ABSTRACT
As cells grow, move, and divide, they must reorganize and rearrange their membranes and cytoskeleton. The F-BAR protein family links cellular membranes with actin cytoskeletal rearrangements in processes including endocytosis, cytokinesis, and cell motility. Here we review emerging information on mechanisms of F-BAR domain oligomerization and membrane binding, and how these activities are coordinated with additional domains to accomplish scaffolding and signaling functions.
KEYWORDS: actin cytoskeleton, cell motility, cytokinesis, endocytosis, F-BAR domain, membrane bending, membrane binding, oligomerization, scaffolding, signaling, tubulation
Introduction
Dynamic cellular processes like motility, endocytosis, and cytokinesis require cells to remodel their membranes in concert with cytoskeleton reorganizations.1 The Bin/Amphiphysin/Rvs (BAR) domain family of proteins is a central player in these processes, acting to link the plasma membrane to the actin cytoskeleton. The BAR protein family is defined by its membrane-binding BAR domain that folds into a dimeric, tightly interwound 6-helix bundle with a curved, crescent-like shape.2,3 Structural studies have determined these domains interact with membranes through the concave face of their crescent-shaped structures.2,3 However, not all BAR domains are shaped the same; multiple structural varieties exist, including classical BAR domains whose membrane-binding face is highly curved (BAR, N-BARs),2 inverse BAR domains whose membrane-binding face bows outward to form a convex curve (I-BARs),4 and Fer/Cip4 homology (FCH) BAR domains that form an elongated, shallow curve (F-BARs).3 In this review we will focus on the banana-shaped F-BAR family. F-BAR domains are accompanied by a variety of other domains in proteins, including SH3, µHD, tyrosine kinase, or GTPase activating domains (GAPs) (Fig. 1). Here we will discuss recent work on F-BAR proteins with an emphasis on integrating F-BAR domain activities like oligomerization with functions of their additional domains in pursuit of a complete understanding of F-BAR protein function in vivo.
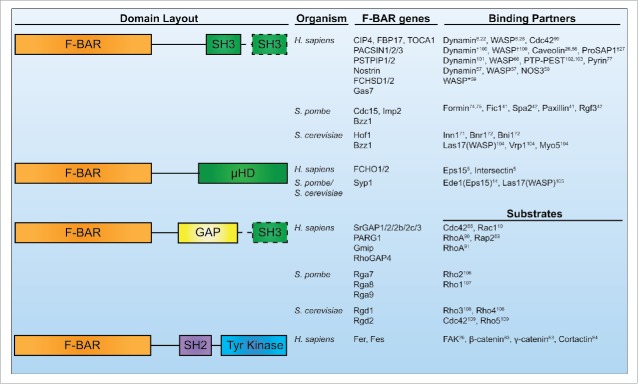
Figure 1.
Human, S. pombe, and S. cerevisiae F-BAR proteins. Conserved domain layouts and select binding partners or substrates of human, S. pombe, and S. cerevisiae F-BAR proteins. Dashed lines indicate the domain is present only in a subset of the listed F-BAR proteins. *Indicates data from Drosophila homologs. †Indicates data from mouse or rat homologs.
F-BAR domain activities
Membrane binding
The crescent-shaped F-BAR domain binds directly to membranes, localizing F-BAR proteins to various sites of action in cells. Emphasizing this important characteristic, membrane binding activity is essential for F-BAR protein function in all cases tested. For instance, point mutations within the FCHo2 and FBP17 F-BARs that specifically disrupt membrane binding prevent the proteins from localizing to the plasma membrane and sites of endocytosis.5-7 Likewise, Schizosaccharomyces pombe Cdc15 or Imp2 membrane binding mutants are not functional.8,9 Proteins lacking the F-BAR domain entirely also fail to properly localize and function: srGAP2ΔF-BAR and PACSINΔF-BAR fail to localize to the plasma membrane,10,11 while Saccharomyces cerevisiae Hof1ΔF-BAR and Syp1ΔF-BAR lose localization to the bud neck12,13 and sites of endocytosis,14 respectively.
F-BAR domains interact with negatively charged membranes primarily through the concave face of their crescent shaped dimers, utilizing multiple positively charged surface residues.3,8,9,15 PACSIN F-BAR domains also contain a unique amphipathic “wedge-loop” that partially inserts into the bilayer16; mutations in this region consequently disrupt membrane binding. The concave orientation of membrane binding is conserved in all F-BARs, though a few variations have been proposed. Under certain conditions in vitro, the FBP17 F-BAR domain associates with membranes through a side face,7 and the Drosophila Nwk F-BAR has also been observed in a side conformation on membranes.17 It is not yet clear if these alternative orientations are important for function or if they occur in vivo; mutations that disrupt this conformation must be tested for functionality in vivo to confirm that a sideways orientation is utilized in cells.
Given that F-BAR domains use positively charged residues for membrane binding, it is not surprising that they are generally capable of binding membranes containing negatively charged phospholipids. While most prefer phosphatidylserine (PS) or various phosphorylated phosphatidylinositol (PIP) lipid head groups, some F-BAR domains including srGAPs,18,19 Drosophila Nwk,17 and S. pombe Cdc15, bind membranes containing multiple species of PIPs.9 Other F-BAR domains display a preference for a certain lipid, though this preference appears less stringent than that of other lipid binding domains such as pleckstrin homology (PH) domains.20 FBP17 and CIP4 prefer PI(4,5)P2 as well as PS,6,21 PACSINs prefers PS,21 Fer prefers phosphatidic acids,22 S. pombe Imp2 prefers PI(4)P,8 and S. cerevisiae Rgd1p prefers PI(4,5)P215. However, in all of these cases the F-BAR domains can bind membranes containing only PS as a negatively charged lipid in vitro.
In healthy eukaryotic cells, PS and PI(4,5)P2 are exclusively located in the inner leaflet of the plasma membrane.23 PS comprises ∼2–10% of the inner leaflet of the PM, while PI(4)P and PI(4,5)P2 are present in trace amounts (≤1 %).23 Therefore preferences for PS and PI(4,5)P2 may be important to direct F-BAR proteins to specific areas enriched in these phospholipids, like the plasma membrane or endocytic sites. Conversely, some evidence suggests this relationship works in the opposite direction; F-BAR proteins may serve to cluster certain plasma membrane lipids into stable micro-domains.24 F-BAR domain-mediated clustering of lipids could be important for generating distinct lipid environments at cellular structures like endocytic sites,25 caveolae,26 or neuronal spines.27 This hypothesis requires further investigation in vivo, perhaps with specific lipid sensors to assay for lipid organization defects in F-BAR membrane-binding mutants.
Oligomerization and membrane bending
Since the earliest characterizations of F-BARs, multiple F-BAR domains were observed not only to bind, but to bend membranes into thin tubules when present at high concentration.6,21,28 Tubulation has been observed when certain F-BAR domains are added to liposomes in vitro, or when overproduced in cultured cells. F-BAR-coated membrane tubules formed in this manner adopt a range of diameters from ∼50 to ∼200 nm,7,8 indicating a degree of flexibility in the F-BAR coat. This heterogeneity initially precluded structural determination of the F-BAR coat; however, Frost and colleagues generated homogenously coated tubules using careful in vitro slow-annealing methods. Using cryo-electron microscopy, the structure of the F-BAR coat upon these tubules was determined,7 revealing that CIP4 F-BAR dimers oligomerize through complex lateral and tip to tip interactions to form a dense coat upon the membrane.7,29 Accordingly, mutations that disrupt oligomerization of F-BAR domains prevent tubulation in overexpression assays.7,8 Computational methods corroborate the oligomerization model of tubulation, as an assembly of oligomerized F-BAR domains can bend a flat membrane into a tubule in molecular dynamics simulations.30 Attractive models proposing F-BAR domains oligomerize upon a membrane in order to collectively induce membrane curvature in endocytosis and other processes have arisen from these structural studies.31-34
Yet, the ability to induce inward-oriented membrane tubules isn't conserved in all F-BAR domains. srGAP F-BAR domains induce tubules of the opposite curvature, outward from the plasma membrane.10,19 This curvature generation is likely accomplished through similar mechanisms as I-BAR proteins, which also oligomerize to collectively bend a membrane.35 Recent evidence suggests even these two varieties of membrane tubulation do not adequately describe the functions of all F-BAR family members. In fact, multiple F-BAR proteins do not tubulate membranes in standard in vitro liposome binding or cultured cell overexpression assays. These include: Fer,6 Fes, RhoGAP4, Gas7, and FCHSD1/2,9 as well as S. pombe Cdc15,9 S. cerevisiae Hof1,15 and Drosophila Nwk.17
It could be argued that the perfect condition (such as a specific lipid composition) has not been discovered to support tubulation of these F-BARs. However, cultured cells contain a variety of membranes with different compositions that overexpressed F-BAR domains can access,36 so this seems an unlikely possibility. Also, multiple compositions mimicking physiological membranes have been tested in vitro, as well as lipid extracts from tissue.9 Synthetic membrane conditions with higher concentrations of PIPs or other lipids depart from a realistic cell-like environment. Further, sufficiently concentrating any protein upon a membrane by adding more binding sites (such as PIPs) is sufficient to induce tubulation through molecular crowding effects.37 A simple explanation for the observed lack of tubulation is certain F-BAR domains do not oligomerize in a manner that confers tubulation activity. And indeed, despite not tubulating membranes, these F-BAR domains do oligomerize.9,17,38-40 Membrane tubulation by F-BAR domains, therefore, appears be one specific consequence of a generally shared ability to oligomerize and simultaneously bind membranes.
One example of an oligomerization mechanism that does not lead to membrane tubulation can be found within the S. pombe Cdc15 F-BAR. EM studies showed this F-BAR domain oligomerizes into linear filaments even in the absence of membrane9 through direct tip to tip electrostatic interactions between F-BAR dimers. Other examples include oligomerization of the Drosophila Nwk F-BAR, which forms short zig-zag structures that can subtly bend and pucker membranes in vitro,17 and lateral contacts between dimers of the Fer and RhoGAP4 F-BAR domains.9 Each case of F-BAR oligomerization studied so far has defined a distinct mechanism of dimer-dimer interaction; F-BAR domains therefore have evolved multiple ways to link together. Further investigation will be necessary to determine the full complement of oligomerization mechanisms used by this protein domain. Targeted mutagenesis of prominent charged surface patches on the tips and sides of F-BAR domains (which could mediate dimer-dimer interactions), and subsequent screening for loss of oligomerization has previously been successful in identifying oligomerization interfaces.9
Oligomerization that does not lead to tubulation nevertheless appears central to F-BAR protein function. Cdc15's linear oligomerization supports a robust avidity (as each repeating F-BAR unit has membrane binding contacts) toward a flat membrane surface. This high avidity membrane binding is critical to accumulate and stabilize Cdc15 at the cell division site9 where it recruits and scaffolds multiple cytokinesis proteins.41,42 Mutations that disrupt F-BAR oligomerization sharply decrease the abundance and increase the turnover of Cdc15 at the division site, which consequently leads to cytokinetic failures.9 Mutations in Fer that block oligomerization compromised its ability to induce lamellipodia formation and enhance cell migration, possibly due to a loss of strong localization to the leading edge membrane.9 Additionally, mutations in the RhoGAP4 F-BAR that disrupt oligomerization compromised RhoGAP4's ability to inhibit cell migration.9 These examples highlight what may be a generally important characteristic of F-BAR domains - their ability to form oligomers on membrane surfaces for the purpose of scaffolding additional protein elements or forming signaling centers.
Surprisingly, the importance of oligomerization for physiological function in the cases of tubulating F-BARs has rarely been directly tested, though it is clear that blocking oligomerization inhibits tubulation of liposomes in vitro and of the plasma membrane in mammalian cell overexpression assays.7 In other words, whether the tubulation activity of F-BAR proteins in vitro is connected to their functions in vivo has not been rigorously established in most cases. To our knowledge, this connection has been tested in only two cases of tubulating F-BARs and different results have been obtained. In the first, it was found that patient derived mutations in FCHO2 that block oligomerization but not membrane binding inhibit FCHO2 recruitment to endocytic sites.5 This may be due to a reduction in membrane binding avidity, similar to the case of Cdc15. In the second, mutations that block oligomerization and tubulation of S. pombe Imp2 have no discernable impact on its localization or function.8 While this is still surprising, the lack of oligomerization may be compensated for by other protein-protein interactions which enforce a local concentration of Imp2 at the division site.42 Clearly though, these examples highlight the value of clarifying the importance of F-BAR domain oligomerization in other F-BAR proteins. As it is now established that F-BAR domains utilize different interaction surfaces to bind one another and oligomerize, this will not be a simple matter of creating homologous mutations, but will require elucidation of each protein's mechanism of oligomerization.
Considering the diversity of oligomerization modes and their functional importance, we propose F-BAR domains in general act as membrane binding, oligomerizing modules that serve to concentrate and stabilize F-BAR proteins at sites of action (Fig. 2). Beyond this generalization, it is likely that different F-BAR domains possess oligomerization interactions that are tailored for distinct functional contexts. When F-BARs are organized by cellular functions, some trends emerge: many endocytic F-BAR domains possess oligomerization interactions that confer binding to or may induce curved membranes (such as FBP17, CIP4, and PACSINs), while F-BARs involved in cytokinesis or cell migration are tuned to bind a relatively “flat” plasma membrane (including Fer, Fes, RhoGAP4, and S. pombe Cdc15) (Fig. 2).
Figure 2.
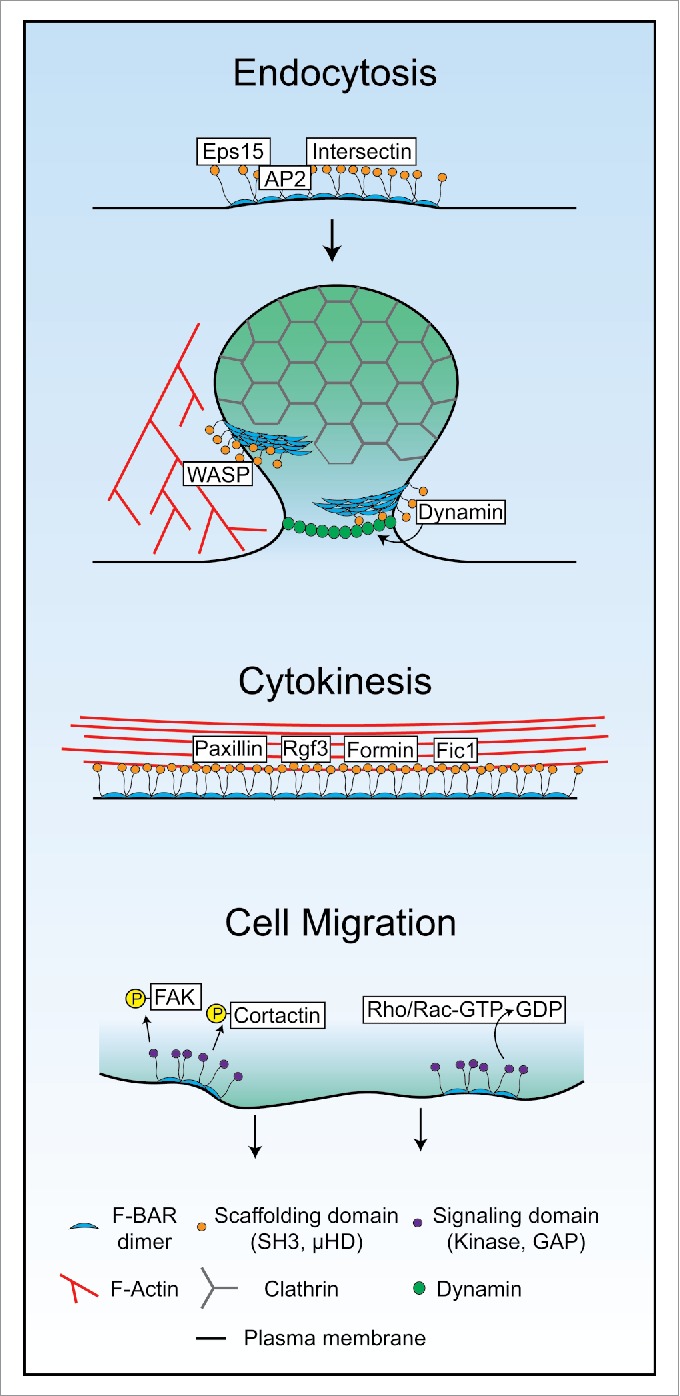
Diverse modes of F-BAR oligomerization in endocytosis, cytokinesis, and cell migration. Schematics of possible modes of F-BAR protein oligomerization, protein recruitment, and signaling in endocytosis, cytokinesis, and cell migration.
Additional domains
F-BAR domain membrane binding and oligomerization are only part of an F-BAR protein's job. Once an F-BAR is bound and oligomerized upon a membrane, it utilizes additional domains to perform scaffolding and signaling functions.
Scaffolding functions
The majority of F-BAR proteins contain either a SH3 or µHD domain that they use to connect with other proteins (Fig. 1). In the cases of F-BAR proteins involved in endocytosis, they recruit partners that in turn have scaffolding and protein recruitment functions. For example, FCHO1/2 F-BAR proteins are two of the first components to localize at incipient sites of endocytosis.5 FCHO2 uses a µHD domain to directly recruit Eps15 and Intersectin,5 and an unstructured middle region to bind and allosterically activate AP2.43,44 In yeast, Syp1 acts similarly; it is present early at sites of endocytosis and recruits Ede1, an Eps15 homolog.14,45
The tubulating activity of FCHO1/2 F-BAR domains3 led to the idea that they might induce the initial membrane curvature early at an endocytic site.5 However, single-molecule imaging experiments suggest that FCHO1/2 “stabilize” the growing bud but do not initiate curvature.46 Efficient recruitment of binding partners and activation of AP2,43,44 aided by clustering from oligomerization,5 may instead explain how FCHO2 acts as a nucleator of clathrin-mediated endocytosis. Indeed, multiple other proteins at endocytic sites are likely responsible for curvature generation, including the FCHO2 binding partner Eps15,47,48 multiple classical BAR domain proteins,33 and the triskelion clathrin coat.49
Slightly later in endocytosis, FBP17 and CIP4 F-BAR proteins bind the budding vesicle.50 FBP17 and CIP4 may contribute to branched actin network formation at the endocytic site through SH3 domain-mediated recruitment of Arp2/3 activators WASP6,51,52 and, for CIP4, WAVE.53 Both FBP17 and CIP4's SH3 domains also recruit the GTPase Dynamin,6,21,28,50 a critical component for vesicle scission54 (Fig. 2). The PACSIN group of F-BAR proteins similarly scaffold WASP and Dynamin and are present in clathrin-mediated endocytosis in certain cells,55 as well as caveolar endocytic sites.26,56 Nostrin also functions at caveolae by recruiting WASP, Dynamin, and a specific substrate, nitric oxide synthase, to regulate its internalization.57,58 Drosophila Nwk proteins correspondingly recruit WASP,59 Dynamin,60 and sorting nexin Snx1661 in neurons to regulate synaptic growth receptor signaling at presynaptic neuromuscular junctions.61,62 Therefore, multiple F-BAR proteins at endocytic sites in different cell types build branched actin networks through recruitment of WASP or WAVE, and assist in vesicle scission through recruitment of Dynamin.
In other cellular processes, F-BAR proteins perform similar scaffolding functions to bridge the membrane to the actin cytoskeleton. As examples, CIP4's binding and recruitment of WASP is also important in regulating lamellipodia during cell migration,63,64 and seems to be a critical component inducing invadopodia in cancer cells.65 In neurons, PACSIN2 interacts with ProSAP1 to form stable membrane-bound structures in neural spines (presumably through oligomerization) which regulate spine organization.27 Furthermore, the PSTPIP1 F-BAR protein scaffolds PTP-PEST phosphatases together with substrates such as WASP66,67 and Abl68 to modulate the actin cytoskeleton.
Model organism studies have also contributed to our understanding of scaffolding F-BAR proteins. In fission yeast, the Cdc15 and Imp2 F-BAR proteins are membrane-bound components of the contractile ring.69,70 Using redundant SH3 domains, these proteins recruit crucial contractile ring proteins including Fic1, Spa2, Rgf3, and Pxl1.41,42 Oligomerization by Cdc15 is critical to localize its partners; oligomerization mutants recruit ∼50% less SH3 binding partners, compromising cytokinesis.9 In S. cerevisiae, the homologous Hof1 F-BAR protein is also important for cytokinesis, recruiting Inn1 through its SH3 domain which activates the chitin synthase necessary for division.71 Hof1's SH3 domain also binds the cytokinetic formin Bnr1,72 which “tunes” the formin's activities.73
While many F-BAR proteins use µHD and SH3 domains for scaffolding functions, in certain cases F-BAR domains also interact directly with other proteins. For example, the S. pombe Cdc15 F-BAR domain directly binds and recruits the formin Cdc12,74,75 which is responsible for F-actin formation in the contractile ring.76 Human PSTPIP1's F-BAR domain interacts with Pyrin77; this interaction activates Pyrin to initiate pyroptosome formation and an inflammatory response.78 It is possible that these two interactions occur simultaneously with membrane binding; the F-BAR domain could bind the membrane on its concave face and a partner on its opposite, cytoplasmic face. In contrast, the PACSIN2 F-BAR domain can interact directly with F-actin filaments in vitro through its concave face79; this interaction excludes simultaneous membrane binding. The cytoplasmic face of F-BAR domains may represent a more generally utilized surface for F-BAR proteins to form linkages with other proteins upon the membrane than currently appreciated.
Based upon much work in the field since their original description, it is clear that many F-BAR family proteins serve as membrane bound scaffolds for a variety of binding partners (Fig. 1). Oligomerization through their F-BAR domains aids scaffolding by locally concentrating the proteins upon membranes. SH3 domains have relatively low affinity (∼1–100 µM) for substrates80; F-BAR oligomerization may therefore help to build a high density network of SH3 or µHDs to strongly link with actin cytoskeletal partners (Fig. 2).
Signaling functions
Other F-BAR proteins contain protein kinase or GAP domains (Fig. 1) and act as signal transducers to the cytoskeleton. Fer and Fes are unique non-receptor tyrosine kinases whose F-BAR domains localize the proteins to the leading edges of migrating cells22 or focal adhesions,81 respectively. Fer and Fes F-BAR domain oligomerization impacts activation of their tyrosine kinase domains through trans-phosphorylation,38,39 similar conceptually to how receptor tyrosine kinase clustering promotes trans-activation. When activated, Fer and Fes phosphorylate several substrates including FAK,82 β- and γ-catenin,83 and cortactin84 to modulate cell-cell and cell-matrix contacts.85
The last class of F-BAR proteins contain GTPase activating domains (GAPs) that promote GTP to GDP catalysis by small GTPases. The most well studied of these in humans is the srGAP group. srGAP1/2/2b-c/3 participate in neural morphogenesis and migration.18,86 srGAP1 is a critical effector of repulsive Slit-Robo signaling; ROBO1 bound to extracellular SLIT2 activates srGAP1s GAP domain at the membrane to specifically inactivate the Cdc42 GTPase, leading to actin cytoskeletal changes that decrease migration toward the SLIT2-displaying cells.86 srGAP2 is important for the biogenesis of neurites,10,87 as well as regulation of Slit-Robo mediated contact inhibition during cell migration through its GAP domain's inactivation of Rac1.88 Additional F-BAR GAP proteins in humans include PARG1, Gmip, and RhoGAP4. PARG1 and Gmip's GAP domains target RhoA,89-91 while RhoGAP4's substrates are unknown. RhoGAP4 functions in inhibiting cell migration,92 but little is known about PARG1 and Gmip cellular function.
Significant future study is required to understand the substrates of signaling F-BARs and integrate this functionally with their F-BAR domain activities.
Future directions
We have discussed the various activities of F-BAR domains (membrane binding, oligomerization, partner binding, signaling); however, these activities are not constitutive in cells, but instead are dynamically regulated. In fact, the membrane and partner binding capacity of many F-BAR domain proteins are autoinhibited and specific activation is required to allow these proteins to carry out their functions (reviewed in Roberts-Galbraith and Gould93). Phosphoregulation is one mechanism that allows for dynamic regulation in line with the short time windows of F-BAR protein activity in dynamic processes. Detailed analyses of phosphoregulation have been carried out for only a few F-BAR proteins such as S. pombe Cdc1540,94 and S. cerevisiae Hof1,12,95 and thus there is the opportunity to learn more about how F-BAR protein function is integrated in signaling networks, particularly in human cells. In other cases, it is argued that a binding partner pries apart an intramolecular interaction to release the F-BAR domain for membrane binding.96,97 Further investigation is necessary to identify the molecular mechanisms of human F-BAR protein spatial and temporal activation.
In certain processes we have described such as endocytosis and cell motility, multiple F-BAR proteins participate simultaneously. In these cases, it is not clear to what extent F-BAR proteins have overlapping or distinct functions. It is possible that multiple F-BAR proteins, through different oligomerization, membrane binding, or partner binding properties, act together or sequentially to coordinate these processes. One approach that could begin to tease this complexity apart is F-BAR domain swapping experiments. These types of experiments will clarify the importance of specific functionalities within different F-BARs such as unique modes of oligomerization or selective membrane binding preferences, and determine whether there is plasticity between different F-BAR domains.8 As discussed above, this will also require elucidation of each F-BAR domains' mechanism of oligomerization.
Finally, most studies of F-BAR proteins to date (and studies of proteins in cultured cells in general) rely upon exogenous expression and overexpression in cultured cells. With the advent of efficient genome editing methods such as CRISPR,98 it is becoming simpler to study endogenous proteins and make mutations of genes at the endogenous locus in human cells. Endogenous mutations and fluorescent tags will refine our knowledge of F-BAR protein localization and functionality in different circumstances. Working in the absence of wildtype protein and with correct endogenous expression levels will further remove many confounding effects of exogenous expression studies, including artificial membrane tubulation by highly concentrated F-BAR proteins.
Abbreviations
- BAR
Bin/Amphiphysin/Rvs domain
- FCH
Fer/CIP4 homology
- F-BAR
FCH-BAR domain
- SH3
SRC homology 3 domain
- µHD
muniscin homology domain
- GAP
GTPase activating domain
- FAK
focal adhesion kinase
- PIP
phosphorylated phosphatidylinositol
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank Dr. Janel Beckley, MariaSanta Mangione, and Chloe Snider for critical reading of the manuscript.
Funding
N.A.M. was supported by AHA fellowship 15PRE21780003. This work was supported by NIH grant GM101035 to K.L.G.
References
- [1].Bezanilla M, Gladfelter AS, Kovar DR, Lee WL. Cytoskeletal dynamics: A view from the membrane. J Cell Biol 2015; 209:329-37; PMID:25963816; http://dx.doi.org/ 10.1083/jcb.201502062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 2004; 303:495-9; PMID:14645856; http://dx.doi.org/ 10.1126/science.1092586 [DOI] [PubMed] [Google Scholar]
- [3].Henne WM, Kent HM, Ford MG, Hegde BG, Daumke O, Butler PJG, Mittal R, Langen R, Evans PR, McMahon HT. Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure 2007; 15:839-52; PMID:17540576; http://dx.doi.org/ 10.1016/j.str.2007.05.002 [DOI] [PubMed] [Google Scholar]
- [4].Millard TH, Bompard G, Heung MY, Dafforn TR, Scott DJ, Machesky LM, Fütterer K. Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53. EMBO 2005; 24:240-50; http://dx.doi.org/ 10.1038/sj.emboj.7600535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Henne WM, Boucrot E, Meinecke M, Evergren E, Vallis Y, Mittal R, McMahon HT. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science 2010; 328:1281-4; PMID:20448150; http://dx.doi.org/ 10.1126/science.1188462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol 2006; 172:269-79; PMID:16418535; http://dx.doi.org/ 10.1083/jcb.200508091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Frost A, Perera R, Roux A, Spasov K, Destaing O, Egelman EH, De Camilli P, Unger VM. Structural basis of membrane invagination by F-BAR domains. Cell 2008; 132:807-17; PMID:18329367; http://dx.doi.org/ 10.1016/j.cell.2007.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].McDonald NA, Takizawa Y, Feoktistova A, Xu P, Ohi MD, Vander Kooi CW, Gould KL. The tubulation activity of a fission yeast F-BAR protein is dispensable for its function in cytokinesis. Cell Rep 2016; 14:534-46; PMID:26776521; http://dx.doi.org/ 10.1016/j.celrep.2015.12.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].McDonald NA, Vander Kooi CW, Ohi MD, Gould KL. Oligomerization but not membrane bending underlies the function of certain f-bar proteins in cell motility and cytokinesis. Dev Cell 2015; 35:725-36; PMID:26702831; http://dx.doi.org/ 10.1016/j.devcel.2015.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell 2009; 138:990-1004; PMID:19737524; http://dx.doi.org/ 10.1016/j.cell.2009.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dharmalingam E, Haeckel A, Pinyol R, Schwintzer L, Koch D, Kessels MM, Qualmann B. F-BAR proteins of the syndapin family shape the plasma membrane and are crucial for neuromorphogenesis. J Neurosci 2009; 29:13315-27; PMID:19846719; http://dx.doi.org/ 10.1523/JNEUROSCI.3973-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Meitinger F, Boehm ME, Hofmann A, Hub B, Zentgraf H, Lehmann WD, Pereira G. Phosphorylation-dependent regulation of the F-BAR protein Hof1 during cytokinesis. Genes Dev 2011; 25:875-88; PMID:21498574; http://dx.doi.org/ 10.1101/gad.622411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Oh Y, Schreiter J, Nishihama R, Wloka C, Bi E. Targeting and functional mechanisms of the cytokinesis-related F-BAR protein Hof1 during the cell cycle. Mol Biol Cell 2013; 24:1305-20; PMID:23468521; http://dx.doi.org/ 10.1091/mbc.E12-11-0804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Reider A, Barker SL, Mishra SK, Im YJ, Maldonado-Báez L, Hurley JH, Traub LM, Wendland B. Syp1 is a conserved endocytic adaptor that contains domains involved in cargo selection and membrane tubulation. EMBO J 2009; 28:3103-16; PMID:19713939; http://dx.doi.org/ 10.1038/emboj.2009.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Moravcevic K, Alvarado D, Ferguson KM, Lemmon MA, Moravcevic K, Alvarado D, Schmitz KR, Kenniston JA, Mendrola JM. Comparison of saccharomyces cerevisiae F-BAR domain structures reveals a conserved inositol phosphate binding site. Structure 2015; 23:252-63; http://dx.doi.org/ 10.1016/j.str.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang Q, Navarro MV, Peng G, Molinelli E, Goh SL, Judson BL, Rajashankar KR, Sondermann H. Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. PNAS 2009; 106:12700-5; PMID:19549836; http://dx.doi.org/ 10.1073/pnas.0902974106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Becalska AN, Kelley CF, Berciu C, Stanishneva-Konovalova TB, Fu X, Wang S, Sokolova OS, Nicastro D, Rodal AA. Formation of membrane ridges and scallops by the F-BAR protein nervous wreck. Mol Biol Cell 2013; 24:2406-18; PMID:23761074; http://dx.doi.org/ 10.1091/mbc.E13-05-0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Carlson BR, Lloyd KE, Kruszewski A, Kim IH, Rodriguiz RM, Heindel C, Faytell M, Dudek SM, Wetsel WC, Soderling SH. WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J Neurosci 2011; 31:2447-60; PMID:21325512; http://dx.doi.org/ 10.1523/JNEUROSCI.4433-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Coutinho-Budd J, Ghukasyan V, Zylka MJ, Polleux F. The F-BAR domains from srGAP1, srGAP2 and srGAP3 regulate membrane deformation differently. J Cell Sci 2012; 125:3390-401; PMID:22467852; http://dx.doi.org/ 10.1242/jcs.098962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol 2008; 9:99-111; PMID:18216767; http://dx.doi.org/ 10.1038/nrm2328 [DOI] [PubMed] [Google Scholar]
- [21].Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell 2005; 9:791-804; PMID:16326391; http://dx.doi.org/ 10.1016/j.devcel.2005.11.005 [DOI] [PubMed] [Google Scholar]
- [22].Itoh T, Hasegawa J, Tsujita K, Kanaho Y, Takenawa T. The tyrosine kinase Fer is a downstream target of the PLD-PA pathway that regulates cell migration. Sci Signal 2009; 2:ra52; PMID:19738202; http://dx.doi.org/ 10.1126/scisignal.2000393 [DOI] [PubMed] [Google Scholar]
- [23].Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006; 443:651-7; PMID:17035995; http://dx.doi.org/ 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- [24].Zhao H, Michelot A, Koskela EV, Tkach V, Stamou D, Drubin DG, Lappalainen P. Membrane-sculpting BAR domains generate stable lipid microdomains. Cell Rep 2013; 4:1213-23; PMID:24055060; http://dx.doi.org/ 10.1016/j.celrep.2013.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Posor Y, Eichhorn-Grunig M, Haucke V. Phosphoinositides in endocytosis. Biochim Biophys Acta 2015; 1851:794-804; PMID:25264171; http://dx.doi.org/ 10.1016/j.bbalip.2014.09.014 [DOI] [PubMed] [Google Scholar]
- [26].Hansen CG, Howard G, Nichols BJ. Pacsin 2 is recruited to caveolae and functions in caveolar biogenesis. J Cell Sci 2011; 124:2777-85; PMID:21807942; http://dx.doi.org/ 10.1242/jcs.084319 [DOI] [PubMed] [Google Scholar]
- [27].Schneider K, Seemann E, Liebmann L, Ahuja R, Koch D, Westermann M, Hübner CA, Kessels MM, Qualmann B. ProSAP1 and membrane nanodomain-associated syndapin i promote postsynapse formation and function. J Cell Biol 2014; 205:197-215; PMID:24751538; http://dx.doi.org/ 10.1083/jcb.201307088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kamioka Y, Fukuhara S, Sawa H, Nagashima K, Masuda M, Matsuda M, Mochizuki N. A novel dynamin-associating molecule, formin-binding protein 17, induces tubular membrane invaginations and participates in endocytosis. J Biol Chem 2004; 279:40091-9; PMID:15252009; http://dx.doi.org/ 10.1074/jbc.M404899200 [DOI] [PubMed] [Google Scholar]
- [29].Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, et al.. Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell 2007; 129:761-72; PMID:17512409; http://dx.doi.org/ 10.1016/j.cell.2007.03.040 [DOI] [PubMed] [Google Scholar]
- [30].Yu H, Schulten K. Membrane sculpting by F-BAR domains studied by molecular dynamics simulations. PLoS Comput Biol 2013; 9:e1002892; PMID:23382665; http://dx.doi.org/ 10.1371/journal.pcbi.1002892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Frost A, Unger VM, De Camilli P. The BAR domain superfamily: membrane-molding macromolecules. Cell 2009; 137:191-6; PMID:19379681; http://dx.doi.org/ 10.1016/j.cell.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mim C, Unger VM. Membrane curvature and its generation by BAR proteins. Trends Biochem Sci 2012; 37:526-33; PMID:23058040; http://dx.doi.org/ 10.1016/j.tibs.2012.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Qualmann B, Koch D, Kessels MM. Let's go bananas: revisiting the endocytic BAR code. EMBO 2011; 30:3501-15; http://dx.doi.org/ 10.1038/emboj.2011.266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Suetsugu S, Toyooka K, Senju Y. Subcellular membrane curvature mediated by the BAR domain superfamily proteins. Semin Cell Dev Biol 2010; 21:340-9; PMID:19963073; http://dx.doi.org/ 10.1016/j.semcdb.2009.12.002 [DOI] [PubMed] [Google Scholar]
- [35].Saarikangas J, Zhao H, Pykäläinen A, Laurinmäki P, Mattila PK, Kinnunen PKJ, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol 2009; 19:95-107; PMID:19150238; http://dx.doi.org/ 10.1016/j.cub.2008.12.029 [DOI] [PubMed] [Google Scholar]
- [36].van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 2008; 9:112-24; PMID:18216768; http://dx.doi.org/ 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, Hayden CC. Membrane bending by protein-protein crowding. Nat Cell Biol 2012; 14:944-9; PMID:22902598; http://dx.doi.org/ 10.1038/ncb2561 [DOI] [PubMed] [Google Scholar]
- [38].Cheng H, Rogers JA, Dunham NA, Smithgall TE. Regulation of c-Fes tyrosine kinase and biological activities by N-terminal coiled-coil oligomerization domains. Mol Cell Biol 1999; 19:8335-43; PMID:10567558; http://dx.doi.org/ 10.1128/MCB.19.12.8335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Craig AW, Zirngibl R, Greer P. Disruption of coiled-coil domains in Fer protein-tyrosine kinase abolishes trimerization but not kinase activation. J Biol Chem 1999; 274:19934-42; PMID:10391941; http://dx.doi.org/ 10.1074/jbc.274.28.19934 [DOI] [PubMed] [Google Scholar]
- [40].Roberts-Galbraith RH, Ohi MD, Ballif BA, Chen JS, McLeod I, McDonald WH, Gygi SP, Yates JR, Gould KL. Dephosphorylation of F-BAR protein Cdc15 modulates its conformation and stimulates its scaffolding activity at the cell division site. Mol Cell 2010; 39:86-99; PMID:20603077; http://dx.doi.org/ 10.1016/j.molcel.2010.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Roberts-Galbraith RH, Chen JS, Wang J, Gould KL. The SH3 domains of two PCH family members cooperate in assembly of the schizosaccharomyces pombe contractile ring. J Cell Biol 2009; 184:113-27; PMID:19139265; http://dx.doi.org/ 10.1083/jcb.200806044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Ren L, Willet AH, Roberts-Galbraith RH, McDonald NA, Feoktistova A, Chen JS, Huang H, Guillen R, Boone C, Sidhu SS, et al.. The Cdc15 and Imp2 SH3 domains cooperatively scaffold a network of proteins that redundantly ensure efficient cell division in fission yeast. Mol Biol Cell 2015; 26:256-69; PMID:25428987; http://dx.doi.org/ 10.1091/mbc.E14-10-1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Umasankar PK, Ma L, Thieman JR, Jha A, Doray B, Watkins SC, Traub LM. A clathrin coat assembly role for the muniscin protein central linker revealed by TALEN-mediated gene editing. Elife 2014; 3:1-33; http://dx.doi.org/ 10.7554/eLife.04137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hollopeter G, Lange JJ, Zhang Y, Vu TN, Gu M, Ailion M, Lambie EJ, Slaughter BD, Unruh JR, Florens L, et al.. The membrane-associated proteins FCHo and SGIP are allosteric activators of the AP2 clathrin adaptor complex. Elife 2014; 3:e03648; http://dx.doi.org/ 10.7554/eLife.03648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Stimpson HEM, Toret CP, Cheng AT, Pauly BS, Drubin DG. Early-Arriving Syp1p and Ede1p function in endocytic site placement and formation in budding yeast. Mol Biol Cell 2009; 20:4640-51; PMID:19776351; http://dx.doi.org/ 10.1091/mbc.E09-05-0429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Cocucci E, Aguet F, Boulant S, Kirchhausen T. The first five seconds in the life of a clathrin-coated pit. Cell 2012; 150:495-507; PMID:22863004; http://dx.doi.org/ 10.1016/j.cell.2012.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Busch DJ, Houser JR, Hayden CC, Sherman MB, Lafer EM, Stachowiak JC. Intrinsically disordered proteins drive membrane curvature. Nat Commun 2015; 6:7875; PMID:26204806; http://dx.doi.org/ 10.1038/ncomms8875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Stachowiak JC, Brodsky FM, Miller EA. A cost–benefit analysis of the physical mechanisms of membrane curvature. Nat Cell Biol 2013; 15:1019-27; PMID:23999615; http://dx.doi.org/ 10.1038/ncb2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ungewickell EJ, Hinrichsen L. Endocytosis: clathrin-mediated membrane budding. Curr Opin Cell Biol 2007; 19:417-25; PMID:17631994; http://dx.doi.org/ 10.1016/j.ceb.2007.05.003 [DOI] [PubMed] [Google Scholar]
- [50].Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol 2011; 9:e1000604; PMID:21445324; http://dx.doi.org/ 10.1371/journal.pbio.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Leibfried A, Fricke R, Morgan MJ, Bogdan S, Bellaiche Y. Drosophila Cip4 and WASp define a branch of the Cdc42-Par6-aPKC pathway regulating E-Cadherin endocytosis. Curr Biol 2008; 18:1639-48; PMID:18976911; http://dx.doi.org/ 10.1016/j.cub.2008.09.063 [DOI] [PubMed] [Google Scholar]
- [52].Hartig SM, Ishikura S, Hicklen RS, Feng Y, Blanchard EG, Voelker KA, Pichot CS, Grange RW, Raphael RM, Klip A, et al.. The F-BAR protein CIP4 promotes GLUT4 endocytosis through bidirectional interactions with N-WASp and Dynamin-2. J Cell Sci 2009; 122:2283-91; PMID:19509061; http://dx.doi.org/ 10.1242/jcs.041343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Fricke R, Gohl C, Dharmalingam E, Grevelhörster A, Zahedi B, Harden N, Kessels M, Qualmann B, Bogdan S. Drosophila Cip4/Toca-1 integrates membrane trafficking and actin dynamics through WASP and SCAR/WAVE7. Curr Biol 2009; 19:1429-37; PMID:19716703; http://dx.doi.org/ 10.1016/j.cub.2009.07.058 [DOI] [PubMed] [Google Scholar]
- [54].Ferguson SM, De Camilli P. Dynamin, a membrane-remodelling GTPase. Nat Rev Mol Cell Biol 2012; 13:75-88; PMID:22233676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].de Kreuk B-J, Anthony EC, Geertss D, Hordijk PL. The F-BAR protein PACSIN2 regulates epidermal growth factor receptor internalization. J Biol Chem 2012; 287:43438-53; PMID:23129763; http://dx.doi.org/ 10.1074/jbc.M112.391078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Senju Y, Itoh Y, Takano K, Hamada S, Suetsugu S. Essential role of PACSIN2/syndapin-II in caveolae membrane sculpting. J Cell Sci 2011; 124:2032-40; PMID:21610094; http://dx.doi.org/ 10.1242/jcs.086264 [DOI] [PubMed] [Google Scholar]
- [57].Icking A, Matt S, Opitz N, Wiesenthal A, Müller-Esterl W, Schilling K. NOSTRIN functions as a homotrimeric adaptor protein facilitating internalization of eNOS. J Cell Sci 2005; 118:5059-69; PMID:16234328; http://dx.doi.org/ 10.1242/jcs.02620 [DOI] [PubMed] [Google Scholar]
- [58].Zimmermann K, Opitz N, Dedio J, Renne C, Muller-Esterl W, Oess S. NOSTRIN: a protein modulating nitric oxide release and subcellular distribution of endothelial nitric oxide synthase. PNAS 2002; 99:17167-72; PMID:12446846; http://dx.doi.org/ 10.1073/pnas.252345399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Coyle IP, Koh YH, Lee WC, Slind J, Fergestad T, Littleton JT, Ganetzky B. Nervous wreck, an SH3 adaptor protein that interacts with Wsp, regulates synaptic growth in drosophila. Neuron 2004; 41:521-34; PMID:14980202; http://dx.doi.org/ 10.1016/S0896-6273(04)00016-9 [DOI] [PubMed] [Google Scholar]
- [60].Rodal AA, Motola-Barnes RN, Littleton JT. Nervous wreck and Cdc42 cooperate to regulate endocytic actin assembly during synaptic growth. J Neurosci 2008; 28:8316-25; PMID:18701694; http://dx.doi.org/ 10.1523/JNEUROSCI.2304-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rodal AA, Blunk AD, Akbergenova Y, Jorquera RA, Buhl LK, Littleton JT. A presynaptic endosomal trafficking pathway controls synaptic growth signaling. J Cell Biol 2011; 193:201-17; PMID:21464232; http://dx.doi.org/ 10.1083/jcb.201009052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].O'Connor-Giles KM, Ho LL, Ganetzky B. Nervous Wreck Interacts with Thickveins and the endocytic machinery to attenuate retrograde BMP signaling during synaptic growth. Neuron 2008; 58:507-18; http://dx.doi.org/ 10.1016/j.neuron.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Saengsawang W, Mitok K, Viesselmann C, Pietila L, Lumbard DC, Corey SJ, Dent EW. The F-BAR protein CIP4 inhibits neurite formation by producing lamellipodial protrusions. Curr Biol 2012; 22:494-501; PMID:22361215; http://dx.doi.org/ 10.1016/j.cub.2012.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Saengsawang W, Taylor KL, Lumbard DC, Mitok K, Price A, Pietila L, Gomez TM, Dent EW. CIP4 coordinates with phospholipids and actin-associated proteins to localize to the protruding edge and produce actin ribs and veils. J Cell Sci 2013; 126:2411-23; PMID:23572514; http://dx.doi.org/ 10.1242/jcs.117473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Pichot CS, Arvanitis C, Hartig SM, Jensen SA, Bechill J, Marzouk S, Yu J, Frost JA, Corey SJ. Cdc42-interacting protein 4 promotes breast cancer cell invasion and formation of invadopodia through activation of N-WASp. Cancer Res 2010; 70:8347-56; PMID:20940394; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-4149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wu Y, Spencer SD, Lasky LA. Tyrosine Phosphorylation Regulates the SH3-mediated binding of the wiskott-aldrich syndrome protein to PSTPIP, a cytoskeletal-associated protein. J Biol Chem 1998; 273:5765-70; PMID:9488710; http://dx.doi.org/ 10.1074/jbc.273.10.5765 [DOI] [PubMed] [Google Scholar]
- [67].Côté JF, Chung PL, Théberge JF, Hallé M, Spencer S, Lasky LA, Tremblay ML. PSTPIP is a substrate of PTP-PEST and serves as a scaffold guiding PTP-PEST toward a specific dephosphorylation of WASP. J Biol Chem 2002; 277:2973-86; http://dx.doi.org/ 10.1074/jbc.M106428200 [DOI] [PubMed] [Google Scholar]
- [68].Cong F, Spencer S, Côté JF, Wu Y, Tremblay ML, Lasky LA, Goff SP. Cytoskeletal protein PSTPIP1 directs the PEST-type protein tyrosine phosphatase to the c-Abl kinase to mediate Abl dephosphorylation. Mol Cell 2000; 6:1413-23; PMID:11163214; http://dx.doi.org/ 10.1016/S1097-2765(00)00138-6 [DOI] [PubMed] [Google Scholar]
- [69].Demeter J, Sazer S. imp2, a new component of the actin ring in the fission yeast. J Cell Biol 1998; 143:415-27; PMID:9786952; http://dx.doi.org/ 10.1083/jcb.143.2.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V. The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell 1995; 82:435-44; PMID:7634333; http://dx.doi.org/ 10.1016/0092-8674(95)90432-8 [DOI] [PubMed] [Google Scholar]
- [71].Nishihama R, Schreiter JH, Onishi M, Vallen EA, Hanna J, Moravcevic K, Lippincott MF, Han H, Lemmon MA, Pringle JR, et al.. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J Cell Biol 2009; 185:995-1012; PMID:19528296; http://dx.doi.org/ 10.1083/jcb.200903125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, Ozaki K, Takai Y. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p: Implication in cytokinesis in Saccharomyces cerevisiae. J Biol Chem 1998; 273:28341-5; PMID:9774458; http://dx.doi.org/ 10.1074/jbc.273.43.28341 [DOI] [PubMed] [Google Scholar]
- [73].Graziano BR, Yu HY, Alioto SL, Eskin JA, Ydenberg CA, Waterman DP, Garabedian M, Goode BL. The F-BAR protein Hof1 tunes formin activity to sculpt actin cables during polarized growth. Mol Biol Cell 2014; 25:1730-43; PMID:24719456; http://dx.doi.org/ 10.1091/mbc.E14-03-0850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Carnahan RH, Gould KL. The PCH family protein, Cdc15p, recruits two F-actin nucleation pathways to coordinate cytokinetic actin ring formation in Schizosaccharomyces pombe. J Cell Biol 2003; 162:851-62; PMID:12939254; http://dx.doi.org/ 10.1083/jcb.200305012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Willet AH, Mcdonald NA, Bohnert KA, Baird MA, Allen JR, Davidson MW, Gould KL. The F-BAR Cdc15 promotes contractile ring formation through the direct recruitment of the formin Cdc12. J Cell Biol 2015; 208:391-9; PMID:25688133; http://dx.doi.org/ 10.1083/jcb.201411097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Chang F, Drubin D, Nurse P. Cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol 1997; 137:169-82; PMID:9105045; http://dx.doi.org/ 10.1083/jcb.137.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Shoham NG, Centola M, Mansfield E, Hull KM, Wood G, Wise CA, Kastner DL. Pyrin binds the PSTPIP1/CD2BP1 protein, defining familial Mediterranean fever and PAPA syndrome as disorders in the same pathway. PNAS 2003; 100:13501-6; PMID:14595024; http://dx.doi.org/ 10.1073/pnas.2135380100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Yu JW, Fernandes-Alnemri T, Datta P, Wu J, Juliana C, Solorzano L, McCormick M, Zhang Z, Alnemri ES. Pyrin Activates the ASC pyroptosome in response to engagement by autoinflammatory PSTPIP1 mutants. Mol Cell 2007; 28:214-27; PMID:17964261; http://dx.doi.org/ 10.1016/j.molcel.2007.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kostan J, Salzer U, Orlova A, Törö I, Hodnik V, Senju Y, Zou J, Schreiner C, Steiner J, Meriläinen J, et al.. Direct interaction of actin filaments with F-BAR protein pacsin2. EMBO Rep 2014; 15:1154-62; PMID:25216944; http://dx.doi.org/ 10.15252/embr.201439267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J 2005; 390:641-53; PMID:16134966; http://dx.doi.org/ 10.1042/BJ20050411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Naba A, Reverdy C, Louvard D, Arpin M. Spatial recruitment and activation of the Fes kinase by ezrin promotes HGF-induced cell scattering. EMBO J 2008; 27:38-50; PMID:18046454; http://dx.doi.org/ 10.1038/sj.emboj.7601943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Oh MA, Choi S, Lee MJ, Choi MC, Lee SA, Ko W, Cance WG, Oh ES, Buday L, Kim SH, et al.. Specific tyrosine phosphorylation of focal adhesion kinase mediated by Fer tyrosine kinase in suspended hepatocytes. Biochim Biophys Acta 2009; 1793:781-91; PMID:19339212; http://dx.doi.org/ 10.1016/j.bbamcr.2009.01.015 [DOI] [PubMed] [Google Scholar]
- [83].Kim L, Wong TW. The cytoplasmic tyrosine kinase FER is associated with the catenin-like substrate pp120 and is activated by growth factors. Mol Cell Biol 1995; 15:4553-61; PMID:7623846; http://dx.doi.org/ 10.1128/MCB.15.8.4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Kim L, Wong TW. Growth factor-dependent phosphorylation of the actin-binding protein cortactin is mediated by the cytoplasmic tyrosine kinase FER. J Biol Chem 1998; 273:23542-8; PMID:9722593; http://dx.doi.org/ 10.1074/jbc.273.36.23542 [DOI] [PubMed] [Google Scholar]
- [85].Greer P. Closing in on the biological functions of Fps/Fes and Fer. Nat Rev Mol Cell Biol 2002; 3:278-89; PMID:11994747; http://dx.doi.org/ 10.1038/nrm783 [DOI] [PubMed] [Google Scholar]
- [86].Wong K, Ren XR, Huang YZ, Xie Y, Liu G, Saito H, Tang H, Wen L, Brady-Kalnay SM, Mei L, et al.. Signal transduction in neuronal migration: Roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell 2001; 107:209-21; PMID:11672528; http://dx.doi.org/ 10.1016/S0092-8674(01)00530-X [DOI] [PubMed] [Google Scholar]
- [87].Charrier C, Joshi K, Coutinho-Budd J, Kim J-E, Lambert N, de Marchena J, Jin WL, Vanderhaeghen P, Ghosh A, Sassa T, et al.. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell 2012; 149:923-35; PMID:22559944; http://dx.doi.org/ 10.1016/j.cell.2012.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Fritz RD, Menshykau D, Martin K, Reimann A, Pontelli V, Pertz O. SrGAP2-Dependent integration of membrane geometry and Slit-Robo-Repulsive cues regulates fibroblast contact inhibition of locomotion. Dev Cell 2015; 35:78-92; PMID:26439400; http://dx.doi.org/ 10.1016/j.devcel.2015.09.002 [DOI] [PubMed] [Google Scholar]
- [89].Myagmar B-E, Umikawa M, Asato T, Taira K, Oshiro M, Hino A, Takei K, Uezato H, Kariya K-I. PARG1, a protein-tyrosine phosphatase-associated RhoGAP, as a putative Rap2 effector. Biochem Biophys Res Commun 2005; 329:1046-52; PMID:15752761; http://dx.doi.org/ 10.1016/j.bbrc.2005.02.069 [DOI] [PubMed] [Google Scholar]
- [90].Saras J, Franzén P, Aspenström P, Hellman U, Gonez LJ, Heldin CH. A novel GTPase-activating protein for Rho interacts with a PDZ domain of the protein-tyrosine phosphatase PTPL1. J Biol Chem 1997; 272:24333-8; PMID:9305890; http://dx.doi.org/ 10.1074/jbc.272.39.24333 [DOI] [PubMed] [Google Scholar]
- [91].Aresta S, de Tand-Heim M-F, Beranger F, de Gunzburg J. A novel Rho GTPase-activating-protein interacts with Gem, a member of the Ras superfamily of GTPases. Biochem J 2002; 367:57-65; PMID:12093360; http://dx.doi.org/ 10.1042/bj20020829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Vogt DL, Gray CD, Young WS, Orellana SA, Malouf AT. ARHGAP4 is a novel RhoGAP that mediates inhibition of cell motility and axon outgrowth. Mol Cell Neurosci 2007; 36:332-42; PMID:17804252; http://dx.doi.org/ 10.1016/j.mcn.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Roberts-Galbraith RH, Gould KL. Setting the F-BAR: Functions and regulation of the F-BAR protein family. Cell Cycle 2010; 9:4091-7; PMID:20948299; http://dx.doi.org/ 10.4161/cc.9.20.13587 [DOI] [PubMed] [Google Scholar]
- [94].Ullal P, McDonald NA, Chen JS, Lo Presti L, Roberts-Galbraith RH, Gould KL, Martin SG. The DYRK-family kinase Pom1 phosphorylates the F-BAR protein Cdc15 to prevent division at cell poles. J Cell Biol 2015; 211:653-68; PMID:26553932; http://dx.doi.org/ 10.1083/jcb.201504073 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [95].Meitinger F, Palani S, Hub B, Pereira G. Dual function of the NDR-kinase Dbf2 in the regulation of the F-BAR protein Hof1 during cytokinesis. Mol Biol Cell 2013; 24:1290-304; PMID:23447700; http://dx.doi.org/ 10.1091/mbc.E12-08-0608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Rao Y, Ma Q, Vahedi-Faridi A, Sundborger A, Pechstein A, Puchkov D, Luo L, Shupliakov O, Saenger W, Haucke V. Molecular basis for SH3 domain regulation of F-BAR-mediated membrane deformation. PNAS 2010; 107:8213-8; PMID:20404169; http://dx.doi.org/ 10.1073/pnas.1003478107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kelley CF, Messelaar EM, Eskin TL, Sokolova OS, Nicastro D, Rodal AA, Kelley CF, Messelaar EM, Eskin TL, Wang S, et al.. Membrane charge directs the outcome of F-BAR domain lipid binding and autoregulation. Cell Rep 2015; 13:2597-609; PMID:26686642; http://dx.doi.org/ 10.1016/j.celrep.2015.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014; 346:1258096-1-9; PMID:25430774; http://dx.doi.org/ 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- [99].Aspenström P. A Cdc42 target protein with homology to the non-kinase domain of FER has a potential role in regulating the actin cytoskeleton. Curr Biol 1997; 7:479-87; PMID:9210375; http://dx.doi.org/ 10.1016/S0960-9822(06)00219-3 [DOI] [PubMed] [Google Scholar]
- [100].Qualmann B, Roos J, DiGregorio PJ, Kelly RB. Syndapin I, a synaptic dynamin-binding protein that associates with the neural Wiskott-Aldrich syndrome protein. Mol Biol Cell 1999; 10:501-13; PMID:9950691; http://dx.doi.org/ 10.1091/mbc.10.2.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Cooper KM, Bennin DA, Huttenlocher A. The PCH family member Proline-Serine-Threonine Phosphatase–interacting Protein 1 Targets to the leukocyte uropod and regulates directed cell migration. Mol Biol Cell 2008; 19:3180-91; PMID:18480402; http://dx.doi.org/ 10.1091/mbc.E08-02-0225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Marcos T, Ruiz-Martin V, de la Puerta ML, Trinidad AG, Rodriguez MDC, de la Fuente MA, Sanchez MC, Alonso A, Bayon Y. Proline-serine-threonine phosphatase interacting protein 1 inhibition of T-cell receptor signaling depends on its SH3 domain. FEBS J 2014; 281:3844-54; PMID:25040622; http://dx.doi.org/ 10.1111/febs.12912 [DOI] [PubMed] [Google Scholar]
- [103].Dowbenko D, Spencer S, Quan C, Lasky LA. Identification of a novel polyproline recognition site in the cytoskeletal associated protein, proline serine threonine phosphatase interacting protein. J Biol Chem 1998; 273:989-96; PMID:9422760; http://dx.doi.org/ 10.1074/jbc.273.2.989 [DOI] [PubMed] [Google Scholar]
- [104].Soulard A, Lechler T, Spiridonov V, Shevchenko A, Shevchenko A, Li R, Winsor B. Saccharomyces cerevisiae Bzz1p is implicated with type I myosins in actin patch polarization and is able to recruit actin-polymerizing machinery in vitro. Mol Cell Biol 2002; 22:7889-906; PMID:12391157; http://dx.doi.org/ 10.1128/MCB.22.22.7889-7906.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Boettner DR, D'Agostino JL, Torres OT, Daugherty-Clarke K, Uygur A, Reider A, Wendland B, Lemmon SK, Goode BL. The F-BAR protein Syp1 negatively regulates WASp-Arp2/3 complex activity during endocytic patch formation. Curr Biol 2009; 19:1979-87; PMID:19962315; http://dx.doi.org/ 10.1016/j.cub.2009.10.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Villar-Tajadura MA, Coll PM, Madrid M, Cansado J, Santos B, Pérez P. Rga2 is a Rho2 GAP that regulates morphogenesis and cell integrity in S. pombe. Mol Microbiol 2008; 70:867-81; PMID:18793338 [DOI] [PubMed] [Google Scholar]
- [107].Yang P, Qyang Y, Bartholomeusz G, Zhou X, Marcus S. The novel Rho GTPase-activating protein family protein, Rga8, provides a potential link between Cdc42/p21-activated kinase and Rho signaling pathways in the fission yeast, Schizosaccharomyces pombe. J Biol Chem 2003; 278:48821-30; PMID:14506270; http://dx.doi.org/ 10.1074/jbc.M306819200 [DOI] [PubMed] [Google Scholar]
- [108].Doignon F, Weinachter C, Roumanie O, Crouzet M. The yeast Rgd1p is a GTPase activating protein of the Rho3 and Rho4 proteins. FEBS Lett 1999; 459:458-62; PMID:10526184; http://dx.doi.org/ 10.1016/S0014-5793(99)01293-4 [DOI] [PubMed] [Google Scholar]
- [109].Roumanie O, Weinachter C, Larrieu I, Crouzet M, Doignon F. Functional characterization of the Bag7, Lrg1 and Rgd2 RhoGAP proteins from saccharomyces cerevisiae. FEBS Lett 2001; 506:149-56; PMID:11591390; http://dx.doi.org/ 10.1016/S0014-5793(01)02906-4 [DOI] [PubMed] [Google Scholar]