Summary
The lack of controllable in vitro models that can recapitulate the features of solid tumors such as Ewing’s sarcoma limits our understanding of the tumor initiation and progression and impedes the development of new therapies. Cancer research still relies of the use of simple cell culture, tumor spheroids, and small animals. Tissue-engineered tumor models are now being grown in vitro to mimic the actual tumors in patients. Recently, we have established a new protocol for bioengineering the Ewing’s sarcoma, by infusing tumor cell aggregates into the human bone engineered from the patient’s mesenchymal stem cells. The bone niche allows crosstalk between the tumor cells, osteoblasts and supporting cells of the bone, extracellular matrix and the tissue microenvironment. The bioreactor platform used in these experiments also allows the implementation of physiologically relevant mechanical signals. Here, we describe a method to build an in vitro model of Ewing’s sarcoma that mimics the key properties of the native tumor and provides the tissue context and physical regulatory signals.
Keywords: human sarcoma, tumor model, cancer research, bioreactor platform, Ewing’s sarcoma
1. Introduction
Predictive models of human tumors are of paramount importance for studies of tumor initiation, progression and remission, identification of therapeutic targets, and development of new therapeutic modalities. Animal models have greatly contributed to our understanding of cancer, but their value in anticipating the effectiveness of treatment strategies in clinical trials remains uncertain [1]. In vitro testing of anticancer drugs typically involves growing cancer cell lines in monolayers on tissue-culture plastic dishes. Despite major progresses, monolayer cultures remain poor predictors of whether a given drug will be safe and ultimately provide clinical benefit [2,3].
Most in vitro models, including tumor spheroids, cancer cells in scaffolds, and small cancer organoids lack the complexity of the native tumor milieu that mediates cancer processes [4,5,6]. Specifically, in the bone microenvironment, osteoblasts, osteoclasts, and mesenchymal stem cells (hMSC) mediate primary tumor growth and metastasis [7]. In addition, mechanical forces generated either from external sources or by cell contractions, play important roles in cancer invasion into the bone [8,9].
Bioengineering methods that have advanced stem cell research and regenerative medicine, are becoming essential tools for cancer research [10]. Importantly, bioengineered tumors can provide cancer cells with a tissue context incorporating the extracellular matrix (ECM), supporting cells and physical signals.
In our effort to introduce substantial improvements over existing 3D models to study bone tumors we have developed a method to engineer in vitro human bone tumors, and to maintain these tumors in culture for prolonged periods of time (weeks to months). We cultured the Ewing’s sarcoma (ES) cell spheroids within tissue engineered human bone. The bone was grown from adult hMSC capable of osteogenic differentiation within native bone ECM serving as a structural scaffold [11]. Additionally we exposed the cancer cells to biophysical stimuli mimicking those normally present within the bone tumor microenvironment, by using a bioreactor capable of providing mechanical compression [12].
We propose a model that allows crosstalk between cancer cells and the key components of the bone tumor environment, such as mechanical forces and native mineralized ECM (Fig. 1). This bioengineered human tumor could dramatically improve upon the current preclinical drug-screening paradigm by providing valuable information about new therapeutic targets and anticancer drug efficacy.
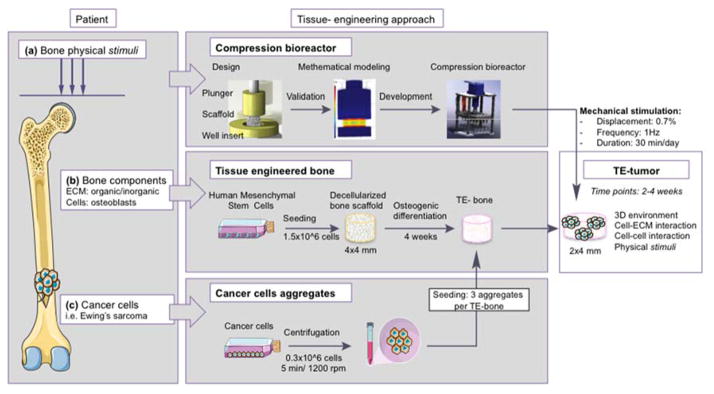
Figure 1. Overall approach to the bioengineered bone tumor in vitro.
The model recapitulates (a) physical stimuli in the patient’s bone; (b) patient’s bone comprising both organic and inorganic extracellular matrix (ECM) and bone cells (osteoblasts); (c) bone tumor compartment with Ewing’s sarcoma cells. In order to mimic the tumor microenvironment and to generate a tissue-engineered tumor (TE-tumor), three cancer cell aggregates were infused into the tissue-engineered bone made of human mesenchymal stem cell derived osteoblasts. Then, the TE-tumor is cultured in a compression bioreactor and stimulated 30 minutes per day under physiological conditions (displacement 0.7%; frequency 1Hz) for 2 and 4 weeks.
2. Materials
2.1.Decellularized bone scaffold (Fig. 2b)
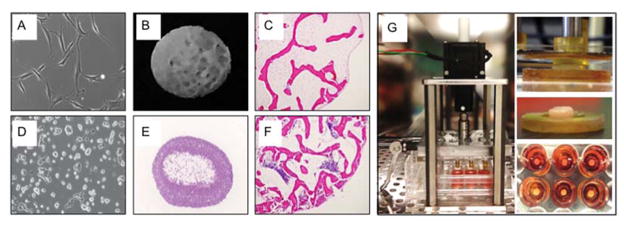
Figure 2. Overview of key methods.
A Human mesenchymal stem cells expanded in culture. B Decellularized bone scaffold used to seed mesenchymal cells and grow bone. C Scaffold seeded with mesenchymal stem cells to engineer bone (H&E stain). D Ewing’s sarcoma cells (RD-ES line), expanded in culture. E Cancer cell aggregate. F Human Ewing’s sarcoma tumor grown in vitro. G Apparatus for mechanical stimulation of engineered tumors.
Prepare all solutions using milli-Q ultrapure water
Driller (Dewat, cat. no DC970K2)
Diamond drill bites (Starlite Industries, cat. no 102045)
Fresh young bovine bone samples (Epiphysis of bovine femur) from a local slaughterhouse (Green Village Packing Co., New Jersey, USA).
1x PBS, 0.1% (w/v) EDTA
-
Decellularization solutions: Prepare 1 L of a 10 mM Tris-HCl (pH.8) as stock solution (see Note 1).
Solution 1: 10 mM Tris-HCl (pH.8), 0.1% (w/v) EDTA
Solution 2: 10 mM Tris-HCl (pH.8), 0.5% (w/v) SDS
DNAse/RNAse solution. Using a syringe, take 5 mL of 10 mM Tris-HCl (pH.8) stock solution and inject it into a vial of DNAse (Life Technologies, cat. no 18068-015). Mix the solution by gentle vortex. Add the DNase solution (5mL) to 35mL of mQ water in a 50mL Falcon tube (total volume =40mL), and incorporate 1 unit/mL RNAse A (Roche Applied Sciences, cat. no. 10109142001) (see Note 2). Mix the DNAse/RNAse solution by vortexing. Keep DNAse/RNAse solution at 4° C until use.
Caliber
Dremel polish rotary tool
2.2.Cells
Purchase Ewing’s sarcoma cell lines from ATCC: RD-ES cell line (ATCC® Number: HTB-166) and SK-N-MC (ATCC® Number: HTB-10); purchase human Mesenchymal Stem cells (hMSC) from Lonza (Catalog number: PT-2501). (Fig. 2a,d)
2.3.Culture Media
Cancer media
Culture the RD-ES cells in ATCC-formulated RPMI-1640 Medium (RPMI) and the SK-N-MC cells in ATCC-formulated Eagle’s Minimum Essential Medium (EMEM). Supplement both media with 10% (v/v) Hyclone FBS and 1% penicillin/streptomycin.
hMSC medium
Basic medium for culturing hMSC consist in DMEM supplemented with 1 ng/ml Fibroblast Growth Factor (FGF), 10% (v/v) Hyclone FBS and 1% penicillin/streptomycin
Osteogenic medium
Prepare hMSC basic medium supplemented with 1 μM dexamethasone, 10 mM β-glycerophosphate and 50 μM ascorbic acid-2-phosphate
2.4.PDMS rings
Polydimethylsiloxane (PDMS) and curing agent (KRAYDEN; cat. no DC2065622)
Glass petri dish
Vacuum
Oven at 60° C
8mm and 4mm biopsy punch (VWR; cat. no 82030-348, cat. no 82030-354)
2.5.TE-Tumor
Non-treated 6-well plates (Nunc cat. no 150239)
100 mm culture dish (Corning; cat. no 353003)
Sterile tweezers
Sterile glass slides (Fisher; cat. no 12-548-5B)
Sterile razor blade (BD Bard-Parker, cat. no 371110)
2.6.Bioreactor: design and parts
The culture module:
-
1
24-well plate (NUNC, cat. no 144530).
-
2
Well-insert: using a CNC milling machine mill a 30×30×3 mm (width x length x thickness) sheet of Ultem (McMaster Carr, cat. no 8685K47) to obtain a disc with a diameter of 15.5 mm and a central hole with a 6 mm in diameter and 1 mm deep. This insert is placed on the bottom of the culture well and allows the exact placement of the scaffold (see Note 3). Make at least 24.
-
3
The plunger: machine a polycarbonate rod (McMaster Car, cat. no 8571K31) to obtain a cylinder that is 8 mm in diameter and 30 mm in height. Make at least 24.
-
4
Lid: machine a polycarbonate bar (McMaster Car, cat. no 8574K321) to obtain a rectangular sheet of 127.89 × 85.60 mm (length x width) dimensions. Drill an 8 mm in diameter and 5 mm deep hole aligned to the center of the culture well. Repeat this for the remaining 23 wells.
-
5
Press fit the plungers into the drilled hole. To make sure all the plungers have the same height, face them down 0.5 mm using the milling machine.
The displacement module:
-
6
Using a shaft clamp (McMaster Car, cat. no 9660T3), connect the lid to a linear actuator controlled by a stepper motor (HaydonKerk 5700 series). The compression load is the result of the vertical motion provided by the linear actuator and the contact between the plungers and the scaffolds.
The control module and user interface:
-
7
The stepper motor is controlled by custom made microcontroller that includes an Arduino Mini Pro (Adafruit, cat. no 1501) and an A4988 stepper motor driver (Pololu, cat. no 1182) which are connected via a USB cable to a PC. The user can control all the experimental parameters form a custom made GUI (general user interface) created using open- source Arduino libraries.
2.7.Bioreactor: Mathematical modeling
To characterize and predict forces generated in the TE-tumor upon exposure to mechanical loading in the bioreactor, run a finite element analysis in COMSOL® Multiphysics 4.2a. This analysis is can identify the stress field generated by a physiological-like loading on the TE-tumor. First, model the 3D geometry of the culture chamber, including the plunger, the well insert and a cylindrical scaffold (4 mm in diameter ×2 mm thick) using the drawing tool in the software. Second, insert the parameters related to the bone’s mechanical properties such as: Young modulus (50 MPa), density (434 kg/m3) and Poisson ratio (0.3). Run a quasi-static analysis that solves a time-dependent problem, assuming the structural mechanics component being static. Finally, solve the Von Mises’s tensor to evaluate the stress field generated in culture chamber.
3. Methods
3.1. TE-Bone
Culture hMSC in basic medium (DMEM supplemented with 10% v/v Hyclone FBS and 1% penicillin/streptomycin) for maintenance and expansion. Sterilize scaffolds in ethanol 70% overnight. Suspend 1.5 ×106 hMSCs (passage 3) into 50 μL of medium and pipette the cells suspension onto the top of a blot-dried scaffolds and allow to percolate through. After 15 min, rotate scaffold 180°, and add 10 μL of medium to prevent the cells from drying out. Repeat this process every 15 min for up to 2 h to facilitate uniform cell distribution. Culture the scaffolds in 6 mL of osteogenic medium for 4 weeks in a non-treated 6-well plate.
3.2. Cancer cell aggregates (Fig. 2e)
Centrifuge 0.3 ×106 Ewing’s sarcoma cells in 4mL of cancer medium in a 15 mL Falcon tube, at 290g for 5min. After centrifugation, culture the aggregates for one week at 37°C in a humidified incubator, 5% CO2 (see Note 4).
3.3. PDMS rings
Measure out 27g of PDMS into a crystal petri dish
Add 3g of curing agent (ratio 9:1)
Gently mix them up for a few minutes with a spatula
Carefully place the petri dish in the vacuum 30min until all the bubbles disappear from the solution.
Once all the bubbles have cleared from the mixture, carefully place the PDMS mold into the oven (see Note 5).
Let the samples bake for 1hour at 60° C (or 24h at room temperature)
Peel the PDMS off the mold using a sharp-tipped knife.
Punch the PDMS using a 8mm biopsy punch for the inner part of the ring and a 4mm biopsy punch for the core
Autoclave the rings before using.
3.4. TE-Tumor (Fig 2c,f)
Add 6mL of cancer medium to 6-well plates (Nunc, cat. no150239). Keep them at 37°C in a humidified incubator containing 5% CO2 until use.
Using sterile tweezers, place one glass slide on a 100 mm dish (see Note 6) and one PDMS ring onto the glass slide. Be sure that the PDMS ring is attached at the slide though the sticky side of the ring.
Harvest the cancer aggregates from the 15mL Falcon tubes using a 5mL pipette. Aspirate about 1mL of medium carrying an aggregate and place it on a100 mm culture dish. Repeat this step until harvesting all the aggregates.
Using sterile tweezers, place a TE-bone in an empty 100 mm dish. Bisect it (axial cut) using a razor blade. Immediately, bring one half to a well of 6-well plate with medium and culture it in the incubator.
Introduce one half of the TE-bone in the PDMS ring attached to the slide. Do not leave space between the bottom of the TE-bone and the crystal slide.
Using a 100μl micropipette, load about 30–50μl of medium containing 3 cancer aggregates onto the TE-Bone placed into the PDMS ring.
Using a tweezers, flip out the ring with the TE-Bone-cancer aggregates. Push gently the TE-bone against the slide and let the aggregates get into the construct. Again, do not leave space between the bottom of the TE-bone and the crystal slide.
Discard the ring and place the TE-tumor in a well of 6-well plate with medium at 37°C in a humidified incubator containing 5% CO2 during 2 weeks.
Change medium biweekly.
3.5. Bioreactor stimulation (Fig. 2g)
Using sterile tweezers place the well inserts at the bottom of a 24-well plate (Nunc, cat. no 144530). Add 2mL of cancer medium to each well. Keep the plate at 37°C in a humidified incubator containing 5% CO2 until use.
Using sterile tweezers transfer the TE-tumor in the 24 well plate. Place the scaffold in the center of the well insert (see Note 7).
Place the 24 well plate in the center of the bioreactor.
Lower the lid and plungers using the controls on the GUI. Make sure the lid covers completely the plate.
Place the bioreactor at 37°C in a humidified incubator containing 5% CO2.
The compression protocol consisted in twenty-four hours of culture in the bioreactor with three loading inputs. The first application consisted of 0.7% of strain (for a 2 mm thick scaffold it equals 14 μm of displacement amplitude), applied using a sinusoidal wave form for the vertical motion at 1 Hz frequency for 1800 loading cycles (equivalent of 30 minutes of stimulation) (see Note 8).
Stimulate sample right after the relocation into the bioreactor, let rest overnight and stimulate again, both in the morning and in the afternoon. (see Note 9)
Acknowledgments
We gratefully acknowledge funding support by the National Institutes of Health (grants UH3EB017103 and EB002520) and the Alfonso Martin Escudero Foundation.
Footnotes
Add 10mL of a 1M Tris-HCl, pH 8, stock solution, (ThermoFisher scientific; 15568-025), to a 1-L graduated cylinder. Mix with 990mL of mQ water and adjust pH using HCl. Store at room temperature.
Close the screw-top caps loosely (Do NOT close all the way so that gas can be exchanged).
Make sure the scaffold is well centered. Use the well insert off-set feature as a reference.
A flat paper or aluminum foil layer placed underneath the petri dish is a good idea just in case the PDMS overflows and sticks to the oven surface.
This protocol uses a 6-well plate format. Amounts may be scaled up or down if using another size format.
You can also use the lid of a 100 mm culture dish.
Make sure the well insert is placed correctly at the bottom of the well.
Various stimulation protocols can be used. It is possible to tune several culture parameters such as frequency, strain and time depending on the user’s need.
Tune stimulation cycles according to experimental requirements.
Disclosure of potential conflicts of interest
The authors indicate no potential conflicts of interest.
References
- 1.van der Worp HB, Howells DW, Sena ES, Porritt MJ, Rewell S, O’Collins V, Macleod MR. Can animal models of disease reliably inform human studies? PLoS medicine. 2010;7(3):e1000245. doi: 10.1371/journal.pmed.1000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voskoglou-Nomikos T, Pater JL, Seymour L. Clinical predictive value of the in vitro cell line, human xenograft, and mouse allograft preclinical cancer models. Clinical cancer research: an official journal of the American Association for Cancer Research. 2003;9(11):4227–4239. [PubMed] [Google Scholar]
- 3.Hutchinson L, Kirk R. High drug attrition rates--where are we going wrong? Nature reviews Clinical oncology. 2011;8(4):189–190. doi: 10.1038/nrclinonc.2011.34. [DOI] [PubMed] [Google Scholar]
- 4.Correia AL, Bissell MJ. The tumor microenvironment is a dominant force in multidrug resistance. Drug resistance updates: reviews and commentaries in antimicrobial and anticancer chemotherapy. 2012;15(1–2):39–49. doi: 10.1016/j.drup.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bissell MJ, Hines WC. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nature medicine. 2011;17(3):320–329. doi: 10.1038/nm.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer cell. 2012;21(3):309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 7.Villasante A, Vunjak-Novakovic G. Bioengineered tumors. Bioengineered. 2015;6(2):73–76. doi: 10.1080/21655979.2015.1011039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch ME, Brooks D, Mohanan S, Lee MJ, Polamraju P, Dent K, Bonassar LJ, van der Meulen MC, Fischbach C. In vivo tibial compression decreases osteolysis and tumor formation in a human metastatic breast cancer model. Journal of bone and mineral research: the official journal of the American Society for Bone and Mineral Research. 2013;28(11):2357–2367. doi: 10.1002/jbmr.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tse JM, Cheng G, Tyrrell JA, Wilcox-Adelman SA, Boucher Y, Jain RK, Munn LL. Mechanical compression drives cancer cells toward invasive phenotype. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(3):911–916. doi: 10.1073/pnas.1118910109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villasante A, Vunjak-Novakovic G. Tissue-engineered models of human tumors for cancer research. Expert opinion on drug discovery. 2015;10(3):257–268. doi: 10.1517/17460441.2015.1009442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villasante A, Marturano-Kruik A, Vunjak-Novakovic G. Bioengineered human tumor within a bone niche. Biomaterials. 2014;35(22):5785–5794. doi: 10.1016/j.biomaterials.2014.03.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marturano-Kruik A, Yeager K, Bach D, Villasante A, Cimetta E, Vunjak-Novakovic G. Mimicking biophysical stimuli within bone tumor microenvironment. Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Annual Conference; 2015; 2015. pp. 3561–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]