Abstract
To evaluate if HIV transmission networks could be elucidated from data collected in a short time frame, 131 HIV-1 pol sequences were analyzed which were generated from treatment-naïve Korean individuals who were sequentially identified over 1 year. A transmission linkage was inferred when there was a genetic distance <1.5% and a total of 16 clusters, involving 39/131 (29.8%), were identified. Younger age and heterosexual exposure were independently related with clustering in the inferred network, which demonstrated that molecular epidemiology with currently generated data (i.e., drug resistance genotypes) can be used to identify local transmission networks, even over a short timeframe.
Keywords: human immunodeficiency virus, molecular epidemiology, cluster analysis, phylogeny
INTRODUCTION
The South Korean HIV epidemic likely started around 1985, and while overall prevalence is still less than 0.1%, it is increasing [KCDC, 2013]. The HIV epidemics in South Korea is characterized as unique Korean Clade B and male preponderance [Kim et al., 2008, 2012]. Subtype B has become majority strain [Kang et al., 1998] soon after the first introduction of HIV in South Korea and monophyletic clustering of Korean Clade B, distinctive from those of other countries, was explained by the slow effective growth rate of HIV infected population in South Korea after early 1980s [Kim et al., 2012]. Besides virologic characteristics, focus of epidemics has gradually shifted from importation of infected individuals from foreign countries and heterosexual exposure (HTS) to currently being concentrated in men who have sex with men (MSM) [Choe, 2007]; however, both HTS and MSM remain important to the current epidemic. [KCDC, 2011].
Since HIV-1 pol sequences that have been generated for surveillance of transmitted drug resistance (TDR) can be used to infer local transmission networks [Smith et al., 2009; Little et al., 2014], we used such available sequence data to determine if HIV sub-networks, that is, clusters, could be identified among antiretroviral therapy (ART) naïve HIV-infected individuals sequentially evaluated over 1 year. If such sub-networks could be identified early, then preventative interventions could be enacted to make a substantial impact locally. [Little et al., 2014].
MATERIALS AND METHODS
The study population consisted of ART naïve HIV-1 infected individuals consecutively enrolled at the National Medical Center, Seoul, South Korea from February 2013 to 2014 as described in a prior study about TDR surveillance [Chin et al., 2015]. The National Medical Center is a 500-bed public hospital which is located in the urban center of Seoul with 10 million population which is about one fifth of total population in South Korea. It is one of the major HIV clinics in South Korea and about 800 HIV-infected individuals were regularly visiting the clinic in 2013 while the total cumulative number of people living with HIV/AIDS in South Korea was 8,662 [KCDC, 2013]. During the study period, retrospective cohort of 131 ART naïve HIV-1 infected Korean individuals of age over 18, mostly newly diagnosed within 1 year before sampling, was constructed whose pre-ART genotypic resistance test result is available as well as pol gene sequence data. HIV-1 genotyping was performed using ViroSeq™ version 2.0 (Abbott Laboratories, Abbott Park, IL), as previously described [Chin et al., 2010]. Sequences with complete protease (amino acids 1–99) and partial reverse transcriptase (amino acids 1–335) genes of the pol region with the length of 1,302 nucleotide were aligned using Bioedit 7.2.5 software and the presence of TDR was determined using the Stanford HIV Drug Resistance Database (Version 7.0) and the World Health Organization HIV Surveillance Drug Resistance Mutation list [Bennett et al., 2009]. In this study, the prevalence of overall TDR was 8.4% and K103N TDR increased significantly (6.1%) compared with prior studies which were performed in South Korea. Sequence data obtained during the study (KM820292-KM820422) were screened for contamination, duplication, and for APOBEC hypermutation and subtyped using the Los Alamos National Laboratory HIV Sequence Database [Rose and Korber, 2000]. Sequence alignment and analysis for network inference was performed using the HyPhy package. [Kosakovsky Pond et al., 2005] Specifically, sequences were analyzed for genetic relatedness via pairwise distance comparison using the Tamura-Nei 93 model. [Wertheim et al., 2014] Since population based sequencing can produce sequences with mixed nucleotide bases, and these mixed bases could hypothetically influence clustering, as can drug resistance mutations, we evaluated four algorithms. These algorithms included: (i) leaving mixed bases unresolved and averaging genetic distance between sequences, (ii) resolving mixed bases for all possibilities before calculating genetic distance between sequences, (iii) stripping drug resistance codons before calculating genetic distance between sequences, and (iv) leaving drug resistance codons in all sequences when calculating genetic distance between sequences. There was no substantive difference in the putative clustering by any of these approaches. A transmission linkage between two sequences was inferred when there was a genetic distance <1.5% between them [Little et al., 2014]. This threshold is the current standard based observations that HIV-1 subtype B pol has an evolutionary rate of 0.7%/year within individuals, and that the expected genetic distance between unrelated HIV-1 pol sequences was >5% [Hightower et al., 2013].
We utilized demographic information, reported date of HIV infection, risk factor, CD4 T cell count, and viral load collected from the medical record. These characteristics were compared between individuals who clustered and those who did not. Categorical variables were compared using the Fisher’s exact test, and continuous variables were evaluated using the Wilcoxon Rank test. Multivariate analysis was performed by logistic regression. Utilization of HIV-1 sequence data and medical record information were approved by the institutional review board.
RESULTS AND DISCUSSION
Among the 131 HIV-1 sequences, most were HIV-1 subtype B (n = 117, 89.3%) followed by CRF01_AE (6.1%). The vast majority of participants were male (94.5%) with a median age of 31 years (IQR 25–40). A reported risk factor for HIV infection was available in 76 cases (58%): 49 were MSM and the other 27 individuals reported HTS. More than half (69.3%) were diagnosed with HIV-1 infection between January 2013 and February 2014, and the median interval between HIV-1 infection diagnosis and sampling was 2.4 months (interquartile range, IQR 0.9–8.8). Presence of TDR was identified in 11 individuals (8.4%). The most common TDR mutation was K103N (8/11, 72.7%), and one individual had resistance mutations to both protease inhibitor (M46L) and non-nucleoside reverse transcriptase inhibitor (K103N).
The HIV network was inferred using all available HIV-1 pol sequences, and a total of 16 clusters, involving 39/131 (29.8%) of the sampled sequences, were identified (Fig. 1). The clusters ranged in size from two to four individuals and all belonged to subtype B strains (39/117, 33.3%). Because there was no clustering case in non-subtype B infections (0/14, P = 0.010), factors associated with clustering was examined only among the 117 subtype B infections. Compared between individuals who clustered (n = 39) and non-clustered (n = 78), younger age at diagnosis of HIV, recent diagnosis of HIV (Jan 2013–Feb 2014), shorter interval form HIV diagnosis to sampling and HTS were associated with clustering (Table I). There was no association with clustering based on gender, CD4 count, or HIV-1 viral load. In multivariate analysis, younger age (per year more than 33.1, OR 0.940, 95%CI 0.903–0.979) and HTS compared with MSM (OR 9.604, 95%CI 2.681–34.404) were independently related with clustering.
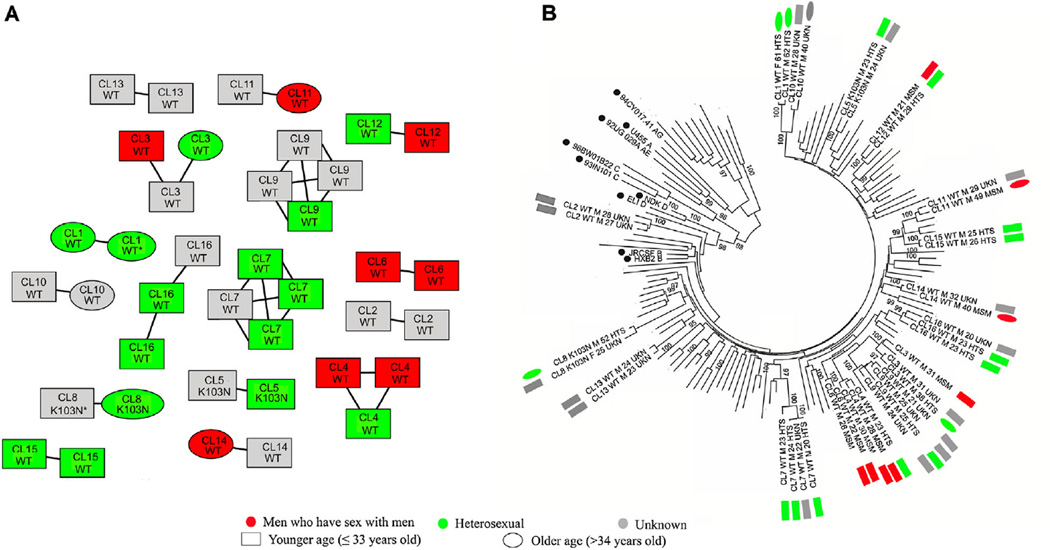
Fig. 1.
Clustering among treatment naïve Korean HIV-1 infected individuals. A: Inferred transmission network of study population; clustering individuals are colored by their reported risk factor in red (men who have sex with men, MSM), green (heterosexual, HTS), or gray (unknown, UKN), respectively. All edges represent a genetic distance ≤1.5% between nodes. Absence (WT: Wild Type) or presence of transmitted drug resistance (TDR) is indicated for each individual. All but two individuals (represented by an *) are male. Younger (≤33 years old) and older individuals are labeled as square and ellipse, respectively. CL represents the abbreviation of cluster number. B: Consistent clustering was observed in phylogenetic tree analysis. Taxon represents cluster number, presence of TDR, gender, age, and reported risk factor. M and F represents the abbreviation of male and female gender, respectively. Sequences with solid circle besides taxon represent reference strains from Los Alamos HIV Database. Phylogenetic tree was built using the Neighbor-joining method generating 1,000 bootstrap sampling, as implemented in MEGA version 6. Value lower than 95% was not displayed.
TABLE I.
Correlates of Clustering Among 117 Individuals With HIV-1 Subtype B Infections
| Clustering (n = 39) |
Not clustering (n = 78) |
P-value | Multivariate OR (95%CI) |
|
|---|---|---|---|---|
| Age at HIV-1 diagnosis (median, year)a | 25 (IQR 23–31) | 32 (IQR 25–45) | 0.007 | 0.940 (0.903–0.979)b |
| Male sex (n, %) | 37 (94.9) | 78 (100) | 0.109 | |
| Risk group (n, %) | 0.001 | |||
| MSM | 8 (20.5) | 35 (44.9) | 1.0 (Ref) | |
| HTS | 15 (38.5) | 9 (11.5) | 9.604 (2.681–34.404) | |
| Unknown | 16 (41.0) | 34 (43.6) | 2.409 (0.856–6.781) | |
| Reported year of HIV diagnosis (n, %)a | 0.036 | 1.516 (0.394–5.844) | ||
| 2010 | 3 (7.7) | 12 (16.0) | ||
| 2011/2012 | 3 (7.7) | 17 (22.7) | ||
| 2013/2014 | 33 (84.6) | 46 (61.3) | ||
| Interval between HIV diagnosis and sampling (median, month)a |
1.87 (IQR 0.5–5.43) | 3.17 (IQR 1.01–22.0) | 0.020 | 0.978 (0.931–1.027) |
| CD4 count (median, cells/mm3) | 270 (IQR 172–412) | 314 (IQR 190–397) | 0.681 | |
| Log10 HIV-1 RNA (median, copies/ml) | 4.37 (IQR 4.10–4.89) | 4.32 (IQR 3.84–4.98) | 0.894 | |
| SDRM (n, %) | 4 (10.3) | 7 (9.0) | 1.000 |
OR, odds ratio; CI, confidence interval; IQR, interquartile range; MSM, men who have sex with men; HTS, heterosexual contact; SDRM, surveillance drug resistance mutation.
Data missing for three participants. HIV diagnosis year was not available in three cases.
The odds ratio is per year over 33.1 which is the mean value of study population.
Younger age is commonly reported contributing factor for viral genetic clustering among HIV-1 infected individuals. However, most studies have identified MSM as contributing factor for clustering contrary to our study [Chalmet et al., 2010; Lubelchek et al., 2015; Poon et al., 2015] and it is an important limitation of our study that HIV infection risk group information was available for only just more than half of study population as well as small sample size of our study. However, our study showed that clustering was relatively more frequent among HTS group than MSM (15/24, 62.5% vs. 8/43, 18.6%), while MSM was more frequent than HTS (43 vs. 24) as a risk factor itself. This unique feature implies that there have been more active transmission networks among HTS groups in South Korea while HIV transmissions among MSM have occurred rather sporadically within limited networks contrary to the study results from other areas. When we redefined the cluster as the group with three and more nodes [Chalmet et al., 2010], the preponderance for clustering of HTS group over MSM was still valid (8/24, 33.3% vs. 3/43, 7.0%).
The cohort of 131 participants in this study, was enrolled over 1 year with no targeting for enrollment based on HIV risk factors or demographics. Genetic distance analysis of available sequence data was able to identify clustering among around a quarter of all participants. While this overall level of clustering is relatively low (likely because of the small numbers of sequences and lack of targeting specific risk populations), this analysis was able to identify a very interesting HIV transmission sub-network predominantly made up of individuals of younger age (OR 0.940 for clustering per year more than 33.1) and risk factor of HTS. In fact, clustering was identified among half of individuals reporting HTS risk. Early identification and characterization of such sub-networks offers a unique opportunity for prevention. This opportunity could be realized if: (i) baseline drug resistance genotyping data were routinely analyzed in real time to define local transmission networks and (ii) individuals identified within these clusters were targeted for early treatment to prevent further transmission.
Acknowledgments
Grant sponsor: Department of Veterans Affairs; Grant sponsor: National Institutes of Health; Grant numbers: AI100665; DA034978; MH083552; MH062512; K01AI110181; AI093163; MH100974; Grant sponsor: James B. Pendleton Charitable Trust; Grant sponsor: University of California, San Diego; Grant sponsor: Center for AIDS Research (CFAR); Grant sponsor: NIH-Funded Program; Grant numbers: P30 AI036214; DA034978
Footnotes
This work was performed at University of California San Diego.
Conflicts of interest: All authors declare no conflict of interest.
AUTHORS’ CONTRIBUTIONS
D. S. coordinated and conducted the analyses and led the manuscript writing; B. C. assisted analyses and manuscript writing; S. M. and J. W. helped establishing the transmission network analysis and assisted with data interpretation; H. S. coordinated study cohort implementation and contributed to interpretation of results; K. K. contributed to interpretation of results. All authors provided significant input into manuscript drafts.
REFERENCES
- Bennett D, Camacho R, Otelea D, Kuritzkes D, Fleury H, Kiuchi M, Heneine W, Kantor R, Jordan M, Schapiro J, Vandamme A, Sandstrom P, Boucher C, van de Vijver D, Rhee S, Liu T, Pillay D, Shafer R. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PLoS ONE. 2009;4:e4724. doi: 10.1371/journal.pone.0004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmet K, Staelens D, Blot S, Dinakis S, Pelgrom J, Plum J, Vogelaers D, Vandekerckhove L, Verhofstede C. Epidemiological study of phylogenetic transmission clusters in a local HIV-1 epidemic reveals distinct differences between subtype B and non-B infections. BMC Infect Dis. 2010;10:262. doi: 10.1186/1471-2334-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin BS, Choi JY, Han Y, Kuang J, Li Y, Han SH, Choi H, Chae YT, Jin SJ, Baek JH, Lim YS, Kim CO, Song YG, Yong D, Li T, Kim JM. Comparison of genotypic resistance mutations in treatment-naive HIV type 1-infected patients in Korea and China. AIDS Res Hum Retroviruses. 2010;26:217–221. doi: 10.1089/aid.2009.0157. [DOI] [PubMed] [Google Scholar]
- Chin BS, Shin HS, Kim G, Wagner GA, Gianella S, Smith DM. Short communication: Increase of HIV-1 K103N transmitted drug resistance and its association with efavirenz use in South Korea. AIDS Res Hum Retroviruses. 2015;31:603–607. doi: 10.1089/aid.2014.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe KW. Epidemiology of HIV/AIDS—Current status, trend and prospect. J Korean Med Assoc. 2007;50:296–302. [Google Scholar]
- Hightower GK, May SJ, Perez-Santiago J, Pacold ME, Wagner GA, Little SJ, Richman DD, Mehta SR, Smith DM, Pond SL. HIV-1 clade B pol evolution following primary infection. PLoS ONE. 2013;8:e68188. doi: 10.1371/journal.pone.0068188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MR, Cho YK, Chun J, Kim YB, Lee I, Lee HJ, Kim SH, Kim YK, Yoon K, Yang JM, Kim JM, Shin YO, Kang C, Lee JS, Choi KW, Kim DG, Fitch WM, Kim S. Phylogenetic analysis of the nef gene reveals a distinctive monophyletic clade in Korean HIV-1 cases. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;17:58–68. doi: 10.1097/00042560-199801010-00009. [DOI] [PubMed] [Google Scholar]
- The Korea Centers for Disease Control and Prevention. Annual Report on the Notified HIV/AIDS in Korea. 2011 [Google Scholar]
- The Korea Centers for Disease Control and Prevention. Annual Report on the Notified HIV/AIDS in Korea. 2013 [Google Scholar]
- Kim GJ, Nam JG, Shin BG, Kee MK, Kim EJ, Lee JS, Kim SS. National survey of prevalent HIV strains: Limited genetic variation of Korean HIV-1 clade B within the population of Korean men who have sex with men. J Acquir Immune Defic Syndr. 2008;48:127–132. doi: 10.1097/QAI.0b013e31816b6ae6. [DOI] [PubMed] [Google Scholar]
- Kim GJ, Yun MR, Koo MJ, Shin BG, Lee JS, Kim SS. Estimating the origin and evolution characteristics for Korean HIV type 1 subtype B using Bayesian phylogenetic analysis. AIDS Res Hum Retroviruses. 2012;28:880–884. doi: 10.1089/aid.2011.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Little SJ, Kosakovsky Pond SL, Anderson CM, Young JA, Wertheim JO, Mehta SR, May S, Smith DM. Using HIV networks to inform real time prevention interventions. PLoS ONE. 2014;9:e98443. doi: 10.1371/journal.pone.0098443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubelchek RJ, Hoehnen SC, Hotton AL, Kincaid SL, Barker DE, French AL. Transmission clustering among newly diagnosed HIV patients in Chicago, 2008 to 2011: Using phylogenetics to expand knowledge of regional HIV transmission patterns. J Acquir Immune Defic Syndr. 2015;68:46–54. doi: 10.1097/QAI.0000000000000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon AF, Joy JB, Woods CK, Shurgold S, Colley G, Brumme CJ, Hogg RS, Montaner JS, Harrigan PR. The impact of clinical, demographic and risk factors on rates of HIV transmission: A population-based phylogenetic analysis in British Columbia, Canada. J Infect Dis. 2015;211:926–935. doi: 10.1093/infdis/jiu560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PP, Korber BT. Detecting hypermutations in viral sequences with an emphasis on G—A hypermutation. Bioinformatics. 2000;16:400–401. doi: 10.1093/bioinformatics/16.4.400. [DOI] [PubMed] [Google Scholar]
- Smith DM, May SJ, Tweeten S, Drumright L, Pacold ME, Kosakovsky Pond SL, Pesano RL, Lie YS, Richman DD, Frost SD, Woelk CH, Little SJ. A public health model for the molecular surveillance of HIV transmission in San Diego, California. AIDS. 2009;23:225–232. doi: 10.1097/QAD.0b013e32831d2a81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Leigh Brown AJ, Hepler NL, Mehta SR, Richman DD, Smith DM, Kosakovsky Pond SL. The global transmission network of HIV-1. J Infect Dis. 2014;209:304–313. doi: 10.1093/infdis/jit524. [DOI] [PMC free article] [PubMed] [Google Scholar]