Abstract
A scientific milestone that has tremendously impacted the cardiac research field has been the discovery and establishment of human-induced pluripotent stem cells (hiPSC). Key to this discovery has been uncovering a viable path in generating human patient and disease-specific cardiac cells to dynamically model and study human cardiac diseases in an in vitro setting. Recent studies have demonstrated that hiPSC-derived cardiomyocytes can be used to model and recapitulate various known disease features in hearts of patient donors harboring genetic-based cardiac diseases. Experimental drugs have also been tested in this setting and shown to alleviate disease phenotypes in hiPSC-derived cardiomyocytes, further paving the way for therapeutic interventions for cardiac disease. Here, we review state-of-the-art methods to generate high-quality hiPSC and differentiate them towards cardiomyocytes as well as the full range of genetic-based cardiac diseases, which have been modeled using hiPSC. We also provide future perspectives on exploiting the potential of hiPSC to compliment existing studies and gain new insights into the mechanisms underlying cardiac disease.
Introduction
Despite important achievements over the last decade, heart disease remains the principal cause of death in developed societies. Intense research efforts have been directed at using human pluripotent stem cells (hiPSC) to invoke cardiac regeneration for heart repair and to model human cardiac development and diseases in vitro (Davis et al., 2011). hPSCs, which include both human-induced pluripotent stem cells (hiPSC) and embryonic stem cells (hESC), have the potential to differentiate into a variety of cell types, including cardiomyocytes. Seminal work using hESC demonstrated that by mimicking the cell-signaling environment during early stages of cardiogenesis, it is possible to differentiate hESC towards beating cardiomyocytes (Mummery et al., 2012). The contribution of hESC research to the cardiac field has been invaluable and their potential for addressing questions of a developmental nature is unmatched; however, their use for modeling human genetic-based diseases has certain limitations. hESC lines are typically isolated from the progeny of people who have no known genetic diseases, and as such genetic manipulation of the genome is required to introduce known mutations causative for specific diseases in order to emulate the genetic defect. In contrast, hiPSC offer unique advantages in that they are patient and disease specific, since they are generated through genetic reprogramming of an affected donor’s cells (dermal skin fibroblasts (Takahashi et al., 2007), adipocytes (Sugii et al., 2010), nucleated blood cells (Loh et al., 2009), and dental pulp (Yan et al., 2010)). In addition to carrying the genetic mutation of interest, these somatic cells can be isolated using relatively non-invasive procedures. Use of hiPSC also circumvents the ethical concerns associated with the use of cells from an embryonic origin. iPSC technology has been rapidly exploited by the biomedical research community as an invaluable tool to recapitulate human disease in a cell culture setting as a means to gain new understanding of underlying molecular mechanisms (Ebert et al., 2009; Israel et al., 2012; Marchetto et al., 2010; Soldner et al., 2009), and more recently the cardiac research field has begun to explore its potential (Davis et al., 2011; Hoekstra et al., 2012; Musunuru et al., 2010). To date, iPSC technology has been used to model several human genetic-based cardiac diseases and systemic diseases involving cardiac defects, which include LEOPARD Syndrome (Carvajal-Vergara et al., 2010), Long QT syndrome (Itzhaki et al., 2011; Lahti et al., 2012; Matsa et al., 2011; Moretti et al., 2010), Timothy Syndrome (Yazawa et al., 2011), Catecholami-nergic Polymorphic Ventricular Tachycardia (Fatima et al., 2011; Itzhaki et al., 2012; Jung et al., 2012; Kujala et al., 2012), familial Dilated Cardiomyopathy (Sun et al., 2012), Arrhythmogenic Cardiomyopathy (Kim et al., 2013; Ma et al., 2012), as well as an overlapping syndrome of a cardiac Na+ channel disease (Davis et al., 2012). Here, we discuss available methods to generate high-quality hiPSC and successfully differentiate them towards the cardiac lineage. We also review recent studies, which have modeled genetic-based cardiac diseases arising from structural, signaling, and electrophysiological abnormalities using iPSC technology, as well as provide perspectives on their contributions to the cardiac field.
iPSC generation
Classic hiPSC reprogramming methods as outlined by Yamanaka and colleagues consisted of retroviral-based gene delivery of oct4, sox2, klf4, and c-myc into human dermal fibroblasts, which triggered dramatic changes in cell morphology and behavior resembling hESC (Takahashi et al., 2007; Takahashi and Yamanaka, 2006). Validation of the pluripotent nature of hiPSC was demonstrated by their ability to express hallmark ESC-specific surface antigens and transcription factors typical of undifferentiated cells (Adewumi et al., 2007). In order to demonstrate the pluripotency of iPSC, several assays have been carried out, indicating their resemblance to hESC, which include their (i) ability to aggregate into spheroid cellular structures containing a mass of pluripotent cells resembling embryoid bodies (EB) (Takahashi et al., 2007), (ii) unguided, spontaneous differentiation of hiPSC in high serum into the three embryonic germ layers (endoderm, mesoderm, and ectoderm) (Takahashi et al., 2007; Takahashi and Yamanaka, 2006), (iii) subcutaneous implantation of hiPSC into mice (teratoma assays) resulted in tumors consisting of cell types and tissues originating from the three embryonic germ layers (Takahashi et al., 2007; Takahashi and Yamanaka, 2006), and (iv) karyotype and microarray analyses of hiPSC further demonstrated the chromosomal stability and genetic (transcript) resemblance of hiPSC to hESC, respectively (Takahashi et al., 2007; Takahashi and Yamanaka, 2006). Demethylation of pluripotency transcription factors (e.g., nanog) promoters in hiPSC also provided evidence of the epigenetic conversion of cells from a somatic to a pluripotent state (Takahashi et al., 2007). Modifications of the original method including the removal of c-myc (Li et al., 2010) and replacement of c-myc and klf4, with lin28 and nanog (Yu et al., 2007), also achieved similar results, with the possible advantage of reducing hiPSC’s oncogenic potential. The production and validation of high-quality hiPSC is critical to disease modeling. Lesser quality cells (i.e., karyotypically abnormal or that are unable to differentiate properly) could produce artifactual phenotypes that are not related to the disease under study. Therefore, iPSC lines should be routinely taken through a panel of pluripotent-based assays to ensure their bona fide status. Additionally, standard approaches towards analyzing results from hiPSC have been to obtain results from at least three independent clones to rule out the possibility that the cell behavior observed was not due to random DNA insertion events. However, rising concerns towards their potential in vivo applications has prompted refinement of hiPSC generation strategies to allow for removal of transgenes after reprogramming, or to avoid integration altogether. These novel strategies could provide an appealing alternative to disease modeling and have been discussed in detail elsewhere (Li and Izpisua Belmonte, 2012). It could also shed light on whether reprogramming through newer methods (e.g., integration-free) is an important factor in producing cells that could qualitatively lead to better disease modeling, when compared to traditional methods.
Differentiation of iPSC towards the cardiac lineage
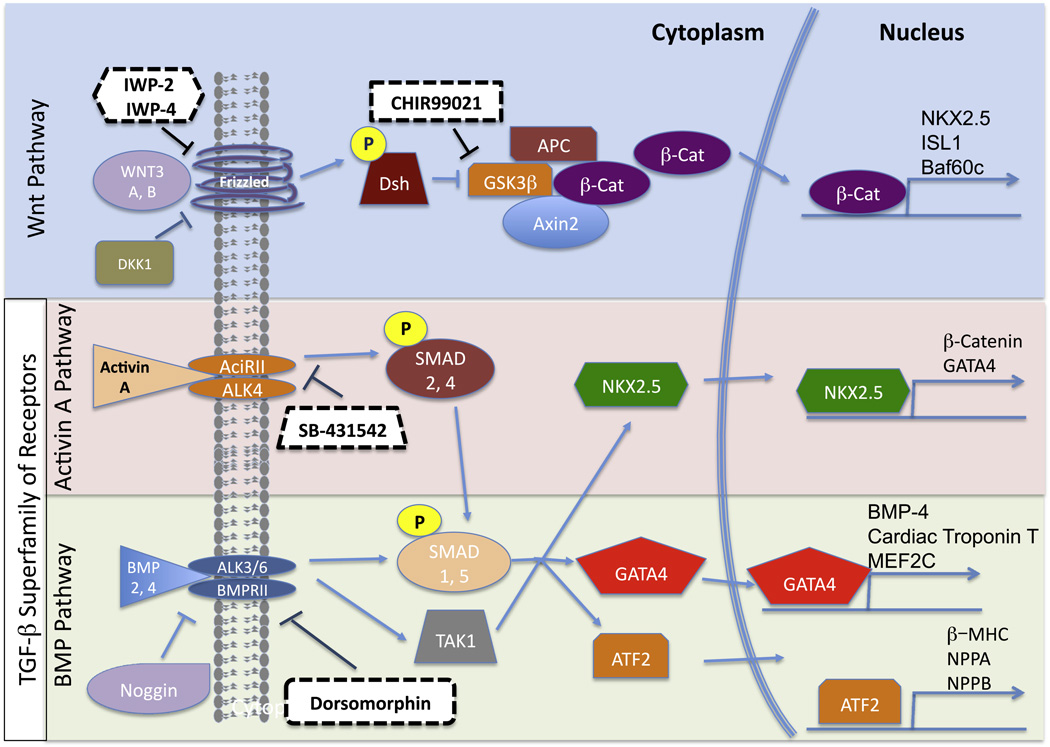
The most reproducible and efficient methods to differentiate hPSC to the cardiogenic path rely on defined and temporal manipulation of the Wnt (Wnt3a, Dickkopf-related protein (DKK)-1) and transforming growth factor (TGF)-β, (activin A, and bone morphogenetic protein (BMP)-4 and −2) signaling pathways (Kattman et al., 2011; Lian et al., 2012b; Paige et al., 2010; Zhang et al., 2012) (Fig. 1). These pathways are essential in the early steps of cardiac development (Davis et al., 2011) and must be utilized in a stage-specific manner to drive cardiac differentiation of hPSC (Mummery et al., 2012). The importance of these pathways is further highlighted as small molecules that inhibit specific downstream targets of these pathways can be used as an alternative to efficiently drive cardiac differentiation of hPSC (Hao et al., 2008; Kattman et al., 2011; Lian et al., 2012a; Minami et al., 2012; Willems et al., 2011) (Fig. 1). A number of cardiac differentiation protocols for hPSC are available, and their advantages and disadvantages have been discussed in greater detail elsewhere (Mummery et al., 2012). For simplicity, we broadly categorize these protocols into monolayer (Lian et al., 2012a; Paige et al., 2010; Zhang et al., 2012), embryoid body (Kattman et al., 2011; Kehat et al., 2001), and “co-culture” (Mummery et al., 2007) strategies, while providing details of efficiencies as well as chemically defined and undefined conditions to drive hPSC towards a cardiogenic path (Table 1). Key to the successful use of these approaches is the need to optimize doses of chemicals used for each cell line (Kattman et al., 2011; Lian et al., 2012b; Mummery et al., 2012; Paige et al., 2010), as well as the growth kinetics and seeding densities of cells prior to EB formation or monolayer differentiation (Lian et al., 2012b). Thus, future studies detailing how these optimizations can be reproducibly achieved will propel the field forward. Importantly, current studies have been based on PSC-derived cardiomyocytes presenting structural and biochemical features reminiscent of early human fetal car-diomyocytes (Beqqali et al., 2006), indicative of immaturity. As a result, this may limit their ability to recapitulate certain aspects of late onset or end-stage disease, where mature and junctionally connected cardiomyocytes are required (Davis et al., 2011). This may be especially pertinent for (i) cardiac channelopathies since the expression and function of various channels (e.g., hERG) are influenced by cardiomyocyte maturity (Kujala et al., 2011) and (ii) certain genetic-based cardiac diseases, such as arrhythmogenic right ventricular cardiomypathy, where the disease develops postnatally and mutations are associated with cell junctional components (Sheikh et al., 2009). Thus, generating hiPSC-derived cardiomyocytes resembling postnatal heart cardiomyocytes will be increasingly important. Interestingly, the majority of the studies that exploit hiPSC to model cardiac disease have used undefined conditions for cardiac differentiation (Table 2). Given the availability and high efficiency of defined protocols to produce cardiomyocytes, it will be of interest to note whether these approaches can more consistently reproduce the range of pathological features in these disease models.
Fig. 1.
A simplified schemata of the Wnt and TGF-β signaling pathways, which include molecular components that are targets to induce cardiac differentiation of PSC. Dashed boxes highlight small molecules that are currently used to efficiently drive cardiac differentiation of PSC.
Table 1.
Overview of the most commonly used PSC cardiac differentiation protocols, including monolayer, EB, and “co- culture” strategies. Time points at which various cytokines and small molecule inhibitors are added are denoted for each protocol along with the reported efficiencies for these protocols.
| Format | Time Points (Days) | Reported Efficiency |
Refs | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Chemically Defined |
Monolayer (2D) |
Basic Monolayer |
D-2 | D-1 | D0 | D1 | D3 | D4 | D5 | D7 | D8 | D14 | ≤90% | Paige et al., 2010 |
|
100ng/ml WNT3A* |
100ng/ml Activin A |
10ng/ml BMP-4 | 200ng/ml DKK-1* | |||||||||||
|
Matrix Sandwich |
D-2 | D-1 | D0 | D1 | D3 | D4 | D5 | D7 | D8 | D14 | ≤98% | Zhang et al., 2012 | ||
| Matrigel Overlay |
Matrigel Overlay 100ng/ml Activin A |
10ng/ml BMP-4 5ng/ml bFGF |
||||||||||||
|
Small Molecules |
D-2 | D-1 | D0 | D1 | D3 | D4 | D5 | D7 | D8 | D14 | ≤98% | Lian et al., 2012a | ||
| 12 M CHIR99021 µg/ml Insulin* |
5 µM IWP-2 or IWP-4 |
|||||||||||||
|
EB (3D) |
Staged, defined |
D-2 | D-1 | D0 | D1 | D3 | D4 | D5 | D7 | D8 | D14 | 30–60% | Kattman et al., 2011 | |
| 0.4ng/ml BMP-4 |
0–12ng/ml Activin A 2.5–20 ng/ml BMP-4 5ng/ml bFGF |
150ng/ml DKK-1 10ng/mlVEGF |
10ng/ml VEGF 5ng/ml bFGF | |||||||||||
|
5.4 M SB-431542* 0.6µM Dorsomorphin* |
||||||||||||||
| Chemically Undefined | Undefined | D-2 | D-1 | D0 | D1 | D3 | D4 | D5 | D7 | D8 | D14 | 0.1–10% | Kehat et al., 2001 | |
| 5–20% FBS or KSR | ||||||||||||||
| Co-culture(2D) | D-2 | D-1 | D0 | D1 | D3 | D4 | D5 | D7 | D8 | D14 | ≤25% | Mummery et al, 2007 | ||
| END2 conditioned medium | ||||||||||||||
D2 and D1 refer to days prior to differentiation. D0-D14 refer to days at time of differentiation.
FBS: Fetal Bovine Serum. KSR: Knockout Serum replacement.
Refers to reported modifications of the original protocols aimed to enhance the cardiogenic potential of PSC.
Table 2.
Overview of current iPSC models of genetic-based cardiac disease including the genetic mutation responsible, the reprogramming and differentiation methods applied, and disease phenotypes observed.
| Disease model | Genetic cause | Source of somatic cells |
Reprogramming strategy |
Cardiac differentiation |
Disease phenotypes observed | Rescue | |
|---|---|---|---|---|---|---|---|
| LEOPARD syndrome (Carvajal-Vergara et al., 2010) |
ptpn11 (SHP2) T468M substitution |
Skin | Retrovirus: oct4, sox2, klf4, and c- myc |
EB, staged, defined |
Increased cell size and sarcomeric organization; Nuclear localization of NFATC4 |
N/A | |
| Long QT 1 (Moretti et al., 2010) | kcnq1 (IKs) R190Q missense mutation |
Skin | Retrovirus: oct4, sox2, klf4, and c- myc |
EB, serum- based |
Catecholamine-induced tachyarrhythmia | Propanolol | |
| Long QT 2 (Itzhaki et al., 2011) | kcnh2/herg (IKr) A614V missense mutation |
Skin | Retrovirus: oct4, sox2, and klf4 |
EB, serum- based |
Prolongation of AP duration | EAD; Triggered arrhythmias |
Nifepidine, Pinacidil, and Ranolazine |
| Long QT 2 (Matsa et al., 2011) | kcnh2/herg (IKr) G1681A point mutation |
Skin | Lentivirus: oct4, sox2, nanog, and lin28 |
EB, serum- based |
Deficient IKr activity | Arrhythmias and EAD triggered by E4031 and isoproterenol |
Propanolol, Nadalol, Nicorandil, and PD118057 |
| Long QT 2 (Lahti et al., 2012) | kcnh2/herg (IKr) R176W missense mutation |
Skin | Retrovirus: oct4, sox2, klf4, and c- myc |
Co-culture with END2 |
Spontaneous arrhythmogenic activity, aggravated by E4031 and sotalol |
N/A | |
| Timothy syndrome (Yazawa et al., 2011) |
cacna1c (CaV1.2) G406R missense mutation |
Skin | Retrovirus: oct4, sox2, klf4, and c- myc |
EB, serum- based |
Irregular contraction, prolonged action potentials, altered Ca2+ influx, and transients in ventricular-like cells |
Roscovitine | |
| Catecholaminergic polymorphic ventricular tachycardia (Itzhaki et al., 2012) |
RYR2 M4109R point mutation |
Skin | Retrovirus: oct4, sox2, and klf4 |
EB, serum- based |
DAD and abnormal Ca2+ handling |
Isoproterenol and forskolin increased frequency and magnitude of AD, also leading to development of triggered activity; Whole-cell Ca2+ transient irregularities, worsened with adrenergic stimulation and Ca2+ overload |
Flecainide, Thapsighargin, and Propanolol |
| Catecholaminergic polymorphic ventricular tachycardia (Fatima et al., 2011) |
ryr2 p.F2483I mutation |
Skin | Retrovirus: oct4, sox2, klf4, and c- myc |
Co-culture with END2 |
Development arrhythmogenic behaviors and DAD following isoproterenol treatment; Higher amplitude and longer duration of spontaneous local Ca2+ releasing events |
N/A | |
| Catecholaminergic polymorphic ventricular tachycardia (Jung et al., 2012) |
RYR2 S406L missense mutation |
Skin | Retrovirus: oct4, sox2, klf4, and c- myc |
EB, serum- based |
DAD and abnormal Ca2+ handling |
Prolongation of Ca2+ sparks | Dantrolene |
| Catecholaminergic polymorphic ventricular tachycardia (Kujala et al., 2012) |
RYR2 P2328S mutation |
Skin | Retrovirus: oct4, sox2, klf4, and c- myc |
Co-culture with END2 cells |
Abnormal Ca2+ signaling and arrhythmias upon catecholaminergic stress; Reduced SR Ca2+ content, indicating leakage of Ca2+ from the SR; DAD during spontaneous beating and in response to adrenaline; EAD during spontaneous beating |
N/A | |
| Familial dilated cardiomyopathy (Sun et al., 2012) |
TNNT2 (cTNT) R173W point mutation |
Adipose tissue | Retrovirus: oct4, sox2, klf4, and c- myc |
EB, staged, defined |
Abnormal sarcomeric structure and contractility, aggravated by β-adrenergic stimulation; Alterations in Ca2+ handling |
Metoprolol and forced overexpression of Serca2a |
|
| Arrhythmogenic right ventricular cardiomyopathy (Ma et al., 2012) |
plakophilin-2 c.1841T.C nucleotide change |
Skin | Retrovirus oct4, sox2, klf4, and c- myc |
EB, serum- based |
Increased lipid accumulation in AC CMs following |
Reduced plakoglobin expression. Larger and denser lipid droplets |
N/A |
| Arrhythmogenic right ventricular cardiomyopathy (Kim et al., 2013) |
plakophilin-2 homozygous c.2484C.T frameshift mutation and c.2013delC frameshift mutation |
Skin | Retrovirus oct4, sox2, klf4, and c- myc |
EB, serum- based |
adipogenic conditions |
Nuclear localization of plakoglobin, reduced β-catenin activity, and increased apoptosis. Metabolic shift from FAO towards glycolysis. Deficient Ca2+ relaxation and reduced serca2a and ncax1 expression |
Forced expression of plakophilin-2. N-acetyl cysteine and ascorbic acid; GW966, T0070907, and GW6471 |
| Overlapping Na+ channel disease syndrome (Davis et al., 2012) |
SCN5A (INa) 1795insD insertion mutation |
Skin | Floxed lentivirus: oct4, sox2, klf4, and c-myc |
Co-culture with END2 cells |
Decrease in peak INa and an increase in persistent INa | N/A | |
EAD = Early afterdepolarizations; DAD = delayed afterdepolarizations; FAO = fatty acid oxidation.
Methods to enrich for hPSC-derived cardiomyocytes
Current cardiac differentiation protocols do not induce hiPSC to differentiate into 100% cardiomyocytes, indicating that non-cardiomyocytes are also included within the pool of cells obtained (Mummery et al., 2012) (Table 1). Hence, both genetic and non-genetic approaches have been used to purify hPSC-derived cardiomyocytes to circumvent limitations in analyzing cardiac muscles within a mixed pool of cells within the context of disease models. Targeting the enhanced green fluorescent protein into the cardiac transcription factor NKX2.5 locus has allowed the purification of hESC-derived cardiac progenitors and cardiomyocytes (Elliott et al., 2011). Non-genetic approaches developed for PSC-derived cardio-myocyte purification include physical separation through percoll gradients (Zhu et al., 2011), metabolic selection using lactate-rich media (Tohyama et al., 2013), and fluorescence-activated cell sorting (FACS) based on mitochondrial content (Hattori et al., 2010) and the expression of signal-regulatory protein alpha (SIRPα) (Dubois et al., 2011).
Modeling cardiac disease using hiPSC-derived cardiomyocytes—what have we learned?
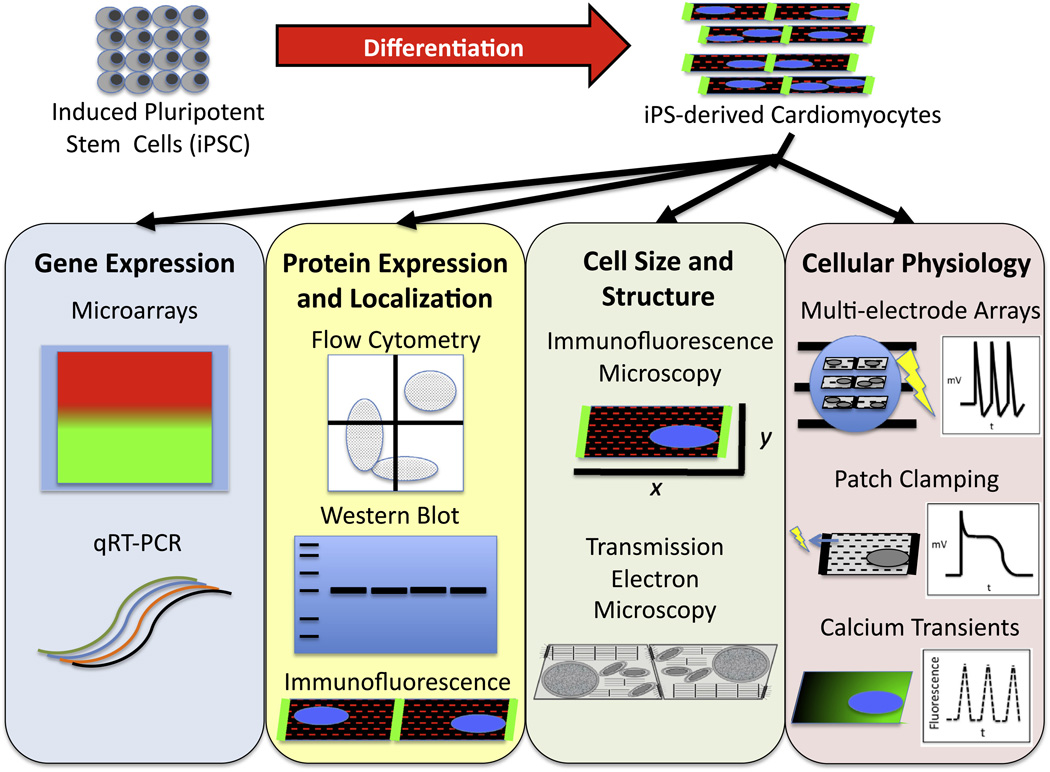
hiPSC have been exploited to model several human genetic-based cardiac diseases that manifest defects in specific cardiac structural components, signaling pathways, and electrophysiological properties (Carvajal-Vergara et al., 2010; Davis et al., 2012; Fatima et al., 2011; Ma et al., 2012; Matsa et al., 2011; Moretti et al., 2010; Sun et al., 2012; Yazawa et al., 2011). Importantly, hiPSC-derived cardiomyocytes can recapitulate specific cardiac muscle defects in vitro observed in the hearts of the same donor patients in vivo (Carvajal-Vergara et al., 2010; Davis et al., 2012; Itzhaki et al., 2011, 2012; Lahti et al., 2012; Ma et al., 2012; Matsa et al., 2011; Moretti et al., 2010; Sun et al., 2012; Yazawa et al., 2011), validating the powerful nature of the technique as an important system to gain mechanistic insights into human cardiac disease. Currently, a wide variety of molecular, cellular, and physiological assays have been adapted to investigate disease phenotypes in hiPSC-derived cardiomyocytes, which are summarized in Fig. 2. hiPSC-derived cardiomyocytes have been used to test experimental drugs, which can revert certain disease features (Itzhaki et al., 2011, 2012; Lahti et al., 2012; Matsa et al., 2011; Moretti et al., 2010; Sun et al., 2012; Yazawa et al., 2011), further setting the stage to use this human model system as a platform for experimental drug screening. Table 2 provides a comprehensive overview of the genetic-based cardiac diseases modeled using hiPSC, including specific methods used for both hiPSC generation and cardiac differentiation as well as key cellular features observed.
Fig. 2.
Schemata of molecular, cellular, and functional techniques used for analysis of disease phenotypes in hiPSC-derived cardiomyocytes.
LEOPARD syndrome
LEOPARD Syndrome (LS) is a complex systemic disease involving a number of features that form its acronym: Lentigines, Electrocardiographic abnormalities, Ocular hypertelorism, Pulmonary valve stenosis, Abnormal genitalia, Retardation of growth, and Deafness (Kontaridis et al., 2006). LS along with Noonan syndrome (NS) is a family of autosomal dominant syndromes caused by germline mutations in components of the Kirsten Murine Sarcoma virus K (KRAS)/v-raf murine sarcoma viral oncogene homolog b1 (RAF)/Mitogen-Activated Protein Kinase Kinase 1 (MEK)/Extracellular Signal-Regulated Kinase (ERK) mitogen-activating protein kinase signaling pathways (Lauriol and Kontaridis, 2011). Although LS and NS mutations result in opposing defects in signaling pathways, patients bearing these mutations display similar cardiac abnormalities (Lauriol and Kontaridis, 2011). However, the mechanisms leading to these similarities have not been completely elucidated. Approximately 85% of LEOPARD Syndrome patients manifest heart defects with the most common life-threatening cardiac manifestation resulting in hypertrophic cardiomyopathy (Gelb and Tartaglia, 2007; Sarkozy et al., 2008). To provide deeper insights into LS, a human hiPSC model was developed using fibroblasts from an affected individual carrying a mutation in ptpn11, which causes a substitution (T468M) in the tyrosine phosphatase SHP2. An unaffected relative donated the control cells used in the study (Carvajal-Vergara et al., 2010). LS hiPSC-derived cardiomyocytes displayed increased cardiomyocyte size and sarcomeric organization as well as preferential nuclear localization of the nuclear factor of activated T cells 4, a transcription factor associated with calcineurin-mediated hypertrophic signaling (Carvajal-Vergara et al., 2010), consistent with a hypertrophic phenotype. Increased phosphorylation of MEK1 and ERK was also detected in LS hiPSC (Carvajal-Vergara et al., 2010). Similar findings have been observed in early developmental stages (larvae) of a Drosophila model of LS (Oishi et al., 2009). However, it remains unclear whether the signaling defects could also be detected in LS hiPSCs-derived cardiomyocytes. This study highlighted the ability for LS hiPSC-derived cardiomyocytes to recapitulate cardiac features found in LS patients, which paved the way towards future studies in establishing hiPSC as a model system to study human genetic-based cardiac diseases.
Long QT syndrome
Long QT syndrome (LQTS) is a congenital cardiac electro-physiological disorder that affects young individuals, a majority of which are asymptomatic. LQTS is caused by aberrantly prolonged cardiac repolarization times (prolonged QT interval as detected via electrocardiogram), leading to increased risk of polymorphic ventricular tachycardia and sudden cardiac death (Kramer and Zimetbaum, 2011). Genetic studies have linked LQTS to mutations in potassium and sodium channels resulting in their loss of function. There are several types of LQTS, which include both inherited (LQT type 1 (LQT1), LQT type 2 (LQT2), LQT type 3 (LQT3)) and acquired forms (drug-induced). LQT1 is the most common form, which arises from loss-of-function mutations in kcnq1, encoding IKs, an adrenergic-sensitive potassium channel in the heart, which has importance during exercise. LQT2 and drug-induced forms arise and are affected from loss-of-function mutations in kcnh2 (also known as hERG), encoding IKr, an important repolarizing potassium current in the heart. LQT3 arises from mutations that disrupt fast inactivation of the cardiac sodium channel SCN5A (Roden, 2008). Considerable attention has focused on using iPSC technology as a means to provide a human cell model system to dissect underlying mechanisms associated with LQTS and to test whether new experimental drugs elicit arrhythmias associated with drug-induced forms of LQTS. Electrophysiological analysis performed on hiPSC-derived cardiomyocytes from LQT1 patients carrying the R190Q mutation in KCNQ1 revealed a prolongation of the duration of the action potential in ventricular-like and atrial-like cells (Moretti et al., 2010). The R190Q–KCNQ1 mutation was shown to dominantly impair the targeting of the channel subunits to the cell membrane due to a trafficking defect (Moretti et al., 2010). Concordantly, reduction in IKs current, as well as altered channel activation and deactivation properties, was observed (Moretti et al., 2010). hiPSC-derived cardiomyocytes from LQT1 patients also displayed increased susceptibility to tachyarrhythmias induced by the β-adrenergic agonist isoproterenol, which could be attenuated with the β-adrenergic blocker, propanolol (Moretti et al., 2010).
At present, three groups have modeled LQT2 using hiPSC (Itzhaki et al., 2011; Lahti et al., 2012; Matsa et al., 2011). Prolongation of action potential duration linked to deficiencies in the potassium channel (IKr) was observed in all three hiPSC LQT2 studies. Administration of the potassium channel openers, nicorandil and pinacidil, had beneficial effects on cells in two of the reported studies (Itzhaki et al., 2011; Matsa et al., 2011). In one study, a hiPSC line from a patient carrying the A614V missense mutation in the KCNH2 gene was generated (Itzhaki et al., 2011). LQT2 hiPSC-derived cardiomyocytes from this patient displayed marked arrhythmogenicity, characterized by early afterdepolarizations (EAD) and triggered arrhythmias (Itzhaki et al., 2011). Interestingly, the calcium channel inhibitor, nifedipine, and the potassium channel opener, pinacidil, and late-sodium channel blocker, ranolazine, were able to rescue the arrhythmias when delivered in short regimens (Itzhaki et al., 2011). Independent studies focused on hiPSC-derived cardiomyocytes from an LQT2 patient and an asymptomatic relative, both carrying the G1681A point mutation in KCNH2 and healthy controls (Matsa et al., 2011). Administration of the IKr blocker E4031 triggered arrhythmias and EAD in LQT2 hiPSC-derived car-diomyocytes, the latter was also observed upon isoproterenol treatment. β-Blockers, propanolol and nadalol (currently included in the patient’s management), were able to reverse the EAD induced by isoproterenol. Nicorandil and PD118057, both experimental potassium channel enhancers, induced action potential shortening, being able to mitigate EAD in some of the experimental conditions (Matsa et al., 2011). Others modeled LQT2 by using hiPSC-derived cardiomyocytes from an asymptomatic 61-year-old male carrier of the R176W missense mutation in KCNH2, with a family history of sudden death at an early age (Lahti et al., 2012). Ventricular-like LQT2 hiPSC-derived cardiomyocytes developed spontaneous arrhythmias, which became more frequent when the cells were exposed to the IKr blocker E4031 and the β-blocker, sotalol (Lahti et al., 2012). Interestingly, potential therapies proposed for LQTS include sodium channel blockers (such as flecainide), potassium channel activators (pinacidil, nicorandil, PD118057), and calcium channel blockers (nifipedine) (Khan and Gowda, 2004). However, non-clinical evaluation of these drugs for their potential to affect cardiac repolarization has revealed that these drugs may also shorten QT interval, which has been suggested to be pro-arrhythmogenic (Shah, 2010). Furthermore, most patients displaying idiopathic ventricular fibrillation show a shortened QT interval (Shah, 2010). These observations highlight the risks and potential consequences of QT interval manipulation, suggesting more tailored therapeutic approaches are required for LQTS. Altogether, these studies indicate that hiPSC-derived cardiomyocytes can accurately recapitulate LQTS phenotypes caused by kncq1 and kcnh2 mutations and can provide insights into drugs that can be further tailored for larger-scale pre-clinical testing.
Timothy syndrome
Timothy Syndrome (TS) is a rare condition that encompasses cardiac electrophysiological defects associated with LQTS, developmental defects including webbed fingers and toes (syndactyly), immune deficiency, intermittent hypoglycemia, cognitive abnormalities, and autism (Splawski et al., 2004). Remarkably, all TS cases result from the CaV1.2 missense mutation G406R, which causes maintained inward Ca2+ currents by complete loss of voltage-dependent channel inactivation (Splawski et al., 2004). TS hiPSC were generated from two patients carrying the G406R mutation in CaV1.2 (Yazawa et al., 2011). TS hiPSC-derived EB contracted at a slower rate (30 bpm) than controls (60 bpm). TS ventricular-like hiPSC-derived cardiomyocytes displayed a delayed inac-tivation of ICa, along with abnormal intracellular Ca2+ handling, reflected by larger and prolonged Ca2+ transients. These effects were reversed by roscovitine, a small molecule that caused an increase in the voltage-dependent inactivation of CaV1.2 (Yazawa et al., 2011). Interestingly, roscovitine also rescued disease phenotypes in hiPSC-derived neurons of TS patients (Pasca et al., 2011). Roscovitine also acts as a cyclin-dependent kinase inhibitor and has been used in clinical trials as an anti-proliferative drug for treatment of advanced cancers; however, high toxicity was observed in some treatment schedules (Le Tourneau et al., 2010). Nevertheless, its beneficial effects in TS hiPSC-derived cardiomyocytes are encouraging. The ability to recapitulate cardiac cell calcium handling defects in TS patients using TS hiPSC highlights their potential use as a tool to identify new pharmacological agents to treat this condition.
Catecholaminergic polymorphic ventricular tachycardia
Catecholaminergic Polymorphic Ventricular Tachycardia (CVPT) is a genetic-based arrhythmogenic disorder, thought to be caused by aberrant Ca2+ storage and release at the sarcoplasmic reticulum (SR), which culminates in tachyar-rhythmias and sudden cardiac death following exercise or emotional stress (Venetucci et al., 2012). Most CPVT patients exhibit mutations in the cardiac ryanodine receptor gene (ryr2) with variable penetrance (Laitinen et al., 2001; Priori et al., 2001). However, in a smaller number of patients, the disease arises from recessive mutations in the cardiac calse-questrin 2 (casq2) gene (Lahat et al., 2001). At present, four CPVT hiPSC models have been reported, and all studies describe that CPVT hiPSC-derived cardiomyocytes exhibit basal defects in Ca2+ handling. These defects became exacerbated by β-adrenergic stimulation, which led to increased susceptibility to delayed afterdepolarizations (DAD) (Fatima et al., 2011; Itzhaki et al., 2012; Jung et al., 2012; Kujala et al., 2012). The first hiPSC model of CPVT analyzed the p.F2483I mutation in RYR2. Interestingly, in this study, the Ca2+ induced Ca2+ release events observed after repolarization were abolished by increasing the cytosolic cAMP levels with forskolin (Fatima et al., 2011). Other studies have generated hiPSC-derived cardiomyocytes from a CPVT patient carrying the M4109R heterozygous point mutation in RYR2 (Itzhaki et al., 2012) and used two small molecules to rescue the CPVT phenotypes observed. One includes flecainide, which is a sodium channel inhibitor proposed to have beneficial effects on CPVT, which is currently in an ongoing clinical trial for CPVT (Faggioni et al., 2012). The other includes thapsigargin, an intracellular calcium releaser acting through inhibition of the Ca2+ pump located in sarcoplasmic and endoplasmic reticulum of cardiomyocytes (Wrzosek et al., 1992). Both drugs eliminated all afterdepolarizations. CPVT hiPSC-derived cardiomyocytes also displayed significant whole-cell Ca2+ transient irregularities, which were worsened by β-adrenergic stimulation and Ca2+ overload, and could be rescued with β-blockers. The threshold of store-overload-induced Ca2+ release was also significantly reduced in the CPVT hiPSC-derived cardiomyocytes (Itzhaki et al., 2012). Further studies modeled CPVT using hiPSC from a patient harboring the novel S406L mutation in RYR2 (Jung et al., 2012). The authors showed that dantrolene, a small molecule that stabilizes the active conformation of skeletal and cardiac RYR, restored Ca2+-handling properties and rescued the arrhythmogenic phenotype of CPVT hiPSC-derived cardiomyocytes. Thus, the underlying arrhythmogenic mechanism caused by the S406L mutation appears to reside in impaired conformational changes within the RYR2 channel (Jung et al., 2012). Independent studies analyzed the RYR2 mutation P2328S in a 25-year-old male CPVT patient (Kujala et al., 2012). Catecholaminergic stress led to abnormal Ca2+ signaling and arrhythmias in CPVT hiPSC-derived cardiomyocytes, which also displayed reduced SR Ca2+ content, indicating leakage from the SR. Adrenaline induced both DAD and EAD during spontaneous beating, recapitulating the ECG abnormalities in patients carrying the same mutation (Kujala et al., 2012). Altogether, the CPVT hiPSC models have replicated known electrophysiological abnormalities seen in patients and have indicated that flecainide, thapsigargin, and dantro-lene could have potential to be explored as a novel therapeutic management for CPVT.
Familial dilated cardiomyopathy
Familial Dilated Cardiomyopathy (fDCM) is associated with a family history of heart failure and death arising from dilation and contractile dysfunction of the left ventricle (Fatkin, 2011). Genetic studies have linked fDCM to mutations in genes encoding sarcomeric, cytoskeletal, mitochondrial, and nuclear membrane proteins, as well as proteins involved in Ca2þ homeostasis (Taylor et al., 2006). fDCM is considered the prevalent cause of heart failure following coronary artery disease and hypertension and is the leading cause for heart transplantation (Harmon et al., 2005). Hence, the need has increased to develop robust human models, such as fDCM hiPSC-derived cardiomyocytes, to provide new insights into the disease. Recently, a comprehensive study was performed using hiPSC-derived cardiomyocytes generated from three generations of a fDCM family, which carried a point mutation in the gene encoding the contractile protein cardiac troponin T (TNNT2) and healthy family members who did not carry the mutation (Sun et al., 2012). fDCM hiPSC-derived cardiomyocytes displayed sarcomeric disorganization and punctate distribution of sarcomeric α-actinin. The effects were exacerbated through β-adrenergic stimulation, which also impaired contraction. In turn, the effects could be rescued with the β1-selective blocker metoprolol (Sun et al., 2012), which is commonly included in the management of fDCM patients (Taylor et al., 2006). Alterations in Ca2þ handling were also observed in fDCM hiPSC-derived cardiomyocytes (Sun et al., 2012). Forced overexpression of sarcoplasmic reticulum Ca2+ATPase (SERCA2a) could also rescue the Ca2+-handling abnormalities as well as increase contractile force in fDCM hiPSC-derived cardiomyocytes (Sun et al., 2012). Altogether, this study demonstrated a faithful recapitulation of cardiac structural and electrophysiological defects observed in fDCM patients, indicating the potential of hiPSC technology to uncover molecular underpinnings of the disease.
Arrhythmogenic right ventricular cardiomyopathy
Arrhythmogenic right ventricular cardiomyopathy (ARVC) is a genetic-based cardiomyopathy initially described by its classic right ventricular involvement and dysfunction (Marcus et al., 1982). However, recent studies have identified non-classical left ventricular prominent forms of the disease (Sen-Chowdhry et al., 2007), leading to a revision of the criteria for diagnosis and included adoption of the broader term arrhythmogenic cardiomyopathy (AC) when describing the disease (Delmar and McKenna, 2010). Characteristic of ARVC is also fibrotic/fatty replacement of the ventricle as well as high frequency of ventricular arrhythmias leading to sudden cardiac death in seemingly healthy young people, including athletes (Saffitz, 2011). ARVC has been termed a desmosomal disease, since 40% of patients carry mutations in desmosomal cell junction components, key components of the intercalated disc involved in the mechanical integrity of the myocardium (Saffitz, 2011; Sheikh et al., 2009). At present, two groups have modeled ARVC arising from mutations in the desmosomal component plakophilin-2 in patients with clinical diagnosis of ARVC (Kim et al., 2013; Ma et al., 2012). The first study by Ma and colleagues examined a 30-year-old male patient carrying a novel heterozygous c.1841T > C nucleotide change mutation in plakophilin-2 and an unaffected age- and sex-matched individual (Ma et al., 2012). Subsequently, Kim and colleagues focused on a 44-year-old female patient harboring a previously reported homozygous c.2484C.T mutation in plakophilin-2 (Awad et al., 2006), as well as an undisclosed aged male patient carrying the c.2013delC mutation in exon 10 of plakophilin-2, which were evaluated with H9 ESC as a control in this study (Kim et al., 2013). Interestingly, in both studies, hiPSC-derived cardiomyocytes from patients carrying plakophilin-2 mutations displayed no basal ARVC phenotype. However, consistent with fatty deposition being observed as a feature of ARVC in human patients (Kimura et al., 2010), ARVC hiPSC-derived cardiomyocytes displayed an increased propensity to develop lipid accumulation when taken through strongly adipogenic conditions after cardiac differentiation (Kim et al., 2013; Ma et al., 2012). Interestingly, in the latter studies, the enhanced lipogenesis observed in ARVC hiPSC-derived cardiomyocytes was linked to a metabolic shift from fatty acid and glucose consumption to predominantly glucose consumption, which was suggested to impact cell viability and induce Ca2+ handling deficiencies (Kim et al., 2013). However, whether these metabolic abnormalities occur in ARVC patients remains to be explored. Nevertheless, the authors were able to successfully reduce the number of lipid droplets found in ARVC-derived cardiomyocytes through lentivirus-mediated expression of wild-type plakophilin in ARVC EB (Kim et al., 2013). Hence, both hiPSC models of ARVC have been able to reproduce the lipid accumulation that is linked to the cardiomyopathy aspect of ARVC seen in a subset of patients. It remains to be explored whether these models can reproduce in greater detail the deadly arrhythmogenic components of ARVC. The suggestion that ARVC may present a metabolic component is also intriguing and deserves further exploration.
Overlapping Na+ channel disease syndrome
hiPSC were also used to model a syndrome characterized by a combination of cardiac abnormalities caused by the heterozygous 1795insD loss-of-function mutation in SCN5A (Davis et al., 2012). While gain-of-function mutations in SCN5A lead to the prolongation of repolarization and AP prolongation seen in LQT3, loss-of-function mutations lead to slower upstroke velocity of the AP, associated with conduction disease and Brugada Syndrome (BrS) (Wilde and Bezzina, 2005). In order to model this disease condition, fibroblasts from a 47-year-old male patient carrying the SCN5A 1795insD mutation and healthy control fibroblasts from a 51-year-old female were isolated. In a knock-in mouse model, the authors had determined that this mutation caused a decrease in peak INa and an increase in persistent INa (Remme et al., 2009). This hiPSC model replicated the same INa abnormalities found in the murine model of this mutation, which were not observed in control hiPSC-derived cardiomyocytes (Davis et al., 2012).
Future perspectives
hiPSC have provided an exciting tool to recapitulate known defects and uncover new defects and mechanisms underlying human cardiac disease. Transcription activator-like effector nucleases (TALENs) (Cermak et al., 2011) and zinc finger nucleases (ZFNs) (Urnov et al., 2005) have been used to genetically repair disease-causing single mutations in PSC (Soldner et al., 2011). Although current hiPSC-derived cardiac disease models have not exploited genetic repair approaches, future use of isogenic repaired lines may provide inarguable validation to phenotypes presented in disease models to link them to the causality of mutations. This may be especially important for some cardiac diseases modeled using hiPSC, since different mutations (some previously shown to be pathogenic and others of unknown pathogenicity) were analyzed for the same disease and in some cases, the distinct biochemical impact of these mutations remains unclear. Thus, future studies should also explore how distinct mutations specifically impact the biochemistry of the molecule to render its pathogenic nature in disease. Furthermore, additional validation of the results obtained from hiPSC-derived cardiomyocytes against myocardial biopsies from the same patients would offer strong support to any in vitro discoveries, especially in the context where new pathways are uncovered. Hence, in order to perform reasonable comparisons between adult tissue and PSC-derived cardiomyocytes, the lack of maturity of the latter needs to be thoroughly addressed. It has been demonstrated that long-term culture could be used as a strategy for maturation (Kamakura et al., 2013); however, such long experimental settings may prove impractical. Tissue engineering approaches have highlighted the importance of the cardiac extracellular matrix (ECM) and milieu in driving cardiac stem cell differentiation and maturation (Thavandiran et al., 2013). Thus, the modulation of the ECM and cell microenvironment constitutes an attractive avenue to promote PSC-derived cardiomyocyte maturation. One example is the use of native cardiac extracellular matrices that can promote cardiac cell junction protein localization indicative of a more mature phenotype (DeQuach et al., 2010). Thus, the combination of tissue engineering technologies with cardiac robust differentiation protocols may help to propel the field forward in being able to evaluate cardiomyocytes at a stage that would be more reflective of postnatal cardiomyocytes, providing a better context to study postnatal onset cardiac diseases. Larger quantities of mature hiPSC-derived cardiomyocytes will also enable large-scale screening approaches to uncover new molecular pathways, compounds, and targets for cardiac disease. Excitingly, experimental drugs as well as drugs that have been used for the patient’s management have shown potential to reverse the disease phenotypes in hiPSC-derived cardiomyocytes in vitro, highlighting the potential for hiPSC as a pre-clinical model for drug testing. As a result, these studies may also advance the future evaluation of the therapeutic potential of these drugs in human clinical trials. In summary, although many challenges and questions remain, the rapid advances in stem cell biology have been quickly incorporated into the cardiac field, bringing great promise to better understand and manage several devastating cardiac diseases that burden the human population.
Acknowledgments
We would like to thank Drs. Jennifer Lowe and Matthew Stroud (University of California-San Diego (UCSD)) for critically reading the manuscript. Funding was provided by the Science and Technology Blasker (Blasker-Rose-Miah Fund) grant from the San Diego Foundation (F.Z.). F.Z. and R.C.L were previously supported by UCSD Cardiovascular Scholarship award. F.Z and R.C.L. are current recipients of the American Heart Association Postdoctoral fellowship. F.S. is supported by grants from the National Institutes of Health and California Institute of Regenerative Medicine.
REFERENCES
- Adewumi O, Aflatoonian B, Ahrlund-Richter L, Amit M, Andrews PW, Beighton G, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nature Biotechnology. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- Awad MM, Dalal D, Tichnell C, James C, Tucker A, Abraham T, et al. Recessive arrhythmogenic right ventricular dysplasia due to novel cryptic splice mutation in PKP2. Human Muta-tation. 2006;27:1157. doi: 10.1002/humu.9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beqqali A, Kloots J, Ward-van Oostwaard D, Mummery C, Passier R. Genome-wide transcriptional profiling of human embryonic stem cells differentiating to cardiomyocytes. Stem Cells. 2006;24:1956–1967. doi: 10.1634/stemcells.2006-0054. [DOI] [PubMed] [Google Scholar]
- Carvajal-Vergara X, Sevilla A, D’Souza SL, Ang YS, Schaniel C, Lee DF, et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–812. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RP, Casini S, van den Berg CW, Hoekstra M, Remme CA, Dambrot C, et al. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation. 2012;125:3079–3091. doi: 10.1161/CIRCULATIONAHA.111.066092. [DOI] [PubMed] [Google Scholar]
- Davis RP, van den Berg CW, Casini S, Braam SR, Mummery CL. Pluripotent stem cell models of cardiac disease and their implication for drug discovery and development. Trends in Molecular Medicine. 2011;17:475–484. doi: 10.1016/j.molmed.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Delmar M, McKenna WJ. The cardiac desmosome and arrhythmogenic cardiomyopathies: from gene to disease. Circ Res. 2010;107:700–714. doi: 10.1161/CIRCRESAHA.110.223412. [DOI] [PubMed] [Google Scholar]
- DeQuach JA, Mezzano V, Miglani A, Lange S, Keller GM, Sheikh F, et al. Simple and high yielding method for preparing tissue specific extracellular matrix coatings for cell culture. PLoS One. 2010;5:e13039. doi: 10.1371/journal.pone.0013039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois NC, Craft AM, Sharma P, Elliott DA, Stanley EG, Elefanty AG, et al. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nature Biotechnology. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF, Jr, Mattis VB, Lorson CL, Thomson JA, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DA, Braam SR, Koutsis K, Ng ES, Jenny R, Lagerqvist EL, et al. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nature Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- Faggioni M, Kryshtal DO, Knollmann BC. Calsequestrin mutations and catecholaminergic polymorphic ventricular tachycardia. Pediatric Cardiology. 2012;33:959–967. doi: 10.1007/s00246-012-0256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima A, Xu G, Shao K, Papadopoulos S, Lehmann M, Arnaiz-Cot JJ, et al. In vitro Modeling of Ryanodine Receptor 2 Dysfunction Using Human Induced Pluripotent Stem Cells. Cellular Physiology and Biochemistry. 2011;28:579–592. doi: 10.1159/000335753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatkin D. Guidelines for the diagnosis and management of familial dilated cardiomyopathy. Heart, Lung and Circulation. 2011;20:691–693. doi: 10.1016/j.hlc.2011.07.008. [DOI] [PubMed] [Google Scholar]
- Gelb BD, Tartaglia M. LEOPARD Syndrome. In: Pagon RA, Adam MP, Bird TD, editors. GeneReviews. Seattle: University of Washington, Seattle; 2007. pp. 1993–2013. [Google Scholar]
- Hao J, Daleo MA, Murphy CK, Yu PB, Ho JN, Hu J, et al. Dorsomorphin, a selective small molecule inhibitor of BMP signaling, promotes cardiomyogenesis in embryonic stem cells. PLoS One. 2008;3:e2904. doi: 10.1371/journal.pone.0002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon WE, McDonald RA, Reyes JD, Bridges ND, Sweet SC, Sommers CM, et al. Pediatric transplantation, 1994–2003. American Journal of Transplantation. 2005;5:887–903. doi: 10.1111/j.1600-6135.2005.00834.x. [DOI] [PubMed] [Google Scholar]
- Hattori F, Chen H, Yamashita H, Tohyama S, Satoh YS, Yuasa S, et al. Nongenetic method for purifying stem cell-derived cardiomyocytes. Nature Methods. 2010;7:61–66. doi: 10.1038/nmeth.1403. [DOI] [PubMed] [Google Scholar]
- Hoekstra M, Mummery CL, Wilde AA, Bezzina CR, Verkerk AO. Induced pluripotent stem cell derived cardiomyocytes as models for cardiac arrhythmias. Frontiers in Physiology. 2012;3:346. doi: 10.3389/fphys.2012.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482:216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzhaki I, Maizels L, Huber I, Gepstein A, Arbel G, Caspi O, et al. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. Journal of the American College of Cardiology. 2012;60:990–1000. doi: 10.1016/j.jacc.2012.02.066. [DOI] [PubMed] [Google Scholar]
- Itzhaki I, Maizels L, Huber I, Zwi-Dantsis L, Caspi O, Winterstern A, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–229. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- Jung CB, Moretti A, Mederos y, Schnitzler M, Iop L, Storch U, et al. Dantrolene rescues arrhythmogenic RYR2 defect in a patient-specific stem cell model of catecholaminergic polymorphic ventricular tachycardia. EMBO Molecular Medicine. 2012;4:180–191. doi: 10.1002/emmm.201100194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakura T, Makiyama T, Sasaki K, Yoshida Y, Wuriyanghai Y, Chen J, et al. Ultrastructural maturation of human-induced pluripotent stem cell-derived cardiomyocytes in a long-term culture. Circulation Journal. 2013;77:1307–1314. doi: 10.1253/circj.cj-12-0987. [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardio-myocytes. Journal of Clinical Investigation. 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IA, Gowda RM. Novel therapeutics for treatment of long-QT syndrome and torsade de pointes. International Journal of Cardiology. 2004;95:1–6. doi: 10.1016/j.ijcard.2003.04.018. [DOI] [PubMed] [Google Scholar]
- Kim C, Wong J, Wen J, Wang S, Wang C, Spiering S, et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494:105–110. doi: 10.1038/nature11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura F, Matsuo Y, Nakajima T, Nishikawa T, Kawamura S, Sannohe S, et al. Myocardial fat at cardiac imaging: how can we differentiate pathologic from physiologic fatty infiltration? Radiographics. 2010;30:1587–1602. doi: 10.1148/rg.306105519. [DOI] [PubMed] [Google Scholar]
- Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. Journal of Biological Chemistry. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- Kramer DB, Zimetbaum PJ. Long-QT syndrome. Cardiology in Review. 2011;19:217–225. doi: 10.1097/CRD.0b013e3182203504. [DOI] [PubMed] [Google Scholar]
- Kujala K, Paavola J, Lahti A, Larsson K, Pekkanen-Mattila M, Viitasalo M, et al. Cell model of catecholaminergic polymorphic ventricular tachycardia reveals early and delayed after-depolarizations. PLoS One. 2012;7:e44660. doi: 10.1371/journal.pone.0044660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujala V, Pekkanen-Mattila M, Aalto-Setälaand K. Human pluripotent stem cell-derived cardiomyocytes: maturity and electrophysiology. In: Kallos MS, editor. Embryonic Stem Cells— Differentiation and Pluripotent Alternatives. Croatia: Intech; 2011. pp. 185–204. [Google Scholar]
- Lahat H, Pras E, Olender T, Avidan N, Ben-Asher E, Man O, et al. A missense mutation in a highly conserved region of CASQ2 is associated with autosomal recessive catecholamine-induced polymorphic ventricular tachycardia in Bedouin families from Israel. American Journal of Human Genetics. 2001;69:1378–1384. doi: 10.1086/324565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti AL, Kujala VJ, Chapman H, Koivisto AP, Pekkanen-Mattila M, Kerkela E, et al. Model for long QT syndrome type 2 using human iPS cells demonstrates arrhythmogenic characteristics in cell culture. Disease Models & Mechanisms. 2012;5:220–230. doi: 10.1242/dmm.008409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen PJ, Brown KM, Piippo K, Swan H, Devaney JM, Brahmb-hatt B, et al. Mutations of the cardiac ryanodine receptor (RyR2) gene in familial polymorphic ventricular tachycardia. Circulation. 2001;103:485–490. doi: 10.1161/01.cir.103.4.485. [DOI] [PubMed] [Google Scholar]
- Lauriol J, Kontaridis MI. PTPN11-associated mutations in the heart: has LEOPARD changed Its RASpots? Trends in Cardiovascular Medicine. 2011;21:97–104. doi: 10.1016/j.tcm.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tourneau C, Faivre S, Laurence V, Delbaldo C, Vera K, Girre V, et al. Phase I evaluation of seliciclib (R-roscovitine), a novel oral cyclin-dependent kinase inhibitor, in patients with advanced malignancies. European Journal of Cancer. 2010;46:3243–3250. doi: 10.1016/j.ejca.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Li M, Izpisua Belmonte JC. No factor left behind: generation of transgene-free induced pluripotent stem cells. American Journal of Stem Cells. 2012;1:75–80. [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhao H, Lan F, Lee A, Chen L, Lin C, et al. Generation of human-induced pluripotent stem cells from gut mesentery-derived cells by ectopic expression of OCT4/SOX2/NANOG. Cellular Reprogramming. 2010;12:237–247. doi: 10.1089/cell.2009.0103. [DOI] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proceedings of the National Academy of Sciences of the United States of America. 2012a;109:E1848–E1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nature Protocols. 2012b;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Agarwal S, Park IH, Urbach A, Huo H, Heffner GC, et al. Generation of induced pluripotent stem cells from human blood. Blood. 2009;113:5476–5479. doi: 10.1182/blood-2009-02-204800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Wei H, Lu J, Ho S, Zhang G, Sun X, et al. Generation of patient-specific induced pluripotent stem cell-derived cardiomyocytes as a cellular model of arrhythmogenic right ventricular cardiomyopathy. European Heart Journal. 2012;34:1122–1133. doi: 10.1093/eurheartj/ehs226. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, et al. A model for neural development and treatment of rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus FI, Fontaine GH, Guiraudon G, Frank R, Laurenceau JL, Malergue C, et al. Right ventricular dysplasia: a report of 24 adult cases. Circulation. 1982;65:384–398. doi: 10.1161/01.cir.65.2.384. [DOI] [PubMed] [Google Scholar]
- Matsa E, Rajamohan D, Dick E, Young L, Mellor I, Staniforth A, et al. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. European Heart Journal. 2011;32:952–962. doi: 10.1093/eurheartj/ehr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami I, Yamada K, Otsuji TG, Yamamoto T, Shen Y, Otsuka S, et al. A small molecule that promotes cardiac differentiation of human pluripotent stem cells under defined, cytokine- and xeno-free conditions. Cell Reports. 2012;2:1448–1460. doi: 10.1016/j.celrep.2012.09.015. [DOI] [PubMed] [Google Scholar]
- Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flugel L, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. New England Journal of Medicine. 2010;363:1397–1409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- Mummery CL, Ward D, Passier R. Differentiation of human embryonic stem cells to cardiomyocytes by coculture with endoderm in serum-free medium. In: Chambers K, editor. Current Protocols in Stem Cell Biology. John Wiley & Sons, Inc; 2007. pp. 2.1–2.14. [DOI] [PubMed] [Google Scholar]
- Mummery CL, Zhang J, Ng ES, Elliott DA, Elefanty AG, Kamp TJ. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circulation Research. 2012;111:344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K, Domian IJ, Chien KR. Stem cell models of cardiac development and disease. Annual Review of Cell and Developmental Biology. 2010;26:667–687. doi: 10.1146/annurev-cellbio-100109-103948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Zhang H, Gault WJ, Wang CJ, Tan CC, Kim IK, et al. Phosphatase-defective LEOPARD syndrome mutations in PTPN11 gene have gain-of-function effects during Drosophila development. Human Molecular Genetics. 2009;18:193–201. doi: 10.1093/hmg/ddn336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Osugi T, Afanasiev OK, Pabon L, Reinecke H, Murry CE. Endogenous Wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS One. 2010;5:e11134. doi: 10.1371/journal.pone.0011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, et al. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nature Medicine. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priori SG, Napolitano C, Tiso N, Memmi M, Vignati G, Bloise R, et al. Mutations in the cardiac ryanodine receptor gene (hRyR2) underlie catecholaminergic polymorphic ventricular tachycardia. Circulation. 2001;103:196–200. doi: 10.1161/01.cir.103.2.196. [DOI] [PubMed] [Google Scholar]
- Remme CA, Scicluna BP, Verkerk AO, Amin AS, van Brunschot S, Beekman L, et al. Genetically determined differences in sodium current characteristics modulate conduction disease severity in mice with cardiac sodium channelopathy. Circ Res. 2009;104:1283–1292. doi: 10.1161/CIRCRESAHA.109.194423. [DOI] [PubMed] [Google Scholar]
- Roden DM. Clinical practice. Long-QT syndrome. New England Journal of Medicine. 2008;358:169–176. doi: 10.1056/NEJMcp0706513. [DOI] [PubMed] [Google Scholar]
- Saffitz JE. Arrhythmogenic cardiomyopathy: advances in diagnosis and disease pathogenesis. Circulation. 2011;124:e390–e392. doi: 10.1161/CIRCULATIONAHA.111.064022. [DOI] [PubMed] [Google Scholar]
- Sarkozy A, Digilio MC, Dallapiccola B. Leopard syndrome. Orphanet Journal of Rare Diseases. 2008;3:13. doi: 10.1186/1750-1172-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen-Chowdhry S, Syrris P, Ward D, Asimaki A, Sevdalis E, McKenna WJ. Clinical and genetic characterization of families with arrhythmogenic right ventricular dysplasia/cardiomyopathy provides novel insights into patterns of disease expression. Circulation. 2007;115:1710–1720. doi: 10.1161/CIRCULATIONAHA.106.660241. [DOI] [PubMed] [Google Scholar]
- Shah RR. Drug-induced QT interval shortening: potential harbinger of proarrhythmia and regulatory perspectives. British Journal of Pharmacology. 2010;159:58–69. doi: 10.1111/j.1476-5381.2009.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh F, Ross RS, Chen J. Cell-cell connection to cardiac disease. Trends in Cardiovascular Medicine. 2009;19:182–190. doi: 10.1016/j.tcm.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Laganiere J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, et al. Generation of isogenic pluripotent stem cells differing exclusively at two early onset Parkinson point mutations. Cell. 2011;146:318–331. doi: 10.1016/j.cell.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, et al. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Sugii S, Kida Y, Kawamura T, Suzuki J, Vassena R, Yin YQ, et al. Human and mouse adipose-derived cells support feeder-independent induction of pluripotent stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3558–3563. doi: 10.1073/pnas.0910172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Yazawa M, Liu J, Han L, Sanchez-Freire V, Abilez OJ, et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Science Translational Medicine. 2012;4 doi: 10.1126/scitranslmed.3003552. 130ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Taylor MR, Carniel E, Mestroni L. Cardiomyopathy, familial dilated. Orphanet Journal of Rare Diseases. 2006;1:27. doi: 10.1186/1750-1172-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thavandiran N, Nunes SS, Xiao Y, Radisic M. Topological and electrical control of cardiac differentiation and assembly. Stem Cell Research & Therapy. 2013;4:14. doi: 10.1186/scrt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardio-myocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Venetucci L, Denegri M, Napolitano C, Priori SG. Inherited calcium channelopathies in the pathophysiology of arrhythmias. Nature Reviews Cardiology. 2012;9:561–575. doi: 10.1038/nrcardio.2012.93. [DOI] [PubMed] [Google Scholar]
- Wilde AA, Bezzina CR. Genetics of cardiac arrhythmias. Heart. 2005;91:1352–1358. doi: 10.1136/hrt.2004.046334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E, Spiering S, Davidovics H, Lanier M, Xia Z, Dawson M, et al. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circulation Research. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzosek A, Schneider H, Grueninger S, Chiesi M. Effect of thapsigargin on cardiac muscle cells. Cell Calcium. 1992;13:281–292. doi: 10.1016/0143-4160(92)90063-x. [DOI] [PubMed] [Google Scholar]
- Yan X, Qin H, Qu C, Tuan RS, Shi S, Huang GT. iPS cells reprogrammed from human mesenchymal-like stem/progenitor cells of dental tissue origin. Stem Cells and Development. 2010;19:469–480. doi: 10.1089/scd.2009.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa M, Hsueh B, Jia X, Pasca AM, Bernstein JA, Hallmayer J, et al. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–234. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circulation Research. 2012;111:1125–1136. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu WZ, Van Biber B, Laflamme MA. Methods for the derivation and use of cardiomyocytes from human pluripotent stem cells. Methods in Molecular Biology. 2011;767:419–431. doi: 10.1007/978-1-61779-201-4_31. [DOI] [PMC free article] [PubMed] [Google Scholar]