Abstract
The thymus is the site of T cell maturation. Notch receptors (Notch1-4) and ligands (DLL1-3 and Jagged1-2) constitute one of several pathways involved in this process. Our data revealed differential constitutive expression of Notch genes and ligands in T lymphocytes and thymic dendritic cells (tDCs), suggesting their participation in human thymocyte maturation. nTreg analyses indicated that the Notch components function in parallel to promote maturation in the thymus.
Keywords: Thymus, Notch, Treg, TDCS
1. Introduction
The thymus is a privileged and indispensable site for the generation and maturation of T cells in vivo, as this microenvironment induces and supports lineage commitment, differentiation, and survival of thymus-seeding cells. Indeed, the thymus is both where the T cell repertoire is generated and where T cells are shaped by positive and negative selection, giving rise to a broad functional MHC-restricted naïve TCR αβ repertoire [1].
T cell progenitors derived from hematopoietic stem cells migrate from the bone marrow and undergo development in the thymic environment to generate the T cell lineage. Notch signaling is one of several pathways involved in promoting T lineage commitment and maturation [2]. Notch receptors and their ligands are a family of trans membrane proteins involved in a variety of cell fate decisions that affect the developmental functions of many organs and systems, including hematopoiesis and the immune system. Mammalian genomes encode four Notch receptors (Notch 1–4, which are homologous to the single Notch receptor in Drosophila) and five ligands (Delta-like 1, 3, and 4 and Jagged 1 and 2, which are homologous to the Delta and Serrate ligands in Drosophila) [3]. The best established function for Notch signaling in the hematopoietic system is the essential role it plays in promoting T lineage commitment and maturation [4].
T lineage specification is mediated exclusively by Notch 1, and induced deletion of Notch 1 in hematopoietic progenitors results in a complete block of T cell development and the ectopic differentiation of immature B cells in the thymus [5]. Notch receptor-ligand interactions communicate signals between neighboring cells via highly conserved signaling mechanism activated through the interaction of the Notch receptor with its cognate ligand [6]. This pathway is initiated when Notch-ligand engagement induces two successive proteolytic cleavages, the second of which is mediated by a presenilin-containing complex exhibiting γ-secretase activity that releases the Notch intracellular domain (NICD). This activated form of Notch translocates to the nucleus where it binds to the CSL protein (also known as RBPJ, an important nuclear mediator of Notch signaling that is similar to the DNA-binding protein found in other species), thereby displacing co-repressors and recruiting co-activators [7].
Each Notch receptor interacts with a specific Notch ligand in a context-dependent manner. Numerous studies have identified transcripts for all Notch ligands in the thymus [8]. Members of the Delta-like family appear to be crucial for T lineage commitment and maturation. A report by Mohtashami and Zúgñiga-Pflücker [9] highlighted the importance of Delta-induced signals by demonstrating that the loss of DLL1 and DLL4 expression led to the inability of fresh, ex vivo thymic stromal monolayers to support T lineage specification following disruption of their three-dimensional organization. In contrast, the inactivation of the DLL1 ligand does not block T cell development, suggesting a redundant role for this and other DLL ligands, such as DLL4 [4]. These data suggest that Notch1 signaling in early thymocyte progenitors is restricted to DLL4 in the thymus [2].
T regulatory cells of the CD4+CD25+FOXP3+phenotype, also known as natural T regulatory cells (nTreg), are generated in the thymus as a specific T cell lineage and are critical for the maintenance of immune homeostasis and the suppression of naturally occurring self-reactive T cells [10]. The generation and maturation of this specific T cell lineage involve particular and complex processes within the thymus, and many signaling pathways participate in these processes. Ou-Yang and colleagues (2008) demonstrated that Notch signaling modulates the Foxp3 promoter through a RBPJ- and HES-1-dependent mechanism (notch target genes with transcriptional repressor functions that negatively regulate gene transcription) [11]; however, further investigations will be required to determine the components of this pathway (in the context of Notch signaling) that are involved in the processes of nTreg commitment, maturation, and differentiation.
Within the thymic medullary region, groups of epithelial cells, termed Hassal׳s corpuscles, have been shown to play a critical role in the maturation of developing thymocytes [12] and the generation of nTreg [13]. Human Hassal׳s corpuscles express thymic stromal lymphopoietin (TLSP), which activates thymic dendritic cells (tDC) to express high levels of CD80 and CD86. Once activated, tDCs induce the proliferation and differentiation of CD4+CD8-CD25-thymic T cells into CD4+CD25+FOXP3+cells, suggesting that tDCs play a crucial role in dendritic cell-mediated secondary positive selection of nTreg [13].
In this study, we investigated the expression of the Notch receptor and ligand genes in immature thymocytes, nTreg and tDCs to evaluate the potential involvement of this pathway in the maturation of T lymphocyte and nTreg cells in the human thymus.
2. Materials and methods
2.1. Patient samples
Thymic tissues were obtained from 10 patients who underwent corrective cardiac surgery at the Hospital do Coração (HCor), SP, Brazil ( mean 3.24 years). Each sample where analyzed in three independent experiments. The patients did not exhibit signs of immunodeficiency. The ethics committees at the Hospital do Coração and the School of Medicine at the University of São Paulo approved this study. Informed consent was obtained from the parents of all children.
2.2. Thymus tissue dissociation, cell isolation, and storage
Thymocytes and tDCs were released from the tissue samples using enzymatic dissociation. The thymus was cut into small fragments and added to 50-mL propylene conical centrifuge tubes. Next, an enzymatic solution (10 mL) containing RPMI pre-warmed to 37 °C, 0.5 mg/mL collagenase A, 0.02 mg/mL DNAse I (Roche Diagnostics, Mannheim, Germany), and 5% FBS was added, and incubation was performed for 10 min at 37 °C under continuous agitation. The digested fragments were homogenized gently and filtered through a plastic sieve with a 70-μM mesh screen (Cell Strainer, BD Falcon, CA, USA) to remove aggregates and stromal material. The resultant cell suspensions were washed twice with wash buffer 1 (50 mL RPMI [Gibco – Life Technologies, Grand Island, NY, USA]) pre-warmed to37 °C, 0.1 mg/mL collagenase A[Roche Diagnostics, Mannheim, Germany], and 0.02 mg/mL DNase I [Roche Diagnostics, Mannheim, Germany]), followed by centrifugation at 540 g for 5 min. Next, the pelleted cells were resuspended in a second wash buffer (50 mL cold PBS, 5 mM EDTA [Sigma Aldrich, Saint Louis, MO, USA], 0.02 mg/mL DNase I, and 5% FBS [Gibco – Life Technologies, USA]) and centrifuged at 540 g for 5 min. The pelleted cells were resuspended immediately in RPMI, and the low-density fraction was collected following centrifugation through Ficoll-Paque (GE Healthcare Bio Science, Uppsala, Sweden) at 540 g for 20 min. The cells were washed twice using RPMI and centrifuged at 1500 rpm for 10 min. The thymic cells were frozen and stored at −80 °C until use.
2.3. Flow cytometry and purification of thymic populations
Thawed cells were re suspended in pre-warmed RPMI, the number of cells per tube was adjusted, and the cells were subjected to fluorescence staining. To characterize cells populations in the human thymus, thymocytes were stained with human anti-CD3 PE-Cy7,-CD4 FITC or APC,-CD8 PE,-CD11cPE and -CD25 PE or FITC antibodies (BD Pharmingen, New Jersey, USA) and the cell populations were sorted into CD3-CD4-CD8-, CD4+CD8+, CD4-CD8+, CD4+CD8-, CD3+CD4+CD8-CD25high, and CD11c+subsets using a FACSAria II flow cytometer (BD Pharmingen, New Jersey, USA). The purity of all sorted cell subsets was greater than 97%.
2.4. RNA purification and preparation of cDNA
After cell sorting, total RNA from the sorted populations was isolated using the RNeasy Plus mini and micro kits (QIAGEN, Hilden, Germany) and was retro transcribed using Sensiscript (QIAGEN, Hilden, Germany) in accordance with the manufacturer׳s instructions.
2.5. Real-time PCR
The NOTCH1, NOTCH2, NOTCH3, DLL1, DLL3,DLL4, JAG1, JAG2, and FOXP3 genes were amplified using specific TaqMan® Gene Expression Assays and SYBR Green fluorescent dye (Applied Biosystems, California, USA) with an ABI 7600 SDS platform (Applied Biosystems, California, USA) in accordance with the manufacturer׳s instructions. The NOTCH4 gene was not evaluated in any of the purified subsets of cells. The GAPDH housekeeping gene was used for normalization. HEK cells were used as a reference for gene expression because they express all evaluated genes (NIH, Bethesda, MD, USA). All the RT-PCR reactions were performed using controls for exogenous contamination and genomic DNA contamination. Gene expression was calculated as 2-ΔCt, which corresponds to the Ct value of the target gene in a population normalized using the Ct value of the housekeeping gene (GAPDH) in the same population and was expressed as the mean±standard error. To perform relative gene analysis, we calculated 2−ΔΔCt in accordance with the method of Livak et al. [14], and the data were expressed as the means±standard errors. The following probes were used: NOTCH1 (Hs01062014), NOTCH2 (Hs01050702), NOTCH3 (Hs01128541), JAG1 (Hs01070032), JAG2 (Hs00171432), DLL1 (Hs00194509), DLL3 (Hs01085096), and FOXP3 (Hs01085834) all from Applied Biosystems, California, USA. The DLL4 primers 5′-CAGAGTGTCGGATATCAGCG-3′ and 5′-CCTGCCTTATACCTCCGTG-3′ (Applied Biosystems, California, USA) were used.
2.6. Statistical analysis
Comparisons of the expression of genes among the evaluated groups were performed using one-way ANOVA, Tukey׳s post-test (to compare all column pairs), and the Bonferroni method (to compare selected pairs of columns). Statistical significance was defined as a p-Value <0.05 using GraphPad Prism software (California, USA).
3. Results
3.1. Characterization of thymic populations and notch receptor and ligand gene expression in developing lymphocytes
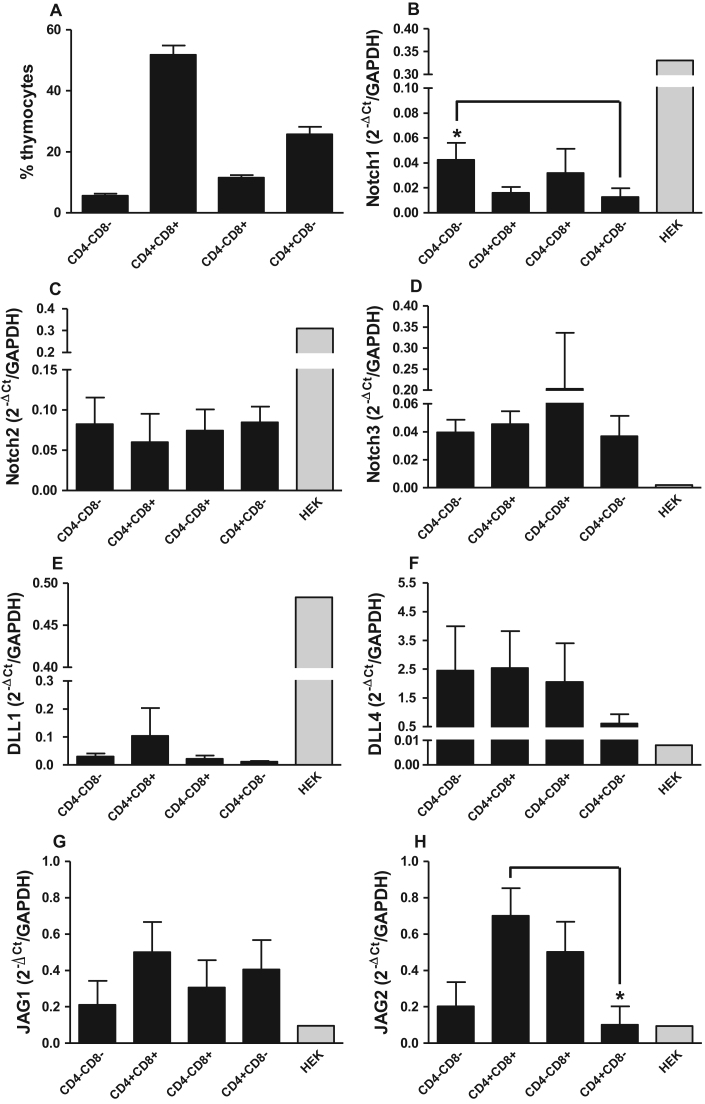
We analyzed suspensions of total thymocytes and characterized the main developmental stages of T cells within the thymus (Fig. 1A). Next, we investigated the expression of the Notch receptor and ligand genes NOTCH1, NOTCH2, NOTCH3, JAG1, JAG2, DLL1, DLL3 and DLL4 in these populations using HEK cells as a reference. All Notch receptors and ligands were expressed in each subset, with the exception of the DLL3 gene (Fig. 1B-H). The NOTCH1 gene was expressed at a higher level in the CD4-CD8-subset than the CD4+CD8-subset (Fig. 1B), and the JAG2 gene exhibited higher expression in the CD4+CD8+subset than the CD4+CD8-cell subset (Fig. 1H). The NOTCH3, DLL1, DLL4, and JAG1 genes exhibited similar expression levels among thymocyte subsets (Fig. 1D-G).
Fig. 1.
Expression of Notch receptor and ligand genes in thymocyte subsets. (A) Total thymocytes were sorted into CD4-CD8-, CD4+CD8+, CD4-CD8+, and CD4+CD8-populations. (B-H) Gene expression of Notch receptors and ligands in total RNA isolated from purified thymocyte subsets and from HEK cells (positive controls). The bars represent the means±SD, *p<0.05 in comparison to an indicated subset (n=10).
3.2. Characterization of nTreg cells and evaluation of notch receptor and ligand gene expression
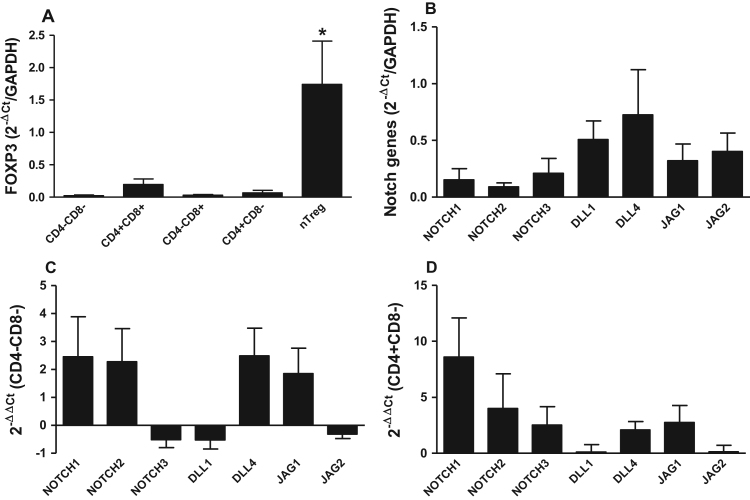
We isolated thymic nTreg cells with the phenotype CD3+CD4+CD8-CD25high; these cells represented 2.16±0.4% of the total thymocytes and expressed the FOXP3 gene at higher levels compared to the other studied populations (Fig. 2A).
Fig. 2.
Expression of FOXP3 and Notch receptor and ligand genes in nTreg. (A) Expression of the FOXP3 gene in total RNA isolated from each subset of thymocytes. (B) Expression of Notch receptor and ligand genes in nTreg. (C) Relative expression of Notch receptor and ligand genes in nTreg cells using the CD4-CD8-subset as a reference. (D) Relative expression of Notch receptor and ligand genes in nTreg using theCD4+CD8-subset as reference. The bars represent the means±SD, *p<0.05 for comparisons between thymocyte subsets (n=10).
After characterizing the nTregs, we evaluated the expression of the NOTCH1,−2, and −3; DLL1,−3 and −4;and JAG1 and −2 genes in this cell subset. The nTreg cells expressed all of these Notch receptor and ligand genes, with the exception of the DLL3 gene. The expression levels of the evaluated Notch receptors and ligands were not significantly different from one another (Fig. 2B). In comparison to the initial stage of thymocyte development (CD4-CD8-phenotype), nTreg cells demonstrated increased expression of the NOTCH1, NOTCH2, DLL4, and JAG1 genes but lower expression of the NOTCH3, DLL1, and JAG2 genes (Fig. 2C). Interestingly, when nTreg cells were compared to cells at the later stage of development with a non-regulatory phenotype (CD4+CD8-), we observed increased expression of the NOTCH1, NOTCH2, NOTCH3, DLL4,and JAG1 genes and similar levels of DLL1 and JAG2 gene expression (Fig. 2D).
3.3. Characterization of notch receptor and ligand expression in tDCs
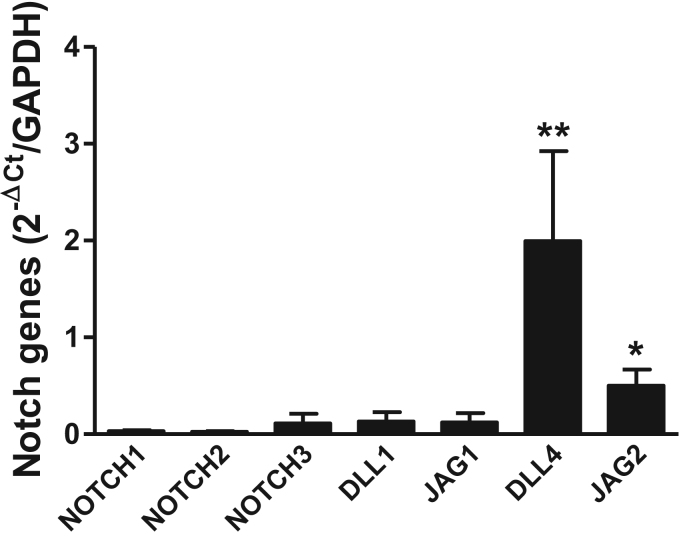
Finally, we characterized tDCs by flow cytometry using the CD11c molecule. After cell sorting, we found that this subset represented 3.1±0.4% of the total thymocyte population. The expression of the DLL4 ligand was higher than those of the other Notch receptor and ligand genes in tDCs (Fig. 3). In addition, the expression of the JAG2 gene was higher than that of the Notch receptor and ligand genes (with the exception of DLL4), and lower levels of NOTCH1, NOTCH3, DLL1, and JAG1 gene expression were detected in tDCs (Fig. 3).
Fig. 3.
Gene expression of Notch receptors and ligands in tDCs. Expression of Notch receptor and ligand genes in tDCs. The bars represent the means±SD (n=10), **p<0.05 for comparisons with all notch receptors and ligands, *p<0.05 for comparisons with all notch receptors and ligands except DLL4 (n=10).
4. Discussion
Notch signaling is involved in regulating the TCR-αβ/γδ lineage decision [15]. Several studies have demonstrated that Notch receptors and ligands are expressed during different phases of T cell maturation in rodents [16], [17], [18], although the situation in humans is less clear. Therefore, we evaluated the expression of Notch receptor and ligand genes in subsets of developing lymphocytes, nTreg cells, and tDC cells in the human thymus.
The frequencies of the lymphocyte subsets at different developmental stages were similar to those previously described in the literature [19], [20]. In the CD4-CD8-subset, we observed an increased level of NOTCH1 gene expression compared to the CD4+CD8-subset, which was consistent with previous observations in mice [3]. In the human thymus, a previous report by Van de Walle et al., 2009 revealed that NOTCH1 was more highly expressed in CD34+cells, with or without CD1 co-expression, compared to TCRαβ+ thymocytes. This result is consistent with our results; our CD4-CD8-subset is equivalent to CD34+thymocytes and our CD4+CD8-subset is included in TCRαβ+ cells. Therefore, NOTCH1 gene expression seems to contribute to T lineage commitment mainly at immature stages, as suggested by Maillard et al. [3].
We observed similar expression of the NOTCH2 gene between TCRαβ+ cells, which differs from published reports in mice demonstrating that this receptor is expressed at higher levels during early stages of maturation, similar to the CD4-CD8-population [21]. Van de Walle et al. previously examined the NOTCH2 gene in human thymus, and also observed similar expression between CD4+CD8+CD3-and TCRαβ+ subsets [22].
Therefore, these data suggest that the NOTCH2 gene is expressed not only in the earlier stages of T cells development but also in mature single positive lymphocytes. Because this receptor is similar to Notch1 [17], our data suggest that NOTCH2 gene expression may be involved in all stages of CD4+and CD8+T cell maturation.
The CD4+CD8-population expressed lower levels of the NOTCH1 gene in comparison to the CD4-CD8-subset, a feature also observed in mice [23]. Nevertheless, in the CD4+CD8-subset, we determined that the JAG2 gene was expressed at a lower level in comparison to CD4+CD8+thymocytes. Studies using a co-culture system of human CD34+CD38-lin- CB cells (uncommitted CB cells) with Jagged2-transduced OP9 stromal cells found that these conditions supported the development of early CD7+CD1+-committed T cell precursors and their further development into CD4+CD8β+double-positive thymocytes and CD3+TCRαβ+ cells [15]. Therefore, our analysis demonstrating constitutive JAG2 gene expression at lower levels among CD4+CD8-lymphocytes compared to CD4+CD8+thymocytes suggests that this gene may be involved in CD4+T cell lineage commitment or may represent a consequence of this type of maturation.
Furthermore, we demonstrated that the DLL1 gene, although at low levels, seems to be expressed constitutively in the CD4+CD8-population of thymocytes in humans; this was not specifically confirmed at the protein level, but it has not been demonstrated previously in mice or humans [2]. This result suggests that this ligand may play an important role in the later stages of T lymphocyte maturation, primarily for CD4+T lymphocyte development.
This study is also the first to demonstrate expression of the DLL4 gene in thymocyte populations, although this ligand has been shown to be expressed preferentially by thymic stromal cells in mice [3], [24]. In mice, the initial steps of T lineage maturation require DLL4 signaling via the Notch1 receptor [25], which, according to our data, may also occur during the later stages of human T lymphocyte development.
Previous reports in the literature have demonstrated the expression of Notch receptor and ligand genes in human nTreg [22]; accordingly, we found that nTreg expressed all the Notch receptor and ligand genes evaluated.
Of the Notch receptors and ligands evaluated in this study, the DLL4 gene appeared to be more highly expressed in nTreg; however, statistical analysis demonstrated that expression of this ligand was not different from that of the other Notch receptor and ligand genes.
When nTreg were compared to the CD4+CD8-population, we observed that NOTCH1 gene expression was increased seven-fold, and NOTCH2 gene expression was increased three-fold in nTreg. These data suggest that nTreg exhibit a significant alteration in NOTCH1 and NOTCH2 gene expression levels during maturation compared to CD4+non-regulatory T cells in the thymus. Notch1 and Notch2 receptors play similar roles during the maturation of developing thymocytes [17], but their roles in nTreg maturation have not been described.
tDCs play a major role in T lymphocyte maturation, principally in the generation of nTreg cells [13], [19]. This role is mediated mainly by cell-to-cell contact; therefore, the expression of Notch receptor and ligand genes in these cells suggests the involvement of the Notch pathway in T lymphocyte maturation in the human thymus.
5. Conclusion
Taken together, our data suggest that changes in the expression of Notch ligands and receptors play a role in the spontaneous lymphocytes maturation in the human thymus. Furthermore, in this study we uncover the possible involvement of Notch signaling in human nTreg maturation.
Conflict of interests
The authors have no conflicts of interest to declare.
Acknowledgments
This study was funded through a Grant from the Laboratory of Medical Investigation – 56, Medical School, University of Sao Paulo, Sao Paulo, Brazil (LIM-56 HC-FMUSP), São Paulo Research Foundation (FAPESP - grant #2012/01248-3) and The National Council for Scientific and Technological Development (CNPq - grant #115603/2015-8). We thank the pediatric cardiac surgery team and surgical center staff at the Hospital do Coração, Associação Sanatorio Sirio (Sao Paulo, SP). We thank Dr. Thomas Arthur Fleisher (NIH, Bethesda, MD, USA) for providing the HEK cell line. We thank Alessandra Pontillo and Maria Notomi Sato for scientific discussions and suggestions regarding this article. Finally, we thank the parents who consented to the research for the realization of this work.
References
- 1.Plum J., De Smedt M., Leclercq G., Taghon T., Kerre T., Vandekerckhove B. Human intrathymic development: a selective approach. Semin. Immunopathol. 2008;30:411–423. doi: 10.1007/s00281-008-0135-2. [DOI] [PubMed] [Google Scholar]
- 2.Ciofani M., Zuniga-Pflucker J.C. The thymus as an inductive site for T lymphopoiesis. Annu. Rev. Cell Dev. Biol. 2007;23:463–493. doi: 10.1146/annurev.cellbio.23.090506.123547. [DOI] [PubMed] [Google Scholar]
- 3.Maillard I., Fang T., Pear W.S. Regulation of lymphoid development, differentiation, and function by the notch pathway. Annu. Rev. Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 4.Yuan J.S., Kousis P.C., Suliman S., Visan I., Guidos C.J. Functions of notch signaling in the immune system: Consensus and controversies. Ann. Rev. Immunol. 2010;28(28):343–365. doi: 10.1146/annurev.immunol.021908.132719. [DOI] [PubMed] [Google Scholar]
- 5.Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H.R., Aguet M. Deficient t cell fate specification in mice with an induced inactivation of notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 6.Bray S.J. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 7.Fiuza U.M., Arias A.M. Cell and molecular biology of notch. J. Endocrinol. 2007;194:459–474. doi: 10.1677/JOE-07-0242. [DOI] [PubMed] [Google Scholar]
- 8.Radtke F., Wilson A., Mancini S.J.C., MacDonald H.R. Notch regulation of lymphocyte development and function. Nat. Immunol. 2004;5:247–253. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 9.Mohtashami M., Zuniga-Pflucker J.C. Three-dimensional architecture of the thymus is required to maintain delta-like expression necessary for inducing t cell development. J. Immunol. 2006;176:S314. doi: 10.4049/jimmunol.176.2.730. [DOI] [PubMed] [Google Scholar]
- 10.Wing K., Sakaguchi S. Regulatory t cells exert checks and balances on self tolerance and autoimmunity. Nat. Immunol. 2010;11:7–13. doi: 10.1038/ni.1818. [DOI] [PubMed] [Google Scholar]
- 11.Ou-Yang H.F., Zhang H.W., Wu C.G., Zhang P., Zhang J., Li J.C., Hou L.H., He F., Ti X.Y., Song L.Q. Notch signaling regulates the foxp3 promoter through rbp-j-and hes1-dependent mechanisms. Mol. Cell Biochem. 2009;320:109–114. doi: 10.1007/s11010-008-9912-4. [DOI] [PubMed] [Google Scholar]
- 12.Senelar R., Escola M.J., Escola R., Serrou B., Serre A. Relationship between hassalls corpuscles and thymocytes fate in guinea-pig fetus. Biomedicine. 1976;24:112–122. [PubMed] [Google Scholar]
- 13.Watanabe N., Wang Y.H., Lee H.K., Ito T., Wang Y.H., Cao W., Liu Y.J. Hassall׳s corpuscles instruct dendritic cells to induce cd4(+)cd25(+) regulatory t cells in human thymus. Nature. 2005;436:1181–1185. doi: 10.1038/nature03886. [DOI] [PubMed] [Google Scholar]
- 14.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Van de Walle I., De Smet G., Gartner M., De Smedt M., Waegemans E., Vandekerckhove B., Leclercq G., Plum J., Aster J.C., Bernstein I.D. Jagged2 acts as a delta-like notch ligand during early hematopoietic cell fate decisions. Blood. 2011;117:4449–4459. doi: 10.1182/blood-2010-06-290049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothenberg E.V. Transcriptional drivers of the t-cell lineage program. Curr. Opin. Immunol. 2012;24:132–138. doi: 10.1016/j.coi.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson P.K., Zuniga-Pflucker J.C. On becoming a t cell, a convergence of factors kick it up a notch along the way. Semin. Immunol. 2011;23:350–359. doi: 10.1016/j.smim.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Radtke F., Fasnacht N., MacDonald H.R. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 19.Tourigny M.R., Mazel S., Burtrum D.B., Petrie H.T. T cell receptor (tcr)-beta gene recombination: Dissociation from cell cycle regulation and developmental progression during t cell ontogeny. J. Exp. Med. 1997;185:1549–1556. doi: 10.1084/jem.185.9.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernandez-Hoyos G., Alberola-Ila J. A notch so simple influence on t cell development. Semin. Cell Dev. Biol. 2003;14:121–125. doi: 10.1016/s1084-9521(02)00180-5. [DOI] [PubMed] [Google Scholar]
- 21.Saito T., Chiba S., Ichikawa M., Kunisato A., Asai T., Shimizu K., Yamaguchi T., Yamamoto G., Seo S., Kumano K. Notch2 is preferentially expressed in mature b cells and indispensable for marginal zone b lineage development. Immunity. 2003;18:675–685. doi: 10.1016/s1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 22.Van de Walle I., De Smet G., De Smedt M., Vandekerckhove B., Leclercq G., Plum J., Taghon T. An early decrease in notch activation is required for human tcr-alphabeta lineage differentiation at the expense of tcr-gammadelta t cells. Blood. 2009;113:2988–2998. doi: 10.1182/blood-2008-06-164871. [DOI] [PubMed] [Google Scholar]
- 23.Parreira L., Neves H., Simoes S. Notch and lymphopoiesis: a view from the microenvironment. Semin. Immunol. 2003;15:81–89. doi: 10.1016/s1044-5323(03)00004-6. [DOI] [PubMed] [Google Scholar]
- 24.Laky K., Fowlkes B.J. Notch signaling in cd4 and cd8 t cell development. Curr. Opin. Immunol. 2008;20:197–202. doi: 10.1016/j.coi.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hozumi K., Mailhos C., Negishi N., Hirano K., Yahata T., Ando K., Zuklys S., Hollander G.A., Shima D.T., Habu S. Delta-like 4 is indispensable in thymic environment specific for t cell development. J. Exp. Med. 2008;205:2507–2513. doi: 10.1084/jem.20080134. [DOI] [PMC free article] [PubMed] [Google Scholar]