Abstract
De novo transcriptome sequencing is a robust method for microRNA (miRNA) target gene prediction, especially for organisms without reference genomes. Following exposure of Megalobrama amblycephala to ammonia (0.1 or 20 mg L−1 ), two cDNA libraries were constructed from the fish gills and sequenced using Illumina HiSeq 2000. Over 90 million reads were generated and de novo assembled into 46, 615 unigenes, which were then extensively annotated by comparing to different protein databases, followed by biochemical pathway prediction. The expression of 2666 unigenes significantly differed; 1961 were up-regulated, while 975 were down-regulated. Among these, 250 unigenes were identified as the targets for 10 conserved and 4 putative novel miRNA families by miRNA target computational prediction. We examined expression of ssa-miRNA-21 and its target genes by real-time quantitative PCR and found agreement with the sequencing data. This study demonstrates the feasibility of identifying miRNA targets by transcriptome analysis. The transcriptome assembly data represent a substantial increase in the genomic resources available for Megalobrama amblycephala and will be useful for gene expression profile analysis and miRNA functional annotation.
Keywords: Megalobrama amblycephala, Ammonia, mRNA, miRNA, Transcriptome
Highlights
-
•
Several differentially expressed genes and miRNA were identified.
-
•
Target prediction indicated that a number of miRNAs involved in immunity.
-
•
The study provides an understanding of molecular mechanisms of ammonia-induced toxicology in fish.
1. Introduction
Next-generation sequencing (NGS)-based RNA sequencing (RNA-seq) methods for transcriptome analysis simultaneously allows acquisition of sequences for gene discovery and also helps identify transcripts involved in specific biological processes. Recent advances in RNA-seq have generated an unprecedented global view of the transcriptome and provide a more efficient method to explore the transcriptional landscape [1], [2]. In addition, the properties of RNA-seq (such as its dynamic range, sensitivity and specificity) also make it ideal for quantitatively analyzing various aspects of gene regulation [3]. Importantly, RNA-seq technologies do not require prior knowledge of the genomic sequence, and compared to traditional sequencing methods, are more practical in terms of time, cost, labor, amount of data produced, data coverage, sensitivity and accuracy [4], [5]. Therefore, they are considered to be efficient and reliable for genome and transcriptome sequencing, and are suitable for the study of non-model organisms such as economically important freshwater fish species.
Blunt snout bream (Megalobrama amblycephala), which accounts for an high proportion of Chinese aquaculture yields, appears prone to disease out-breaks when exposed to high temperatures and elevated ammonia concentrations [6]. Based on this finding, we predicted that M. amblycephala is relatively sensitive to ammonia exposure. This represents an important concern as ammonia is one of major environmental pollutants in fish culture, especially in recirculation systems [7], [8], [9], [10], [11]. While ammonia can be removed by biological filtration or water exchange, a transient, sudden and rapid increase in ammonia levels may be detrimental to fish [12]. Excessive ammonia can cause fish growth reduction [13], [14], tissue erosion and degeneration [15], [16], as well as immune suppression and high mortality [17]. However, the molecular mechanisms involved in ammonia detoxification are still unclear.
MicroRNAs (miRNAs) are 20–22-nt non-coding RNAs that play important roles in post-transcriptional gene regulation. In animal cells, miRNAs regulate their targets by translational inhibition and mRNA destabilization [18], and may also play a role in the stress response [19], [20], [21], [22]. The growing body of literature showing alterations in miRNA profiles in response to environmental and endogenous exposures indicates that miRNAs may play important roles as stress regulators. Therefore, dissecting their biological function may be useful in understanding the molecular mechanisms involved in ammonia-induced toxicity in M. amblycephala. As identification of the biological functions of these miRNAs requires target prediction, in this study, we aimed to construct a more powerful transcriptome dataset for target identification.
Although two parallel M. amblycephala expressed sequence tag analyses have already been conducted using certain tissues [23], [24], the data generated here represent the first effort to characterize ammonia exposure-mediated changes in the M. amblycephala transcriptome. The two cDNA libraries from the ammonia treatment group and control group used for our miRNA analysis were constructed and sequenced with Illumina HiSeq 2000. The obtained reads were assembled into transcripts and annotated by BLAST analysis against various databases before screening for differentially expressed genes, followed by miRNA target prediction. Our work will provide an approach to identify genes targeted by miRNA and to characterize their functional/regulatory networks to increase our understanding of ammonia-induced toxicology in M. amblycephala.
2. Materials and methods
2.1. Ethics statement
All fish handling was conducted in accordance with the Guidelines on the Care and Use of Animals for Scientific Purposes set up by the Institutional Animal Care and Use Committee (IACUC) of the Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences (Wuxi, China).
2.2. Experimental animals
We obtained 300 healthy, similarly sized (mean weight: 15.16±1.24 g) M. amblycephala juveniles from the Freshwater Fisheries Research Center, Chinese Academy of Fishery Sciences, China. The fish were immediately transferred to the aquatic laboratory and held in three 500-L fiberglass tanks (N=100 fish/tank). During acclimation, each tank was supplied with pre-aerated municipal water (pH 7.85±0.08; chloride, 15–18 mg L−1; dissolved oxygen, 5.16–6.53 mg L−1; total ammonia<0.05 mg L−1; and baseline nitrite, <0.1 mg L−1) and maintained at 20±1 °C with natural light and photoperiod. Fish were provided a commercial pelleted diet twice daily at a ration of 3% total body weight. The water exchange rate was 33% per day, and the fecal matter was removed daily from the aerated tanks.
2.3. Sample collection
Based on the LC50 estimate established in a previous study from our group [25], the fish were subjected to a 48-h ammonia exposure using two concentrations of ammonia: 0.1 mg L−1 (control) and 20 mg L−1 (test), with three replicate tanks for each concentration (20 fish per tank). The ambient ammonia concentration for each group was adjusted to the required value by adding a stock solution of total ammonia-nitrogen (TAN). After a 48-h exposure to ammonia, three fish were collected randomly from each tank and anesthetized with MS-222 (Sigma-Aldrich, St Louis, MO, USA) to obtain gill-pooling samples that were stored at −80 °C until RNA extraction for RNA-seq analysis.
2.4. cDNA library construction and sequencing
Total RNA was obtained from the fish gills using a total RNA purification kit (LC Sciences, Houston, TX, USA) and was further purified using the TruSeq RNA LT Sample Prep Kit v2 (Illumina, San Diego, CA, USA) according to the manufacturer’s protocol. Oligo-dT beads were used to obtain poly (A+) mRNA from a total RNA pool consisting of equal quantities of total RNA from two sample types (the control and treatment groups). Purified mRNAs were then fragmented using divalent cations under elevated temperatures, and then converted to dsDNA by two rounds of cDNA synthesis using reverse transcriptase and DNA polymerase I. After the end repair process, DNA fragments were ligated with adaptor oligos [26]. The ligated products were amplified (15 PCR cycles) to generate an RNA-seq library. cDNA sequencing was performed using a Genome Analyzer IIx (Illumina).
2.5. Data processing, assembly and functional annotation
Raw data generated by Illumina sequencing were preprocessed to remove the adaptor sequences and any ambiguous or low-quality reads. Subsequently, de novo assembly of the clean reads was performed using the assembly program Trinity [27], [28]. First, for fast and efficient transcript assembly, the short reads were assembled into high-coverage contigs that could not be extended farther in either direction using a k-mer-based approach. Then, the related contigs were clustered, and a de Bruijn graph was constructed for each cluster. Finally, in the context of the corresponding de Bruijn graph and all plausible transcripts, alternatively spliced isoforms and transcripts were derived.
All assembled transcripts were compared with publicly available databases including Nr (NCBI non-redundant protein sequences), KOG/COG (Clusters of Orthologous Groups of proteins) [29], Swiss-Prot (a manually annotated and reviewed protein sequence database) [30], KO (KEGG ortholog database) [31], Pfam (protein family) [32] and GO (Gene Ontology) (http://www.geneontology. org/). The Nr, KOG/COG, Swiss-Prot and KO databases used BLASTx analysis with a cut-off E-value of 10−5, while Pfam used Hmmerscan and GO used Blast2GO [33]. The best BLAST hits from all BLAST results were parsed for a homology-based functional annotation. For the nr annotations, the Blast2GO program was used to obtain GO annotations of unique assembled transcripts to describe biological processes, molecular functions and cellular components.
2.6. Differential gene expression profiling
The expression abundance for each assembled transcript was measured using the reads per kilobase per million mapped reads (RPKM) values. All reads were mapped onto the non-redundant set of transcripts to quantify the abundance of assembled transcripts. Bowtie was used for read mapping and applied for RPKM-based expression measurement. The expression levels for each read for the two different ammonia concentrations (0.1 mg/L and 20 mg/L) were calculated using the number of reads with a specific match. When comparing the two samples, a minimum two-fold difference in log2 expression was required to indicate differential expression.
2.7. miRNA target prediction
miRNA-seq analysis was conducted in the same biological samples as the mRNA-seq as described above. Small RNA libraries were constructed using a Small RNA Cloning Kit (TaKaRa). RNA was purified by polyacrylamide gel electrophoresis (PAGE) to enrich for molecules in the 17–27 nt range, and then was ligated with 5ʹ and 3ʹ adapters. The resulting samples were used as templates for cDNA synthesis, followed by PCR amplification. The obtained libraries were subjected to sequencing using the Illumina sequencing-by-synthesis technology. After the run, image analysis, sequencing quality evaluation and data production summarization were performed using the Illumina/Solexa pipeline. All small RNA data has been deposited into the NCBI Sequence Read Archive (Database ID: SRA322742). The sequencing data was pretreated to discard low-quality reads, 3ʹ-adaptor reads, 5ʹ-adaptor contaminants and sequences shorter than 18 nucleotides. After trimming the 3ʹ adaptor sequence, sequence tags were mapped onto the transcriptome of M. amblycephala using Bowtie. Any small RNAs that were exact matches to the transcriptome of M. amblycephala were used from further analysis. The mapped reads were compared to the miRBase (19.0) to annotate conserved miRNAs. To predict novel miRNAs, miREvo [34] and miRDeep2 [35] were used.
The miRanda toolbox was utilized for computational identification of differentially expressed miRNA targets [36], using the complementary region between miRNAs and mRNAs and factoring in the thermodynamic stability of the miRNA-mRNA duplex. All the mRNAs used for target prediction came from the differentially expressed unigenes obtained above. Regions of complementarity between the miRNA and the 3׳UTR of the mRNA were examined using a dynamic programming algorithm of the miRanda toolbox and the scores, which were based on sequence complementarity as well as minimum free energy of RNA duplex, were calculated using the Vienna RNA package [37]. All detected targets with scores and energies lower than the threshold parameters of S>90 (single-residue pair scores) and ΔG <−17 kcal/mol (minimum free energy) were selected as potential targets.
2.8. Real-time quantitative PCR (RT-qPCR) validation
The sequencing results were validated by RT-qPCR using the One Step PrimeScript miRNA cDNA Synthesis Kit (TaKaRa) for miRNA, the PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Shiga, Japan) for mRNA and SYBR Premix Ex Taq II (2x) (TaKaRa) for RT-qPCR, according to the manufacturer protocols. The following primers were used: For ssa-miR-21b, [F: 5ʹ-AGCGGCGGTGAGTAT TACTTC-3ʹ, R: 5ʹ-AGCGGCGGTGAGTATTACTTC-3ʹ]; interleukin-10 (IL-10), (F: 5ʹ-AAGGAGCTCCGTTCTG CATAC-3ʹ, R:5ʹ-AGTCGATGGGTGTTTTCGGG-3ʹ); ferritin (FRIS) (F: 5ʹ-CCGAAATCCGCCAGAACTAC-3ʹ, R: 5ʹ-GCTTATCGGCAT GCTCTCTC-3ʹ); and C-C chemokine receptor type 9 (CCR9) (F: 5ʹ-CACACTTCAC AAACCGCCTG-3ʹ, R: 5ʹ-AGCCATAGATGGAATCGGCG-3ʹ). Unless specified, the primers were purchased from Sangon Biotech (Shanghai, China) and RT-qPCR quantification was carried out using the PRISM® 7900HT Real-Time PCR System (Life Technologies/Applied Biosystems, Foster City, CA, USA). To normalize expression values, U6 snRNA for miRNA and β-actin for mRNA were used as housekeeping controls [38], [39]. Expression levels were quantitatively analyzed using the 2−ΔΔCT method [40]. One-way ANOVA tests were performed using SPSS 17.0 to determine significant differences. Each experiment was repeated in triplicate.
3. Results and discussion
3.1. De novo assemblies and annotation of unigenes
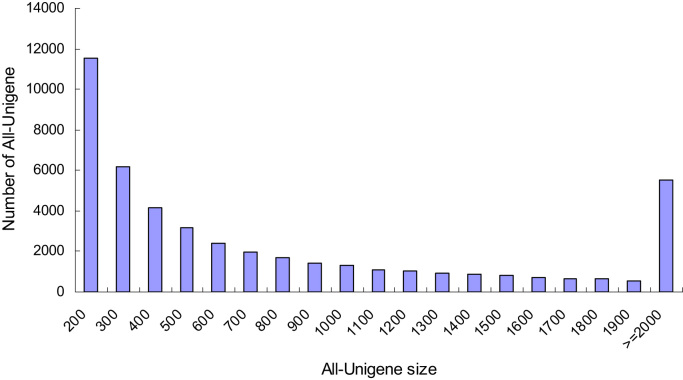
We generated 90.87 million raw reads by sequencing, and submitted this raw data to the NCBI Short Read Archive under the accession numbers SRX1021887 and SRX1021888. After trimming, 89.43 million clean reads corresponding to 10.87 GB clean bases (Table 1) remained, which were then assembled using the de novo assembly program Trinity [27]. These short reads were further assembled into 46,615 unigenes, which had an average length of 939 bp (Table 2). The size distribution of these transcripts ranged from 201 to 16,157 bp, of which 24,715 were larger than 2000 bp. Fig. 1.
Table 1.
Bluntnose black bream transcriptome sequencing results.
| Sample | Raw data | Valid data | Valid ratio (%) | Q20 percentage (%) | ||
|---|---|---|---|---|---|---|
| Read | Base | Read | Base | |||
| CG | 44,578,630 | 4,457,863,000 | 44,103,206 | 4,410,320,600 | 98.93 | 95.24 |
| TG | 46,295,912 | 4,629,591,200 | 45,331,332 | 4,533,133,200 | 98.88 | 95.42 |
CG: Gills of Blunt nose black bream without ammonia exposure; TG: gills of Blunt nose black bream under ammonia exposure 48 h.
Table 2.
cDNA library sequencing results.
| Parameter | Number |
|---|---|
| Number of unigene | 46,615 |
| Total bases of unigene (bp) | 43,774,977 |
| Number of unigene ≥500 bp | 24,715 |
| Mean length of unigenes (bp) | 939 |
| N50 | 1593 |
| Maximal length of unigenes (bp) | 16,157 |
Fig. 1.
Size distribution of the assembled unigenes in the sequenced cDNA library.
3.2. Annotation of predicted proteins
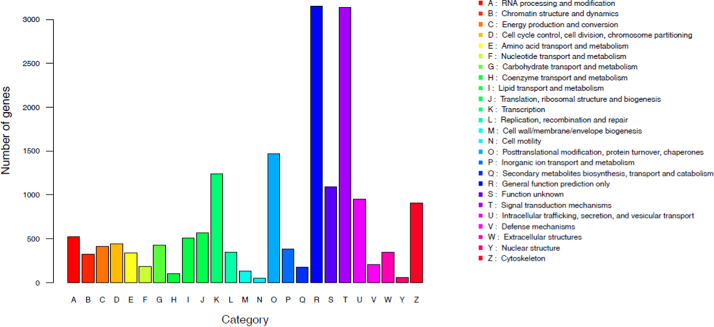
A total of 46,615 unigenes (50.02% of the transcripts) matched known genes corresponding to 25,180 annotated proteins (Table 3, Table S1). An additional functional annotation of the unigenes of M. amblycephala was performed by searching for putative orthologs and paralogs within the KOG database. A total of 46,615 unigenes (37.46%) were assigned to 26 eukaryotic orthologous groups (Fig. 2). The category “General function prediction only,” under which 3,151 unigenes (18.04% of 17,462 unigenes) could be grouped was the largest, followed by the categories “Signal transduction mechanisms” (3140, 17.98%), “post-translational modification, protein turnover, chaperone” (1468, 8.41%) and “Transcription” (1238, 7.10%).
Table 3.
BLAST analysis of non-redundant unigenes against public databases.
| Database | Number of annotated unigenes | Percentage of annoted unigenes (%) |
|---|---|---|
| Nr | 25,180 | 50.02 |
| Pfam | 18,803 | 40.34 |
| Swiss-prot | 19,997 | 40.90 |
| KO | 14,385 | 30.86 |
| KOG | 18,789 | 40.31 |
| GO | 17,773 | 38.13 |
Nr: NCBI non-redundant protein sequences, Pfam: Protein family, Swiss-Prot: A manually annotated and reviewed protein sequence database, KO: KEGG Ortholog database, KOG: Clusters of Orthologous Groups of proteins and GO: Gene Ontology.
Fig. 2.
KOG annotations. of the identified M. amblycephala unigenes.
3.3. GO annotation and KEGG pathway analyses
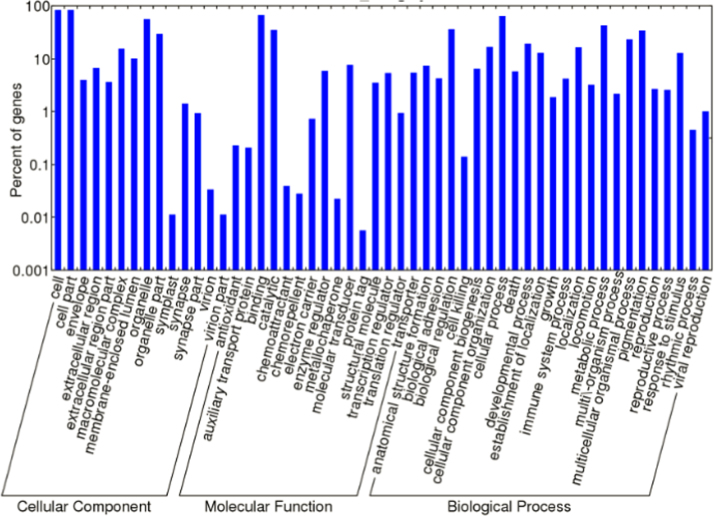
After GO annotation, M. amblycephala transcripts could be assigned to three categories: biological processes, molecular functions and cellular components (Fig. 3). Searching against the Kyoto Encyclopedia of Genes and Genomes Pathway database (KEGG) revealed that 18,780 unigenes could be matched to 148 KEGG pathways. The most represented pathways in hierarchy 2 were the “Cell Communication” (1185 unigenes) and the “Immune System” (1140 unigenes) pathways (Table 4). Multiple pathways and processes related to immune function were also identified, such as the “Toll-like receptor signaling” pathway [41], the “chemokine signaling” pathway [42] and “Complement and coagulation cascades” [43]. This suggests that exposure of M. amblycephala to higher concentrations of ammonia affects various processes involved in immune activation and functioning.
Fig. 3.
Level 2 GO term distributions for the biological process, cellular component and molecular function categories.
Table 4.
Top 10 list of the gene number of pathway.
| Pathway hierarchy 2 | Unigene number |
|---|---|
| Signal transduction | 1502 |
| Cell communication | 1185 |
| Immune system | 1140 |
| Signaling molecules and interaction | 802 |
| Endocrine system | 707 |
| Cancers | 704 |
| Carbohydrate metabolism | 542 |
| Amino acid metabolism | 485 |
| Nervous system | 450 |
| Lipid metabolism | 407 |
3.4. Differential gene expression in response to ammonia exposure
Based on the transcriptome sequence data, two differential gene expression (DGE) libraries representing the control and treatment groups were constructed to identify the differentially expressed unigenes. After removing low-quality reads, 44,103,206 and 45,331,332 clean reads were generated, respectively, for the control and treatment libraries (Table 1). Among these clean reads, 36,138,990.41 readcounts from the control group and 37,738,333.89 readcounts from the treatment group could be mapped to unigenes.
Analysis revealed that 2666 genes showed significantly different expression when comparing the M. amblycephala in two different ammonia concentrations (0.1 mg/L and 20 mg/L). Of these, 1691 were up-regulated and 975 were down-regulated in the control samples compared to the treatment samples (Fig. 4 and Table S2). GO enrichment analysis of the DEGs indicated that these genes were significantly enriched in oxidation–reduction processes (biological processes), integral to membrane (cellular components) and protein binding (molecular functions) (Table S3). Pathway enrichment analysis found the DEGs to be mainly enriched in complement and coagulation cascades (Table S4). Additionally, the findings of this study were consistent with well-established reports of heat shock protein upregulation in response to a wide range of environmental stressors: 70-kilodalton heat shock protein (HSP70), HSP 27 and HSP90 were significantly up-regulated in M. amblycephala following ammonia exposure. HSP are also potent activators of the innate immune system [44], [45]. To inhibit innate immunity and ensure successful infection, pathogens have evolved mechanisms involving host miRNA-mediated down-regulation of heat shock protein expression. Interestingly, ammonia exposure resulted in down-regulation of alpha-2-macroglobulin, complement components and C-type lectin, all of which play important roles in the innate immune response [46], [47], [48], [49]. These findings suggest that exposure to ammonia may impair immune responses in fish. Thus, prolonged ammonia exposure may have population consequences stemming from a reduced the ability to resist disease following increases in the ammonia levels.
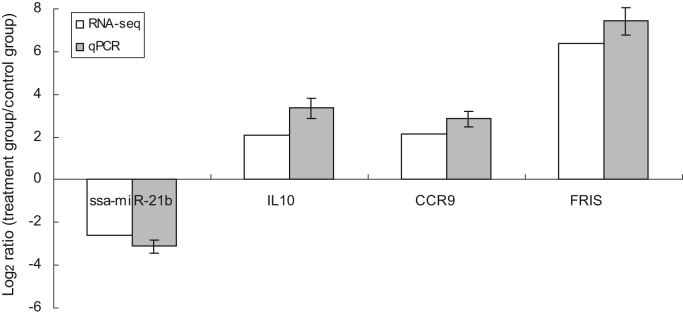
Fig. 4.
qRT-PCR validation of transcriptome sequencing results for the ammonia exposure group vs. the control group. One down-regulated miRNA (ssa-miR-21b) and three up-regulated genes (JNK1, CCR9 and FRIS) were identified.
3.5. miRNA target prediction
The identification of miRNAs and their targets is important in understanding their physiological and functional roles. To probe the targets of differentially expressed miRNAs following ammonia exposure in fish, we produced small RNA libraries from the gill samples of control or ammonia-exposed M. amblycephala and subjected these to Illumina deep sequencing. Small RNA deep sequencing data were aligned with miRBase 18.0 to search for known miRNAs with complete matches, that is, to narrow down potential targets to those differentially expressed miRNAs whose expression was inversely related to that of the mRNAs. This approach increases the strength to enable discovery of the true target genes and functions affected by miRNA dysregulation. In total, among the differentially expressed target genes, the 250 genes identified were differentially expressed in the opposite direction in the target tissue. These genes were found to be the targets of 10 conserved and 4 putative novel miRNA families, including ssa-miR-142a-5p, ssa-miR-21b-3p, aca-miR-125a-5p, ssa-miR-199a-3p, ccr-let-7i, aca-miR-184, aca-miR-214-3p and dre-miR-144-3p (Table S5). In particular, our analysis revealed that many of the most highly expressed miRNAs in the treatment group were those playing a role in the immune response.
As key players in the response to ammonia, the genes targeted by miR-21, a miRNA down-regulated in the treatment group, were further analyzed in the present study. miR-21 was predicted to target 23 differentially expressed M. amblycephala genes, including protein kinase, interleukin-10 (IL-10), FRIS and CCR9 (Table S6). Many of these play important roles in the immune response: IL-10, which was initially identified in the supernatants of Con-A-stimulated T cells based on its ability to inhibit the synthesis of pro-inflammatory cytokines, has been shown to be indispensable for the regulation of inflammation and other immune processes [50]. Chemokine receptors such as CCR9 act as the main receptors of chemokines, which are responsible for chemotaxis and are released at the sites of infection, inflammation and injury [51], [52]. Ferritin, which is encoded by FRIS, is likely to play a role in iron sequestration and protection against oxidative stress [53]. The increased mRNA levels of JNK1, FRIS, and CCR9 in the ammonia treatment group samples support our present finding where miR-21 is significantly down-regulated following ammonia exposure. We validated the sequencing data by examining miR-21, JNK1, FRIS and CCR9 expression by RT-qPCR in the same samples, and found agreement (Fig. 5).
Fig. 5.
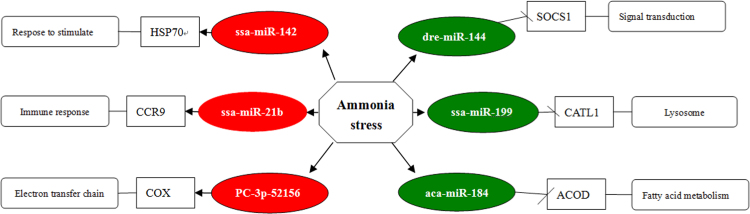
MicroRNA-gene network analysis. Depiction of gene targets of the differentially expressed miRNA: miRNAs identified as being differentially expressed following M. amblycephala ammonia exposure were assembled in a network based on GO annotations. The ovals represent the miRNAs (red, down-regulated; green, up-regulated), while the rectangles represent the target genes.
In addition, some members of the let-7 family such as let-7a-e and i were also expressed abundantly in the two libraries. Previous studies have shown that the let-7 family can regulate the expression of major cytokine-inducible proteins in response to microbial challenge in mammals [54], [55]. Taken together, our results suggest that miRNAs might play a key role in the regulation of immune-related gene expression in fish exposed to ammonia.
miRNAs play important roles in gene regulation by pairing to protein-encoding mRNAs to direct their post-transcriptional repression [56], [57]. The identification of the genes targeted by various miRNAs is an important step in understanding their physiological role and in this study, we provide an overview of miRNA-mediated gene regulation in M. amblycephala following ammonia exposure (Fig. 5). Most miRNA-associated computational methods comprise the prediction of miRNA genes and their targets, and an increasing number of computational algorithms and web-based resources such as like miRanda [37], TargetScan [58], RNAhybrid [59] and PicTar [60] have been developed to aid miRNA research, However, animal miRNA targets are difficult to predict by bioinformatics methods since miRNA: mRNA duplexes often contain several mismatches, gaps and G:U base pairs in many positions, thus limiting the maximum length of contiguous sequences of matched nucleotides [61]. As alternatives, several studies have suggested that simultaneous expression profiling [62] or inverse expression relationship analysis between miRNAs and mRNAs [63] are effective strategies for more reliably identifying miRNA-target mRNA pairs in large sets of transcriptome experiments [64], [65]. To decrease the false positive rates in this study, RNA-seq and miRNA-seq experiments were conducted using the same biological samples. Likewise, to downsize the number of putative target genes, the miRNA targets were predicted from only among the differentially expressed genes.
4. Conclusions
In this study, we used a high-throughput sequencing approach to characterize the transcriptome of M. amblycephala, a species for which little genomic data are available. By mapping DGE tags to the assembled transcriptome, a large number of candidate genes involved in response to ammonia-induced stress were identified. This study strongly indicates that miRNA is a critical factor in determining mRNA abundance and regulation during ammonia, further study will focus on the experimental validation of the miRNAs of interest, using in vitro experimental knockdown or over-expression of candidate miRNAs and mRNAs to provide an enhanced understanding of ammonia-induced toxicity mechanisms in M. amblycephala.
Acknowledgments
We thank Chen Zhang (LC-Bio Co. Ltd.) for their help in sequencing and data analysis. This study was financially supported by the Chinese Academy of Fishery Sciences, the China Central Governmental Research Institutional Basic Special Research Project from Public Welfare Fund (2015C06XK01), the Natural Science Foundation of Jiangsu Pvovince, China (SBK2015020058), the Modern Agriculture Industrial Technology System Special Project-the National Technology System for Conventional Freshwater Fish Industries (CARS-46), and the National “Twelfth Five-Year” Plan for Science & Technology Support (2012BAD25B07).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.rinim.2016.03.001.
Appendix A. Supplementary material
Supplementary material
References
- 1.Huang da, Sherman Lempicki. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucl. Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Pan, Liu De novo transcriptome sequencing of radish (Raph anus sativus L.) and analysis of major genes involved in glucosinolate metabolism. BMC Genom. 2013;14:836. doi: 10.1186/1471-2164-14-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graveley Molecular biology: power sequencing. Nature. 2008;453(7199):1197–1198. doi: 10.1038/4531197b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozsolak Kapranov, Foissac Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143(6):1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhardwaj Chauhan, Swarnkar Comprehensive transcriptomic study on horse gram (Macrotyloma uniflorum): de novo assembly, functional characterization and comparative analysis in relation to drought stress. BMC Genom. 2013;14:647. doi: 10.1186/1471-2164-14-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Liao, Yuan Pathological observation of bacterial septicemia in Megalobrama amblycephala. J. Southwest. Univ. 2006;28(3):483–490. in chinese. [Google Scholar]
- 7.Dosdat Ruyet, Coves Effect of chronic exposure to ammonia on growth, food utilization and metabolism of the European sea bass (Dicentrarchus labrax) Aquat. Living. Resour. 2003;16(6):509–520. [Google Scholar]
- 8.Foss Imsland, Roth Effects of chronic and periodic exposure to ammonia on growth and blood physiology in juvenile turbot (Scophthalmus maximus) Aquaculture. 2009;296(1–2):45–50. [Google Scholar]
- 9.Lemarie Dosdat, Coves Effect of chronic ammonia exposure on growth of European seabass (Dicentrarchus labrax) juveniles. Aquaculture. 2004;229(1–4):471–491. [Google Scholar]
- 10.Person-Le Ruyet Lamers, Roux Long term ammonia exposure of turbot: effects of plasma parameters. J. Fish. Biol. 2003;62(4):879–894. [Google Scholar]
- 11.Gonçalvesa P.áscoaa, Nevesb The inhibitory effect of environmental ammonia on Danio rerio LPS induced acute phase response. Dev. Comput. Immunol. 2012;36(2):279–288. doi: 10.1016/j.dci.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 12.Foss Siikavuopio, Saether Effect of chronic ammonia exposure on growth in juvenile Atlantic cod. Aquaculture. 2004;237(1–4):179–189. [Google Scholar]
- 13.Atwood Tomasso, Ronan Brain monoamine concentrations as predictors of growth inhibition in channel catfish exposed to ammonia. J. Aquat. Anim. Health. 2000;12:69–73. doi: 10.1577/1548-8667(2000)012<0069:BMCAPO>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 14.El-Shafai El-Gohary, Van Der Steen Nasr. Chronic ammonia toxicity to duckweed-fed tilapia (Oreochromis niloticus) Aquaculture. 2004;232(1–4):117–127. [Google Scholar]
- 15.Benli K.öksalb, Özkulc Sublethal ammonia exposure of Nile tilapia (Oreochromis niloticus L.): effects on gill, liver and kidney histology. Chemosphere. 2008;72(9):1355–1358. doi: 10.1016/j.chemosphere.2008.04.037. [DOI] [PubMed] [Google Scholar]
- 16.Dong Zhang, Qin Acute ammonia toxicity and gill morphological changes of Japanese flounder Paralichthys olivaceus in normal versus supersaturated oxygen. Aquac. Res. 2013;44(11):1752–1759. [Google Scholar]
- 17.Qin Fast, Kai Lethal effects of ammonia and pH on snakehead (Channa striatus) J. World. Aquac. Soc. 1997;28(1):87–90. [Google Scholar]
- 18.Bushati Cohen. MicroRNA functions. Annu. Rev. Cell. Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 19.Hou Wang, Baccarelli Environmental chemicals and microRNAs. Mutat. Res. 2011;714(1–2):105–112. doi: 10.1016/j.mrfmmm.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lema Cunningham. MicroRNAs and their implications in toxicological research. Toxicol. Lett. 2010;198(2):100–105. doi: 10.1016/j.toxlet.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 21.Paul Kim, Park Impact of miRNA deregulation on mRNA expression profiles in response to environmental toxicant, nonylphenol. Mol. Cell. Toxicol. 2011;7(3):259–269. [Google Scholar]
- 22.Yokoi Nakajima. Toxicological implications of modulation of gene expression by microRNAs. Toxicol. Sci. 2011;123(1):1–14. doi: 10.1093/toxsci/kfr168. [DOI] [PubMed] [Google Scholar]
- 23.Gao Luo, Liu Transcriptome analysis and SSR/SNP markers information of the blunt snout bream (Megalobrama amblycephala) Plos. One. 2012;7:e42637. doi: 10.1371/journal.pone.0042637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tran Gao, Zhao Transcriptome analysis and microsatellite discovery in the blunt snout bream (Megalobrama amblycephala) after challenge with Aeromonas hydrophila. Fish. Shellfish. Immunol. 2015;45(1):72–82. doi: 10.1016/j.fsi.2015.01.034. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Sun, Ge Acute effects of ammonia exposure on histopathology of gill, liver and kidney in juvenile Megalobrama amblycephala and the post-exposure recovery. J. Fish. China. 2015;39(2):233–244. In Chinese. [Google Scholar]
- 26.Pruitt Tatusova, Maglott NCBI reference sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabherr Haas, Yassour Fulllength transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haas Papanicolaou, Yassour De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koonin Fedorova, Jackson A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5(2):R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boeckmann Bairoch, Apweiler The SWISS-PROT protein knowledge base and its supplement TrEMBL in 2003. Nucl. Acids Res. 2003;31(1):365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa KEGG: kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Punta Coggill, Eberhardt The Pfam protein families database. Nucl. Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Götz García-G.ómez, Terol High-throughput functional annotation and data mining with the Blast2GO suite. Nucl. Acids Res. 2008;36(10):3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen Shen, Shi miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012;13:140. doi: 10.1186/1471-2105-13-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Friedlander Mackowiak, Chen miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucl. Acids Res. 2012;40(1):37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.John Enright, Aravin Human microRNA targets. Plos. Biol. 2005;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enright John, Gaul MicroRNA targets in drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mi Wei, Zhou Identification and profiling of sex-biased microRNAs from sea urchin Strongylocentrotus nudus gonad by Solexa deep sequencing. Com. Biochem. Phys. D. 2014;10:1–8. doi: 10.1016/j.cbd.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Sun Ge, Xuan Nitrite-induced hepatotoxicity in bluntsnout bream (Megalobrama amblycephala): the mechanistic insight from transcriptome to physiology analysis. Environ. Toxicol. Pharmacol. 2014;37(1):55–65. doi: 10.1016/j.etap.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 40.Livak Schmittgen. Analysis of relative gene expression data using real-time quantitative PCR and the 2−△△CT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Akira Takeda. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 42.Rot von Andrian. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu. Rev. Immunol. 2004;22:891–928. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]
- 43.Liu Li, Fu Two novel homologs of simple C-type lectin in grass carp (Ctenopharyngodon idellus): potential role in immune response to bacteria. Fish. Shellfish. Immunol. 2011;31(6):765–773. doi: 10.1016/j.fsi.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 44.Moseley Stress proteins and the immune response. Immunopharmacology. 2000;48(3):299–302. doi: 10.1016/s0162-3109(00)00227-7. [DOI] [PubMed] [Google Scholar]
- 45.Tsan Gao. Heat shock protein and innate immunity. Cell. Mol. Immunol. 2004;1(4):274–279. [PubMed] [Google Scholar]
- 46.Lin Chen, Mana Modulation of innate immunity and gene expressions in white shrimp Litopenaeus vannamei following long-term starvation and re-feeding. Results Immunol. 2012;2:148–156. doi: 10.1016/j.rinim.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chuang Liu, Hung Purification, characterization and molecular cloning of alpha-2-macroglobulin in cobia, Rachycentron canadum. Fish. Shellfish Immunol. 2014;41(2):346–355. doi: 10.1016/j.fsi.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 48.Bathige Whang, Umasuthan Three complement component 1q genes from rock bream, Oplegnathus fasciatus: genome characterization and potential role in immune response against bacterial and viral infections. Fish. Shellfish Immunol. 2013;35(5):1442–1454. doi: 10.1016/j.fsi.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 49.Saraiva Grund, Komegae Nattectin a fish C-type lectin drives Th1 responses in vivo: licenses macrophages to differentiate into cells exhibiting typical DC function. Int. Immunopharmacol. 2011;11(10):1546–1556. doi: 10.1016/j.intimp.2011.05.012. [DOI] [PubMed] [Google Scholar]
- 50.Grayfera Belosevica, Miodrag Identification and molecular characterization of the interleukin-10 receptor 1 of the zebrafish (Danio rerio) and the goldfish (Carassius auratus L.) Dev. Comp. Immunol. 2012;36(2):408–417. doi: 10.1016/j.dci.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Galindo-Villegas Mulero, García-Alcazar Recombinant TNFα as oral vaccine adjuvant protects European sea bass against vibriosis: insights into the role of the CCL25/CCR9 axis. Fish. Shellfish Immunol. 2013;35(4):1260–1271. doi: 10.1016/j.fsi.2013.07.046. [DOI] [PubMed] [Google Scholar]
- 52.Zhu Wang, Ren Characterization of the CCR3 and CCR9 genes in miiuy croaker and different selection pressures imposed on different domains between mammals and teleosts. Dev. Comp. Immunol. 2013;41(4):631–643. doi: 10.1016/j.dci.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 53.Zheng Hu, Sun Identification and analysis of a Scophthalmus maximus ferritin that is regulated at transcription level by oxidative stress and bacterial infection. Comp. Biochem. Phys. B. 2010;156(3):222–228. doi: 10.1016/j.cbpb.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 54.Schulte Eulalio, Mollenkopf Analysis of the host microRNA response to salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011;30(10):1977–1989. doi: 10.1038/emboj.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voinnet Micro-balancing innate immunity to salmonella. EMBO J. 2011;30(10):1877–1879. doi: 10.1038/emboj.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krol Loedige, Filipowicz The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 57.Bartel MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lewis Shih, Jones-Rhoades Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 59.Rehmsmeier Steffen, Hochsmann Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10(10):1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krek Grun, Poy Combinatorial microRNA target predictions. Nat. Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 61.Stark Brennecke, Russell Identification of drosophila microRNA targets. Plos. Biol. 2003;1(3):e60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang Babak, Corson Using expression profiling data to identify human microRNA targets. Nat. Methods. 2007;4(12):1045–1049. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- 63.Gennarino Sardiello, Avellino MicroRNA target prediction by expression analysis of host genes. Genome Res. 2009;19(3):481–490. doi: 10.1101/gr.084129.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Iterson Bervoets, de Meijer Integrated analysis of microRNA and mRNA expression: adding biological significance to microRNA target predictions. Nucl. Acids Res. 2013;41(15):e146. doi: 10.1093/nar/gkt525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muniategui Pey, Plane Joint analysis of miRNA and mRNA expression data. Brief. Bioinform. 2013;14(3):263–278. doi: 10.1093/bib/bbs028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material