Abstract
Purpose
The ability to test for and detect prostate cancer with minimal invasiveness has the potential to reduce unnecessary prostate biopsies. This study was conducted as part of a clinical investigation for the development of an OligoFISH® probe panel for more accurate detection of prostate cancer.
Materials and methods
One hundred eligible male patients undergoing transrectal ultrasound biopsies were enrolled in the study. After undergoing digital rectal examination with pressure, voided urine was collected in sufficient volume to prepare at least two slides using ThinPrep. Probe panels were tested on the slides, and 500 cells were scored when possible. From the 100 patients recruited, 85 had more than 300 cells scored and were included in the clinical performance calculations.
Results
Chromosomes Y, 7, 10, 20, 6, 8, 16, and 18 were polysomic in most prostate carcinoma cases. Of these eight chromosomes, chromosomes 7, 16, 18, and 20 were identified as having the highest clinical performance as a fluorescence in situ hybridization test and used to manufacture the fluorescence in situ hybridization probe panels. The OligoFISH® probes performed with 100% analytical specificity. When the OligoFISH® probes were compared with the biopsy results for each individual, the test results highly correlated with positive and negative prostate biopsy pathology findings, supporting their high specificity and accuracy. Probes for chromosomes 7, 16, 18, and 20 showed in the receiver operator characteristics analysis an area under the curve of 0.83, with an accuracy of 81% in predicting the biopsy result.
Conclusion
This investigation demonstrates the ease of use with high specificity, high predictive value, and accuracy in identifying prostate cancer in voided urine after digital rectal examination with pressure. The test is likely to have positive impact on clinical practice and advance approaches to the detection of prostate cancer. Further evaluation is warranted.
Keywords: prostate cancer detection, OligoFISH®, oncology, PSA screening
Introduction
Prostate cancer is the most common nondermatologic malignancy of males in the United States. In 2015, there were approximately 221,000 new cases of prostate cancer, and more than 27,000 of the men died.1 Approximately 2.9 million American men are living with prostate cancer.2 Lifetime risk of developing prostate cancer is 14% (2010–2012 data1), while lifetime risk of dying from the disease is 2.8% or 1 in 38.3 Risk factors include age (>75% of prostate cancer diagnosed in men >65); African-American and Caribbean-African ancestry (1.6 times more likely to develop prostate cancer; 2.4 times more likely to die compared to Caucasians); geography; positive family history for prostate cancer; mutations of BRCA1 or BRCA2 genes and presence of hereditary nonpolyposis colorectal cancer, or HNPCC/Lynd Syndrome; high red meat or high-fat dairy consumption; obesity; smoking, workplace exposures; and prostatitis.4–7
Prostate-specific antigen (PSA) testing with risk stratification based on serum levels forms the basis for early detection of prostate cancer.8 However, given falsely elevated PSA levels occurring in common clinical conditions9 and that transrectal biopsies, which only sample a small part of the gland, result in a high rate of false negatives,4,10,11 considerable debate and disagreement exist around PSA screening. The most recent American Urological Association (AUA) guidelines state that PSA screening should be done every 2 years or more with greatest benefit for men aged 55–69 years, whereas the US Prevention Services Task Force position is that mortality reduction is marginal, even in the “optimal age range of 55–69 years”12 and that the “potential harms do not outweigh the benefits.”12 In addition, these interventions are associated with significant health care costs. For the Medicare population aged >65 years, PSA screening expenditures and follow-up associated interventions, including prostate biopsy, were $125 Billions in 2010, and estimated to exceed $158 Billions (in 2010 dollars) annually by 2020.13,14 Expanding health care expenditures and on-going uncertainties related to available early prostate cancer screening approaches support evaluation of new methods including DNA analyses, molecular urine testing, and fluorescence in situ hybridization (FISH).15–17
FISH uses principles of high affinity molecular or biological targets, minimizing nonspecific cellular uptake and retention where tumor cell levels are low or difficult to detect by standard methodologies.18–23 Whole chromosome gains are the most frequently identified change in malignant cells, and extensive research validates aneuploidy as a biomarker of cancer.23 Research on these OligoFISH® probes is enabled through the use of the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer, whereby highly sensitive and specific probes for both high- and low-grade tumors can be developed.24,25
The goal of this study was to develop a noninvasive prostate cancer detection method that would predict the findings from prostate cancer using OligoFISH® probes for aneuploidy detection in male volunteers undergoing prostate evaluation in voided urine after a digital rectal examination (DRE) with pressure.
Materials and methods
Sample collection and OligoFISH® procedure
Patients undergoing prostate cancer surveillance by scheduled DRE with pressure and biopsy were recruited for the study. The study protocol was approved by the Western Independent Review Board. All subjects were informed of the procedures and risks of the study and provided informed consent.
On the day of biopsy, the urologist performed a DRE with pressure. Prostate lobes were stroked through the rectum three times with approximately 1 cm depression. The lobes were stroked from base to apex and from lateral to median line including a final stroke on median line from base to apex to empty prostatic fluid into the urethra. Subjects were asked to provide 40–80 mL of voided urine, including first stream. Preservative was added to the urine (2:1 urine:preservative) to prevent bacterial growth and preserve cells and DNA (PreservCyt; Hologic, Inc., Bedford, MA, USA). Samples were stored at 4°C for no longer than 48 hours before urothelial cells were washed in CytoLyt (Hologic, Inc. Bedford, MA, USA.), and the slides were prepared. Urothelial slides were prepared using ThinPrep machines (Hologic, Inc.) following manufacturer’s instructions. Slides were partially digested in IsoThermal Protease (Cellay, Inc., Cambridge, MA, USA) at room temperature for 15 minutes, followed by 5-minute incubation in Detergent Solution (Cellay, Inc.). Cells were lightly fixed for 5 minutes in 1% formaldehyde and rinsed in phosphate-buffered saline. Cellular DNA was denatured in IsoThermal Denaturing Solution (Cellay, Inc.) at room temperature for 10 minutes. Slides were dehydrated in 85% and 100% alcohol for 1 minute each and air dried. Slides were hybridized with 3 µL of OligoFISH® probe mix (chromosomes Y. 6, 8 and 10), coverslips applied, and incubated for 10 minutes at 37°C. Slides were washed in 2× SSC (saline sodium citrate) under agitation to float off the coverslips and then incubated in IsoThermal Wash Solution (2× SSC and Isothermal Denaturing Reagent) for 5 minutes at room temperature to remove unbound or nonspecifically hybridized probes. Slides were rinsed in 2× SSC and mounted with Antifade with 4′,6-diamidino-2-phenylindole dihydrochloride and analyzed with an epifluorescence microscope. Each specimen was scored for 500 cells. The only cells excluded from analysis were the easily recognizable poly-morph nucleated white blood cells and sperm cells.
Once the slides were scored, they were washed in 2× SSC 0.1% sodium dodecyl sulfate to remove coverslips and Antifade and denatured again for 5 minutes to strip the probes. Slides were then hybridized with the second probe panel for the detection of chromosomes 7, 16, 18, and 20. Five hundred cells were scored for each sample.
Determining the number of prostate cells in urine samples
Proportion of prostatic cells in urine samples of the first ten subjects was obtained through cytology to determine the percentage of prostate cells. Up to 500 cells were scored and categorized as prostatic or urothelial using nuclear:cytoplasmic ratio. Potential prostate cells were identified by their small size compared to urothelial, high ratio nucleus/cytoplasm, and eccentric position of the nucleus. Cells with polylobulated nucleus, presumed to be white blood cells, were excluded as were identifiable sperm cells.
The samples were processed using this methodology, and following these morphological criteria, 500 cells were classified as belonging to the urothelium or to the prostate.
Analytical performance of the four chromosome OligoFISH® probe panel
Analytical performance of the OligoFISH® panels was validated based on schema previously described for hybridizing probes.18 In brief, the probe panel was validated against normal blood cells in metaphase from five chromosomally normal individuals to determine analytical specificity and sensitivity.
Clinical performance
Comparison of OligoFISH® prostate test and the pathology results
By comparing biopsy pathology results for the presence of cancer to the OligoFISH® results, the true positives, true negatives, false positives, and false negatives were determined, and the clinical performance of the OligoFISH® assay was calculated. Because the data do not follow a Gaussian distribution, the interval at 95% confidence was calculated using the Clopper–Pearson method.26
Receiver operator characteristics (ROC) curves
By varying the normal cut-off (NCO) from zero polysomic cells to the maximum percentage observed, we were able to calculate true positives, true negatives, false positives, false negatives, sensitivity, specificity, 1-specificity, positive predictive value (PPV), negative predictive value (NPV), accuracy, and Youden’s index (J) on every cut-off value. This calculation was performed at every cut-off value for each one of the two 4 chromosome panels, as well as for the individual chromosomes. Sensitivity against 1–specificity was plotted, and the area under the curve (AUC) was calculated using Prism 6 for Windows (version 6.02; GraphPad Software, Inc., La Jolla, CA, USA). For every probe panel as well as the individual chromosomes, the Youden’s index was used to determine the best NCO. AUC was used to determine the power of differentiating between benign and cancer of each one of the probe panels and the chromosomes.
Results
One hundred subjects were enrolled in the study. Mean age was 68.8 years (standard deviation [SD] ±8.4); 21% were African American, non-Hispanic and the remainder were Caucasian, non-Hispanic. Voided urine samples after DRE with pressure were collected from each patient, and 500 cells were scored in each sample when possible.
Analytical performance of the four chromosome OligoFISH® probe panel
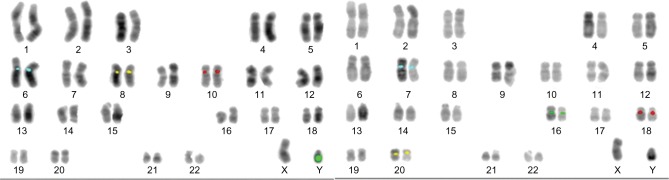
OligoFISH® probes performed with 100% analytical specificity and >98% analytical sensitivity (Table 1), consistent with what the probes are designed to provide as a rapid test that targets only specific parts of the chromosome and exceeding the recommendations by the American College of Genetic Medicine27 as well as most traditional FISH probes. Figure 1 illustrates the correct location of both OligoFISH® probe panels.
Table 1.
Analytical sensitivity of OligoFISH® probes
| Chromosome probe | Fluorochrome | Analytical sensitivity (%) | Margin of error, 95% (%) |
|---|---|---|---|
| Yq12 | Green | 99.5 | 0.95 |
| 6 | Aqua | 99.5 | 0.95 |
| 8 | Gold | 99.5 | 0.95 |
| 10 | Red | 99.1 | 1.30 |
| 7 | Aqua | 99.0 | 1.38 |
| 16 | Green | 99.0 | 1.38 |
| 18 | Red | 98.5 | 1.7 |
| 20 | Gold | 98.0 | 1.9 |
Figure 1.
Examples of the metaphases fully karyotyped using reverse DAPI.
Notes: Left karyotype: chromosome 6 centromere labeled in aqua, chromosome 8 centromere labeled in gold, chromosome 10 centromere labeled in red and chromosome Y heterochromatin labeled in green. Right karyotype: chromosome 7 centromere labeled in aqua, chromosome 16 centromere labeled in green, chromosome 18 centromere labeled in red and chromosome 20 centromere labeled in gold.
Abbreviation: DAPI, diamidino-2-phenylindole dihydrochloride.
Presence of prostate cells in urine samples
Urine samples were successfully collected after DRE with pressure. When 500 cells were scored and classified as prostatic or not according to their morphology, an average of 65%±9.1% prostatic cells was found; the range was 40.0%–83%. Urine samples collected without DRE with pressure were found to contain almost no prostatic epithelial cells (results not shown), demonstrating that DRE with pressure is crucial for performing the test.
Clinical performance
When possible, 500 cells were scored on each slide after the first hybridization and again after the second hybridization. If less than 300 cells were scored, the subject was excluded from the calculations; this reduced our sample from 100 subjects to 85. The results for each kit were combined for each subject to determine the clinical validity of the eight chromosomes together.
The clinical performance and ROC data are listed in Table 2. ROC curves were calculated by varying the cut-off value from clinical specificity =1 (100%) and sensitivity =0 to specificity =0 and sensitivity =1. The values of true positive rates (clinical sensitivity) were plotted against false-positive rates (1–specificity), and the AUC was calculated using GraphPad Prism 6 software. The highest AUC was obtained when the combination of chromosomes 7, 16, 18, and 20 was used with a value of 0.83, corresponding to a very good test for discriminating between benign and cancerous pathological result (Figure 2 and Table 3).
Table 2.
OligoFISH® panel performance results
| Performance measure | % | Margin of error 95% |
|---|---|---|
| Sensitivity | 72 | 53–86 |
| Specificity | 92 | 74–99 |
| PPV | 92 | 74–99 |
| NPV | 72 | 53–86 |
| Accuracy | 81 | 68–90 |
Abbreviations: PPV, positive predictive value; NPV, negative predictive value.
Figure 2.
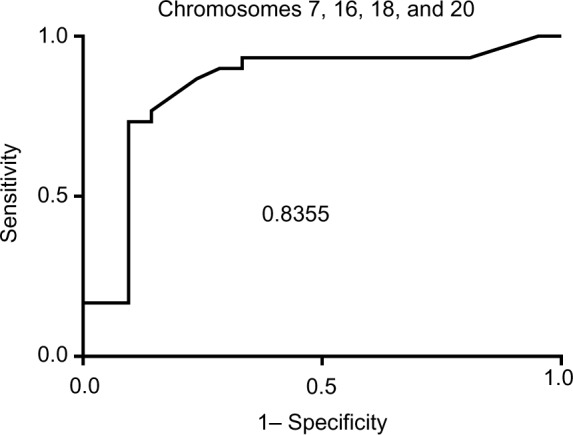
ROC and AUC for chromosome panel for chromosomes 7, 16, 18, and 20.
Abbreviations: ROC, receiver operator characteristics; AUC, area under the curve.
Table 3.
ROC AUC for each chromosome and chromosome panels
| Chromosomes | AUC |
|---|---|
| 6, Y, 10 and 8 | 0.79 |
| 6 | 0.69 |
| Y | 0.70 |
| 10 | 0.75 |
| 8 | 0.73 |
| 7, 16, 18, and 20 | 0.83 |
| 7 | 0.76 |
| 16 | 0.74 |
| 18 | 0.75 |
| 20 | 0.74 |
Abbreviations: ROC, receiver operator characteristics; AUC, area under the curve.
Best cut-off was determined by plotting sensitivity and specificity at all the possible cut-offs. The best cut-off point was defined as the cut-off where sensitivity and specificity intersect. Clinical sensitivity, clinical specificity, PPV, NPV, and accuracy were calculated as previously described.
The OligoFISH® probe panels showed high specificity (92%), PPV (92%), and accuracy (81%) for the detection of chromosomes 7, 16, 18, and 20, which are associated with prostate cancer.
Cancer vs benign
Pathological results of the OligoFISH® test are listed in Table 4. The test correlated highly with positive and negative pathological results. Out of 37 positive OligoFISH® tests, 34 (92%) were cancerous by pathology testing. Of the 48 negative OligoFISH® tests, 13 (27%) were cancerous by pathology testing. These data demonstrate the high correlation of the test with pathology findings. As shown in Figure 3, the collection of prostate cells in urine following DRE with pressure resulted in markedly increased average percentage of cells with chromosome gains in cancer patients compared to those with benign and low-grade lesions, increasing cellular material for testing and improving test validity through sufficient sample size collection. Figure 3 depicts prostatic cells using the OligoFISH® probe. Arrows show cells with trisomy and tetrasomy 6 signals in Aqua.
Table 4.
Pathological results of OligoFISH® probes
| Results of OligoFISH® test | Pathological results
|
||
|---|---|---|---|
| Cancer | Benign | Total | |
| Positive | 34 | 3 | 37 |
| Negative | 13 | 34 | 48 |
| Total | 47 | 37 | 85 |
Figure 3.
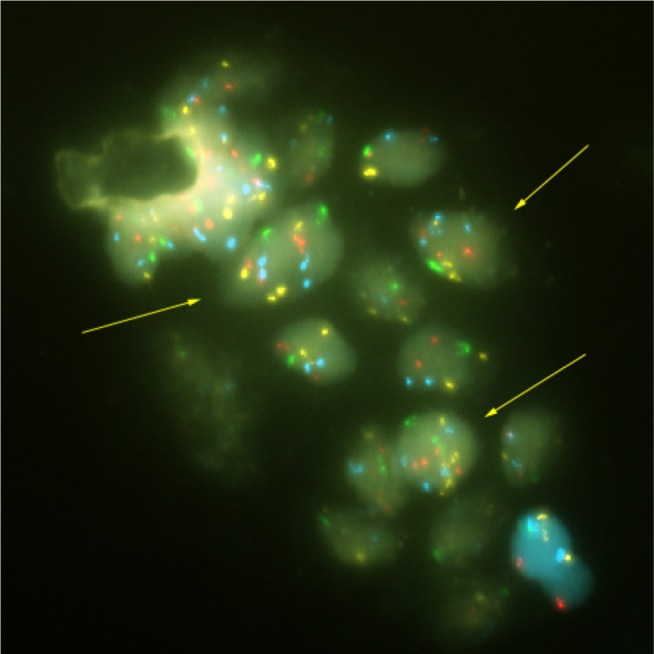
Prostatic cells released in the urine.
Note: Arrows show cells with trisomy and tetrasomy 6 signal in aqua.
Discussion
Despite decades of surveillance, research, and outreach, screening for prostate cancer remains a significant public health priority as well as an ongoing debate. There is agreement among urologists and other health care groups that epidemiologic, clinical, and economic data support the need for more effective, noninvasive, or minimally invasive screening and diagnostic approaches.8,12 This study demonstrated that OligoFISH® has the appropriate level of reproducible specificity and sensitivity needed for accurate early detection of prostate cancer using minimally invasive procedures. Prostate cell collection in urine collected following DRE with pressure significantly increased the prostate cell harvest that allowed for sufficient cell collection to test the OligoFISH® probe panel. The proportion of prostate cells found in the urine samples tested varied from 40% to 83% after DRE with pressure. This allowed for the collection of a sufficient number of high-quality prostate cells to perform the test. Urine samples collected without prior DRE with pressure were found to contain almost no prostate cells, demonstrating that DRE with pressure is crucial for performing the test. These probe panels were developed based on chromosomes identified through the use of Mitelman’s database for aneuploidy as a biomarker of prostate cancer.24 This enables the creation of a very powerful probe panel for prostate cancer detection. Specifically, chromosomes 7, 16, 18, and 20 were definitively found to be most correlated with prostate cancer.
Since we looked to score between 300 and 500 cells, 15 patients had to be excluded in this exploratory phase. With an NCO value at 3%, all positive patients became positive in the first 100 cells scored. In the final test, probably 100 cells will be enough and would avoid test failure from low cellularity.
The protocol for the OligoFISH® probes was relatively uncomplicated and streamlined. Most of the OligoFISH® tests were conducted at room temperature, with minimal methodological complexity, in a single day. Tissue samples were voided urine and can be obtained with minimal invasiveness, as DRE with pressure can reliably produce the needed concentration of prostate cells, both cancerous and benign. The probes are designed to provide a rapid test that targets only specific chromosomal regions. This was validated, as the OligoFISH® probes performed with 100% analytical specificity and high sensitivity.
In addition, one of the most important outcomes of this study was high correlation between the OligoFISH® results and pathology findings. This demonstrates that a chromosomal target within malignant cells, isolated from voided urine and not from a prostate biopsy, can detect the presence or absence of malignancy with high sensitivity and specificity. Thus, OligoFISH® probe prostate cancer screening could be done as part of routine DREs with minimal invasiveness, making routine screening more accessible, more accepted by patients and clinicians, and cost-effective. In addition, if OligoFISH® was used periodically to monitor patients with indolent prostate cancer, changes in percentage of abnormal cells could signal progression in the disease.
Table 5 compares available data for currently available tests: tissue biopsy-based epigenetic multiplex polymerase chain reaction (PCR) (ConfirmMDx; MDxHealth, Irvine, CA, USA), and PCA3 gene assay (Progensa; Hologic, Inc.), with those for the OligoFISH® probe. ConfirmMDx has high NPV, which means it is most reliable if the patient has a negative biopsy. However, despite having an excellent NPV, ConfirmMDx’s PPV is only 33%, resulting in high false positives. Similar tradeoffs occur with the Progensa test. Although PCA3 shows a high specificity, its sensitivity is 41%, which means it is the NPV that assists physicians in deciding if a new biopsy is required after initial negative biopsy. In contrast, OligoFISH® test possesses both high specificity and high sensitivity, maximizing the test’s accuracy.
Table 5.
Clinical performance comparison of prostate biomarkers
| OligoFISH® Chromosome 7, 16, 18, 20 | Tissue Biopsy-Based Epigenetic Multiplex PCR Assay (ConfirmMDxa) | PCA3 Gene Assay (Progensab) | |
|---|---|---|---|
| Tissue Source | Urine | Prostate Tissue via Biopsy | Urine |
| Sensitivity % | 72 | 95 | 42 |
| (CI 95%) | (53–86) | (76–100) | (36–48) |
| Specificity % | 92 | 38 | 91 |
| (CI 95%) | (74–99) | (27–51) | (87–94) |
| PPV % | 92 | 33 | 80 |
| (CI 95%) | (74–99) | (22–47) | (72–86) |
| NPV % | 72 | 96 | N/A |
| (CI 95%) | (53–86) | (80–100) |
Most urologists have been trained in prostate massages or DRE with pressure. The use of the OligoFISH® test is extremely easy compared to other FISH tests. The probes hybridize in 10 minutes, allowing results in a record time; most pathologists are trained in scoring FISH results. Most of the pathology laboratories already offer FISH tests, and incorporating a new one should be very easy. Regarding the cost of the test in Michigan, the full procedure of a transrectal ultrasound needle biopsy is being reimbursed by Centers for Medicare and Medicate Services at $1,377, while the OligoFISH® procedure is reimbursed at $700. Since a high number of prostate biopsies result in a benign diagnosis, this test could avoid these procedures, which results not only in an economical benefit but also improved clinical outcomes, since biopsies can result in extra complications, due to the procedure.
One limitation of a FISH test scoring polysomic cells to detect cancer is that aneuploidy is a very general marker for human carcinomas. When obtaining the urine, cells from other organs can be collected as well (including bladder or kidneys). If a patient suspected for prostate cancer has bladder or kidney cancer, this could result in a positive test independent of prostate cancer status.
Conclusion
The OligoFISH® probes used in this study showed high clinical performance. Larger clinical trials are warranted to demonstrate that the use of this rapid, simple, and minimally invasive OligoFISH® test could significantly improve prostate cancer detection, avoiding unnecessary prostate biopsies.
Acknowledgments
We would like to thank Marissa Seligman for her editorial work.
Footnotes
Disclosure
All the authors from Cellay are current or past employees. Joan Aurich-Costa is a stock holder of Cellay. The authors report no other conflicts of interest in this work.
References
- 1.National Cancer Institute: Surveillance, Epidemiology and End Results Program SEER Stat Fact Sheets-Prostate Cancer. [Accessed February 5, 2016]. Available from: http://seer.cancer.gov/statfacts/html/prost.html.
- 2.Centers for Disease Control and Prevention Prostate cancer rates by rate and ethnicity. [Accessed February 5, 2016]. Available from: http://www.cdc.gov/cancer/prostate/statistics/race.htm.
- 3.American Cancer Society Lifetime probability of dying from prostate cancer. [Accessed February 5, 2016]. Available from: http://www.cancer.org/cancer/cancerbasics/lifetime-probability-of-developing-or-dying-from-cancer.
- 4.Pienta KJ, Esper PS. Risk factors for prostate cancer. Ann Intern Med. 1993;118(10):793–803. doi: 10.7326/0003-4819-118-10-199305150-00007. [DOI] [PubMed] [Google Scholar]
- 5.American Cancer Society What are the risk factors for prostate cancer. [Accessed February 5, 2016]. Available from: http://www.cancer.org/cancer/prostatecancer/detailedguide/prostate-cancer-risk-factors.
- 6.Taksler GB, Keating NL, Cutler DM. Explaining racial differences in prostate cancer mortality. Cancer. 2012;118:4280–4289. doi: 10.1002/cncr.27379. [DOI] [PubMed] [Google Scholar]
- 7.Zenka D. African Americans at higher risk for prostate cancer. [Accessed February 5, 2016]. Available from: http://www.pcf.org/site/c.leJRIROrEpH/b.8368629/k.D6C1/African_Americans_At_Higher_Risk_for_Prostate_Cancer.htm.
- 8.American Urological Association Guidelines on Prostate Cancer: detection of prostate cancer. [Accessed February 5, 2016]. Available from: https://www.auanet.org/education/guidelines/prostate-cancer-detection.cfm.
- 9.Barry MJ. Clinical practice. Prostate-specific-antigen testing for early diagnosis of prostate cancer. N Engl J Med. 2001;344(18):1373–1377. doi: 10.1056/NEJM200105033441806. [DOI] [PubMed] [Google Scholar]
- 10.Daneshgari F, Taylor GD, Miller GJ, Crawford ED. Computer simulation of the probability of detecting low volume carcinoma of the prostate with six random systematic core biopsies. Urology. 1995;45(4):604–609. doi: 10.1016/S0090-4295(99)80051-X. [DOI] [PubMed] [Google Scholar]
- 11.Hull GW, Rabbani F, Abbas F, Wheeler TM, Kattan MW, Scardino PT. Cancer control with radical prostatectomy alone in 1,000 consecutive patients. J Urol. 2002;167:528–534. doi: 10.1016/S0022-5347(01)69079-7. [DOI] [PubMed] [Google Scholar]
- 12.US Prevention Services 9,13 Force Prostate cancer screening. [Accessed February 5, 2016]. Available from: http://www.uspreventiveservicestaskforce.org/Page/Document/UpdateSummaryFinal/prostate-cancer-screening.
- 13.Ma X, Wang R, Long JB, et al. The cost implications of prostate cancer screening in the Medicare population. Cancer. 2014;120(1):96–102. doi: 10.1002/cncr.28373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Cancer Institute Cancer costs projected to reach at least $158 billion in 2020. [Accessed February 5, 2016]. Available from: http://www.cancer.gov/news-events/pressreleases/2011/CostCancer2020.
- 15.Prenser JR, Rubin MA, Wei JT, Chinnalyan AM. Beyond PSA: the next generation of prostate cancer biomarkers. Sci Transl Med. 2012;4(127):127rv3. doi: 10.1126/scitranslmed.3003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tosoian J, Loeb S. PSA and beyond: the past, present, and future of investigative biomarkers for prostate cancer. ScientificWorldJournal. 2010;10:1919–1931. doi: 10.1100/tsw.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Duin M, van Marion R, Vissers K, et al. High-resolution array comparative genomic hybridization of chromosome arm 8q: evaluation of genetic progression markers for prostate cancer. Genes Chromosomes Cancer. 2005;44:438–449. doi: 10.1002/gcc.20259. [DOI] [PubMed] [Google Scholar]
- 18.Aurich-Costa J, Zamecheck L, et al. Oligo fluorescence in situ hybridization (Oligo-Fish), a new strategy for enumerating chromosomes in interphase nuclei. Fertil Steril. 2007;88:S86. [Google Scholar]
- 19.Aurich-Costa J, Ng C, Selvaggio S, Colls P, Bradley S. Oligonucleotide (ODN) fluorescence in situ hybridization (Oligo-FISH) and conventional FISH allow enumeration of 24 chromosomes in 6 successive hybridizations performed in a single day. Fertil Steril. 2009;92:S50. [Google Scholar]
- 20.Zhang S, Zhang HS, Cordon-Cardo C, et al. Selection of tumor antigens as targets for immune attack using immunohistochemistry: II. Blood group-related antigens. Int J Cancer. 1997;73:50–56. doi: 10.1002/(sici)1097-0215(19970926)73:1<50::aid-ijc9>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 21.Sokolova I, Halling KC, Jenkins RB, et al. The development of multitarget, multicolor fluorescence in situ hybridization assay for the detection of urothelial carcinoma in urine. J Mol Diagn. 2000;2:116–123. doi: 10.1016/S1525-1578(10)60625-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui J, Deubler DA, Rohr LR, et al. Chromosome 7 abnormalities in prostate cancer detected by dual-color fluorescence in situ hybridization. Cancer Genet Cytogenet. 1998;107:51–60. doi: 10.1016/s0165-4608(98)00074-0. [DOI] [PubMed] [Google Scholar]
- 23.French M. Chromosomal biomarkers of genomic instability relevant to cancer. Drug Discov Today. 2007;7:1128–1137. doi: 10.1016/s1359-6446(02)02502-3. [DOI] [PubMed] [Google Scholar]
- 24.Mitelman F, Johansson B, Mertens F. Mitelman F, Johansson B, Mertens F, editors. Mitelman Database of chromosome aberrations and gene fusions in cancer. 2011. [Accessed May 24, 2016]. Available from: http://cgap.nci.nih.gov/Chromosomes/Mitelman.
- 25.Reddy KS, Tripod J. Contemplations on preclinical validation of fluorescence in situ hybridization probe assay for paraffin-embedded tissues in hematologic disorders. Cancer Genet Cytogenet. 2008;183:1–5. doi: 10.1016/j.cancergencyto.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Clooper C, Pearson S. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26:404–413. [Google Scholar]
- 27.Relm H, Bale SJ, Bayrak-Toydemir P, et al. Working Group of the American College of Medical Genetics and Genomics Laboratory Quality Assurance Committee ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–747. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trock BJ, Brotzman MJ, Mangold LA, et al. Evaluation of GSTP1 and APC methylation as indicators for repeat biopsy in a high-risk cohort of men with negative initial prostate biopsies. BJU Int. 2012;110(1):56–62. doi: 10.1111/j.1464-410X.2011.10718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei JT, Feng Z, Partin AW, et al. Can Urinary PCA3 Supplement PSA in the Early Detection of Prostate Cancer? J Clin Oncol. 2014;32(36):4066–4072. doi: 10.1200/JCO.2013.52.8505. [DOI] [PMC free article] [PubMed] [Google Scholar]