300-3; PMID:10894778; http://dx.doi.org/ 10.1126/science.289.5477.300 [DOI] [PubMed] [Google Scholar]
- [12].Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol 2003; 161:281-94; PMID:12707311; http://dx.doi.org/ 10.1083/jcb.200208092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nasmyth K, Haering CH. Cohesin: Its roles and mechanisms. Annu Rev Genet 2009; 43:525-58; PMID:19886810; http://dx.doi.org/ 10.1146/annurev-genet-102108-134233 [DOI] [PubMed] [Google Scholar]
- [14].Howard-Till RA, Lukaszewicz A, Novatchkova M, Loidl J. A single cohesin complex performs mitotic and meiotic functions in the protist Tetrahymena. PLoS Genetics 2013; 9:e1003418; PMID:23555314; http://dx.doi.org/ 10.1371/journal.pgen.1003418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Signon L. New insights into the regulation of anaphase by mitotic cyclins in budding yeast. Cell Cycle 2014; 10:1655-68; http://dx.doi.org/ 10.4161/cc.10.10.15632 [DOI] [PubMed] [Google Scholar]
- [16].Surana U, Robitsch H, Price C, Schuster T, Fitch I, Futcher AB, Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell 1991; 65:145-61; PMID:1849457; http://dx.doi.org/ 10.1016/0092-8674(91)90416-V [DOI] [PubMed] [Google Scholar]
- [17].Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 2013; 140:3079-93; PMID:23861057; http://dx.doi.org/ 10.1242/dev.091744 [DOI] [PubMed] [Google Scholar]
- [18].Nugent JH, Alfa CE, Young T, Hyams JS. Conserved structural motifs in cyclins identified by sequence analysis. J Cell Sci 1991; 99:669-74; PMID:1834684 [DOI] [PubMed] [Google Scholar]
- [19].Stover NA, Rice JD. Distinct cyclin genes define each stage of ciliate conjugation. Cell Cycle 2014; 10:1699-701; PMID:NOT_FOUND; http://dx.doi.org/ 10.4161/cc.10.10.15548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].La H, Li J, Ji Z, Cheng Y, Li X, Jiang S, Venkatesh PN, Ramachandran S. Genome-wide analysis of cyclin family in rice (Oryza Sativa L.). Mol Genet Genomics 2006; 275:374-86; PMID:16435118; http://dx.doi.org/ 10.1007/s00438-005-0093-5 [DOI] [PubMed] [Google Scholar]
- [21].Hu X, Cheng X, Jiang H, Zhu S, Cheng B, Xiang Y. Genome-wide analysis of cyclins in maize (Zea mays). Genetics Mol Res 2010; 9:1490-503; http://dx.doi.org/ 10.4238/vol9-3gmr861 [DOI] [PubMed] [Google Scholar]
- [22].Wang G, Kong H, Sun Y, Zhang X, Zhang W, Altman N, DePamphilis CW, Ma H. Genome-wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin-like proteins. Plant Physiol 2004; 135:1084-99; PMID:15208425; http://dx.doi.org/ 10.1104/pp.104.040436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Orias E, Cervantes MD, Hamilton EP. Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res Microbiol 2011; 162:578-86; PMID:21624459; http://dx.doi.org/ 10.1016/j.resmic.2011.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Martindale DW, Allis CD, Bruns PJ. Conjugation in Tetrahymena thermophila: A temporal analysis of cytological stages. Exp Cell Res 1982; 140:227-36; PMID:7106201; http://dx.doi.org/ 10.1016/0014-4827(82)90172-0 [DOI] [PubMed] [Google Scholar]
- [25].Mochizuki K, Novatchkova M, Loidl J. DNA double-strand breaks, but not crossovers, are required for the reorganization of meiotic nuclei in Tetrahymena. J Cell Sci 2008; 121:2148-58; PMID:18522989; http://dx.doi.org/ 10.1242/jcs.031799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lukaszewicz A, Shodhan A, Loidl J. Exo1 and Mre11 execute meiotic DSB end resection in the protist Tetrahymena. DNA Repair 2015; 35:137-43; PMID:26519827; http://dx.doi.org/ 10.1016/j.dnarep.2015.08.005 [DOI] [PubMed] [Google Scholar]
- [27].Shodhan A, Lukaszewicz A, Novatchkova M, Loidl J. Msh4 and Msh5 function in SC-independent chiasma formation during the streamlined meiosis of Tetrahymena. Genetics 2014; 198:983-93; PMID:25217051; http://dx.doi.org/ 10.1534/genetics.114.169698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Loidl J, Scherthan H. Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J Cell Sci 2004; 117:5791-801; PMID:15522890; http://dx.doi.org/ 10.1242/jcs.01504 [DOI] [PubMed] [Google Scholar]
- [29].Howard-Till RA, Lukaszewicz A, Loidl J. The recombinases Rad51 and Dmc1 play distinct roles in DNA break repair and recombination partner choice in the meiosis of Tetrahymena. PLoS Genetics 2011; 7:193-202;; http://dx.doi.org/ 10.1371/journal.pgen.1001359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Orias E, Hamilton EP, Orias JD. Tetrahymena as a laboratory organism: Useful strains, cell culture, and cell line maintenance. Methods Cell Biol 2000; 62:189-211; PMID:10503191; http://dx.doi.org/ 10.1016/S0091-679X(08)61530-7 [DOI] [PubMed] [Google Scholar]
- [31].Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One 2009; 4:e4429; PMID:19204800; http://dx.doi.org/ 10.1371/journal.pone.0004429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Xiong J, Lu X, Lu Y, Zeng H, Yuan D, Feng L, Chang Y, Bowen J, Gorovsky M, Fu C, et al.. Tetrahymena Gene Expression Database (TGED): A resource of microarray data and co-expression analyses for Tetrahymena. Sci China Life Sci 2011; 54:65-7; PMID:21253873; http://dx.doi.org/ 10.1007/s11427-010-4114-1 [DOI] [PubMed] [Google Scholar]
- [33].Xiong J, Lu X, Zhou Z, Chang Y, Yuan D, Tian M, Zhou Z, Wang L, Fu C, Orias E, et al.. Transcriptome Analysis of the Model Protozoan, Tetrahymena thermophila, Using Deep RNA Sequencing. PLoS One 2012; 7:e30630; PMID:22347391; http://dx.doi.org/ 10.1371/journal.pone.0030630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Huang D, Friesen H, Andrews B. Pho85, a multifunctional cyclin-dependent protein kinase in budding yeast. Mol Microbiol 2007; 66:303-14; PMID:17850263; http://dx.doi.org/ 10.1111/j.1365-2958.2007.05914.x [DOI] [PubMed] [Google Scholar]
- [35].Nguyen TB, Manova K, Capodieci P, Lindon C, Bottega S, Wang XY, Refik-Rogers J, Pines J, Wolgemuth DJ, Koff A. Characterization and expression of mammalian cyclin b3, a prepachytene meiotic cyclin. J Biol Chem 2002; 277:41960-9; PMID:12185076; http://dx.doi.org/ 10.1074/jbc.M203951200 [DOI] [PubMed] [Google Scholar]
- [36].Ozaki Y, Saito K, Shinya M, Kawasaki T, Sakai N. Evaluation of Sycp3, Plzf and Cyclin B3 expression and suitability as spermatogonia and spermatocyte markers in zebrafish. Gene Expr Patterns 2011; 11:309-15; PMID:21402175; http://dx.doi.org/ 10.1016/j.gep.2011.03.002 [DOI] [PubMed] [Google Scholar]
- [37].Zhang T, Qi ST, Huang L, Ma XS, Ouyang YC, Hou Y, Shen W, Schatten H, Sun QY. Cyclin B3 controls anaphase onset independent of spindle assembly checkpoint in meiotic oocytes. Cell Cycle 2015; 14:2648-54; PMID:26125114; http://dx.doi.org/ 10.1080/15384101.2015.1064567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nanney DL. Nucleo-Cytoplasmic interaction during conjugation in tetrahymena. Biological Bulletin 1953; 105:133-48;; http://dx.doi.org/ 10.2307/1538562 [DOI] [Google Scholar]
- [39].Gaertig J, Cole ES. The role of cortical geometry in the nuclear development of Tetrahymena thermophila. J Eukaryot Microbiol 2000; 47:590-6; PMID:11128713; http://dx.doi.org/ 10.1111/j.1550-7408.2000.tb00095.x [DOI] [PubMed] [Google Scholar]
- [40].Moore AT, Rankin KE, von Dassow G, Peris L, Wagenbach M, Ovechkina Y, Andrieux A, Job D, Wordeman L. MCAK associates with the tips of polymerizing microtubules. J Cell Biol 2005; 169:391-7; PMID:15883193; http://dx.doi.org/ 10.1083/jcb.200411089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sun SC, Kim NH. Spindle assembly checkpoint and its regulators in meiosis. Hum Reprod Update 2012; 18:60-72; PMID:22086113; http://dx.doi.org/ 10.1093/humupd/dmr044 [DOI] [PubMed] [Google Scholar]
- [42].Mochizuki K. High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene 2008; 425:79-83; PMID:18775482; http://dx.doi.org/ 10.1016/j.gene.2008.08.007 [DOI] [PubMed] [Google Scholar]
- [43].Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, et al.. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res 2015; 43:D213-21; PMID:25428371; http://dx.doi.org/ 10.1093/nar/gku1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389-402; PMID:9254694; http://dx.doi.org/ 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al.. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23:2947-8; PMID:17846036; http://dx.doi.org/ 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- [46].Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30:2725-9; PMID:24132122; http://dx.doi.org/ 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 2007; 23:127-8; PMID:17050570; http://dx.doi.org/ 10.1093/bioinformatics/btl529 [DOI] [PubMed] [Google Scholar]
- [48].Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 2011; 39:W475-8; PMID:21470960; http://dx.doi.org/ 10.1093/nar/gkr201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 2014; 42:W320-4; PMID:24753421; http://dx.doi.org/ 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mar JC, Wells CA, Quackenbush J. Defining an informativeness metric for clustering gene expression data. Bioinformatics 2011; 27:1094-100; PMID:21330289; http://dx.doi.org/ 10.1093/bioinformatics/btr074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wu Z, Wang X, Zhang X. Using non-uniform read distribution models to improve isoform expression inference in RNA-Seq. Bioinformatics 2011; 27:502-8; PMID:21169371; http://dx.doi.org/ 10.1093/bioinformatics/btq696 [DOI] [PubMed] [Google Scholar]
- [52].Kim D, Salzberg SL. TopHat-Fusion: An algorithm for discovery of novel fusion transcripts. Genome Biol 2011; 12:R72; PMID:21835007; http://dx.doi.org/ 10.1186/gb-2011-12-8-r72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012; 7:562-78; PMID:22383036; http://dx.doi.org/ 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Team RC. R: A Language and Environment for Statistical Computing Vienna, Austria: R Foundation for Statistical Computing, 2015 [Google Scholar]
- [55].Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al.. Gene Ontology: Tool for the unification of biology. Nat Genet 2000; 25:25-9; PMID:10802651; http://dx.doi.org/ 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005; 21:3448-9; PMID:15972284; http://dx.doi.org/ 10.1093/bioinformatics/bti551 [DOI] [PubMed] [Google Scholar]
- [57].Lukaszewicz A, Howard-Till RA, Novatchkova M, Mochizuki K, Loidl J. MRE11 and COM1/SAE2 are required for double-strand break repair and efficient chromosome pairing during meiosis of the protist Tetrahymena. Chromosoma 2010; 119:505-18; PMID:20422424; http://dx.doi.org/ 10.1007/s00412-010-0274-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.