ABSTRACT
Transcription factor SOX2 is multiple phosphorylated. However, the kinase and the timing regulating SOX2 phosphorylation remains poorly understood. Here we reported mitotic phosphorylation of SOX2 by Aurora kinase A (AURKA). AURKA inhibitors (VX680, Aurora kinase Inhibitor I) but not PLK1 inhibitors (BI2536, CBB2001) eliminate the mitotic phosphorylation of SOX2. Consistently, siRNA inhibition of AURKA can eliminate mitotic SOX2 phosphorylation. Ser220 and Ser251 are two sites that identified for mitotic phosphorylation on SOX2. Moreover, SOX2 mutants (S220A and S251A) can promote SOX2 induced OCT4 re-expression in differentiated cells. These findings reveal a novel regulation mechanism of SOX2 phosphorylation mediated by AURKA in mitosis and its function in stem cell pluripotency maintenance in cancer cells.
KEYWORDS: AURKA, OCT4, phosphorylation, PA-1 cell, SOX2
Introduction
Transcription factor SOX2 plays critical role in the central regulatory hub of stem cell pluripotency and self-renewal. Expression of SOX2 in self-renewing progenitor cells can inhibit neuronal differentiations.1 SOX2, combining with other factors OCT4, KLF4 and c-Myc, is sufficient to reprogram differentiated cells to induced pluripotency stem cells (iPS).2,3 The balance of SOX2 expression is strictly regulated, small increases (two fold or less) in SOX2 protein can trigger the differentiation of embryonic stem cell (ESC) into neuroectoderm, mesoderm, and trophectoderm but not endoderm,4 while knocking down of SOX2 in ESC promotes their differentiation into trophectoderm-like cells.5 But engineered ESC (i-OSKM-ESC), which express OCT4, SOX2, KLF4 and c-Myc from an inducible transgene, do not differentiate when each of these four factors is elevated ∼2-fold for at least 5 passages.6 Thus the expression changes of the proteins do not appear to be the main contributor.6 Comparing interactomes of SOX2 in ESC and ESC beginning to differentiate, it indicates that the SOX2-interactomes changes dramatically within 24 hours.7 It is unclear why the SOX2-interactome changes so rapidly. The attractive changes are the post-translational modifications of interacting proteins. For example, one hour after human ESC initiate differentiation, their phosphoproteome changes by ∼50%.8
SOX2 activity and expression is regulated by multiple post-translational modifications, including ubiquitination, methylation, phosphorylation and SUMOylation (SUMO, small ubiquitin-related modifier 1, an ubiquitin-like protein that can covalently bind to target proteins as monomer or a lysine-linked polymer as a kind of post-transcriptional modification). Mouse SOX2 is SUMOylated at lysine 247.9 Sumoylation of SOX2 inhibits nanog expression and SUMOylation of SOX2 by Pias3 impairs its interaction with OCT4.10 CARM1 methylates SOX2 at Arg113, enhances SOX2 self-association and facilitates SOX2-mediated transactivation.11 SETD7 monomethylates SOX2 at Lys119, which induces SOX2 ubiquitination and degradation.12 PKCι-mediated SOX2 phosphorylation at Thr118 is required for HHAT promoter occupancy and maintenance of a stem cell like phenotype in lung squamous cell carcinoma.13 AKT1 also phosphorylates SOX2 at Thr118, but this phosphorylation antagonizes K119me by SETD7 to stabilize SOX2.12 SOX2 is also phosphorylated by Cyclin-dependent kinase (CDK), CDK-mediated SOX2 phosphorylation at S39 and S253 is required for establishing the pluripotent state during reprogramming but is dispensable for ESC maintenance.14 Although it is reported that SOX2 is phosphorylated in human ESCs, little is known about the biological significance of SOX2 phosphorylation in mitosis, which contains a series of events concerning chromatin organization, condensation and separation. In addition, mitosis normally accompanies with rich modifications of cellular proteins and most of their functions remain unknown.
Among cell cycle regulatory kinases, Aurora kinase A is a key regulator of centrosome duplication and spindle assembly. It is a cell cycle oscillated kinase which presents activity mainly in G2 and M phase, along with obvious centrosome and spindle localization.15,16 AURKA regulates mitotic events through phosphorylating the substrates to cause protein structure transformation, activity switch on/off, as well as cellular translocation. AURKA wildly presents rich expression in kinds of tumors and is considered as target for inhibitors development and cancer therapy.17
In this study, we used human ovarian cancer PA-1 cells, a rapid proliferating cell lines with rich SOX2 and OCT4 expression,18 to study the regulatory connection of cell cycle and SOX2. Exactly, we discovered that SOX2 is heavily modified in mitosis. The mitotic SOX2 were multiple phosphorylated and AURKA is the kinase that responsible for mitotic SOX2 phosphorylation. Ser220 and Ser251 are the two sites showed to be phosphorylated on SOX2 in mitosis. Finally, we proved that AURKA directly regulated the mitotic phosphorylation of SOX2, which is important for cancer stem-cell like cell maintenance.
Results
SOX2 is highly modified in M phase
Stemness factors SOX2, OCT4 and nanog are strictly regulated and normally expressed in embryonic stage. However, the high expression of SOX2 and OCT4 was frequently found in many cancers and showed a significant association with poor prognosis and disease-free survival.13,18-21 Understanding the mechanism of how stemness factors are regulated is of great significance for cancer pathogenesis and clinical therapy. To uncover the relationship between cell cycle oscillation and stemness maintenance such as OCT4 and SOX2 expression, we arrested PA-1 cells with nocodazole (50 ng/mL) and detected SOX2 using specific antibody. We discovered that SOX2 was highly modified and presented a slow shift band distinguish from its main bands in SDS-PAGE gel (Fig. 1A). Moreover, the slow shift band is mitotic specific and decayed completely within one hour in the mitotic releasing cells (Fig. 1C). These results are consistent with that observed in other SOX2 expressing cell lines F9, H520 and MCF7 (Fig. 1B).
Figure 1.
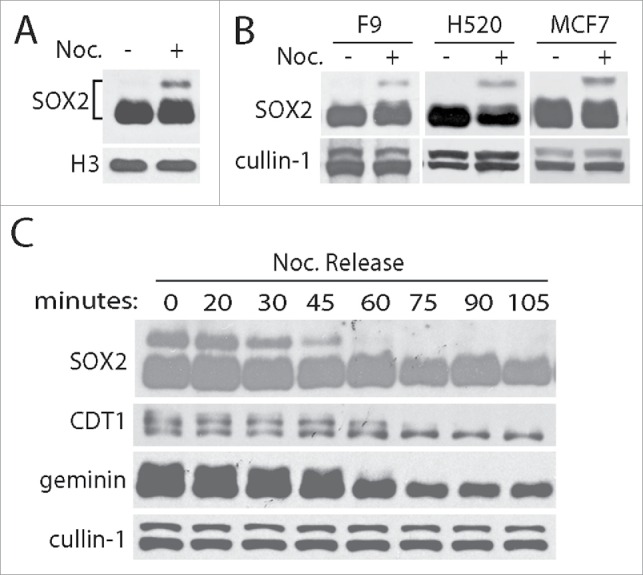
SOX2 is specific modified in mitosis. (A) SOX2 was obviously modified in mitosis. In nocodazole (Noc., 50 ng/mL for 12 hours) treated human ovarian PA-1 cells, SOX2 exerts a slow-migration band observed by western blotting. Histone H3 was blotted as loading control. (B) Mitotic specific modifications of SOX2 can similarly observed in F9, H520 and MCF7 cell lines. (C) The modified SOX2 decayed along with the exit of mitosis. Mitotic PA-1 cells (arrested by nocodazole for 12 hours) were released for 0, 20, 30, 45, 60, 75, 90, 105 minutes. Cell cycle regulation and mitotic modified proteins CDT1 and geminin were examined for cell phase control and unchanged cullin-1 was blotted as loading control.
AURKA regulates mitotic modifications of SOX2
To explore what modifications formed on SOX2 in mitosis, we incubated the mitotic arrest PA-1 cells with small kinase inhibitors separately. Surprisingly, we found that inhibition of AURKA kinase with inhibitory chemical VX680 completely abolished the shift band of SOX2. Consistently, the pan Aurora kinase inhibitor ZM447439 also exerted partial inhibition on mitotic SOX2 modifications. Besides, in the parallel experiments using Aurora kinase Inhibitor I, a novel specific inhibitor to AURKA, we confirmed that inhibition of AURKA activity is capable of attenuating mitotic modification of SOX2. However, inhibition of PLK1, an interrelated kinase at the same period, with specific inhibitors BI2536 or CBB2001 did not alter the mitotic modifications of SOX2. Similarly, pan CDKs inhibitor roscovitine also did not exert obvious effect on mitotic SOX2 modification (Fig. 2A). Thus, AURKA is the most likely kinase that regulates the mitotic modifications of SOX2.
Figure 2.
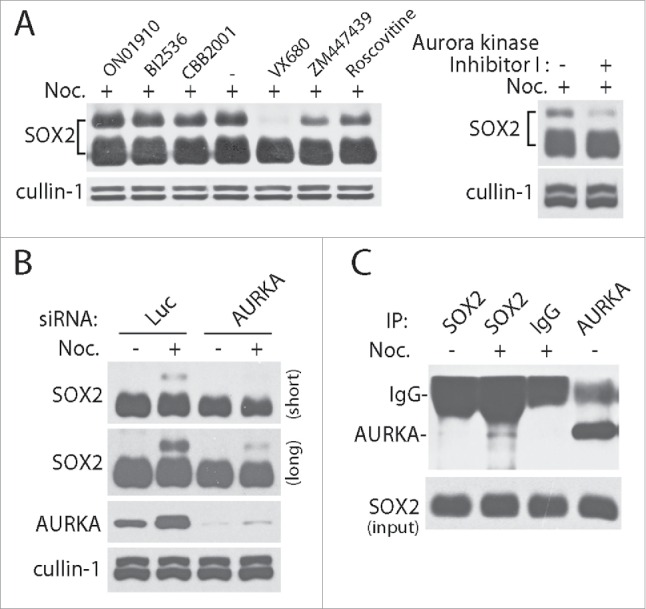
AURKA regulates the mitotic modification of SOX2. (A) Mitotic modification of SOX2 was sensitive to AURKA inhibition. PA-1 cells were treated with nocodazole for 12 hours and further incubated with chemical inhibitors for 30 minutes as indicated. ON01910, BI2536 and CBB2001 are PLK1 inhibitors, VX680 and Aurora kinase inhibitor I are AURKA inhibitors, ZM447439 is a pan Aurora kinase inhibitor. Roscovitine is a pan CDKs inhibitor. (B) siRNA knockdown of AURKA abolished the mitotic modification of SOX2. PA-1 cells were transfected with siRNAs specific to Luciferase (Luc, as negative control) and AURKA for 48 hours, and then incubated with DMSO or nocodazole (Noc.) for 12 hours. Both short and long exposure results were showed. (C) AURKA interacts with SOX2 directly in M phase. PA-1 cells were treated with nocodazole (Noc.) or DMSO for 12 hours, and the protein complexes in the lysates were co-immunoprecipitated (co-IP) with antibodies.
To further confirm the regulatory role of AURKA kinase on SOX2, we designed small interference RNA to knock down AURKA expression in PA-1 cells. As expected, siRNA disruption of AURKA expression inhibited the mitotic modifications of SOX2 (Fig. 2B). On this basis, we further performed protein co-immunoprecipitation using specific antibodies to detect the interaction between SOX2 with AURKA. Both normal and mitotic arrested PA-1 cells were harvested and lysis for the experiments. Consistently, AURKA interacts with SOX2 and their interactions were enhanced in mitosis (Fig. 2C).
SOX2 is multiple phosphorylated in mitosis
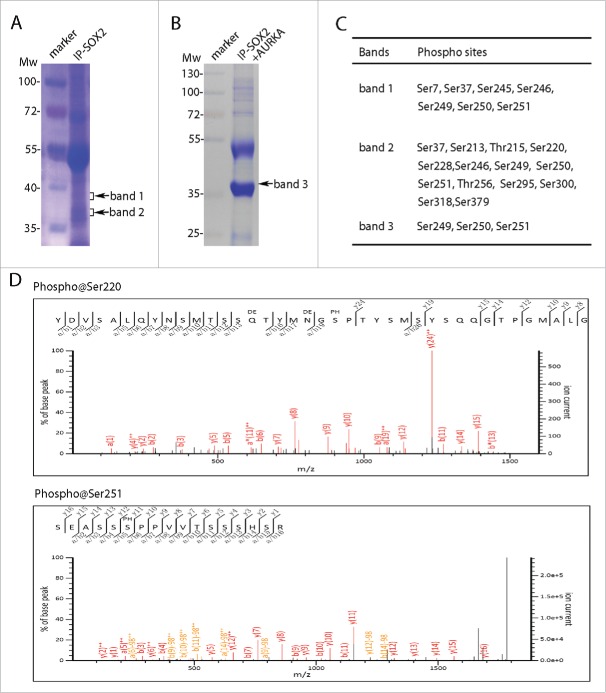
In order to verify the types and sites of mitotic SOX2 modifications, we performed mass spectrometry analysis to identify the modifications of mitotic SOX2. Firstly, PA-1 cells were arrested in mitosis upon nocodazole incubation, and then cell lysate was subjected to affinity purification of SOX2 protein using antibodies. Immunoprecipitated SOX2 proteins complex were separated by SDS-PAGE and viewed by coomassie blue G250 staining (Fig. 3A). In order to precisely locate the SOX2 containing bands, we matched the gel with the parallel performed western blotting using SOX2 specific antibodies. Therefore, we obtained two protein bands (band 1 and band 2, Fig. 3A). Then protein bands containing SOX2 were subjected to mass spectrometry analysis according to the protocol (refer to Materials and Methods for mass spectrometry sample preparation, peptide spectrum mapping, database searching and phosphorylation sites analysis). We successfully identified SOX2 protein in both band 1 and 2, along with multiple phosphorylation sites. In addition, we performed in vitro kinase reaction on SOX2 using active AURKA kinase (Fig. 3B). The reaction products were SDS-PAGE separated, coomassie blue stained, SOX2-containing band located and collected for mass spectrometry analysis as indicated previously. Consistently, we also identified SOX2 protein as well as the phosphorylation sites (Fig. 3C). From these results, we successfully identified multiple phosphorylation sites on SOX2 (Fig. 3C and D).
Figure 3.
SOX2 was multiple phosphorylated in M phase. (A) SOX2 protein was separated from mitotic PA-1 cells using SOX2 specific antibody. SDS-PAGE separated proteins were stained and extracted for mass spectrometry analysis. (B) Coomassie brilliant blue staining of SOX2 protein in AURKA kinase assay. PA-1 cells were incubated with VX680 (0.5 μM) for 5 hours and then immunoprecipitated using SOX2 specific antibody. SOX2 proteins were subjected to in vitro kinase reaction with AURKA. Reaction products were SDS-PAGE separated and analyzed by mass spectrometry. (C) Confirmed phosphorylation sites in SOX2 proteins from band 1, band 2 in (A), and band 3 in (B). (D) Two representative spectrums of SOX2 phosphorylations on sites Ser220 and Ser251.
Ser220 and Ser251 are the two sites required for mitotic SOX2 phosphorylation
With successfully identify of multiple phospho-sites of SOX2, we intended to functional validating the key sites responding for mitotic SOX2 phosphorylation. Site directed mutations were obtained on plasmid containing wild type SOX2 open reading frame and transiently expressed in PA-1 cells as exogenous SOX2 (HA-SOX2). Similarly, mitotic PA-1 cells were harvested and protein gel blotting for SOX2. Both endogenous and exogenous expressed wild type SOX2 showed mitotic phosphorylations. While for the mutants, only S220A and S251A but not others (including S37A, T116A, S228A, S246A, S249A, and S250A) can completely abolished the mitotic phosphorylation of SOX2 (Fig. 4). Moreover, we also confirmed the results in HA-SOX2 stably expressed PA-1 cells, which were selected by puromycin. The above results suggested that Ser220 and Ser251 are the two sites requiring for mitotic phosphorylations of SOX2.
Figure 4.
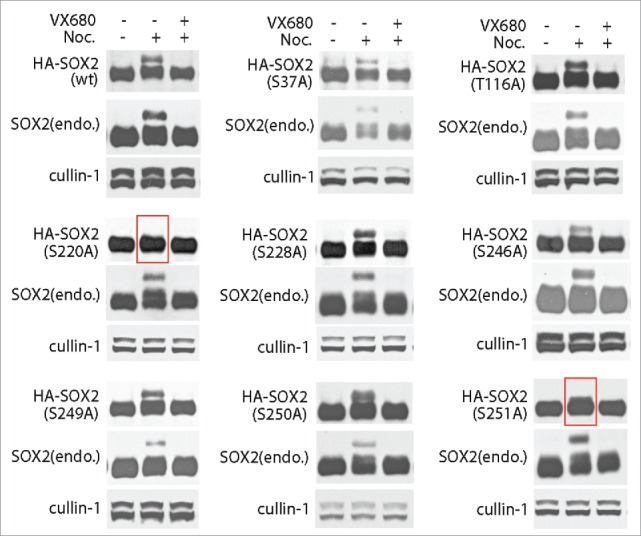
Ser220 and Ser251 are the critical sites for M phase specific modifications of SOX2. PA-1 cells expressing HA tagged wild type or mutants SOX2 (HA-SOX2) were incubated with DMSO or nocodazole for 12 hours to arrest cells at mitosis. VX680 were added (for 30 minutes) for AURKA inhibition. AURKA inhibition can inhibit the mitotic phosphorylation of SOX2. Consistently, mutations on Ser220 and Ser251 can also completely abolish the mitotic phosphorylation of SOX2. For other mutants, including S37A, T116A, S228A, S246A, S249A, and S250A, they did not show any effect on the mitotic phosphorylation of SOX2.
SOX2 mutants (S220A and S251A) are capable of cell reprogramming and induced OCT4 re-expression in differentiated cells
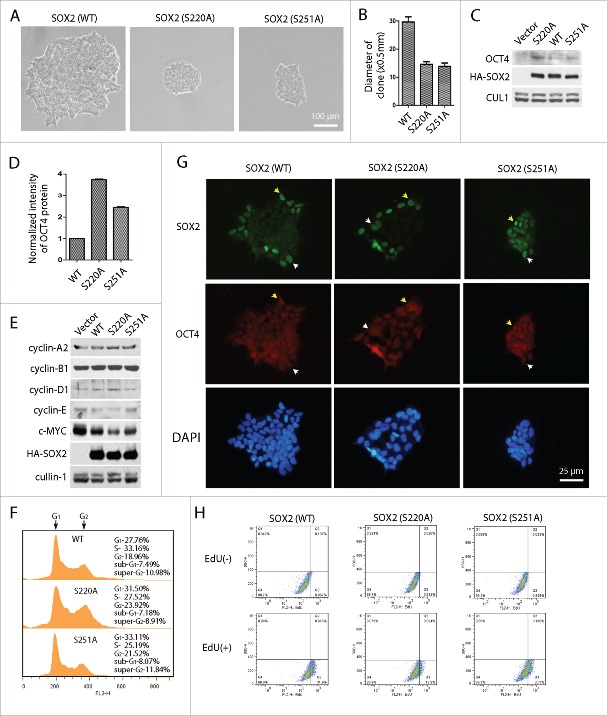
To further explore the role of phosphorylated SOX2, we performed clone formation experiments using somatic 293 cells. Wild type SOX2 and S220A and S251A mutants of SOX2 were respectively expressed in 293 cells. Positive cells were selected under puromycin and diluted into single cell for clone formation. Clones were imaged on day 6. Cells expressing wild type SOX2 showed larger clones and rapid proliferation. In contrast, cells expressing SOX2 mutants (S220A and S251A) presented much smaller clones (Fig. 5A and B). More interestingly, we found OCT4 re-expression in forced SOX2 expressing cells. Both mutants showed higher expression of OCT4 than that of wild type SOX2. It should be noted that the control cells with vector transfection did not show any expression of OCT4 (Fig. 5C and D). In order to elucidate whether SOX2 expression altered the cell cycle state, we detected cell cycle regulator as well as c-Myc in SOX2 expressing cells. C-Myc is a transcription factor that non-specifically binds to DNA and activates the transcription of growth related genes. Thus down regulation of c-Myc is relating to reduced cell proliferation.22 We found that c-Myc and cyclin-E are both obviously down regulated in SOX2 S220A and S251A mutants expressing-clones, while cyclin-A2, cyclin-B1 and cyclin-D1 are slightly changed (Fig. 5E). FACS analysis also confirmed a slight cell cycle arrest in S220A and S251A clones (Fig. 5F). Interestingly, the established clones did not express SOX2 in all cells, only partial of them expressed both SOX2 and OCT4, or even only expressed SOX2 but not OCT4 (Fig. 5G). EdU incorporation experiments also suggest that clones expressing SOX2 mutants (especially S220A) showed decelerated cell proliferation (Fig. 5H). These phenotypes are consistent with the characters of stem cells of slow proliferation rate and asymmetrical division. These results indicated that phosphorylation at Ser220 and Ser251 on SOX2 mediated by AURKA may be the key event involving in stem cell reprogramming and pluripotency maintenance.
Figure 5.
SOX2 mutants S220A and S251A promote SOX2 induced differentiated cells reprogramming and OCT4 expression. (A) Single-cell clones of 293 cells expressing wild type (WT) SOX2 and point mutated (S220A or S251A) SOX2. 293 cells were infected by lentivirus expressing SOX2, and selected under 2 μg/mL puromycin treatment. SOX2-expressing 293 cells were distributed by serial dilution into single cells. Clones were expanded and imaged at day 6. (B) Diameter of the clones in (A) were measured and calculated. Data presented as mean ±SD of 10 clones each. (C) Lysates of the clones in (A) were analyzed by protein gel blotting to detect the expression of OCT4. (D) The expressions of OCT4 in (C) were measured and quantified by Gel-Pro Analyzer. Mean ±SD of three independent experiments. (E) Cell cycle regulators were examined. Cyclin-E and c-Myc showed remarkably down regulation in S220A or S251A clones. (F) Cell cycle analyses of the SOX2 expressing clones. (G) Immunofluorescence imaging of SOX2 expressing clones showed regional expression of SOX2 and OCT4. Some cells expressed both SOX2 and OCT4 (arrow head in yellow); while more cells only expressed SOX2 (arrow head in white). (H) EdU incorporation experiments assayed by FACS suggested the decelerated cell proliferation in S220A clones.
Discussion
SOX2 is a key component of the transcriptional network that maintains stem cell self-renew and differentiation. Over-expression of SOX2 in C3H10T1/2 cells inhibited osteoblast differentiation.23 SOX2 also confers cancer cells with pluripotency property that more sensitive to LSD1 inhibition and this could be a promising strategy for cancer therapy.24 Moreover, differentiated cells can be reprogrammed by combinations of SOX2 with small molecules or even merely with small compounds.24-26 However, the underlying mechanisms by which SOX2 contributes to self-renewal or reprogramming processes remain unknown. The post-translational modifications are considered as important events during ESCs early differentiation.8 AKT phosphorylated SOX2 at Thr118, which enhances the transcriptional activity of SOX2 in ESCs and enables more efficient induction of iPSCs.27 Additional phosphor-sites were identified on SOX2 mediated by CDK at Ser39 and Ser253, which is required for establishing the pluripotent state during reprogramming.14 In this study, we examined the mitotic SOX2 protein and identified rich phospho-sites which contain potential regulatory information. We found SOX2 is highly phosphorylated in mitosis. The phosphorylation of SOX2 in mitosis is regulated by AURKA kinase, but not other cell cycle regulators such as PLK1 or CDKs. AURKA directly interacted with SOX2 and phosphorylated SOX2. Indeed, Ser220 and Ser251 are the two key sites for mitotic SOX2 phosphorylation and present sensitivity to the kinase activity of AURKA. Either inhibition of AURKA kinase activity using specific inhibitors or knockdown of AURKA by siRNA can abolish the mitotic phosphorylation of SOX2. Both solid tumor and cultured cells were heterogeneous, the number of cells, which are named stem-cell like cell and capable of differentiated into kinds of other cells, is limited (Fig. 6). We presumed that the population of stem cell is regulated through phosphorylated SOX2 mediated by AURKA. When disrupt the mitotic modification of SOX2, either by mutating SOX2 or inactivating AURKA, the ratio of stem-cell like cells was increased (Fig. 6). Indeed, we discovered that SOX2 mutants S220A and S251A remarkably changed the cell state and reprogrammed differentiated cells, evoked the re-expression of OCT4 (Fig. 5). This may partially explain why AURKA kinase inhibitors developed as anti-tumor drugs were failed in clinical trial. Thus our discoveries will contribute to elucidating the features of stem-cell like cells in cancer and cancer therapy.
Figure 6.

Schematic model displays the mitotic regulation of SOX2 by AURKA is critical for cancer stem-cell like cell maintenance. Both solid tumor and cultured cells were heterogeneous and the number of stem-cell like cells is limited. It is proposed here that the ratio of stem-cell like cells is regulated through phosphorylation on SOX2 mediated by AURKA. AURKA phosphorylates SOX2 and induce the M phase specific modification of SOX2. Mitotic phosphorylation of SOX2 by AURKA restricts the ratio of stem-cell like cells. When disrupt the mitotic modification of SOX2, either by mutating SOX2 or inactivating AURKA, the ratio of stem-cell like cells increased.
Materials and methods
Cell culture
Human ovarian carcinoma PA-1, breast adenocarcinoma MCF7, lung carcinoma H520, embryonic kidney fibroblast 293 and mouse teratocarcinoma F9 cells were obtained from American Type Cell Collection (ATCC). F9, MCF7 and 293 were cultured with Dulbecco's Modified Eagle Medium (DMEM); PA-1 cultured with Minimum Essential Medium (MEM), H520 cultured with Roswell Park Memorial Institute (RPMI) 1640 medium. All medium was supplemented with 10% fetal bovine serum and MCF7 with additional 0.01 mg/mL human recombinant insulin (#I3536, Sigma-Aldrich Co. LLC, http://www.sigmaaldrich.com/catalog/product/sigma/i3536?lang=zhandregion=CN).
Chemicals and small interference RNA
AURKA inhibitor I (http://www.selleck.cn/products/AURKA-Inhibitor-I.html,28 VX680 (also know as Tozasertib or MK-0457; http://www.selleck.cn/products/VX-680(MK-0457).html) and BI2536 (http://www.selleck.cn/products/BI-2536.html) were from Selleck Chemicals,17 nocodazole (https://www.sigmaaldrich.com/catalog/product/sigma/m1404?lang=zhandregion=CN) and roscovitine (http://www.sigmaaldrich.com/catalog/product/sigma/r7772?lang=zhandregion=CN) were obtained from Sigma-Aldrich Inc., ON01910 (also know as Rigosertib) was from Selleck Chemicals; http://www.selleck.cn/products/ON-01910.html), ZM447439 was from Santa Cruz (http://www.scbt.com/datasheet-200696-zm-447439.html). CBB2001 was synthesized and used as described.29 The concentration of chemical inhibitors used for treatment of the cells is: AURKA inhibitor I, 25 nM, VX-680, 0.5 μM, BI2536 0.1 μM, roscovitine 5 μM, ON10910, 5 μM, ZM447439, 5 μM, CBB2001, 1 μM, and nocodazole 50 ng/mL. Active AURKA kinase was expressed and purified as described previously.29 Small interference RNA targeting human AURKA is: CCACUGAAUAACACCCAAA, and SOX2 3′-UTR is: UGCCGAGAAUCCAUGUAUA. DharmaFECT 1 Transfection Reagent (#T-2001-03, Thermo Fisher Scientific Inc.) was used to knock down the expression of target proteins, according to the protocol provided by the manufactures.
Site-directed mutation
Wild type plasmid pMSCVpuro-SOX2 (Clontech Laboratories, Inc.) was used as a PCR template for site-directed mutation using specific primers. Then PCR products were incubated with DpnI (#1235B, Takara Biotechnology (Dalian) Co., Ltd.) to remove the template from the mutated DNA, and transfected into competent E.coli cell DH5α to obtain the clone. Positive clones were sequenced to confirm the success of mutation.
Flow cytometry analysis
About 500,000 cells were trypsin harvested and resuspended in 200 μL PBS, followed by propidium iodide staining (25 μg/mL, 0.02% Triton-X100), incubation at 37 °C for 30 minutes. Dilute the cells ten times in PBS and immediately analyzed by BD FACSCalibur. Raw data were analyzed by FlowJo 7.6.1(FlowJo, LLC; http://www.flowjo.com). For 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay, Click-iT EdU Alexa Fluor 488 Flow Cytometry Assay Kit were purchased (Thermo Fisher Scientific Inc., https://www.thermofisher.com/order/catalog/product/C10425) and performed according to the protocol.
Real-time quantitative PCR (RT-qPCR)
Total cellular RNA was extracted using RNAiso Plus (#9109, Takara Biotechnology (Dalian) Co., Ltd., http://www.takara.com.cn/ProductShow.aspx?m=20141220151857153056andpro ductID=20141226160653343219), then reverse transcribed to cDNA by Reverse Transcriptase M-MLV (RNase H-) (#2641A, Takara Biotechnology (Dalian) Co., Ltd.). The relative mRNA levels of the target genes were quantified by SYBR Fast qPCR Mix (#RR430S, Takara Biotechnology (Dalian) Co., Ltd. http://www.takara.com.cn/Product.aspx?m=20141215102926123157 #p) in a CFX Connect Real-Time PCR Detection System (#1855200, Bio-Rad Laboratories, Inc.). β-actin was quantified as control.
Immunofluorescence
Cells were seeded on cover slides in 35-mm dish for 5 d to allow clones formation. After fixed with paraformaldehyde (4%) for 15 min at room temperature and permeabilized for 5 min on ice, cells were rinsed with PBS and incubated with the primary antibodies overnight at 4 °C. After rinsing in PBS to remove the unbound antibodies, cells were incubated with FITC- or TRITC- conjugated secondary antibodies for 1 h at room temperature and mounted with mowiol (#81381, Sigma-Aldrich, Co. LLC.) containing 1 μg/mL 4′, 6-diamidino-2-phenylindole dihydrochloride (DAPI) (#D8417, Sigma-Aldrich, Co. LLC.) to stain the DNA. Microscopic images were captured on an OLYMPUS IX73 fluorescence microscope.
Western blotting and antibodies
Proteins were separated by SDS-PAGE and transferred to nitrocellular membrane, blocked one hour with 3% non-fat milk and incubated with primary antibody overnight at 4 °C. After rinsed three times in TBS (with 0.2% Tween-20) and incubated with the HRP conjugated secondary antibody for one hour at room temperature, the membrane was developed onto the X-ray film. Antibodies anti SOX2 (A301-741, http://www.bethyl.com/product/A301-741A/SOX2_Antibody?r eferrer=search_default) was from Bethyl Laboratories.Inc., AURKA (#4718) from Cell Signaling Technology (http://www.cellsignal.com/products/primary-antibodies/AURKA-aik-1g4-rabbit-mab/4718?N=4294956287&Ntt=Aurora+Aand fromPage=plp). OCT4 (#sc-5279) was from Santa Cruz Biotechnology. Inc. (http://www.scbt.com/datasheet-5279-oct-3-4-c-10-antibody.html). Anti-HA-tag (AB104-02) from TIANGEN Biotech (Beijing) Co., Ltd, (http://www.tiangen.com/?product Show/t1/8/id/197.html) and anti-cullin1 was laboratory made using purified antigen as described.21,30 The intensity of protein bands were measured by Gel-Pro Analyzer software (Media Cybernetics, Inc.). Three independent experiments were analyzed to obtain the average values that were used to plot histograms.
Co-Immunoprecipitation
Cells were lysed at 4 °C for 15 minutes in lysis buffer (0.5 % NP-40, 50 mM Tris-HCl pH 7.5, 150 mM NaCl, protease and phosphotase inhibitors), and centrifuged twice at 4 °C for 15 minutes. The supernatant was incubated with 1 μg primary antibodies for 3 hours on rotator at 4 °C, and further incubated with 25 μL protein-A sepharose beads for 1 hour to pull down the protein complexes. Beads-protein complexes were rinsed in lysis buffer for five times to remove unspecific bounds.
LC-MS/MS sample preparation and analysis
Proteins were separated by SDS-PAGE (Hoefer SE400 Vertical Electrophoresis System, 18×16 cm), stained with coomassie blue G250. The protein bands were cut, destained, and dehydrated by acetonitrile. Protein was digested by Trypsin Gold (V5280, Promega Biotech Co., Ltd; http://www.promega.com/resources/msds/msdss/v5000/v5111/?cs=y) in gel at 37 °C for 14 hours. Peptides were extracted from the gel with 70% acetonitrile (with 0.02% trifluoroacetic acid) and vacuum-dry concentrated. Peptides were dissolved in liquid chromatography (LC) buffer (95% H2O, 5% acetonitrile, 0.1% formic acid) thoroughly and ready for mass spectrometry analyzing by AB SCIEX TripleTOF 5600.
Phosphorylation sites identification
Raw data in .wiff files of mass spectrums generated from AB SCIEX TripleTOF 5600 were processed to generating peaks and searching Human uniprot-all.fasta database (Proteome ID=UP000005640) by ProteinPilot 5.0 software (with Paragon Algorithm 5.0.0.0, 4767) to create a project. “group” file, which contain the annotated spectrums and proteins identified. Phospho-peptides and phosphor-sites localization were performed using the Mascot Server (Matrix Science. Ltd; http://www.matrixscience.com/cgi/search_form.pl?FORMVER =2&SEARCH=MIS). Briefly, the neutral loss of phosphate (H3PO4, 98 Da, or HPO3, 80 Da) from the intact peptide can be the characteristic indicator to the identity of the phosphorylated residue. During mass spectrum mapping, intact peptide is observed in a full scan by mass spectrometry (MS or MS1). Then intact peptides are fragmented by colliding them with an inert gas. The fragmented ions are also detected (MS/MS, or MS2). Phospho-site localization is realized through observation of ions that can distinguish between possible phosphorylatable residues. If a strong neutral loss of 98 Da is observed, the intact peptide is expected phsophorylating on serine or theronine (pS or pT). Mascot provides an algorithm for phosphorylation site mapping, spectrum depicting and scoring. Normally, the neutral losses of phosphate in corresponding fragmented ions are clearly displayed in the spectrum.31,32
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
D.D. Q. and F. L. conceived this project, designed the experiments, performed experiments, analyzed and organized the data, wrote the manuscript. Q.Q. W and M. Y performed the experiments. R.F. L. designed the experiments, performed experiments, and wrote the manuscript. S.M. L. implemented the mass spectrometry experiments, analyzed the data, and reviewed the manuscript.
Funding
We thank grants from National Natural Science Foundation of China (NSFC) (21402167, 21133002), Natural Science Foundation of Guangdong Province (2014A030313779) and the Peking University Shenzhen Graduate School (Key State Laboratory of Chemical Genomics open project fellowship program).
References
- [1].Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat Neurosci 2003; 6:1162-8; PMID:14517545; http://dx.doi.org/ 10.1038/nn1131 [DOI] [PubMed] [Google Scholar]
- [2].Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature 2007; 448:313-7; PMID:17554338; http://dx.doi.org/ 10.1038/nature05934 [DOI] [PubMed] [Google Scholar]
- [3].Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131:861-72; PMID:18035408; http://dx.doi.org/ 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- [4].Kopp JL, Ormsbee BD, Desler M, Rizzino A. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells 2008; 26:903-11; PMID:18238855; http://dx.doi.org/ 10.1634/stemcells.2007-0951 [DOI] [PubMed] [Google Scholar]
- [5].Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P, et al.. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol 2005; 25:6031-46; PMID:15988017; http://dx.doi.org/ 10.1128/MCB.25.14.6031-6046.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gao Z, Cox JL, Gilmore JM, Ormsbee BD, Mallanna SK, Washburn MP, Rizzino A. Determination of protein interactome of transcription factor Sox2 in embryonic stem cells engineered for inducible expression of four reprogramming factors. J Biol Chem 2012; 287:11384-97; PMID:22334693; http://dx.doi.org/ 10.1074/jbc.M111.320143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mallanna SK, Ormsbee BD, Iacovino M, Gilmore JM, Cox JL, Kyba M, Washburn MP, Rizzino A. Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells 2010; 28:1715-27; PMID:20687156; http://dx.doi.org/ 10.1002/stem.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Van Hoof D, Munoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, Mummery CL, Krijgsveld J. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 2009; 5:214-26; PMID:19664995; http://dx.doi.org/ 10.1016/j.stem.2009.05.021 [DOI] [PubMed] [Google Scholar]
- [9].Tsuruzoe S, Ishihara K, Uchimura Y, Watanabe S, Sekita Y, Aoto T, Saitoh H, Yuasa Y, Niwa H, Kawasuji M, et al.. Inhibition of DNA binding of Sox2 by the SUMO conjugation. Biochem Biophys Res Commun 2006; 351:920-6; PMID:17097055; http://dx.doi.org/ 10.1016/j.bbrc.2006.10.130 [DOI] [PubMed] [Google Scholar]
- [10].Wu Y, Guo Z, Wu H, Wang X, Yang L, Shi X, Du J, Tang B, Li W, Zhang Y. SUMOylation represses Nanog expression via modulating transcription factors Oct4 and Sox2. PLoS One 2012; 7:e39606; PMID:22745796; http://dx.doi.org/ 10.1371/journal.pone.0039606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhao HY, Zhang YJ, Dai H, Zhang Y, Shen YF. CARM1 mediates modulation of Sox2. PLoS One 2011; 6:e27026; PMID:22046437; http://dx.doi.org/ 10.1371/journal.pone.0027026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fang L, Zhang L, Wei W, Jin X, Wang P, Tong Y, Li J, Du JX, Wong J. A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol Cell 2014; 55:537-51; PMID:25042802; http://dx.doi.org/ 10.1016/j.molcel.2014.06.018 [DOI] [PubMed] [Google Scholar]
- [13].Justilien V, Walsh MP, Ali SA, Thompson EA, Murray NR, Fields AP. The PRKCI and Sox2 oncogenes are coamplified and cooperate to activate Hedgehog signaling in lung squamous cell carcinoma. Cancer Cell 2014; 25:139-51; PMID:24525231; http://dx.doi.org/ 10.1016/j.ccr.2014.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ouyang J, Yu W, Liu J, Zhang N, Florens L, Chen J, Liu H, Washburn M, Pei D, Xie T. Cyclin-Dependent Kinase-mediated Sox2 phosphorylation enhances the ability of Sox2 to establish the pluripotent State. J Biol Chem 2015; 290:22782-94; PMID:26139602; http://dx.doi.org/ 10.1074/jbc.M115.658195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res 2007; 5:1-10; PMID:17259342; http://dx.doi.org/ 10.1158/1541-7786.MCR-06-0208 [DOI] [PubMed] [Google Scholar]
- [16].Fu J, Bian M, Liu J, Jiang Q, Zhang C. A single amino acid change converts Aurora-A into Aurora-B-like kinase in terms of partner specificity and cellular function. Proc Natl Acad Sci U S A 2009; 106:6939-44; PMID:19357306; http://dx.doi.org/ 10.1073/pnas.0900833106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA, Demur C, Hercend T, Diu-Hercend A, et al.. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med 2014; 10:262-7; http://dx.doi.org/ 10.1038/nm1003 [DOI] [PubMed] [Google Scholar]
- [18].Yin F, Lan R, Zhang X, Zhu L, Chen F, Xu Z, Liu Y, Ye T, Sun H, Lu F, et al.. LSD1 regulates pluripotency of embryonic stem/carcinoma cells through histone deacetylase 1-mediated deacetylation of histone H4 at lysine 16. Mol Cell Biol 2014; 34:158-79; PMID:24190971; http://dx.doi.org/ 10.1128/MCB.00631-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Du L, Yang Y, Xiao X, Wang C, Zhang X, Wang L, Li W, Zheng G, Wang S, Dong Z. Sox2 nuclear expression is closely associated with poor prognosis in patients with histologically node-negative oral tongue squamous cell carcinoma. Oral Oncol 2011; 47:709-13; PMID:21689966; http://dx.doi.org/ 10.1016/j.oraloncology.2011.05.017 [DOI] [PubMed] [Google Scholar]
- [20].Zhang X, Lu F, Wang J, Yin F, Xu Z, Qi D, Wu X, Cao Y, Liang W, Liu Y, et al.. Pluripotent stem cell protein Sox2 confers sensitivity to LSD1 inhibition in cancer cells. Cell Rep 2014; 5:445-57; http://dx.doi.org/ 10.1016/j.celrep.2013.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R, Liu Y, Ward D, Quan J, Ye T, et al.. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res 2011; 71:7238-49; PMID:21975933; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-0896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Taira N, Mimoto R, Kurata M, Yamaguchi T, Kitagawa M, Miki Y, Yoshida K. DYRK2 priming phosphorylation of c-Jun and c-Myc modulates cell cycle progression in human cancer cells. J Clin Invest 2012; 122:859-72; PMID:22307329; http://dx.doi.org/ 10.1172/JCI60818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ding D, Xu H, Liang Q, Xu L, Zhao Y, Wang Y. Overexpression of Sox2 in C3H10T1/2 cells inhibits osteoblast differentiation through Wnt and MAPK signalling pathways. Int Orthop 2011; 36:1087-94; PMID:22012572; http://dx.doi.org/ 10.1007/s00264-011-1368-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, et al.. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 2009; 5:491-503; PMID:19818703; http://dx.doi.org/ 10.1016/j.stem.2009.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell 2008; 3:568-74; PMID:18983970; http://dx.doi.org/ 10.1016/j.stem.2008.10.004 [DOI] [PubMed] [Google Scholar]
- [26].Li X, Zuo X, Jing J, Ma Y, Wang J, Liu D, Zhu J, Du X, Xiong L, Du Y, et al.. Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell 2015; 17:195-203; PMID:26253201; http://dx.doi.org/ 10.1016/j.stem.2015.06.003 [DOI] [PubMed] [Google Scholar]
- [27].Jeong CH, Cho YY, Kim MO, Kim SH, Cho EJ, Lee SY, Jeon YJ, Lee KY, Yao K, Keum YS, et al.. Phosphorylation of Sox2 cooperates in reprogramming to pluripotent stem cells. Stem Cells 2010; 28:2141-50; PMID:20945330; http://dx.doi.org/ 10.1002/stem.540 [DOI] [PubMed] [Google Scholar]
- [28].Aliagas-Martin I, Burdick D, Corson L, Dotson J, Drummond J, Fields C, Huang OW, Hunsaker T, Kleinheinz T, Krueger E, et al.. A class of 2, 4-bisanilinopyrimidine Aurora A inhibitors with unusually high selectivity against Aurora B. J Med Chem 2009; 52:3300-7; PMID:19402633; http://dx.doi.org/ 10.1021/jm9000314 [DOI] [PubMed] [Google Scholar]
- [29].Lan R, Lin G, Yin F, Xu J, Zhang X, Wang J, Wang Y, Gong J, Ding YH, Yang Z, et al.. Dissecting the phenotypes of Plk1 inhibition in cancer cells using novel kinase inhibitory chemical CBB2001. Lab Invest 2012; 92:1503-14; PMID:22890557; http://dx.doi.org/ 10.1038/labinvest.2012.114 [DOI] [PubMed] [Google Scholar]
- [30].Qi D, Wang Q, Li H, Zhang T, Lan R, Kwong D, Wong W, Wong K, Li S, Lu F. SILAC-based quantitative proteomics identified lysosome as a fast response target to PDT agent Gd-N induced oxidative stress in human ovarian cancer IGROV1 cells. Mol Bio Syst 2015; 11:3059-67 [DOI] [PubMed] [Google Scholar]
- [31].Dephoure N, Gould KL, Gygi SP, Kellogg DR. Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Mol Biol Cell 2013; 24:535-42; PMID:23447708; http://dx.doi.org/ 10.1091/mbc.E12-09-0677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Savitski MM, Lemeer S, Boesche M, Lang M, Mathieson T, Bantscheff M. and Kuster B. Confident phosphorylation site localization using the Mascot Delta Score. Mol Cell Proteomics 2011; 10:M110.003830; PMID:21057138; http://dx.doi.org/ 10.1074/mcp.M110.003830 [DOI] [PMC free article] [PubMed] [Google Scholar]