ABSTRACT
FXR1 belongs to a family of RNA-binding proteins that play critical roles in post-transcriptional regulation of gene expression in immunity, development and cancer. FXR1 is associated with regulation of specific mRNAs in myocytes and macrophages. In quiescent cells (> 24 h of extended serum-starvation, ∼30-48 h or more), a spliced isoform of FXR1, FXR1a, promotes translation of the cytokine TNFα, independent of the effects of RNA levels. Here we examined the role of FXR1 in THP1 human monocytic leukemic cells that were grown in serum, as well as in early (24 h) serum-starvation conditions that demonstrates differences in gene expression mechanisms and is distinct from quiescent (> 24 h extended serum-starvation) cells. Global RNA profiling, conducted to investigate the role of FXR1 on mRNA levels, revealed that FXR1 affects levels of specific mRNAs in serum-grown and in early 24 h serum-starvation conditions. FXR1 decreases levels of several mRNAs, including as previously identified, CDKN1A (p21CIP1 or p21) mRNA in serum-grown cells. Interestingly, we find that FXR1 positively regulates mRNA levels of specific cytokines and chemokines in serum-grown and in early 24 h serum-starvation conditions. These include IL1β and CCL2 that control cell migration. Accordingly, depletion and overexpression of FXR1 decreased and increased levels of CCL2 mRNA. Consistent with the reduced levels of IL1β, CCL2 and other chemokines upon FXR1 depletion, our data reveal that depletion of FXR1 decreases the ability of these cells to induce cell migration of neighboring monocytic cells. These data reveal a new role of FXR1 in controlling induction of monocyte migration.
KEYWORDS: cell migration, chemokines, FXR1, gene expression, monocyte, mRNA
Introduction
RNA binding proteins play critical roles in post-transcriptional regulation of gene expression in immunity, development and cancer.1,2 The RNA binding protein, Fragile-X-Mental-Retardation-syndrome-Related protein 1 (FXR1)3-5 is overexpressed and associated with poor clinical outcomes in multiple cancers.6 FXR1 is similar to Fragile-X-Mental Retardation Protein 1 (FMR1),4,7-14 and is implicated at multiple levels of post-transcriptional control, including translation, mRNA stability and transport.7-10, 12-13, 15-18 FXR1 is associated with negative regulation of specific growth factor and cytokine mRNAs in myocytes5,19-22 and macrophages,16 which can control development, cell differentiation and cell state specific functions. However, our previous studies demonstrated that a spliced isoform of FXR1, FXR1a, promotes specific mRNA translation independent of RNA levels, in association with an altered microRNP (microRNA-protein complex), in distinct conditions, such as oocytes and quiescent (> 24 h, 30–48 h extended serum-starved) mammalian cells.17,23-25 In cells that are induced to quiescence by extended serum-starvation (> 24 h), FXR1a isoform promotes translation of Tumor Necrosis Factor α (TNFα) cytokine23 (independent of RNA level changes), which can regulate monocyte cell state, signaling and differentiation, and tumors.26-32 In early (24 h) serum-starved human THP1 acute monocytic leukemic cells, our data recently revealed global changes in gene expression mechanisms33 that are distinct from those in quiescent (extended > 24 h serum-starved) cells. Cytokines like TNFα are transcribed and detected in these conditions; however, the role of FXR1 in regulating specific mRNA levels and expression in serum grown and in early 24 h serum-starved THP1 cells remains to be outlined. Given the effect of FXR1 on cell differentiation5,20-22 and cancer6 and its regulation of TNFα cytokine mRNA expression in quiescent, extended serum-starved cells, we examined the role of FXR1 in regulating mRNA levels in serum grown cells and in early (24 h) serum-starved monocytic leukemic cells.
Here we find that FXR1 is required for regulating RNA levels and thereby, expression of chemokines and cytokines that induce cell migration—IL1β and CCL2.34-39 Consistently, we find that FXR1 is required for the ability of monocytes to induce cell migration of neighboring cells. These data reveal a new role of FXR1 in controlling induction of cell migration via regulation of mRNA levels and thereby, gene expression in monocytic leukemic cells—with implications for monocyte functions and cell signaling in immune/inflammatory response, and in cancer.
Results
Global transcriptome profiling reveals that FXR1 depletion affects the levels of distinct mRNAs in THP1 cells
To investigate the role of FXR1, we created stable THP1 monocytic cell lines that inducibly expressed40 a control shRNA (shCtrl) or a specific shRNA to knock down FXR1 (shFXR1). FXR1 upregulates the cytokine, TNFα, at the translation level (independent of RNA levels) in quiescent cells induced by extended serum-starvation (30-48 h serum-starvation). Since TNFα mRNA is transcribed and detectable in early (24 h) serum-starved cells17,23 where gene expression mechanisms are altered33 and distinct from those in quiescent (extended > 24 h serum-starved) cells, we examined the effect of FXR1 on mRNA levels in early (24 h) serum-starved as well as in serum-grown THP1 human monocytic leukemic cells. The cells were induced with doxycycline to express the shRNA for 3 d and further grown in serum media or serum-starved for 24 h. Western blot analysis showed that FXR1 was effectively depleted in both conditions at the protein level and RNA levels (Fig. 1A–C).
Figure 1.
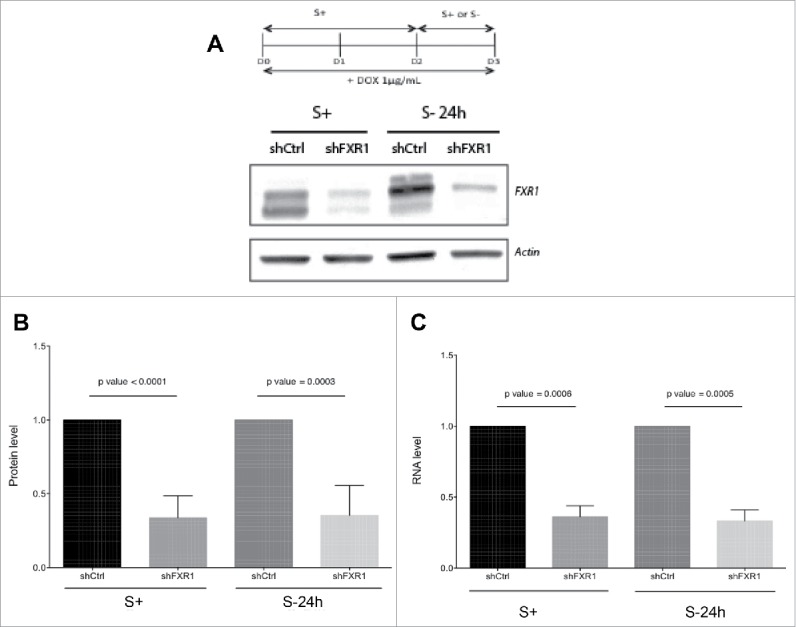
FXR1 knockdown in serum grown and serum-starved THP1 monocytic leukemic cells. (A) Western blot analysis of FXR1 in THP1 stable cell lines expressing control shRNA (shCtrl) or shRNA against FXR1 (shFXR1) after 3 d of doxycycline induction in serum containing media (S+) or serum-free media for 24 h (S- 24 h). (B) Quantitation of multiple Western blots using image J. The data represented are the mean and SD values of 6 experiments. p-values were calculated by one tailed paired t test. (C) RNA level after knockdown of FXR1 evaluated by qRT-PCR. The data represent the mean and SD of 5 experiments p-values were calculated by one-tailed paired t-test.
FXR1 can regulate mRNA levels16 apart from translation,23 and could thereby, affect monocyte functions. We therefore, investigated the mRNAs that are affected by FXR1 depletion in serum grown and 24 h serum-starved cells. Total RNA from shCtrl and shFXR1 cells, grown in serum containing media or serum-starved for 24 h, were subject to global mRNA profiling using Affymetrix GeneChip Human Gene ST 2.0 Arrays (Fig. 2A, Table S1). Profiling results were validated by qRT-PCR and revealed decreased FXR1 levels as well as altered levels of distinct mRNAs in serum-grown and serum-starved cells (Fig. 2B-C, S1, Table S2A-B). An increase in CDKN1A (p21CIP1 or p21) mRNA levels is observed in serum-grown cells upon FXR1 knockdown (Fig. 2B, S+), which is in agreement with previously published data demonstrating CDKN1A increase upon FXR1 depletion.22 These data reveal that distinct mRNAs are regulated in the absence of FXR1 in THP1 cells.
Figure 2.
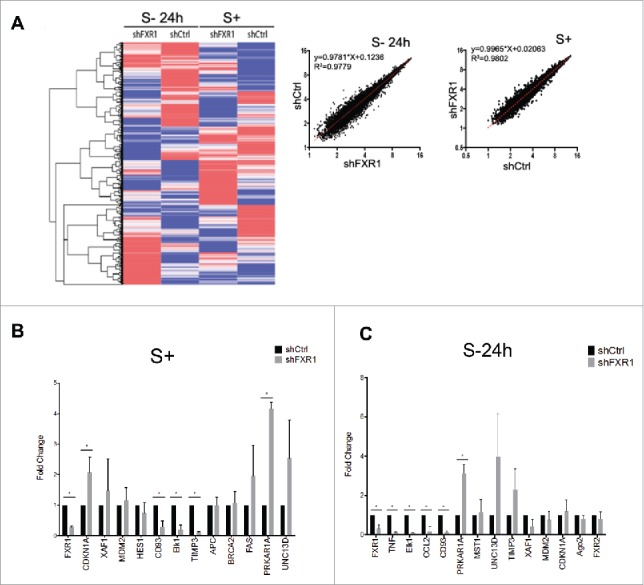
Global gene expression profiling of FXR1 regulated mRNAs by microarray analysis in control and FXR1 knockdown THP1 monocytes. (A) (Left) Heatmap analysis of microarray data (Table S1) from THP1 control (shCtrl) and FXR1 knockdown (shFXR1) cells grown in serum containing media (S+) or serum-starved for 24 h (S-24 h). Heatmap representation and clustering (Euclidean) was done using GENE-E software. (Right) The logarithmic value of individual RNAs in FXR1 knockdown cells (shFXR1) was plotted against those values in control cells (shCtrl), grown in serum containing media (S+) or serum-starved for 24 h (S-24 h). R square and curve equations were calculated using linear regression model with prism6. (B-C) Fold change values from qRT-PCR of a subset of targets identified from the microarray from serum grown (S+) and serum-starvation (S- 24h) conditions (Tables S2A-B). The data plotted are the mean and SD of 4 biological replicates. Statistical significance was determined with t test comparison using the Holm-Sidak method.
Depletion of FXR1 leads to decreased cytokine and chemokine mRNAs and reduced gene expression of cell migration regulators, IL1β and CCL2
Gene ontology (GO) analysis of the microarray data revealed that immune genes, including several cytokines and chemokines, were significantly affected by the decrease of FXR1 (Fig. S2, 2C, 3A–B, Table S3). These effects were more clearly observed in 24 h serum-starved cells where many of the cytokine and chemokine mRNAs are transcribed and detectable in comparison to serum-grown cells. The cytokines and chemokines affected by FXR1, include genes such as Interleukin 1β (Il1β), chemokine (C-C motif) ligand 2 (CCL2) and CCL3 (Fig. 3A–B, Table S3) that affect cell migration and invasion,34-39 suggesting that FXR1 regulates distinct pathways. The chemokine, CCL2 mRNA, is detectable in serum-grown as well as 24 h serum-starved cells and was observed to decrease upon FXR1 knockdown (Fig. 3C). If FXR1 is required for the expression of these chemokines, then CCL2 mRNA should be upregulated upon overexpression of FXR1. Consistently, we find that CCL2 mRNA is significantly increased in THP1 cells that stably overexpress FXR1a, in both serum-grown and 24 h serum-starved conditions (Fig. 3D–E, FXR1a ox). These data support that FXR1 regulates the levels and expression of specific cytokines and chemokines.
Figure 3.
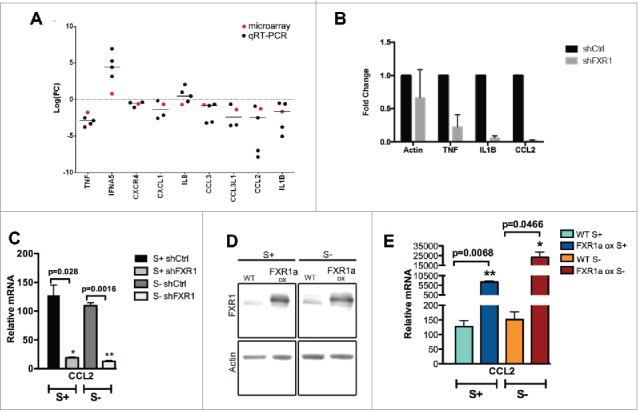
Immune response genes, including cytokines and chemokines, are regulated by FXR1. (A) Gene Ontology (GO) analysis for differentially expressed genes was performed using the DAVID tool, as previously conducted.33 Immune response associated genes in serum-starved cells (S-24 h) are observed to be significantly affected by FXR1 depletion (Fig. S2, Tables S2B, S3). Analysis of RNA expression for a subset of mRNAs related to immune response in serum-starved FXR1 depleted cells compared to serum-starved shCtrl cells: the log base 2 of the fold change values for qRT-PCR (black dot) and microarray (red dot) are plotted. (B) qRT-PCR analysis of select cytokine and chemokine mRNAs in serum-starved cells showing the fold decrease upon FXR1 knockdown. (C) qRT-PCR analysis of CCL2 mRNA normalized to tRNA-lys RNA levels in control (shCtrl) and FXR1 knockdown (shFXR1) cells grown in serum (S+) and 24 h serum-starvation (S-) conditions. The average of 3 technical replicates is shown with SEM as error bars. p-values were calculated by 2-tailed paired t-test. (D) Western blot analysis of FXR1 in THP1 stable cell lines without (WT) or with FXR1a constitutive overexpression (FXR1a ox), grown in serum (S+) and 24 h serum-starvation (S- 24 h) conditions. (E) qRT-PCR analysis of CCL2 mRNA normalized to tRNA-lys RNA levels in control (WT) and FXR1 overexpression (FXR1a ox) cells grown in serum (S+) and 24 h serum-starvation (S-) conditions. The average of 3 technical replicates is shown with SEM as error bars. p-values were calculated by 2-tailed paired t-test.
Il1β mRNA and protein levels are decreased upon FXR1 depletion
The transcript of the cytokine, TNFα, is more easily detectable in 24 h serum-starved THP1 cells where TNFα is induced compared to serum grown cells.17,23 TNFα mRNA levels decreased in 24 h serum-starved THP1 cells upon FXR1 depletion (Fig. 3A–B, Table S3). The cytokine, Il1β mRNA, is also not significantly detectable in serum-grown cells but is detected in 24 h serum-starved THP1 cells. Like TNFα, Il1β mRNA levels also decreased in the absence of FXR1, as observed from the microarray analysis, and as validated by qRT-PCR (Tables S1, S2B, S3, Fig. 3B). Consistent with the regulation of Il1β mRNA levels, Western blot analysis revealed that Il1β protein levels are present in wildtype 24 h serum-starved cells but are significantly decreased upon FXR1 depletion (Fig. 4A). TNFα was concurrently tested—as a positive control that is regulated by FXR1—and consistently revealed a significant decrease in TNFα protein levels in FXR1 depleted cells by Western blot analysis (Fig. 4B). These data suggest that Il1β cytokine mRNA levels and thereby, Il1β protein levels, are regulated by FXR1 in 24 h serum-starved THP1 cells.
Figure 4.
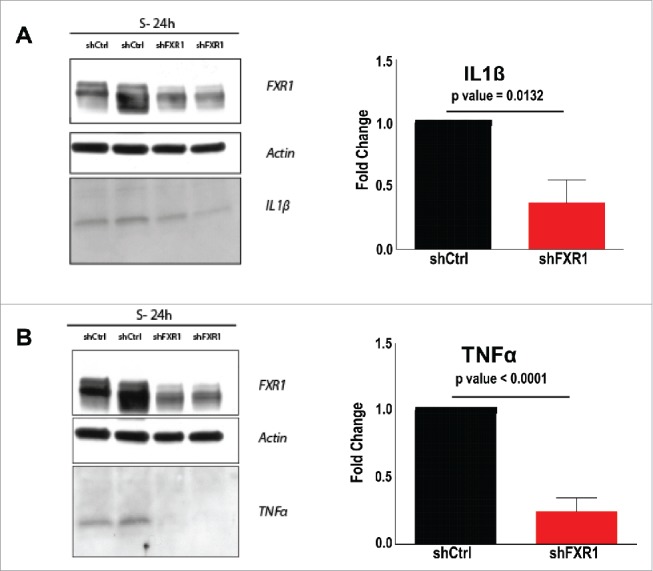
Il1β levels are decreased upon FXR1 knockdown. (A-B) Western blot analysis of IL1β and TNFα upon FXR1 depletion. Cell lysate and medium from 24 h serum-starved cells were concentrated using Strataclean resin and the eluates were analyzed by Western blotting. Western blot analysis of (A) IL1β and (B) TNFα, with actin as loading control. The average of 3 replicates is shown with SEM as error bars. p-values were calculated by 2-tailed unpaired t-test.
FXR1 depletion decreases the ability of monocytes to induce cell migration of neighboring cells
FXR1 affects differentiation5,19-21 and macrophage physiology.16 Moreover, FXR1 depletion resulted in altered expression of TNFα23 and other cytokine and chemokine mRNAs16 such as IL1β and CCL2 (Figs. 3–4) that are involved in monocyte functions such as neighboring cell migration. We therefore tested whether FXR1 affects induction of cell migration by THP1 monocytic cells. Cell migration assays were performed in transwells using wildtype and FXR1 knockdown cells to test the migration of another THP1 monocyte cell line that stably expresses GFP for visualization.41 The top chambers of the transwells were filled with equal numbers of a THP1 cell line that stably expresses GFP. Equal numbers of 24 h serum-starved control shCtrl cells or shFXR1 cells (that do not express GFP) were placed in the bottom chambers supplemented with 10% serum. These cells were tested for their ability to produce chemokines to promote migration of the GFP expressing cells from the top chamber into the bottom. As shown in Figs. 5A–C, FXR1 depletion severely reduced cell migration, compared to shCtrl cells. The decreased cytokine and chemokine mRNAs, observed in FXR1 knockdown cells (Fig. 3), are critical for cell migration,34-39 explaining the decreased induction of cell migration observed in FXR1 depleted cells (Fig. 5A–C). These data support that FXR1 regulates the induction of cell migration of THP1 monocytes.
Figure 5.
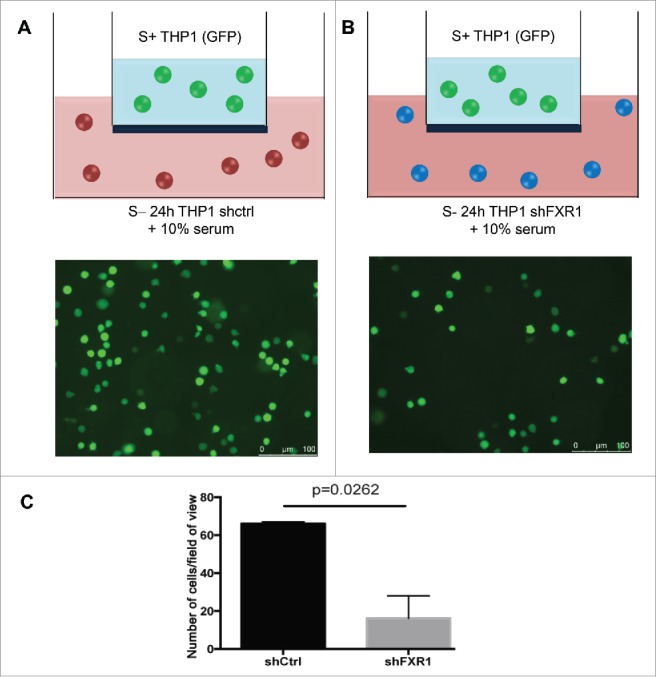
FXR1 knockdown leads to decreased induction of cell migration by THP1 monocytes Cell migration assays were performed in transwells to test the migration of a THP1 monocytic cell line that stably expresses GFP for visualization.41 Cell migration assay was performed using THP1 cell lines expressing (A) control shRNA (shCtrl) or (B) shRNA against FXR1 (shFXR1), which were grown in serum-starvation conditions (S-24 h), and then placed in the bottom chambers and supplemented with 10% serum. The top chamber was filled with a THP1 monocyte cell line that stably expresses GFP. (C) Graph representing cell migration count using imageJ of GFP positive cells that migrated through the membrane filter in (A-B). p-values were calculated by one-tailed unpaired t-test.
Discussion
FXR1 is an RNA binding protein that plays important roles in differentiation, development and cancer.5,6, 16,19-22, 24 Our previous data demonstrated a role for an isoform of FXR1 in quiescent (extended > 24 h serum-starved) cells, in upregulating translation (independent of RNA level changes) of a critical cytokine, TNFα17 that can promote monocyte functions, signaling and differentiation.26,28-31 In early (24 h) serum-starved THP1 acute monocytic leukemic cells, our data revealed altered gene expression mechanisms33 that are distinct from those in quiescent (extended > 24 h serum-starved) cells. Cytokines like TNFα are transcribed and detected in these conditions; however, the role of FXR1 in regulating specific mRNA levels and expression in serum grown and in early (24 h) serum-starved THP1 cells remained to be explored. We therefore, examined the role of FXR1 in regulating mRNA levels in monocytic leukemic cells in these conditions. We find that FXR1 regulates the mRNA levels of a number of genes—including promoting the RNA levels and thereby, expression of specific cytokines and chemokines that are associated with inducing cell migration.34-39 Consistently, we find that FXR1 depletion reduces the ability of such monocytes to induce cell migration of neighboring cells, revealing a new role for this RNA binding protein and one consequence of its role in regulation of gene expression.
FXR1 is an RNA binding protein implicated at multiple levels of post-transcriptional control, including translation, mRNA stability and transport.7-10, 12-13,15-18 FXR1 has been found to affect mRNA levels and thereby, regulate gene expression.6,16, 18,22 CDKN1A mRNA was previously demonstrated to be downregulated by FXR122; accordingly, our microarray data also showed that depletion of FXR1 in serum-grown cells promotes CDKN1A mRNA levels (Fig. 2B, S+, Table S2A). However, our previous data reveal that FXR1 can also promote expression of specific genes, such as TNFα in quiescent (extended > 24 h serum-starved) cells17 and Myt1 mRNA in oocytes.24 Additionally, recent publications reveal that FXR1 promotes expression of specific mRNAs such as Epithelial cell transforming 2 (ECT2) in lung cancer cells.6 Consistently, we find that FXR1 is also required to promote the RNA levels and thereby, expression of specific mRNAs in monocytic leukemic THP1 cells in serum grown and 24 h serum-starved conditions (Figs. 2B–C, 3A–B).
Our previous data demonstrated a role for FXR1 in promoting expression of the cytokine, TNFα, in cells induced to quiescence by extended serum-starvation (30-48 h, > 24 h of serum-starvation), at the translation level, independent of changes in RNA levels.17 Interestingly, our microarray data revealed several genes that are upregulated at the RNA level by FXR1 and consequently decreased upon its depletion in both serum grown cells, as well as in early (24 h) serum-starved cells (Fig. 2B–C, 3A–B, Tables S1, S2A-B), where many of these mRNAs are induced and transcribed. Early (24 h) serum-starved cells are distinct from quiescent (> 24 h extended serum-starved) cells and represent a transitional stage where the cells decide to enter quiescence, or return to proliferation, or enter other cell states such as differentiation. Consistently, we previously found that global gene expression mechanisms are altered in early (24 h) serum-starved cells in a manner distinct from that observed in quiescent (> 24 h extended serum-starved) cells.33 The identified regulation—with FXR1 promoting specific mRNA levels in early (24 h) serum-starvation conditions, where many of these RNAs are induced and transcribed—may serve as an early response/initial regulatory mechanism, prior to regulation of specifically increased mRNAs by different mechanisms (translation upregulation independent of RNA level changes) upon subsequent extended serum-starvation/entry into quiescence. Accordingly, TNFα mRNA levels and thereby, protein levels, are increased in early (24 h) serum-starved cells by FXR1—and then transitionally upregulated, independent of mRNA levels, in extended > (24 h) serum-starved, quiescent cells.17 These distinct mechanisms—regulation of mRNA and thereby protein levels in early (24 h) serum-starved cells, and regulation of translation/protein levels independent of mRNA level changes in quiescent (extended >24 h serum-starved) cells—may provide alternative means of upregulating specific gene expression, and at potentially different levels, as required in these distinct conditions.
These mRNAs not only include the previously identified TNFα17 but also other cytokines like IL1β, and chemokines like CCL2 (Fig. 3, S2, Table S3). The cytokines and chemokines that are regulated by FXR1 control cell migration and invasion,34-39 indicating the specificity of FXR1 in regulating distinct pathways. Apart from this role of FXR1 in promoting the expression of TNFα and IL1β cytokines in serum-starved THP1 monocytes, FXR1 has been shown previously to downregulate the expression of these cytokines in LPS stimulated mouse macrophages,16 indicating that FXR1 can differentially regulate gene expression in distinct conditions to elicit specific gene expression outcomes.
In FXR1 depleted cells, the downregulated levels of cytokines such as IL1β mRNA (Fig. 3B) is also accompanied by decreased levels of IL1β protein (Fig. 4A), indicating reduced levels of gene expression due to diminished mRNA levels, and subsequently, decreased protein levels. One possible mechanism could be the involvement of conserved elements such as 3′-UTR AU-rich elements, since FXR1 is known to interact with AU-rich elements that are present not only in TNFα mRNA but also in IL1β, CCL2 and other chemokine and cytokine mRNAs.16,17, 42,43 Alternative mechanisms including competition with other RNA regulators, other sequence elements in the 3′-UTRs of these specific mRNAs, mRNA export, and regulation at the translation level may be additionally involved and cannot be ruled out. The regulation of these genes by FXR1 could be indirect—via regulation of mRNA levels and translation by other unknown effectors that may in turn be controlled directly by FXR1 and remain to be elucidated.
FXR1 affects critical cellular functions in development, differentiation, immunity and cancer.5,6 16,19-22, 24 Deregulation of FXR1 levels affects cell growth and cancer via control over cell cycle and growth factor gene expression.6,22 FXR1 also affects the gene expression of immune modulators at the post-transcriptional level.16,17 Our data reveal that FXR1 is required for upregulated mRNA levels and thereby, expression of specific chemokines and cytokines, such as CCL2 and IL1β that induce cell migration and play critical roles in invasion, cell-to-cell communication, differentiation, and inflammatory response.34-39 Consequently, depletion of FXR1 decreases such chemokines and cytokines, and reduces induction of cell migration by FXR1 depleted monocytes. These data reveal a new consequence for the role of FXR1 in gene expression regulation, in controlling the induction of cell migration by monocytic cells, which plays important roles in immune cell functions and cell-cell communications in inflammation and in cancer.
Methods
Cell culture
THP1 monocytic leukemic cell line was cultured in RPMI 1640 (Life Technologies), supplemented with 10% fetal bovine serum (Life Technologies), 100 nM penicillin/streptomycin (Life Technologies), 2mM L-glutamine (Life Technologies). HEK293T cell line was cultivated in DMEM (Life Technologies) supplemented with 10% fetal bovine serum (life technologies), 100 nM penicillin/streptomycin (Life Technologies), 2mM L-glutamine (Life Technologies). In S- (24 h) conditions the cells are cultivated for (24 h) in medium without serum23; both conditions were maintained at 5% CO2 and 37°C.
Lentiviral transduction
Lentiviruses were produced in 293T packaging cells with FXR1 shRNA V3THS_340198 (used in all figures), V2THS_18038, and pTRIPZ vectors (GE Dharmacon, Open Biosystems) as conducted previously.33 Supernatants were collected every (24 h) on 3 consecutive days starting 24 h after transfection, and viral particles were concentrated by centrifugation at 15,000 rpm for 2 h at 4°C. Approximately 10,000 THP1 cells were seeded (per well) in a 96-well culture plate and infected with 10 μl of concentrated virus in the presence of polybrene (7 μg/ml). The cells were spun down at 1000g for 30 minutes at 37°C. The medium was replaced and cells were transferred to a bigger plate. After 3 d of culture, 1μg/mL of puromycin was added to the culture and the medium was changed every 48 h.
Cell migration assay
Cell migration assays were performed as described41 to test the migration of a THP1 monocyte cell line that stably expresses GFP for visualization. Transwell membranes (Sigma, 8 mm pore size) were washed with PBS and pre-incubated with serum-free RPMI media for 30 min at 37°C. After removing media, equal numbers of THP1 cells that stably express GFP were added to the upper chambers. Following serum starvation for 24 h, equal numbers of wild-type or FXR1 knockdown THP1 cells were plated in the lower chambers and supplemented with 10% serum. The number of migrated THP1 cells expressing GFP in the lower chambers was measured using the imageJ software.
Microarray
Total RNA was extracted with 3 volumes of TRIzol (Invitrogen) and cleaned using RNeasy mini kit (Qiagen). The synthesized cDNA probe (WT Expression Kit; Ambion) was hybridized to GeneChip Human Gene ST 2.0 Array (Affymetrix) by the Partners Healthcare Center for Personalized Genetic Medicine Microarray facility. A 1.5-fold change in microarray expression was used as the cutoff to determine differentially regulated genes (Tables S1-3, Figs 2–3). Gene Ontology (GO) analysis for differentially expressed genes was performed with the DAVID tool as previously conducted.33
Plasmids
pTRIPZ constructs40 against control and FXR1 (V2THS_18042; V2THS_18038; V3THS_340198) were obtained from Open Biosystems, Thermo. pTRIPZ vector, which expresses an shRNA with miR30a pri-cursor-miR sequences, was used as control (shCtrl). FXR1a was previously described in reference 17 (FXR1a is labeled according to reference 3; GenBank: BC028983.1, which is also called transcript variant 2 or isoform b in GenBank, Accession version: NM_001013438.2).
Western blot analysis of cytokines
THP1 cells were grown in medium containing serum and 1 μg/ml doxycycline for 2 d and then switched to serum-starvation conditions with 1 μg/ml doxycycline for 24 h. The cells were harvested by centrifugation, resuspended in lysis buffer and snap frozen in dry ice. The lysates were sonicated for 10 min (Bioruptor high intensity with 30 s on and 30 s off). After 10 min of centrifugation at 10000 rpm at 4°C, the protein concentration of the supernatant was measure using Bradford assay. The culture media (500 μl) that was separated from the centrifuged cells was saved and incubated with 50 μl of Strataclean resin (Stratagene) for 20 min at 4°C to bind the secreted proteins, including cytokines and chemokines in the culture media. The resin was washed 2 times with PBS and the proteins were eluted by boiling the resin for 10 min in 1x SDS loading buffer. After 2 min of centrifugation at 5000 g, the supernatant (which constitutes the secreted cytokines and chemokines present in the culture media) was combined with the cell lysate and analyzed by SDS-PAGE.
qRT-PCR analysis
RNA was extracted using Trizol (Invitrogen) according to the manufacturer's protocol. cDNA synthesis was performed using Random Primers (Invitrogen) using M-MULV reverse transcriptase (NEB). Quantitative real-time PCR (qRT-PCR) was performed using primers (IL1β-for: TACCTGTCCTGCGTGTTGAA; IL1β-rev: TCTTTGGGTAATTTTTGGGATCT; CCL2-for: AGTCTCTGCCGCCCTTCT; CCL2-rev: GTGACTGGGGCATTGATTG; FXR1-for: AGCTGCGACAGATTGGTTCT; FXR1-rev: TCAGAGGGGTTAGACAGCTCA; IFNA5-for: TGTATGATGCAGGAGGTTGG; IFNA5-rev: TCACAGTCAGGATAGAGTCCACA; CXCR4-for: GGATATAATGAAGTCACTATGGGAAAA; CXCR4-rev: GGGCACAAGAGAATTAATGTAGAAT; CXCL1-for: TCCTGCATCCCCCATAGTTA; CXCL1-rev: CTTCAGGAACAGCCACCAGT; IL8-for: GAGCACTCCATAAGGCACAAA; IL8-rev: ATGGTTCCTTCCGGTGGT; CCL3-for: GGCTCTCTGCAACCAGTTCT; CCL3-rev: AATCTGCCGGGAGGTGTAG; CCL3L1-for: CTCCAAGCCCAGTGTCATC; CCL3L1-rev: GAAGCTTCTGGACCCCTCA; TNFα and tRNA-lys primers as described previously23) following the manufacturer's directions (Applied Biosystems) on a lightcycler 480 (Roche).
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the Partners Healthcare Center for Personalized Genetic Medicine Microarray facility for microarray analysis, JH Lee for constructing the overexpression FXR1 plasmid, S.I.A. Bukhari, A. Classon and C.J. Wilusz for critical reading, assistance and advice.
Author contributions
OLT created the cell lines and conducted the majority of the experiments. SK validated some of the chemokines and contributed Fig. 3C–E as well as contributed in preparing all the figures for publication. SL conducted the cell migration assay in Fig. 5. MA-S and SST performed the corrections for the revised manuscript. SV planned the experiments and prepared the manuscript. All authors reviewed the manuscript.
Ethics statement
No human patients or samples are involved. The procedures are ethically approved by MGH Safety Board which serves as the representative of the US Ethics committee and are in accordance with the Helsinki Declaration of 1975.
Funding
This work was supported by MGH start-up funds, MGH Interim Support, V Foundation, the Leukemia & Lymphoma Society New Idea Award and GM100202 from NIGMS awarded to SV. SL is supported by the Fund for Medical Discovery postdoctoral fellowship.
References
- [1].Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 2007; 8:113-26; PMID:17245413; http://dx.doi.org/ 10.1038/nrm2104 [DOI] [PubMed] [Google Scholar]
- [2].Groppo R, Richter JD. Translational control from head to tail. Curr Opin Cell Biol 2009; 21:444-51; PMID:19285851; http://dx.doi.org/ 10.1016/j.ceb.2009.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kirkpatrick LL, McIlwain KA, Nelson DL. Alternative splicing in the murine and human FXR1 genes. Genomics 1999; 59:193-202; PMID:10409431; http://dx.doi.org/ 10.1006/geno.1999.5868 [DOI] [PubMed] [Google Scholar]
- [4].Kirkpatrick LL, McIlwain KA, Nelson DL. Comparative genomic sequence analysis of the FXR gene family: FMR1, FXR1, and FXR2. Genomics 2001; 78:169-77; PMID:11735223; http://dx.doi.org/ 10.1006/geno.2001.6667 [DOI] [PubMed] [Google Scholar]
- [5].Mientjes EJ, Willemsen R, Kirkpatrick LL, Nieuwenhuizen IM, Hoogeveen-Westerveld M, Verweij M, Reis S, Bardoni B, Hoogeveen AT, Oostra BA, et al.. Fxr1 knockout mice show a striated muscle phenotype: implications for Fxr1p function in vivo. Hum Mol Genet 2004; 13:1291-302; PMID:15128702; http://dx.doi.org/ 10.1093/hmg/ddh150 [DOI] [PubMed] [Google Scholar]
- [6].Qian J, Hassanein M, Hoeksema MD, Harris BK, Zou Y, Chen H, Lu P, Eisenberg R, Wang J, Espinosa A, et al.. The RNA binding protein FXR1 is a new driver in the 3q26-29 amplicon and predicts poor prognosis in human cancers. Proc Natl Acad Sci USA 2015; 112:3469-74; PMID:25733852; http://dx.doi.org/ 10.1073/pnas.1421975112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Siomi MC, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol Cell Biol 1996; 16:3825-32; PMID:8668200; http://dx.doi.org/ 10.1128/MCB.16.7.3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al.. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 2001; 107:477-87; PMID:11719188; http://dx.doi.org/ 10.1016/S0092-8674(01)00568-2 [DOI] [PubMed] [Google Scholar]
- [9].Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci 2004; 24:7272-6; PMID:15317853; http://dx.doi.org/ 10.1523/JNEUROSCI.2306-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci 2004; 7:113-7; PMID:14703574; http://dx.doi.org/ 10.1038/nn1174 [DOI] [PubMed] [Google Scholar]
- [11].Darnell JC, Fraser CE, Mostovetsky O, Darnell RB. Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs. Hum Mol Genet 2009; 18:3164-3177; PMID:19487368; http://dx.doi.org/ 10.1093/hmg/ddp255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zang JB, Nosyreva ED, Spencer CM, Volk LJ, Musunuru K, Zhong R, Stone EF, Yuva-Paylor LA, Huber KM, Paylor R, et al.. A mouse model of the human Fragile X syndrome I304N mutation. PLoS Genet 2009; 5:e1000758; PMID:20011099; http://dx.doi.org/ 10.1371/journal.pgen.1000758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al.. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011; 146:247-61; PMID:21784246; http://dx.doi.org/ 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Phan AT, Kuryavyi V, Darnell JC, Serganov A, Majumdar A, Ilin S, Raslin T, Polonskaia A, Chen C, Clain D, et al.. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat Struct Mol Biol 2011; 18:796-804; PMID:21642970; http://dx.doi.org/ 10.1038/nsmb.2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ceman S, O'Donnell WT, Reed M, Patton S, Pohl J, Warren ST. Phosphorylation influences the translation state of FMRP-associated polyribosomes. Hum Mol Genet 2003; 12:3295-305; PMID:14570712; http://dx.doi.org/ 10.1093/hmg/ddg350 [DOI] [PubMed] [Google Scholar]
- [16].Garnon J, Lachance C, Di Marco S, Hel Z, Marion D, Ruiz MC, Newkirk MM, Khandjian EW, Radzioch D. Fragile X-related protein FXR1P regulates proinflammatory cytokine tumor necrosis factor expression at the post-transcriptional level. J Biol Chem 2005; 280:5750-63; PMID:15548538; http://dx.doi.org/ 10.1074/jbc.M401988200 [DOI] [PubMed] [Google Scholar]
- [17].Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 2007; 128:1105-18; PMID:17382880; http://dx.doi.org/ 10.1016/j.cell.2007.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ascano M Jr, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al.. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 2012; 492:382-86; PMID:23235829; http://dx.doi.org/ 10.1038/nature11737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Khandjian EW, Bardoni B, Corbin F, Sittler A, Giroux S, Heitz D, Tremblay S, Pinset C, Montarras D, Rousseau F, et al.. Novel isoforms of the fragile X related protein FXR1P are expressed during myogenesis. Hum Mol Genet 1998; 7:2121-8; PMID:9817930; http://dx.doi.org/ 10.1093/hmg/7.13.2121 [DOI] [PubMed] [Google Scholar]
- [20].Dube M, Huot ME, Khandjian EW. Muscle specific fragile X related protein 1 isoforms are sequestered in the nucleus of undifferentiated myoblast. BMC Genet 2000; 1:4; PMID:11178106; http://dx.doi.org/ 10.1186/1471-2156-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huot ME, Bisson N, Davidovic L, Mazroui R, Labelle Y, Moss T, Khandjian EW. The RNA-binding protein fragile X-related 1 regulates somite formation in Xenopus laevis. Mol Biol Cell 2005; 16:4350-61; PMID:16000371; http://dx.doi.org/ 10.1091/mbc.E05-04-0304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Davidovic L, Durand N, Khalfallah O, Tabet R, Barbry P, Mari B, Sacconi S, Moine H, Bardoni B. A novel role for the RNA-binding protein FXR1P in myoblasts cell-cycle progression by modulating p21/Cdkn1a/Cip1/Waf1 mRNA stability. PLoS Genet 2013; 9:e1003367; PMID:23555284; http://dx.doi.org/ 10.1371/journal.pgen.1003367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Truesdell SS, Mortensen RD, Seo M, Schroeder JC, Lee JH, LeTonqueze O, Vasudevan S. MicroRNA-mediated mRNA Translation Activation in Quiescent Cells and Oocytes Involves Recruitment of a Nuclear microRNP. Sci Rep 2012; 2:842; PMID:23150790; http://dx.doi.org/ 10.1038/srep00842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mortensen RD, Serra M, Steitz JA, Vasudevan S. Posttranscriptional activation of gene expression in Xenopus laevis oocytes by microRNA-protein complexes (microRNPs). Proc Natl Acad Sci USA 2011; 108:8281-6; PMID:21536868; http://dx.doi.org/ 10.1073/pnas.1105401108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science 2007; 318:1931-4; PMID:18048652; http://dx.doi.org/ 10.1126/science.1149460 [DOI] [PubMed] [Google Scholar]
- [26].Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, Schenkman DI, Gilkeson GS, Broxmeyer HE, Haynes B, et al.. A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 1996; 4:445-54; PMID:8630730; http://dx.doi.org/ 10.1016/S1074-7613(00)80411-2 [DOI] [PubMed] [Google Scholar]
- [27].Prewitt TW, Matthews W, Chaudhri G, Pogrebniak HW, Pass HI. Tumor necrosis factor induces doxorubicin resistance to lung cancer cells in vitro. J Thorac Cardiovasc Surg 1994; 107:43-9; PMID:8283917 [PubMed] [Google Scholar]
- [28].Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 1999; 10:387-98; PMID:10204494; http://dx.doi.org/ 10.1016/S1074-7613(00)80038-2 [DOI] [PubMed] [Google Scholar]
- [29].Cain BS, Harken AH, Meldrum DR. Therapeutic strategies to reduce TNF-alpha mediated cardiac contractile depression following ischemia and reperfusion. J Mol Cell Cardiol 1999; 31:931-47; PMID:10336835; http://dx.doi.org/ 10.1006/jmcc.1999.0924 [DOI] [PubMed] [Google Scholar]
- [30].Rothman N, Skibola CF, Wang SS, Morgan G, Lan Q, Smith MT, Spinelli JJ, Willett E, De Sanjose S, Cocco P, et al.. Genetic variation in TNF and IL10 and risk of non-Hodgkin lymphoma: a report from the InterLymph Consortium. Lancet Oncol 2006; 7:27-38; PMID:16389181; http://dx.doi.org/ 10.1016/S1470-2045(05)70434-4 [DOI] [PubMed] [Google Scholar]
- [31].Grimm D, Wehland M, Pietsch J, Infanger M, Bauer J. Drugs interfering with apoptosis in breast cancer. Curr Pharm Des 2011; 17:272-83; PMID:21348828; http://dx.doi.org/ 10.2174/138161211795049723 [DOI] [PubMed] [Google Scholar]
- [32].Leibovich SJ, Polverini PJ, Shepard HM, Wiseman DM, Shively V, Nuseir N. Macrophage-induced angiogenesis is mediated by tumour necrosis factor-alpha. Nature 1987; 329:630-2; PMID:2443857; http://dx.doi.org/ 10.1038/329630a0 [DOI] [PubMed] [Google Scholar]
- [33].Lee S, Truesdell SS, Bukhari SI, Lee JH, LeTonqueze O, Vasudevan S. Upregulation of eIF5B controls cell-cycle arrest and specific developmental stages. Proc Natl Acad Sci USA 2014; 111:E4315-E4322; PMID:25261552; http://dx.doi.org/ 10.1073/pnas.1320477111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pomorski P, Watson JM, Haskill S, Jacobson KA. How adhesion, migration, and cytoplasmic calcium transients influence interleukin-1beta mRNA stabilization in human monocytes. Cell Motil Cytoskeleton 2004; 57:143-57; PMID:14743348; http://dx.doi.org/ 10.1002/cm.10159 [DOI] [PubMed] [Google Scholar]
- [35].Imhof BA, Aurrand-Lions M. Adhesion mechanisms regulating the migration of monocytes. Nat Rev Immunol 2004; 4:432-44; PMID:15173832; http://dx.doi.org/ 10.1038/nri1375 [DOI] [PubMed] [Google Scholar]
- [36].Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol 2005; 5:953-64; PMID:16322748; http://dx.doi.org/ 10.1038/nri1733 [DOI] [PubMed] [Google Scholar]
- [37].Dirkx AE, Oude Egbrink MG, Wagstaff J, Griffioen AW. Monocyte/macrophage infiltration in tumors: modulators of angiogenesis. J Leukoc Biol 2006; 80:1183-96; PMID:16997855; http://dx.doi.org/ 10.1189/jlb.0905495 [DOI] [PubMed] [Google Scholar]
- [38].Hume DA. Differentiation and heterogeneity in the mononuclear phagocyte system. Mucosal Immunol 2008; 1:432-41; PMID:19079210; http://dx.doi.org/ 10.1038/mi.2008.36 [DOI] [PubMed] [Google Scholar]
- [39].Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol 2014; 14:392-404; PMID:24854589; http://dx.doi.org/ 10.1038/nri3671 [DOI] [PubMed] [Google Scholar]
- [40].Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, Lowe SW. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet 2005; 37:1289-95; PMID:16200064 [DOI] [PubMed] [Google Scholar]
- [41].McSherry EA, Brennan K, Hudson L, Hill AD, Hopkins AM. Breast cancer cell migration is regulated through junctional adhesion molecule-A-mediated activation of Rap1 GTPase. Breast Cancer Res 2011; 13:R31; PMID:21429211; http://dx.doi.org/ 10.1186/bcr2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res 2006; 34:D111-4; PMID:16381826; http://dx.doi.org/ 10.1093/nar/gkj052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Khabar KS. Post-transcriptional control during chronic inflammation and cancer: a focus on AU-rich elements. Cell Mol Life Sci 2010; 67:2937-55; PMID:20495997; http://dx.doi.org/ 10.1007/s00018-010-0383-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


