Abstract
RNA sequencing (RNA-seq) has revolutionary roles in transcriptome identification and quantification of different types of tissues and cells in many organisms. Although numerous RNA-seq data derived from many types of human tissues and cell lines, little is known on the transcriptome repertoire of human aortic valve. In this study, we sequenced the total RNA prepared from two calcified human aortic valves and reported the whole transcriptome of human aortic valve. Integrating RNA-seq data of 13 human tissues from Human Body Map 2 Project, we constructed a transcriptome repertoire of human tissues, including 19,505 protein-coding genes and 4,948 long intergenic noncoding RNAs (lincRNAs). Among them, 263 lincRNAs were identified as novel noncoding transcripts in our data. By comparing transcriptome data among different human tissues, we observed substantial tissue specificity of RNA transcripts, both protein-coding genes and lincRNAs, in human aortic valve. Further analysis revealed that aortic valve-specific lincRNAs were more likely to be recently derived from repetitive elements in the primate lineage, but were less likely to be conserved at the nucleotide level. Expression profiling analysis showed significant lower expression levels of aortic valve-specific protein-coding genes and lincRNA genes, when compared with genes that were universally expressed in various tissues. Isoform-level expression analysis also showed that a majority of mRNA genes had a major isoform expressed in the human aortic valve. To our knowledge, this is the first comparative transcriptome analysis between human aortic valve and other human tissues. Our results are helpful to understand the transcriptome diversity of human tissues and the underlying mechanisms that drive tissue specificity of protein-coding genes and lincRNAs in human aortic valve.
Keywords: transcriptome, long noncoding RNAs, protein-coding genes, aortic valve, tissue specificity
Introduction
Transcriptome is the complete set of transcribed transcripts in a cell, including mRNAs, noncoding RNAs, and small RNAs. As a revolutionary tool for transcriptome analysis,1 RNA sequencing (RNA-seq) has been widely applied to determine the repertoire of RNA transcripts,2 the different isoform structures of a gene,3 and differentially expressed transcripts under various conditions.4 These studies have gained important insights into the functional elements of the genome, the constituents of different cells and tissues, and the molecular mechanisms of development and disease.5
Among the large amount of transcriptome studies, human transcriptome is one of the most explored, and thus well-understood, transcript repertoires in the past several years. In an earlier work, Wu et al.6 interrogated the transcribed loci in 420 ENCyclopedia Of DNA Elements (ENCODE) regions using rapid amplification of cDNA ends (RACE) sequencing, and they concluded that much of the human genomes are transcribed. Using RNA-seq, Wang et al.7 and Sultan et al.8 sequenced diverse human tissue and cell line transcriptomes. They confirmed extensive transcription of human genome in different cell types and revealed that more than 90% of human genes are under regulation of alternative splicing.7,8 Further analysis found that human transcriptome is dominated by one transcript per protein-coding gene, which hinted that not all the transcripts contributing to transcriptome diversity are equally likely to contribute to protein diversity.9 Recent advances in human transcriptome studies include deep sequencing of the whole transcriptome repertoires in different human cell lines and tissues.10–12 In addition to protein-coding genes, which cover only 3% of the whole human genome, long intergenic noncoding RNAs (lincRNAs) are a class of noncoding RNAs that do not encode proteins.13 LincRNAs are transcribed from most human genomic regions and have important roles in regulating gene expression.14 Cabili et al.15 collected ∼4 billion RNA-seq reads from 24 human tissues and cell types and cataloged more than 8,000 lincRNAs. Using a de novo transcriptome assembly method, Hangauer et al.16 identified tens of thousands more putative lincRNAs in the human transcriptome. A recent study14 curated 7,256 RNA-seq libraries from human tumors, normal tissues, and cell lines, yielding a catalog of 58,648 human lincRNAs in the human transcriptome. Comparative transcriptome analysis among human tissues and between human beings and other mammals revealed tissue-specific expression of human protein-coding transcripts and lincRNAs in evolutionary conserved tissues.17,18 Furthermore, Genotype-Tissue Expression (GTEx) project19 investigated transcriptome variations across human tissues and individuals. They found stable transcriptional signatures in tissues, and the signatures are dominated by a small number of genes.19 All these studies have brought us a much comprehensive portrait of human transcriptome, and finally a detailed annotation of human transcripts.20
Although previous studies have sequenced transcriptomes of many human tissues, most of them are tissues that can be easily acquired from clinical samples,12,21 such as the brain, lung, breast, heart, liver, and testis. In order to gain insights into the transcriptome diversity of human tissues, transcriptome components of some other tissues still need to be addressed. Some attempts were performed in the past several years, such as transcriptome analysis of human retina,22 testis,23 and kidney.24 These studies identified tissue-specific transcripts that are complementary to known human transcripts, and some of them are related to tissue physiology and function. Here, we reported a transcriptome analysis of another less investigated human tissue, aortic valve. Aortic valve is a kind of semilu-nar valve of the heart, and its open and close can modulate the pressure of the left ventricle.25 The dysfunction of aortic valve ranges from aortic stenosis, aortic regurgitation, aortic aneurysms, and aortic dissection.26 We sequenced the whole repertoire of both protein-coding transcripts and lincRNAs from a diseased bicuspid aortic valve (BAV) and a calcified tricuspid aortic valve (TAV). Next, we compared human aortic valve transcriptome with those of 16 human tissues from Human Body Map 2 project15 and identified aortic valve- specific protein-coding genes and lincRNAs. We further analyzed isoform structures and their expression of each protein-coding gene in the human aortic valve. Finally, we compared the characteristic features of aortic valve-specific lincRNAs to those of lincRNAs that are universally expressed in human tissues.
Materials and Methods
Sample collection
We collected two calcified human aortic valve samples during operation, one from a BAV patient and the other from a TAV patient. The baseline characteristics of these two patients are listed in Table 1. These two samples were obtained from the Jiangsu Province People’s Hospital with informed consent from all patients concerned. This study was considered by the Ethics Committee of the Jiangsu Province People’s Hospital, and the requirement of ethics approval was waived. The research complied with the principles of the Declaration of Helsinki.
Table 1.
Baseline characteristics of the two patients used in this study.
| cHARAcTeRiSTicS | TAV | bAV |
|---|---|---|
| Sex | Male | Male |
| Age at operation | 53 | 49 |
| Body surface area (m2) | 1.78 | 1.62 |
| Predominant valve dysfunction | Moderate to severe AR | Moderate AS |
| Valve morphology | N/A | R-L fusion |
| Leaflet calcification | + | ++ |
Abbreviations: TAV, tricuspid aortic valve; BAV, bicuspid aortic valve; predominant valve dysfunction: AR, aortic regurgitation; AS, aortic stenosis; valve morphology: R–L fusion, functionally bicuspid due to fusion of right and left coronary leaflets; N/A, not applicable; leaflet calcification: ++ for heavily calcified leaflets (>2/3 leaflet area), + for mild calcification (<1/3 leaflet area).
Library preparation and high-throughput sequencing
Total RNA was obtained from BAV and TAV samples using TRIzol (Invitrogen) according to manufacturer’s protocol. Genomic DNA was removed using DNase (New England Biolabs), and RNA purity was assessed using the NanoDrop 2000. Each RNA sample had an A260:A280 ratio above 1.8 and A260:A230 ratio above 2.2. The total RNA was subjected to ribosomal RNA depletion according to the manufacturer’s protocol of RiboMinus kit. Next, RNA was fragmented into 200 base pairs (bps) using the RNA fragmentation kit (Ambion) and quantified with NanoDrop. The first cDNA strand was synthesized using random hexamer primers, and the second cDNA strand was synthesized where dUTP was used instead of dTTP. In this step, actinomycin D was used to increase strand specificity by inhibiting second-strand cDNA synthesis. At 15 °C, 0.5 µL of actinomycin D solution (120 ng/µL), 0.5 µL of RNaseOUT (40 units/µL, Invitrogen), and 0.5 µL of SuperScript III polymerase (200 units/µL, Invitrogen) were added to the reaction. Then, Ethidium bromide (20 µL; 10 mM Tris-Cl, pH 8.5, Qiagen) was added to the reaction, and the Deoxy-ribonucleotide Triphosphates were removed by purification of the first strand mixture on a self-made 200 µL G-50 gel filtration spin-column equilibrated with 1 mM Tris-Cl, pH 7.0. After the second strand synthesis and DNA fragmentation process, the sequencing libraries were further constructed by following the manufacturer’s instructions (Illumina). Fragments of 300–400 bps were recovered and purified, and then enriched by Polymerase Chain Reaction for 15 cycles. Each library was loaded into one lane of the Illumina HiSeq 2500 for 2 × 100 bps pair-end (PE) sequencing.
In order to investigate the tissue specificity of aortic valve transcriptome, we downloaded transcriptome data of 16 human tissues from NCBI Gene Expression Omnibus (GEO) under accession GSE30611 (Supplementary Table 1).15 This data set is part of the Human Body Map 2 project, and each tissue had both single-end (SE) and PE poly-adenylation (polyA)-enriched libraries. Since aortic valve transcriptome contains transcripts without polyA tail at its 3′ end, we should remove these transcripts before comparing aortic valve transcriptomes against transcriptomes from the Human Body Map 2 project.15 Therefore, we also downloaded a data set of human polyA sites from NCBI GEO under accession GSM747477.15
Construction of human transcriptome from RNA-seq data
We used FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to control the quality of sequencing data. At an initial filtering step, we discarded low quality reads, including reads that have adaptors, reads with more than 10% unknown bases, and reads that have more than 50% bases with quality value less than 5. Then, the clean reads were mapped to the human reference genome (hg19) by TopHat27 using “-G” option and Ensembl gene annotation (ftp://ftp.ensembl.org/pub/release-78/gtf/homo_sapiens). Here, “-G” option was used to supply the Ensembl gene model as a GTF formatted file for TopHat mapping. Then, the mapped RNA-seq reads were de novo assembled separately for each sample using Cufflinks.28 Finally, the assembled transcriptomes of each sample were merged to an integrated transcriptome of human tissues using Cuffmerge.28
To construct a transcriptome repertoire in human tissues, the merged transcriptome was compared with known human gene annotation using Cuffcompare.28 For transcripts that have different genomic locations against the reference gene set, we identified a reliable catalog of novel lincRNAs. First, we carried out a six-step identification process to detect a set of putative novel lincRNAs: (1) we extracted transcripts within a class code of “u” in the tracking rows from Cuffcompare output, which was interpreted as unknown intergenic transcript and may contain novel lincRNAs; (2) we used in-house Perl scripts to exclude transcripts smaller than 200 bps; (3) we aligned the remaining transcript to Ensemble known lincRNA using blastN29 with a cutoff e-value of 1e−10 to remove some transcript segments from known human linc-RNAs; (4) we used TransDecoder (http://transdecoder.sourceforge.net/) to identify putative open reading frame (ORF) in each transcript, and removed transcripts have putative ORFs longer than 300 bps30; (5) we aligned the remaining transcripts to NCBI nonredundant (nr) protein database to eliminate transcripts with potential protein-coding ability (E-value ≤1 × 10−5); (6) we used the Coding-Potential Assessment Tool31 to estimate the coding potential of each transcript and removed transcripts that had higher coding probability. As suggested by Wang et al.31, the optimal cutoff of human coding probability was 0.364, which gave the highest sensitivity and specificity. After that, we discarded single-exon transcripts and lowly expressed multiexon transcripts. The expression level was determined by read counts, and only transcripts that have read counts larger than ten in at least one sample were retained. Since the differences of sequencing libraries between aortic valve samples and other human tissue samples, we further removed novel lincRNA candidates that did not overlap or end within 100 bps of human polyA sites in the two aortic valve samples. For transcripts that overlapped to known human gene annotations, we filtered known linc-RNAs by removing lowly expressed lincRNA transcripts (read counts less than ten) in all samples and lincRNAs annotated as level 3 (automatically annotated loci) in GENCODE.20 As for known protein-coding genes, we constructed the stringent set of mRNA transcripts by excluding lowly expressed transcripts (read counts less than ten) in all tissue samples.
The flow chart of transcriptome construction is shown in Figure 1.
Figure 1.
A schematic computational workflow of RNA-seq data in constructing a stringent repertoire of human RNA transcripts.
Expression quantification and normalization of human transcripts in all tissue samples
Based on the TopHat alignment BAM file, Cuffnorm28 was used to estimate and quantify gene expression separately for each RNA-seq data with default parameters, yielding raw read count and expression abundance for each of the protein-coding genes and lincRNA genes across all human samples. Gene expression was measured in fragments per kilobase of exon per million reads mapped (FPKM). FPKM calls were log10-normalized (after addition of ε = 0.001). We also measured isoform abundances of each gene using RSEM,32 which requires the short read alignments of RNA-seq data to the human genome. We used a threshold of five mapped reads to consider a transcript as expressed. Major isoform of a gene was defined as the isoform with the highest expression level within a gene. The fraction of the mRNA pool can be explained as the ratio of the sum of FPKM for major isoforms and the sum of FPKM for all the isoforms.
Tissue specificity score of protein-coding genes and lincRNAs
To evaluate the tissue specificity of a gene, we calculated a tissue specificity score as suggested by Cabili et al.15 This score is an entropy-based measure that quantifies the distance of a given transcript’s expression vector to a predefined expression vector that represents the extreme case of only being transcribed in one tissue.15 We started with an average FPKM value in each tissue, and this expression vector was transformed to a normalized density vector with values between 0 and 1. The tissue specificity score was calculated for each tissue, and the maximum value across all human tissues was set to be the tissue specificity score of the gene. We chose tissue specificity score of 0.4 as the cutoff in differentiating the tissue-specific gene and the tissue universal gene. Genes with tissue specificity score larger than 0.4 were defined as tissue-specific genes. Since the aortic valve tissues were obtained from two patients with different aortic valve diseases, we only retained genes that are not differentially expressed between BAV and TAV samples for tissue specificity analysis. We used edgeR33 to identify differentially expressed genes by pair wise comparison.
Gene Ontology and KEGG enrichment analysis of aortic valve-specific genes
For each aortic valve-specific mRNA gene, we performed Gene Ontology (GO) enrichment analysis of aortic-specific genes, including aortic valve-specific lincRNA-related protein-coding genes and aortic valve-specific protein-coding genes, using Go:TermFinder.34 We also performed KEGG pathway enrichment analysis using the KEGG Orthology-Based Annotation System (KOBAS).35
Conservation score
To assess the conservation level of each lincRNA and protein-coding gene, SIte-specific PHY-logenetic analysis (SiPhy)36 was run on the 46-way alignment available from UCSC genome browser.37 The omega value from SiPhy calculation36 was used to evaluate nucleotide sequence conservation throughout this study.
Evolutionary dynamics
We used pairwise alignments from UCSC genome browser37 to analyze the evolutionary dynamics of RNA transcripts. To this end, we downloaded the chain files between human beings and chimpanzee, rhesus, mouse, rat, cow, and the chain files with nonhuman species as reference. LiftOver facilities were chosen to map genomic positions from human beings to another species. If a region was covered, we then mapped the putative orthologous region back to the human genome and tested if the mapping is reciprocal. The orthologous fractions shown in Figure 4D were estimated using the proportion of the locus that can be mapped reciprocally to the total locus.
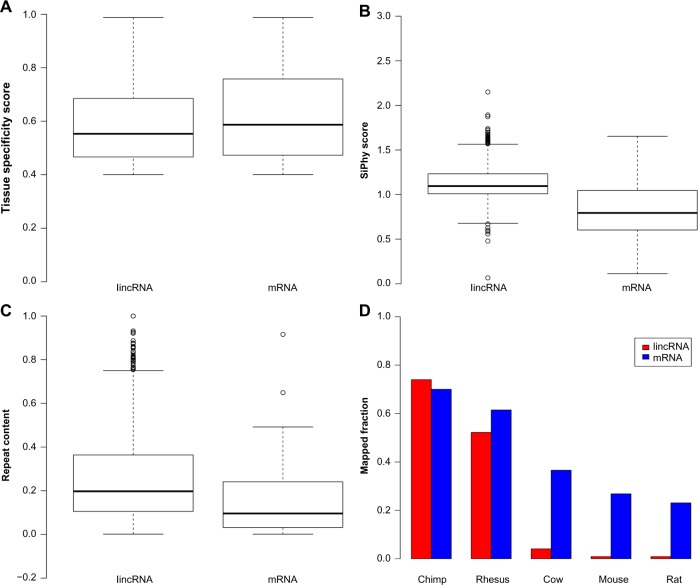
Figure 4.
Comparison of aortic valve-specific mRNAs and lincRNAs: (A) tissue specificity score; (B) SiPhy score; (C) repeat content; and (d) fraction of homologous sequences in chimp, rhesus, cow, mouse, and rat.
Repeat content
Repeat annotation of human genome was downloaded from UCSC genome browser.37 To quantify the repeat content of each transcript, we calculated the ratio of repetitive elements in its exonic region.
Statistical analysis
In our analysis, we used Mann–Whitney two-tailed test to compare two distributions. To group human tissues based on expression values of protein-coding genes and lincRNAs, we performed hierarchical cluster analysis and principal component analysis. All these analyses were performed by R platform (http://www.r-project.org/).
Results
Transcriptome repertoire of human aortic valve
We collected two human aortic valve samples from a diseased BAV patient and a calcium TAV patient. Total RNAs were purified from these two samples, and a ribo-depleted library was constructed for each sample. Next, these two libraries were sequenced on Illumina HiSeq 2500 platform. In total, high-throughput sequencing created a mean of 112 million PE reads for each sample (Supplementary Table 1), and the sequencing quality is high with Q30 values larger than 92.05% for both samples. The raw RNA-seq data of two human aortic valve samples were freely accessible upon request. To construct a repertoire of human transcripts in human beings and compare transcriptome among human tissues, we also downloaded RNA-seq data of 16 human tissues from Human Body Map 2 Project,15 which include one SE reads data and one PE reads data for each tissue (Supplementary Table 1). RNA-seq data from both aortic valve samples and Human Body Map 2 Project were used for downstream bioinformatic analysis. For each RNA-seq data, we mapped short reads to the human genome using known gene annotation. Based on mapping results, we quantified the expression of known protein-coding genes, transcript isoforms, and lincRNAs using in-house pipeline (see Materials and methods section, Fig. 1). To identify novel linc-RNAs that were not annotated in current databases, we de novo assembled transcriptome data from each separate sample and filtered transcripts that are possible false positives (see Materials and methods section, Fig. 1). Finally, we constructed a stringent set of human RNA transcripts, including 19,505 protein-coding genes and 4,948 lincRNAs. Among them, 263 lincRNAs were putative novel lincRNAs identified from our dataset (Fig. 1).
Tissue-specific analysis of aortic valve transcriptome at the gene level
We quantified the expression of protein-coding genes from each RNA-seq data and compared gene expression profiles among human tissues. Hierarchal clustering analysis of expression value of 19,505 protein-coding genes showed tissue-based expression patterns of protein-coding genes in human beings (Fig. 2A). Samples from the same human tissue were clustered together in the histogram (Fig. 2A). In particular, aortic valves were clustered with human testis and brain with regard to protein-coding gene expression, which were located at the leftmost side of the histogram (Fig. 2A). Principal component analysis of expression values of protein-coding genes in these human tissues showed similar pattern (Supplementary Fig. 1). Differential expression analysis showed that 18,178 protein-coding genes and 3,094 lincRNAs had similar expression in two human aortic valves. This set of protein-coding genes and lincRNAs was used to represent human aortic valve transcriptome for tissue specificity analysis. We calculated tissue specificity score for each gene in all 17 human tissues and counted the number of tissue-specific protein-coding genes based on tissue specificity score (Fig. 2C). Aortic valve had 454 tissue-specific protein-coding genes, which was only smaller than brain and testis (Fig. 2C). KEGG enrichment analysis revealed that genes in some aortic valve function-related molecular pathways, such as RIG-I-like receptor signaling pathway and Jak-STAT signaling pathway, were significantly enriched in aortic valve-specific mRNA gene set (Supplementary Table 2). GO enrichment analysis also suggested that genes in some functional categories, including G-protein coupled receptor activity, were enriched in aortic valve-specific protein-coding gene set (Supplementary Table 2). Moreover, we compiled the top 100 genes that have the highest expression level in the human aortic valve. Aortic valve function-related molecular pathways, such as complement and coagulation cascades, were significantly enriched in this gene set (Supplementary Table 3).
Figure 2.
Hierarchical clustering of (A) protein-coding and (B) lincRNA genes based on expression values in two aortic valve samples and 32 human tissue samples from Human Body Map 2 Project. The number of tissue-specific (C) protein-coding and (d) lincRNA genes in each human tissue was also shown.
Isoform-level analysis of human transcriptome
The diversified repertoire of mammalian transcripts is mainly driven by isoform structure, which is a general characteristic of mammalian genes and regulated by alternative splicing. To understand the isoform structure in human aortic valve, we estimated the expression level of all isoforms of each gene in the aortic valve and assigned isoforms as major isoforms or minor isoforms based on their relative expression (see Materials and methods section). We observed that more than 60% of protein-coding genes in human aortic valve had more than one isoform. However, most of these multi-isoform genes expressed only one major isoform in the aortic valve (Fig. 3A). The expression level of major isoforms in human aortic valve was significantly higher than that of minor isoforms (P-value < 2.2e−16; Figure 3B). The expression level of major isoforms accounted more than 60% of the whole gene expression, while the second abundant isoform of each gene only explained less than 20% of the whole transcript repertoire (Fig. 3C).
Figure 3.
Isoform-level expression analysis of human mRNA genes in aortic valve. Isoforms of each gene were ranked by their expression level, and all isoforms were grouped based on its rank. We showed (A) the relative transcript abundance of isoforms in each rank group; (B) the histogram of expression level of major isoforms and minor isoforms; and (C) percentage of mRNA transcripts in each rank group.
Tissue-specific analysis of lincRNAs in human tissues
Other than protein-coding genes, lincRNAs are another set of abundant RNA transcripts that are expressed in human tissues. In comparison to protein-coding genes, lincRNAs in aortic valves had smaller number of exons and shorter transcript length (Supplementary Fig. 2). Next, we performed lincRNA expression comparison among all human tissues by hierarchal clustering analysis. We observed tissue-specific expression of 4,948 lincRNAs in human tissues, where samples from the same tissue were grouped together in the histogram (Fig. 2B). Interestingly, aortic valve samples were also grouped with human testis samples at the right side of the histogram (Fig. 2B). Principal component analysis of expression values of lincRNAs in these human tissues showed a similar pattern (Supplementary Fig. 3). Tissue specificity analysis of human lincRNAs showed that aortic valve had 725 tissue-specific lincRNAs (Fig. 2D). Similar to protein-coding genes (Fig. 2C), testis, aortic valve, and brain were the top three tissues with the highest tissue specificity of linc-RNAs (Fig. 2D).
Characteristics of aortic valve-specific transcripts
In previous sections, we observed substantial tissue specificity of both protein-coding genes (Fig. 2C) and lincRNA genes (Fig. 2D) in human aortic valves. To understand the basic characteristics of aortic valve-specific RNA transcripts, we first compared several features of aortic valve-specific lincR-NAs with those of aortic valve-specific mRNAs. We found a low expression level of both aortic valve-specific mRNAs and lincRNAs, and their expression values were statistically similar (P-value > 0.1). The tissue specificity of aortic valve-specific lincRNAs was statistically smaller than that of aortic-specific protein-coding genes (Fig. 4A, P-value < 0.0017). Aortic valve-specific lincRNAs had smaller sequence conservation (higher SiPhy score) than those of aortic valve- specific mRNAs (Fig. 4B; P-value < 2.2e−16). Further analysis showed that most aortic valve-specific lincRNAs were more likely to be recently evolved in primate lineage, while aortic valve-specific protein-coding genes were gradually evolved along the process of mammalian evolution (Fig. 4D). We also investigated the contribution of repetitive elements to the original of aortic valve-specific lincRNAs and mRNAs. We found that repeat content was significantly higher in tissue-specific lincRNAs (Fig. 4C; P-value < 1.017e−09).
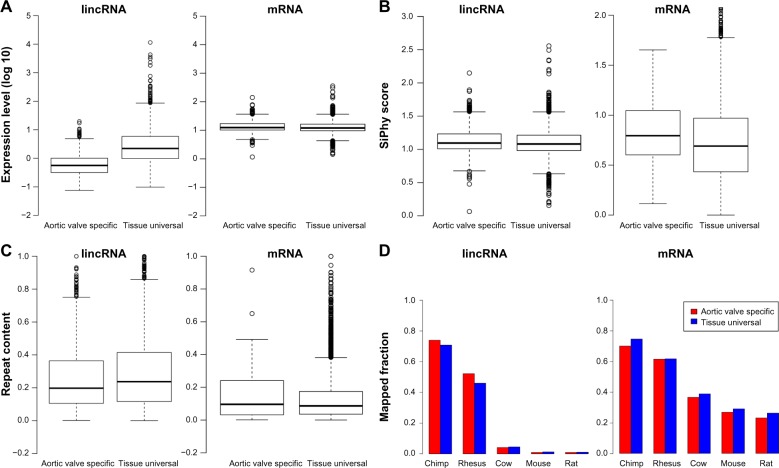
Next, we compiled a set of RNA transcripts that were universally expressed in all human tissues (see Materials and methods section) and compared the features of aortic valve-specific RNA transcripts with those of tissue-universal transcripts. We observed that both aortic valve-specific mRNAs (P-value < 2.2e−16) and lincRNAs (P-value < 2.2e−16) had significantly smaller expression levels than tissue-universal transcripts (Fig. 5A). SiPhy score was significantly smaller in universally expressed transcripts than those in aortic valve-specific transcripts (Fig. 5B, P-value < 8.325e−13 for mRNAs and P-value < 0.007946 for lincRNAs), which suggested sequences of universally expressed transcripts were under higher selective pressure in keeping their biological function. In general, aortic valve-specific mRNAs and lincRNAs were less conserved, comparing to transcripts that were universally expressed in human tissues (Fig. 5D). For repeat content in the transcripts, we found higher repeat content in aortic valve-specific lincRNAs than tissue-universal lincRNAs (Fig. 5C; P-value < 0.00289). But, repeat content in aortic valve-specific mRNAs was statistically equal to that in tissue-universal mRNAs (Fig. 5C; P-value > 0.1).
Figure 5.
Comparison of aortic valve-specific transcripts against tissue-universal transcripts: (A) expression level; (B) SiPhy score; (C) repeat content; and (d) fraction of homologous sequences in chimp, rhesus, cow, mouse, and rat.
Discussion
In this study, we reported a transcriptome analysis of human aortic valve samples and its comparison with other human tissues. Our main aim was to identify aortic valve-specific transcripts, both protein-coding genes and lincRNAs, in human aortic valve. This is different from a previous study that also analyzed the transcriptome of human aortic valves.38 In order to ascertain the underlying mechanism of valvular degeneration, Padang et al.38 compared the transcriptome of valve tissues from patients with diseased BAV and calcified TAV. They identified several differentially expressed mRNA genes in different patient groups, including genes involving Notch1-signaling pathway. In contrast, we constructed a transcriptome repertoire of human aortic valve, including protein-coding genes and lincRNAs. Other than known mRNAs and lincRNAs in current public database, we computationally predicted 263 putative novel lincRNAs with strict filtering settings, although wet laboratory experiments are needed to validate these novel lincRNAs. Some previous studies have integrated various sources of human transcriptomes from different tissues and cell lines.12,14,19,39 Our study complemented these studies with transcriptomes of two human aortic valves and constructed a new repertoire of human transcriptome. In comparison to a recently published human lincRNA repertoire – mitranscriptome,14 73 of 263 (28%) putative novel lincRNAs were included in mitranscriptome. The remaining 190 putative lincRNAs were more likely to be lincRNAs specifically expressed in human aortic valve. We found that most protein-coding genes have one major transcript expressed at significantly higher level than others (Fig. 3A), and the major transcripts contributed more than 60% to the total mRNA in aortic valve (Fig. 3C). This suggested that the transcriptome of aortic valve from protein-coding loci was dominated by one transcript per gene, which was consistent with the previous findings observed in 16 other human tissues.9 When comparing to transcriptomes of other human tissues, we found a set of mRNAs and lincRNAs that were specifically expressed in aortic valve. Finally, we analyzed the characteristics of these aortic valve-specific transcripts, including sequence conservation, evolutionary dynamics, repeat content, etc. It is noteworthy to mention that the aortic valve transcriptome we constructed was from diseased patients, which might be somewhat different from a normal human aortic valve. For example, transcripts that were downregulated in both TAV and BAV will be missed in our aortic valve transcriptome.
Previous studies have found that human testis and brain are the top two human tissues with the highest tissue specificity.14,19,21,23,40 Djureinovic et al.23 compared mRNAs in 27 human tissues and found that human testis had the largest number of tissue-specific mRNAs. Yeo et al.40 found that human brain and testis had the highest level of alternative splicing events. Melé et al.12 observed that tissue-specific transcripts were exclusive to testis, and most were lincRNAs. Consistent with these previous findings, we observed high tissue specificity of both mRNAs and lincRNAs in human testis and brain (Fig. 2C and D). Interestingly, our results showed that aortic valve had substantial tissue specificity of protein-coding genes and lincRNAs, which was similar to testis and brain (Fig. 2C and D). Although the exact reason why aortic valve had such high tissue specificity is unknown, it may be explained by its unique cell component and biological function. For example, LUM is a representative gene among the top 100 genes that is highly expressed in human aortic valve, which encodes a member of the small leucinerich proteoglycan family, including biglycan, decorin, fibromodulin, epiphycan, keratocan, and osteoglycin. These molecules are closely related to the physical characteristics and function of aortic valve.15 In these bifunctional molecules, the protein moiety binds collagen fibrils and the highly charged hydrophilic glycosaminoglycans regulate interfibrillar spacings. Lumican, as the major keratan sulfate proteoglycan of the cornea, is also distributed in interstitial collagenous matrices throughout the body.41 Lumican may regulate collagen fibril organization and circumferential growth, corneal transparency, and epithelial cell migration and tissue repair.42,43
We found that aortic valve-specific mRNAs and linc-RNAs had similar but low expression level (Fig. 5A), which was significantly smaller than transcripts that are universally expressed in most human tissues (Fig. 5A). This suggested that tissue specificity of aortic valve was mainly determined by a set of lowly expressed RNA transcripts, rather than by some genes that were only highly expressed in the aortic valve. One possible reason is that highly expressed genes in aortic valve are more likely to perform basic biological functions that are important to all cell types. Therefore, highly expressed genes should have less likelihood to be tissue specific in aortic valve. Another probability is that highly tissue-specific genes tend to evolve rapidly at the gene sequence level but slowly at the expression profile level.44 In our results, we also observed higher sequence divergence in aortic valve-specific mRNAs and lincRNAs than that of tissue-universal transcripts (Fig. 5B). On the other hand, evolutionary dynamics analysis showed that aortic valve-specific mRNAs and lincRNAs were evolved more recently in human genome than tissue-universal genes (Fig. 5D). These results suggested that both gene sequence evolution and recent origination of RNA transcripts contributed to the transcriptome specificity of aortic valve.
Unlike protein-coding genes, lincRNAs are a set of long noncoding RNAs that play diverse regulatory roles in human development. Aortic valve-specific lincRNAs were more likely to be recently evolved in primate lineage, while aortic valve-specific mRNAs were gradually gained along the evolutionary history of human genome (Fig. 5D). In addition, sequence conservation of lincRNAs was also significantly smaller than protein-coding genes (Fig. 5B). These results are consistent with the findings in several recent studies that analyzed different human tissues.12,18,45 Melé et al.12 observed that most tissue-specific transcripts were lincRNAs that expressed in testis. Hezroni et al.45 and Cabili et al.15 showed that linc RNA exons were under less negative selective pressures and evolved faster. Washietl et al.18 characterized human lincRNA expression patterns in nine tissues across six mammalian species and found that ∼20% of human lincRNAs were hominid-specific lincRNAs that were more tissue specific and faster evolving within the human lineage. All these characteristics of lincRNAs suggested that gene regulators, such as linc RNAs, were more important to human tissue specificity and functional diversification. As a result, evolutionary changes in gene expression may account for most phenotypic differences in human evolution.46 Moreover, previous studies showed that new lincRNAs are partly originated by exonization of repetitive elements.45 In our results, we observed higher repetitive content in aortic valve-specific lincRNAs than that in protein-coding genes and tissue-universal lincRNAs (Fig. 5C). This confirmed that repetitive elements might also play important roles in creating aortic valve-specific lincRNAs.
Supplementary Material
Supplementary Table 1. Sequencing library and number of sequenced reads from each human tissue sample.
Supplementary Table 2. KEGG and GO enrichment analysis of aortic valve-specific protein-coding genes. Aortic valve-specific protein-coding genes are also listed.
Supplementary Table 3. KEGG and GO enrichment analysis of the top 100 genes that have highest expression level in human aortic valves.
Supplementary Figure 1. Principal component analysis of protein-coding genes based on the expression values in the two aortic valve samples and 32 human tissue samples from Human Body Map 2 Project.
Supplementary Figure 2. Comparison of (A) exon number and (B) gene length between mRNA genes and lincRNA genes.
Supplementary Figure 3. Principal component analysis of lincRNAs based on the expression values in the two aortic valve samples and 32 human tissue samples from Human Body Map 2 Project.
Footnotes
ACADEMIC EDITOR: Jike Cui, Associate Editor
PEER REVIEW: Four peer reviewers contributed to the peer review report. Reviewers’ reports totaled 951 words, excluding any confidential comments to the academic editor.
FUNDING: This work was supported in part by Jiangsu Provincial Department of Education (Grants CXZZ11_0726), the Priority Academic Program of Jiangsu Higher Education Institutions (Grants JX10231801), and the projects BK20151590 of the National Natural Science Foundation of Jiangsu Province. The authors confirm that the funders had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Ying Wang and Tong Yu, two of the authors of the manuscript, are currently affiliated with Nanjing Decode Genomics Biotechnology Corporation. This does not alter the authors’ adherence to all the journal policies on sharing data and materials.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
JW, LC and YS conceived and designed the experiments. TY and Wanjun G analyzed the data. JW and YW wrote the first draft of the manuscript. Weidong G, BN, and HS contributed to the writing of the manuscript. All authors agree with manuscript results and conclusions. LC and YS made critical revisions and approved final version as corresponding authors. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Wang Z, Gerstein M, Snyder M. RNA-seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan MH, Au KF, Yablonovitch AL, et al. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res. 2013;23:201–16. doi: 10.1101/gr.141424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–31. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trapnell C, Roberts A, Goff L, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pickrell JK, Marioni JC, Pai AA, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–72. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu JQ, Du J, Rozowsky J, et al. Systematic analysis of transcribed loci in ENCODE regions using RACE sequencing reveals extensive transcription in the human genome. Genome Biol. 2008;39(1):R3. doi: 10.1186/gb-2008-9-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–6. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sultan M, Schulz MH, Richard H, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 9.Gonzàlez-Porta M, Frankish A, Rung J, et al. Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. 2013;14(7):R70. doi: 10.1186/gb-2013-14-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–8. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klijn C, Durinck S, Stawiski EW, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotech. 2015;33:306–12. doi: 10.1038/nbt.3080. [DOI] [PubMed] [Google Scholar]
- 12.Melé M, Ferreira PG, Reverter F, et al. GTEx Consortium The human transcriptome across tissues and individuals. Science. 2015;348:660–5. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerstein MB, Joel R, Koon-Kiu Y, et al. Comparative analysis of the transcriptome across distant species. Nature. 2014;512:445–8. doi: 10.1038/nature13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014;24:616–28. doi: 10.1101/gr.165035.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ardlie KG, Deluca DS, Segre AV, et al. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang HJ, Kawasawa YI, Cheng F, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–9. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farkas MH, Grant GR, White JA, et al. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genomics. 2013;14:486. doi: 10.1186/1471-2164-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Djureinovic D, Fagerberg L, Hallström B, et al. The human testis-specific proteome defined by transcriptomics and antibody-based profiling. Mol Hum Reprod. 2014;20:476488. doi: 10.1093/molehr/gau018. [DOI] [PubMed] [Google Scholar]
- 24.Habuka M, Fagerberg L, Hallström BM, et al. The kidney transcriptome and proteome defined by transcriptomics and antibody-based profiling. PLoS One. 2014;9(12):e116125. doi: 10.1371/journal.pone.0116125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutcheson JD, Aikawa E, Merryman WD. Potential drug targets for calcific aortic valve disease. Nat Rev Cardiol. 2014;11:218–31. doi: 10.1038/nrcardio.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siu SC, Silversides CK. Bicuspid aortic valve disease. J Am Coll Cardiol. 2010;55:2789–800. doi: 10.1016/j.jacc.2009.12.068. [DOI] [PubMed] [Google Scholar]
- 27.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Frith MC, Forrest AR, Nourbakhsh E, et al. The abundance of short proteins in the mammalian proteome. PLoS Genet. 2006;2:e52. doi: 10.1371/journal.pgen.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyle EI, Weng S, Gollub J, et al. GO:TermFinder–open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–5. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie C, Mao X, Huang J, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–22. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009;25:i54–62. doi: 10.1093/bioinformatics/btp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karolchik D, Baertsch R, Diekhans M, et al. University of California Santa Cruz The UCSC genome browser database. Nucleic Acids Res. 2003;31:51–4. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padang R, Bagnall RD, Tsoutsman T, Bannon PG, Semsarian C. Comparative transcriptome profiling in human bicuspid aortic valve disease using RNA-sequencing. Physiol Genomics. 2015;47(3):75–87. doi: 10.1152/physiolgenomics.00115.2014. [DOI] [PubMed] [Google Scholar]
- 39.Linn F, Björn MH, Per O, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chakravarti S. Functions of lumican and fibromodulin: lessons from knockout mice. Glycoconj J. 2002;19:287–93. doi: 10.1023/A:1025348417078. [DOI] [PubMed] [Google Scholar]
- 42.Yamanaka O, Yuan Y, Coulson-Thomas VJ, et al. Lumican binds ALK5 to promote epithelium wound healing. PLoS One. 2013;8:e82730. doi: 10.1371/journal.pone.0082730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engebretsen KVT, Lunde IG, Strand ME, et al. Lumican is increased in experimental and clinical heart failure, and its production by cardiac fibroblasts is induced by mechanical and proinflammatory stimuli. FEBS J. 2013;280:2382–98. doi: 10.1111/febs.12235. [DOI] [PubMed] [Google Scholar]
- 44.Liao B-Y, Zhang J. Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol Biol Evol. 2006;23:1119–28. doi: 10.1093/molbev/msj119. [DOI] [PubMed] [Google Scholar]
- 45.Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Reports. 2015;11:1110–22. doi: 10.1016/j.celrep.2015.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khaitovich P, Enard W, Lachmann M, Pääbo S. Evolution of primate gene expression. Nat Rev Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Sequencing library and number of sequenced reads from each human tissue sample.
Supplementary Table 2. KEGG and GO enrichment analysis of aortic valve-specific protein-coding genes. Aortic valve-specific protein-coding genes are also listed.
Supplementary Table 3. KEGG and GO enrichment analysis of the top 100 genes that have highest expression level in human aortic valves.
Supplementary Figure 1. Principal component analysis of protein-coding genes based on the expression values in the two aortic valve samples and 32 human tissue samples from Human Body Map 2 Project.
Supplementary Figure 2. Comparison of (A) exon number and (B) gene length between mRNA genes and lincRNA genes.
Supplementary Figure 3. Principal component analysis of lincRNAs based on the expression values in the two aortic valve samples and 32 human tissue samples from Human Body Map 2 Project.