Abstract
Antibiotics remain essential tools in the control of infectious diseases. With the emergence of new diseases, resistant forms of diseases such as tuberculosis and malaria, as well as the emergence of multidrug-resistant bacteria, it has become essential to develop novel antibiotics. Development of the existing antibiotics involved three strategies, including discovery of new target sites, modification of existing antibiotic structures, and the identification of new resources for novel antibiotics. Marine microorganisms have clearly become an essential new resource in the discovery of new antibiotic leads.
Keywords: Antibiotics, Bacillus subtilis, Escherichia coli, marine fungi, marine microorganisms, resistance, Staphylococcus aureus
Introduction
An antibiotic is a chemical substance produced by microorganisms, which has the capacity to inhibit the growth and even to destroy bacteria and other microorganisms in low concentration [1]. This definition distinguishes between chemicals produced by microorganisms and antimicrobial compounds synthesized in the laboratory. To date the term antibiotic is used to describe both of these groups of antimicrobial agents.
The history of antibiotics began with Alexander Fleming’s discovery of penicillin in 1929 and since then thousands of antibiotics have been isolated and characterized. Many antibiotics are used commercially or are useful in medicine for activities other than their antibiotic activity. Antibiotics are used as enzyme inhibitors, antitumor, immunosuppressive, hypocholesterolemic and antiparasitic agents, in addition to their applications as antibiotics.
Antibiotics have remained unrivaled in their control of infectious bacteria and fungi for the last 60 years. Ongoing research focuses on the discovery of new or novel antibiotics and further modifications to existing antibiotics are still in progress. There are several important reasons why the discovery and development of antibiotics with novel structural classes are particularly important, including the development of resistant bacteria and other pathogens. Not all antibiotics have activity against all microorganisms. In addition, every antibiotic which is released for clinical application has a limited shelf-life. This means that antibiotics will no longer be active against bacteria at some point in time [2•]. Some microorganisms are naturally resistant to antibiotics and antibiotic resistance can be an inherent property of a microorganism, or can be acquired. Microorganisms may have an inherent resistance to an antibiotic because, for example, the microorganism may lack the target an antibiotic inhibits, be impermeable to antibiotics, be able to alter antibiotics to an inactive form or may modify the target of the antibiotics, or be able to pump out an antibiotic entering the cell [3•].
Bacteria have a high capacity to mutate and exchange genetic material with other microorganisms and, as a result, the development of resistant bacteria has become a serious concern. The development of resistance to existing antibiotic classes by pathogenic bacteria is in direct correlation with the level of utilization of the antibiotics to treat them, and now different types of antibiotics are required to treat the same bacterial infection. For example, treatment for an infection by Staphylococcus aureus was changed from penicillin to methicillin, and now vancomycin is required to treat this infection.
Antibiotic resistance can be achieved by horizontal transfer of resistant genes (carried by plasmids or transposons), by recombination of foreign DNA into a chromosome, or by mutation in different chromosomal loci. Mutations in different loci produce a change in MIC values and stable maintenance of heterogeneous antibiotic resistance expression classes in bacterial populations [4]. Resistance to antifolate drugs by the malaria parasite is the result of point mutations in the substrate binding site of target enzymes. There are seven identified point mutations occurring in the dihydrofolate reductase genes which are associated with reduced drug binding capacity in resistant strains of Plasmodium falciparum [5]. Mycobacterium tuberculosis resistance to rifampin arises due to mutation in the β-subunit of RNA polymerase encoded by the gene rpoB [6]. Resistance to macrolide antibiotics involves modification of the ribosome, specifically methylation of an adenine residue in domain V of the 23S rRNA [7].
There are now many examples of antibiotic-resistant bacteria that are of concern such as methicillin-resistant S aureus (MRSA), vancomycin-resistant enterococci (VRE), multidrug resistance in M tuberculosis and multidrug-resistant Gram-negative bacteria. Other examples of resistance include penicillin resistance in Neisseria gonorrhoeae, pneumococcal infections that are resistant to macrolide therapy and drug-resistant meningococci and Haemophilus influenzae [8].
The toxicity of current antibiotics limits their application, thus providing another reason why the development of novel antibiotics is of importance. Many existing antibiotics have toxicity at or near to their therapeutic dose. Amphotericin B is used in the treatment of serious disseminated dimorphic fungal infection and infections caused by Blastomyces, Candida, Cryptococcus and Histoplasma spp. Unfortunately, amphothericin B causes nephrotoxicity, reduction of renal blood flow, nausea, vomiting and anorexia. Griseofulvin causes hepatotoxicity and gastrointestinal distress but is used for the treatment of certain dermatophyte infections caused by Epidermophyton, Microsporum and Tricophyton spp [9].
The development of infectious conditions has risen significantly as a result of increasingly widespread use of therapies that depress the immune system. The rise has also been due to the frequent and often indiscriminate use of broad-spectrum antibacterial agents and the common use of indwelling intravenous devices combined with the advent of chronic immunosuppressive viral infections such as AIDS. These factors have intensified the need to search for new, safer and more efficacious antibiotics [10]. A changing patient population, including immunocompromised cases, an elderly population and patients under aggressive chemotherapy, has generated increasing sensitivity to infectious diseases and spurred a resurgent interest in the search for new antibiotics [11].
Finally, increasing concerns that terrorists will use infectious diseases as weapons of mass destruction has had an impact on our need for new antibiotics, and as a result, novel antibiotics are needed to combat infections agents, eg, Bacillus anthracis, Clostridium botulinum and Yersinia pestis [12].
The emergence of new infectious diseases, the re-emergence of resistant forms of well known diseases such as tuberculosis and malaria, and the emergence of antibiotic-resistant bacteria have made the development of new antibiotics essential. Different approaches and strategies can be utilized in the search for novel antibiotics and include the discovery of new target sites, structural modification of currently available antibiotics, and sourcing of antibiotic producers. The first and second strategies have been discussed in a review by Schmidt [13•]. This review will emphasize the exploration of new sources for the production of novel antibiotics, with a focus on marine-derived microorganisms as a final frontier for the discovery and development of novel antibiotics.
There are many natural resources that can be explored as sources of antibiotics, such as rare actinomycetes [14], endophytes [15–17], myxobacteria [18] and marine microorganisms [19–21]. Many antibiotic researchers believe that a vast number of new antibiotics will be discovered if other groups of microorganisms are examined.
Marine microorganisms as resources of antibiotics
The oceans are a resource of biodiversity that greatly exceeds that of terrestrial environments. The oceans cover 71% of the earth’s surface and 80% of life on the planet is found under the ocean surface. In addition, the marine environment provides 95% of habitat space on the planet and the oceans offer tremendous diversity with two-thirds of phyla exclusively or dominantly marine organisms [22]. The oceans have been a valuable resource for a unique and large group of structurally diverse natural products that are mainly accumulated in invertebrates, such as sponges, tunicates, bryozoans and mollusks [23–25]. Different ecological areas have been investigated as potential sources of microbial diversity. Therefore, the biological diversity will be reflected by chemical diversity of metabolites [26].
The marine environment also provides a tremendous biodiversity of microorganisms. Microorganisms play an important role in all of the major element cycles in the ocean and are involved in many ecological phenomena. Marine microorganisms have developed unique metabolites and physiological capabilities that not only ensure survival in extreme habitats, but also offer the potential for production of metabolites not observed in terrestrial microorganisms [27]. The isolation of antimicrobial metabolites from bacteria collected from sponges suggested that these bacteria may play a role in a defense mechanism of these invertebrates [28,29].
Terrestrial microorganisms have proved to be rich sources for bioactive antibiotics and the exploration of terrestrial microorganisms and, in particular, the Actinomycetes gave rise to the discovery of many new species and novel bioactive compounds. The expectation to identify novel, diverse and bioactive metabolites from marine microorganisms is based on the following reasons:
The diversity of microorganisms is certain to yield diversity in the production of secondary metabolites.
The production and diversity of microbial metabolites can be regulated by modification of culture conditions.
Most bioactive compounds are produced by a specific strain rather than a taxonomically defined species of microorganisms. Therefore, new bioactive compounds can be obtained by the cultivation of isolated strains without prior taxonomic classification.
Only a small quantity of plant or animal tissue is required to cultivate marine microorganisms, thus protecting macroorganisms from over sampling.
A preservation of natural habitat is possible since once microbes are cultured and preserved, recollection is not required.
Culturable marine microorganisms will provide an adequate supply of bioactive compounds required for scientific and economic development.
Microorganisms are more readily manipulated than their macro counterparts [30•,31].
The most significant obstacles which have hindered the development of drugs from marine natural products are the supply issues. The majority of promising marine compounds have complex structures, rich in centers of asymmetry, limiting the large-scale preparation of antibiotics by chemical synthesis [32•,33•]. In addition, the yield from harvesting and isolating from macroorganisms may be quite low because the macroorganism is rare or the active compound is a minor component. Since it is impossible to collect unlimited quantities of marine animal or plant material, the isolation and cultivation of symbiotic microorganisms are of great importance for a sufficient and constant supply of bioactive compounds. Symbiotic marine microorganisms which can be cultivated will facilitate future development of marine natural product leads if the compound can be produced by an associated microorganism [34•].
The biology of the bacteria-sponge relationship has elicited considerable interest. The responsibility of bacteria for defense strategies of sponges and the production of chemical products by the sponge-associated bacteria are important observations for the exploitation of natural substances [35,36••,37]. Several metabolites have been identified from bacterial cultures isolated from sponges. For example, diketopiperazine, previously reported as a metabolite of the sponge Tedania ignis, is produced by the Micrococcus sp isolated from T ignis [38].
Marine microorganisms can be isolated from seawater, sediment, animate and inanimate surfaces and the internal space of invertebrate animals. The surface of marine organisms is more nutrient rich than sea water and sediment, and numerous bacterial strains can be observed on the surface of marine plants and animals [39,40••,41•].
Antibiotics from marine bacteria
Bacteria in the marine environment are diverse, and more than 40% of sponge weight may contain bacteria. Marine bacteria are considered as an emerging source of novel bioactive metabolites with respect to their existence, diversity and function in the marine environment [42••,43]. Here, we report promising antibiotics derived from marine bacteria.
Massetolides
Bacterial isolate MK90e85 was obtained from the marine leafy red alga and produced massetolides A (Figure 1) to D, while a tube worm-derived strain (MK91CC8) produced massetolides E to H. Both isolates belong to the Pseudomonas genus based on fatty acid analysis. Massetolide A exhibited promising antimicrobial activity against M tuberculosis and Mycobacterium avium intracellulare with MIC values of 5 to 10 μg/ml and 2.5 to 5 μg/ml, respectively. No activity was demonstrated against human pathogenic bacteria, including Escherichia coli and S aureus. A single intraperitoneal injection of 10 mg/kg of massetolide A was non-toxic to mice [44].
Figure 1.
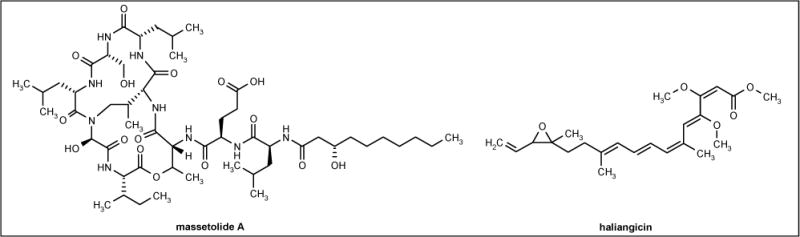
The structures of massetolide A and haliangicin.
Haliangicin
Haliangicin (Figure 1) is a β-methoxyacrylate antibiotic, which was isolated from the culture broth of a marine myxobacterium Haliangium luteum. Haliangicin demonstrated potent activities against filamentous fungi such as Aspergillus niger AJ 117374, Aspergillus fumigatus AJ 117190, Botrytis cinerea AJ 117140, Fusarium sp AJ 117167 and Mucor hiemalis AJ 117374 with MIC values of 3.1 to 12.5 μg/ml. The compound also showed potency against oomycete fungi Phythium ultimum IFO 32210 and Saprolegnia parasitica IFO 8978, which were insensitive to amphotericin B and nystatin [45].
Macrolactins
Macrolactins (Figure 2) were reported as secondary metabolites from an unidentified bacteria isolated from a deep sea sediment sample from California, USA [46], Bacillus sp isolated from a macroalga Schizymenia dubyi [47] and Bacillus sp SC 026 identified from a sediment sample around Sichang Island, Thailand [48]. Gustafson et al reported that macrolactin A inhibited herpes simplex type 1 virus strain LL (IC50 = 5.0 μg/ml) as well as type II virus strain G (IC50 = 5.0 μg/ml) and HIV (IC50 = 10 μg/ml) [46]. The MIC values of macrolactin A against Bacillus subtilis and S aureus were 20 and 5 μg/disk, respectively. Nagao et al reported that macrolactin A, G, H, I, L and M were more effective against S aureus (MIC = 10 ppm) than B subtilis (MIC = 60 ppm). Macrolactin J was the most active with MIC values of 5 and 30 μg/ml against S aureus and B subtilis, respectively, while macrolactins F and K were considered inactive. This lack of antibacterial activity was due to the presence of ketone carbonyl at C(15) and suggested that the hydroxyl group at C(15) was necessary for antibacterial activity.
Figure 2.
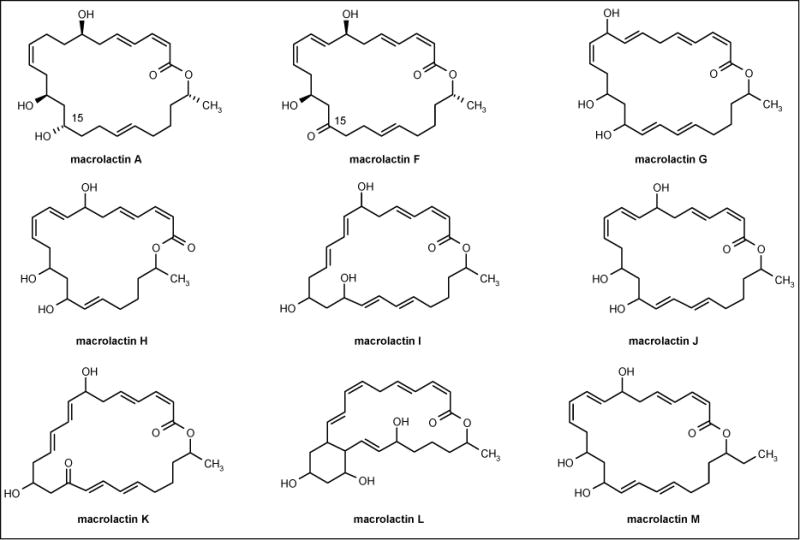
The structures of macrolactins.
Bacillus laterosporus antibiotics
Bacterium PNG-276 tentatively identified as Bacillus laterosporus produced the cyclic decapeptide antibiotics loloatin A to D (SeaTek Marine Biotechnology Inc; Figure 3) [49], the cationic peptide bogorol A (Figure 3) [50], basiliskamide A and B (Figure 3), and tupuseleiamides A and B [51]. Loloatin A to D inhibited MRSA, vancomycin-resistant Enterococcus faecalis, vancomycin-resistant Enterococcus faecium, penicillin-resistant Streptococcus pneumoniae, as well as Candida albicans with MIC values from 0.5 to 8 μg/ml. Bogorol A displayed potent activity against both MRSA (MIC = 2 μg/ml) and VRE (MIC = 10 μg/ml) but exhibited no activity against C albicans and drug-resistant P aeruginosa. Basiliskamide A and B showed activity against C albicans with MIC values of 1.0 and 3.1 μg/ml, respectively. In addition, the MIC values of basiliskamide A and B against A fumigatus were 2.5 and 5.0 μg/ml, respectively. In vitro cytotoxicity studies comparing basiliskamide A with amphotericin B revealed that basiliskamide A produced minimal toxicity at 100 μg/ml and no cytopathic effect, while amphotericin B produced a cytopathic effect at 12.5 μg/ml.
Figure 3.
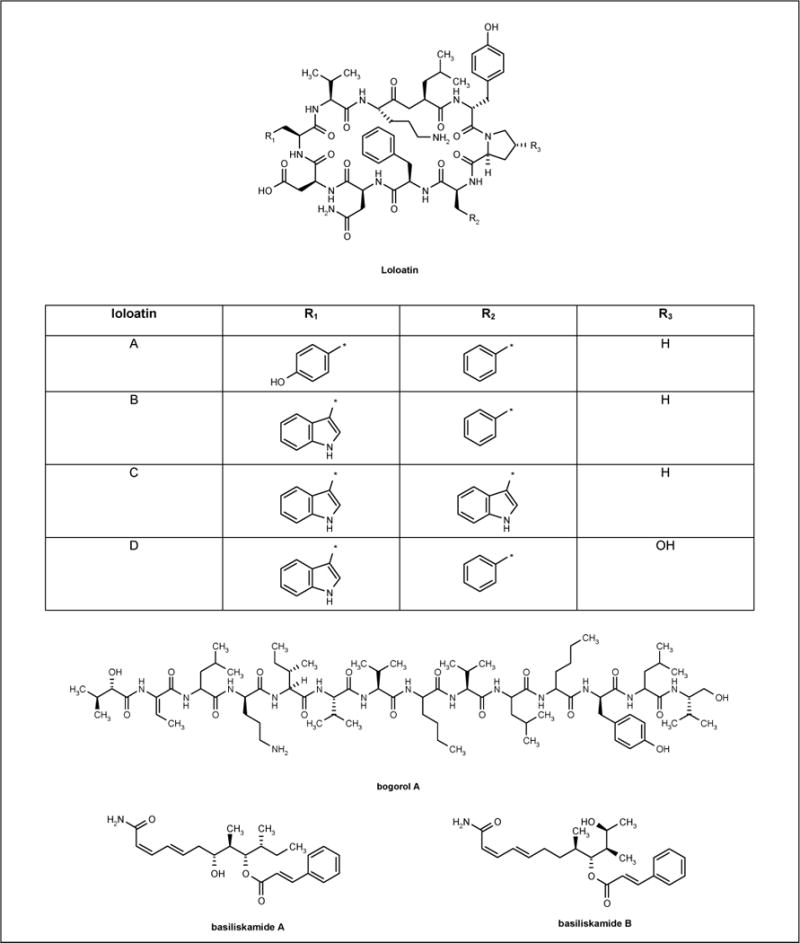
The structure of antibiotics isolated from Bacillus laterosporus.
Tetrocarcins
Arisostatin A and B (Figure 4) are new members of the tetroarcin class of antibiotics and were isolated from the cultured broth of the actinomycetes strain TP-AO316, which was identified as a Micromonospora sp strain isolated from a sea water sample collected in Toyama Bay, Japan. Arisostatins exhibited antibacterial activity against Gram-positive bacteria but no activity against Gram-negative bacteria and yeast [52]. The MIC values are shown in Table 1.
Figure 4.
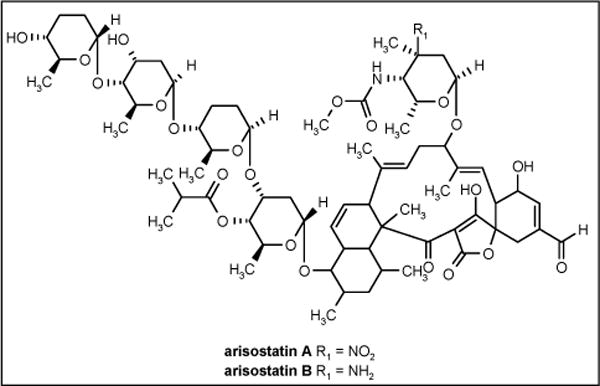
The structures of arisostatins.
Table 1.
The MIC values of arisostatins against Gram-positive bacteria.
| Arisostatin A (μg/ml) | Arisostatin B (μg/ml) | |
|---|---|---|
| Staphylococcus aureus 209P JC-1 | 100 | 50 |
| Bacillus subtilis ATCC 6633 | 0.39 | 25 |
| Micrococcus luteus ATCC 9341 | 12.5 | 3.1 |
Lomaiviticins
The antitumor antibiotics lomaiviticins A and B (Figure 5) were isolated from halophilic strain LL-371366 and identified as the new species Micromonospora lomaivitiensis. The strain was isolated from the inner core of the ascidian Polysyncraton lithostrotum. In a plate assay, the antibiotics demonstrated potent activity against S aureus and E faecium with MIC values of 6 to 25 ng/spot [53].
Figure 5.
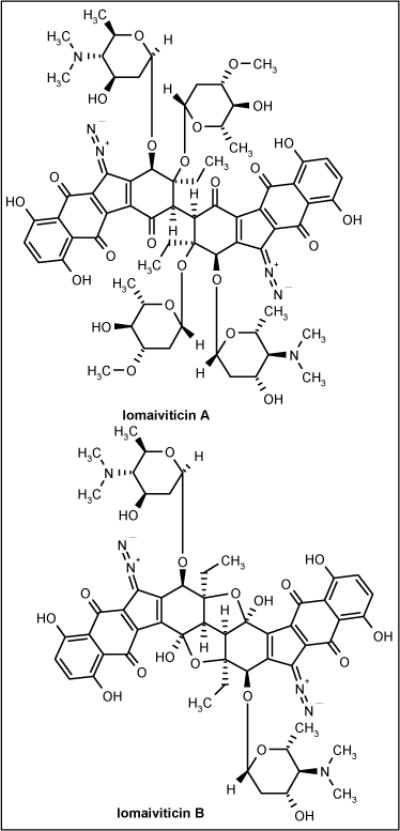
The structures of lomaiviticins.
Pelagiomicins
The new marine bacterium Pelagiobacter variabilis was isolated from macroalga Pocockiella variegata collected at Palau. This bacterium is halophilic, Gram-negative and produced a series of compounds called pelagiomicin A (Figure 6) to C. Pelagiomicin A was active against Gram-positive and -negative bacteria (Table 2) but was inactive against yeast. The pelagiomicins are the third example containing griseoluteic acid. The other two were griseoluteins and senacarcin produced by terrestrial Streptomyces sp, and this raises interesting questions regarding the propagation of secondary metabolite genes [54].
Figure 6.
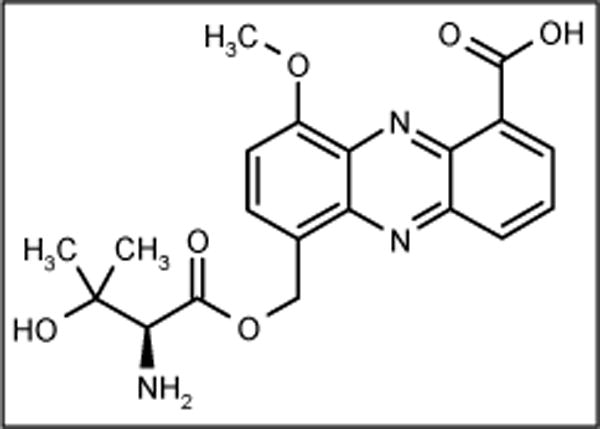
The structure of pelagiomicin A.
Table 2.
MIC values of pelagiomicin A against Gram-positive and -negative bacteria.
| Bacteria | MIC (μg/ml) |
|---|---|
| Staphylococcus aureus ATCC 6538P | 2.6 |
| Enterococcus hirae ATCC 10541 | 0.16 |
| Bacillus subtilis # 10707 | 0.16 |
| Klebsiella pneumoniae ATCC 10031 | 0.16 |
| Escherichia coli ATCC 26 | 1.3 |
| Pseudomonas aeruginosa Bin H#1 | 5.2 |
| Proteus vulgaris ATCC 6897 | < 0.04 |
| Shigella sonnei ATCC 9290 | 1.3 |
Thiocoraline
A new depsipeptide, thiocoraline (PharmaMar SA; Figure 7), was isolated from marine Micromonospora strain L-13-ACM2-092 identified as Micromonospora marina. The strain was obtained from a soft coral collected in the Indian Ocean. Thiocoraline showed potent antibiotic activity against Gram-positive bacteria S aureus, B subtilis and Micrococcus luteus with MIC values of 0.05, 0.05 and 0.03 μg/ml, respectively, while demonstrating low activity against Gram-negative bacteria. This antibiotic inhibits RNA synthesis more specifically than DNA synthesis [55].
Figure 7.
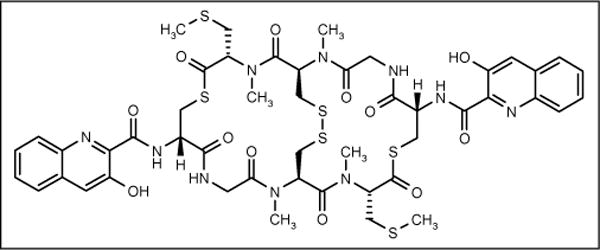
The structure of thiocoraline.
Parimycin
Marine actinomycete strain B8652, found in the sediment of the Laguna de Terminos at the Gulf of Mexico, produced the antibiotic parimycin (Figure 8). The antibiotic showed moderate activity against E coli, B subtilis and S aureus at concentrations of > 20 μg [56].
Figure 8.
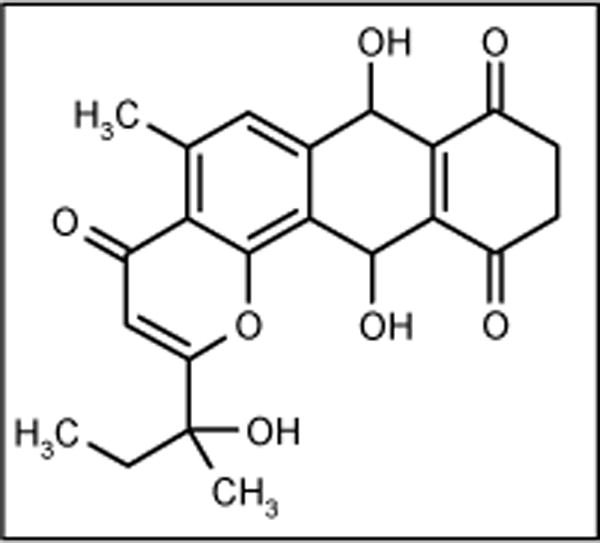
The structure of parimycin.
IB-00208
A polycyclic xanthone IB-00208 (Figure 9) was isolated from strain BL-42-PO13-046 identified as Actinomadura. This compound showed good antibiotic activity against Gram-positive bacteria, but poor activity against Gram-negative bacteria. The MIC values against B subtilis ATCC 6051, S aureus ATCC 6538P and M luteus ATCC 9341 were 1.4, 1.4 and 0.09 nM, respectively [57,58].
Figure 9.
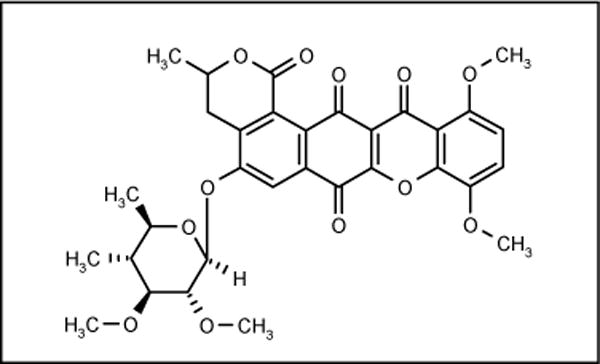
The structure of IB-00208.
YM-266183 and YM-266184
Bacillus cereus QN 03323 was isolated from a marine sponge Halicondria japonica, collected at Hoshisuna Beach, Irimoto Island, Japan and produced novel thiopeptide antibiotics YM-266183 (Figure 10; Yamanouchi Pharmaceutical Co Ltd) and YM-266184 (Figure 10; Yamanouchi Pharmaceutical Co Ltd) along with thiocillin I and II. Both compounds were active against Gram-positive bacteria, including MRSA (MIC = 0.78 and 0.39 μg/ml, respectively), VRE (MIC = 0.025 μg/ml) and methicillin-resistant Staphylococcus epidermidis (MRSE) (MIC = 1.56 and 0.2 μg/ml, respectively), but inactive against Gram-negative bacteria, fungi and yeast. The activities against MRSA, MRSE and VRE were comparable to vancomycin [59,60].
Figure 10.
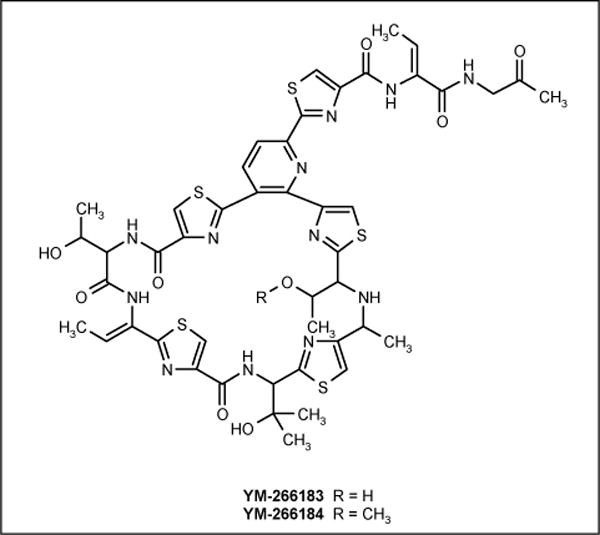
The structures of YM-266183 and YM-266184.
Chalcomycins
Marine Streptomyces sp B7064 was isolated from a mangrove sediment near Pohoiki, Hawaii and was reported to produce new types of macrolide antibiotics chalcomycin A and B (Figure 11). The compounds showed activity against B subtilis, E coli and S aureus with MIC values of 6.25, 750 and 0.39 μg/ml, respectively [61].
Figure 11.
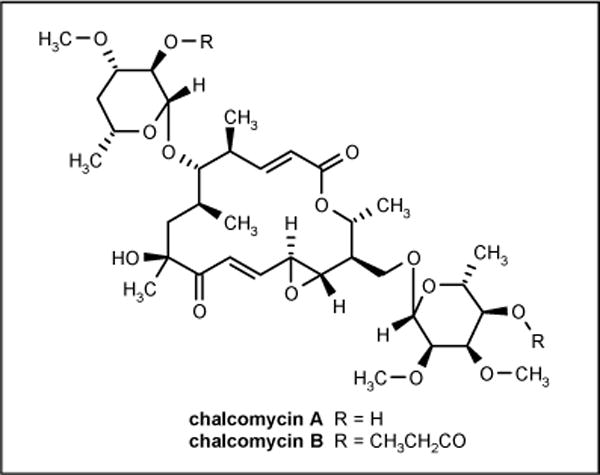
The structures of chalcomycins.
Antibiotics from marine fungi
Terrestrial fungi have proven a rich source of pharmaceutically interesting secondary metabolites, and it has been postulated that marine fungi will also become an important source of secondary metabolites. Higher marine fungi are distributed in littoral and deep water. Their geographic distribution is largely a function of temperature and salinity requirements. These fungi are morphologically different and have different growth requirement from terrestrial fungi. The diversity, biological activity and secondary metabolite production of fungi associated with marine sponges were investigated in order to assess the potential of these fungi for the production of novel biologically active compounds [62,63].
Zopfiellamides
Two novel antimicrobial compounds zopfiellamide A and B (Figure 12) were isolated from a submerged culture Zopfiella latipes. The strain was isolated from a sediment sample from the Indian Ocean near New Delhi. Zopfiellamide A was reported to be active against Arthrobacter citreus, Bacillus brevis, B subtilis, B licheniformis, Corynebacterium insidiosum, M luteus, Mycobacterium phlei and Streptomyces sp with MIC values between 2 and 10 μg/ml; zopfiellamide B was 5-fold less active [64].
Figure 12.
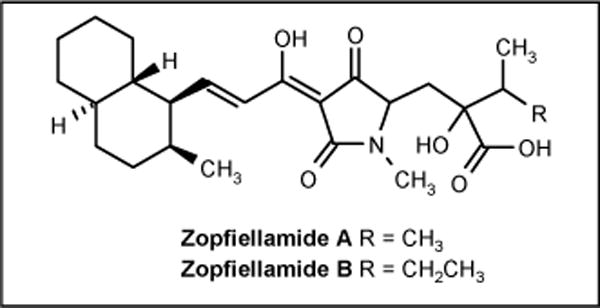
The structures of zopfiellamides.
Pestalone
Pestalone (Figure 13) was obtained by mixed fermentation of the fungus Pestalotia sp and an unidentified antibiotic-resistant marine bacteria. The fungus was isolated from the surface of the brown alga Rosenvingea sp collected at the Bahamas Island. Interestingly, pestalone was not produced when the fungus and bacteria were cultured separately. This fact suggests that the antibiotic is produced in response to antagonism. It is possible that the pathogens responsible for the biosynthesis of certain compounds are regulated by factors elicited by one microbe and detected by another. The compound showed activity against MRSA (MIC = 37 ng/ml) and VRE (MIC = 78 ng/ml) [65].
Figure 13.
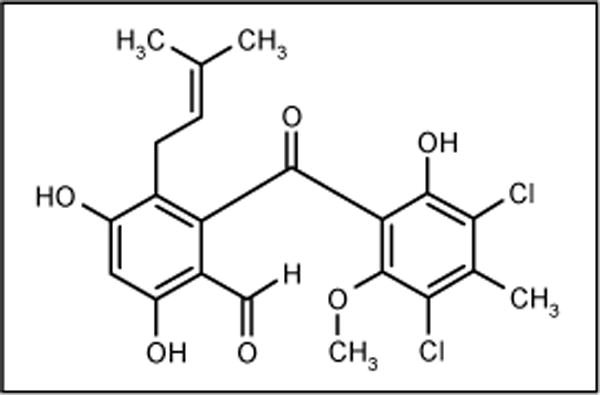
The structure of pestalone.
Halorosellinic acid
A culture broth of marine fungus Halorosellina oceanica BCC 5149 collected in Thailand was the source of an ophiobolane sesterpene halorosellinic acid (Figure 14). The compound exhibited moderate antimalarial activity against Plasmodium falciparum with an IC50 value of 13 μg/ml but weak antimicrobial activity [66].
Figure 14.
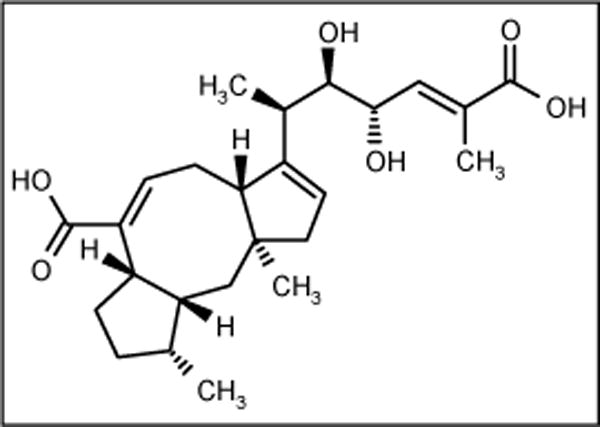
The structure of halorosellinic acid.
Trichodermamides
Two new modified dipeptides trichodermamides A and B (Figure 15) were isolated from marine-derived fungi CNL 910 and CNK 266, identified as Trichoderma virens. The fungus CNL 910 was isolated from the well-known ascidian Didemnum molle collected in Papua New Guinea, while the fungus CNK 266 was isolated from the surface of the green alga of the genus Halimeda. Trichodermamide B exhibited moderate antimicrobial activity against MRSA, VRE and amphotericin-resistant C albicans with MIC values of 15 μg/ml for all three strains, while trichodermamide A was completely inactive suggesting that the chlorine atom at C(5) is essential for activity [67].
Figure 15.
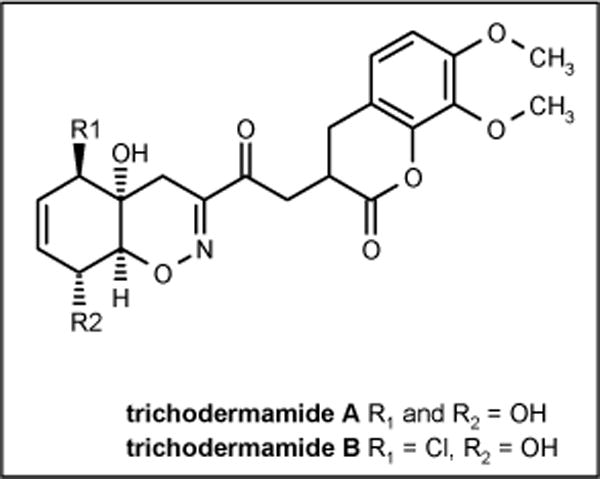
The structures of trichodermamides.
Varixanthone
The fungus Emericella variecolor isolated from a sponge collected in Venezuelan waters of the Caribbean produced varixanthone (Figure 16) with activity against E coli, Proteus sp, B subtilis and S aureus (MIC = 12.5 μg/ml in all cases) [68].
Figure 16.
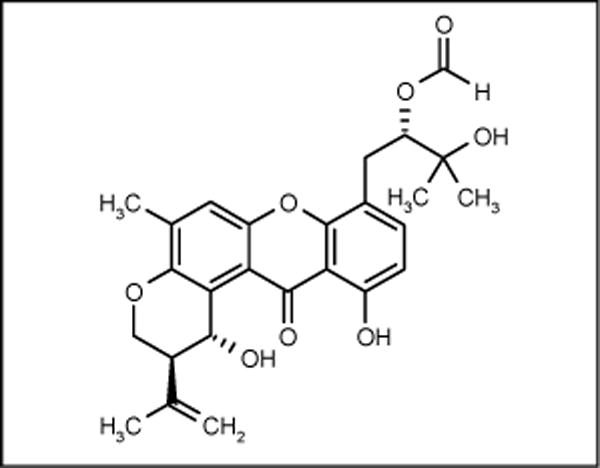
The structure of varixanthone.
YM-202204
Marine fungus Phoma sp Q60596 was isolated from the sponge Halicondria japonica collected at Hoshisuna beach, Okinawa, Japan. The fungus produced the antifungal antibiotic YM-202204 (Figure 17; Yamanouchi Pharmaceutical Co Ltd) which showed activity against C albicans ATCC 10231, Cryptococcus neoformans TIMM0362, A fumigatus TIMM1776 and Saccharomyces cerevisae YFC805 with IC80 values of 6.25, 1.56, 12.5 and 1.56 μg/ml, respectively. The compound has activity as an inhibitor of glycosyl-phosphatidyl inositol (GPI)-anchoring. Since GPI-anchoring protein is essential for the growth of yeast and fungi, this may be a unique target for antifungal chemotherapy [69].
Figure 17.
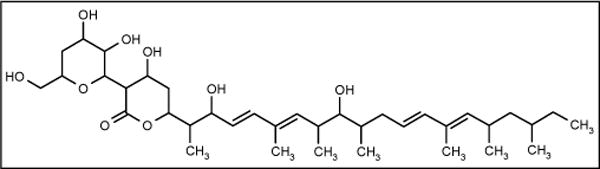
The structure of YM-202204.
Future prospects
Novel antibiotics from marine microorganisms are attracting attention because of the growing demand for antibiotics to combat infectious diseases. Many unusual antibiotic classes with different mechanisms of action from existing antibiotics have been discovered. These require further development of fermentation processes since their yields from natural sources are quite low. Fenical recommended 100-liter fermentation of marine bacteria to yield sufficient quantities of the target compound for analysis [70•].
Bioprocess intensification focusing on optimizing fermentation of marine microorganisms will become an important strategy for improving the yields [71••]. For industrial purposes, marine microorganisms should have properties related to current industrial microorganisms, such as the production of the metabolite of interest, availability in pure culture, genetic stability and culture on a large-scale, stability over long periods, rapid growth and production of the desired product quickly, ability to grow in inexpensive liquid media, ease of separation between media and biomass, and finally the microorganisms should be amenable to genetic manipulation.
Many technologies that have been successfully applied to terrestrial microorganisms can be applied to marine microorganisms in order to achieve overproduction of the desired product. Random mutagenesis and selection techniques are common for strain improvement [72].
Recent advances in molecular biology, knowledge of biosynthetic pathways and biosynthetic gene clusters offer many possibilities in strain improvement, such as increasing gene dosage by introduction of additional copies of genes encoding the pathway, increasing gene expression by introduction of regulatory genes in high copy numbers and gene replacement technologies [73–76]. Further understanding of antibiotic biosynthesis pathways will increasingly permit reprogramming of antibiotic biosynthesis, resulting in more desirable molecular structures, simplified production patterns and enhanced product yield [77].
Conclusion
Marine microorganisms provide a wide range of diversity in chemical structure. The activity of compounds derived from marine microorganisms is comparable with existing antibiotics and can address the problem of resistance and reemergence in infectious diseases. In addition to the molecular novelty and bioactivity, marine microbes present a secure and renewable supply of targeted metabolites for scientific enquiry and commercial development.
Rapid developments in molecular biology, genetic engineering and fermentation technologies provides the capacity to solve the problem of renewable supply associated with marine natural products as well as uncultivable and unexplored marine microorganisms. Considerable effort will be required before the potential of the marine microorganism is fully realized.
References
- 1.Demain AL. Pharmaceutically active secondary metabolites of microorganisms. Appl Microbiol Biotechnol. 1999;52(4):455–463. doi: 10.1007/s002530051546. [DOI] [PubMed] [Google Scholar]
- 2•.Walsh C. Where will new antibiotics come from? Nat Rev Microbiol. 2003;1(1):65–70. doi: 10.1038/nrmicro727. Review detailing the discovery and innovation of antibiotics. [DOI] [PubMed] [Google Scholar]
- 3•.Amabile-Cuevas C. New antibiotics and new resistance. Am Sci. 2003;91:138–149. Review detailing the mechanisms of antibiotic resistance. [Google Scholar]
- 4.Martinez JL, Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother. 2000;44(7):1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phllips RS. Current status of malaria and potential for control. Clin Microbiol Rev. 2001;14(1):208–226. doi: 10.1128/CMR.14.1.208-226.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillespie SH. Evolution of drug resistance in Mycobacterium tuberculosis: Clinical and molecular perspective. Antimicrob Agents Chemother. 2002;46(2):267–274. doi: 10.1128/AAC.46.2.267-274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vester B, Douthwaite S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother. 2001;45(1):1–12. doi: 10.1128/AAC.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell AD. Antibiotic and biocide resistance in bacteria: Introduction. J Appl Microbiol Symposium Suppl. 2002;92:1S–3S. [PubMed] [Google Scholar]
- 9.Gupte M, Kulkarni P, Ganguli BN. Antifungal antibiotics. Appl Microbiol Biotechnol. 2002;58(1):46–57. doi: 10.1007/s002530100822. [DOI] [PubMed] [Google Scholar]
- 10.Ghannoum MA, Rice LB. Antifungal agents: Mode of action, mechanism of resistance and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev. 1999;12(4):501–517. doi: 10.1128/cmr.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosamond J, Allsop A. Harnessing the power of the genome in the search for new antibiotics. Science. 2000;287(5460):1973–1976. doi: 10.1126/science.287.5460.1973. [DOI] [PubMed] [Google Scholar]
- 12.Atlas RM. Bioterrorism and biodefence research: Changing the focus of microbiology. Nat Rev Microbiol. 2003;1(1):70–74. doi: 10.1038/nrmicro728. [DOI] [PubMed] [Google Scholar]
- 13•.Schmidt FR. The challenge of multidrug resistance: Actual strategies in the development of novel antibacterials. Appl Microbiol Biotechnol. 2004;63(4):335–343. doi: 10.1007/s00253-003-1344-1. Mini-review detailing three strategies in the development of antibiotics. [DOI] [PubMed] [Google Scholar]
- 14.Lazzarini A, Cavaletti L, Toppo G, Marinelli F. Rare actinomycetes as potential producers of new antibiotics. Antonie van Leeuwenhoek. 2000;78(3–4):399–405. [PubMed] [Google Scholar]
- 15.Tan RX, Zou WX. Endophytes: Rich sources of functional metabolites. Nat Prod Rep. 2001;18(4):448–459. doi: 10.1039/b100918o. [DOI] [PubMed] [Google Scholar]
- 16.Kongsaeree P, Prabpai S, Sriubolmas N, Vongvein C, Wiyakrutta S. Antimalarial dihydroisocoumarins produced by Geotrichum sp an endophytic fungus of Crassocephalum crepidioides. J Nat Prod. 2003;66(5):709–711. doi: 10.1021/np0205598. [DOI] [PubMed] [Google Scholar]
- 17.Wagenaar MM, Corwin J, Strobel G, Clardy J. Three new cytochalasins produced by an endophytic fungus in the genus Rhinocladiella. J Nat Prod. 2000;63(12):1692–1695. doi: 10.1021/np0002942. [DOI] [PubMed] [Google Scholar]
- 18.Reichenbach H, Hofle G. Biologically active secondary metabolites from myxobacteria. Biotechnol Adv. 1993;11(2):219–277. doi: 10.1016/0734-9750(93)90042-l. [DOI] [PubMed] [Google Scholar]
- 19.Haefner B. Drugs from the deep: Marine natural products as drug candidates. Drug Disc Today. 2003;8(12):536–544. doi: 10.1016/s1359-6446(03)02713-2. [DOI] [PubMed] [Google Scholar]
- 20.Hentschel U. Natural product from marine microorganisms. ChemBioChem. 2000;1(11):1151–1154. doi: 10.1002/1439-7633(20021104)3:11<1151::AID-CBIC1151>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 21.Proksch P, Edrada RA, Ebel R. Drugs from the seas - current status and microbiological implications. Appl Microbiol Biotechnol. 2002;59(2–3):125–134. doi: 10.1007/s00253-002-1006-8. [DOI] [PubMed] [Google Scholar]
- 22.Patrzykat A, Douglas SE. Gone gene fishing: How to catch a novel marine antimicrobials. Trends Biotechnol. 2003;21(8):362–369. doi: 10.1016/S0167-7799(03)00145-8. [DOI] [PubMed] [Google Scholar]
- 23.Blunt JW, Copp BR, Munro MHG, Northcote PT, Prinsep MR. Marine natural products. Nat Prod Rep. 2003;20(1):1–48. doi: 10.1039/b207130b. [DOI] [PubMed] [Google Scholar]
- 24.Faulkner DJ. Marine natural products. Nat Prod Rep. 2002;19(1):1–48. doi: 10.1039/b009029h. [DOI] [PubMed] [Google Scholar]
- 25.Faulkner DJ. Marine natural products. Nat Prod Rep. 2001;18(1):1–49. doi: 10.1039/b006897g. [DOI] [PubMed] [Google Scholar]
- 26.Ditullio D, Rocheleau N, Talbot MK, Millett W, McAlpine J, Robakiewicz P. Strain diversity within a microbial collection. J Antibiot. 2002;55(10):899–903. doi: 10.7164/antibiotics.55.899. [DOI] [PubMed] [Google Scholar]
- 27.Garson MJ. Ecological perspectives on marine natural product biosynthesis. In: McClintok JB, Baker BJ, editors. Marine Chemical Ecology. Springer-Verlag; Heidelberg, Germany: 2001. pp. 71–114. [Google Scholar]
- 28.Unson MD, Faulkner DJ. Cyanobacterial symbiont biosynthesis of chlorinated metabolite from Dysedia herbacea (Porifera) Experentia. 1993;49:349–353. [Google Scholar]
- 29.Pietra F. Secondary metabolites from marine microorganisms: Bacteria, protozoa, algae and fungi. Achievements and prospects. Nat Prod Rep. 1997;14(5):453–464. doi: 10.1039/np9971400453. [DOI] [PubMed] [Google Scholar]
- 30•.Okami Y. The search for bioactive metabolites from marine bacteria. J Mar Biotechnol. 1993;1:59–65. Mini-review describing factors that influence the production of secondary metabolites from marine bacteria. [Google Scholar]
- 31.Konig GM, Wright AD. Trends in marine biotechnology. In: Grabley S, Thiericke R, editors. Drug Discovery from Nature. Springer-Verlag Telos; Heidelberg, Germany: 2000. pp. 180–185. [Google Scholar]
- 32•.Davidson BS. New dimensions in natural products research: Cultured marine microorganisms. Curr Opin Biotechnol. 1995;6(3):284–291. Review of the marine compounds isolated from marine microorganisms. [Google Scholar]
- 33•.Pomponi SA. The bioprocess - technological potential of the sea. J Biotechnol. 1999;70(1–3):5–13. The challenges and prospects of marine natural products are reviewed. [Google Scholar]
- 34•.Kobayashi J, Ishibashi M. Bioactive metabolites of symbiotic marine microorganisms. Chem Rev. 1993;93(5):1763–1769. This review describes the collection of bioactive metabolite derived from symbiotic or associated bacteria. [Google Scholar]
- 35.Imhoff JF, Stohr R. Sponge-associated bacteria: General overview and special aspects of bacteria associated with Halicondria panacea. In: Muller WEG, editor. Sponge (Porifera): Progress in Molecular and Subcellular Biology. Springer-Verlag; Berlin, Germany: 2003. pp. 35–57. [DOI] [PubMed] [Google Scholar]
- 36••.Faulkner DJ, Harper MK, Haygood MG, Salomon CE, Schmidt EW. Symbiotic bacteria in sponges: Sources of bioactive substance. In: Fusetani N, editor. Drugs from the Sea. S Karger AG; Basel, Switzerland: 2000. pp. 107–119. A general overview on the importance of symbiotic bacteria as sources of marine bioactive compounds. [Google Scholar]
- 37.Haygood MG, Schmidt EW, Davidson SK, Faulkner DJ. Microbial symbionts of marine invertebrates: Opportunities for microbial biotechnology. J Mol Microbiol Biotechnol. 1999;1(1):33–43. [PubMed] [Google Scholar]
- 38.Stierle AC, Cardellina JH, 2nd, Singleton FL. A marine Micrococcus produces metabolites ascribed to the sponge Tedania ignis. Experientia. 1988;44(11–12):1021. doi: 10.1007/BF01939910. [DOI] [PubMed] [Google Scholar]
- 39.Bultel-Ponce V, Berge J-P, Debitus C, Nicolas J-L, Guyot M. Metabolites from the sponge-associated bacterium Pseudomonas species. Mar Biotechnol. 1999;1(4):384–390. doi: 10.1007/pl00011792. [DOI] [PubMed] [Google Scholar]
- 40••.Hentschel U, Fieseler L, Wehrl M, Gernert C, Steinert M, Hacker J, Horn M. Microbial diversity of marine sponges. In: Muller WEG, editor. Sponge (Porifera): Progress in Molecular and Subcellular Biology. Springer-Verlag; Berlin, Germany: 2003. pp. 59–88. A review detailing the microbial diversity of marine sponge; three examples are included. [DOI] [PubMed] [Google Scholar]
- 41•.Webster NS, Wilson KJ, Blackall LL, Hill RT. Phylogenetic diversity of bacteria associated with the marine sponge Rhopaloeides odorabile. Appl Environ Microbiol. 2001;67(1):434–444. doi: 10.1128/AEM.67.1.434-444.2001. This paper reports data regarding the diversity of bacteria associated with the marine sponge Rhopaloides odorabile. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.Fenical W, Jensen P. Marine microorganisms and drug discovery: Current status and future potential. In: Fusetani NS, editor. Drugs from the Sea. Karger AG; Switzerland: 2000. pp. 6–29. Review of the importance of marine microorganisms and the current status of drug discovery. [Google Scholar]
- 43.Austin B. Novel pharmaceutical compounds from marine bacteria. J Appl Bacteriol. 1989;67(5):461–470. doi: 10.1111/j.1365-2672.1989.tb02517.x. [DOI] [PubMed] [Google Scholar]
- 44.Gerard J, Lloyd R, Barsby T, Haden P, Kelly MT, Andersen RJ. Massetolides A–H, antimycobacterial cyclic depsipeptides produced by two pseudomonads isolated from marine habitats. J Nat Prod. 1997;60(3):223–229. doi: 10.1021/np9606456. [DOI] [PubMed] [Google Scholar]
- 45.Fudou R, Iizuka T, Yamanaka S. Haliangicin, a novel antifungal metabolite produced by marine myxobacterium. 1. Fermentation and biological characteristics. J Antibiot. 2001;54(2):149–152. doi: 10.7164/antibiotics.54.149. [DOI] [PubMed] [Google Scholar]
- 46.Gustafson K, Roman M, Fenical W. The macrolactins, a novel class of antiviral and cytotoxic macrolide from a deep sea marine bacterium. J Am Chem Soc. 1989;111(19):7519–7524. [Google Scholar]
- 47.Nagao T, Adachi K, Sakai M, Nishijima M, Sano H. Novel macrolactins as antibiotic lactones from a marine bacterium. J Antibiot. 2001;54(4):333–339. doi: 10.7164/antibiotics.54.333. [DOI] [PubMed] [Google Scholar]
- 48.Jaruchoktaweechai C, Suwanborirux K, Tanasupawatt S, Kittakoop P, Menasveta P. New macrolactins from marine Bacillus sp Sc026. J Nat Prod. 2000;63(7):984–986. doi: 10.1021/np990605c. [DOI] [PubMed] [Google Scholar]
- 49.Gerard JM, Haden P, Kelly MT, Andersen RJ. Loloatins A-D, cyclic decapeptide antibiotics produced in culture by a tropical marine bacterium. J Nat Prod. 1999;62(1):80–85. doi: 10.1021/np980219f. [DOI] [PubMed] [Google Scholar]
- 50.Barsby T, Kelly MT, Gagne SM, Andersen RJ. Bogorol A produced in culture by a marine Bacillus sp reveal a novel template for cationic peptide antibiotics. Org Lett. 2001;3(3):437–440. doi: 10.1021/ol006942q. [DOI] [PubMed] [Google Scholar]
- 51.Barsby T, Kelly MT, Andersen RJ. Tupuseleiamides and basiliskamides, new acyldipeptides and antifungal polyketides produced in culture by a Bacillus laterosporus isolate obtained from tropical marine habitat. J Nat Prod. 2002;65(10):1447–1451. doi: 10.1021/np0201321. [DOI] [PubMed] [Google Scholar]
- 52.Furumai T, Takagi K, Igarashi Y, Saito N, Oki T. Arisostatins A and B, new members of tetrocarcin class of antibiotics from Micromonospora sp TP-A0316. I. Taxonomy, fermentation, isolation and biological properties. J Antibiot. 2000;53(3):227–232. doi: 10.7164/antibiotics.53.227. [DOI] [PubMed] [Google Scholar]
- 53.He H, Ding W-D, Bernan VS, Richardson AD, Ireland CM, Greenstein M, Ellestad GA, Carter GT. Lomaiviticins A and B, potent antitumor antibiotics from Micromonospora lomaivitiensis. J Am Chem Soc. 2001;123(22):5362–5363. doi: 10.1021/ja010129o. [DOI] [PubMed] [Google Scholar]
- 54.Imamura N, Nishijima M, Takadera T, Adachi K, Sakai M, Sano H. New anticancer antibiotics pelagiomicins, produced by a new marine bacterium Pelagiobacter variabilis. J Antibiot. 1997;50(1):8–12. doi: 10.7164/antibiotics.50.8. [DOI] [PubMed] [Google Scholar]
- 55.Romero F, Espliego F, Perez Baz J, Garcia Quesada T, Gravalos D, de la Calle F, Fernandez-Puentes JL. Thiocoraline, a new depsipeptide with antitumor activity produced by marine Micromonospora. I. Taxonomy, fermentation, isolation, and biological activities. J Antibiot. 1997;50(9):734–737. doi: 10.7164/antibiotics.50.734. [DOI] [PubMed] [Google Scholar]
- 56.Maskey RP, Helmke E, Fiebig H-H, Laatsch H. Parimycin: Isolation and structure elucidation of novel cytotoxic 2,3-dihydroquinizarin analogue of γ-indomycinone from a marine Streptomycete isolate. J Antibiot. 2002;55(12):1031–1035. doi: 10.7164/antibiotics.55.1031. [DOI] [PubMed] [Google Scholar]
- 57.Malet-Cascon L, Romero F, Espeliego-Vazquez F, Gravalos D, Fernandez-Puentes JL. IB-00208, a new cytotoxic polycyclic xanthone produced by a marine-derived Actinomadura. I. Isolation of the strain, taxonomy and biological activities. J Antibiot. 2003;56(3):219–225. doi: 10.7164/antibiotics.56.219. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez JC, Fernandez-Puentes JL, Baz JP, Canedo LM. IB-00208, a new cytotoxic polycyclic xanthone produced by marine derived Actinomadura. II. Isolation, physico-chemical properties and structure determination. J Antibiot. 2003;56(3):318–321. doi: 10.7164/antibiotics.56.318. [DOI] [PubMed] [Google Scholar]
- 59.Nagai K, Kamigiri K, Arao N, Suzumura K, Kawano Y, Yamaoka M, Zhang H, Watanabe M, Suzuki K. YM-266183 and YM-266184, novel thiopeptide antibiotics produced by Bacillus cereus isolated from a marine sponge. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological properties. J Antibiot. 2003;56(2):123–128. doi: 10.7164/antibiotics.56.123. [DOI] [PubMed] [Google Scholar]
- 60.Suzumura K, Yokoi T, Funatsu M, Nagai K, Tanaka K, Zhang H, Suzuki K. YM-266183 and YM-266184, novel thiopeptide antibiotics produced by Bacillus cereus isolated from a marine sponge. II. Structure elucidation. J Antibiot. 2003;56(2):129–134. doi: 10.7164/antibiotics.56.129. [DOI] [PubMed] [Google Scholar]
- 61.Asolkar RN, Maskey RP, Helmke E, Laatsch H. Chalcomycin B, a new macrolide antibiotic from the marine isolate Streptomyces sp B7064. J Antibiot. 2002;55(10):893–898. doi: 10.7164/antibiotics.55.893. [DOI] [PubMed] [Google Scholar]
- 62.Holler U, Wright AD, Matthee GF, Konig GM, Draeger S, Aust H-J, Schulz B. Fungi from marine sponges: Diversity, biological activity and secondary metabolites. Mycol Res. 2000;104(11):1354–1365. [Google Scholar]
- 63.Cuomo V, Palomba I, Perretti A, Guerriero A, D’Ambrosio M, Pietra F. Antimicrobial activities from marine fungi. J Mar Biotechnol. 1995;2:199–204. [Google Scholar]
- 64.Daferner M, Anke T, Sterner O. Zopfiellamide A and B, antimicrobial pyrolidinone derivatives from marine fungus Zopfiella latipes. Tetrahedron. 2002;58(39):7781–7784. [Google Scholar]
- 65.Cueto M, Jensen PR, Kauffman C, Fenical W, Lobkovsky E, Clardy J. Pestalone, a new antibiotic produced by a marine fungus in response to bacterial challenge. J Nat Prod. 2001;64(11):1444–1446. doi: 10.1021/np0102713. [DOI] [PubMed] [Google Scholar]
- 66.Chinworrungsee M, Kittakoop P, Isaka M, Rungrod A, Tanticharoen M, Thebtaranonthy Y. Antimalarial halorosellinic acid from the marine fungus Halorosellina oceanica. Bioorg Med Chem Lett. 2001;11(15):1965–1969. doi: 10.1016/s0960-894x(01)00327-4. [DOI] [PubMed] [Google Scholar]
- 67.Garo E, Starks CM, Jensen PR, Fenical W, Lobkovsky E, Clardy J. Tricodermamides A and B, cytotoxic modified dipeptides from the marine-derived fungus Trichoderma virens. J Nat Prod. 2003;66(3):423–426. doi: 10.1021/np0204390. [DOI] [PubMed] [Google Scholar]
- 68.Malmstrom J, Christophersen C, Barrero AF, Oltra JE, Justicia J, Rosales A. Bioactive metabolites from a marine-derived strain of fungus Emericella variecolor. J Nat Prod. 2002;65(3):364–367. doi: 10.1021/np0103214. [DOI] [PubMed] [Google Scholar]
- 69.Nagai K, Kamigiri K, Matsumoto H, Kawano Y, Yamaoka M, Shimoi H, Watanabe M, Suzuki K. YM-202204, a new antifungal antibiotic produced by marine fungus Phoma sp. J Antibiot. 2002;55(12):1036–1041. doi: 10.7164/antibiotics.55.1036. [DOI] [PubMed] [Google Scholar]
- 70•.Fenical W. Chemical studies of marine bacteria: Developing a new resource. Chem Rev. 1993;93:1673–1683. Chemical approaches to isolate marine microorganisms are described. [Google Scholar]
- 71••.Marwick JD, Wright PC, Burgess JG. Bioprocess intensification for production of novel marine bacterial antibiotics through bioreactor operation and design. Mar Biotechnol. 1999;1(5):495–507. doi: 10.1007/pl00011806. This review describes the design requirements for a bioreactor for the production of antibiotics from marine bacteria. [DOI] [PubMed] [Google Scholar]
- 72.Parekh S, Vinci VA, Strobel RJ. Improvement of microbial strains and fermentation processes. Appl Microbiol Biotechnol. 2000;54(3):287–301. doi: 10.1007/s002530000403. [DOI] [PubMed] [Google Scholar]
- 73.Paradkar AS, Mosher RH, Anders C, Griffin A, Griffin J, Hughes C, Greaves P, Barton B, Jensen E. Applications of gene replacement technology to Streptomyces clavuligerus strain development for clavulanic acid production. Appl Environ Microbiol. 2001;67(5):2292–2297. doi: 10.1128/AEM.67.5.2292-2297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu H, Ochi K. Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistant mutations. Appl Environ Microbiol. 2001;67(4):1885–1892. doi: 10.1128/AEM.67.4.1885-1892.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Butler MJ, Takano E, Bruheim P, Jovetic S, Marinelli F, Bibb MJ. Deletion of scbA enhances antibiotic production in Streptomyces lividans. Appl Microbiol Biotechnol. 2003;61(5–6):512–516. doi: 10.1007/s00253-003-1277-8. [DOI] [PubMed] [Google Scholar]
- 76.Butler MJ, Bruheim P, Jovetic S, Marinelli F, Postma PW, Bibb MJ. Engineering of primary carbon metabolism for improved antibiotic production in Streptomyces lividans. Appl Environ Microbiol. 2002;68(10):4731–4739. doi: 10.1128/AEM.68.10.4731-4739.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Agust PR, Yu TW, Floss HG. Molecular biological aspect of antibiotic biosynthesis. In: Thiericke Grabley S., editor. Drug Discovery from Nature. Springer-Verlag Telos; Heidelberg, Germany: 2000. pp. 215–230. [Google Scholar]


