Abstract
Pure natural products isolated from marine sponges, algae, and cyanobacteria were examined for antioxidant activity using a 2,2-diphenyl-1-picrylhydrazyl radical (DPPH) solution-based chemical assay and a 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA) cellular-based assay. The DCFH system detects only antioxidants that penetrate cellular membranes. Potent antioxidants were identified and the results from each system compared. The algal metabolites cymopol (1), avrainvilleol (3), and fragilamide (4), and the invertebrate constituent puupehenone (5) showed strong antioxidant activity in both systems. Several compounds were active in the DPPH assay but significantly less active in the DCFH system. The green algal metabolite 7-hydroxycymopol (2) was isolated from Cymopolia barbata and its structure determined. Compound 2 was significantly less active in the DCFH system than cymopol (1). The sponge metabolites (1S)-(+)-curcuphenol (6), aaptamine (7), isoaaptamine (8), and curcudiol (9) and the cyanobacterial pigment scytonemin (10) showed strong antioxidant activity in the DPPH assay, but were relatively inactive in the DCFH system. Thus, cellular uptake dramatically affects the potential significance of antioxidants discovered using only the DPPH assay. The apparent “proantioxidants” hormothamnione A diacetate (11) and Laurencia monomer diacetate (12) require metabolic activation for antioxidant activity. Significant advantages are achieved using both a solution- and cellular-based assay to discover new antioxidants.
Reactive oxygen species (ROS) and oxidative stress play an important role in the etiology and progression of major human degenerative diseases.1 This realization has sparked great interest in substances that act as endogenous and exogenous antioxidants. Many solution-based chemical antioxidant assay systems, such as the 2,2-diphenyl-1-picrylhydrazyl radical (DPPH•) assay, have been reported.2 However, it is also important to evaluate the effects of antioxidants within living cells. Fluorescent technology has made it possible to evaluate antioxidants in living cells using specific probes such as 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA).3 This cell-based fluorescent method is useful to directly examine the ability of natural products to penetrate cell membranes and inhibit ROS in living human cells.
Marine organisms have proven to be rich sources of structurally novel and biologically active secondary metabolites.4 These natural products have served as important chemical prototypes for the discovery of new agents for use in the treatment of disease,5 probes in molecular pharmacology,6 and agrochemicals for pest management.7 However, relatively few attempts have been made to explore this vast resource of structurally unique chemistry for new antioxidant prototypes. Marine antioxidant research has largely focused on the antioxidant effects of crude extracts.8 The previously characterized marine antioxidant substances are mainly chemicals that are structurally related to plant-derived antioxidants.9 These marine antioxidants include pigments such as chlorophylls10 and carotenoids11 and tocopherol derivatives such as vitamin E and related isoprenoids.12 Alternatively, certain phenolic substances produced by marine algae13 and UV-absorbing mycosporine-like amino acids found in marine microalgae and invertebrates have been shown to have antioxidant activity.14
Many studies of plant-derived antioxidants have examined the reduction potential or radical-scavenging effects of natural products in solution-based or TLC-based DPPH autographic chemical assays.2,15 Researchers have recently begun to look at the antioxidant effects of natural products in living systems.3 A cell-based method to directly examine the ability of natural products to penetrate living human cells and inhibit reactive oxygen species (ROS)-catalyzed oxidation was used to evaluate marine natural products for their ability to scavenge exogenous ROS induced by TPA (12-O-tetradecanoylphorbol 13-acetate) in HL-60 cells.
Results and Discussion
In the present study 130 structurally diverse pure marine natural products, isolated from the research efforts of three marine natural products research groups, were assembled and evaluated in both antioxidant evaluation systems. This panel of natural products included metabolites produced by marine Chlorophyta (green algae), Cnidaria (i.e., soft corals), Chrysophyta (golden-brown algae), Cyanobacteria (blue-green algae), Mollusca (i.e., sea hares), Phaeophyta (brown algae), Porifera (sponges), and Rhodophyta (red algae). This assemblage of secondary metabolites represented a significant level of structural-chemical diversity. Examples of the substances evaluated include the following classes of marine natural products: alkaloids (aplysinopsins, araguspongines, 1H-benzo[de]1,6-naphthyridines, bromopyrroles, bromotyrosines, brominated indole alkaloids, makaluvamines), lipopeptides (cyclic and linear, malyngamides, microcolins), furocoumarins, oxylipins (eicosanoids, prostanoids, carbocyclic and heterocyclic), peptides (simple and cyclic depsipeptides), phenolics (simple, polyphenolics, polyhalogenated, styrylchromones, quinones and hyroquinones), and terpenoids (oxygenated C10–C30, polyhalogenated C10–C20, cyclic, linear, cembranoids, steroids, and terpene quinones).
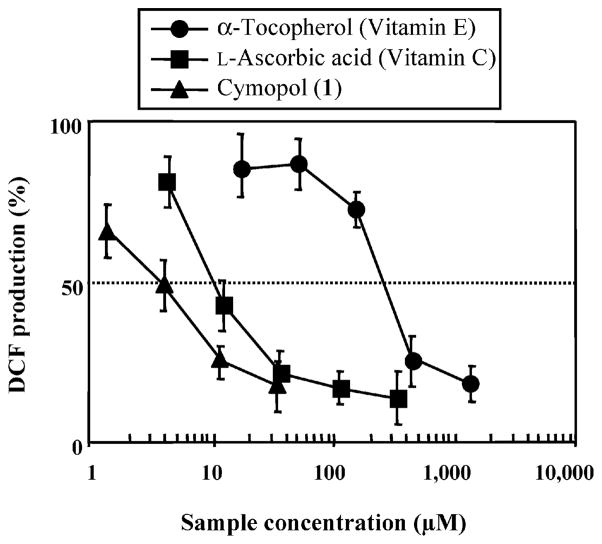
Our findings have demonstrated the potential of marine natural products to act as potent antioxidants. By using the solution-based chemical assay along with the cellular-based assay we were able to compare the results in both systems. Three interesting findings were demonstrated by this approach. Antioxidant compounds were discovered that displayed activity in both assay systems (Table 1). These include compounds such as the marine algal compounds cymopol16 (1), 7-hydroxycymopol17 (2), avrainvilleol18 (3), and fragilamide19 (4), and invertebrate metabolites such as puupehenone20 (5). These compounds not only act as antioxidants in solution-based antioxidant assays but can also be taken up by living cells and maintain their activity. Typical dose–response curves obtained for 1 and antioxidant standards α-tocopherol and ascorbic acid on TPA-stimulated hydrogen peroxide DCFH-DA oxidation in HL-60 cells are shown in Figure 1.
Table 1.
Marine Natural Products Found to Have Antioxidant Activity
| compound | sources (ref) | DPPHa activity | DCFH-DAb IC50 (μM) |
|---|---|---|---|
| cymopol (1) | Cymopolia barbata16 | strong | 4.0 |
| 7-hydroxycymopol (2) | Cymopolia barbata17 | strong | >14.6 |
| avrainvilleol (3) | Avrainvillia spp.18 | strong | 6.1 |
| aragilamide (4) | Martensia fragilis19 | moderate | 11 |
| puupehenone (5) | Hyrtios spp.,20 and other species | strong | 27 |
| (1S)-(+)-curcuphenol (6) | Didiscus spp.,22 and other species | moderate | 209 |
| aaptamine (7) | Aaptos aaptos,23 and other species | strong | >55 |
| isoaaptamine (8) | Aaptos aaptos,24 and other species | strong | >55 |
| (1S)-curcudiol (9) | Didiscus spp.,22 and other species | moderate | not active |
| scytonemin (10) | Scytonema spp.,25 and other species | moderate | >23 |
| hormothamnione diacetate (11) | Chrysophaeum taylori26 | not active | 18.3 |
| Laurencia monomer diacetate (12) | Laurencia spectabolis27 | not active | 49 |
| hormothamnione (13) | Chrysophaeum taylori28 | moderate | >31 |
| carotene (α,β-mixture) | weak | >58 | |
| α-tocopherol (vitamin E) | strong | 255 | |
| ascorbic acid (vitamin C) | strong | 9.7 |
TLC-based 1,1-diphenyl-2-picrylhydrazyl radical scavenger antioxidant assay.
Cell-based 2′,7′-dichlorodihydrofluorescein diacetate antioxidant bioassay.
Figure 1.
Inhibition of cymopol (1), L-ascorbic acid, and α-tocopherol on DCFH-DA oxidation by TPA-stimulated hydrogen peroxide in HL-60 cells. The data are presented as percentages of inhibitions based upon measurement of fluorescence at 530 nm. Values are means of three independent determinations ± SD. Statistical analysis was performed using the Student’s t-test.
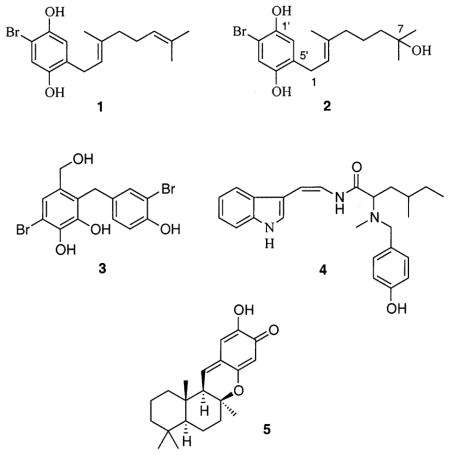
Compound 2 was isolated from a CH2Cl2–MeOH (1:1, v/v) extract of a Jamaican collection of Cymopolia barbata (L.) Lamouroux (Dasycladaceae). Compound 2 has not previously been reported to be a natural product. However, 2 has been reported as an intermediate in the synthesis of 7-hydroxycymoprolone diacetate [4′-bromo-2′,5′-diacetaoxyphenyl)-E-(2,6-dimethyl-6-hydroxyhept-2-enyl] ketone.17 Few spectroscopic data were available to positively confirm the structure of 2 from the original synthetic work. Estrada and co-workers reported only low-field strength 1H NMR data (60 or 90 MHz) and no 13C NMR spectrum from which to make an exact identification. A methoxylated analogue of 2 (3′-methoxy-7-hydroxycymopol) has recently been isolated from a Cuban collection of C. barbata.21 Since this is the first report of 2 from a natural source, the isolation, structure elucidation, and essential spectroscopic/spectrometric data are reported herein.
A detailed analysis of 13C NMR, 1H, DEPT-135, and HRESIMS data of compound 2 gave the molecular formula of C16H23BrO3 (molecular weight 342), with five degrees of unsaturation. The 13C and DEPT-135° NMR spectra indicated the presence of three methyl groups, four methylenes, three methines, and six quaternary carbons, providing a partial molecular formula of C16H20 and mass of 212 Da. The 13C and 1H NMR spectra were quite similar to those of 1 except for the methylene carbon resonance (43.27 ppm) at position C-6, the quaternary carbon resonance (71.87 ppm) at position C-7, and two fewer sp2 carbon resonances. The 1H–13C HMBC spectrum showed the connectivity of the hydroxyl-bearing carbon C-7 (71.87 ppm) to the proton resonances for the methylene at position H-6 (1.49 ppm) and the geminal methyl group resonances (1.24 ppm). The methylene carbon (43.27 ppm) at position C-6 was coupled to the methyl proton resonances (1.24 ppm) at positions H3-8 and C-7-Me and also with the methylene proton resonance (2.07 ppm) at position H-4. The 1H–1H spin system (1H–1H COSY) established the connectivity of the methylene resonances at positions H-4, H-5, and H-6. The methylene carbon (39.97 ppm) at position C-4 was coupled to the methine proton resonance (5.29 ppm) at position H-2 and methyl protons (1.72 ppm) at position C-3-Me and also with the methylene proton (1.51 ppm) at position H-5. The quaternary carbon at C-3 (138.46 ppm) showed long-range1H–13C couplings (1H–13C HMBC) with the methylene protons at positions H-1, H-4 and the protons of the methyl group attached to C-3. While 1 showed strong antioxidant activity in both assay systems, the increase in polarity associated with the 7-hydroxyl moiety significantly decreased the activity of 2 when examined using the cell-based antioxidant system.
Other marine natural products were shown to be active in the chemical assay, but had no significant activity inside cells. These include the sponge metabolites (1S)-(+)-curcuphenol22 (6), aaptamine23 (7), isoaaptamine24 (8), and curcudiol22 (9) and the cyanobacterial UV-sunscreen pigment known as scytonemin25 (10). This suggests that these compounds do not enter the cells due to poor cellular uptake or reduced medium solubility, or perhaps lack the capacity to quench 2′,7′-dichlorofluorescein (DCF) fluorescence inside the cell.
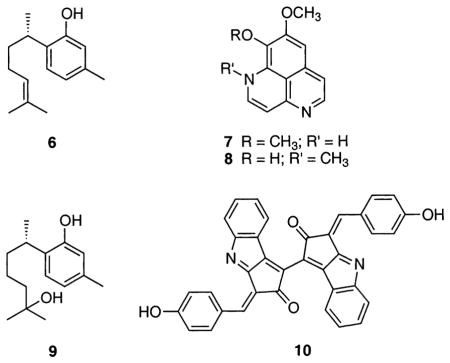
Finally, several semisynthetic marine natural product derivatives showed no activity in the solution-based assay, but showed good activity in the cellular-based assay. These include the acetylated semisynthetic styrylchromone derivative hormothamnione A diacetate26 (11) and the acetylated red algal metabolite simply referred to as Laurencia monomer diacetate27 (12). This apparent “cellular activation” is most likely due to the presence of acetylated hydroxyl groups, which upon uptake by the cell, are hydrolyzed by cellular esterases to yield free hydroxyl derivatives, which can serve as proton donors. The natural product hormothamnione A (free phenol-OH groups)28 (13), isolated from the marine chrysophyte Chrysophaeum taylori, was also evaluated in both systems. Hormothamnione A (13) displayed only mild antioxidant activity in the solution-based chemical assay, but did not show significant activity inside the cells. It is believed that hormothamnione A diacetate (11) and the Laurencia monomer diacetate (12) act as metabolically activated antioxidants or “proantioxidants” and are more lipophilic and better absorbed into living cells.
Experimental Section
General Experimental Procedures
The UV spectrum was obtained using a Hewlett-Packard 8453 spectrophotometer. The IR spectrum was obtained using an AATI Mattson Genesis Series FTIR. NMR spectra (1H, 13C, COSY, HMQC, and HMBC) of 2 were recorded in CDCl3 on a Bruker DRX 500 spectrometer operating at 500 MHz for 1H and 125 MHz for 13C, running gradients and using residual solvent peaks as internal references. Structures of other marine natural products examined were either determined as part of the original referenced research or confirmed by matching NMR spectra and MS data with that of authentic standards. The HREIMS data were acquired on a Finnigan-MAT 95 mass spectrometer (University of Minnesota Department of Chemistry, MS Service Laboratory), and the HRESIMS data were acquired on a Bruker BioAPEX 30es instrument (NCNPR, University of Mississippi). DPPH (1,1-diphenyl-2-picrylhydrazyl) was purchased from Sigma Chemical Co.
Algal Material
The marine green alga C. barbata (DNJ.074) was collected at Drax Cove, Jamaica, in July 1998. This sample was frozen and shipped to our (D.G.N.) laboratory storage facility (−20 °C) for storage prior to extraction. A voucher sample was placed in the UM Herbarium (Department of Biology, University of Mississippi).
Extraction and Isolation of 7-Hydroxycymopol (2) from Cymopolia barbata
Frozen C. barbata was lyophilized (286.34 g) and exhaustively extracted with 50% CH2Cl2 in MeOH (v/v) and dried under vacuum to yield 22.38 g of lipid extract. Analysis by 2D-TLC revealed this extract to be chemically rich with a variety of UV-absorbing secondary metabolites that produced charred products upon treatment with ethanolic H2SO4 (heat). A portion of crude lipid extract (5.90 g) was fractionated by Si gel vacuum-liquid chromatography with a hexanes–EtOAc–MeOH gradient. The column fraction (300 mg) that eluted with 60% EtOAc in hexanes from the Si gel vacuum-liquid chromatography was separated by Sephadex LH-20 chromatography [50% CH2Cl2 in MeOH (v/v)]. Fractions 15–18 were combined (110 mg), and a portion (11 mg) was chromatographed by NP-HPLC (Prodigy Si gel, 5 μm, 4.6 × 250 mm, 30% EtOAc in hexanes (v/v), 2.0 mL/min, photodiode array detection monitored at 254 nm) to yield 2 (6.6 mg, 1.1% yield).
7-Hydroxycymopol (2)
oil; UV (MeOH) λmax (log ε) 296 (3.50) nm; IR (film) νmax 2915, 1705, 1595, 1490, 1203 cm−1; 1H NMR (CDCl3, 500 MHz) δ 1.24 (6H, s, C-8 and C-7-Me), 1.49 (2H, m, H-6), 1.51 (2H, m, H-5), 1.72 (3H, s, C-3-Me), 2.07 (2H, m, H-4), 3.28 (2H, d, J = 7.2 Hz, H-1), 5.29 (1H, t, J = 7.2 Hz, H-2), 6.81 (1H, s, H-6′), 6.94 (1H, s, H-3′); 13C NMR (CDCl3, 125 MHz) δ 16.34 (CH3, C-3-Me), 22.61 (CH2, C-5), 29.06 (CH2, C-1), 29.36 (C, C-7-Me), 29.36 (C, C-8), 39.97 (CH2, C-4), 43.27 (CH2, C-6), 71.87 (C, C-7), 106.88 (C, C2′), 116.94 (CH, C-6′), 118.85 (CH, C-3′), 121.55 (CH, C-3′), 129.04 (C, C-5′), 138.46 (C, C-3), 146.51 (C, C-1′), 148.42 (C, C-4′); HRESIMS m/z 341.0728 [M – H]− (calcd for C16H2BrO3, 341.0753).
TLC Autographic Assay for DPPH Radical-Scavenging Effect
Si gel GF plates (10 × 20 cm; 250 μm; Uniplate) were scored to create small individual squares that could accommodate up to 96 samples of test compounds prepared in 96-well microplates. Pure compounds (prepared in 96-well plates) were dissolved in DMSO at a concentration of 2.0 mg/mL, 4.0 μg of each was applied in the form of a spot (4–5 mm in diameter) using a multichannel pipet, and the residual DMSO was removed under a vacuum (15–20 min). The radical-scavenging effects of the marine natural products selected were detected on the TLC plates, using a spray reagent composed of a 0.2% (w/v) solution of 1,1-diphenyl-2-picrylhydrazyl radical (DPPH) in MeOH.9,10 Plates were observed 30 min after spraying. Active compounds are observed as yellow spots against a purple background. Relative radical-scavenging activity was assigned as “strong” (compounds that produced an intense bright yellow zone), “medium” (compounds that produced a clear yellow spot), “weak” (compounds that produce only a weakly visible yellow spot), or “not active” (compounds that produced no sign of any yellow spot).
Microplate Assay for the Detection of Oxidative Products with DCFH-DA
This method is based on a fluorimetric assay described by Rosenkranz and co-workers.3b Promyelocytic HL-60 cells (1 × 106 cells/mL, ATCC) were suspended in RPMI 1640 medium with 10% FBS and antibiotics at 37 °C in 5% CO2–95% air. A 125 μL aliquot of the cell suspension was added to a well of a 96-well plate. After treatment with different concentrations of the test materials for 30 min, cells were stimulated with 100 ng/mL 12-O-tetradecanoylphorbol 13-acetate (TPA, Sigma) for 30 min. Then 2′,7′-dichlorodihydrofluorescein diacetate (5 μg/mL) (DCFH-DA, Molecular Probes) was added to the cells, and they were incubated for another 15 min. DCFH-DA is a nonfluorescent probe that diffuses into cells. Cytoplasmic esterases hydrolyze DCFH-DA to 2′,7′-dichlorodihydrofluorescein (DCFH). Reactive oxygen species (ROS) generated within HL-60 cells oxidize DCFH to the fluorescent dye 2′,7′-dichlorofluorescein (DCF). The ability of the test materials to inhibit exogenous cytoplasmic ROS-catalyzed oxidation of DCFH in HL-60 cells was measured by TPA-treated control incubations with and without the test materials. Levels of DCF were measured using a CytoFluor 2350 Fluorescence Measurement System (Millipore) with excitation wavelength at 485 nm (bandwidth 20 nm) and emission at 530 nm (bandwidth 25 nm).
Acknowledgments
The Natural Resource Conservation Authority, Jamaica, and Discovery Bay Marine Laboratory are gratefully acknowledged for assistance with sample collections. This work was supported in part by the National Center for Natural Products Research (NCNPR) and Research Institute of Pharmaceutical Sciences (RIPS) [University of Mississippi], and the USDA Agricultural Research Service Specific Cooperative Agreement # 58-6408-2-0009. Efforts at OSU were supported by NIH CA52955.
References and Notes
- 1.(a) Breimer LH. Mol Carcinog. 1990;3:188–197. doi: 10.1002/mc.2940030405. [DOI] [PubMed] [Google Scholar]; (b) Cerutti PA. Science. 1985;227:375–380. doi: 10.1126/science.2981433. [DOI] [PubMed] [Google Scholar]
- 2.(a) Brand-Williams W, Cuvelier ME, Berset C. Food Sci Technol. 1995;28:25–30. [Google Scholar]; (b) Cuendet M, Hostettmann K, Potterat O, Dyatmyko W. Helv Chim Acta. 1997;80:1144–1152. [Google Scholar]; (c) Calis I, Kuruuzum A, Demirezer OL, Sticher O, Ganci W, Ruedi P. J Nat Prod. 1999;62:1101–1105. doi: 10.1021/np9900674. [DOI] [PubMed] [Google Scholar]
- 3.(a) Bass DA, Parce JW, Dechatelet LR, Szejda P, Seeds MC, Thomas M. J Immunol. 1983;130:1910–1917. [PubMed] [Google Scholar]; (b) Rosenkranz AR, Schmaldienst S, Stuhlmeier KM, Chen W, Knapp W, Zlabinger GJ. J Immunol Methods. 1992;156:39–45. doi: 10.1016/0022-1759(92)90008-h. [DOI] [PubMed] [Google Scholar]; (c) Lee SK, Qing WG, Mar W, Luyengi L, Mehta RG, Kawanishi K, Fong HHS, Beecher CWW, Kinghorn AD, Pezzuto JM. J Biol Chem. 1998;273:19829–19833. doi: 10.1074/jbc.273.31.19829. [DOI] [PubMed] [Google Scholar]; (d) Chung HS, Chang LC, Lee SK, Shamon LA, van Breemen RB, Mehta RG, Farnsworth NR, Pezzuto JM, Kinghorn AD. J Agric Food Chem. 1999;47:36–41. doi: 10.1021/jf980784o. [DOI] [PubMed] [Google Scholar]; (e) Rajbhandari I, Takamatsu S, Nagle DG. J Nat Prod. 2001;64:693–695. doi: 10.1021/np0101346. [DOI] [PubMed] [Google Scholar]; (f) Bedir E, Tatli II, Khan RA, Zhao J, Takamatsu S, Walker LA, Goldman P, Khan I. J Agric Food Chem. 2002;50:3150–3155. doi: 10.1021/jf011682s. [DOI] [PubMed] [Google Scholar]
- 4.Capon RJ. Eur J Org Chem. 2001:633–645.and references therein. Scheuer PJ. J Chem Educ. 1999;76:1075–1079.and references therein. Cragg GM, Newman DJ, Snader KM. J Nat Prod. 1997;60:52–60. doi: 10.1021/np9604893.
- 5.Blunden G. In: Pharmacognosy. 15. Evans WC, editor. W. B. Saunders; New York: 2002. pp. 115–124. and references therein Fusetani N, editor. Drugs from the Sea. S. Karger AG; New York: 2000. p. 158.and references therein.
- 6.(a) Tremblay JF. Chem Eng News. 2000 Jan 3;:14–15. [Google Scholar]; (b) Coxon A, Bestor TH. Chem Biol. 1995;2:119–121. doi: 10.1016/1074-5521(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 7.El Sayed KA, Dunbar DC, Perry TL, Wilkins SP, Hamann MT. J Agric Food Chem. 1997;45:2735–2739.and references therein. Schrader KK, Nagle DG, Wedge DE. In: Advances in Microbial Toxin Research and its Biotechnological Exploitation. Upadhyay RK, editor. Kluwer Acedemic/Plenum; New York: 2002. pp. 171–195.and references therein.
- 8.(a) Nakagawa H, Murata M, Tachibana K, Shiba T. Phycol Res. 1998;46(Suppl):9–12. [Google Scholar]; (b) Guzmán S, Gato A, Calleja JM. Phytother Res. 2001;15:224–230. doi: 10.1002/ptr.715. [DOI] [PubMed] [Google Scholar]
- 9.(a) Potterat O. Curr Org Chem. 1997;1:415–440. [Google Scholar]; (b) Son BW, Kim JC, Choi HD, Kang JS. Arch Pharm Res. 2002;25:77–79. doi: 10.1007/BF02975266. [DOI] [PubMed] [Google Scholar]
- 10.(a) Watanabe N, Yamamoto K, Ihshikawa H, Yagi A, Sakata K, Brinen LS, Clardy J. J Nat Prod. 1993;56:305–317. [Google Scholar]; (b) Cahyana AH, Shuto Y, Kinoshita Y. Biosci Biotech Biochem. 1992;56:1533–1535. doi: 10.1271/bbb.56.958. [DOI] [PubMed] [Google Scholar]; (c) Yamamoto K, Sakata K, Watanabe N, Yagi A, Brinen LS, Clardy J. Tetrahedron Lett. 1992;33:2587–2588. [Google Scholar]
- 11.(a) Choi JS, Lee WK, Cho YJ, Kim DS, Kim A, Chung HY, Jung JH, Im KS, Choi WC, Choi HD, Son BW. Nat Prod Sci. 2000;6:122–125. [Google Scholar]; (b) Miki W, Otaki N, Yokoyama A, Izumida H, Shimidzu N. Experientia. 1994;50:684–686. [Google Scholar]; (c) Miki W. Pure Appl Chem. 1991;63:141–146. [Google Scholar]
- 12.(a) Kobayashi MJ. Chem Res (S) 1994:494–495. [Google Scholar]; (b) Yamamoto Y, Fujisawa A, Hara A, Dunlap WC. Proc Natl Acad Sci USA. 2001;98:13144–13148. doi: 10.1073/pnas.241024298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujimoto K, Kaneda T. Hydrobiologia. 1984;116/117:111–113. [Google Scholar]
- 14.(a) Dunlap WC, Shick JM, Yamamoto Y. Redox Rep. 1999;4:301–306. doi: 10.1179/135100099101535142. [DOI] [PubMed] [Google Scholar]; (b) Dunlap WC, Yamamoto Y. Comp Biochem Physiol. 1995;112B:105–114. [Google Scholar]
- 15.(a) Cavin A, Potterat O, Wolfender JL, Hostettmann K, Dyatmyko W. J Nat Prod. 1998;61:1497–1501. doi: 10.1021/np980203p. [DOI] [PubMed] [Google Scholar]; (b) Masuda T, Matsumura H, Oyama Y, Takeda Y, Jitoe A, Kida A, Hidaka K. J Nat Prod. 1998;61:609–613. doi: 10.1021/np970555g. [DOI] [PubMed] [Google Scholar]; (c) Hwang BY, Kim HS, Lee JH, Hong YS, Ro JS, Lee KS, Lee JJ. J Nat Prod. 2001;64:82–84. doi: 10.1021/np000251l. [DOI] [PubMed] [Google Scholar]; (d) Ohtani II, Gotoh N, Tanaka J, Higa T, Gyamfi MA, Aniya Y. J Nat Prod. 2000;63:676–679. doi: 10.1021/np990396w. [DOI] [PubMed] [Google Scholar]; (e) Westenburg HE, Lee KJ, Lee SK, Fong HHS, van Breemen RB, Pezzuto JM, Kinghorn AD. J Nat Prod. 2000;63:1696–1698. doi: 10.1021/np000292h. [DOI] [PubMed] [Google Scholar]
- 16.Högberg H-E, Thompson RH, King TJ. J Chem Soc, Perkin Trans 1. 1976:1697–1701. [Google Scholar]
- 17.Estrada DM, Martin JD, Pérez C. J Nat Prod. 1987;50:735–737. [Google Scholar]
- 18.Sun HH, Paul VJ, Fenical W. Phytochemistry. 1983;22:743–745. [Google Scholar]
- 19.Kirkup MP, Moore RE. Tetrahedron Lett. 1983;24:2087–2090. [Google Scholar]
- 20.Ravi BN, Perzanowski HP, Ross RA, Erdman TR, Scheuer PJ, Finer J, Clardy J. Pure Appl Chem. 1979;51:1893–1900. [Google Scholar]
- 21.Dorta E, Darias J, San Martín A, Cueto M. J Nat Prod. 2002;65:329–333. doi: 10.1021/np010418q. [DOI] [PubMed] [Google Scholar]
- 22.(a) McEnroe FJ, Fenical W. J Nat Prod. 1978;34:1661–1664. [Google Scholar]; (b) Wright AE, Pomponi SA, McConnell OJ, Komoto S, McCarthy PJ. J Nat Prod. 1987;50:976–978. [Google Scholar]; (c) El Sayed KA, Yousaf M, Hamann MT, Avery MA, Kelly M, Wipf P. J Nat Prod. 2002;65:1547–1553. doi: 10.1021/np020213x. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura H, Kobayashi J, Ohizumi Y, Hirata Y. Tetrahedron Lett. 1982;23:5555–5558. [Google Scholar]
- 24.Shen YC, Lin TT, Sheu JH, Duh CY. J Nat Prod. 1999;62:1264–1267. doi: 10.1021/np990156g. [DOI] [PubMed] [Google Scholar]
- 25.Proteau P, Gerwick WH, Garcia-Pichel F, Castenholz R. Experientia. 1993;49:825–829. doi: 10.1007/BF01923559. [DOI] [PubMed] [Google Scholar]
- 26.(a) Gerwick WH, Lopez A, van Duyne GD, Clardy J, Ortiz W, Baez A. Tetrahedron Lett. 1986;27:1979–1982. [Google Scholar]; (b) Ayyangar NR, Khan RA, Deshpande VH. Tetrahedron Lett. 1988;29:2347–2348. [Google Scholar]
- 27.Bernart MW, Gerwick WH, Corcoran EE, Lee AY, Clardy J. Phytochemistry. 1992;31:1273–1276. [Google Scholar]
- 28.Gerwick WH, Lopez A, van Duyne GD, Clardy J, Ortiz W, Baez A. Tetrahedron Lett. 1986;27:1979–1982. [Google Scholar]