Abstract
Only a few studies indentified the significance of circulating microRNAs in blood as a predictive biomarker for chemoresistance in esophageal squamous cell carcinoma (ESCC). In this study, we tested whether oncogenic miR-21 promoted chemoresistance in ESCC and served as a biomarker for predicting chemoresistance in plasma of patients with ESCC. All consecutive patients underwent the preoperative chemotherapy regimen (JCOG9907 trial) with cisplatin plus 5-fluorouracil. As a result, pretreatment plasma concentrations of miR-21 were significantly higher in ESCC patients with a low histopathological response than in those with a high histopathological response (P = 0.0416). Multivariate analysis revealed that a high pretreatment plasma concentration of miR-21 was an independent risk factor of chemoresistance (p = 0.0150; Odds Ratio 9.95 (range: 1.56-63.4)). The expression of miR-21 was also significantly higher in pretreatment ESCC tissues with a low histopathological response than in those with a high histopathological response (P = 0.0409). In vitro, although the growth of KYSE 170 ESCC cells transfected with the control mimics was markedly inhibited by the 5-fluorouracil or cisplatin treatment, the inhibitory effects of 5-FU (P < 0.05) or cisplatin (P < 0.05) were significantly reduced in KYSE170 cells that overexpressed miR-21. Taken together, the overexpression of miR-21 contributed to chemoresistance and circulating miR-21 in plasma of patients with ESCC could be a useful biomarker for predicting chemoresistance.
Keywords: Esophageal cancer, microRNA, plasma, biomarker, chemoresistance, liquid biopsy
Introduction
Esophageal carcinoma is the sixth leading cause of cancer-related deaths worldwide [1]. Although there are two distinctive histological types of esophageal carcinoma, esophageal adenocarcinoma and esophageal squamous cell carcinoma (ESCC), ESCC is the predominant histological type globally [2], particularly in Asian countries. ESCC accounts for approximately 90% of esophageal carcinomas [3] and remains one of the most aggressive carcinomas of the gastrointestinal tract. Although surgical techniques, perioperative management, and perioperative chemo- and/or radiotherapy regimens have greatly progressed, ESCC continues to present with an extremely poor prognosis.
Chemotherapy is an important component in the treatment paradigm for ESCC. To improve the prognosis of patients with ESCC, combination therapies of preoperative chemotherapy or chemoradiotherapy followed by surgery have been developed and widely practiced worldwide [4,5]. However, both intrinsic and acquired drug resistance remains a major clinical obstacle to successful treatment [6]. Only certain patients receive the benefit of shrinkage in tumor mass and repression. Otherwise, the resistance of cancer cells to chemotherapeutic agents may result in progression of the disease and the subsequent metastasis of cancer cells [7,8]. There is currently no validated sensitivity and/or resistance predictive factor available in clinical settings and the mechanisms involved in cancer cell chemoresistance remain largely unknown. Therefore, a better understanding of drug resistance mechanisms and the detection of clinically relevant biomarkers for predicting chemoresistance are needed in order to improve the survival of patients with ESCC.
MicroRNAs, which are small non-coding RNAs, regulate the translation of specific protein-coding genes. The altered expression of microRNAs has been associated with several diseases, and tumor microRNAs have been shown to play a critical role in carcinogenesis and the development of various types of cancer [9-12]. Several studies recently reported that microRNAs were detected in plasma/serum [9,13-15]. Tumor-derived microRNAs are resistant to endogenous ribonuclease activity because these may bind to proteins, such as the Argonaute 2 protein and high-density lipoprotein [16,17], or may be packaged by secretory particles such as exosomes in plasma/serum [18-21]. Therefore, microRNAs can be present in a very stable form [21,22] and the expression levels of blood microRNAs were shown to be reproducible and consistent among individuals [13,21]. Moreover, secretory vesicles, which include specific microRNAs, can function as intercellular transmitters. Secreted microRNAs from donor cells can be transferred to and function in recipient cells [23-25].
Recent studies demonstrated that specific microRNAs in tumor tissues were involved in regulating drug resistance [26-28]. However, it remains unknown whether plasma microRNA levels are useful biomarkers for ESCC patients who receive chemotherapy. In the present study, we focused on the most famous and promising oncogenic microRNA, miR-21, because previous studies already confirmed that the expression of miR-21 in several types of cancer tissues could be used to predict chemoresistance (Supplementary Table 1) [29-46]. However, only a few studies have investigated the clinical significance of circulating miR-21 in the plasma/serum of patients with cancers for predicting chemoresistance [40,47,48]. These findings prompted us to determine the biological and clinicopathological significance of miR-21 chemoresistance in ESCC.
Concerning miR-21 in ESCC, our and other groups have already clarified the potential utility of circulating miR-21 in plasma/serum in clinical applications to detect cancer, monitor tumors, and predict prognosis [49-51]. However, the relationship between circulating miR-21 levels in the plasma/serum of patients with ESCC and chemoresistance has yet to be determined. In the present study, we clearly demonstrated that plasma miR-21 levels were useful for predicting chemoresistance in preoperative chemotherapy. Our results demonstrated that plasma miR-21 levels may contribute to clinical decision making for ESCC treatments to a clinically satisfactory degree.
Materials and methods
Patients and samples
This study was approved by the Institutional Review Board of Kyoto Prefectural University of Medicine, and each subject provided written informed consent. Between March 2010 and May 2012, 37 consecutive pretreatment plasma samples and tissue specimens were collected from consecutive ESCC patients who received preoperative chemotherapy and then underwent curative esophagectomy at the Kyoto Prefectural University of Medicine. All patients were pathologically diagnosed with ESCC. Tumor stages were assessed according to the 7th Union of International Control of Cancer (UICC)/TNM classification [52].
As a control, plasma was collected from 20 healthy volunteers. These healthy volunteers included medical personnel and patients with benign diseases such as cholelithiasis. These patients underwent medical examinations and were found not to have any esophageal or other cancerous disease. Peripheral blood (7 ml) was obtained from each patient before neoadjuvant chemotherapy and from the healthy volunteers. Immediately after being collected, cell-free nucleic acids were isolated from the blood samples using a 3-spin protocol (1500 rpm for 30 min, 3000 rpm for 5 min, and 4500 rpm for 5 min) to prevent contamination by cellular nucleic acids. Plasma samples were stored at - 80°C until further processing.
Twenty ESCC specimens were collected from primary ESCC tumors, and five normal esophageal tissue specimens of the abdominal esophagus were collected from patients undergoing total gastrectomy for gastric cancer; these patients were selected for the normal specimens in order to strictly compare molecular expression between ESCC tissue and normal esophageal tissue from patients without ESCC because the non-cancerous esophageal tissues of ESCC patients may still exhibit dysplasia or potentially be cancerous tissue, although differences in molecular expression may be proven between cancerous and non-cancerous tissues in paired samples. The resected specimens were fixed in formalin and embedded in paraffin for pathological diagnoses. The macroscopic and microscopic classification of tumors was based on the UICC/TMN staging system [52].
All patients underwent the same preoperative chemotherapy regimen (JCOG9907 regimen) with cisplatin plus 5-fluorouracil, which was repeated twice every 3 weeks. A dose of 80 mg/m2 cisplatin was administered by an intravenous drip infusion for 2 h on day 1; 5-fluorouracil was administered at a dose of 800 mg/m2 by continuous infusion on days 1 through 5 [4].
Evaluation of responses to chemotherapy
In the present study, we used the histopathological response grade of tumors as an indication of the chemotherapeutic effect because previous studies suggested that the histopathological response correlated more strongly with clinical outcomes or survival than the clinical response grade in ESCC [53-55]. The degree of histopathological tumor regression in surgical specimens was classified into 5 categories according to the 10th guidelines of the Japan Esophageal Society [56]. The percentage of viable residual tumor cells within the total cancerous tissue was assessed as follows: Grade 3, no viable residual tumor cells; Grade 2, less than 1/3 residual tumor cells; Grade 1b, 1/3 to 2/3 residual tumor cells; Grade 1a, more than 2/3 residual tumor cells; Grade 0, no significant response to chemotherapy [56]. The number of patients with each histopathological grade was one patient with Grade 3, 6 patients with Grade 2, 6 patients with Grade 1b, 23 patients with Grade 1a, and one patient with Grade 0, respectively. In the present study, we allocated 13 patients with Grade 3, 2, 1b and 24 patients with Grade 1a, 0 into the low and high histopathological response groups, respectively.
RNA extraction
Plasma total RNA was extracted from 400 μl of plasma using the mirVana PARIS Kit (Ambion, Austin, TX, USA) and finally eluted into 100 μl of preheated (95°C) Elution Solution according to the manufacturer’s protocol. Using the formalin-fixed paraffin-embedded tissues, tissue total RNA was extracted from four 15-μm-thick slices of tissue (total 60 μm in thickness) using the RecoverAll Total Nucleic Acid Isolation Kit (Ambion, Austin, TX, USA) and then eluted into 60 μl of Elution Solution according to the manufacturer’s protocol. Total RNA was extracted from the freshly frozen tissue samples or cell lines using TRIzol Reagent (Invitrogen, San Diego, CA, USA), and small-sized RNAs were isolated using a mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA), both according to the manufacturer’s instructions.
Protocol for the detection of miR-21
The amounts of microRNAs in the plasma samples were quantified by qRT-PCR using human TaqMan MicroRNA Assay Kits (Applied Biosystems, Foster City, CA, USA). The reverse transcription reaction was carried out using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) in 15 μl solution containing 5 μl of RNA extract, 0.15 μl of 100 mM dNTPs, 1 μl of MultiScribe reverse transcriptase (50 U/μl), 1.5 μl of 10× reverse transcription buffer, 0.19 μl of RNase inhibitor (20 U/μl), 1 μl of gene-specific primer (has-miR-21, Assay ID: 000397 and RNU6B, Assay ID: 001093), and 4.16 μl of nuclease-free water. The reaction mixtures for cDNA synthesis were incubated at 16°C for 30 min, at 42°C for 30 min, and at 85°C for 5 min and then held at 4°C. A total of 1.33 μl of cDNA solution was then amplified using 10 μl of TaqMan 2× Universal PCR Master Mix with no AmpErase UNG reagent (Applied Biosystems), 1 μl of a gene-specific primer/probe, and 7.67 μl of nuclease-free water in a final volume of 20 μl. Quantitative PCR was run on a 7300 Real-time PCR system (Applied Biosystems), and the reaction mixtures were incubated at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, and 60°C for 1 min. Cycle threshold (Ct) values were calculated using SDS 1.4 software (Applied Biosystems).
Plasma microRNA levels were calculated on a standard curve constructed using synthetic microRNAs from the mirVana miRNA Reference Panel (Ambion, Austin, TX, USA). Standard reference microRNAs were amplified by RT-PCR of a 10-fold serial dilution of the mirVana miRNA Reference Panel. The linearity of quantitative RT-PCR was confirmed between the logarithm of the amount of input microRNA and Ct values for a range of concentrations (1 fmol-0.0001 fmol) of each synthetic microRNA (Supplementary Figure 1). Although plasma microRNAs levels have previously been determined by comparing with internal control microRNAs [57], it remains controversial as to which microRNAs are suitable as internal controls for plasma assays. Therefore, we confirmed a linear correlation between the logarithm of the amount of input synthetic microRNA and the Ct value using real-time PCR, as well as the feasibility of extracting total RNA and amplifying specific microRNA in plasma samples. On the basis of these findings, we utilized the absolute concentration to measure plasma microRNA in this study.
The expression of microRNAs from tissue and cell line samples was normalized using the 2-ΔΔCT method relative to U6 small nuclear RNA (RNU6B). ΔCt was calculated by subtracting the Ct values of RNU6B from those of the microRNAs of interest. ΔΔCt was then calculated by subtracting ΔCt of normal tissue from ΔCt of ESCC tissues. The change in gene expression was calculated using the equation 2-ΔΔCt [58]. Using several housekeeping genes as internal controls was more appropriate for the study in cancer tissues and cell lines; however, we used U6 only as an internal control for the of expression miR-21 in cancer tissues and cell lines, as described in our previous study [50].
ESCC cell line and culture
We selected the KYSE170 cell line, which was established from the surgically resected tumors of ESCC [59] and presented stable cell growth and comparatively low expression of miR-21. The KYSE170 cell line was cultured in Roswell Park Memorial Institute-1640 medium (Sigma, St. Louis, MO, USA) in 50 mL/L carbon dioxide at 37°C in a humidified chamber.
Oligonucleotide transfection
To overexpress miR-21, the miR-21 mimic (MC10206) or control mimic microRNA (mirVana miRNA mimic Negative Control #1) selected from the mirVana miRNA mimic panel (Ambion, Austin, TX, USA) was transfected into cells (50 μM) using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s instructions. The overexpression of miR-21 was confirmed by qRT-PCR using human TaqMan MicroRNA Assay Kits (Applied Biosystems, Foster City, CA, USA).
Cell viability assays
To assess chemoresistance to the 5-fluorouracil (5-FU) plus cisplatin treatment, oligonucleotide-transfected cells were plated onto a 24-well plate (3x104 cells/ml) and incubated overnight under normal culture conditions. The cells were then incubated with various concentrations of 5-FU (1, 4, 16, 64 or 256 μM) or cisplatin (1, 2, 4, or 16 μM) for the KYSE170 cells. These cells were subjected to the WST-8 assay 72 h after the 5-FU treatment and 48 h after the cisplatin treatment. The number of viable cells was determined with a Cell Counting Kit (Dojindo Molecular Technologies, Inc., Gaithersburg, MD), which counted the number of living cells using WST-8.
Statistical analysis
Discrete variables were compared using the Chi-squared test or Fisher’s exact probability test and continuous variables were compared using the Mann-Whitney U-test or Student’s t-test. The area under the receiver operating characteristic (ROC) curve (AUC) was used to assess the feasibility of using plasma miR-21 levels as a diagnostic tool to predict the chemoresistance of ESCC. The Youden index was used to determine the cut-off value for plasma microRNA levels [60]. Multivariate logistic regression analysis was performed to identify independent risk factors associated with chemoresistance. Multivariate odds ratios are presented with 95% confidence intervals.
Results
Study design to develop a novel biomarker for chemoresistance in ESCC using plasma microRNA
This study was divided into four parts: (1) Evaluation of whether pretreatment plasma miR-21 levels could independently predict chemoresistance. (2) Demonstration of higher miR-21 levels in the ESCC tissues and plasma of patients with chemoresistance. (3) Demonstration that miR-21 was associated with chemoresistance to the 5-FU or cisplatin treatment in vitro (Figure 1).
Figure 1.
Study design to develop a novel biomarker for chemoresistance in ESCC using plasma microRNA.
Comparison of circulating miR-21 levels according to pathological response grades to preoperative chemotherapy in the plasma of patients with ESCC
To evaluate the appropriateness of this plasma assay, we first amplified a 10-fold serial dilution of the mirVana miRNA Reference Panel using the real-time RT-PCR assay. The linearity of quantitative RT-PCR was confirmed from the concentrations of 1 fmol to 0.0001 fmol of each synthetic miR-21 (R2 = 0.9968) between the logarithm of the amount of input microRNAs and the Ct values (Supplementary Figure 1).
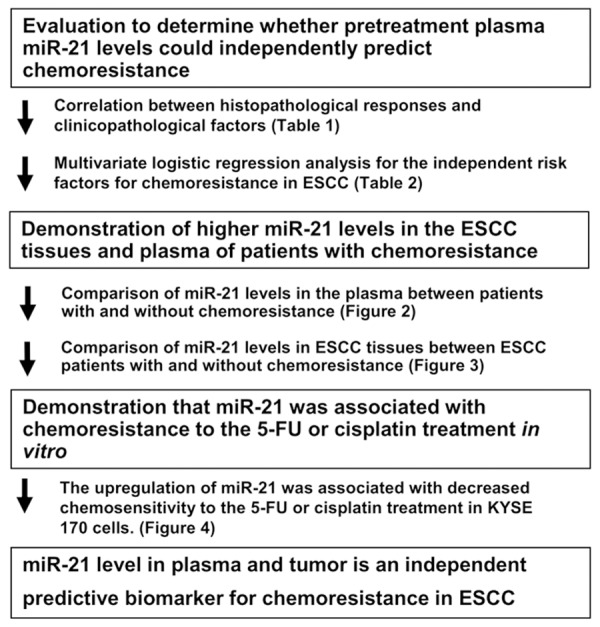
We hypothesized that the higher expression of miR-21 in primary ESCC tissues would influence the expression of miR-21 in the plasma of ESCC patients, and this may be associated with chemoresistance. The results obtained showed that the plasma concentrations of miR-21 were significantly higher in ESCC patients with a low pathological response (Grade 0 or 1a) than in those with a high pathological response (Grade 1b, 2 or 3) (P = 0.0416). Plasma miR-21 levels were significantly higher in both the low and high response groups than in the healthy volunteers (P = 0.0029 and P = 0.0433, respectively) (Figure 2A). These results clearly indicated that the histopathological response grade may be inversely correlated with the expression of miR-21 in the plasma of patients with ESCC.
Figure 2.
Comparison of circulating miR-21 levels according to the pathological response grades to preoperative chemotherapy in the plasma of patients with ESCC. A. The plasma concentrations of miR-21 were significantly higher in ESCC patients with a low pathological response (Grade 0 or 1a) than in those with a high pathological response (Grade 1b, 2 or 3) (P = 0.0416). Plasma miR-21 levels were significantly higher in both the low and high response groups than in healthy volunteers (P = 0.0029 and P = 0.0433, respectively). B. A representation of the data using a ROC plot showed a strong separation between the low and high pathological response groups, with an AUC of 0.6794. An optimal cut-off point was indicated at 0.8154 amol/μl with a sensitivity of 92.3%, specificity of 54.2%, and accuracy of 67.6%.
Determination of the cut-off value for the plasma miR-21 level to predict chemosensitivity and the relationship between the histopathological response to chemotherapy and clinicopathological factors
A representation of the data using a ROC plot showed a strong separation between the low and high pathological response groups, with an AUC of 0.6794 (Figure 2B). In this model, an optimal cut-off point was indicated at 0.8154 amol/μl with a sensitivity of 54.2%, specificity of 92.3%, and accuracy of 67.6%. We then examined the relationship between the histopathological response to chemotherapy and clinicopathological factors in ESCC patients. Patient characteristics with respect to age, sex, venous invasion, lymphatic invasion, clinical and pathological T stage, clinical and pathological N stage, and disease stage are shown in Table 1. Factors such as lymphatic invasion (P = 0.0170) and pretreatment plasma miR-21 levels (P = 0.0165) correlated with the histopathological response (Table 1). Multivariate logistic regression analysis revealed that a high pretreatment plasma concentration of miR-21 was an independent risk factor for chemoresistance (p = 0.0150; Odds Ratio 9.95 (range: 1.56-63.4)) (Table 2).
Table 1.
Correlation between the histopathological responses and clinicopathological features
| Histopathological responses | ||||
|---|---|---|---|---|
|
|
||||
| n | Low (n = 24) | High (n = 13) | a p value | |
| Sex | ||||
| Male | 30 | 20 (83%) | 10 (77%) | |
| Female | 7 | 4 (17%) | 3 (23%) | 0.6779 |
| Age | ||||
| < 65 | 19 | 12 (50%) | 7 (54%) | |
| ≥ 65 | 18 | 12 (50%) | 6 (46%) | 0.9036 |
| Lymphatic invasion | ||||
| Negative | 18 | 8 (33%) | 10 (77%) | |
| Positive | 19 | 16 (67%) | 3 (23%) | 0.0170 |
| Venous invasion | ||||
| Negative | 24 | 14 (58%) | 10 (77%) | |
| Positive | 13 | 10 (42%) | 3 (23%) | 0.3051 |
| Depth of invasion | ||||
| cT0-T2 | 9 | 6 (25%) | 3 (23%) | |
| cT3-T4 | 28 | 18 (75%) | 10 (77%) | 1.0000 |
| Depth of invasion | ||||
| pT0-T2 | 18 | 10 (42%) | 8 (62%) | |
| pT3-T4 | 19 | 14 (58%) | 5 (38%) | 0.4179 |
| Lymph node mestasis | ||||
| cN0 | 9 | 6 (25%) | 3 (23%) | |
| cN1-3 | 28 | 18 (75%) | 10 (77%) | 1.0000 |
| Lymph node mestasis | ||||
| pN0 | 12 | 6 (25%) | 6 (46%) | |
| pN1-N3 | 25 | 18 (75%) | 7 (54%) | 0.3449 |
| pStage | ||||
| I/II | 18 | 9 (37%) | 9 (69%) | |
| III/III | 19 | 15 (63%) | 4 (31%) | 0.1338 |
| Pretreatment plasma miR-21 | ||||
| Low | 21 | 10 (42%) | 11 (85%) | |
| High | 16 | 14 (58%) | 2 (15%) | 0.0165 |
p values are from the x2 or Fisher’s exact test and were significant at 0.05.
Table 2.
Multivariable logistic regression analysis for chemoresistance in ESCC using pretreatment factors
| Variables | Multivariate analysisa | |||
|---|---|---|---|---|
|
| ||||
| ORb | 95% CIc | p valued | ||
| Sex | Male vs. female | 2.688 | 0.322-22.22 | 0.3605 |
| Age (years old) | 65 ≤ vs. < 65 | 1.333 | 0.287-6.189 | 0.7140 |
| Depth of invasion | cT3-T4 vs. cT0-T2 | 0.681 | 0.092-4.694 | 0.6965 |
| Lymph node metastasis | cN1-N3 vs. cN0 | 0.701 | 0.116-4.237 | 0.6986 |
| Pretreatment plasma miR-21 | High vs. Low | 9.953 | 1.562-63.42 | 0.0150 |
Multivariate analysis was performed using logistic regression analysis;
OR: Odds ratio;
CI: confidence interval;
p value < 0.05 was significant.
Comparison of miR-21 levels according to the histopathological response grades to preoperative chemotherapy in primary ESCC tissues
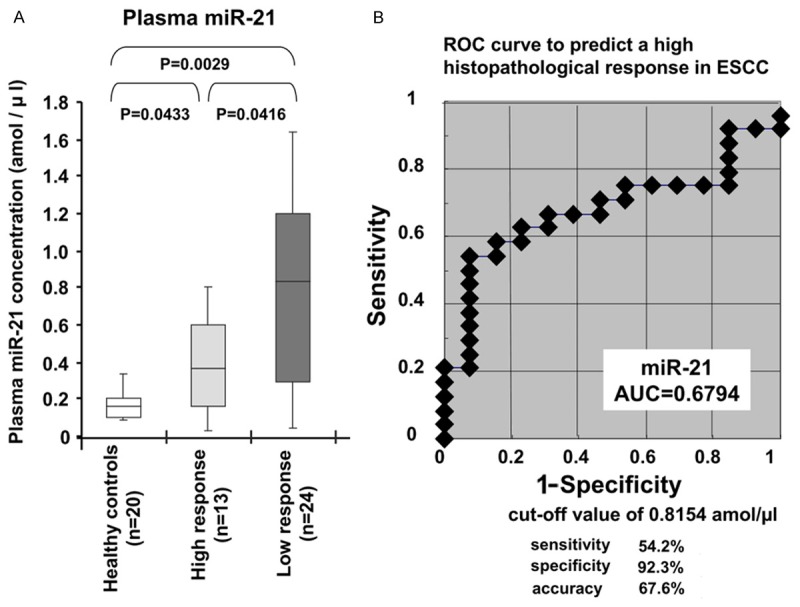
To confirm the high expression of miR-21 in pretreatment ESCC tissues of patients with a low histopathological response, we investigated whether the histopathological response grade to chemotherapy was associated with the expression of miR-21 in primary ESCC tissues. we used 20 pretreatment biopsy tissues of ESCC derived from 10 consecutive patients with a high histopathological response and 10 consecutive patients with a low histopathological response to chemotherapy using qRT-PCR. The results are shown after normalization to the expression of RNU6B. As a result, the expression of miR-21 was slightly higher in the ESCC tissues of patients with a low histopathological response (Grade 0 or 1a) than in those with a high histopathological response (Grade 1b, 2 or 3) (P = 0.0409) (Figure 3). These results indicated that the histopathological response grade inversely correlated with the expression of miR-21 in pretreatment ESCC tissues as well as plasma.
Figure 3.
Comparison of miR-21 levels according to the histopathological response grades to preoperative chemotherapy in primary ESCC tissues. The expression of miR-21 was slightly higher in the ESCC tissues of patients with a low histopathological response (Grade 0 or 1a) than in those with a high histopathological response (Grade 1b, 2 or 3) (P = 0.0409). The upper and lower limits of the boxes and lines inside the boxes indicate the 75th and 25th percentiles and the median, respectively. The upper and lower horizontal bars denote the 90th and 10th percentiles, respectively.
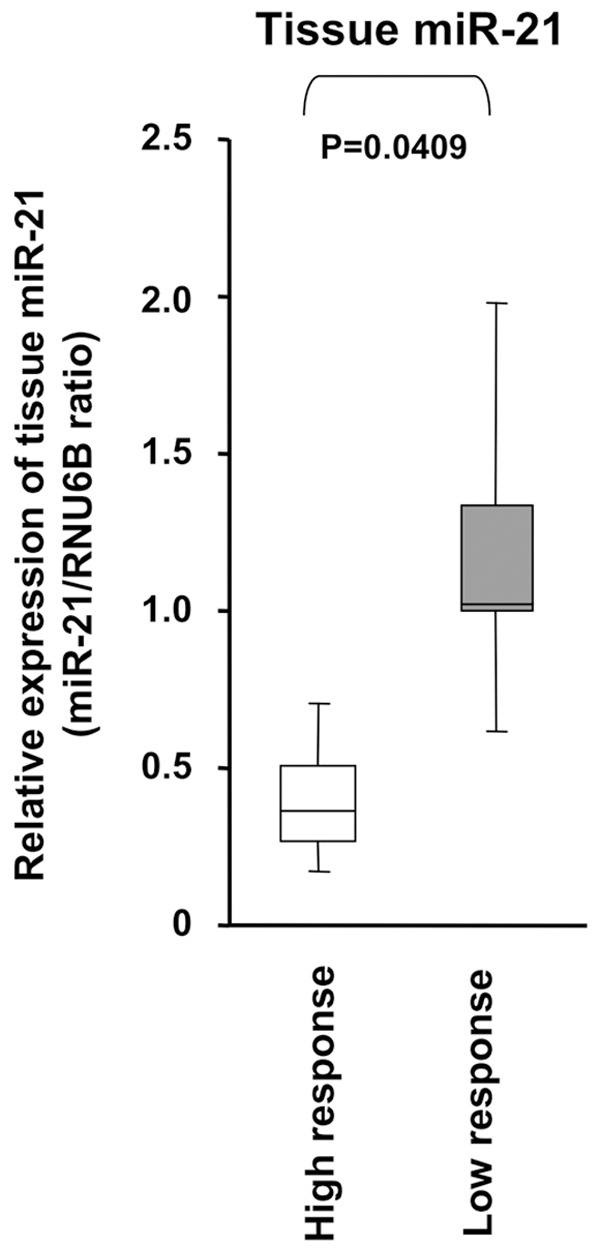
The upregulation of miR-21 was associated with decreased chemosensitivity to the 5-FU or cisplatin treatment in the KYSE 170 cell line
To determine the effects of the overexpression of miR-21 on the chemoresistance of cancer cells to the 5-FU or cisplatin treatment, ESCC cells were transfected with miR-21 mimics to increase the expression of miR-21 (Figure 4). After confirming the overexpression of miR-21 following transfection of the miR-21 mimic (Figure 4A), the transfected cells were then treated with increasing concentrations of 5-FU or cisplatin, and cell growth was measured using the CCK-8 assay. As shown in Figure 4B and 4C, the growth of KYSE170 cells transfected with the control mimics was markedly inhibited by the 5-FU or cisplatin treatment, while the inhibitory effect of 5-FU (P < 0.05) or cisplatin (P < 0.05) was significantly reduced in miR-21-transfected KYSE170 cells.
Figure 4.
The upregulation of miR-21 was associated with decreased chemosensitivity to the 5-FU or cisplatin treatment in the KYSE 170 cell line. A. The KYSE170 cell line was selected because of its stable cell growth and comparatively low expression of miR-21. B. After confirming the overexpression of miR-21 following transfection of the miR-21 mimic in KYSE170 cells. C. The growth of KYSE170 cells transfected with the control mimics was markedly inhibited by the 5-FU or cisplatin treatment, while the inhibitory effect of 5-FU or cisplatin was significantly reduced in the miR-21-transfected KYSE170 cells.
Discussion
Evidence to show that circulating microRNA levels in the blood may be useful for decision-making in the treatment of patients with cancers is increasing [9,13-15]. However, most studies have focused on microRNA levels in the blood as diagnostic, monitoring, and predicting prognostic markers for cancers [15,49-51,61-67]. Several studies recently demonstrated that microRNA levels in the plasma/serum were useful for predicting the response to chemotherapy in several types of cancer [40,47,48,68-73] (Supplementary Table 2). Regarding ESCC, serum miR-200c levels were reported to be associated with chemoresistance in ESCC [70], and the relationship between plasma microRNAs and chemosensitivity or resistance in ESCC has yet fully to be examined. Therefore, we hypothesized that novel plasma microRNAs may be associated with sensitivity or resistance to chemotherapy in patients with ESCC.
We focused on plasma miR-21 concentrations to verify this hypothesis because we previously demonstrated its significance in making diagnoses, monitoring tumor dynamics, and predicting the prognoses of patients with ESCC [49,50]. miR-21 is the most famous oncomir that is associated with resistance to chemotherapeutic agents. As shown in Supplementary Table 1, the dysregulation of miR-21 was identified as a predictor of tumor responses to several cytotoxic chemotherapeutic agents, such as cisplatin in nasopharyngeal cancer [42], oral cancer [31,43], head and neck squamous cell carcinoma (HNSCC) [29], and ovarian cancer [5], gemcitabine in pancreatic cancer [40] and cholangiocarcinoma [33], doxorubicin in lung cancer [39] and bladder cancer [36], 5-fluorouracil in hepatocellular carcinoma [37] and colorectal cancer [38], tamoxifen [35], faslodex, and topotecan [41] in breast cancer, docetaxel [34] and staurosporine [44] in prostate cancer, VM-26 in glioblastoma [32], and arabiosylcytosine in leukemia [45]. However, the relationship between miR-21 and the chemoresistance of ESCC has not yet been examined either in vitro or in vivo. In the present study, we clearly demonstrated that pretreatment plasma concentrations of miR-21 could predict histopathological responses to neoadjuvant chemotherapy in patients with ESCC. This is the first study to demonstrate that plasma miR-21 levels are potentially useful markers for predicting the responses of patients with ESCC to chemotherapy. Moreover, we clarified that the overexpression of miR-21 in ESCC cells was strongly associated with 5-FU and cisplatin chemoresistance in vitro. Although the detailed molecular mechanisms of chemoresistance associated with miR-21 remain unknown in the present study, PDCD4 and PTEN may also be related to chemoresistance in ESCC, as shown in previous studies regarding the oncogenesis of miR-21 in ESCC [74,75] and chemoresistance associated with 5-FU or cisplatin in other cancers [29,37,42,46].
Concerning biomarkers to predict the chemoresistance of ESCC, until now, there have been few comparative biomarkers in ESCC, particularly in blood-based microRNA. In the present study, the sensitivity, specificity, and accuracy by which ESCC patients with histopathological chemoresistance were detected were 54.2%, 92.3%, and 67.6%, respectively (Figure 2B). A previous study reported that the sensitivity, specificity, and accuracy of detecting ESCC patients with clinical chemoresistance on computed tomography using serum miR-200c were 68.0%, 61.5%, and 64.1% [69]. However, the meaning of these data differs depending on whether the definition of chemoresistance is a histopathological or clinical response and whether the blood sample used is plasma or serum. Therefore, further studies are needed to evaluate candidate microRNAs including novel candidates using the same definition of chemoresistance and same kind of blood sample.
The advantages and disadvantages of the different approaches used to examine exosomal microRNAs [76] or free circulating microRNAs [49-51] currently remain unclear. Arroyo JD et al reported that more than 70% of microRNAs in the blood bound to plasma proteins while the others bound to plasma proteins and/or were packaged in secretory particles such as exosomes [16]. Therefore, we have mainly focused on the circulating microRNAs that bind to plasma proteins. However, the significance of exosomal microRNAs in the blood is currently being investigated at our institute and will be reported on in the near future.
The non-invasive plasma assay used to predict chemosensitivity or resistance with specific microRNAs may be extremely beneficial for clinical applications in various cancers because of its perspective technical simplicity, rapidity, and reliability. However, evaluating chemoresistance or sensitivity using molecular markers in clinical settings to date has only been possible in patients with tumor tissue that could be obtained from resected tumor specimens or biopsies [77,78]. In the near future, we may be able to utilize circulating microRNAs in plasma/serum and predict chemosensitivity or resistance using a so-called ‘liquid biopsy’, even in patients with micrometastasis and distant metastasis, the tumor tissue of which cannot be obtained easily [79].
The so-called ‘bench-to-bedside’ medicine has been desired for a long time. A non-invasive assay using circulating nucleic acids has opened up a new and very interesting field for the evaluation of chemosensitivity or resistance as well as the diagnosis and monitoring of cancer patients. Nevertheless, many issues must be addressed before these findings can be translated into a clinically useful screening strategy for ESCC patients. We will prospectively confirm the usefulness of plasma miR-21 in predicting chemoresistance in a large number of patients. Further studies are warranted to detect more sensitive biomarkers among plasma microRNAs for the chemoresistance or sensitivity of ESCC for translation into clinical settings. These issues are also currently being evaluated in high-throughput platforms, such as genome-wide array-based approach, digital PCR-based approach and next-generation sequencing, using large candidate numbers of microRNA and will be reported in the near future.
Supporting Information
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Cook MB. Non-acid reflux: the missing link between gastric atrophy and esophageal squamous cell carcinoma? Am J Gastroenterol. 2011;106:1930–2. doi: 10.1038/ajg.2011.288. [DOI] [PubMed] [Google Scholar]
- 3.Hiyama T, Yoshihara M, Tanaka S, Chayama K. Genetic polymorphisms and esophageal cancer risk. Int J Cancer. 2007;121:1643–58. doi: 10.1002/ijc.23044. [DOI] [PubMed] [Google Scholar]
- 4.Ando N, Kato H, Igaki H, Shinoda M, Ozawa S, Shimizu H, Nakamura T, Yabusaki H, Aoyama N, Kurita A, Ikeda K, Kanda T, Tsujinaka T, Nakamura K, Fukuda H. A randomized trial comparing postoperative adjuvant chemotherapy with cisplatin and 5-fluorouracil versus preoperative chemotherapy for localized advanced squamous cell carcinoma of the thoracic esophagus (JCOG9907) Ann Surg Oncol. 2012;19:68–74. doi: 10.1245/s10434-011-2049-9. [DOI] [PubMed] [Google Scholar]
- 5.Gebski V, Burmeister B, Smithers BM, Foo K, Zalcberg J, Simes J. Survival benefits from neoadjuvant chemoradiotherapy or chemotherapy in oesophageal carcinoma: a meta-analysis. Lancet Oncol. 2007;8:226–34. doi: 10.1016/S1470-2045(07)70039-6. [DOI] [PubMed] [Google Scholar]
- 6.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–34. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 7.Broxterman HJ, Gotink KJ, Verheul HM. Understanding the causes of multidrug resistance in cancer: a comparison of doxorubicin and sunitinib. Drug Resist Updat. 2009;12:114–26. doi: 10.1016/j.drup.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Fojo T. Multiple paths to a drug resistance phenotype: mutations, translocations, deletions and amplification of coding genes or promoter regions, epigenetic changes and microRNAs. Drug Resist Updat. 2007;10:59–67. doi: 10.1016/j.drup.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 14.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 15.Ichikawa D, Komatsu S, Konishi H, Otsuji E. Circulating microRNA in digestive tract cancers. Gastroenterology. 2012;142:1074–8. e1. doi: 10.1053/j.gastro.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Hasselmann DO, Rappl G, Tilgen W, Reinhold U. Extracellular tyrosinase mRNA within apoptotic bodies is protected from degradation in human serum. Clin Chem. 2001;47:1488–9. [PubMed] [Google Scholar]
- 20.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–52. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu H, Fan GC. Extracellular/circulating microRNAs and their potential role in cardiovascular disease. Am J Cardiovasc Dis. 2011;1:138–49. [PMC free article] [PubMed] [Google Scholar]
- 23.Rechavi O, Erlich Y, Amram H, Flomenblit L, Karginov FV, Goldstein I, Hannon GJ, Kloog Y. Cell contact-dependent acquisition of cellular and viral nonautonomously encoded small RNAs. Genes Dev. 2009;23:1971–9. doi: 10.1101/gad.1789609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 26.Du L, Pertsemlidis A. microRNA regulation of cell viability and drug sensitivity in lung cancer. Expert Opin Biol Ther. 2012;12:1221–39. doi: 10.1517/14712598.2012.697149. [DOI] [PubMed] [Google Scholar]
- 27.Sarkar FH, Li Y, Wang Z, Kong D, Ali S. Implication of microRNAs in drug resistance for designing novel cancer therapy. Drug Resist Updat. 2010;13:57–66. doi: 10.1016/j.drup.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Li Y, Ahmad A, Azmi AS, Kong D, Banerjee S, Sarkar FH. Targeting miRNAs involved in cancer stem cell and EMT regulation: An emerging concept in overcoming drug resistance. Drug Resist Updat. 2010;13:109–18. doi: 10.1016/j.drup.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bourguignon LY, Earle C, Wong G, Spevak CC, Krueger K. Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene. 2012;31:149–60. doi: 10.1038/onc.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa PM, Cardoso AL, Nobrega C, Pereira de Almeida LF, Bruce JN, Canoll P, Pedroso de Lima MC. MicroRNA-21 silencing enhances the cytotoxic effect of the antiangiogenic drug sunitinib in glioblastoma. Hum Mol Genet. 2013;22:904–18. doi: 10.1093/hmg/dds496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawakita A, Yanamoto S, Yamada S, Naruse T, Takahashi H, Kawasaki G, Umeda M. MicroRNA-21 promotes oral cancer invasion via the Wnt/beta-catenin pathway by targeting DKK2. Pathol Oncol Res. 2014;20:253–61. doi: 10.1007/s12253-013-9689-y. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Li W, Yang Y, Lu Y, He C, Hu G, Liu H, Chen J, He J, Yu H. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009;1286:13–8. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 33.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, Schmittgen TD, Patel T. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–29. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 34.Shi GH, Ye DW, Yao XD, Zhang SL, Dai B, Zhang HL, Shen YJ, Zhu Y, Zhu YP, Xiao WJ, Ma CG. Involvement of microRNA-21 in mediating chemoresistance to docetaxel in androgen-independent prostate cancer PC3 cells. Acta Pharmacol Sin. 2010;31:867–73. doi: 10.1038/aps.2010.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 36.Tao J, Lu Q, Wu D, Li P, Xu B, Qing W, Wang M, Zhang Z, Zhang W. microRNA-21 modulates cell proliferation and sensitivity to doxorubicin in bladder cancer cells. Oncol Rep. 2011;25:1721–9. doi: 10.3892/or.2011.1245. [DOI] [PubMed] [Google Scholar]
- 37.Tomimaru Y, Eguchi H, Nagano H, Wada H, Tomokuni A, Kobayashi S, Marubashi S, Takeda Y, Tanemura M, Umeshita K, Doki Y, Mori M. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-alpha/5-fluorouracil in hepatocellular carcinoma cells. Br J Cancer. 2010;103:1617–26. doi: 10.1038/sj.bjc.6605958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valeri N, Gasparini P, Braconi C, Paone A, Lovat F, Fabbri M, Sumani KM, Alder H, Amadori D, Patel T, Nuovo GJ, Fishel R, Croce CM. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2) Proc Natl Acad Sci U S A. 2010;107:21098–103. doi: 10.1073/pnas.1015541107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang K, Li PF. Foxo3a regulates apoptosis by negatively targeting miR-21. J Biol Chem. 2010;285:16958–66. doi: 10.1074/jbc.M109.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang P, Zhuang L, Zhang J, Fan J, Luo J, Chen H, Wang K, Liu L, Chen Z, Meng Z. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol Oncol. 2013;7:334–45. doi: 10.1016/j.molonc.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37:2584–95. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang GD, Huang TJ, Peng LX, Yang CF, Liu RY, Huang HB, Chu QQ, Yang HJ, Huang JL, Zhu ZY, Qian CN, Huang BJ. Epstein-Barr Virus_Encoded LMP1 upregulates microRNA-21 to promote the resistance of nasopharyngeal carcinoma cells to cisplatin-induced Apoptosis by suppressing PDCD4 and Fas-L. PLoS One. 2013;8:e78355. doi: 10.1371/journal.pone.0078355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu ZW, Zhong LP, Ji T, Zhang P, Chen WT, Zhang CP. MicroRNAs contribute to the chemoresistance of cisplatin in tongue squamous cell carcinoma lines. Oral Oncol. 2010;46:317–22. doi: 10.1016/j.oraloncology.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 44.Li T, Li D, Sha J, Sun P, Huang Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun. 2009;383:280–5. doi: 10.1016/j.bbrc.2009.03.077. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Zhu X, Gu J, Hu H, Dong D, Yao J, Lin C, Fei J. Anti-miR-21 oligonucleotide enhances chemosensitivity of leukemic HL60 cells to arabinosylcytosine by inducing apoptosis. Hematology. 2010;15:215–21. doi: 10.1179/102453310X12647083620840. [DOI] [PubMed] [Google Scholar]
- 46.Liu S, Fang Y, Shen H, Xu W, Li H. Berberine sensitizes ovarian cancer cells to cisplatin through miR-21/PDCD4 axis. Acta Biochim Biophys Sin (Shanghai) 2013;45:756–62. doi: 10.1093/abbs/gmt075. [DOI] [PubMed] [Google Scholar]
- 47.Wei J, Gao W, Zhu CJ, Liu YQ, Mei Z, Cheng T, Shu YQ. Identification of plasma microRNA-21 as a biomarker for early detection and chemosensitivity of non-small cell lung cancer. Chin J Cancer. 2011;30:407–14. doi: 10.5732/cjc.010.10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan J, Chen L, Chen X, Sun W, Zhou X. Identification of serum microRNA-21 as a biomarker for chemosensitivity and prognosis in human osteosarcoma. J Int Med Res. 2012;40:2090–7. doi: 10.1177/030006051204000606. [DOI] [PubMed] [Google Scholar]
- 49.Komatsu S, Ichikawa D, Takeshita H, Konishi H, Nagata H, Hirajima S, Kawaguchi T, Arita T, Shiozaki A, Fujiwara H, Okamoto K, Otsuji E. Prognostic impact of circulating miR-21 and miR-375 in plasma of patients with esophageal squamous cell carcinoma. Expert Opin Biol Ther. 2012;12(Suppl 1):S53–9. doi: 10.1517/14712598.2012.681373. [DOI] [PubMed] [Google Scholar]
- 50.Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H, Shiozaki A, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2011;105:104–11. doi: 10.1038/bjc.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurashige J, Kamohara H, Watanabe M, Tanaka Y, Kinoshita K, Saito S, Hiyoshi Y, Iwatsuki M, Baba Y, Baba H. Serum microRNA-21 is a novel biomarker in patients with esophageal squamous cell carcinoma. J Surg Oncol. 2012;106:188–92. doi: 10.1002/jso.23064. [DOI] [PubMed] [Google Scholar]
- 52.Sobin L, Gospodarowicz M, Wittekind C, editors; Cancer IUA, editor. TNM Classification of Malignant Tumours seventh edition. New York: Wiley-Blackwell; 2009. [Google Scholar]
- 53.Ajani JA, Walsh G, Komaki R, Morris J, Swisher SG, Putnam JB Jr, Lynch PM, Wu TT, Smythe R, Vaporciyan A, Faust J, Cohen DS, Nivers R, Roth JA. Preoperative induction of CPT-11 and cisplatin chemotherapy followed by chemoradiotherapy in patients with locoregional carcinoma of the esophagus or gastroesophageal junction. Cancer. 2004;100:2347–54. doi: 10.1002/cncr.20284. [DOI] [PubMed] [Google Scholar]
- 54.Yano M, Takachi K, Doki Y, Miyashiro I, Kishi K, Noura S, Eguchi H, Yamada T, Ohue M, Ohigashi H, Sasaki Y, Ishikawa O, Imaoka S. Preoperative chemotherapy for clinically node-positive patients with squamous cell carcinoma of the esophagus. Dis Esophagus. 2006;19:158–63. doi: 10.1111/j.1442-2050.2006.00558.x. [DOI] [PubMed] [Google Scholar]
- 55.Francis AM, Sepesi B, Correa AM, Blum MA, Erasmus JJ, Lee JH, Maru DM, Mehran RJ, Rice DC, Roth JA, Vaporciyan AA, Walsh GL, Welsh JW, Swisher SG, Hofstetter WL. The influence of histopathologic tumor viability on long-term survival and recurrence rates following neoadjuvant therapy for esophageal adenocarcinoma. Ann Surg. 2013;258:500–7. doi: 10.1097/SLA.0b013e3182a196f4. [DOI] [PubMed] [Google Scholar]
- 56.Kuwano H, Nishimura Y, Oyama T, Kato H, Kitagawa Y, Kusano M, Shimada H, Takiuchi H, Toh Y, Doki Y, Naomoto Y, Matsubara H, Miyazaki T, Muto M, Yanagisawa A. Guidelines for Diagnosis and Treatment of Carcinoma of the Esophagus April 2012 edited by the Japan Esophageal Society. Esophagus. 2015;12:1–30. doi: 10.1007/s10388-014-0465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375–81. doi: 10.1136/gut.2008.167817. [DOI] [PubMed] [Google Scholar]
- 58.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 59.Shimada Y, Imamura M, Wagata T, Yamaguchi N, Tobe T. Characterization of 21 newly established esophageal cancer cell lines. Cancer. 1992;69:277–84. doi: 10.1002/1097-0142(19920115)69:2<277::aid-cncr2820690202>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 60.Akobeng AK. Understanding diagnostic tests 3: Receiver operating characteristic curves. Acta Paediatr. 2007;96:644–7. doi: 10.1111/j.1651-2227.2006.00178.x. [DOI] [PubMed] [Google Scholar]
- 61.Hirajima S, Komatsu S, Ichikawa D, Takeshita H, Konishi H, Shiozaki A, Morimura R, Tsujiura M, Nagata H, Kawaguchi T, Arita T, Kubota T, Fujiwara H, Okamoto K, Otsuji E. Clinical impact of circulating miR-18a in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:1822–9. doi: 10.1038/bjc.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kawaguchi T, Komatsu S, Ichikawa D, Morimura R, Tsujiura M, Konishi H, Takeshita H, Nagata H, Arita T, Hirajima S, Shiozaki A, Ikoma H, Okamoto K, Ochiai T, Taniguchi H, Otsuji E. Clinical impact of circulating miR-221 in plasma of patients with pancreatic cancer. Br J Cancer. 2013;108:361–9. doi: 10.1038/bjc.2012.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Komatsu S, Ichikawa D, Tsujiura M, Konishi H, Takeshita H, Nagata H, Kawaguchi T, Hirajima S, Arita T, Shiozaki A, Kubota T, Fujiwara H, Okamoto K, Otsuji E. Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res. 2013;33:271–6. [PubMed] [Google Scholar]
- 64.Konishi H, Ichikawa D, Komatsu S, Shiozaki A, Tsujiura M, Takeshita H, Morimura R, Nagata H, Arita T, Kawaguchi T, Hirashima S, Fujiwara H, Okamoto K, Otsuji E. Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. Br J Cancer. 2012;106:740–7. doi: 10.1038/bjc.2011.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morimura R, Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Nagata H, Konishi H, Shiozaki A, Ikoma H, Okamoto K, Ochiai T, Taniguchi H, Otsuji E. Novel diagnostic value of circulating miR-18a in plasma of patients with pancreatic cancer. Br J Cancer. 2011;105:1733–40. doi: 10.1038/bjc.2011.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, Komatsu A, Jitsukawa M, Matsubara H. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:644–52. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi K, Fujiwara H, Okamoto K, Otsuji E. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–9. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen J, Wang W, Zhang Y, Chen Y, Hu T. Predicting distant metastasis and chemoresistance using plasma miRNAs. Med Oncol. 2014;31:799. doi: 10.1007/s12032-013-0799-x. [DOI] [PubMed] [Google Scholar]
- 69.Jung EJ, Santarpia L, Kim J, Esteva FJ, Moretti E, Buzdar AU, Di Leo A, Le XF, Bast RC Jr, Park ST, Pusztai L, Calin GA. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer. 2012;118:2603–14. doi: 10.1002/cncr.26565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanaka K, Miyata H, Yamasaki M, Sugimura K, Takahashi T, Kurokawa Y, Nakajima K, Takiguchi S, Mori M, Doki Y. Circulating miR-200c levels significantly predict response to chemotherapy and prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. Ann Surg Oncol. 2013;20(Suppl 3):S607–15. doi: 10.1245/s10434-013-3093-4. [DOI] [PubMed] [Google Scholar]
- 71.Wang H, Tan G, Dong L, Cheng L, Li K, Wang Z, Luo H. Circulating MiR-125b as a marker predicting chemoresistance in breast cancer. PLoS One. 2012;7:e34210. doi: 10.1371/journal.pone.0034210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang J, Zhang K, Bi M, Jiao X, Zhang D, Dong Q. Circulating microRNA expressions in colorectal cancer as predictors of response to chemotherapy. Anticancer Drugs. 2014;25:346–52. doi: 10.1097/CAD.0000000000000049. [DOI] [PubMed] [Google Scholar]
- 73.Zhao R, Wu J, Jia W, Gong C, Yu F, Ren Z, Chen K, He J, Su F. Plasma miR-221 as a predictive biomarker for chemoresistance in breast cancer patients who previously received neoadjuvant chemotherapy. Onkologie. 2011;34:675–80. doi: 10.1159/000334552. [DOI] [PubMed] [Google Scholar]
- 74.Hiyoshi Y, Kamohara H, Karashima R, Sato N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M, Baba H. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res. 2009;15:1915–22. doi: 10.1158/1078-0432.CCR-08-2545. [DOI] [PubMed] [Google Scholar]
- 75.Ma WJ, Lv GD, Tuersun A, Liu Q, Liu H, Zheng ST, Huang CG, Feng JG, Wang X, Lin RY, Sheyhidin I, Lu XM. Role of microRNA-21 and effect on PTEN in Kazakh’s esophageal squamous cell carcinoma. Mol Biol Rep. 2011;38:3253–60. doi: 10.1007/s11033-010-0480-9. [DOI] [PubMed] [Google Scholar]
- 76.Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, Watanabe M, Baba H. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119:1159–67. doi: 10.1002/cncr.27895. [DOI] [PubMed] [Google Scholar]
- 77.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, Aprile G, Kulikov E, Hill J, Lehle M, Ruschoff J, Kang YK. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–97. doi: 10.1016/S0140-6736(10)61121-X. [DOI] [PubMed] [Google Scholar]
- 78.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 79.Diaz LA Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, Kinzler KW, Oliner KS, Vogelstein B. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–40. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


