Abstract
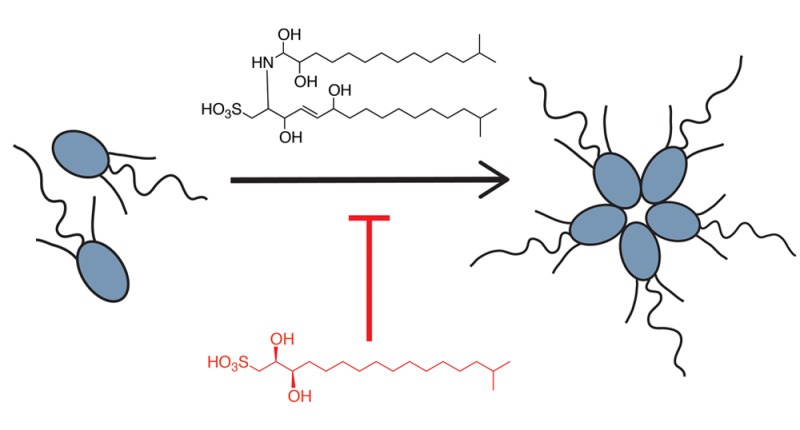
The choanoflagellate Salpingoeca rosetta is a microbial marine eukaryote that can switch between unicellular and multicellular states. As one of the closest living relatives of animals, this organism has become a model for understanding how multicellularity evolved in the animal lineage. Previously our laboratories isolated and synthesized a bacterially produced sulfonolipid that induces S. rosetta to form multicellular “rosettes.” In this study, we report the identification of a bacterially produced inhibitor of rosettes (IOR-1) as well as the total synthesis of this molecule and all of its stereoisomers. Our results confirm the previously noted specificity and potency of rosette-modulating molecules, expand our understanding of the complex chemical ecology between choanoflagellates and rosette-inducing bacteria, and provide a synthetic probe template for conducting further mechanistic studies on the emergence of multicellularity.
Choanoflagellates are motile microbial eukaryotes that reside in aquatic environments and feed on bacteria. Much like the collar cells of sponges, these microscopic organisms use a single apical flagellum to sweep surrounding bacteria into their actin-rich collar, where the bacteria are phagocytosed.1 Choanoflagellates, which are the closest living relatives of animals, express diverse genes, such as C-type lectins, cadherins, and tyrosine kinases, that are known to regulate multicellular processes in animals.2−5 While predominately unicellular, several species of choanoflagellate, including Salpingoeca rosetta, alternate between unicellular and multicellular states. In an embryogenesis-like process, the multicellular form, known as a “rosette,” arises through multiple rounds of cell division in which the sister cells do not completely separate from each other (see the Supporting Information for an image of a rosette).6,7 While full mechanistic understanding of rosette development is yet to be achieved, further study of the transition to multicellularity in this ancient organism could provide meaningful insights into how multicellularity evolved in the animal lineage.
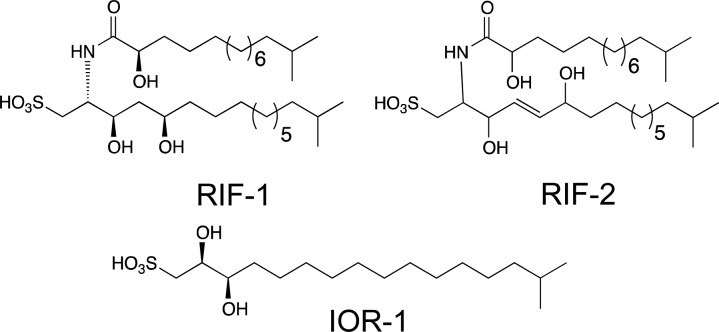
We previously showed that the transition between the unicellular form and the multicellular rosette is induced by a sulfonolipid produced by Algoriphagus machipongonensis (“Algoriphagus” for short), a marine bacterium originally coisolated with S. rosetta that serves as prey for the choanoflagellate.8,9 Subsequent synthesis of the inducing molecule, termed rosette-inducing factor-1 (RIF-1), revealed the absolute configuration of the molecule as well as the strict stereochemical requirements for activity (Figure 1).10
Figure 1.
Previously isolated rosette-inducing molecules RIF-1 and RIF-2 and inhibitor of rosettes (IOR-1).
However, while RIF-1 could induce a small percentage of cells to form rosettes, the activity of RIF-1 alone did not faithfully recapitulate the activity observed with live bacteria or conditioned medium. Additionally, we noted apparent fluctuations in the activity of isolated (natural) RIF-1, as well as sphingolipid-enriched extracts, leading us to hypothesize that Algoriphagus produces additional choanoflagellate-modulating molecules that could serve as alternative inducers, synergists, or possibly even inhibitors. In this report, we describe the isolation and synthesis of a bacterially produced sulfonate-containing lipid that inhibits sulfonolipid-induced rosette formation in S. rosetta.
We performed a chloroform/methanol extraction on the cell pellet of Algoriphagus and fractionated the extract by reversed-phase (C-18) HPLC using a broad elution range in order to expand our search beyond sulfonolipids.8 We then tested each fraction in combination with inducers of rosette development to determine whether any of the fractions contained molecules with inhibitory activity. As inducers we used either a sulfonolipid-enriched fraction (RIF-mix) that elicits high levels of rosette formation (with up to 30% of cells in rosettes) or a purified sulfonolipid, RIF-2, a close structural analogue of RIF-1 whose complete stereostructure remains to be fully elucidated (Woznica and Cantley et al., submitted; Figure 1). We identified two adjacent fractions that reduced rosette formation when treated in combination with either RIF-mix or RIF-2.
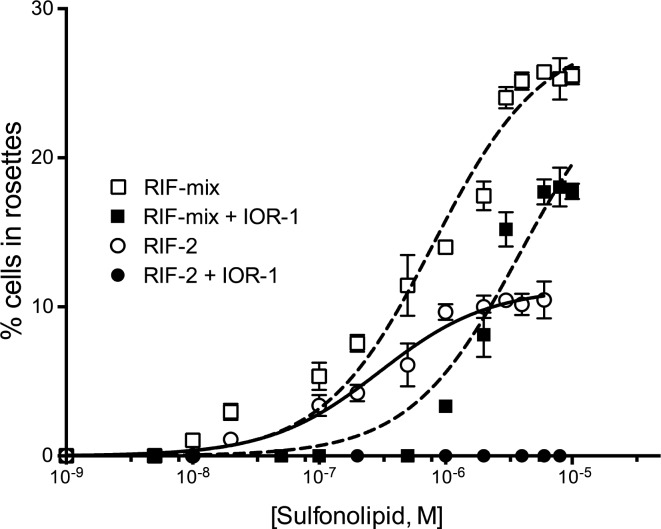
High-resolution mass spectrometry revealed that both fractions predominately contained a molecule with a mass of [M – H] 351.2216 Da, matching a predicted formula of C17H35O5S. One- and two-dimensional NMR experiments (Figures S1–S6) permitted us to propose a planar structure for this molecule, which we have named inhibitor of rosettes (IOR-1) (Figure 1). IOR-1 is optically active ([α]D22 = +24, c 0.125, MeOH), and its absolute configuration was ultimately determined through synthesis as described below. Dose–response curves using purified IOR-1 showed an optimal inhibitory concentration of 2.5 nM (Figure 2), which corresponds with our observation of IOR-1’s single-digit-nanomolar concentration in Algoriphagus-conditioned medium (Supporting Information, Methods).
Figure 2.
Cotreatment of IOR-1 (2.5 nM) with RIF-2 and RIF-mix. Graphs were generated using GraphPad Prism 6 statistical software. The rosette induction data were analyzed using a one-site (specific binding) model.
We were intrigued by the structure of IOR-1 for several reasons. It resembles the capnine base found in bacterially produced sulfonolipids, especially in that it contains the sulfonic acid headgroup present in the previously identified RIF-1 and RIF-2. As capnine bases, like the analogous sphingoid bases, are biosynthetically derived from amino acids, the 2-position typically has an −NH2 substituent, so the −OH group at this position on IOR-1 is a notable modification. In general little is known about capnines, and while they have been postulated to facilitate bacterial gliding, their functions are not well understood and their distribution is quite limited.11,12 The more common class of sphingosine bases (or lysosphingolipids) act through G protein-coupled receptors to modulate diverse biological processes including triggering apoptosis and mediating inflammation.13−15 The structural similarity between IOR-1 and these signaling molecules suggests that they may also share functional similarities.
We synthesized IOR-1 both to establish its absolute stereostructure and to determine whether it shared the same strict stereochemical requirements seen in RIF-1. Additionally, the relatively simpler synthesis of IOR-1 compared with that of RIF-1 makes IOR-based probes potentially valuable tools for identifying the host targets of rosette-modulating molecules.
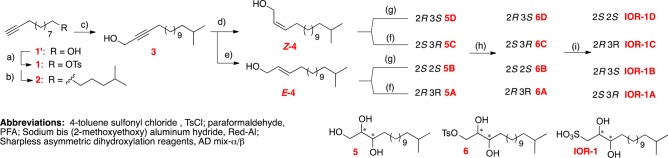
As we needed to access all four possible configurations of the hydroxyl groups at C2 and C3, we reasoned that we could reduce an alkyl chain ending in a propargylic alcohol to either the corresponding cis- or trans-alkene and then perform Sharpless asymmetric dihydroxylations on both alkenes using either the α or β mix to yield all four stereoisomers. In a final step, the sulfonic acid moiety could be added to each purified stereoisomer through nucleophilic substitution.
To reach 15-methylhexadec-2-yn-1-ol (3), we started with commercially available 10-undecyn-1-ol (1′). We elongated the acyl chain and added the isopropyl tail through a Grignard reaction with isopentyl-MgBr in the presence of Li2CuCl4 to yield 2 in a manner similar to that described previously.16 Propargylic alcohol 3 was obtained through acetylide formation and subsequent nucleophilic addition to paraformaldehyde.17 At this stage our synthetic strategy diverged to obtain both the cis- and trans-alkene. We used Lindlar’s catalyst to reduce alkyne 3 to the cis-alkene Z-4 in the presence of H2 and Red-Al to obtain the trans-alkene E-4. These reductions were achieved in acceptable yields of 74% and 70% respectively. From this branch point we could access each diol configuration pattern through asymmetric bishydroxylation using the Sharpless reagents (AD mix-α and AD mix-β) in the presence of methanesulfonamide, which afforded yields in the 70–80% range.18 This stage proved suitable to purify the diols via chiral chromatography, yielding enantiopure 6A, 6B, 6C, and 6D (Scheme 1; also see Figures S7–S23).
Scheme 1. Synthesis of IOR-1 Stereoisomers A–D.
Conditions: (a) TsCl, pyridine, CH2Cl2, 4 °C, 10 h. (b) Isopentyl-MgBr, THF, Li2CuCl4 (cat.), 0 °C to RT, overnight, 42% over two steps. (c) THF, n-BuLi, PFA, 0 °C to RT, 2.5 h, 66%. (d) Lindlar’s catalyst, MeOH, H2, RT, overnight, 74%. (e) Red-Al, ether, 0 °C to RT, overnight, 70%. (f) AD mix-β, H2O, t-BuOH, methanesulfonamide, 0 °C, 6–24 h, 77–87%. (g) AD mix-α, H2O, t-BuOH, methanesulfonamide, 0 °C, 6–24 h, 73–82%. (h) TsCl, pyridine, DCM, 4 °C, 10 h. Note: at this step, tosylated compounds were purified by chiral HPLC to give the pure enantiomers. (i) Na2SO3, H2O, EtOH, 62 °C, overnight, 14–20%.
Although we previously introduced the sulfonic acid moiety of RIF-1 through a Mitsonobu reaction using thioacetic acid followed by oxidation, with tosylates 6A-D in hand a simple nucleophilic displacement strategy at this position was more efficient. While substitution with thioacetic acid and subsequent oxidation yielded IOR-1, side-product formation frustrated the final purification. Addition of sodium sulfite in a heated biphasic solution of water and ethanol yielded fewer side products and, while giving a low yield of the final product (14–20%), allowed a much simpler purification process and higher overall conversion.19
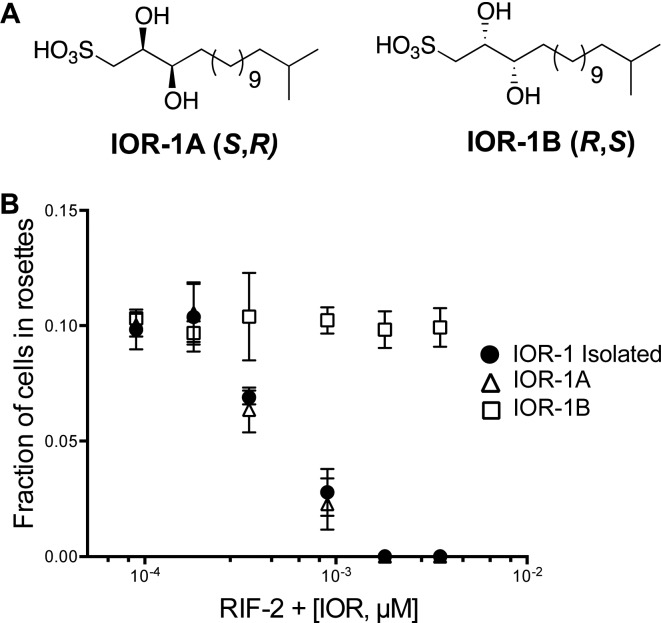
The 1H NMR spectra of compounds IOR-1A and IOR-1B were identical to that of isolated IOR-1 (Figures S24–S26), whereas compounds IOR-1C and IOR-1D exhibited different chemical shifts of protons at positions C1, C2, and C3. Determination of the optical rotations for these molecules revealed matching signs and values for IOR-1A and IOR-1 (Supporting Information, Methods), suggesting that IOR-1A is likely a match to the isolated molecule.
To verify the activity and specificity of IOR-1, we tested each of the synthetic stereoisomers in our rosette inhibition assay. Full dose–response curves revealed almost identical activity for IOR-1A compared to the isolated inhibitor, whereas IOR-1B displayed no activity (Figure 3); unsurprisingly, IOR-1C and IOR-1D were also inactive (Figure S27). Given both the spectroscopic and biological data, we were able to determine the absolute configuration of IOR-1 as 2S,3R. Significantly, only one stereoisomer of the inhibitor is active, reprising the theme that these molecules interact in a highly specific manner with their target. We further validated this specificity by testing a handful of commercially available IOR-1 analogues, and none were active at concentrations ranging from 0.1 ng/mL to 1 μg/mL (Figure S28).
Figure 3.
(A) Structures of the synthetic compounds IOR-1A and IOR-1B. (B) Comparison of dose–response curves of IOR-1A, IOR-1B, and isolated IOR-1. RIF-2 treated at 2 μM.
Furthermore, this specificity suggests that IOR-1 (IOR-1A) is an appropriate starting point for the development of a bioaffinity probe that could be used to investigate the choanoflagellate target and mechanism of rosette-modulating molecules. Its straightforward synthesis and scalability allows for quick access to modified versions of IOR-1, and its potency (2.5 nM) would minimize the likelihood of nonspecific interactions even if the probes were of somewhat lower potency. Our synthetic route also supplies us with inactive stereoisomers of IOR-1, which can serve as useful negative controls for target identification.
The assignment of the hydroxyls of IOR-1 in the syn configuration was unexpected; we had predicted that the hydroxyls would have the same relative configuration as the 2-amine and 3-hydroxyl groups observed in RIF-1, which is by far the most common stereochemistry for sulfonolipids and sphingolipids.13 While not unprecendented, it is quite rare for capnine bases to exhibit the syn configuration, and biosynthesis of the syn-diol has not been reported.20,21 Exploration of the fully annotated genome of Algoriphagus confirmed the presence of a number of transaminases, which could invert the configuration of the hydroxyl group at C2 during conversion from an amino group (Figures S29 and S30).22 As the biosynthesis of IOR-1 clearly has components that are distinct from the known sulfonolipids (cf. RIF-1), this molecule is unlikely to be either a degradation product or a precursor to the more standard sphingolipids and sulfonolipids. Further investigation into the biosynthesis and regulation of IOR-1 are ongoing and will be of great interest in understanding the ecological context in which these molecules are produced.
From an ecological perspective, the isolation and characterization of IOR-1 raises a number of interesting questions about the choanoflagellate–bacterium predator–prey relationship. The isolation of both an inducer and an inhibitor from the same bacterium highlights the complexity of the relationship between Algoriphagus and S. rosetta. Our current hypothesis is that rosette formation improves bacterial prey capture by choanoflagellates, and if this is true, production of factors that attenuate rosette-colony formation would confer an apparent benefit to the producing bacteria.23 Understanding how IOR-1 and RIFs are produced and regulated should begin to reveal how Algoriphagus could use both sets of molecules to manipulate its predators. More generally, examining the complex phenotypic effects triggered by these bacterially produced small molecules will increase our understanding of the role of bacteria in the evolution of multicellular organisms.
In summary, we have isolated, characterized, and synthesized an atypical sulfonolipid that potently inhibits the conversion from a unicellular to a multicellular morphology in choanoflagellates. Through synthesis we were able to confirm that this lipid has the rare syn-diol configuration and that the 2S,3R stereochemistry is necessary for activity. The discovery of this molecule reveals that the chemical interaction between choanoflagellates and rosette-inducing bacteria is more complex than previously imagined and argues that further investigation is warranted. Finally, IOR-1 provides a starting point for pathway identification in this important model system.
Acknowledgments
This work was funded by the NIH (GM099533). C.B. was supported by a postdoctoral fellowship from the German National Academy of Science Leopoldina (LPDS2011-2). N.K. is an investigator in the Howard Hughes Medical Institute and a Senior Scholar in the Integrated Microbial Biodiversity Program of the Canadian Institute for Advanced Research. We thank Furong Sun at the University of Illinois at Urbana–Champaign Mass Spectrometry Laboratory. We also thank Drs. D. Dietrich and P. Stallforth for consultations on synthetic strategy and manuscript preparation.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b01190.
Synthesis and isolation procedures, structure characterization, and bioassay data (PDF)
Author Contributions
∥ A.M.C. and A.W. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Hibberd D. J. J. Cell Sci. 1975, 17, 191–219. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I.; Roger A. J.; Burger G.; Gray M. W.; Lang B. F. Mol. Biol. Evol. 2008, 25, 664–672. 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek T. B. H.; Gringhuis S. I. Nat. Rev. Immunol. 2009, 9, 465–479. 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G.; Young S. L.; Miller W. T.; Zhai Y. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 9674–9679. 10.1073/pnas.0801314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough S. R.; Chen Z.; Kramer E.; Zeng Q.; Young S.; Robertson H. M.; Begovic E.; Richter D. J.; Russ C.; Westbrook M. J.; Manning G.; Lang B. F.; Haas B.; Nusbaum C.; King N. Genome Biol. 2013, 14, R15. 10.1186/gb-2013-14-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel M. J.; Alegado R. A.; Fairclough S. R.; Levin T. C.; Nichols S. A.; McDonald K.; King N. Dev. Biol. 2011, 357, 73–82. 10.1016/j.ydbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough S. R.; Dayel M. J.; King N. Curr. Biol. 2010, 20, R875–R876. 10.1016/j.cub.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegado R. A.; Brown L. W.; Cao S.; Dermenjian R. K.; Zuzow R.; Fairclough S. R.; Clardy J.; King N. eLife 2012, 1, e00013. 10.7554/eLife.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegado R. A.; Grabenstatter J. D.; Zuzow R.; Morris A.; Huang S. Y.; Summons R. E.; King N. Int. J. Syst. Evol. Microbiol. 2013, 63, 163–168. 10.1099/ijs.0.038646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemelmanns C.; Woznica A.; Alegado R. A.; Cantley A. M.; King N.; Clardy J. J. Am. Chem. Soc. 2014, 136, 10210–10213. 10.1021/ja5046692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godchaux W. III; Leadbetter E. R. J. Biol. Chem. 1984, 259, 2982–2990. [PubMed] [Google Scholar]
- Abbanat D. R.; Godchaux W. III; Leadbetter E. R. Arch. Microbiol. 1988, 149, 358–364. 10.1007/BF00411656. [DOI] [Google Scholar]
- Merrill A. H. J. Biol. Chem. 2002, 277, 25843–25846. 10.1074/jbc.R200009200. [DOI] [PubMed] [Google Scholar]
- Salcedo M.; Cuevas C.; Alonso J. L.; Otero G.; Faircloth G.; Fernandez-Sousa J. M.; Avila J.; Wandosell F. Apoptosis 2007, 12, 395–409. 10.1007/s10495-006-0573-z. [DOI] [PubMed] [Google Scholar]
- Hannun Y. A.; Bell R. M. Science 1987, 235, 670–674. 10.1126/science.3101176. [DOI] [PubMed] [Google Scholar]
- Takikawa H.; Nozawa D.; Kayo A.; Muto S. E.; Mori K. J. Chem. Soc., Perkin Trans. 1 1999, (17), 2467–2477. 10.1039/a904258j. [DOI] [Google Scholar]
- Romuald C.; Cazals G.; Enjalbal C.; Coutrot F. Org. Lett. 2013, 15, 184–187. 10.1021/ol303186j. [DOI] [PubMed] [Google Scholar]
- Sharpless K. B.; Amberg W.; Bennani Y. L.; Crispino G. A.; Hartung J.; Jeong K. S.; Kwong H. L.; Morikawa K.; Wang Z. M. J. Org. Chem. 1992, 57, 2768–2771. 10.1021/jo00036a003. [DOI] [Google Scholar]
- Sacoman J. L.; Hollingsworth R. I. Carbohydr. Res. 2011, 346, 2294–2299. 10.1016/j.carres.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Molinski T. F.; Biegelmeyer R.; Stout E. P.; Wang X.; Frota M. L. C. Jr.; Henriques A. T. J. Nat. Prod. 2013, 76, 374–381. 10.1021/np300744y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruett S. T.; Bushnev A.; Hagedorn K.; Adiga M.; Haynes C. A.; Sullards M. C.; Liotta D. C.; Merrill A. H. J. Lipid Res. 2008, 49, 1621–1639. 10.1194/jlr.R800012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. H. J. Bacteriol. 1984, 159, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper M.; Dayel M. J.; Pepper R. E.; Koehl M. A. R. Phys. Rev. Lett. 2013, 110, 228104. 10.1103/PhysRevLett.110.228104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.