Abstract
The majority of therapeutics target membrane proteins, accessible on the surface of cells, to alter cellular signaling. Cells use membrane proteins to transduce signals into cells, transport ions and molecules, bind the cell to a surface or substrate, and catalyze reactions. Newly devised technologies allow us to drug conventionally “undruggable” regions of membrane proteins, enabling modulation of protein–protein, protein–lipid, and protein–nucleic acid interactions. In this review, we survey the state of the art in high-throughput screening and rational design in drug discovery, and we evaluate the advances in biological understanding and technological capacity that will drive pharmacotherapy forward against unorthodox membrane protein targets.
Keywords: transmembrane domains, drug discovery, high-throughput screening, rational design, curvature sensing
1. MEMBRANE PROTEINS: CHALLENGES AND OPPORTUNITIES
Contemporary medicine is unrecognizable without pharmaceuticals and biologics. The entire enterprise of drug discovery rests on the selective binding of the drug molecule to its target. New approaches in biomedical research and development are usually slow to take hold, and drug discovery has thus far been plagued by the so-called streetlight effect—that is, scientists have been looking for new targets where looking is easiest. The shadowy areas of this street are the “undruggable” targets that have proven too difficult to target by the standard modus operandi. Enzymes, transporters, ion channels, and receptors are all common membrane protein (MP) drug targets; virtually all therapeutics bind proteins within solvated regions outside the membrane.
Whereas MPs make up ~23% of the human proteome (1), an analysis performed 10 years ago by Overington et al. (2) concluded that MPs constitute more than 60% of current drug targets. A few categories of targets are highly overrepresented; more than one-third of small-molecule drugs target proteins from the G protein–coupled receptor (GPCR) superfamily to inhibit or activate signal transduction (3). In the past decade, there has been a push to (a) find new drug targets and (b) create new classes of agents, but the number of disease-associated targets is limited and will eventually be exhausted (4). However, is it reasonable to expect that these new agents track already discovered drug–target interactions? A hallmark of druggability is the requirement for a solvent-accessible hydrophobic pocket (5), often the active site of an enzyme in the case of orthosteric drugs (6). The first major challenge to this dogma came from the success of therapeutic monoclonal antibodies, which function by specifically binding an extracellular epitope on the surface of an MP with high affinity. Monoclonal antibodies can bind to receptors or their ligands to modulate signaling, or they can deliver conjugated drugs to individual cell types on the basis of differences in MP surface expression. Still, drug design rests on a core assumption that there are no specific interactions within the membrane that can be exploited for drug development. In light of new evidence, this view is becoming increasingly doubtful. Transmembrane domains (TMDs) are not simply passive membrane-spanning anchors for MPs; rather, they play active roles in oligomerization and specifically drive protein–protein interactions (PPIs) within the plasma membrane. In this review, we attempt to reframe the idea of druggability by discussing a new model that includes anti-TMD peptides and small molecules. The dearth of solved three-dimensional MP structures has been a barrier to rational drug design, but advances in structural biology have led to new opportunities. Here we appraise the strategies used to discover potential therapeutics that interact with MP TMDs, by (a) considering the interactions between membranes and MPs, (b) examining biological understanding of the cell membrane, and (c) analyzing new technologies used to investigate TMD-mediated signal transduction, in order to bring new MP targets into the light (Figure 1). We focus on the challenges and opportunities surrounding various therapeutic modalities, including small molecules, peptides, and peptidomimetics, with an emphasis on cell surface MPs and the plasma membrane. We refer readers interested in other aspects of drug discovery to excellent reviews of chemical genetics (7), antibiotics targeting bacterial proteins (8), targeting of PPIs with synthetic agents (9–11), drugging of GPCRs based on structural motifs that differ between GPCR families (12–14), and general drug design strategies for targeting GPCRs (15).
Figure 1.
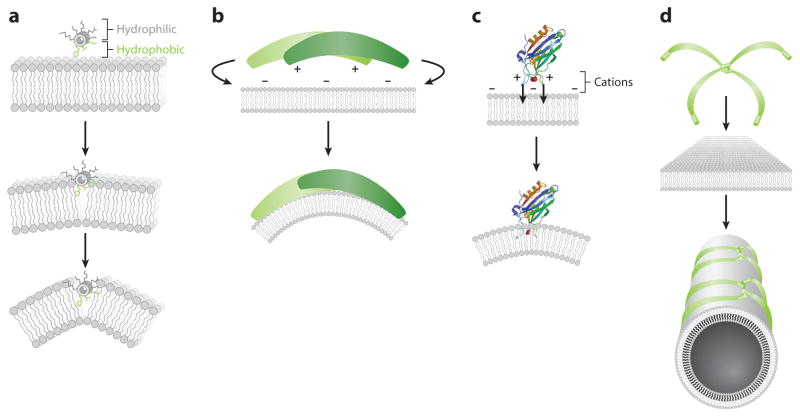
Uses of exogenous chemical probes to investigate cell membranes and membrane proteins (MPs). (a) MP transmembrane domain (TMD) structure–function relationships can be investigated without crystallizing full-length MPs. (b) TMD structures also enable rational design of anti-TMD peptides and small molecules. (c) Curvature-sensing peptides and proteins can be used to sense curved membranes, such as those found on small, highly curved extracellular vesicles. (d) Modulating membrane protein–protein and protein–lipid interactions also offers an opportunity to understand the fine-tuning of the immune response in response to pattern recognition receptor activation, with applications in cancer immunotherapy. (e) Conjugating an environment-sensitive fluorophore to peptide probes provides a convenient readout for interaction with the membrane. (f) Computational advances have improved predictions of TMD–TMD interactions.
2. MEMBRANE PROTEINS EMERGING FROM “UNDRUGGABLE” TARGETS
2.1. Structural Basis for Targeting Membrane Proteins
Major advances in structural biology have facilitated the analyses of many previously inaccessible MP targets, helping to overcome a major hurdle in targeting MPs—the lack of high-resolution three-dimensional structures. Less than 1% of all solved protein crystal structures are MPs (16), but as more MP complex structures are solved, structure–function studies and structure-based design of drugs targeting MPs will become more feasible. Near-atomic-level resolution of transmembrane protein structures by cryoelectron microscopy (cryo-EM) (17), advances in X-ray crystallography such as femtosecond- or even attosecond-timescale pulse lasers (18), and solid-state nuclear magnetic resonance (NMR) in lipid bilayers (19) are advancing membrane structural biology. New MP structures will undoubtedly lead to functional insights and enable rational design to target many previously undruggable MPs.
2.1.1. Cryoelectron microscopy for membrane protein structures
For larger, symmetric MP complexes, cryo-EM can now achieve near-atomic resolution using single-particle analysis. New direct electron detectors lead to less noisy images, and improved image-processing algorithms such as ReliOn (Regularised Likelihood Optimisation) employ a Bayesian framework to handle the dramatically increased amounts of data (17). The synergy between these developments allow researchers to build near-complete atomic models de novo at the level of amino acid side chains, despite the relatively smooth nature of macromolecular structures (17).
A prominent example of an MP complex structure determined by cryo-EM is the γ-secretase complex. γ-Secretase is a multisubunit intramembrane protease that can generate the amyloid-β plaques that accumulate in the brain of patients with Alzheimer disease and are hypothesized to cause this disease. TMDs are known mostly for their roles in anchoring MPs, in connecting extracellular and intracellular domains, and in providing selective permeability to channel proteins; however, a few TMDs have proteolytic capabilities (20). More than 25% of γ-secretase substrates contain Gx3G TMD dimerization motifs (21), including the amyloid precursor protein (22). Recent research also suggests a role for oligomerization (23) or helix-destabilizing amino acids within TMDs facilitating cleavage by γ-secretase (24). The structural basis of substrate recognition within the membrane remained a mystery until the Shi group (25) used cryo-EM both to derive the structure of the entire γ-secretase complex at a 4.32-Å resolution and to unambiguously assign all TMDs. Cryo-EM will continue to find use in structural studies of large multiprotein membrane complexes, and will prove especially useful for determining TMD structures.
2.1.2. NMR spectroscopy for membrane proteins
Obtaining high-quality crystals amenable for X-ray crystallography is still a limiting factor for solving MP structures by this technique, so in many cases NMR is advantageous for structural determination. Solid-state NMR has become an important tool for MP complex studies, characterizing the structures of MP complexes and TMD PPIs in liquid crystalline lipid bilayers (19). NMR of MPs yields high structural resolution; multidimensional magic-angle-spinning correlation NMR measures structural constraints of MPs in lipid bilayers and provides information about torsion angles, interatomic distances, orientation, and insertion depth (26). More recently, MPs have been studied in their native environment through the use of on-cell NMR to investigate conformations of MPs in live cells (27). Live on-cell NMR relies on isotope-labeled residues, for example, by expressing proteins in mammalian cell lines grown in isotopically labeled media. Mammalian expression may give low yields and require large amounts of labeled media, so a sensible alternative for investigating smaller MPs is to express them in Escherichia coli, purify them, and then introduce them into mammalian cells. Researchers confirmed the possibility of live on-cell NMR to characterize receptor–ligand interactions between peptides and cell surface proteins by using saturation transfer double-difference NMR to study binding of the pentapeptide ligand cyclo(Arg–Gly–Asp–D-Phe–Val) to αIIbβ3 integrin in intact platelets, where the affinity was substantially enhanced compared to integrins in liposomes (28). With tiny amounts of receptor protein (25 pmol, which was achieved with 108 cells, or ~150,000 receptors per cell), these investigators characterized structural changes occurring during binding in live cells, demonstrating the importance of studying MPs in native environments.
NMR has also been used to study MPs in living cancer cells using two-dimensional transferred nuclear Overhauser effect spectroscopy NMR (29). Two MPs involved in angiogenesis, cluster of differentiation (CD)13 and αvβ3 integrin, bind to another cyclic RGD peptide called cilengitide. The authors compared NMR spectra of these MPs bound to cilengitide from normal and β3-knockdown cells to decrease αvβ3 integrin expression (29). Thus, MP structural studies can be feasibly carried out in live cells using NMR spectroscopy, providing more relevant MP structures and illuminating MP structure–function relationships in native environments.
2.1.3. Structural features of TMD–TMD complexes
PPIs were classically considered undruggable because of their large interaction faces. A landmark study by Clackson & Wells (30) on the structure of human growth hormone and its receptor (hGH–hGHR) first brought this view of “nonspecificity” into question. Although ~30 side-chain residues from each protein make contact, more than three-quarters of the binding energy comes from a central hydrophobic region dominated by two tryptophan residues (30). Thus, PPIs can function through hot spots, small and complementary sets of contact residues that maintain binding affinity. This discovery first legitimized the idea of designing small-molecule inhibitors of PPIs by finding hot-spot residues and designing drugs to specifically bind these hot spots and inhibit PPIs.
Hydrophobic matching governs the insertion, orientation, and depth of TMDs within lipid bilayers. A difference between the hydrophobic length of the TMD and the hydrophobic thickness of the bilayer causes hydrophobic mismatch, resulting in helical tilting of TMDs that would otherwise be too long to be completely contained within the hydrophobic lipid environment and would expose hydrophobic residues to the hydrophilic environment outside the membrane. During MP folding and association, PPIs can occur in solvated regions and within the membrane between TMDs. Various motifs discovered in TMDs determine whether helices cross in a left- or right-handed fashion (e.g., GASRight and GASLeft motifs) and can be oriented in either a parallel or antiparallel helix-packing geometry (31, 32). Additionally, TMD helices must undergo conformational changes to transduce signals across the membrane. Whether TMDs accomplish transduction through motions such as translation, piston-like motion, or rotation parallel or perpendicular to the membrane remains unknown for most MPs (33). Several conserved motifs within TMDs are involved in specific, and possibly switchable, PPIs (34). Furthermore, some TMD motifs facilitate specific protein–lipid interactions that may control oligomerization of TMD helices. The head groups of lipids bind membrane-adjacent regions of TMDs through electrostatic interactions, and lipid tails bind hydrophobic residues on the TMD helix surfaces via van der Waals interactions (35).
Known conserved TMD–TMD interaction motifs include Gx3G (more broadly, small–x3–small), leucine and glycine zippers, polar–xx–polar/aromatic–xx–aromatic, and serine/threonine-rich motifs (34). Interactions between polar side chains in membranes enable interhelical hydrogen bonds that are stronger in the membrane than in solution (36). The low dielectric constant within the membrane means that carboxylic groups are protonated and have two polar side-chain atoms capable of forming hydrogen bonds. Despite the high thermodynamic cost of transferring polar side chains from water into the membrane, newer models based on known structures explain how polar residues with long side chains can be found in the membrane by “snorkeling” toward the solvent interface (37). In the case of arginine, snorkeling allows the guanidinium group to avoid the hydrophobic core, where the energetic cost of desolvation is highest, and instead interact with negatively charged phospholipid head groups (37). However, bulky residues can cause steric clashes that disrupt dimerization if they are located at the interacting surface.
Nonpolar interactions feature in other TMD PPIs, as exemplified by the π-stacking responsible for aromatic–xx–aromatic motif interactions (34). Several closely related structural motifs, including Gx3G, small–x3–small, and glycine zippers, permit extensive van der Waals interactions at the TMD–TMD interface, as the Gx3G motif forms an extensive and nearly flat contact surface that docks against ridges in the neighboring helix to form a tightly packed GASRight motif (31, 32). Weak interhelical hydrogen bonding between a CαH and a carbonyl group may also contribute to these TMD–TMD interactions (36). The TMD glycine zipper motif has either a (G,A,S)xxxGxxxG or GxxxGxxx(G,S,T) sequence within at least one of the associating TMDs (38). The minimal entropic penalty required to bury a small residue may explain the presence of glycine, alanine, and serine at the TMD–TMD interface (38). The exposure of backbone atoms enables weakly polar backbone hydrogen bonds, and small residues may permit a closer approach of helices to maximize packing interactions (38). The Langosch group (39) discovered the leucine zipper, known to facilitate helix–helix interactions in soluble proteins, in TMD helix–helix structures by, where it forms a knobs-into-holes side-chain packing.
The most-studied TMD interaction to date is the interaction between the two subunits of the single-pass MP glycophorin A (GpA), an erythrocyte sialoglycoprotein. The possible involvement of TMD interactions in GpA was suggested as far back as 1976 by the Marchesi group (40, 41). During studies of erythrocyte MPs, these researchers found that GpA homodimers were unusually resistant to sodium dodecyl sulfate (SDS). They synthesized a peptide derived from the GpA TMD sequence, which they used to prevent subunits from interacting after heat-induced dissociation. In the 1990s, interest in TMD structures reemerged after Manolios et al. (42) discovered that the T cell receptor (TCR)–CD3 complex associates through subunit TMDs. This interaction relies on hydrogen bonding between basic residues in the TCR TMD and acidic residues in the CD3 TMDs to form three-helix bundles.
Many PPIs rely on complementary shape and electrostatic interactions; how, then, could flat hydrophobic TMDs associate with any specificity? One answer came from research done by the Langosch and Engelman laboratories on what would come to be known as the Gx3G motif (43). These investigators studied specific interactions between TMDs in chimeric GpA constructs by using denaturing gel electrophoresis and circular dichroism spectral analysis after deletion mutagenesis (44). A conservative valine-to-leucine mutation disrupted dimerization, implicating sterics and specifically glycine residues as important for GpA TMD–TMD interactions (44). Computational prediction of the dimer based on mutagenesis data predicted a right-handed interaction at a −30° angle, stabilized by an interhelical threonine–threonine hydrogen bond (45). In the first published use of the ToxR system, Langosch et al. (46) investigated GpA in a living cell membrane, enabling future studies of TMD dimerization by directed evolution.
The first reported structure of the GpA TMD dimer was found by solution NMR in aqueous detergent micelles (47), and several significant differences from the previous computational predictions emerged. The interhelical crossing angle of the packed interface was at a steeper −40° angle, and threonine–threonine interhelical hydrogen bonding between the two helices was not required for interaction. Mutagenesis of glycine residues also led to steric clashes that disrupted dimerization. Thus, specific TMD–TMD interactions might actually be stabilized solely by van der Waals interactions along the length of the transmembrane interface. Drawing on the above-described study by Langosch et al. (46), Russ & Engelman (48) used directed evolution of a transmembrane library in a modified ToxR system called TOXCAT, selecting against noninteracting TMDs and ultimately finding that more than 80% of isolates contained a TMD Gx3G motif. The Langosch group (49) performed its own directed evolution study using a high-diversity library coding for TMD peptides; they observed that the resulting high-affinity sequences were strongly enriched with tryptophan residues, further implicating hot-spot residues in TMD–TMD interactions.
2.1.4. Toll-like receptor structures
Another integral MP family of cell surface receptors with potential as drug targets, the Toll-like receptor (TLR) family, is currently undergoing TMD structure–function studies. TLRs are pattern recognition receptors (PRRs) used by sentinel cells of the innate immune system to detect nonself patterns, either microbe- or damage-associated molecular patterns (MAMPs or DAMPs) from microbes, viruses, and necrotic cells, in order to initiate an inflammatory response to extracellular ligands. Activated TLRs transduce signals through cytosolic Toll/interleukin-1 receptor homology (TIR) domains to form a helical signaling complex (50) termed the myddosome, which recruits kinases, activating transcription factors including nuclear factor κB (NF-κB), leading to expression of proinflammatory cytokines. This process is followed by delayed anti-inflammatory cytokine production.
TLR4 forms a complex with myeloid differentiation protein 2 (MD-2) and CD14 to sense lipopolysaccharide (LPS) bound to LPS-binding protein (LBP). Like GpA, TLRs are single-pass MPs that function as dimers. Structures exist for TLR ectodomains and TIR domains, but no full-length structures have yet been solved. The Lee lab solved ectodomain structures for TLR2 in a ligand-bound complex with TLR1 (51) and TLR6 (52), as well as the structure for the eritoran-bound TLR4–MD-2 complex (53); the Davies group solved the TLR3–dsRNA structure (54); the Wilson group (55) crystallized and characterized flagellin-bound TLR5 ; and the Shimizu lab was the first to solve crystal structures for ectodomains of apo- and liganded TLR8 (56) and TLR9 (57). The Wilson (58) and Shimizu groups (59) also solved the structure of the nonsignaling TLR4 homolog radioprotective 105 (RP105) in complex with the MD-2 homolog MD-1. Interestingly, although the complex is homologous to TLR4–MD-2, RP105–MD-1 forms a 2:2 homodimer upon binding an endogenous lipid, assembling in an unusual head-to-head arrangement to inhibit TLR4 signaling by preventing TLR4 homodimerization (58, 59). Subsequently, the Wilson group discovered that flagellin-bound TLR5 homodimers also assemble into a symmetric 2:2 tail-to-tail complex in a similar organization to TLR1/2 and TLR3 bound to ligand (55).
Crystal structures of both unliganded and liganded TLR8 ectodomains indicate that conformational changes after ligand binding include both a ring rotation and a hinge motion, bringing together the C termini of the ectodomains; structures of TLR9 with activating and inhibitory DNA show how ligand binding might induce TLR oligomerization after Z-loop processing. Agonist binding is thought to bring TLR extracellular C-terminal regions into juxtaposition, allowing intracellular TIR domains to initiate signaling cascades. TLR7–9 are believed to be expressed as preformed dimers (56), but to date no crystal structures have been solved for full-length TLRs, including TMDs. Knowledge of these structures would enable rational design in order to target TLR extracellular domains, but decades after TLRs were accepted as key PRRs in the immune response, the mechanism by which TLR TMDs transduce signals across the membrane are still unknown.
How could TMD–TMD interactions regulate the signaling of dimeric MPs? Dimerization of MP subunits can be driven by TMDs, such as in the case of integrins, where switchable TMD PPIs stabilize the inactive conformation (32). Other activation mechanisms may be possible: Ligand binds to monomer, leading to dimerization and activation, or ligand binds to preformed dimer, leading to conformational change that relieves autoinhibition, in turn causing signal transduction across the membrane (32). Nishiya & DeFranco (60) expressed constructs in primary bone marrow–derived macrophages from TLR4−/− mice, where the ectodomain of TLR4 was fused with the TMD and cytoplasmic domains of TLR1–9. Some chimeras were expressed on the cell surface and were capable of signaling to produce the proinflammatory cytokine tumor necrosis factor (TNF) in response to the TLR4 ligand LPS, indicating that the transmembrane or cytosolic domains were responsible for subcellular localization and signaling. Constitutive activation occurred in N-terminal deletion variants of TLR4, suggesting that the ectodomains may be autoinhibitory (61). Previously, Yin and colleagues (23) used circular dichroism and Förster resonance energy transfer (FRET) to show that peptides derived from TLR TMDs can oligomerize in micelles, and they employed a ToxR assay to demonstrate that they also oligomerize in E. coli membranes. TLR TMD functions in live cell membranes are an active area of investigation that may determine how TLRs mediate signal transduction.
2.2. Novel Biological Insights Revealed
Membrane shape (e.g., curvature) and composition have become fast-growing areas of drug discovery and targeting. Protein–lipid interactions regulate MP clustering (62, 63), lipid raft interactions (64), cell–cell signaling (65, 66), and membrane curvature (67–69). Cell organelles rely on regulated membrane curvature for proper function, and many different proteins sense or induce the curvature required for intracellular functions such as the dynamic motions of the endoplasmic reticulum (ER), vesicular trafficking, endocytosis, and exocytosis.
2.2.1. Extracellular vesicles
A remarkable development in the cell signaling field is the discovery that secretion of bilayered membrane vesicles, conserved from bacteria to humans, enables biomolecular cargoes to be ferried between cells (70, 71). Isolated extracellular vesicles (EVs) contain bioactive lipids (72), RNA (73), and proteins (74) that can function in recipient cells. Mammalian EVs include both exosomes and microvesicles (70, 71). Although there is still no universally agreed-upon definition for these vesicles, exosomes are conventionally described as ~30–100-nm membrane-derived bilayer vesicles. The term exosome is meant to stand in contrast with the term microvesicle, which describes larger (~100–1,000-nm) vesicles that bud directly from the plasma membrane; however, these terms more accurately describe how vesicles are isolated, rather than any biological property (70). Perhaps due to this distinction, mechanisms of sorting cargoes into EVs are only beginning to be understood (75). Furthermore, the mechanism of EV uptake is still unknown and may depend on the cell of origin, the recipient cell, or EV size.
EVs were first investigated for possible use as nonimmunogenic delivery vehicles for cancer vaccines (76). Although questions about the level of purity and absolute quantification of biomolecules are a constant refrain, EVs are attracting great interest in cell–cell communication research. EVs are also recognized for their functions within tumor microenvironments, namely promoting angiogenesis and metastasis. The van Rheenen group (77) recently devised a reporter for EV cargo delivery based on the LoxP–Cre system; using intravital imaging of transplanted tumors in mice, these authors observed that EVs released by malignant cells altered less-malignant cells, enhancing their migratory and metastatic capacities. These findings demonstrate EV-mediated long-range cell–cell communication in vivo.
Validation of EVs as cancer biomarkers for diagnostic purposes comes from a study by Kalluri and colleagues (78). These investigators used mass spectrometry analyses to characterize EVs and found that the presence of glypican-1 on the surface of isolated EVs could distinguish pancreatic cancer patients from both healthy subjects and patients with benign pancreatic disease with absolute specificity and sensitivity.
2.2.2. Molecular mechanisms of curvature sensing
Several mechanisms of protein–lipid interactions allow peptides and proteins to sense and bind curved membranes (Figure 2). Lipid packing is a physical parameter dependent on both the individual lipid geometry and the global membrane curvature; lipid-packing defects arise from a mismatch between these components, leading to transient low-density regions in one leaflet of a lipid bilayer. Amphipathic α-helices containing an Arf GTPase–activating protein 1 lipid-packing sensor (ALPS) motif bind highly curved membranes through the hydrophobic effect; at the same time, bulky hydrophobic side chains (phenylalanine, leucine, tryptophan) on the hydrophobic face of the helix insert into transient lipid-packing defects (Figure 2a), stabilizing these defects and allowing diverse proteins to sense membrane curvature (68). In the contrasting example of α-synuclein, the intrinsically disordered protein also forms an amphipathic α-helix upon interaction with the membrane, but electrostatic interactions are responsible for its membrane curvature sensing. The membrane-adsorbing helical face of α-synuclein contains the small residues valine, alanine, and threonine, but these are flanked by positively charged lysine residues that interact with negatively charged lipid head groups and glutamic acid residues point away from the membrane (69). Proteins can also sense curvature by forming a complementary shape to the curved membrane (Figure 2b). Bin–Amphiphysin–Rvs (BAR) domains form crescent-shaped coiled-coil homodimers with positive residues in the concave face, leading to Coulombic attraction; the concavity of the domain matches the curvature of the membrane and stabilizes the curvature of complementary shape (79). Another mechanism for membrane curvature sensing relies on electrostatic interactions to facilitate the insertion of hydrophobic loops into curved membranes (Figure 2c). For example, the synaptic vesicle–localized Ca2+ sensor synaptotagmin-1 (Syt-1) synchronizes neurotransmitter release during Ca2+-evoked synaptic vesicle fusion. Syt-1 assists in vesicle fusion by bending membranes in a Ca2+-dependent manner with its C2 domains. Ca2+ ions form a complex between membrane-penetrating loops in the C2A and C2B domains and anionic lipid head groups, allowing the loops to insert ~2 nm into the hydrophobic core of the plasma membrane in response to Ca2+ signaling and, ultimately, curve the membrane (80). Oligomerization and scaffolding can also improve sensing of curved membranes (Figure 2d), as typified by the oligomeric networks formed by endophilin at high concentrations on membrane surfaces. This process allows BAR domains to scaffold membranes through higher-order interactions (81).
Figure 2.
Strategies for lipid sensing and curvature targeting. Highly curved membranes contain lipid-packing defects, which are transient low-density regions resulting from a mismatch between individual lipid geometry and global membrane curvature. (a) In hydrophobic insertion, large hydrophobic residues (phenylalanine, leucine, tryptophan) can insert into transient lipid-packing defects in the membrane, stabilizing curvature. (b) In shape-based sensing, shape complementarity between a concave, cationic protein surface and a convex, anionic membrane stabilizes interactions such as the interaction of a Bin–Amphiphysin–Rvs (BAR) domain with a membrane. (c) Electrostatic insertions by metalloproteins use metal ions to coordinate with lipid head groups. In the case of the Ca2+-binding C2B domain of Syt-1 (Protein Data Bank code: 1UOW), Ca2+ ions form a complex between membrane-penetrating loops and anionic lipid head groups, allowing loops to insert ~2 nm into the membrane. (d) Multivalent clustering and oligomerization can also scaffold proteins around membrane curvature.
Proteins may use more than one of these mechanisms, as BAR domains appear to utilize hydrophobic insertions and oligomerization in addition to their complementary shape–based mechanism in membrane interactions (81). Deeper hydrophobic insertions can induce strong bending, as illustrated by reticulons in the peripheral ER and caveolins in the plasma membrane. Rather than sensing curvature, oligomers of these proteins directly cause and stabilize positive curvature as a result of two short hairpin TMDs that do not completely span the bilayer, forming a wedge shape to increase the surface area of the outer membrane leaflet (82). Regulation of membrane curvature is especially important in the ER, which has an elaborate, dynamic morphology that allows ER tubules to appose and signal to other organelles (83). While proteins shape the membrane, the converse is also true, as MP function can be regulated by the lipid environment and membrane shape (84). Disentangling the abilities of different MPs to sense, stabilize, or induce membrane curvature remains a challenge for the future.
2.2.3. Context-specific functions of Toll-like receptor signaling in the immune system
The context in which TLR signaling occurs is essential for understanding the immune response. Advances in immunology and oncoimmunology have already expanded our understanding of the capabilities of TLRs to function in a variety of contexts beyond those originally appreciated, which may become important in personalized therapy. New research on the role played by inflammation and immunity in eliminating early tumors has revealed that the connections between the immune system and tumorigenesis are more substantial than previously thought. For example, the contribution of TLR4 to the success of radiotherapy and chemotherapy treatment by recognizing the DAMP high-mobility-group box 1 (HMGB1) (85) shows that the TLR-activated immune response protects against tumors, not just infection. Recent research into the microbiome has also begun to implicate TLRs in symbiosis (86, 87). However, this understanding is still incomplete, so newly discovered TLR agonists and antagonists could prove useful as probes for understanding how combinations of TLR activation alter the immune response (Figure 3).
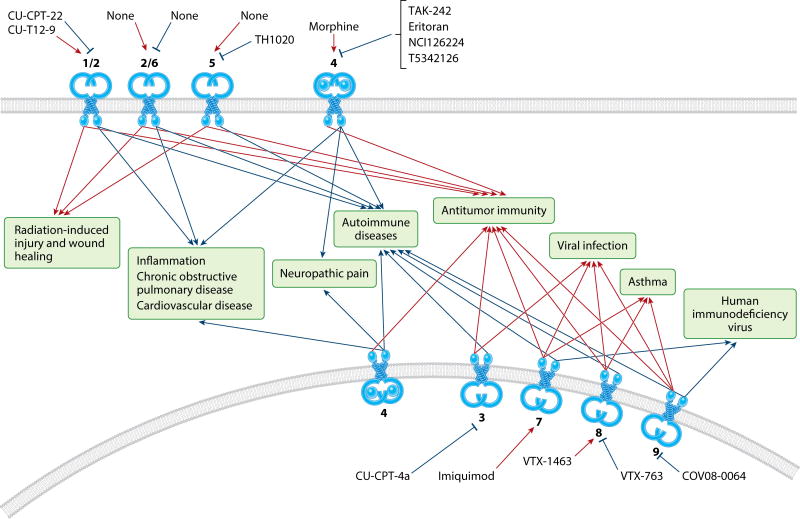
Figure 3.
Selective small molecule and peptide immunomodulators of the Toll-like receptor (TLR) family. TLR agonists and antagonists provide the ability to activate or inhibit the immune response. Agonists are currently being investigated to strengthen the anticancer and antiviral immune response, but TLR antagonists have received the most attention for inflammatory and autoimmune diseases. However, other areas of therapeutic intervention using modulators of TLR signaling continue to be explored. Agonists and antagonists under investigation for targeting TLRs have been reviewed elsewhere (88, 89). This illustration is not meant to be exhaustive but rather to demonstrate the feasibility of using TLR family members as small-molecule drug targets.
Small-molecule and peptide TLR agonists and antagonists are currently being investigated as therapeutics for multiple indications, including inflammatory diseases, autoimmunity, viral infection, and wound healing (88–90). The membrane environment is also important to consider, as protein–lipid interactions regulate TLR function. Knockdowns of TLRs and mass spectrometry–based membrane lipidomics exposed a lipid coregulatory network and TLR–plasma membrane feedback loops (91). TLR signaling causes immune cells to change their plasma membrane composition by modulating sphingolipid metabolism and higher-level functional organization of membrane lipids, ultimately altering TLR trafficking and signaling. Sphingolipid metabolism modulated LPS-induced interleukin (IL)-6 release by altering TLR4 trafficking. Knockdown of acid sphingomyelinase or addition of certain ceramides (e.g., N–C18:0(OH)–Cer or N–C8:0(2H)–Cer) increased TLR4 signaling by altering the abundance of various lipids, thereby increasing surface TLR4 expression.
2.3. Technological Developments
New technologies are enabling investigations into previously inaccessible MP interactions in the areas of novel probing methods, fluorescent biosensors, engineered proteins and peptides, and computational simulations and designs.
2.3.1. Investigation of TMD interactions in membranes
The ToxR system is a two-hybrid assay that has become a standard way to investigate TMD–TMD interactions within a biological membrane. First developed by the Langosch group (43, 46) to investigate GpA TMDs, the Gx3G motif is essential for TMD–TMD interactions. ToxR is a single-pass, inner membrane–spanning, dimerization-dependent transcriptional activator derived from Vibrio cholerae. By replacing the ToxR TMDs with any TMDs of interest, one can measure the strength of TMD interactions. Constructs are expressed in E. coli; N-terminal maltose binding protein localizes to the periplasmic space (fused with the TMD of interest, which localizes to the inner membrane), and the ToxR subunits localize in the cytoplasm. Upon TMD dimerization, the ToxR subunits dimerize and gain the ability to bind the ctx promoter, leading to reporter gene expression. Commonly used reporters include (a) chloramphenicol acetyltransferase (CAT), in which expression provides resistance to chloramphenicol to select for TMD sequences favoring interaction (named TOXCAT by the Engelman group); (b) lacZ coding for β-galactosidase, which hydrolyzes added o-nitrophenyl-β-galactoside to produce o-nitrophenolate, quantifiable by colorimetry; and (c) firefly luciferase reporter expression, which can be quantified using a luminometer (92). It is possible to express two different TMD fusion constructs with two different TMDs to measure heterodimerization by fusing one TMD to a functional ToxR subunit while fusing another TMD to a dominant negative ToxR mutant. Upon TMD-driven heterodimerization, CAT expression decreases, leading to a corresponding decrease in bacterial growth. Although the ToxR system was initially developed for studies of single-pass MPs, Yin and colleagues (93) have also extended it to studies of MPs with multiple TMDs.
2.3.2. Rational design of peptidomimetic and self-organizing probes
By nature, linear peptides are flexible and capable of sampling many conformational states while free in solution. Once bound, these peptides lose rotational and translational freedom. Folded proteins are more constrained and offer guidance for how to overcome this entropic penalty: Increase rigidity and decrease conformational entropy through peptide stapling or cyclization. This procedure works beyond simply increasing the affinity of protein–peptide interactions. As an elegant example, stapled peptides use hydrocarbon linkages to connect amino acid residues, with architectures linking position i with i+3, i+4, or i+7 to bridge one or two helical turns, thereby stabilizing the α-helical conformation (94). Hydrocarbon linkages stabilize α-helicity to enhance uptake and shift equilibrium toward a protein-bound state by reducing the entropic cost of binding (95). Intriguingly, it is possible to go beyond hydrophobic peptides to create synthetic transmembrane assemblies composed of small molecules that undergo supramolecular self-organization within a membrane. Bhosale et al. (96) created biomimetic assemblies of fluorophore scaffolds that oligomerize by transmembrane π-stacking and span a lipid bilayer. These assembles constituted a synthetic photosystem that can produce proton gradients and be converted into an ion channel upon ligand intercalation within the membrane, creating a new paradigm for rational design of multifunctional small-molecule oligomers within a membrane. Small-molecule scaffolds may provide another means of drugging TMDs.
2.3.3. Computational design
Computational design of anti-TMD peptides (97, 98) facilitates development of specific probes that complement antibody-based methods not applicable to TMD regions of MPs. Yin et al. (97) developed a computational strategy, computed helical antimembrane protein (CHAMP), to rationally design peptides that specifically recognize transmembrane helices. From a database of helix pairs from MP structures (31), the small–x3–small TMD motif in both αIIb and αv integrin subunits suggested the preferred mode of interaction. This allowed computationally determined template backbones, based on the parallel GASRight motif, to maximize geometric complementarity between helices by use of a Monte Carlo repacking algorithm to consider different combinations of side chains in low-energy rotamers. Finally, to complete the design, residues at each location of the CHAMP peptide were selected using a residue-based membrane insertion potential model (37).
Wang & Barth (99) employed a similar computational approach to predict TMD structures. They developed an algorithm called EFDOCK-TM, which was implemented in RosettaMembrane. These authors used a direct interaction score to find strongly coevolving residue pairs at predicted contact sites at the interacting interface between TMD helices, then performed TMD helical docking simulations under evolutionary constraints. When overlaid with known NMR structures, the predictions made by EFDOCK-TM achieved atomic accuracy for right-handed TMD homodimers and near-atomic accuracy for left-handed TMD dimers; they also achieved similar accuracy for both a tetramer and a pentamer (99). This algorithm’s success is due to the inclusion of evolutionary conservation information and TMD-adjacent regions in its predictions.
3. APPROACHES TO TARGETING MEMBRANE PROTEIN INTERACTIONS
Approaches to identifying new therapeutics fall into two categories: (a) high-throughput screening (HTS), which is a biomedical brute-force attack to find hits from large libraries, and (b) rational design, which relies on structural knowledge to create molecules.
3.1. High-Throughput Screening
3.1.1. Advantages and challenges
When the genetic basis of a disease is unknown or polygenic or protein structures have not been solved, screening may be a better approach than structural design. In HTS, compounds are screened for activity and off-target binding. Several kinds of HTS are used, including phenotypic screening, target-based screening, and in silico screening (100, 101) of commercial vendor or natural product (102) libraries. Automation allows large chemical libraries to be screened rapidly and quickly identify hits. For instance, the Yin group used HTS to identify inhibitors of TLR interactions (103–105) and other integral MP interactions (106).
Although protein–RNA interactions had previously been considered undruggable, Yin and colleagues have reported selective, small-molecule TLR3 inhibitors. They identified hit compounds from in silico HTS by using the 1.2 million–compound Enamine database; several of these compounds had a surprising scaffold of a D–amino acid conjugated with an aromatic substituent (103). These hits were synthesized and tested in cell-based assays, followed by optimization of synthetic routes and structure–activity relationship studies. Biophysical confirmation of TLR3 binding came with fluorescence anisotropy assays, which demonstrated competitive binding between the final lead compounds CU-CPT-4a and rhodamine-labeled polyinosinic:polycytidylic acid, a synthetic dsRNA analog and a TLR3 agonist, respectively. Later, the Uematsu and Akira groups (107) showed that this compound ameliorates radiation-induced gastrointestinal syndrome in mice after acute γ-ray exposure.
Parkinson’s disease and diffuse Lewy body disease are synucleinopathies, or prion-like accumulations of misfolded oligomeric α-synuclein in the CNS. These oligomers play a central role in the development of a chronic neuroinflammatory milieu and ultimately neurodegeneration; some reports suggest that neuron-released oligomers are DAMPs capable of activating TLR signaling in microglia (108). The Yin lab (105) discovered a small molecule through HTS, CU-CPT-22, that selectively inhibits the TLR1/2 association, providing a useful tool to study the role of this PPI in cell signaling and disease pathology. Recently, CU-CPT-22 was tested for the ability to inhibit the TLR1/2-dependent inflammatory response caused by higher-order α-synuclein oligomers. As measured by NF-κB reporter translocation and TNF secretion in primary mouse microglia, CU-CPT-22 attenuated the overactive microglial response. This finding demonstrates that TLR1/2 is a druggable target to treat neuroinflammation in synucleinopathies (109).
These studies highlight the need for further research into the role of TLR signaling in neuroinflammatory diseases, as well as the therapeutic potential of TLR inhibitors. Similar methods have led to the discovery of an agonist for the same receptors (110), demonstrating that screening for both agonists and antagonists can be fruitful.
Membrane PPIs have also been considered undruggable because of assumptions about nonspecificity and large, flat interfaces. Consider the expression of latent membrane protein 1 (LMP-1) expression during Epstein–Barr virus (EBV) infection. LMP-1 resembles the TNF receptor superfamily member CD40 but homotrimerizes through TMD–TMD interactions, leading to constitutive signaling though its C-terminal domain to activate and immortalize B cells during the latent stage of infection (111). EBV, also known as human herpesvirus 4, is ubiquitous, infecting more than 90% of the population. EBV infection is responsible for many B cell lymphomas (112), and infection is associated with multiple autoimmune diseases (113). Yin and colleagues (111) discovered that a residue in the fifth TMD (TM5) of LMP-1 drives homotrimerization through a hot spot formed by hydrogen-bond interactions among asparagine residues, as well as peptides that interfere with this interaction. The interaction of pentamidine with this TMD was investigated through the ToxR assay, tryptophan fluorescence, coumarin fluorescence dequenching, and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The authors screened a small-molecule library for inhibitors of LMP-1 TM5 interactions, and they identified a compound (NSC 295242) that inhibits oncogenic signaling of LMP-1 by binding to the hot spot and preventing homotrimerization (106). These studies demonstrate the technical feasibility of drugging TMD PPIs using small-molecule agents.
3.1.2. Traptamers
Similar in concept to nucleic acid aptamers, peptide aptamers are proteins built with a randomized sequence within a standard scaffold. Transmembrane peptide aptamers, also known as traptamers, offer a screening-based strategy to find agonists or antagonists of MP TMDs. The E5 protein of bovine papillomavirus, at 44 amino acids, is the smallest known oncoprotein and a known TMD interacting sequence (114). E5 expressed by virally infected cells forms disulfide-linked dimers, then binds to the TMD of platelet-derived growth factor β receptor (PDGFβR), a receptor tyrosine kinase (RTK), to cause dimerization and trans-autophosphorylation. Although PDGFβR is a plasma MP, E5 remains in the ER and Golgi apparatus and activates PDGFβR signaling (115). Experiments by DiMaio and colleagues (115) using TOXCAT with a scaffold based on E5 and internal randomized hydrophobic sequences identified novel proteins that activated PDGFβR signaling to transform cells. The same scaffold was also used to screen for TMD peptides to activate the erythropoietin receptor (EpoR), a type I cytokine receptor, thereby pinpointing sequences capable of selectively activating human EpoR without activating either PDGFβR or murine EpoR (116). EpoR is preformed as a homodimer, and upon activation it changes conformation to activate downstream signaling in erythroid cells, suggesting that EpoR TMDs are capable of forming switchable PPIs to allow anti-TMD peptides to activate signaling. The common drug targeting EpoR, recombinant erythropoietin (produced by Amgen/Janssen), has historically been the highest-expenditure drug by Medicare. The same scaffold-based screening approach also led these researchers to a peptide that inhibited C–C chemokine receptor type 5 (CCR5) expression, preventing HIV entry into T cells (117).
3.2. Rational Design
Computational small-molecule (118) and protein designs (119) take into account structure, thermodynamics (120), and PPI hot spots (30). A major advance in the rational design of novel agents, the design of peptides that target TMD protein–protein (97, 98) and protein–lipid interactions (121–123), illustrates the power of novel drug discovery approaches.
3.2.1. Advantages and challenges
In cases where the one-drug, one-target paradigm holds, rational design aims to maximize selectivity, which can help off-target effects and toxicity (the latter being one of the main causes of failure during clinical trials for small molecules). For protein-based therapeutics, toxicity failures are less common than lack of efficacy. Rational design avoids using expensive libraries and time-consuming validation of hits on target that are often nonspecific or false positives. Designers of peptides and peptidomimetics, such as D-peptides (124), β-peptides (125), and foldamers (126), must consider a number of important factors. For example, a major challenge in peptide-based therapeutics is the entropic cost associated with binding. To overcome this entropic penalty, a peptide can be stapled or cyclized, and thereby stabilized, to increase its affinity for the target. Peptides targeting protein TMDs face additional challenges: They must be soluble in both aqueous and hydrophobic phases, insert into the membrane in the correct orientation, fold, anchor, and finally interact specifically with their TMD target. Peptides and peptidomimetics have a few advantages over larger recombinant proteins, including resistance to protease digestion, reduced immunogenicity, and lower production costs. However, a major issue with peptide drugs is their limited half-life. Peptide drugs specific for MP extracellular domains have generally proven much less effective than monoclonal antibodies because of their rapid elimination, although peptidomimetics and chemical modifications may lengthen half-life (127).
Particularly illustrative of the functional role of TMD helices is that the TMDs of mechanosensitive channels interact with the membrane to sense membrane tension through hydrophobic matching (128). Studies of MP function in bacterial membranes are limited models for physiological function in native membranes, but bacterial MP functions can certainly carry over during expression in mammalian cells. For example, Kralj et al. (129) repurposed a microbial rhodopsin to function as a genetically encoded voltage indicator when expressed in mammalian cell membranes, which enabled recording of individual action potentials in cultured neurons expressing the fluorescent biosensor at a subcellular spatial resolution and submillisecond temporal resolution.
3.2.2. Anti-TMD peptides
On the basis of analyses of TCR and CD3 assembly, Manolios et al. (130) discovered that charged amino acids within the lipid bilayer are critical for stable interactions, and they demonstrated that synthesized peptide analogs of the TCR and CD3 TMDs that peptides derived from the TCR α-chain TMD inhibited T cell activation, as measured by IL-2 secretion and proliferation, without activating B cells, 131). Interestingly, these peptides also inhibited natural killer (NK) cell activation. TMD-derived peptides were extended to GPCRs with studies by Hebert et al. (132), who discovered that β2-adrenergic receptors dimerized and that peptides derived from the sixth TMD could inhibit receptor dimerization and activation. Ng et al. (133) showed that this inhibition was possible with other neurotransmitter GPCRs; using immunoblotting, they showed that dopamine receptor D2 homodimerization in Sf9 cell membranes was specifically blocked with a TMD-derived peptide. Tarasova et al. (134) investigated the structure–activity relationship of multiple GPCR TMD–derived peptides and discovered the importance of charged residues immediately adjacent to the transmembrane residues in order to achieve potent inhibitors.
The ability of TMD peptides to alter signal transduction was first demonstrated with a rationally designed peptide based on CD2 and known α-chain interactions between immunoglobulin E (IgE) and the high-affinity IgE receptor (FcεRI) (135). Both L- and retroenantiomeric D-peptides were cyclized by an intrachain disulfide bond, leaving them constrained yet moderately flexible. In agreement with the hot-spot residue hypothesis, these anti-TMD peptides were able to inhibit IgE signaling in mast cells, preventing degranulation in response to dinitrophenyl–human serum albumin challenge, as monitored by β-hexosaminidase release. Binding was also measured by circular dichroism spectroscopy and surface plasmon resonance, further proof that structure-based design of small constrained peptides could inhibit PPIs, in this case with relevance in type I hypersensitivity.
The CHAMP approach was employed to design and synthesize anti-TMD peptides binding αIIbβ3 and αVβ3 integrins to activate signaling in micelles, bacteria, and ultimately mammalian cell membranes (97). The peptides bound with high affinity in micelles, with a equilibrium dissociation constant (Kd) of 0.32±0.05 μM for αIIb TMD and anti-αIIb, or 3.2 ± 0.5 × 10−4 in mole fraction units of peptide to detergent. The dominant negative TOXCAT assay in E. coli showed that the anti-TMD peptides were specific for their targets. Upon addition to mammalian cell membranes, the rationally designed anti-αv peptide induced platelet aggregation in a dose-dependent manner. The success of this approach was due in part to exploiting known TMD–TMD PPI motifs, as the interaction face between the anti-TMD peptide and target TMD helices resembled a glycine zipper motif. The CHAMP methodology was later extended to a β-peptide foldamer targeting integrins (β-CHAMP), which allowed the DeGrado group (136) to target the Gx3G motif on the αIIb TMD by first positioning a poly(homoglycine) sequence to find the optimal backbone, using a grid search and the CHARM force field, and then optimizing van der Waals contacts with the target TMD.
RTKs are single-pass transmembrane proteins that signal as dimers. Mutations and aberrant activity cause many different developmental disorders and diseases, including several types of cancer (137). In most cases, ligand binding to the extracellular domain induces receptor dimerization and signals through trans-autophosphorylation of intracellular tyrosine kinase domains, which phosphorylate downstream signaling proteins. The Lemmon group (138) used TOXCAT and mutagenesis to show that TMDs from the class I RTK ErbB receptor associate due to Gx3G motifs. Thereafter, Bennasroune et al. (139) used expression vectors coding for ErbB TMD peptides, as well as liposome-delivered synthetic TMD peptides, to inhibit receptor dimerization and extracellular signal–related kinase 1/2 signaling. TMD-derived peptides have been used as RTK activators for the class II RTK insulin receptor (140). Of course, other hydrophobic peptides can activate RTK signaling, as shown by the DiMaio lab (115), which showed that transmembrane peptides produced by directed evolution activate the class III RTK PDGFβR.
TLR TMDs specifically interact; for example the TLR2 TMD specifically interacts with TMDs derived from known interacting partners of TLR2, a finding that suggests the possibility of modulating TLR signaling by TMD-targeting peptides (23). Although the mechanism of these interactions is not fully understood, small–x3–small motifs are found in the TMDs of TLR2, TLR7, and TLR9 (23). Through the use of a ToxR assay, a TLR2 TMD–derived peptide was synthesized and confirmed to interact with TLR2 in membrane mimetic environments, in cultured macrophages, and finally in a murine acute sepsis–like model in which decreased cytokine production and increased survival were observed in response to challenge by the TLR2 agonist lipoteichoic acid when mice were administered 5 mg/kg of the TLR2 TMD peptide (141).
Rational design and molecular modeling will improve with the increasing number of transmembrane protein crystal structures and our improving understanding of TMD interactions with other MPs and lipids. Thus far, the fundamental challenge in designing anti-TMD therapeutics has been poor understanding of TMD interaction specificity. However, as more structural motifs are uncovered, chemical biologists will be able to design molecular probes to validate these interactions.
3.2.3. Curvature-sensing peptides
Rational design has recently been used to create peptides that may be able to target interactions between proteins and curved membranes. The Chapman group (80) described the ability of Syt-1 to bend membranes in a Ca2+-dependent manner with loops within its C2B domain, which insert ~2 nm into the hydrophobic core of membranes to sense Ca2+ signaling and allow for synchronized synaptic vesicle fusion and neurotransmitter release in neurons. As mentioned in Section 2.3.2, above, peptide cyclization has become a common approach to decrease the entropic cost of binding. Combining biological and thermodynamic information, Yin and colleagues (121) derived a fragment from the membrane-penetrating loop of Syt-1 and created a synthetic peptide, which was cyclized by copper-catalyzed “click” chemistry to approximately match the size of the native loop. By labeling the peptide with the solvatochromic fluorophore nitrobenzoxadiazole (NBD), the investigators were able to use the peptide as a fluorescent biosensor for curvature, and cyclizing these peptides enabled them to selectively recognize highly curved membranes in liposomes and small EVs.
Another curvature-sensing peptide, MARCKS-ED, was derived from the effector domain (ED; residues 151–175) of myristoylated alanine-rich C-kinase substrate (MARCKS). MARCKS functions in cells by sequestering the negatively charged phospholipid PI(4,5)P2 into a region on the inner leaflet by Coulombic interactions between its ED and the plasma membrane. It responds to local increases in calcium concentration via removal from the membrane by Ca2+-bound calmodulin, releasing PI(4,5)P2 to freely diffuse throughout the membrane (142). The McLaughlin group found that MARCKS-ED peptides could bind liposomes (143) and that phosphorylation, ionic strength, and calmodulin could all reverse this interaction (144). The Cafiso lab (145) used electron paramagnetic spectroscopy (EPR) to investigate the interaction of cysteine-substituted spin-labeled MARCKS-ED with spin-labeled proxyl-PI(4,5)P2 in vesicles, which indicated that MARCKS-ED binds to membranes and that the subsequent sequestration of negatively charged lipids is mediated by electrostatic interactions. However, magic-angle spinning NMR in lipid bilayers highlighted the depth of the phenylalanine side chains within the lipid bilayer, suggesting that the penetration of the aromatic phenyl rings into the acyl chain region of the membrane could contribute to membrane-binding energy (146). In a further complication, studies with radiolabeled and fluorescence-labeled peptides indicated that MARCKS-ED binds to membranes without developing any secondary structure (147).
The previously discovered BAR domain relies on coiled-coil bundles, and the more recently recognized ALPS motif relies on intrinsically unfolded sequences that form amphipathic α-helices for their membrane curvature–sensing mechanisms. So, does MARCKS contain an unrecognized motif for membrane curvature sensing? MARCKS-ED senses negatively charged, highly curved membranes, such as those found on EVs, and NBD-labeled peptide binds to apoptotic cells in C. elegans, indicating the possibility of employing MARCKS-ED as a fluorescent biosensor by binding exposed phosphatidylserine in vivo (123). The combination of the curvature and lipid composition of EVs can cause lipid-packing defects (148), and computational analyses show that these defects influence binding (149) for both L- and D-MARCKS-ED (150), hinting at shared mechanisms with other curvature sensors. In a similar fashion to naturally occurring biopolymers, the affinity of membrane-binding bradykinin was increased by covalently attaching peptides to form multivalent clusters (122), adding evidence that oligomerization is a mechanism for membrane curvature sensing (Figure 2). New insights into the structural basis of membrane curvature recognition are still necessary for rationally designed curvature-sensing drugs, but the era of membrane curvature research is only beginning.
4. PERSPECTIVES
Targeting TLRs has the potential to steer the immune response in cancer immunotherapy, and drugging TMDs represents a novel approach. Unsuccessful trials for TLR agonists administered systemically have led to redesigns with local administration, and current trials using TLR ligands in combination with blockade of anti-inflammatory cytokines may prove more successful (151). The identification of anti-TMD drugs may rely on the discovery of small-molecule probes, such as the selective TLR1/2 and TLR3 agonists that have increased the set of selective TLR modulators (103, 152). These molecules may be further developed as drugs while also serving dual roles as probes for discovery of new modulators of TLR signaling, including anti-TMD drugs. Additionally, cell-based assays for identifying hits rely mostly on genetically encoded reporters that provide a readout for downstream signaling, but specificity is rarely assured by this approach because transcription factors integrate signals from different receptors. One may foresee development of new biosensors to detect dimerization for a more direct look at how individual receptors are activated, including the conformational changes of TMDs resulting from ligand binding.
Disruptive new platforms are key to developing novel therapeutics targeting MPs. Technologically advanced top-down mass spectrometry, cryo-EM, HTS, and other instrumentation will become de rigueur in drug discovery. Bioengineers have made major advances in developing both protein- and cell-based therapeutics to specifically bind the extracellular surfaces of MPs. Nevertheless, we believe that small molecules and peptides drugging transmembrane helices are the next therapeutic frontier. Our knowledge of MP structure is rapidly improving, and tremendous “omics” databases, increasingly at the single-cell level, are a treasure trove for bioinformaticians. Moreover, innovative instruments and novel biosensors have facilitated biological research, yet the present challenge is to apply the full potential of these technologies to drug discovery by rethinking druggability. Recently discovered protein motifs that exploit membrane protein–protein and protein–lipid interactions epitomize the promise of unexplored binding sites, and the existence of several anti-TMD peptides and small molecules that take advantage of these sites serve as proof-of-mechanism studies for drugging therapeutic targets implicated in metastatic, inflammatory, neurological, and metabolic diseases. The recent conclusion of the National Institutes of Health Molecular Libraries Program, which discovered 375 small-molecule probes, many of which were first in class (153), will help identify targets that are inaccessible to protein-based therapeutics. Still, the membrane proteome is vast, with more than 10,000 unique proteins (1). We believe that developing novel TMD peptide probes with high affinity and specificity is necessary to shed light on the structure–function relationship of MP TMDs in the pursuit of new druggable regions (154). A new category of targets awaits exploration by new modalities; rapid advances in drug discovery may ultimately pivot on the ability to drug the “undruggable.”
Figure 4.
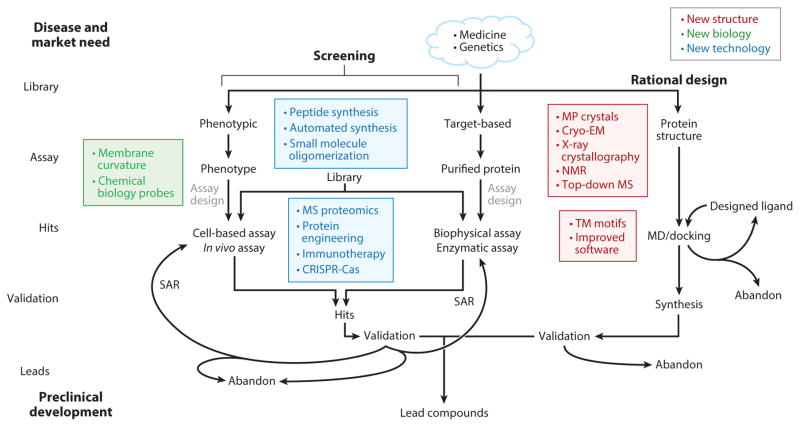
Advances affecting the drug discovery work flow. High-throughput screening and rational design are two contrasting approaches to drug discovery. Improved membrane protein structural information (red), improved biological understanding of membranes (green), and new technologies (blue) affect different segments of the discovery pipeline. MD: molecular dynamics. SAR: structure-activity relationship.
Figure 5.
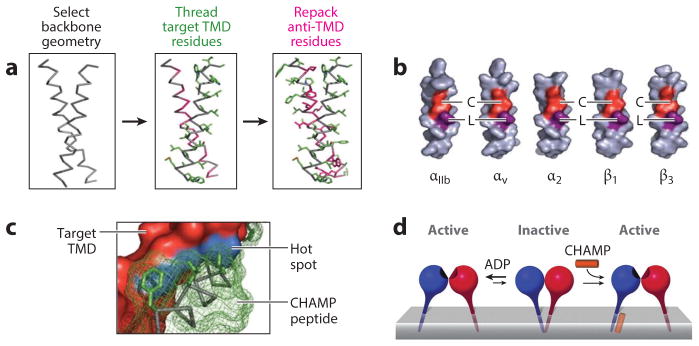
Rational design of anti–transmembrane domain (TMD) peptides. (a) In the initial peptide design, a backbone geometry is first selected from existing structures that contain motifs found in the TMD target; amino acid residues from the target TMD are then added to the backbone (green); and finally a side chain–repacking algorithm is run on the computed helical antimembrane protein (CHAMP) peptide (pink). (b) Sequence motifs are illustrated on target integrin TMD idealized conformations, with common small sequences (red) and a common leucine (purple). (c) A tightly packing interface between the CHAMP peptide (green), the integrin TMD (red), and the hot spot (blue) is predicted. (d) Integrin activation by an anti-TMD peptide is explained by a model indicating the effect of the anti-TMD peptide in shifting the equilibrium of integrin subunits towards the active state. Modified from Reference 97.
Summary Points.
It is technically feasible to modulate MP function by selectively drugging TMD protein–protein and protein–lipid interactions.
Both HTS and rational design approaches have produced leads that challenge the conventional definition of druggability.
Chemical biology probes and fluorescent biosensors are attractive research tools to study membrane curvature and lipid composition.
Unconventional drug–target interactions are providing new directions for drug discovery.
Future Issues.
Using a variety of tactics to uncover additional full-length MP structures will help explain general principles of TMD structure–function relationships.
Hydrophobic peptide delivery remains a challenge for therapeutic use and will likely require advances in drug delivery systems for further preclinical development.
Optimization of peptidomimetics to maximize pharmacological stability will provide another advantage to the drug modality.
Curvature-sensing peptides could find broader use in selectively binding EVs and other curved membranes for further analysis.
Understanding the interrelationship between MPs and the membrane environment may reveal new forms of cellular regulation.
Acknowledgments
We thank the National Institutes of Health (NIH R01GM103843 and R01GM101279) for financial support. A.F. is supported by the Signaling and Cellular Regulation Training Program (NIH T32GM08759). We thank James I. Godfroy III for proofreading the manuscript.
Glossary
- MP
membrane protein
- GPCR
G protein–coupled receptor
- Druggability
a term used to evaluate the likelihood of being able to modify the function of a target protein with a drug that binds the target with high affinity
- TMD
transmembrane domain
- PPI
protein–protein interaction
- Cryoelectron microscopy (cryo-EM)
transmission electron microscopy of samples frozen in a native environment, preserving specimens without fixation, staining, or crystallization
- NMR
nuclear magnetic resonance
- Gx3G motif (also known as the GxxxG or GXXXG motif)
consists of small residues (glycine, alanine, or serine) separated by three residues and stabilizes transmembrane helix interactions
- Protein–lipid interaction
noncovalent interaction between MPs and lipids that may shape the membrane or alter MP function
- TLR
Toll-like receptor
- FRET
Förster resonance energy transfer
- EV
extracellular vesicle
- HTS
high-throughput screening
- In silico screening
process in which a virtual library of small molecules or peptides undergoes simulated docking with a target to identify hits
- Protein–nucleic acid interaction
interaction utilized by conventionally “undruggable” DNA- or RNA-binding proteins or by pattern recognition receptors that sense nucleic acids
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Hang Yin, Email: Hubert.Yin@colorado.edu.
Aaron D. Flynn, Email: Aaron.Flynn@colorado.edu.
LITERATURE CITED
- 1.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, et al. Tissue-based map of the human proteome. Science. 2015;347:6220. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 2.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–96. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 3.Rask-Andersen M, Almén MS, Schiöth HB. Trends in the exploitation of novel drug targets. Nat Rev Drug Discov. 2011;10:579–90. doi: 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]
- 4.Rask-Andersen M, Masuram S, Schiöth HB. The druggable genome: Evaluation of drug targets in clinical trials suggests major shifts in molecular class and indication. Annu Rev Pharmacol Toxicol. 2014;54:9–26. doi: 10.1146/annurev-pharmtox-011613-135943. [DOI] [PubMed] [Google Scholar]
- 5.Cheng AC, Coleman RG, Smyth KT, Cao Q, Soulard P, et al. Structure-based maximal affinity model predicts small-molecule druggability. Nat Biotechnol. 2007;25:71–75. doi: 10.1038/nbt1273. [DOI] [PubMed] [Google Scholar]
- 6.Lahti JL, Tang GW, Capriotti E, Liu T, Altman RB. Bioinformatics and variability in drug response: a protein structural perspective. J R Soc Interface. 2012;9:1409–37. doi: 10.1098/rsif.2011.0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schenone M, Dančík V, Wagner BK, Clemons PA. Target identification and mechanism of action in chemical biology and drug discovery. Nat Chem Biol. 2013;9:232–40. doi: 10.1038/nchembio.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright PM, Seiple IB, Myers AG. The evolving role of chemical synthesis in antibacterial drug discovery. Angew Chem Int Ed Engl. 2014;53:8840–69. doi: 10.1002/anie.201310843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin H, Hamilton AD. Strategies for targeting protein–protein interactions with synthetic agents. Angew Chem Int Ed Engl. 2005;44:4130–63. doi: 10.1002/anie.200461786. [DOI] [PubMed] [Google Scholar]
- 10.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature. 2007;450:1001–9. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 11.Azzarito V, Long K, Murphy NS, Wilson AJ. Inhibition of α-helix-mediated protein–protein interactions using designed molecules. Nat Chem. 2013;5:161–73. doi: 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]
- 12.Lagerström MC, Schiöth HB. Structural diversity of G protein–coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–57. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 13.Rosenbaum DM, Rasmussen SGF, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–63. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–94. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 15.Klabunde T, Hessler G. Drug design strategies for targeting G-protein-coupled receptors. ChemBioChem. 2002;3:928–44. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 16.White SH. Membrane Proteins of Known 3D Structure. 2015 http://blanco.biomol.uci.edu/mpstruc/
- 17.Bai X, McMullan G, Scheres SH. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 2014;40(1):49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Miao J, Ishikawa T, Robinson IK, Murnane MM. Beyond crystallography: diffractive imaging using coherent X-ray light sources. Science. 2015;348:530–35. doi: 10.1126/science.aaa1394. [DOI] [PubMed] [Google Scholar]
- 19.Miao Y, Cross TA. Solid state NMR and protein–protein interactions in membranes. Curr Opin Struct Biol. 2013;23:919–28. doi: 10.1016/j.sbi.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis. Cell. 2000;100:391–98. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 21.Beel AJ, Sanders CR. Substrate specificity of γ-secretase and other intramembrane proteases. Cell Mol Life Sci. 2008;65:1311–34. doi: 10.1007/s00018-008-7462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munter L-M, Voigt P, Harmeier A, Kaden D, Gottschalk KE, et al. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Aβ42. EMBO J. 2007;26:1702–12. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Godfroy JI, Roostan M, Moroz YS, Korendovych IV, Yin H. Isolated Toll-like receptor transmembrane domains are capable of oligomerization. PLOS ONE. 2012;7:e48875. doi: 10.1371/journal.pone.0048875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langosch D, Scharnagl C, Steiner H, Lemberg MK. Understanding intramembrane proteolysis: from protein dynamics to reaction kinetics. Trends Biochem Sci. 2015;40:318–27. doi: 10.1016/j.tibs.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Sun L, Zhao L, Yang G, Yan C, Zhou R, et al. Structural basis of human γ-secretase assembly. PNAS. 2015;112:6003–8. doi: 10.1073/pnas.1506242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong M, Zhang Y, Hu F. Membrane protein structure and dynamics from NMR spectroscopy. Annu Rev Phys Chem. 2012;63:1–24. doi: 10.1146/annurev-physchem-032511-143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freedberg DI, Selenko P. Live cell NMR. Annu Rev Biophys. 2014;43:171–92. doi: 10.1146/annurev-biophys-051013-023136. [DOI] [PubMed] [Google Scholar]
- 28.Claasen B, Axmann M, Meinecke R, Meyer B. Direct observation of ligand binding to membrane proteins in living cells by a saturation transfer double difference (STDD) NMR spectroscopy method shows a significantly higher affinity of integrin αIIbβ3 in native platelets than in liposomes. J Am Chem Soc. 2005;127:916–19. doi: 10.1021/ja044434w. [DOI] [PubMed] [Google Scholar]
- 29.Mari S, Invernizzi C, Spitaleri A, Alberici L, Ghitti M, et al. 2D TR-NOESY experiments interrogate and rank ligand–receptor interactions in living human cancer cells. Angew Chem Int Ed Engl. 2010;49:1071–74. doi: 10.1002/anie.200905941. [DOI] [PubMed] [Google Scholar]
- 30.Clackson T, Wells J. A hot spot of binding energy in a hormone–receptor interface. Science. 1995;267:383–86. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 31.Walters RFS, DeGrado WF. Helix-packing motifs in membrane proteins. PNAS. 2006;103:13658–63. doi: 10.1073/pnas.0605878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore DT, Berger BW, DeGrado WF. Protein–protein interactions in the membrane: sequence, structural, and biological motifs. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matthews EE, Zoonens M, Engelman DM. Dynamic helix interactions in transmembrane signaling. Cell. 2006;127:447–50. doi: 10.1016/j.cell.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 34.Fink A, Sal-Man N, Gerber D, Shai Y. Transmembrane domains interactions within the membrane milieu: principles, advances and challenges. Biochim Biophys Acta. 2012;1818:974–83. doi: 10.1016/j.bbamem.2011.11.029. [DOI] [PubMed] [Google Scholar]
- 35.Stangl M, Schneider D. Functional competition within a membrane: lipid recognition vs. transmembrane helix oligomerization. Biochim Biophys Acta. 2015;1848:1886–96. doi: 10.1016/j.bbamem.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 36.Senes A, Ubarretxena-Belandia I, Engelman DM. The Cα–H···O hydrogen bond: a determinant of stability and specificity in transmembrane helix interactions. PNAS. 2001;98:9056–61. doi: 10.1073/pnas.161280798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senes A, Chadi DC, Law PB, Walters RFS, Nanda V, Degrado WF. Ez, a depth-dependent potential for assessing the energies of insertion of amino acid side-chains into membranes: derivation and applications to determining the orientation of transmembrane and interfacial helices. J Mol Biol. 2007;366:436–48. doi: 10.1016/j.jmb.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 38.Kim S, Jeon T-J, Oberai A, Yang D, Schmidt JJ, Bowie JU. Transmembrane glycine zippers: physiological and pathological roles in membrane proteins. PNAS. 2005;102:14278–83. doi: 10.1073/pnas.0501234102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurezka R, Laage R, Brosig B, Langosch D. A heptad motif of leucine residues found in membrane proteins can drive self-assembly of artificial transmembrane segments. J Biol Chem. 1999;274:9265–70. doi: 10.1074/jbc.274.14.9265. [DOI] [PubMed] [Google Scholar]
- 40.Furthmayr H, Marchesi VT. Subunit structure of human erythrocyte glycophorin A. Biochemistry. 1976;15:1137–44. doi: 10.1021/bi00650a028. [DOI] [PubMed] [Google Scholar]
- 41.Furthmayr H, Galardy RE, Tomita M, Marchesi VT. The intramembranous segment of human erythrocyte glycophorin A. Arch Biochem Biophys. 1978;185:21–29. doi: 10.1016/0003-9861(78)90139-x. [DOI] [PubMed] [Google Scholar]
- 42.Manolios N, Bonifacino J, Klausner R. Transmembrane helical interactions and the assembly of the T cell receptor complex. Science. 1990;249:274–77. doi: 10.1126/science.2142801. [DOI] [PubMed] [Google Scholar]
- 43.Brosig B, Langosch D. The dimerization motif of the glycophorin A transmembrane segment in membranes: importance of glycine residues. Protein Sci. 1998;7:1052–56. doi: 10.1002/pro.5560070423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemmon MA, Flanagan JM, Hunt JF, Adair BD, Bormann BJ, et al. Glycophorin A dimerization is driven by specific interactions between transmembrane α-helices. J Biol Chem. 1992;267:7683–89. [PubMed] [Google Scholar]
- 45.Adams PD, Engelman DM, Brünger AT. Improved prediction for the structure of the dimeric transmembrane domain of glycophorin A obtained through global searching. Proteins. 1996;26:257–61. doi: 10.1002/(SICI)1097-0134(199611)26:3<257::AID-PROT2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 46.Langosch D, Brosig B, Kolmar H, Fritz HJ. Dimerisation of the glycophorin A transmembrane segment in membranes probed with the ToxR transcription activator. J Mol Biol. 1996;263:525–30. doi: 10.1006/jmbi.1996.0595. [DOI] [PubMed] [Google Scholar]
- 47.MacKenzie KR. A transmembrane helix dimer: structure and implications. Science. 1997;276:131–33. doi: 10.1126/science.276.5309.131. [DOI] [PubMed] [Google Scholar]
- 48.Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix–helix association. J Mol Biol. 2000;296:911–19. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 49.Ridder A, Skupjen P, Unterreitmeier S, Langosch D. Tryptophan supports interaction of transmembrane helices. J Mol Biol. 2005;354:894–902. doi: 10.1016/j.jmb.2005.09.084. [DOI] [PubMed] [Google Scholar]
- 50.Lin S-C, Lo Y-C, Wu H. Helical assembly in the MyD88–IRAK4–IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–90. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, et al. Crystal structure of the TLR1–TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–82. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Kang JY, Nan X, Jin MS, Youn S-J, Ryu YH, et al. Recognition of lipopeptide patterns by Toll-like receptor 2–Toll-like receptor 6 heterodimer. Immunity. 2009;31:873–84. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 53.Kim HM, Park BS, Kim J-I, Kim SE, Lee J, et al. Crystal structure of the TLR4–MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–17. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 54.Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, et al. Structural basis of Toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–81. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon S, Kurnasov O, Natarajan V, Hong M, Gudkov AV, et al. Structural basis of TLR5–flagellin recognition and signaling. Science. 2012;335(6070):859–64. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanji H, Ohto U, Shibata T, Miyake K, Shimizu T. Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands. Science. 2013;339:1426–29. doi: 10.1126/science.1229159. [DOI] [PubMed] [Google Scholar]
- 57.Ohto U, Shibata T, Tanji H, Ishida H, Krayukhina E, et al. Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature. 2015;520:702–5. doi: 10.1038/nature14138. [DOI] [PubMed] [Google Scholar]
- 58.Yoon S, Hong M, Wilson IA. An unusual dimeric structure and assembly for TLR4 regulator RP105–MD-1. Nat Struct Mol Biol. 2011;18:1028–35. doi: 10.1038/nsmb.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohto U, Miyake K, Shimizu T. Crystal structures of mouse and human RP105/MD-1 complexes reveal unique dimer organization of the Toll-like receptor family. J Mol Biol. 2011;413:815–25. doi: 10.1016/j.jmb.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 60.Nishiya T, DeFranco AL. Ligand-regulated chimeric receptor approach reveals distinctive subcellular localization and signaling properties of the Toll-like receptors. J Biol Chem. 2004;279:19008–17. doi: 10.1074/jbc.M311618200. [DOI] [PubMed] [Google Scholar]
- 61.Panter G, Jerala R. The ectodomain of the Toll-like receptor 4 prevents constitutive receptor activation. J Biol Chem. 2011;286:23334–44. doi: 10.1074/jbc.M110.205419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van den Bogaart G, Meyenberg K, Risselada HJ, Amin H, Willig KI, et al. Membrane protein sequestering by ionic protein–lipid interactions. Nature. 2011;479:552–55. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aimon S, Callan-Jones A, Berthaud A, Pinot M, Toombes GES, Bassereau P. Membrane shape modulates transmembrane protein distribution. Dev Cell. 2014;28:212–18. doi: 10.1016/j.devcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 65.Yang H, Chen Y-Z, Zhang Y, Wang X, Zhao X, et al. A lysine-rich motif in the phosphatidylserine receptor PSR-1 mediates recognition and removal of apoptotic cells. Nat Commun. 2015;6:5717. doi: 10.1038/ncomms6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Neumann B, Coakley S, Giordano-Santini R, Linton C, Lee ES, et al. EFF-1-mediated regenerative axonal fusion requires components of the apoptotic pathway. Nature. 2015;517:219–22. doi: 10.1038/nature14102. [DOI] [PubMed] [Google Scholar]
- 67.McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–96. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 68.Drin G, Casella J-F, Gautier R, Boehmer T, Schwartz TU, Antonny B. A general amphipathic α-helical motif for sensing membrane curvature. Nat Struct Mol Biol. 2007;14:138–46. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 69.Antonny B. Mechanisms of membrane curvature sensing. Annu Rev Biochem. 2011;80:101–23. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 70.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–89. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 71.Kastelowitz N, Yin H. Exosomes and microvesicles: identification and targeting by particle size and lipid chemical probes. ChemBioChem. 2014;15:923–28. doi: 10.1002/cbic.201400043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Subra C, Laulagnier K, Perret B, Record M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie. 2007;89:205–12. doi: 10.1016/j.biochi.2006.10.014. [DOI] [PubMed] [Google Scholar]
- 73.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–76. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen B, Wu N, Yang J-M, Gould SJ. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J Biol Chem. 2011;286:14383–95. doi: 10.1074/jbc.M110.208660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, et al. Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell derived exosomes. Nat Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 77.Zomer A, Maynard C, Verweij FJ, Kamermans A, Schäfer R, et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161:1046–57. doi: 10.1016/j.cell.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–82. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gallop JL, Jao CC, Kent HM, Butler PJG, Evans PR, et al. Mechanism of endophilin N-BAR domain–mediated membrane curvature. EMBO J. 2006;25:2898–910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hui E, Johnson CP, Yao J, Dunning FM, Chapman ER. Synaptotagmin-mediated bending of the target membrane is a critical step in Ca2+-regulated fusion. Cell. 2009;138:709–21. doi: 10.1016/j.cell.2009.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Isas JM, Ambroso MR, Hegde PB, Langen J, Langen R. Tubulation by amphiphysin requires concentration-dependent switching from wedging to scaffolding. Structure. 2015;23:873–81. doi: 10.1016/j.str.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zurek N, Sparks L, Voeltz G. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 2011;12:28–41. doi: 10.1111/j.1600-0854.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Westrate LM, Lee JE, Prinz WA, Voeltz GK. Form follows function: the importance of endoplasmic reticulum shape. Annu Rev Biochem. 2015;84:791–811. doi: 10.1146/annurev-biochem-072711-163501. [DOI] [PubMed] [Google Scholar]
- 84.Phillips R, Ursell T, Wiggins P, Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–85. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, et al. Toll-like receptor 4–dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050–59. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 86.Round JL, Lee SM, Li J, Tran G, Jabri B, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–77. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–27. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 88.Hennessy EJ, Parker AE, O’Neill LAJ. Targeting Toll-like receptors: emerging therapeutics? Nat Rev Drug Discov. 2010;9:293–307. doi: 10.1038/nrd3203. [DOI] [PubMed] [Google Scholar]
- 89.Wang X, Smith C, Yin H. Targeting Toll-like receptors with small molecule agents. Chem Soc Rev. 2013;42:4859–66. doi: 10.1039/c3cs60039d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hoque R, Farooq A, Malik A, Trawick BN, Berberich DW, et al. A novel small-molecule enantiomeric analogue of traditional (−)-morphinans has specific TLR9 antagonist properties and reduces sterile inflammation-induced organ damage. J Immunol. 2013;190:4297–4304. doi: 10.4049/jimmunol.1202184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Köberlin MS, Snijder B, Heinz LX, Baumann CL, Fauster A, et al. A conserved circular network of coregulated lipids modulates innate immune responses. Cell. 2015;162:170–83. doi: 10.1016/j.cell.2015.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roth L, Nasarre C, Dirrig-Grosch S, Aunis D, Crémel G, et al. Transmembrane domain interactions control biological functions of neuropilin-1. Mol Biol Cell. 2008;19:646–54. doi: 10.1091/mbc.E07-06-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Joce C, Wiener AA, Yin H. Multi-Tox: application of the ToxR-transcriptional reporter assay to the study of multi-pass protein transmembrane domain oligomerization. Biochim Biophys Acta. 2011;1808:2948–53. doi: 10.1016/j.bbamem.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cromm PM, Spiegel J, Grossmann TN. Hydrocarbon stapled peptides as modulators of biological function. ACS Chem Biol. 2015;10:1362–75. doi: 10.1021/cb501020r. [DOI] [PubMed] [Google Scholar]
- 95.London N, Movshovitz-Attias D, Schueler-Furman O. The structural basis of peptide–protein binding strategies. Structure. 2010;18:188–99. doi: 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 96.Bhosale S, Sisson AL, Talukdar P, Fürstenberg A, Banerji N, et al. Photoproduction of proton gradients with pi-stacked fluorophore scaffolds in lipid bilayers. Science. 2006;313:84–86. doi: 10.1126/science.1126524. [DOI] [PubMed] [Google Scholar]
- 97.Yin H, Slusky JS, Berger BW, Walters RS, Vilaire G, et al. Computational design of peptides that target transmembrane helices. Science. 2007;315:1817–22. doi: 10.1126/science.1136782. [DOI] [PubMed] [Google Scholar]
- 98.Caputo GA, Litvinov RI, Li W, Bennett JS, Degrado WF, Yin H. Computationally designed peptide inhibitors of protein–protein interactions in membranes. Biochemistry. 2008;47:8600–6. doi: 10.1021/bi800687h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang Y, Barth P. Evolutionary-guided de novo structure prediction of self-associated transmembrane helical proteins with near-atomic accuracy. Nat Commun. 2015;6:7196. doi: 10.1038/ncomms8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shoichet BK, Kobilka BK. Structure-based drug screening for G-protein-coupled receptors. Trends Pharmacol Sci. 2012;33:268–72. doi: 10.1016/j.tips.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lounkine E, Keiser MJ, Whitebread S, Mikhailov D, Hamon J, et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012;486:361–67. doi: 10.1038/nature11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Harvey AL, Edrada-Ebel R, Quinn RJ. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov. 2015;14:111–29. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 103.Cheng K, Wang X, Yin H. Small-molecule inhibitors of the TLR3/dsRNA complex. J Am Chem Soc. 2011;133:3764–67. doi: 10.1021/ja111312h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang S, Cheng K, Wang X, Yin H. Selection, synthesis, and anti-inflammatory evaluation of the arylidene malonate derivatives as TLR4 signaling inhibitors. Bioorg Med Chem. 2012;20:6073–79. doi: 10.1016/j.bmc.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheng K, Wang X, Zhang S, Yin H. Discovery of small-molecule inhibitors of the TLR1/TLR2 complex. Angew Chem. 2012;124:12412–15. doi: 10.1002/anie.201204910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang X, Saludes JP, Zhao TX, Csakai A, Fiorini Z, et al. Targeting the lateral interactions of transmembrane domain 5 of Epstein–Barr virus latent membrane protein 1. Biochim Biophys Acta. 2012;1818:2282–89. doi: 10.1016/j.bbamem.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Takemura N, Kawasaki T, Kunisawa J, Sato S, Lamichhane A, et al. Blockade of TLR3 protects mice from lethal radiation-induced gastrointestinal syndrome. Nat Commun. 2014;5:3492. doi: 10.1038/ncomms4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim C, Ho D-H, Suk J-E, You S, Michael S, et al. Neuron-released oligomeric α-synuclein is an endogenous agonist of TLR2 for paracrine activation of microglia. Nat Commun. 2013;4:1562. doi: 10.1038/ncomms2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Daniele SG, Béraud D, Davenport C, Cheng K, Yin H, Maguire-Zeiss KA. Activation of MyD88-dependent TLR1/2 signaling by misfolded α-synuclein, a protein linked to neurodegenerative disorders. Sci Signal. 2015;8:ra45. doi: 10.1126/scisignal.2005965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cheng K, Gao M, Godfroy JI, Brown PN, Kastelowitz N, Yin H. Specific activation of the TLR1–TLR2 heterodimer by small-molecule agonists. Sci Adv. 2015;1:e1400139. doi: 10.1126/sciadv.1400139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sammond DW, Joce C, Takeshita R, McQuate SE, Ghosh N, et al. Transmembrane peptides used to investigate the homo-oligomeric interface and binding hotspot of latent membrane protein 1. Biopolymers. 2011;95:772–84. doi: 10.1002/bip.21672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Young LS, Rickinson AB. Epstein–Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–68. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 113.Küppers R. Mechanisms of B cell lymphoma pathogenesis. Nat Rev Cancer. 2005;5:251–62. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 114.Talbert-Slagle K, DiMaio D. The bovine papillomavirus E5 protein and the PDGF β receptor: It takes two to tango. Virology. 2009;384:345–51. doi: 10.1016/j.virol.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Freeman-Cook LL, Dixon AM, Frank JB, Xia Y, Ely L, et al. Selection and characterization of small random transmembrane proteins that bind and activate the platelet-derived growth factor β receptor. J Mol Biol. 2004;338:907–20. doi: 10.1016/j.jmb.2004.03.044. [DOI] [PubMed] [Google Scholar]
- 116.Cammett TJ, Jun SJ, Cohen EB, Barrera FN, Engelman DM, Dimaio D. Construction and genetic selection of small transmembrane proteins that activate the human erythropoietin receptor. PNAS. 2010;107:3447–52. doi: 10.1073/pnas.0915057107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scheideman EH, Marlatt SA, Xie Y, Hu Y, Sutton RE, DiMaio D. Transmembrane protein aptamers that inhibit CCR5 expression and HIV coreceptor function. J Virol. 2012;86:10281–92. doi: 10.1128/JVI.00910-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tinberg CE, Khare SD, Dou J, Doyle L, Nelson JW, et al. Computational design of ligand-binding proteins with high affinity and selectivity. Nature. 2013;501:212–16. doi: 10.1038/nature12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Samish I, MacDermaid CM, Perez-Aguilar JM, Saven JG. Theoretical and computational protein design. Annu Rev Phys Chem. 2011;62:129–49. doi: 10.1146/annurev-physchem-032210-103509. [DOI] [PubMed] [Google Scholar]
- 120.Chaires JB. Calorimetry and thermodynamics in drug design. Annu Rev Biophys. 2008;37:135–51. doi: 10.1146/annurev.biophys.36.040306.132812. [DOI] [PubMed] [Google Scholar]
- 121.Saludes JP, Morton LA, Ghosh N, Beninson LA, Chapman ER, et al. Detection of highly curved membrane surfaces using a cyclic peptide derived from synaptotagmin-I. ACS Chem Biol. 2012;7:1629–35. doi: 10.1021/cb3002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Saludes JP, Morton LA, Coulup SK, Fiorini Z, Cook BM, et al. Multivalency amplifies the selection and affinity of bradykinin-derived peptides for lipid nanovesicles. Mol Biosyst. 2013;9:2005–9. doi: 10.1039/c3mb70109c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Morton LA, Yang H, Saludes JP, Fiorini Z, Beninson L, et al. MARCKS-ED peptide as a curvature and lipid sensor. ACS Chem Biol. 2013;8:218–25. doi: 10.1021/cb300429e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao L, Lu W. Mirror image proteins. Curr Opin Chem Biol. 2014;22:56–61. doi: 10.1016/j.cbpa.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kritzer JA, Stephens OM, Guarracino DA, Reznik SK, Schepartz A. β-Peptides as inhibitors of protein–protein interactions. Bioorg Med Chem. 2005;13:11–16. doi: 10.1016/j.bmc.2004.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Horne WS, Gellman SH. Foldamers with heterogeneous backbones. Acc Chem Res. 2008;41:1399–408. doi: 10.1021/ar800009n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Werle M, Bernkop-Schnürch A. Strategies to improve plasma half life time of peptide and protein drugs. Amino Acids. 2006;30:351–67. doi: 10.1007/s00726-005-0289-3. [DOI] [PubMed] [Google Scholar]
- 128.Haswell ES, Phillips R, Rees DC. Mechanosensitive channels: What can they do and how do they do it? Structure. 2011;19:1356–69. doi: 10.1016/j.str.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods. 2012;9:90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Manolios N, Collier S, Taylor J, Pollard J, Harrison LC, Bender V. T cell antigen receptor transmembrane peptides modulate T cell function and T cell–mediated disease. Nat Med. 1997;3:84–88. doi: 10.1038/nm0197-84. [DOI] [PubMed] [Google Scholar]
- 131.Huynh NT, Ffrench RA, Boadle RA, Manolios N. Transmembrane T-cell receptor peptides inhibit B- and natural killer–cell function. Immunology. 2003;108:458–64. doi: 10.1046/j.1365-2567.2003.01614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hebert TE, Moffett S, Morello J-P, Loisel TP, Bichet DG, et al. A peptide derived from a 2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation. J Biol Chem. 1996;271:16384–92. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 133.Ng GY, O’Dowd BF, Lee SP, Chung HT, Brann MR, et al. Dopamine D2 receptor dimers and receptor-blocking peptides. Biochem Biophys Res Commun. 1996;227:200–4. doi: 10.1006/bbrc.1996.1489. [DOI] [PubMed] [Google Scholar]
- 134.Tarasova NI, Rice WG, Michejda CJ. Inhibition of G-protein-coupled receptor function by disruption of transmembrane domain interactions. J Biol Chem. 1999;274:34911–15. doi: 10.1074/jbc.274.49.34911. [DOI] [PubMed] [Google Scholar]
- 135.McDonnell JM, Beavil AJ, Mackay GA, Jameson BA, Korngold R, et al. Structure based design and characterization of peptides that inhibit IgE binding to its high-affinity receptor. Nat Struct Biol. 1996;3:419–26. doi: 10.1038/nsb0596-419. [DOI] [PubMed] [Google Scholar]
- 136.Shandler SJ, Korendovych IV, Moore DT, Smith-Dupont KB, Streu CN, et al. Computational design of a β-peptide that targets transmembrane helices. J Am Chem Soc. 2011;133:12378–81. doi: 10.1021/ja204215f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mendrola JM, Berger MB, King MC, Lemmon MA. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J Biol Chem. 2002;277:4704–12. doi: 10.1074/jbc.M108681200. [DOI] [PubMed] [Google Scholar]
- 139.Bennasroune A, Fickova M, Gardin A, Dirrig-Grosch S, Aunis D, et al. Transmembrane peptides as inhibitors of ErbB receptor signaling. Mol Biol Cell. 2004;15:3464–74. doi: 10.1091/mbc.E03-10-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee J, Miyazaki M, Romeo GR, Shoelson SE. Insulin receptor activation with transmembrane domain ligands. J Biol Chem. 2014;289:19769–77. doi: 10.1074/jbc.M114.578641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fink A, Reuven EM, Arnusch CJ, Shmuel-Galia L, Antonovsky N, Shai Y. Assembly of the TLR2/6 transmembrane domains is essential for activation and is a target for prevention of sepsis. J Immunol. 2013;190:6410–22. doi: 10.4049/jimmunol.1202033. [DOI] [PubMed] [Google Scholar]
- 142.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–11. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 143.Kim J, Blackshear PJ, Johnson JD, McLaughlin S. Phosphorylation reverses the membrane association of peptides that correspond to the basic domains of MARCKS and neuromodulin. Biophys J. 1994;67:227–37. doi: 10.1016/S0006-3495(94)80473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kim J, Shishido T, Jiang X, Aderem A, McLaughlin S. Phosphorylation, high ionic strength, and calmodulin reverse the binding of MARCKS to phospholipid vesicles. J Biol Chem. 1994;269:28214–19. [PubMed] [Google Scholar]
- 145.Rauch ME, Ferguson CG, Prestwich GD, Cafiso DS. Myristoylated alanine-rich C kinase substrate (MARCKS) sequesters spin-labeled phosphatidylinositol 4,5-bisphosphate in lipid bilayers. J Biol Chem. 2002;277:14068–76. doi: 10.1074/jbc.M109572200. [DOI] [PubMed] [Google Scholar]
- 146.Zhang W, Crocker E, McLaughlin S, Smith SO. Binding of peptides with basic and aromatic residues to bilayer membranes: Phenylalanine in the myristoylated alanine-rich C kinase substrate effector domain penetrates into the hydrophobic core of the bilayer. J Biol Chem. 2003;278:21459–66. doi: 10.1074/jbc.M301652200. [DOI] [PubMed] [Google Scholar]
- 147.Wang J, Gambhir A, Hangyás-Mihályné G, Murray D, Golebiewska U, McLaughlin S. Lateral sequestration of phosphatidylinositol 4,5-bisphosphate by the basic effector domain of myristoylated alanine-rich C kinase substrate is due to nonspecific electrostatic interactions. J Biol Chem. 2002;277:34401–12. doi: 10.1074/jbc.M203954200. [DOI] [PubMed] [Google Scholar]
- 148.Vanni S, Hirose H, Barelli H, Antonny B, Gautier R. A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat Commun. 2014;5:4916. doi: 10.1038/ncomms5916. [DOI] [PubMed] [Google Scholar]
- 149.Morton LA, Tamura R, de Jesus AJ, Espinoza A, Yin H. Biophysical investigations with MARCKS-ED: dissecting the molecular mechanism of its curvature sensing behaviors. Biochim Biophys Acta. 2014;1838:3137–44. doi: 10.1016/j.bbamem.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yan L, de Jesus AJ, Tamura R, Li V, Cheng K, Yin H. Curvature sensing MARCKS-ED peptides bind to membranes in a stereo-independent manner. J Pept Sci. 2015;21:577–85. doi: 10.1002/psc.2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lu H. TLR agonists for cancer immunotherapy: tipping the balance between the immune stimulatory and inhibitory effects. Front Immunol. 2014;5:83. doi: 10.3389/fimmu.2014.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cheng K, Wang X, Zhang S, Yin H. Discovery of small-molecule inhibitors of the TLR1/TLR2 complex. Angew Chem Int Ed Engl. 2012;51:12246–49. doi: 10.1002/anie.201204910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schreiber SL, Kotz JD, Li M, Aubé J, Austin CP, et al. Advancing biological understanding and therapeutics discovery with small-molecule probes. Cell. 2015;161:1252–65. doi: 10.1016/j.cell.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yin H. Exogenous agents that target transmembrane domains of proteins. Angew Chem Int Ed Engl. 2008;47:2744–52. doi: 10.1002/anie.200704780. [DOI] [PubMed] [Google Scholar]