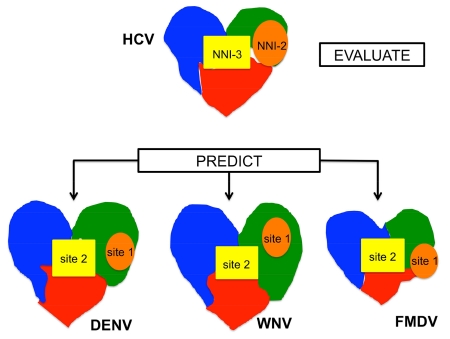
Graphical abstract
1. Introduction
Over the past two decades, there have been many advances in biochemical and biophysical techniques that allow us to identify the proteins responsible for human diseases. An important factor in treating many of these diseases is identifying sites on the corresponding proteins where small molecules may bind in order to modulate protein function. Unfortunately, sometimes it is difficult to determine the three-dimensional structure and location of binding sites with experimental techniques such as X-ray crystallography(Hassell, An et al. 2007). In such scenarios computational methods may be of significant utility. Over the past decade we have seen an increase in the number of computational methods available to predict protein binding sites. Many of these methods can be used in combination with experimental studies as a validation tool or to guide experimental design. With these considerations in mind, we have assessed the ability of four Ligand Binding Site Predictors (LBSPs) to predict known binding sites within the Hepatitis C virus (HCV) polymerase. The most successful LBSP was then used to identify novel binding sites on the surfaces of three structurally related but less well-studied viral polymerases.
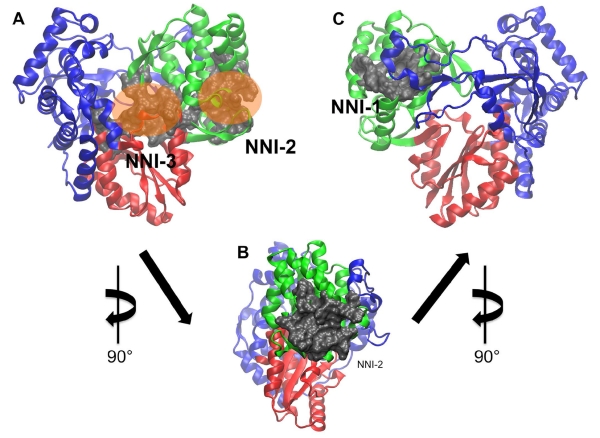
Viral polymerases are crucial players in the life cycle of viruses and are often validated targets for therapeutics(De Clercq 2005, Sesmero and Thorpe 2015). Consequently, our studies have broad implications in suggesting new locations on viral polymerases that can be targeted by small molecules and thus new therapeutic strategies for viral infections. The HCV polymerase is of particular interest, as it displays multiple binding sites for different classes of small molecule inhibitors. Thus, it serves as a useful and interesting model system with which to test the efficacy of binding site prediction algorithms. HCV infection continues to be a global health concern, affecting approximately 200 million people worldwide(Seff and Hoofnagle 2003, Beaulieu 2009). The therapeutic landscape for HCV infection has improved significantly in recent years with the introduction of new and more effective therapies(Lawitz, Mangia et al. 2013, Isaac, Christudas et al. 2015, Keating 2015). However, these therapies continue to have limitations including high cost. The HCV polymerase is one of the key enzyme targets of small molecule therapeutics approved to treat HCV infection and is an ongoing focus of drug discovery efforts. In addition to the active site, the enzyme possesses four allosteric sites that can be targeted for enzyme inhibition(Beaulieu 2009, Li, Tatlock et al. 2009). The HCV polymerase possesses the canonical “right hand” structure common to viral polymerases, consisting of the fingers, thumb and palm subdomains. However, it resembles a “closed hand” rather than the “open hand” configuration frequently observed in other viral polymerases. The active site and two allosteric sites are found within the palm domain, while the remaining two allosteric sites are located in the thumb domain (figure 1). A range of small molecules with diverse chemical properties have been discovered and optimized to target these sites(Condon 2005, Beaulieu 2007, Burton and Everson 2009). Active site inhibitors are either nucleoside inhibitors (NIs) or pyrophosphate analogs, while those that target the allosteric pockets are termed nonnucleoside inhibitors (NNIs)(Condon 2005, Beaulieu 2007, Burton and Everson 2009). Consequently, the allosteric binding sites are referred to as NNI pockets (figure 1).
Figure 1.
Allosteric sites in the HCV polymerase used to evaluate the LBSPs. Protein residues are shown in ribbon representation while residues lining the binding pockets are shown in grey surface representation. Panel A shows a frontal view of the NNI-2 and NNI-3 sites, which are differentiated by the positions of orange translucent spheres. Panel B is a magnified lateral view of the NNI-2 site rotated 90° about the y-axis. Panel C is a rear view of the protein depicting the NNI-1 pocket after an additional 90° rotation about the y-axis. The thumb, palm and fingers domains are shown in green, red and blue respectively.
The strong structural similarities shared by the polymerases of HCV and other viruses suggest that other viral polymerases may exhibit allosteric sites analogous to those observed in the HCV enzyme. If so, it may be possible to identify novel small molecules that could inhibit these enzymes in a similar manner to that achieved for HCV. This is particularly important for viruses that do not currently have therapeutic options available. In this study, we examined polymerases of Dengue (DENV), West Nile (WNV) and Foot-and-mouth disease (FMDV) viruses in order to predict the location of potential allosteric binding sites. These diseases have no or limited treatment options available (Malet, Massé et al. 2008, Noble, Chen et al. 2010) and have become increasingly prevalent, cause significant mortality, morbidity or economic cost. The first major obstacle encountered in performing a study such as this is that there is not much biochemical or structural information identifying allosteric sites within these polymerases. This makes the problem well suited for the application of computational tools to predict novel binding sites within these proteins. In doing so, we have placed emphasis on allosteric sites for two main reasons: i) active sites are well conserved across many viral families, thus predicting active site binding is not anticipated to be difficult and ii) in drug discovery for polymerases, allosteric sites tend to be unique to the virus. Thus, inhibitors that bind at these sites may reduce nonspecific side effects that can lead to host cellular toxicities. We believe that the HCV polymerase is an excellent model to evaluate the effectiveness of ligand binding site predictor (LBSP) tools in predicting allosteric sites due to the wealth of structural and biochemical data available describing the interactions between allosteric inhibitors and this enzyme. Additionally, we suggest that one can use existing information available for the HCV polymerase to make meaningful inferences about the locations of allosteric sites in the DENV, WNV and FMDV polymerases due to the strong structural and functional similarities among these enzymes.
Four different LBSPs were employed to determine which tool performed best at predicting allosteric binding sites within the HCV polymerase. Our target sites are: NNI-1 and NNI-2 located in the thumb and NNI-3 found in the palm (figure 1). Because the two allosteric sites found within the palm domain (NNI-3, NNI-4) largely overlap, we treated these as a single site, using the residues for NNI-3 as a proxy for both palm allosteric sites given that NNI-3 spans the palm domain and thumb-fingers junction. Additionally, our previous studies have shown that an NNI-3 inhibitor is able to interact with diverse protein residues throughout the palm and thumb domains, including residues that are typically considered part of the NNI-4 pocket. This observation suggests that the NNI-3 and NNI-4 sites are not distinct binding locations but instead are different regions within a single, large binding pocket in the palm domain (Brown and Thorpe 2015). Targeting these three allosteric sites with diverse residue composition, size, shape and location allows for a robust evaluation of the LBSPs in identifying allosteric binding sites in viral polymerases. The tools evaluated were: FTSite (Ngan, Hall et al. 2012), QsiteFinder (Laurie and Jackson 2005), LISE (Xie and Hwang 2012) and Ligsitecsc (Huang and Schroeder 2006). For this study, we focused on allosteric sites in the HCV polymerase because these pockets have been validated as drug targets and structures of the corresponding protein-ligand complexes have been extensively characterized, providing an effective means to test our hypothesis. In contrast, there is much less structural and biochemical information available for allosteric sites on other viral polymerases.
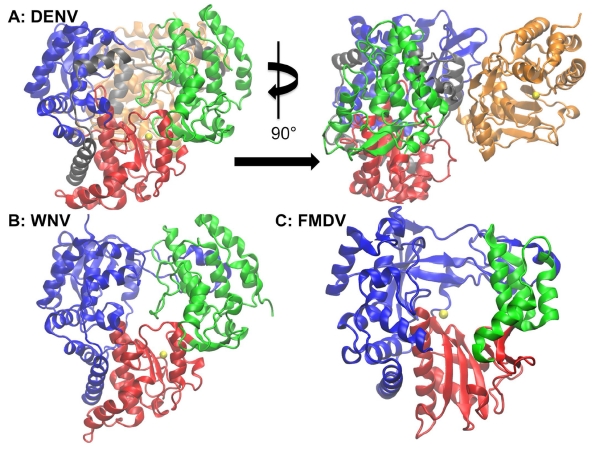
DENV and WNV are both clinically important viruses that, like HCV, are members of the flaviviridae family (figure 2). Like HCV, both DENV and WNV possess positive-sense single-stranded RNA genomes. They are considered emerging pathogens whose outbreaks may lead to fatal outcomes(Malet, Egloff et al. 2007). About 50% of the world’s population across 128 countries is at risk for DENV infection(Brady, Gething et al. 2012, Bhatt, Gething et al. 2013). As of 2010, there were an estimated 390 million DENV infection events globally, of which about 96 million possess some form of clinical severity(Bhatt, Gething et al. 2013). WNV has also significantly increased in global prevalence since an outbreak in 1999 in which the virus was introduced to the United States and Europe(G 2004, Brandler and Tangy 2013). WNV epidemics from 1999 to 2006 resulted in 23,925 human disease cases and 946 deaths in the United States alone(Malet, Egloff et al. 2007) and this virus continues to induce additional mortality(Brandler and Tangy 2013). The polymerases of DENV, WNV and HCV are alike in their global protein folds(Yap, Xu et al. 2007) (figures 1 and 2). However, the full-length polymerases of DENV and WNV are larger than that of HCV, possessing a methyltransferase (MTase) domain in addition to the RNA-dependent RNA polymerase domain(Zhou, Ray et al. 2007, Malet, Massé et al. 2008, Noble, Chen et al. 2010).
Figure 2.
Polymerases of DENV, WNV and FMDV. The enzyme subdomains are colored as in Figure 1: this color scheme is maintained throughout the remaining figures. The left image in panel A shows the frontal view of the DENV polymerase with a magnesium ion (yellow) present in the active site while the right image depicts the DENV enzyme rotated 90° about the y-axis to allow for the methyltransferase (MTase) domain (colored in gold) to be visible. Panel C is the WNV polymerase with a magnesium ion (yellow) in the active site. While WNV also possesses an MTase domain, a full-length structure containing both the polymerase and MTase domains has not yet been solved. Finally, panel C shows the FMDV with a catalytic manganese ion (yellow).
There have been efforts to discover and develop vaccines and antiviral therapies against DENV and WNV, but these have thus far been unsuccessful(Gu, Ouzunov et al. 2006, Malet, Massé et al. 2008, Chappell, Stoermer et al. 2008, Noble, Chen et al. 2010). Currently, there are several vaccine candidates in clinical trials for DENV (Blaney, Matro et al. 2005, Thisyakorn and Thisyakorn 2014), although an approved vaccine is not yet available. In addition, inhibitors have been identified that target various enzymes critical to the life cycle of DENV(Lim, Wang et al. 2013, Schwartz, Halloran et al. 2015, Smith, Lim et al. 2015). An adenosine analog (an active site inhibitor) as well as an N-sulfonylanthranilic acid NNI (thought to bind at the entrance of the template channel) of the DENV polymerase(Yin, Chen et al. 2009) have been identified, though these small molecules have not progressed to clinical trials. To date, there are at least 5 vaccine candidates for WNV that have entered clinical trials, with a trial for one of the most promising candidates beginning as recently as July 2015(Brandler and Tangy 2013, Pinto, Richner et al. 2013, Petersen 2014, Yamshchikov 2015). There have also been several studies identifying small molecule inhibitors of the WNV polymerase(Jordan, Briese et al. 2000, Gu, Ouzunov et al. 2006, Furuta, Gowen et al. 2015). Unfortunately, these small molecules are primarily broad spectrum active site antivirals that may result in nonspecific targeting of host proteins or fail during in vivo experiments(Jordan, Briese et al. 2000, Furuta, Gowen et al. 2015).
FMDV is a member of the Aphthovirus genus in the picornaviridae family and infects cloven-hoofed domestic animals such as cattle in addition to more than 40 wild animal species(Grubman and Baxt 2004). The cost of a FMDV outbreak in the UK in 2001 has been valued at 6 billion pounds sterling and it is estimated that an epidemic of similar proportions would cost the United States about 100 billion dollars(Ferrer-Orta, Arias et al. 2004). Due to an extremely high mutation rate during replication, there are about 65 subtypes of FMDV. This results in high antigenic diversity and makes it difficult to have a universal vaccine(Domingo, Baranowksi et al. 2002, Durk, Singh et al. 2010). The continued threat of costly outbreaks and lack of a universal vaccine warrants continued drug discovery efforts for FMDV. The FMDV polymerase serves as a challenging test of whether known structural information about the HCV polymerase can be used to garner useful insight into the properties of an enzyme with more significant dissimilarities in genome and structure than DENV or WNV. FMDV possesses a positive sense single strand RNA genome like HCV, DENV and WNV, as well as a polymerase with similar overall architecture to the corresponding enzymes from these viruses. However, there are several etiological and structural differences. The FMDV polymerase is the smallest of the four enzymes, possessing less than 500 residues, and has not been reported to contain an MTase domain(Ferrer-Orta, Arias et al. 2004). Unlike DENV and WNV, there has been an experimental study identifying and confirming the presence of an allosteric inhibitor binding site located in the fingers domain of the FMDV polymerase, proximal to, but not overlapping with the active site(Durk, Singh et al. 2010). While there is no crystal structure containing a ligand bound to this site, mutagenesis has revealed the residues critical for inhibitor binding. Thus, this allosteric site provides another opportunity to test the robustness of the selected LBSP.
An obstacle limiting clinical success of allosteric inhibitors for these viral polymerases is reduced potency with regard to in vivo cellular assays relative to that measured for in vitro studies(Noble, Chen et al. 2010). One of the underlying causes for this observation is that polymerase regions outside of the active site and palm domains are subject to reduced evolutionary constraints to maintain their amino acid composition, as they are not directly involved in catalysis. Consequently, there is a lower barrier for the emergence of resistance as a result of amino acid substitutions. This situation is exacerbated by the fact that these polymerases do not possess proofreading ability. Thus, during infection the virus typically exists as a collection of quasi-species, each with slightly different genetic composition, providing a fertile platform for the development of drug resistance. Nonetheless, it has been shown that applying multiple allosteric inhibitors in combination may diminish this problem(Noble, Chen et al. 2010). Recent simulation studies from our group have begun to elucidate the molecular mechanisms underlying the enhanced inhibition obtained when multiple NNIs are presented to the HCV polymerase(Brown and Thorpe 2015). This knowledge may facilitate future efforts to employ similar strategies in other viral polymerases such as those of DENV, WNV or FMDV.
The results of our present study indicate that LISE is an accurate tool for identifying known allosteric sites in the HCV polymerase. Given the structural similarities of many viral polymerases, this finding suggests that LISE may also be successful in identifying allosteric binding sites in other viral polymerases, especially those that are closely related to the HCV enzyme. Thus, we employed LISE to predict binding sites in the polymerases from DENV, WNV and FMDV. In all three enzymes we observed that there are putative allosteric sites in the palm and thumb domains that are structurally similar to pockets within similar regions in the HCV polymerase, suggesting that the structural commonalities of the enzymes result in allosteric pockets in similar locations on the enzyme surface. Moreover, the geometry and chemical composition of these putative sites are comparable among the different enzymes, suggesting that the common structural features may be associated with similar ability to bind small molecules. In turn, this may translate to similar functional properties in the polymerases, including an ability to be inhibited by allosteric ligands.
2. Materials and Methods
2.1 Description of LBSPs
FTSite
The FTSite algorithm is an energy-based tool developed on the basis that ligand-binding sites will readily accommodate small organic molecules of various shapes and polarity(Ngan, Hall et al. 2012). FTSite employs sixteen organic probes(Kozakov, Hall et al. 2011) to detect sites of favorable protein-ligand interactions. Use of such diverse probes is thought to increase the likelihood of predicting a broad range of ligand-binding pockets. The algorithm docks the probes to the query protein then identifies a collection of favorable docked poses. These poses are subsequently minimized with the CHARMM force field(Brooks, Bruccoleri et al. 1983) using the implicit solvent method Analytic Continuum Electrostatics (ACE) and the Newton-Raphson algorithm(Brenke, Kozakov et al. 2009). The minimized poses are then clustered based on their energies and the six most energetically favored clusters associated with each probe are retained. The next step is to identify overlap between protein regions predicted by different probes and classify these as consensus clusters. These consensus clusters are then ranked according to the number of non-bonded interactions between the protein and all the probes in the cluster. One potential limitation of FTSite is that the use of organic probes can hinder the identification of ligand binding sites on a protein surface. Unlike large cavities, surface binding sites are often shallow and require rearranging of atoms to reduce steric hindrance when accommodating a ligand. The static protein structures employed in most protein-ligand docking experiments preclude rearrangements within the local environment of such pockets to accommodate the probe(s). This in turn provides a potential bias towards ligand binding sites within cavities of proteins where steric hindrances are reduced(Ngan, Hall et al. 2012).
QsiteFinder
Like FTSite, QsiteFinder is an energy-based tool. However, QsiteFinder differs from the former in that it employs only methyl probe(s) to predict the most favorable ligand-binding sites(Laurie and Jackson 2005). QsiteFinder encompasses a number of separate components. First, the LigandSeek module separates the ligand and protein coordinates. The protein coordinates are rotated around their geometric center to better align the protein to the grid, thereby reducing grid points. The next step consists of docking the methyl probe(s) with the query protein. The LIGGRID module calculates the non-bonded interaction energy of the docked probe-protein poses using GRID force parameters(Jackson 2002). Poses with the most favorable energies are retained and subsequently clustered if they are within 1 Å of each other. Finally, the coordinates are rotated back into their original orientation and a putative ligand-binding site is predicted if 25% or higher of predicted probe clusters overlap at a given location. One known caveat of applying this method is that predictions are generally more accurate for ligand bound conformations than unbound conformations. In Qsite Finder’s initial evaluation by Laurie et al. found that even subtle changes in protein structure caused by ligand binding could alter existing binding sites, leading to a drop in precision (Laurie and Jackson 2005). However, we note that ligand induced conformational changes may impact the performance of all of these algorithms since they each employ static protein structures as inputs.
LISE
LISE is a geometric and propensity based algorithm conceptually derived from the docking score function MotifScore(Xie and Hwang 2010). MotifScore works by searching a protein-ligand complex for specific networks of interaction motifs consisting of 3 protein and 2 ligand atoms thought to occur primarily at ligand binding sites. It then orients the ligand into the best position to maximize these interactions(Xie and Hwang 2010). These interaction motifs are termed protein triangles within LISE and are associated with a binding site enrichment factor, which denotes the likelihood of these triangles occurring at a ligand-binding site. The enrichment factor is the ratio of the probabilities of a motif occurring at a specific site relative to the probability of its occurrence throughout the entire protein. Protein triangles with binding site enrichment factors less than one are eliminated. The enrichment factors of the retained protein triangles are used in conjunction with a weighting factor of 1.7 and conservation scores taken directly from PSI-BLAST’s position specific scoring matrix to calculate the triangle scores. A three-dimensional grid at 1 Å intervals is then created around the protein and each grid point denoted as protein occupied or empty based on a specific distance threshold. If a grid point is labeled empty, then a grid point score is computed as the sum of all triangle scores within 4 Å of the grid point. The next step is to calculate the sphere score, which is the sum of all the grid point scores centered within 11 Å of each empty grid point. Finally, the sphere scores are ranked with the top predicted site being associated with the highest value. One limitation of LISE is that it omits protein triangles with enrichment factors that are less than one, which most often are on the protein surface. On the other hand, the statistical propensity approach that LISE employs negates the need for tedious energy calculations such as those used in FTsite and QsiteFinder.
LIGSITEcsc
LIGSITEcsc is a geometry-based algorithm developed using surface-solvent-surface interactions to predict putative binding sites. The first step of this algorithm is to project the query protein onto a three-dimensional grid and align the protein along the xyz axes. After alignment, each grid point is denoted as one of the following: i) protein if it contains a protein atom within 1.6 Å of the grid point; ii) surface (defined using a combination of the protein vdW surface and solvent accessible surface computed using the Connolly algorithm(Connolly 1983)); and iii) solvent (all remaining volume). The next step involves scanning in all direction to identify surface-solvent-surface events, which are characterized as a sequence of protein grid points that are bounded by surface grid points with solvent grid points in between. Next, if a solvent grid point is part of five surface-solvent-surface interactions it is marked as a potential ligand-binding pocket. These pockets are clustered if they are within 3.0 Å of each other. Finally, the clusters are ranked according to the number of grid points in each. One drawback of LIGSITEcsc is that the use of the Connolly Surface restricts the prediction of interior ligand-binding sites within channels and crevices of the protein. Nonetheless, it is also the use of the Connolly surface that allows this tool to consistently identify surface binding sites.
Selection of LSBPs
There are a plethora of LBSPs described in literature, each with its own unique advantages and limitations (see table 1 for a brief summary of the pros and cons of the LBSPs used in this study). However, each of the programs described in this work has already been extensively validated by the original authors on a large set of standardized protein-ligand complexes, allowing for fair comparisons among the methods(Laurie and Jackson 2005, Huang and Schroeder 2006, Ngan, Hall et al. 2012, Xie and Hwang 2012). Our goal in selecting this specific collection of tools was to use programs that have been shown to perform well for a large assortment of protein-ligand complexes and that employ a diverse set of physical and chemical principles to predict ligand-binding sites. More specifically, one reason LIGSITEcsc was selected was because it originated from and performed better than LIGSITE(Hendlich, Rippmann et al. 1997), which is a geometric-based approach that is frequently used as a reference for comparison for newer LBSPs. In contrast, Qsite Finder and FTSite are energy-based methods that are reported to also display superior performance to LIGSITE. LISE differs from the other three tools in that it combines both statistical and geometric computations to make predictions. This program demonstrated superior performance to Qsite Finder and LIGSITEcsc on the Protein Ligand Database dataset (Puvanendrampillai and Mitchell 2003, Huang and Schroeder 2006, Xie and Hwang 2012). It is quite possible that a different collection of LBSPs could perform equally well, or perhaps even better if the right combination can be fortuitously identified. However, our main point is not that these particular LBSPs must necessarily be used, but that the general approach we describe is of utility in predicting unknown ligand binding sites. In principle it can be adapted for any combination of LBSPs that meets the needs of a particular study.
Table 1.
Features of LBSPs Evaluated
| FTSite | Qsite Finder | LISE | LIGSITEcsc | |
|---|---|---|---|---|
|
Type of
Algorithm |
Energy | Energy | Geometric & Propensity |
Geometric |
| Chemical Probe | 16 organic probes |
Methyl probe(s) | none | none |
| Pro(s) | diverse array of probes |
prediction of surface sites |
negates the need for any energy consideration |
prediction of surface sites |
| Con(s) | organic probes cause bias towards predicting large cavities |
predictions more accurate for holo structures than apo structures |
surface sites can be under predicted |
Connolly Surface restricts the prediction of interior sites |
2.2 Evaluation of LBSPs
Identification of known Polymerase Binding Sites
As mentioned previously, our primary mode of evaluating LBSPs was to detect NNI-1, NNI-2 and NNI-3 sites of the HCV polymerase. Since these sites have been previously identified via x-ray crystallography, we used the Ligand Explorer utility of PDB.org and the Visual Molecular Dynamics (VMD) program to locate all protein residues within 8 Å from the ligands. This distance threshold allowed us to capture long-range interactions between protein residues and the ligands that are consistent with x-ray crystallography and biochemical data. These include water mediated protein-ligand hydrogen bonds important to the potency of some NNIs (e.g. NNI-2 ligands) that are not captured using shorter distance cutoffs such as 4 Å.
We note that during the initial development and testing of several LBSPs including FTSite, LISE and LIGSITEcsc, a successful hit is defined when at least one point of a predicted binding site (usually the geometric center) is within 4 Å of any ligand atom. However, such a criterion cannot be applied in cases where the binding site is unknown. In addition, a cutoff this short would omit some of the long-range interactions noted above that are important for binding of NNIs. Consequently, we employ different criteria to assess success in this study. First, we focus on identifying residues that comprise a ligand-binding site rather than on identifying the location of ligand atoms themselves. Since the residues around experimentally confirmed ligand binding pockets are known, we can compare these known residues to the residues surrounding the predicted site. In the case of LISE or LIGSITEcsc that output the geometric center of a predicted site, a cutoff of 8 Å was employed to identify the protein residues surrounding the predicted site. This allowed us to compute the overlap between the known and predicted binding pocket residues. The percentage of overlap is defined as the total number of predicted pocket residues in common with the known pocket residues, divided by the total number of known pocket residues and multiplied by 100. Rather than providing a geometric center, FTSite and Qsite Finder directly list the residues anticipated to comprise each predicted binding site. Thus, a distance cutoff was not required for the latter tools.
We chose an overlap of 50% or greater as denoting a successful hit. This precision threshold ensures that at least half of the residues comprising a binding pocket are identified within the predicted site. When we compare binding site residues described in the literature to those identified using an 8 Å cutoff from the center of mass of each ligand, we observed an overlap of 56% or more. This observation motivates our decision to use a threshold of 50% or greater to denote a successful hit in the other systems employed in this study. Note that this overlap criterion is significantly more stringent than that employed in the development of LBSP tools such as Qsite Finder and LISE, which can be as low as 25%. Another benefit of defining successful hits based on identifying binding pocket residues is that this criterion can be readily extended to cases where the binding site location is unknown. In such cases we can anticipate that residues within 8 Å of predicted binding site locations have a probability of being part of an actual ligand binding site that is at least as high as the precision threshold (i.e. 50%).
For additional validation, we used LISE to predict known allosteric sites on the FMDV and Coxsackievirus (CSV) polymerases. FMDV and CSV have known allosteric sites in the fingers domain and are both members of the picornaviridae viral family. As these viruses are more distantly related to HCV than DENV or WNV, they provide a more stringent test of the ability of LISE to perform well for diverse viral polymerases.
Structure Preparation
We used three protein data bank (PDB) structures that are representative of the target sites in the HCV polymerase. These structures are 2BRL(NNI-1), 2WHO(NNI-2) and 3CO9(NNI-3). We prepared each crystal structure by deleting chains B and higher (if the structure was solved with multiple chains), ions, water molecules and bound ligands. The free protein structures were individually submitted to online servers for each of the four LBSPs. Deletion of each ligand exposes its binding pocket and allows the tools to use only the protein structure to predict putative binding sites, as is the case with unknown query structures.
In using LISE to search for allosteric sites within the DENV, WNV, FMDV and CSV polymerases, we employed three different structures for each viral enzyme. We used protein structures that did not possess any small molecule inhibitors or for which bound ligands were deleted before submission to the server. The DENV query structures have PDB ID’s 4V0R, 2J7W and 2J7U, while for WNV we used the 2HCN, 2HCS and 2HFZ coordinates. For FMDV we used coordinates 1U09, 2F8E and 2D7S while for CSV we used 3DDK, 4Y2A and 4WFZ. Finally, for these systems we only considered and further evaluated predicted sites that occurred in at least 2 out of the 3 query structures for a given enzyme.
Analysis
We conducted both direct and cross comparisons to evaluate each tool’s performance in predicting the target sites in the HCV and FMDV polymerases. The purpose of direct comparisons is to evaluate how well each tool is able to predict each target site when the query protein was previously solved with a ligand in that particular binding pocket. For the direct comparisons, we compared the residues of each predicted site to the residues of the known binding pocket of the query protein. For example, all the predicted sites for the NNI-1 allosteric pocket from the four LBSP tools were compared to the known 2BRL NNI-1 site. In contrast, we performed cross comparisons to evaluate the robustness of each tool. Specifically, we wanted to determine which LBSP was the best at predicting the largest number of known sites regardless of query structure. For cross comparisons, we compared all putative sites to the known binding sites. Using the example above, all predicted pockets for the 2BRL query structure (containing an NNI-1 site) were compared to known NNI-2 and NNI-3 sites. Note that since there is only one elucidated allosteric site in the FMDV and CSV enzymes, there were no cross comparisons for these systems. Finally, electrostatic potentials were evaluated for protein residues comprising the surface of each predicted binding site. The electrostatic potential maps were generated by using the PDB2PQR web server(Dolinsky, Neilsen et al. 2004, Dolinsky, Czodrowski et al. 2007) to convert the respective PDB files to PQR format containing the necessary charge distributions. The PQR files were then employed to calculate the electrostatic potentials with the APBS program(Baker, Sept et al. 2001).
3. Results and Discussion
3.1.1. LISE Effectively Predicted Allosteric Sites in the HCV Polymerase
LISE performed well in predicting each of the target sites within the HCV polymerase in direct comparisons (Table 2). In contrast, both Qsite Finder and LIGSITEcsc were able to identify only the NNI-1 and NNI-2 target sites. FTSite proved to be the least successful, as it only identified the NNI-3 pocket. Although the cross comparisons suggest that both LISE and Qsite Finder were able to predict all of the target sites when every query structure was considered, overall LISE identified the target sites more consistently (tables S1, S2).
Table 2.
Percent Overlap for Direct Comparisons of the Top Predicted Site(s) and Query Site for Each LBSP
| PDB ID of Query Structures: |
Percent Overlap for Top Predicted Sites‡ for Each LBSP | |||
|---|---|---|---|---|
| FTSite | Qsite Finder | LISE | LIGSITECSC | |
| 2BRL (NNI-1) | <50% | 88% | 81% | 81% |
| 2WHO (NNI-2) | <50% | 75% | 66% | 75% |
| 3CO9 (NNI-3) | 50% | <50% | 63% | <50% |
FTSite only gave predictions for the top 3 sites while the other tools gave the top 10 putative sites.
One of our initial hypotheses was that, due to the diverse array of organic probes used in FTSite [Ngan, Bioinformatics 2011], this program would outperform the other tools. However, as mentioned above it failed to predict locations of the NNI-1 and NNI-2 sites. Furthermore, the percent overlap was barely at the threshold cutoff for the NNI-3 pocket and was significantly lower than those observed for LISE and Qsite Finder (table 2, S1, S2 and S3). Both NNI-1 and NNI-2 sites are located outside the large central cavity found in the polymerase palm domain. Thus, this observation is consistent with the known limitation (noted in section 2.1 Methods) that this tool tends not to identify surface sites. Results from the cross comparisons for FTSite also confirmed the tendency for FTSite to predict interior cavities (table S3). Specifically, all the sites predicted by FTSite in table 2 are located within the central cavity of the protein regardless of input structure. Thus, we conclude that FTSite possesses a strong bias for predicting binding sites that are contained within large interior cavities and would not be ideal for predicting external surface sites. This may limit the utility of this program in a protein for which there is no a priori knowledge regarding the binding site location.
As stated above, both Qsite Finder and LIGSITEcsc were able to identify only the NNI-1 and NNI-2 target sites in the direct comparisons (table 2). Both of these sites can be found on the surface of the NS5B polymerase. In addition, both sites are located within the thumb domain of NS5B and are predominantly hydrophobic. As we noted previously, Qsite Finder utilizes methyl probes(Laurie and Jackson 2005) for binding site predictions, which may help to explain why it primarily identified hydrophobic pockets. Despite a bias towards predicting hydrophobic target sites, Qsite Finder was also able to identify other putative sites within different domains of the NS5B protein. While these sites did not correspond to known allosteric sites within NS5B, this observation suggests that Qsite Finder has less bias for predicting pockets within the central cavity of the enzyme compared to FTSite (which only identified sites within this central cavity). An interesting observation was that Qsite Finder was able to predict the NNI-3 site when 2BRL and 2WHO were input structures but not when the corresponding NNI-3 structure 3CO9 was the query. This finding may suggests that Qsite Finder is particularly sensitive to the conformational differences that exist among the query structures. Thus, the predictions from Qsite Finder may be more susceptible to being negatively impacted by the occurrence of ligand induced protein conformational changes (see section 2.1). Unlike the hydrophobic probes employed by Qsite Finder, the geometric criteria employed by LIGSITEcsc are anticipated to bias the program towards identifying surface cavities at the expense of detecting deeper cavities and channels within the protein. Our results confirm this expectation, as LIGSITEcsc only detected the NNI-1 and NNI-2 sites (table 2).
LISE, in comparison to the other three tools, made more consistent predictions across all input queries. The only exception to this observation is the NNI-1 site, which was only detected when 2BRL was used as a query. However, we note that the other LBSPs that were able to predict NNI-1 also were only successful using the 2BRL structure. Unlike FTSite, Qsite Finder and LIGSITEcsc that exhibit biases towards identifying either internal, hydrophobic or surface target sites, LISE is able to accurately predict all site categories. The statistical propensity approach employed in LISE eliminates limitations that may be associated with energy-based methods such as the need to extensively sample the protein structure with probes or the longer search times typically required in order to perform energy calculations(Xie and Hwang 2010, Gu, Qiu et al. 2012). One potential caveat to note in employing LISE is that the calculation of the triangle score often results in the omission of over 70% of surface triangle motifs(Xie and Hwang 2012), which may hinder the prediction of surface ligand-binding sites such as the NNI-1 or NNI-2 target sites. However, LISE was still able to identify both NNI-1 and NNI-2 sites, suggesting that surface sites that are true binding pockets are not removed based on the triangle score.
It is interesting that the NNI-1 site was only predicted when the 2BRL structure was used as an input query. One possible reason for this observation is that this allosteric site only exists when a ligand is present. The ligand binding pocket in 2BRL lies in a cleft between the thumb and fingers domains(Di Marco, Volpari et al. 2005). In the absence of a ligand this cleft is completely filled by a flexible loop (residues 9 – 41) from the fingers domain. Thus, this binding pocket is formed via displacing this loop. In this case, one would anticipate that when the ligand is absent the LBSPs would not be able to predict the presence of this site. Results from the cross comparisons support this hypothesis (see Tables S1 to S4). This finding suggests that the inability to predict ligand-induced binding sites may be a general limitation of LBSPs. This is not surprising given that these tools generally employ static protein structures for their predictions. A consequence of this occurrence is that natively occurring variations in the protein structures employed for the queries may influence the type and number of functionally relevant sites predicted for a given system. Thus, no LBSP had the same target sites predicted across all the query structures. This result probably is due to the fact that the three HCV polymerase query structures used for validation represent, to some degree, the structural variability that one might expect to occur across a solution ensemble. Thus, the three binding sites likely display different characteristics in each structure. LISE displayed the most consistent performance, as the NNI-2 and NNI-3 sites were predicted for all three of the query structures (tables 2 and S1). The noticeably better performance of LISE for the HCV polymerase validates its use to predict polymerase binding sites in the less studied DENV, WNV and FMDV.
Finally, we note that when using these LBSPs it is important to consider their high false positive rates in predicting target sites: every query structure had predictions that were not among any of the known sites. In Supporting Material we provide Tables S5-S7 that display the false positive rate of LISE for each of the HCV query structures and Table S8 that summarizes the average false positive rate for each query. We see that LISE displays a fairly high false positive rate for individual query structures (80-85%). The high rate of false positives for individual structures may be related to the inherent structural variability reflected in these structures and sensitivity of LBSPs to this variability. However, the likelihood that a given prediction is a false positive can be reduced significantly if one employs multiple query structures of a given protein and looks for agreement among the putative binding sites in DENV, WNV and FMDV. Thus, we compare the predictions among the different query structures and choose sites that represent the consensus result. Specifically, as mentioned in section 2.2 we only considered and further evaluated predicted sites that occurred in at least 2 out of the 3 query structures for DENV, WNV and FMDV. We find that these consensus solutions are more likely to identify functionally relevant pockets and exhibit a reduced propensity to incorrectly identify residues as being part of a binding site (see Table S9). This consideration was the primary reason we employed a minimum of three X-ray crystallographic structures as queries for each of the viral polymerases for which the locations of allosteric sites are not necessarily known. In support of this approach, we note that the thumb and palm sites identified in this study for DENV were recently validated by computational and biochemical experiments(Yokokawa, Nilar et al. 2016). This finding will be discussed in greater detail in section 3.2.4.
3.1.2. LISE Predicts Known Allosteric Sites in FMDV and CSV
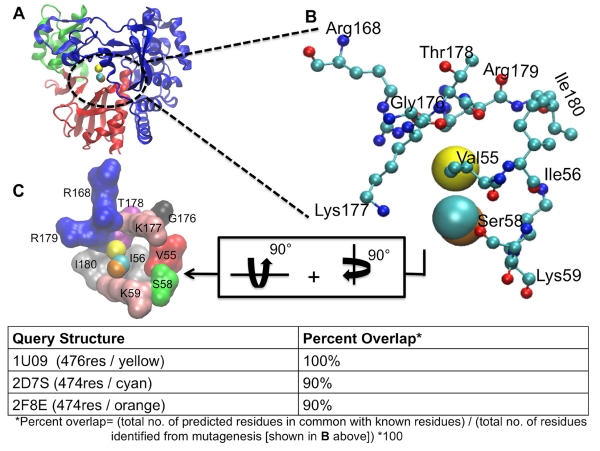
LISE was able to predict the allosteric site in FMDV previously identified by Durk et al. (Durk, Singh et al. 2010). The ligand 5D9 binds to this pocket, which is located in the fingers domain proximal to the NTP binding site (figure 3). Successful matches were observed with all three input structures. Specifically, comparisons of residues making up the predicted LISE sites and known residues elucidated by mutagenesis resulted in a 100% overlap for the 1U09 query structure and 90% overlap for 2D7S and 2F8E (figure 3). Furthermore, each of the LISE matches identified Lys59 and Lys177, which have been shown to be critical for inhibitor binding through the mutagenesis studies by Durk et al. noted above. Another test of the robustness of LISE was its ability to predict the presence of the magnesium ions binding site in the FMDV polymerase, identifying each of the residues (Asp238, Asp240, Asp339, Val239, Ile340 and Thr384) that interact with the magnesium ions (Ferrer-Orta, Arias et al. 2004). Specifically, all of these residues were found in the fourth most highly ranked sites for both the 2D7S and 2F8E query structures while 5 of the residues were identified as the part of the seventh most highly ranked site for the 1U09 input.
Figure 3.
LISE accurately predicts the site of a known allosteric inhibitor of the FMDV polymerase. Panel A shows the rear of the enzyme with the LISE predicted spheres from the 3 query structures in van der Waals (VDW) representation (1U09 in yellow, 2F8E in orange and 2D7S in cyan). Panel B is a magnified view of A showing only the residues (in CPK representation) that have been biochemically established to be critical for ligand binding at this site and the LISE predicted spheres (results for query structure 1U09 in yellow, 2F8E in orange and 2D7S in cyan). Panel C displays the residues making up this site depicted in surface representation after 90° rotations about both the y and x axes. Each residue is separately colored and labeled.
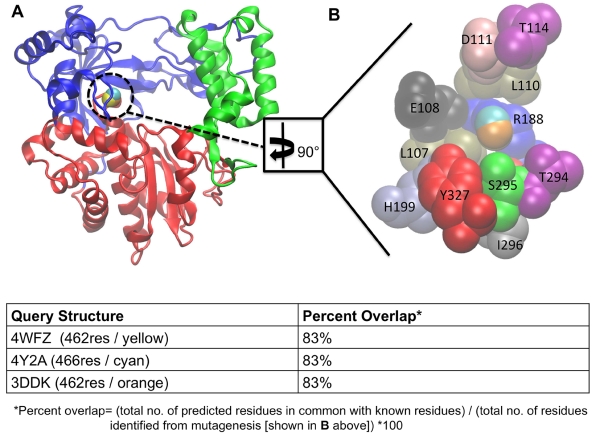
LISE was also able to identify a known allosteric site in the polymerase of CSV (Figure 4) (Van Der Linden, Vives-Adrián et al. 2015). CSV can cause a number of ailments including rashes, upper respiratory tract infections and meningitis. While CSV is not our primary focus, the ability of LISE to identify the known CSV allosteric site provides additional support regarding the predictive power of LISE for viral polymerases. This success is further evidence that LISE can capably identify ligand binding sites in diverse viral polymerases. As we will show below, LISE not only performed exceptionally well at predicting previously identified sites, but also predicted new cavities in DENV, WNV and FMDV that have counterparts on the HCV polymerase.
Figure 4.
LISE accurately predicts the site of a known allosteric inhibitor of the CSV polymerase. Panel A shows a frontal view of the enzyme with the LISE predicted spheres from the three query structures in van der Waals (VDW) representation (4WYZ in yellow, 3DDK in orange and 4Y2A in cyan). Panel B is a magnified view of A after a 90° rotation about the y axis showing only the residues (in VDW representation) comprising the ligand binding site and the LISE predicted spheres (results for query structure 4WFZ in yellow, 3DDK in orange and 4Y2A in cyan). Each residue is colored according to its name and is individually labeled. Note: Y195 (in red), which is behind S295 (in green) is the only residue not fully visible in the current view.
3.2 HCV-like Allosteric Pockets Exist in DENV, WNV and FMDV
3.2.1. DENV
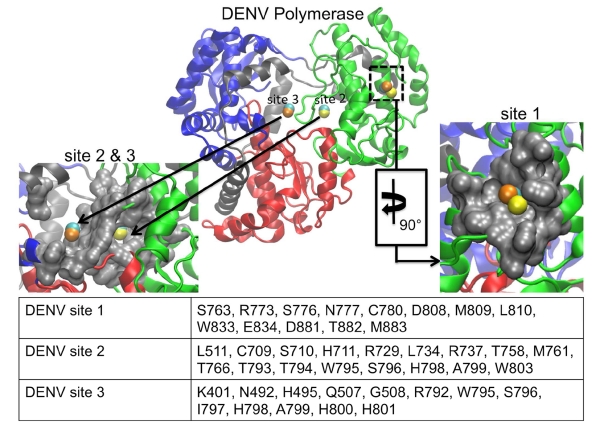
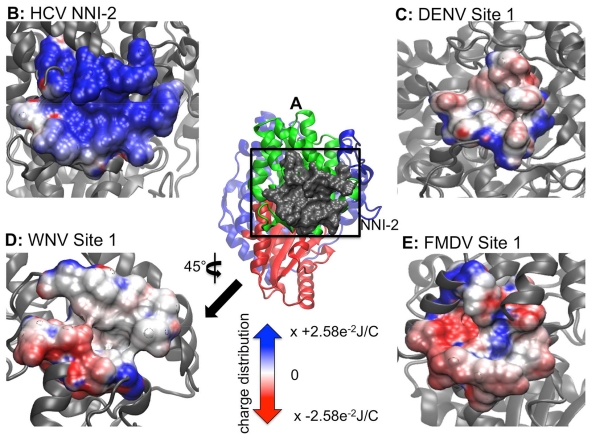
LISE predicted three putative sites in the DENV polymerase that each had a similar counterpart in the HCV polymerase (figure 5). Malet and coworkers had previously used the CASTp(Dundas, Ouyang et al. 2006) and PASS(Brady and Stouten 2000) LBSP tools to predict two cavities in the DENV polymerase and 5 cavities in the WNV enzyme(Malet, Massé et al. 2008). These two pockets in the DENV enzyme were subsequently tested for functional utility via mutagenesis studies. One residue in cavity A (Lys756) and four in cavity B (Leu328, Lys330, Trp859, Ile863) were identified as being crucial for viral replication(Zou, Chen et al. 2011). We compared our LISE predictions with the residues comprising cavities A and B as listed in their work. LISE identified cavity A in the thumb domain for each of the 3 DENV query structures. Specifically, one of the top ten predicted sites (site 1 in figure 5) for each of 4V0R, 2J7U and 2J7W were matches for cavity A. Overlap values based on the residues provided in Malet et. al. (Malet, Massé et al. 2008) were at or above our 50% threshold (50% for 4V0R, 58% for 2J7U and 67% for 2J7W). We believe site 1 in DENV possesses some similarities to the NNI-2 site within the HCV polymerase. Visually, both sites are found on the lateral surface of the thumb domain in similar locations. This region of the enzyme is displayed in panel B of figure 2; further references to the lateral enzyme surface should be understood to denote this location. The DENV site 1 is slightly smaller and is located slightly higher in the thumb domain than the NNI-2 pocket. Thus, only the residues lying at the bottom of the DENV site 1 coincide with the top portion of where the NNI-2 site would be located in the HCV polymerase (figures 1 and 5). Nonetheless, both sites are similarly hydrophobic(Ontoria, Rydberg et al. 2009) and posses a deep grove that runs their entire length. However, they differ in that NNI-2 has more positive charge than site 1 in DENV (figure 6). Although the predicted site 1 did not encompass the critical residue Lys756 identified by Zou et al.(Zou, Chen et al. 2011), this site warrants further biochemical evaluation as the initial study by Zou et al. only mutated 7 of the residues making up the initially predicted site and our current findings include additional residues that may also be important for ligand binding.
Figure 5.
LISE predicted sites in the DENV polymerase. The center image depicts the DENV enzyme with LISE predicted spheres (4V0R in yellow, 2J7U in orange, 2J7W in cyan). The left image is a magnified surface view of both of the predicted sites 2 and 3 located in the palm domain that share structural and chemical similarities with the HCV NNI-3 pocket. The right image is an enlarged surface view of the predicted site 1, which shares structural and chemical features with the HCV NNI-2 pocket, after 90° rotation about the y axis. The table lists residues in common among the LISE predicted sites.
Figure 6.
Electrostatic potential maps of HCV NNI-2 and sites 1 of DENV, WNV and FMDV highlighting chemical and structural features. Panel A is a lateral view of the HCV polymerase depicting the NNI-2 pocket and the reference used to orient the position of the images from other enzymes. Panels B – E depicts the electrostatic potential of the NNI-2/site 1 in HCV, DENV, WNV and FMDV respectively. The perspective in D has been rotated 45° about the y-axis. Regions of blue and red indicate positively and negatively charged surface regions respectively. The potentials are expressed as multiples of 2.58×10−2 Joules per Coulomb (J/C). The color and unit scales are maintained for the remaining electrostatic potential maps.
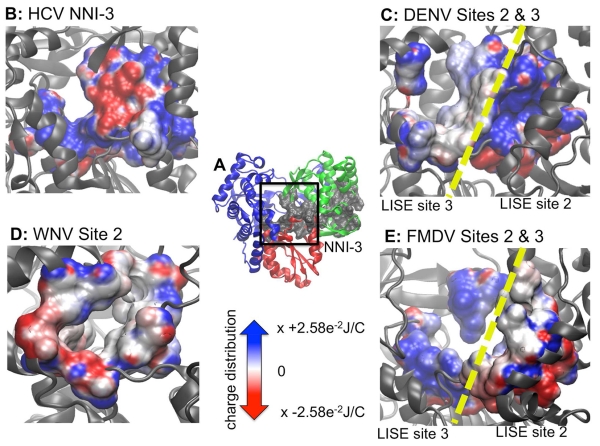
We note that significant differences in structure may contribute to variations in the DENV site predictions. The DENV 4V0R structure is the full-length protein including not only the polymerase domain, but also the methyltransferase (MTase) domain. The MTase has been shown to be involved in the formation of the RNA cap, which plays a role in regulating gene expression(Zhou, Ray et al. 2007, Malet, Massé et al. 2008, Dong, Fink et al. 2014). LISE analysis of full-length 4V0R indicates an accumulation of putative sites at the interface of the polymerase and MTase domain. However, these domains are connected by a flexible linker and it is likely that the structure of the domain interface is highly variable, making it difficult to target this region using structure-based drug discovery efforts. The result obtained using the 4V0R structure also contrast with those obtained for structures containing only the polymerase domain (2J7U and 2J7W). The latter display predicted binding sites in the various subdomains throughout the enzyme, in agreement with predictions made for WNV and FMDV (see below). We hypothesize that in the 4V0R structure, predicted sites outside of the polymerase/MTase interface exhibit a lower rank (i.e. below the top ten) due to the multitude of predictions within the domain interface. Consequently, we decided to focus on DENV structures containing only the polymerase domain to increase the likelihood that consensus sites in readily targeted regions of the enzyme would be identified. Nonetheless, the occurrence of site 1 in both 2J7U and 2J7W suggests that it represents a realistic binding site location for small molecules. It is possible that this site could be targeted with known NNI-2 chemical scaffolds with altered functional moieties that are more chemically appropriate for this site. Furthermore, we note that LISE was also able to predict a zinc-binding site that is known to be located at the base of the thumb domain at the rear of the enzyme. This gives us high confidence that LISE can make accurate binding site predictions for the DENV polymerase. All four residues (H712, H714, C728 and C847) that coordinate with zinc in this location were identified as highly ranked sites for 2J7U and 2J7W. The inability of LISE to identify a zinc-binding site for the full-length 4V0R structure suggests that presence of the MTase domain may preclude the identification of other viable sites within the polymerase domain. LISE also identified two slightly different sites within the central cavity of the protein that share some structural similarities with the HCV NNI-3 pocket (figure 5). When DENV sites 2 and 3 are combined they display significant structural similarity to the NNI-3 site, covering a large portion in the middle of the enzyme. Both also contain a mixture of hydrophobic and hydrophilic residues similar to NNI-3. However, a comparison of the electrostatic potential maps of NNI-3 and DENV sites 2 and 3 demonstrates that the electrostatic potential differs from that present in NNI-3 (figure 7). In a similar manner to suggestions that have been proposed for NNI-3 inhibitors of HCV polymerase(Brown and Thorpe 2015, Davis, Brown et al. 2015), ligands that bind within the region of the predicted DENV site 3 may block portions of the template channel, preventing the RNA template from accessing the active site and disrupting RNA replication. Site 3 is also in close proximity to the G-loop, while both sites 2 and 3 are located close to the priming loop. The G-loop, located in the fingers subdomain (DENV residues 405-418; WNV residues 407-420), typically protrudes towards the active site of Flavivirus polymerases(Yap, Xu et al. 2007, Malet, Egloff et al. 2007, Malet, Massé et al. 2008). Because its location corresponds to the C terminus of the HCV polymerase, the G-loop is hypothesize to play a role in preventing both RNA and incoming NTPs from binding to the enzyme(Malet, Massé et al. 2008). Also protruding towards the active site, the priming loop in the thumb subdomain (DENV residues 792-804; WNV residues 796-809) is thought to stabilize the de novo initiation complex(Malet, Massé et al. 2008). Small molecules binding within these locations may disrupt changes in the conformation of the priming loop that are needed for the enzyme to shift to the elongation stage of replication(Malet, Massé et al. 2008).
Figure 7.
Electrostatic potential maps of HCV NNI-3 and predicted palm sites in DENV, WNV and FMDV highlighting chemical and structural features. Panel A is a frontal view of the HCV NNI-3 pocket and the reference used to orient the position of the images from other enzymes. Panels B – E depicts the electrostatic potential of the known or predicted palm allosteric sites in HCV, DENV, WNV and FMDV respectively. Both DENV and FMDV had two separate predicted sites within the palm domain. However, since they are in similar locations both are displayed simultaneously with a yellow dashed line showing the interface between each pair of sites.
3.2.2. WNV
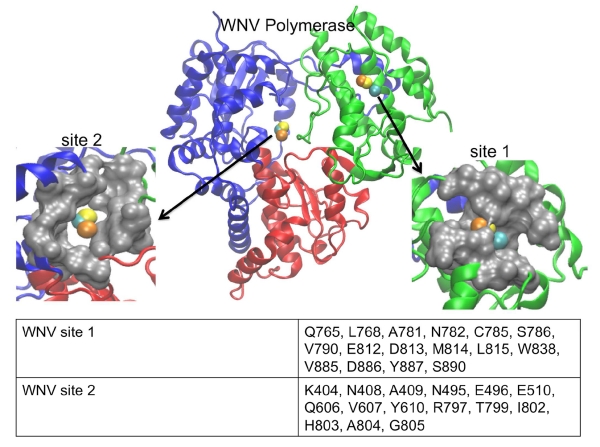
In a similar manner to its performance with DENV, LISE was able to predict two sites in the WNV polymerase that share structural features with two allosteric pockets in the HCV polymerase (figure 8). Cavities A and B that were identified in DENV by Malet et al. (and discussed above) are similar to 2 of the 5 sites that were predicted for the WNV polymerase in the same study. However, we did not observe any matches (i.e. > 50% threshold) between our LISE predictions and residues listed for cavities A and B by Malet et al.(Malet, Massé et al. 2008). We believe that one reason for the lack of a match may be due to differences in the input structures used by Malet et al. and in our work. While we used crystallographic coordinates 2HCN, 2HCS and 2HFZ to make our predictions, it is unclear whether Malet et. al used an X-ray or modeled structure for their studies. As we have noted above, site predictions can vary appreciably based on the query structure. Even though we did not observe matches to residues surrounding the previously predicted cavities by Malet et al., we were encouraged by the similarities between sites 1 and 2 (figure 8) and the NNI-2 and NNI-3 pockets in the HCV polymerase. Specifically, all of the query structures predicted sites 1 and 2 shown in figure 8. Because of the location of site 1 several angstroms away from the expected site of NNI-2, we think this pocket may be novel. In addition, the electrostatic potential map of the WNV site 1 (while being almost opposite to that of NNI-2 in the HCV polymerase: see figures 6B and 6D), is very similar to site 1 predicted in the DENV enzyme. Despite the difference in charge distribution, we observe a significant number of residues within the WNV site 1 that exhibit hydrophobicity comparable to that observed for the NNI-2 pocket. Furthermore, the WNV site 1 possesses a groove that spans the entire cavity, much like that observed in HCV and DENV (figures 6, 8), which may allow similarly shaped ligands to bind. Thus, it may be possible to target site 1 pockets of DENV and WNV with small molecule inhibitors that possess chemical scaffolds that are similar to known HCV NNI-2 ligands (although containing distinct functional groups to match the electrostatic properties of each site). The predicted WNV site 2 is similar to NNI-3 and consists of a mixture of polar and charged residues. It occupies a large fraction of the middle of the enzyme in a similar manner to the corresponding cavity in the HCV polymerase (figures 7B, 7D and 8). Although the WNV site 2 is more enclosed than the HCV NNI-3, it may be possible to target this site with established NNI-3 allosteric inhibitors (due to the chemical similarities shown in figures 7B and 6D) or to at least use such ligands as a starting point for developing WNV inhibitors.
Figure 8.
LISE predicted sites in the WNV polymerase. The center image depicts the WNV enzyme with LISE predicted VDW spheres in the palm and thumb domains (2HCN in yellow, 2HCS in orange, 2HFZ in cyan). The left image is a magnified surface view of the predicted site 2 located in the palm domain that is structurally similar to the HCV NNI-3 pocket. The right image is an enlarged surface view of the predicted site 1, which is structurally similar to the HCV NNI-2 pocket. The table in the lower panel lists the consensus residues making up each LISE predicted site.
3.2.3 FMDV
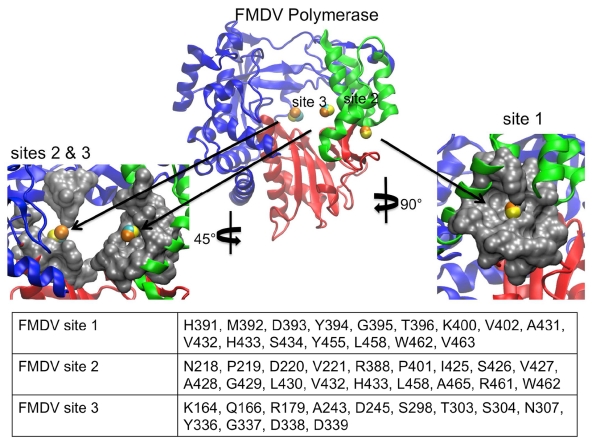
Like DENV and WNV, LISE predicted sites in the FMDV polymerase that share similar locations and electrostatic properties with the HCV NNI-2 and NNI-3 pockets (figures 6, 7 and 9). In a similar manner to DENV and WNV, the predicted site 1 is not as large as NNI-2 but also possesses a groove that spans the majority of its length. It is located on a region of the enzyme surface that is similar to NNI-2 (figure 9). However, it differs from NNI-2 in that it is located at the junction of the thumb and palm domains and includes residues from each domain whereas NNI-2 contains only thumb residues. Sites 2 and 3 when combined occupy a location in the central cavity of the enzyme much like NNI-3. Also, sites 2 and 3 display a ratio of positive and negative charges that is similar to that present in the HCV NNI-3 site (figure 7). These results further support our hypothesis that structural information for one viral polymerase may be used to garner new insights regarding another, even if they possess considerable structural differences. Our findings suggest that despite the genetic and structural differences between the FMDV and HCV polymerases, allosteric sites in the two enzymes have similar geometric and chemical properties. Thus, it may be possible to target these sites with allosteric inhibitors that are similarly shaped.
Figure 9.
LISE predicted sites in the FMDV polymerase. The center image depicts the FMDV enzyme LISE predicted spheres in the palm and thumb domains (1U09 in yellow, 2F8E in orange and 2D7S in cyan). The left image is a magnified surface view of both of the predicted sites 2 and 3 located in the palm domain that share structural similarities with the HCV NNI-3 pocket. The right image is an enlarged surface view of the predicted site 1 which is structurally similar to the HCV NNI-2 pocket. The table lists the consensus residues making up each LISE predicted site.
3.2.4 Potential Functional Significance of Putative Allosteric Sites
Biochemical experiments combined with computational studies provide insight into how inhibitors bound to the HCV NNI-2 and NNI-3 pockets achieve allosteric inhibition. Both NNI-2 and NNI-3 inhibitors have been shown to disrupt an early stage of replication that precedes elongation (Ruebsam, Webber et al. 2008, Ontoria, Rydberg et al. 2009). Molecular simulations by our group suggest that inhibitors of the NNI-2 and NNI-3 pockets generally restrict protein fluctuations and disrupt motions observed in the free enzyme (Davis and Thorpe 2013, Brown and Thorpe 2015, Davis, Brown et al. 2015). In addition, both type of inhibitors altered correlated motions within enzyme functional motifs in a manner that may disfavor the normal cycle of replication. Specifically, these functional regions are thought to play roles in binding of template, the nascent 3’-end or nascent RNA duplex (Moustafa, Shen et al. 2011, Sesmero and Thorpe 2015). Thus, the NNI-2 and NNI-3 allosteric pockets may communicate with residues involved in the binding of template and newly synthesized RNA strands as well as those required to transition to the elongation stage of genome replication. Consequently, the putative allosteric sites identified in the current work may share these functional properties and allow the enzymes to be inhibited in a similar manner to the HCV polymerase.
Recent studies of the DENV polymerase using both experimental and computational techniques support the existence of novel binding sites that are consistent with our predictions. Manvar and coworkers used in vitro screens and SAR analysis to identify a small molecule scaffold that had inhibitory activity against serotype 2 of the DENV polymerase (Manvar, Küçükgüzel et al. 2015). Fluorescence experiments confirmed that three of the ligands with this scaffold interacted with the polymerase, while docking studies suggest that the ligand with the most favorable affinity binds to a site in the thumb domain (Manvar, Küçükgüzel et al. 2015). Six of the ten residues identified to interact with this ligand are found within site 1 identified by LISE in our current work (see figure 5). This result supports the utility of our approach in identifying functionally relevant allosteric sites. Furthermore, in another recent investigation, Yokokawa and coworkers employed fragment-based screening and structure-based drug design to identify a novel non-nucleoside inhibitor of the DENV polymerase that binds to a specific site in the palm domain (Yokokawa, Nilar et al. 2016). Visual inspection of the initial identified fragment by Yokokawa et al. puts this site in the same location as our identified DENV site 2 (figure 5). After drug design efforts and biochemical studies, residues Arg729, Thr794, Trp795, Ser796, His800 and Gln802 were shown to be key interaction points within the binding pocket for a novel small molecule inhibitor (Yokokawa, Nilar et al. 2016). Four of these six residues were also identified in our present work as DENV site 2 (figure 5), further verifying the validity of our approach. Taken together, the studies by Manvar et al. and Yokokawa et al. suggest that the allosteric sites predicted in our work for the DENV polymerase are valid inhibitor binding sites. This finding makes it more likely that the sites we identified for the WNV and FMDV polymerases also support inhibitor binding. Thus, we believe it would be worthwhile to explore these predictions in greater detail for drug discovery.
4. Conclusions
Our results suggest that it is possible to predict allosteric sites for viral polymerases that are structurally similar to the HCV polymerase. Thus, we were able to identify experimentally elucidated allosteric sites within the HCV, FMDV, CSV and DENV polymerases. Given that the overall architecture of these enzymes is relatively common, this general approach may prove useful in identifying allosteric sites for a significant number of other viral polymerases.
Even though we demonstrate that our approach seems to result in valid site predictions for the viral polymerases studied, there are certain caveats that should be considered when LBSPs are used. As discussed in section 3.1, LBSPs are not ideal for predicting ligand-induced sites, as there is no built-in feature within these tools that accounts for the dynamic nature of proteins in sampling various conformations. Thus, there is no way to investigate the presence of sites that only exist within distinct structural states unless one has those states available. To minimize the impact of conformational variation on site prediction, we recommend using as many input structures as feasible and looking for consensus among the predicted sites. In this way one can have greater confidence that the results have converged to describe a realistic binding site when its location is unknown. We believe that these consensus sites, such as those identified above for the DENV, WNV and FMDV polymerases, have a higher probability of being functionally relevant. While we consider predicted sites for the WNV and FMDV enzymes to be putative in lieu of biochemical studies to definitively establish their existence, they likely represent good starting points for future drug discovery efforts targeting these enzymes.
Our studies suggest that the structural similarities of viral polymerases are associated with similar cavities on the protein surface that could be targeted with small molecules. We identified putative allosteric sites in DENV, WNV and FMDV that possess varying degrees of similarity to the NNI-2 and NNI-3 pockets in the HCV polymerase. Even though the chemical environment of the NNI-2 site in HCV polymerase differs from that of site 1 in DENV, WNV and FMDV, their similar shapes suggest that this pocket is a conserved evolutionary feature. Consequently, this location may have functional significance such as interacting with RNA strands during viral replication or interacting with other viral or host proteins. Thus, it is possible that the activities of the different enzymes can be modulated in similar ways when allosteric ligands bind at these locations and that allosteric inhibition is a shared feature of these polymerases. Moreover, in light of their structural similarities it may be possible to identify new allosteric inhibitors for one viral enzyme by using the known allosteric inhibitors of another enzyme as a starting point. For example, it may be enough to simply change the appropriate functional groups on known chemical scaffolds. We also observe that every enzyme possesses a predicted site within the palm domain that is much like the HCV polymerase NNI-3 pocket with regard to its structural and chemical environment. Thus, it may be possible to target these locations in the different enzymes with very similar small molecules. Finally, we emphasize that the strategy employed here of evaluating LBSPs on a well-studied enzyme to predict sites in less studied enzymes may facilitate the identification of novel binding sites. Ultimately this knowledge can assist in the process of discovering new drugs that target these enzymes, especially in cases for which efficacious and cost-effective treatments are not widely available.
Supplementary Material
Highlights.
The HCV polymerase was used to evaluate 4 ligand binding site predictors
LISE was best at predicting known allosteric sites in the HCV polymerase
LISE predicted allosteric sites in the polymerases of DENV, WNV and FMDV
Structural similarities of viral polymerases may include allosteric binding sites
Acknowledgements
Jodian A. Brown was funded by NIH F31 pre-doctoral grant (GM-106958). We wish to thank Paul Zhong-ru Xie for his technical assistance regarding the LISE program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baker NA, et al. Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu PL. Non-nucleoside inhibitors of the HCV NS5B polymerase: progress in the discovery and development of novel agents for the treatment of HCV infections. Current Opinion in Investigational Drugs. 2007;8(8):614–634. [PubMed] [Google Scholar]
- Beaulieu PL. Recent advances in the development of NS5B polymerase inhibitors for the treatment of hepatitis C virus infection. Expert Opinion on Therapeutic Patents. 2009;19(2):145–164. doi: 10.1517/13543770802672598. [DOI] [PubMed] [Google Scholar]
- Bhatt S, et al. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaney JE, et al. Recombinant, Live-Attenuated Tetravalent Dengue Virus Vaccine Formulations Induce a Balanced, Broad, and Protective Neutralizing Antibody Response against Each of the Four Serotypes in Rhesus Monkeys. Journal of Virology. 2005;79(9):5516–5528. doi: 10.1128/JVI.79.9.5516-5528.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady GP, Stouten PFW. Fast Prediction and Visualization of Protein Binding Pockets With PASS. Journal of Computer-Aided Molecular Design. 2000;14(4):383–401. doi: 10.1023/a:1008124202956. [DOI] [PubMed] [Google Scholar]
- Brady OJ, et al. Refining theGlobal Spatial Limits of Dengue Virus Transmission by Evidence-Based Consensus. PLOS Neglected Tropical Diseases. 2012;6(8) doi: 10.1371/journal.pntd.0001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandler S, Tangy F. Vaccines in Development against West Nile Virus. Viruses. 2013;5(10):2384–2409. doi: 10.3390/v5102384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenke R, et al. Fragment-based identification of druggable ‘hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics. 2009;25(5):621–627. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR, et al. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. Journal of Computational Chemistry. 1983;4(2):187–217. [Google Scholar]
- Brown JA, Thorpe IF. Dual Allosteric Inhibitors Jointly Modulate Protein Structure and Dynamics in the Hepatitis C Virus Polymerase. Biochemistry. 2015;54(26):4131–4141. doi: 10.1021/acs.biochem.5b00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton JR, Everson GT. HCV NS5B polymerase inhibitors. Clinical Liver Disease. 2009;13(3):453–465. doi: 10.1016/j.cld.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Chappell KJ, et al. West Nile Virus NS2B/NS3 protease as an antiviral target. Current Medicinal Chemistry. 2008;15(27):2771–2784. doi: 10.2174/092986708786242804. [DOI] [PubMed] [Google Scholar]
- Condon SM. Allosteric Inhibitors of Hepatitis C NS5B RNA-dependent RNA Polymerase. Current Medicinal Chemistry. 2005;4(2):99–110. [Google Scholar]
- Connolly ML. Analytical molecular surface calculation. Journal of Applied Crystallography. 1983;16(5):548–558. [Google Scholar]
- Davis BC, et al. Allosteric Inhibitors Have Distinct Effects, but Also Common Modes of Action, in the HCV Polymerase. Biophysical Journal. 2015;108(7):1785–1795. doi: 10.1016/j.bpj.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BC, Thorpe IF. Thumb inhibitor binding eliminates functionally important dynamics in the hepatitis C virus RNA polymerase. Proteins: Structure, Function and Bioinformatics. 2013;81(1):40–52. doi: 10.1002/prot.24154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Recent highlights in the development of new antiviral drugs. Current Opinion in Microbiology. 2005;8(5):552–560. doi: 10.1016/j.mib.2005.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marco S, et al. Interdomain communication in hepatitis C virus polymerase abolished by small molecule inhibitors bound to a novel allosteric site. Journal of Biological Chemistry. 2005;280(33):29765–29770. doi: 10.1074/jbc.M505423200. [DOI] [PubMed] [Google Scholar]
- Dolinsky TJ, et al. PDB2PQR: Expanding and updgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Research. 2007;35(suppl 2):W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, et al. PDB2PQR: an automated pipeline for the setup, execution, and analysis of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Research. 2004;32(suppl 2):W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E, et al. Foot and mouth disease virus. Comparative Immunology, Microbiology and Infectious Diseases. 2002;25(5-6):297–308. doi: 10.1016/s0147-9571(02)00027-9. [DOI] [PubMed] [Google Scholar]
- Dong H, et al. Flavivirus RNA methylation. Journal of General Virology. 2014;95(Pt 4):763–768. doi: 10.1099/vir.0.062208-0. [DOI] [PubMed] [Google Scholar]
- Dundas J, et al. CASTp: computed atas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Research. 2006;34(suppl 2):W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durk RC, et al. Inhibitors of Foot and Mouth Disase Virus Targeting a Novel Pocket of the RNA-dependent RNA Polymerase. PLoS ONE. 2010;5(12):e15049. doi: 10.1371/journal.pone.0015049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Orta C, et al. Structure of Foot-and-Mouth Disease Virus RNA-dependent RNA Polymerase and Its Complex with a Template-Primer RNA. Journal of Biological Chemistry. 2004;279(45):47212–47221. doi: 10.1074/jbc.M405465200. [DOI] [PubMed] [Google Scholar]
- Furuta Y, et al. Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Research. 2015;100(2):446–454. doi: 10.1016/j.antiviral.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- G P. West Nile virus. Prevalence and significance as a zoonotic pathogen. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 2004;47(7):653–660. doi: 10.1007/s00103-004-0864-x. [DOI] [PubMed] [Google Scholar]
- Grubman MJ, Baxt B. Foot-and-Mouth Disease. Clinical Microbiology Reviews. 2004;17(2):465–493. doi: 10.1128/CMR.17.2.465-493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B, et al. Discovery of small molecule inhibitors of West Nile virus using a high-throughput sub-genomic replicon screen. Antiviral Research. 2006;70(2):39–50. doi: 10.1016/j.antiviral.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Gu J, et al. Identification of Ligand Binding Pockets for Unbound State Proteins Using Local Residue References International Conference on Artificial Intelligence and Embedded Systems; Singapore. 2012.pp. 14–18. [Google Scholar]
- Hassell AM, et al. Crystallization of protein-ligand complexes. Acta Crystallographica Section D: Biological Crystallography. 2007;63(1):72–79. doi: 10.1107/S0907444906047020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendlich M, et al. LIGSITE: automatic and efficient detection of potential small molecule-binding sites in proteins. Journal of Molecular Graphics and Modelling. 1997;15(6):359–363. doi: 10.1016/s1093-3263(98)00002-3. [DOI] [PubMed] [Google Scholar]
- Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Structural Biology. 2006;6(1):19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac NM, et al. Sofosbuvir: New and Promising Treatment for Hepatitis C Virus Infection- A Review. WORLD JOURNAL OF PHARMACY AND PHARMACEUTICAL SCIENCES. 2015;4(9):318–326. [Google Scholar]
- Jackson RM. Q-fit: a probabilistic method for docking molecular fragments by sampling low energy conformational space. Journal of Computer-Aided Molecular Design. 2002;16(1):43–57. doi: 10.1023/a:1016307520660. [DOI] [PubMed] [Google Scholar]
- Jordan I, et al. Ribavirin Inhibits West Nile Virus Replication and Cytopathic Effect in Neural Cells. The Journal of Infectious Diseases. 2000;182(4):1214–1217. doi: 10.1086/315847. [DOI] [PubMed] [Google Scholar]
- Keating GM. Ledipasvir/Sofosbuvir: A Review of Its Use in Chronic Hepatitis C. Drugs. 2015;75(6):675–685. doi: 10.1007/s40265-015-0381-2. [DOI] [PubMed] [Google Scholar]
- Kozakov D, et al. Structural conservation of druggable hot spots in protein-protein interfaces. Proceedings of the National Academy of Sciences. 2011;108(33):13528–13533. doi: 10.1073/pnas.1101835108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie AT, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21(9):1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- Lawitz E, et al. Sofosbuvir for Previously Untreated Chronic Hepatitis C Infection. New England Journal of Medicine. 2013;368(20):1878–1887. doi: 10.1056/NEJMoa1214853. [DOI] [PubMed] [Google Scholar]
- Li H, et al. Discovery of (R)-6-Cyclopentyl-6-(2-(2,6- diethylpyridin-4-yl)ethyl)-3-((5,7-dimethyl- [1,2,4]triazolo[1,5-a]pyrimidin-2-yl)methyl)-4- hydroxy-5,6-dihydropyran-2-one (PF-00868554) as a Potent and Orally Available Hepatitis C Virus Polymerase Inhibitor. Journal of Medicinal Chemistry. 2009;52(5):1255–1258. doi: 10.1021/jm8014537. [DOI] [PubMed] [Google Scholar]
- Lim S, et al. Ten Years of Dengue Drug Discovery: Progress and Prospects. Antiviral Research. 2013;100(2):500–519. doi: 10.1016/j.antiviral.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Malet H, et al. Crystal Structure of the RNA Polymerase Domain of the West Nile Virus Non-structural Protein 5. Journal of Biological Chemistry. 2007;282(14):10678–10689. doi: 10.1074/jbc.M607273200. [DOI] [PubMed] [Google Scholar]
- Malet H, et al. The flavivirus polymerase as a target for drug discovery. Antiviral Research. 2008;80(1):23–35. doi: 10.1016/j.antiviral.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Manvar D, et al. Discovery of conjugated thiazolidinone-thiadiazole scaffold as anti-dengue virus polymerase inhibitors. Biochemical and Biophysical Research Communications. 2015;469(3):743–747. doi: 10.1016/j.bbrc.2015.12.042. [DOI] [PubMed] [Google Scholar]
- Moustafa IM, et al. Molecular Dynamics Simulations of Viral RNA Polymerases Link Conserved and Correlated Motions of Functional Elements to Fidelity. Journal of Molecular Biology. 2011;410(1):159–181. doi: 10.1016/j.jmb.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan CH, et al. FTSite: high accuracy detection of ligand binding sites on unbound protein structures. Bioinformatics. 2012;28(2):286–287. doi: 10.1093/bioinformatics/btr651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble CG, et al. Strategies for development of dengue virus inhibitors. Antiviral Research. 2010;85(3):450–462. doi: 10.1016/j.antiviral.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Ontoria JM, et al. Identification and Biological Evaluation of a Series of 1H-Benzo[de]isoquinoline-1,3(2H)-diones as Hepatitis C Virus NS5B Polymerase Inhibitors. Journal of Medicinal Chemistry. 2009;52(16):5217–5227. doi: 10.1021/jm900517t. [DOI] [PubMed] [Google Scholar]
- Petersen LR. West Nile Virus: From Africa to Europe, America, and Beyond. Springer; Netherlands: 2014. [Google Scholar]
- Pinto AK, et al. A Hydrogen Peroxide-Inactivated Virus Vaccine Elicits Humoral and Cellular Immunity and Protects against Lethal West Nile Virus Infection in Aged Mice. Journal of Virology. 2013;87(4):1926–1936. doi: 10.1128/JVI.02903-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puvanendrampillai D, Mitchell JBO. Protein Ligand Database (PLD): additional understanding of the nature and specificity of protein-ligand complexes. Bioinformatics. 2003;19(14):1856–1857. doi: 10.1093/bioinformatics/btg243. [DOI] [PubMed] [Google Scholar]
- Ruebsam F, et al. Pyrrolo[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase. Bioorganic & Medicinal Chemistry Letters. 2008;18(12):3616–3621. doi: 10.1016/j.bmcl.2008.04.066. [DOI] [PubMed] [Google Scholar]
- Schwartz LM, et al. The dengue vaccine pipeline: Implications for the future of dengue control. Vaccine. 2015;33(29):3293–3298. doi: 10.1016/j.vaccine.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seff LB, Hoofnagle JH. National Institutes of Health Consensus Development Conference: Management of Hepatitis C: 2002. Hepatology. 2003;36(supplement 5B):s1–s2. doi: 10.1053/jhep.2002.36992. [DOI] [PubMed] [Google Scholar]
- Sesmero E, Thorpe IF. Using the Hepatitis C Virus RNA-Dependent RNA Polymerase as a Model to Understand Viral Polymerase Structure, Function and Dynamics. Viruses. 2015;7(7):3974–3994. doi: 10.3390/v7072808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TM, et al. Identifying Initiation and Elongation Inhibitors of Dengue Virus RNA Polymerase in a High-Throughput Lead-Finding Campaign. Journal of Biomolecular Screening. 2015;20(1):153–163. doi: 10.1177/1087057114551141. [DOI] [PubMed] [Google Scholar]
- Thisyakorn U, Thisyakorn C. Latest developments and future directions in dengue vaccines. Therapeutic Advances in Vaccines. 2014;2(1):3–9. doi: 10.1177/2051013613507862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Linden L, et al. The RNA template channel of the RNA-dependent RNA polymerase as a target for development of antiviral therapy of multiple genera within a virus family. PLoS Pathogens. 2015;11(3):e1004733. doi: 10.1371/journal.ppat.1004733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie ZR, Hwang M-J. An interaction-motif-based scoring function for protein-ligand docking. BMC Bioinformatics. 2010;11(298):1–16. doi: 10.1186/1471-2105-11-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie ZR, Hwang M-J. Ligand-binding site prediction using ligand-interacting and binding site-enriched protein triangles. Bioinformatics. 2012;28(12):1579–1585. doi: 10.1093/bioinformatics/bts182. [DOI] [PubMed] [Google Scholar]
- Yamshchikov V. Development of human live attenuated West Nile infectious DNA vaccine: Conceptual design of the vaccine candidate. Virology Journal. 2015;484:59–68. doi: 10.1016/j.virol.2015.04.027. [DOI] [PubMed] [Google Scholar]
- Yap TL, et al. Crystal Structure of the Dengue Virus RNA-Dependent RNA Polymerase Catalytic Domain at 1.85-Angstrom Resolution. Journal of Virology. 2007;81(9):4753–4765. doi: 10.1128/JVI.02283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, et al. An adenosine nucleoside inhibitor of dengue virus. Proceedings of the National Academy of Sciences. 2009;106(48):20435–20439. doi: 10.1073/pnas.0907010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokokawa F, et al. Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design. Journal of Medicinal Chemistry. 2016;59(8):3935–3952. doi: 10.1021/acs.jmedchem.6b00143. [DOI] [PubMed] [Google Scholar]
- Zhou Y, et al. Structure and Function of Flavivirus NS5 Methyltransferase. Journal of Virology. 2007;81(8):3891–3903. doi: 10.1128/JVI.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou G, et al. Functional Analysis of Two Cavities in Flavivirus NS5 Polymerase. Journal of Biological Chemistry. 2011;286(16):14362–14372. doi: 10.1074/jbc.M110.214189. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.