Abstract
Women persistently infected with human papillomavirus (HPV) type 16 are at high risk for development of cervical intraepithelial neoplasia grade 3 or cervical cancer (CIN3+). We aimed to identify biomarkers for progression to CIN3+ in women with persistent HPV16 infection. In this prospective study, 11,088 women aged 20-29 years were enrolled during 1991-1993, and re-invited for a second visit two years later. Cervical cytology samples obtained at both visits were tested for HPV DNA by Hybrid Capture 2 (HC2), and HC2-positive samples were genotyped by INNO-LiPA. The cohort was followed for up to 19 years via a national pathology register. To identify markers for progression to CIN3+, we performed microarray analysis on RNA extracted from cervical swabs of 30 women with persistent HPV16-infection and 11 HPV-negative women. Six genes were selected and validated by quantitative PCR. Three genes were subsequently validated within a different and large group of women from the same cohort. Secondly, Kaplan-Meier and Cox-regression analyses were used to investigate whether expression levels of those three genes predict progression to CIN3+. We found that high transcript levels of TMEM45A, SERPINB5 and p16INK4a at baseline were associated with increased risk of CIN3+ during follow-up. The hazard ratios of CIN3+ per 10-fold increase in baseline expression level were 1.6 (95% CI: 1.1-2.3) for TMEM45A, 1.6 (95% CI: 1.1-2.5) for p16INK4a, and 1.8 (95% CI: 1.2-2.7) for SERPINB5. In conclusion, high mRNA expression levels of TMEM45A, SERPINB5 and p16INK4a were associated with increased risk of CIN3+ in persistently HPV16-infected women.
Keywords: Cervical cancer, CIN3+, HPV16, biomarker, TMEM45A, SERPINB5, p16INK4A
Introduction
Of all high-risk HPV types, HPV16 has the greatest carcinogenic potential [1,2], and women with a persistent HPV16 infection are at high risk of subsequent cervical intraepithelial neoplasia grade 3 or cervical cancer (CIN3+) [1,3,4]. Nevertheless, not all women with a persistent HPV16 infection develop CIN3+, and little is known about the reasons why some women progress to CIN3+ while others do not.
The identification of differentially expressed genes before progression of a persistent HPV16 infection to CIN3+ might help to clarify the molecular mechanisms underlying progression.
Previous studies have thus far identified the viral protein L1 [5], cellular markers such as transglutaminase enzyme type 2 [6], p16INK4a/Ki-67 [7] and SERPINB5 [8,9] as well as specific miRNAs including miR-375, miR-218 or miR-34a [10-12] as potential biomarkers for the progression to CIN3+. However, these studies were of cross-sectional design and compared expression levels of low-grade versus high-grade cervical lesions. Only p16INK4a has been the subject of prospective studies, and a high expression level of p16INK4a-protein was found to be associated with an increased risk of progression to high-grade CIN or cervical cancer. Its predictive value was determined for its protein levels by performing immunohistochemistry staining of biopsy [13-17] or cytology samples [18].
In the present cohort study with up to 19 years of follow-up, we aimed at identifying differentially expressed genes that mark the progression to CIN3+ in women with a persistent HPV16 infection at baseline as this is the most prevalent HR type infection.
Gene expression microarray analysis was used to identify a set of potential candidate markers for progression. Analysis of SERPINB5 and p16INK4a mRNA levels were included based on the potential predictive value of these two markers described in previous reports.
Material and methods
Study cohort
The present study is based on the Danish HPV cohort, which has previously been described in detail [1,19]. Briefly, 11,088 Danish women aged 20-29 years from the general female population of Copenhagen were enrolled during 1991-1993. Two years later, the women were re-invited for a second visit, and a total of 8,656 women participated. At both visits, a Pap-smear was taken, and endo-ectocervical cell material was obtained for HPV testing. Cervical samples were placed in phosphate-buffered saline (with 0.05% methiolate), stored at -80°C, and tested by Hybrid Capture 2 (HC2) (Qiagen, Hilden, Germany). HPV-positive samples were genotyped by INNO-LiPA Genotyping (Innogenetics, Ghent, Belgium) as previously described [1,19].
The cohort was subsequently followed via the Danish national Pathology Data Bank [20], which enabled us to obtain normal and abnormal cervical diagnoses of all participating women. Abnormal cervical diagnoses were reported either according to the dysplasia nomenclature (atypia, mild dysplasia, moderate dysplasia, severe dysplasia, carcinoma in situ, and cancer), the Bethesda nomenclature for cytological diagnoses, or the CIN nomenclature for histological diagnoses. In our analyses, histological diagnoses of severe dysplasia, carcinoma in situ (including adenocarcinoma in situ), CIN3 or cancer (including adenocarcinoma) were categorized as CIN3+. We defined baseline as the date of the second study visit.
Consent and approval
Before entering the study, all participants were informed verbally and in writing about the study, and all participants signed a written informed consent. The study was approved by the national Scientific Ethics Committee and the national Data Protection Board.
RNA extraction
RNA was extracted from cervical swab samples obtained at the second study visit. Extraction was done using the RNeasy Mini Kit (Qiagen, Hilden, Germany), and the RNA was stored at -80°C. RNA quality was determined using an Agilent Bioanalyzer 2100.
Microarray analysis
RNA amplification and hybridization to Affymetrix Human Genome U133 Plus 2.0 arrays were performed at the Microarray Facility Tübingen following standard Affymetrix procedures. The final data analysis was performed by Genedata AG, Basel, as follows: The quality control of the data files from the Human Genome U133 Plus 2.0 arrays was performed using Refiner Array from Genedata AG. Expression values for all probe sets were obtained by applying the GeneChip Robust Multi-array Average condensing method (GC-RMA) [21]. No microarray showed local areas of significantly increased or decreased intensities, and no systematic biases (defects, gradient, distortion, background) were found. All transcriptional profiles have been submitted to the GEO database at NCBI (Accession number: GSE75132).
Quantitative real-time PCR (qPCR)
Primer pairs for the respective biomarker genes were designed using PRIMER3 plus software [22] (Table 1). RNA was reverse transcribed using the Reverse Transcription kit from Qiagen (Hilden, Germany). RNA concentration was determined by NanoDrop and absolute RNA amount used for cDNA synthesis ranged from 10 pg to 1 µg cDNA. 5 µl sample cDNA was subjected to qPCR (Roche Light Cycler 480, Roche Diagnostics) using 1x Light Cycler 480 SYBR green I Master Mix (Roche Diagnostics), 0.3 µM primer and PCR grade H2O. Specificity was verified by melting curve analysis. The transcript levels of each gene, calculated using standard curves, were normalized to PGK1 transcript levels to calculate relative mRNA amounts.
Table 1.
Oligonucleotide primers for quantitative real-time PCR
| Gene Symbol | ID | Primer | Sequence |
|---|---|---|---|
| TMEM45A | 219410_at | Forward | AGTTGGATGCCCACACTATG |
| Reverse | AGTAGCAAGCCCAGTAACCTTG | ||
| TNFAIP6 | 206025_s_at | Forward | TTGAAGATGACCCAGGTTGC |
| Reverse | CATCTGGAAGCTCATCTCCAC | ||
| CD44 | 204490_s_at | Forward | AAGGTGGAGCAAACACAACC |
| Reverse | GCTTTTTCTTCTGCCCACAC | ||
| CCRL2 | 211434_s_at | Forward | GGCGCGGAAATTTGTCTAAG |
| Reverse | CCAGGGTTTGGAGTTTGATG | ||
| SERPINB5 | 204855_at | Forward | GTTGCCGGTTCATGGATTAC |
| Reverse | GCATGTCAAGGAAGAGATGG | ||
| PGK 1 | Forward | CTGTGGGGGTATTTGAATGG | |
| Reverse | CTTCCAGGAGCTCCAAACTG | ||
| p16INK4a | Qiagen QuantiTect Primer Assay | ||
| (#QT00089964) for CDKN2A |
Statistical analysis
The logarithmized gene expression values from the microarray analyses were normalized by locally weighted scatterplot smoothing (LOWESS) [23] and by shifting the medians of each experiment to a value of 10,000. In order to eliminate possible batch effects, expression values were divided by the medians of the corresponding HPV-negative control group. Significantly deregulated genes were selected using t-tests for independent samples and Wilcoxon rank sum tests. Statistics were computed based on the logarithmized and normalized expression values. The p-value, a permutation q-value (100 repeats) and the Benjamini-Hochberg q-value (false discovery rate, FDR) [24] were derived. In addition, we calculated the fold change of the means of the groups.
When analyzing whether the identified genes could predict progression to CIN3+ among persistently HPV16-infected women, we estimated the cumulative incidence of CIN3+ during follow-up using the Kaplan-Meier method. For this analysis, gene expression values were dichotomized at the median. Furthermore, we used Cox proportional hazard models to estimate the hazard ratio (HR) with corresponding 95% confidence intervals (CIs) of progression to CIN3+ during follow-up according to gene expression levels at baseline. In the Cox models, we used continuous log-transformed gene expression values as well as categorical gene expression values (dichotomized at the median) as the independent variable. Hazard ratios are given for both, categorization at the median and for 10-fold increase (continuous scale). The analysis was adjusted for age at baseline. In both the Kaplan-Meier and the Cox regression analyses, time to event was measured from baseline (i.e. date of the second study visit) to date of CIN3+-diagnosis. Repeating the Cox regression using the mid-point between date of diagnosis and date of last examination without CIN3+ did not alter our conclusions.
Results
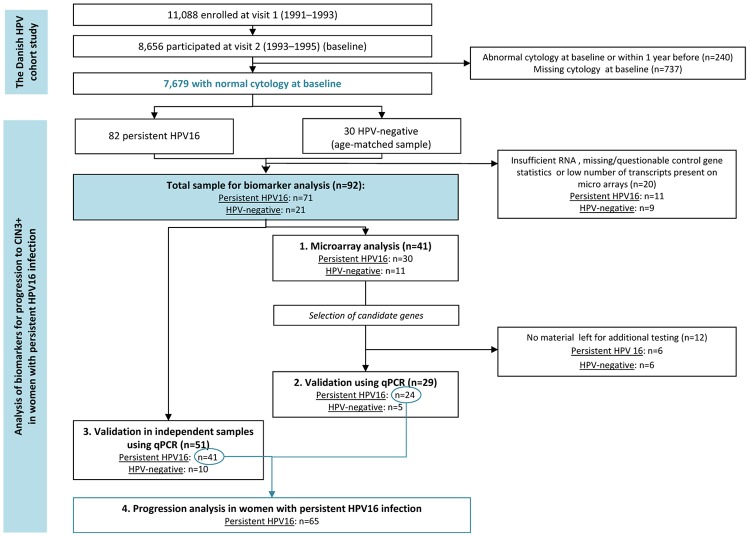
Figure 1 illustrates the design of the present analysis. First, we selected those women from the original cohort who had normal cervical cytology at baseline and no abnormal smears within one year before baseline (n=7,679). Of these, we identified 82 women with a persistent HPV16 infection at the second study visit and at least one cervical examination in the Pathology Databank during follow-up. Persistent HPV16 infection was defined as testing positive for HPV16 DNA by the INNO-LiPA test at both study visits 2 years apart. The median age of the 82 women was 26 years (range 22-32 years). As a control group for the discovery set for the gene expression analysis, we selected an age-matched sample of 30 women who were negative for HPV DNA by HC2 at both study visits and who did not develop cervical abnormalities during follow-up (henceforth referred to as HPV-negative women). The purpose of the control group was to validate the analysis undertaken by identifying genes differentially expressed in women with persistent HPV16 infection compared with HPV-negative women, as a proof of principle as the samples were collected under suboptimal conditions for the later RNA analysis. A total of 20 women (11 women with persistent HPV16 infection and 9 HPV-negative women) were excluded from all analyses due to insufficient RNA, missing/questionable control gene statistics or low number of transcripts (< 10%) present on microarrays. Thus, the final study population included 71 women with persistent HPV16 infection, and 21 women who were HPV-negative at baseline (Figure 1).
Figure 1.
Overview of study design. CIN, Cervical intraepithelial neoplasia; qPCR, quantitative real-time PCR; HPV, Human papillomavirus.
Microarray analysis
Based on RNA quality analysis, samples with similar RNA integrity numbers were selected for microarray experiments. The microarray analysis included a total of 41 samples (30 of the 71 samples from women with persistent HPV16 infection, and 11 of the 21 samples from HPV-negative women; Figure 1).
T-test analysis of the microarray data resulted in 3,177 altered transcript levels at p-values < 0.01, an FDR of 0.18, and a permutation q-value of 0.17. A total of 1,519 genes showed fold changes of > 2 or < 0.5. Of these, 1,090 were down-regulated, and 429 were up-regulated in persistently HPV16-infected women compared with HPV-negative women. The Wilcoxon rank sum test confirmed the total number of deregulated genes, with 1,430 genes showing fold changes of > 2 or < 0.5 (975 down-regulated and 455 up-regulated genes). The overlap between the 3,177 transcripts from the t-tests and the 3,177 transcripts from the Wilcoxon tests counted 2,596 transcripts, and the overlap between the 1,519 and 1,430 transcripts with fold changes of > 2 or < 0.5 comprised 1,366 transcripts.
From this pool of significantly deregulated genes, we selected one up-regulated gene (TMEM45A) and three down-regulated genes (TNFAIP6, CD44 and CCRL2) for validation by qPCR. The selection was performed by degree of statistical significance (all p-values < 0.0001) and the extent of deregulation (ratio of means ranging from 0.04-3.63) (Table 2). Moreover, p16INK4a and SERPINB5 were selected for further analyses based on previous findings [8,9,13,18]. A description of the ontology of the selected genes is provided in Table 2.
Table 2.
Microarray results and description of genes selected for further analysis
| Microarray results | |||||
|---|---|---|---|---|---|
|
|
|||||
| Persistently HPV16-infected versus HPV-negative women | |||||
| Gene symbol | Chromosome | Gene description | Gene ontology | T-test (p-value) | Ratio of means1 |
| TMEM45A | 3q12.2 | Transmembrane protein 45A | Integral to membrane | < 0.0001 | 3.63 |
| TNFAIP6 | 2q23.2 | Tumor necrosis factor, alpha-induced protein 6 | Inflammatory response, cell adhesion, signal transduction, cell-cell signaling | < 0.0001 | 0.05 |
| CD44 | 11p13 | CD44 antigen (homing function and Indian blood group system) | Regulation of cell growth, inflammatory response, cell adhesion, cell migration | < 0.0001 | 0.14 |
| CCRL2 | 3p21 | Chemokine (C-C motif) receptor-like 2 | Chemotaxis, signal transduction, receptor activity | < 0.00001 | 0.04 |
| p16INK4a2 | 9p21 | Cyclin-dependent kinase inhibitor 2A | Cell cycle checkpoint, regulation of cyclin-dependent protein kinase activity, G1/S transition of mitotic cell cycle, DNA fragmentation involved in apoptosis, transcription | 0.5193 | 0.93 |
| SERPINB52 | 18q21.3 | Serpin peptidase inhibitor, clade B (ovalbumin), member 5 | Cell motion, serine-type endopeptidase inhibitor activity, protein binding | 0.0144 | 2.69 |
Ratio of means was computed as “mean expression value in persistently HPV16-infected women”/“mean expression value in HPV-negative women”;
Included in analysis based on à priori hypotheses.
Validation by qPCR
In order to confirm the differential expression of the selected genes, we performed two validation assays using qPCR. The first qPCR validation was performed on the non-amplified original RNA samples previously included in the microarray analysis.
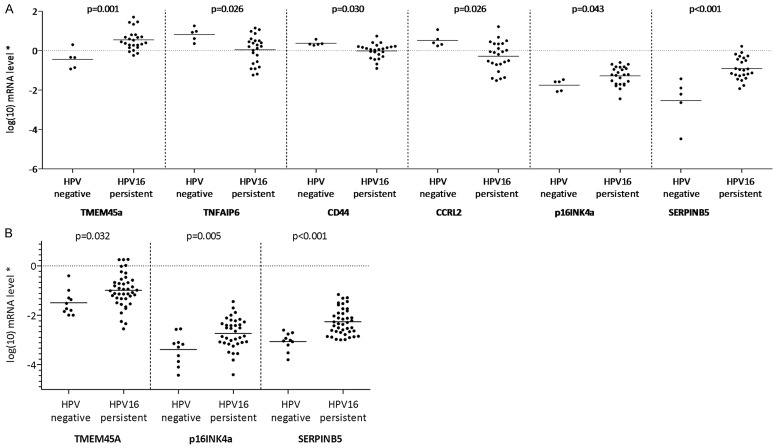
Due to insufficient material, only 29 samples (n=24 persistently HPV16-infected women, n=5 HPV-negative women) could be used for this analysis (Figure 1). Figure 2A presents the results of the first qPCR validation. This validation confirmed the results of the microarray data, showing that the expression of TMEM45A was increased in persistently HPV16-infected women compared with HPV-negative women (p=0.001), while the expression of TNFAIP6, CD44 and CCRL2 was reduced (p=0.026, p=0.030 and p=0.026, respectively). Furthermore, the analysis showed that p16INK4a and SERPINB5 were overexpressed in women with a persistent HPV16 infection compared with HPV-negative women (p=0.043 and p < 0.001, respectively).
Figure 2.
Validation of microarray results by quantitative PCR. (A) on a subset of samples previously included in microarrays and (B) on independent samples not previously included in microarrays. Log-transformed (log10) mRNA levels in 24 and 41 women with persistent HPV16 infection, compared to 5 and 10 HPV-negative women, respectively. P-values refer to T-test for independent samples using logarithmic transformed gene expression values. Solid line indicates mean of log-transformed transcript levels; *Normalized to PGK1 transcript levels; HPV, Human papillomavirus.
The results of the second qPCR validation was performed on a large group of 51 independent samples not previously included in the microarray analysis, comprising 41 women with persistent HPV16 infection and 10 HPV-negative women (Figure 1). Due to limited RNA amounts, this validation was performed only for the transcripts of the three most promising genes, including TMEM45A, p16INK4a and SERPINB5. TMEM45A and SERPINB5 were selected because they showed the most significant difference between HPV-negative and persistently HPV16-infected women in the first validation analysis (p=0.001 and p < 0.001, respectively, Figure 2A). P16INK4a was selected due to a previous report demonstrating that overexpression of this marker might be predictive for the risk of incident high-grade cervical lesions in HPV-positive women [18]. Figure 2B presents the results of the second qPCR validation. This analysis confirmed significantly higher mRNA transcript levels for TMEM45A (p=0.032), p16INK4a (p=0.005) and SERPINB5 (p < 0.001) in women with persistent HPV16 infection compared with HPV-negative women (Figure 2B).
Prediction of progression to CIN3+
Following the validation analyses, we examined whether the transcript levels of the three selected genes were predictive for progression to CIN3+ during follow-up in women with persistent HPV16 infection at baseline. In this analysis, we included women with persistent HPV16 infection at baseline from the first (n=24) and the second (n=41) validation analyses, resulting in a total of 65 women for the progression analysis (Figure 1). Of these 65 persistently HPV-16 infected women, 29 women developed CIN3, and 2 developed cancer during up to 19.3 years of follow-up, resulting in 31 (53.4%) cases of CIN3+.
Figure 3A-C show the cumulative incidence of CIN3+ during follow-up according to baseline expression levels of TMEM45A, p16INK4a and SERPINB5. For all three genes, high (≥ median) transcript levels at baseline were associated with a higher cumulative incidence of CIN3+ during follow-up than low (< median) transcript levels. Furthermore, the Kaplan-Meier curves indicated that TMEM45A or SERPINB5 transcript levels allowed a more pronounced separation of progressors versus non-progressors than p16INK4a transcript levels.
Figure 3.
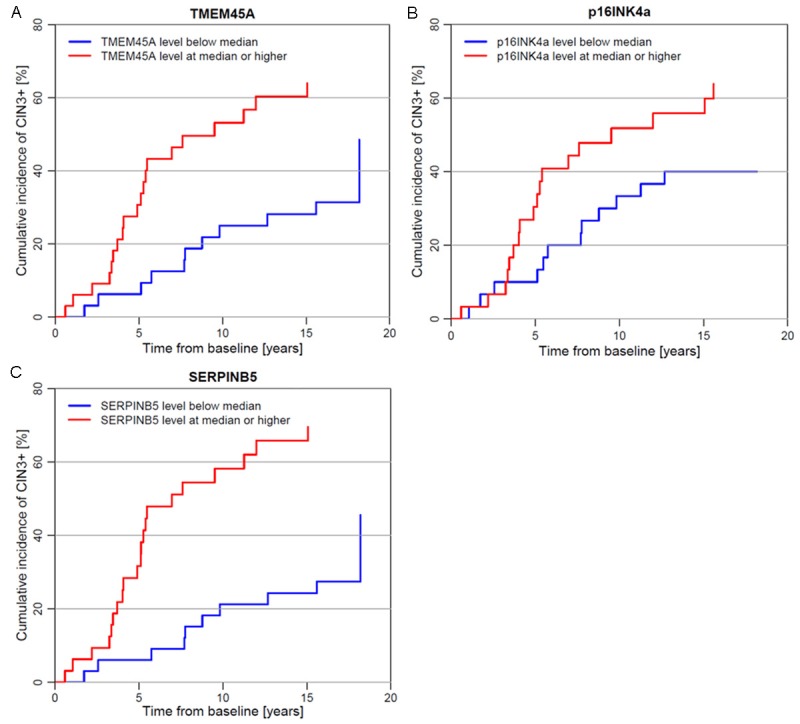
Cumulative incidence of CIN3+ during follow-up in 65 women with persistent HPV16 infection at baseline, according to expression level of TMEM45A (A), p16INK4a (B), and SERPINB5 (C) at baseline. CIN3+, Cervical intraepithelial neoplasia grade 3 or worse; HPV, Human papillomavirus.
Finally, Table 3 shows the age-adjusted hazard ratios (HRs) of CIN3+ in women with persistent HPV16 infection at baseline, according to baseline gene expression levels. For TMEM45A and SERPINB5, baseline transcript levels above versus below the median were significantly associated with higher CIN3+ risk during follow-up (TMEM45A: HR=3.1, 95% CI: 1.4-6.7; SERPINB5: HR=4.2, 95% CI: 1.9-9.3). The same pattern, although not statistically significant, was observed for p16INK4a (HR=1.8, 95% CI: 0.9-3.7). Assuming a log-linear association between gene expression levels and the hazard of CIN3+, all three genes showed a 60-80% increased hazard of CIN3+ per 10-fold increase in the expression level (TMEM45A: HR=1.6, 95% CI: 1.1-2.3; SERPINB5: HR=1.8, 95% CI: 1.2-2.7; p16INK4a: HR=1.6, 95% CI: 1.1-2.5).
Table 3.
Hazard ratios of CIN3+ during up to 19 years of follow-up in 65 women with persistent HPV16 infection at baseline, according to expression level of TMEM45A, p16INK4a and SERPINB5 as measured by quantitative real-time PCR at baseline
| Main analysis | Sensitivity analysis using midpoint of time interval | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Expression level | CIN3+ | Age-adjusted HR of CIN3+ | (95% CI) | Age-adjusted HR of CIN3+ | (95% CI) | |||
|
|
||||||||
| N | n | (%) | ||||||
| TMEM45A | < Median | 32 | 11 | (34.4) | 1.0 | (ref) | 1.0 | (ref) |
| ≥ Median | 33 | 20 | (60.6) | 3.1 | (1.4-6.7) | 3.6 | (1.7-7.9) | |
| Continuous (per 10-fold increase) | 65 | 31 | (47.7) | 1.6 | (1.1-2.3) | 1.2 | (1.1-1.3) | |
| p16INK4a* | < Median | 30 | 13 | (43.3) | 1.0 | (ref) | 1.0 | (ref) |
| ≥ Median | 30 | 18 | (60.0) | 1.8 | (0.9-3.7) | 2.2 | (1.0-4.5) | |
| Continuous (per 10-fold increase) | 60 | 31 | (51.7) | 1.6 | (1.1-2.5) | 1.2 | (1.1-1.4) | |
| SERPINB5 | < Median | 33 | 10 | (30.3) | 1.0 | (ref) | 1.0 | (ref) |
| ≥ Median | 32 | 21 | (65.6) | 4.2 | (1.9-9.3) | 4.8 | (2.2-10.7) | |
| Continuous (per 10-fold increase) | 65 | 31 | (47.7) | 1.8 | (1.2-2.7) | 1.2 | (1.1-1.3) | |
Five women with missing p16INK4a values were excluded.
CI, Confidence interval; CIN3+, Cervical intraepithelial neoplasia grade 3 or worse; HPV, Human papillomavirus; HR, Hazard ratio; (ref), reference group.
Discussion
In the present study, we employed microarray analysis and qPCR to cervical cytology samples of a population-based cohort which has been followed for up to 19 years with virtually no loss to follow-up. We found that in women with normal cytology and persistent HPV16 infection at baseline, increased mRNA expression of TMEM45A, SERPINB5 and p16INK4a was associated with an increased risk of progression to CIN3+ during follow-up.
The TMEM45A (transmembrane protein 45A; also known as DERP7, DNAPTP4 or FLJ10134) gene belongs to the family of uncharacterized predicted transmembrane proteins and contains the Pfam domain DUF716 (domain of unknown function) which is associated with viral infection in plants (InterPro IPR006904). The function of TMEM45A remains unknown. It has been shown that TMEM45A is a hypoxia inducible gene [25,26] with anti-apoptotic functions under genotoxic stress [27], and the expression of TMEM45A in cancer cells suppressed progression of ductual carcinoma into invasive breast cancer [28]. TMEM45A is highly expressed in human keratinocytes [29], and it was reported to be associated with keratinocyte differentiation [30,31]. Up-regulation of TMEM45A expression was found in psoriasis and actinic keratosis [31,32]. TMEM45A has previously been described as a marker for survival of breast cancer patients. Patients with high expression within the tumor tissue had a significantly lower relapse-free survival than those with a low expression [27]. Almost 90% of all cervical cancers reveal a gain of chromosome 3q, which carries the TMEM45A gene [33]. It therefore seems plausible that early overexpression of TMEM45A might constitute a marker for progression to high-grade cervical lesions in high-risk HPV-infected women.
In contrast SERPINB5 or Maspin (mammary serine protease inhibitor) has previously been reported to be differentially expressed in certain types of cancer. A reduced expression was found in prostate cancer [8], breast cancer [34] and gastric carcinoma [35], which supports a tumor suppressor role of SERPINB5 in tumor progression. However, increased expression of SERPINB5 was observed in ovarian cancer [36], breast cancer [37], gastric adenocarcinoma [38], colorectal [39,40], pancreatic [41] and gallbladder cancer [42], suggesting that overexpression is involved in tumor progression in these tissues. Previously published data also showed that SERPINB5 might play a role in disease progression from in situ to invasive cervical cancer [9], and Liu et al. reported that nuclear localization of SERPINB5 might be a marker for prognosis of cervical cancer patients [8]. This is consistent with our results, showing that a higher expression of SERPINB5 in persistently HPV16-infected women with normal cytology has the potential to predict progression to CIN3+.
In contrast to TMEM45A and SERPINB5, immunohistological staining for p16INK4a expression is already being used as a tool for managing patients with atypical squamous cells of undetermined significance (ASC-US) or low-grade squamous intraepithelial lesion (LSIL) cytology results and for triaging HPV-positive women [43]. The p16INK4a protein is a cyclin-dependent kinase (CDK) inhibitor, which is induced by cellular stress including the presence of the papillomavirus oncogene E7. Because of an epigenetic derepression mediated by E7, HPV-associated tumors express high levels of p16INK4a, which justifies its use as a marker for neoplastic lesions [44]. A recent prospective analysis investigated the relation between p16INK4a protein positivity in cytology specimens and the subsequent risk of CIN2+ during 3 years of follow-up [18]. In accordance with our results, this study demonstrates that p16INK4a overexpression is a predictive marker for the development of CIN2+ or worse in women positive for HPV irrespective of the type.
A main strength of our study is the initial use of microarray analysis as an unbiased approach to identify a set of cellular genes deregulated in women with persistent HPV16 infection. This allowed us to combine a genome-wide search for new potential progression markers with previously published data on potential markers. Furthermore, the follow-up via the nationwide pathology register enabled an extended follow-up period with virtually no loss. Lastly, because HPV-status was measured at two time points, we were able to identify women with a persistent HPV16-infection (HPV16-positive at both visits). Therefore, we could focus our analysis on those women who are at highest risk for the development of CIN3+ [1,45].
A limitation of our study is that although the Danish HPV cohort study included more than 11,000 women, there were relatively few CIN3+-cases in the study population for the present analysis (n=31). Therefore, we were unable to evaluate progression to CIN3 and cervical cancer separately. Furthermore, cervical swabs were stored in a buffer not suitable for RNA storage, which led to a certain degree of RNA degradation. Due to limited RNA amounts, we could not include all of the potential genetic markers identified by microarrays in the analysis of the risk for progression to CIN3+.
In conclusion, we found that high expression levels of TMEM45A, SERPINB5 and p16INK4a mRNA in persistently HPV16-infected women with normal cervical cytology increases the risk of future CIN3+. Further investigation is needed to clarify the roles of TMEM45A and SERPINB5 in disease development, which might ultimately help to assess the potential of those markers for risk stratification as well as to a better understanding of the biological processes causing progression to CIN3+ in some, but not all persistent HPV16 infections.
Acknowledgements
This work was supported by the Mermaid project (MERMAID-2) and the European commission (INCA).
Disclosure of conflict of interest
TT has received a travel grant from Sanofi Pasteur MSD. CM has received support for conference participation and speaker’s fees from Sanofi Pasteur MSD. SKK has received speaker’s and advisory board fees and research grants through her institution from Sanofi Pasteur MSD and Merck. TI received speaker honoraria from Hologic GmbH, Becton Dickinson Diagnostics GmbH and Sanofi Pasteur MSD; and his institution received an unconditional research grant from Hologic GmbH and Becton Dickinson Diagnostics GmbH.
References
- 1.Kjaer SK, Frederiksen K, Munk C, Iftner T. Long-term absolute risk of cervical intraepithelial neoplasia grade 3 or worse following human papillomavirus infection: role of persistence. J Natl Cancer Inst. 2010;102:1478–1488. doi: 10.1093/jnci/djq356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munoz N, Castellsague X, de Gonzalez AB, Gissmann L. Chapter 1: HPV in the etiology of human cancer. Vaccine. 2006;24(Suppl 3) doi: 10.1016/j.vaccine.2006.05.115. S3/1-10. [DOI] [PubMed] [Google Scholar]
- 3.Schiffman M, Herrero R, Desalle R, Hildesheim A, Wacholder S, Rodriguez AC, Bratti MC, Sherman ME, Morales J, Guillen D, Alfaro M, Hutchinson M, Wright TC, Solomon D, Chen Z, Schussler J, Castle PE, Burk RD. The carcinogenicity of human papillomavirus types reflects viral evolution. Virology. 2005;337:76–84. doi: 10.1016/j.virol.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Castle PE, Rodriguez AC, Burk RD, Herrero R, Wacholder S, Alfaro M, Morales J, Guillen D, Sherman ME, Solomon D, Schiffman M Proyecto Epidemiológico Guanacaste (PEG) Group. Short term persistence of human papillomavirus and risk of cervical precancer and cancer: population based cohort study. BMJ. 2009;339:b2569. doi: 10.1136/bmj.b2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarmadi S, Izadi-mood N, Pourlashkari M, Yarandi F, Sanii S. HPV L1 capsid protein expression in squamous intraepithelial lesions of cervix uteri and its relevance to disease outcome. Arch Gynecol Obstet. 2012;285:779–784. doi: 10.1007/s00404-011-2010-y. [DOI] [PubMed] [Google Scholar]
- 6.Del Nonno F, Pisani G, Visca P, Signore F, Grillo LR, Baiocchini A, Garbuglia AR, Sepe S, Piacentini M, Falasca L. Role and predictive strength of transglutaminase type 2 expression in premalignant lesions of the cervix. Mod Pathol. 2011;24:855–865. doi: 10.1038/modpathol.2011.40. [DOI] [PubMed] [Google Scholar]
- 7.Reuschenbach M, Seiz M, von Knebel Doeberitz C, Vinokurova S, Duwe A, Ridder R, Sartor H, Kommoss F, Schmidt D, von Knebel Doeberitz M. Evaluation of cervical cone biopsies for coexpression of p16INK4a and Ki-67 in epithelial cells. Int J Cancer. 2012;130:388–394. doi: 10.1002/ijc.26017. [DOI] [PubMed] [Google Scholar]
- 8.Liu Z, Shi Y, Meng W, Liu Y, Yang K, Wu S, Peng Z. Expression and localization of maspin in cervical cancer and its role in tumor progression and lymphangiogenesis. Arch Gynecol Obstet. 2014;289:373–382. doi: 10.1007/s00404-013-2988-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu C, Quddus MR, Sung CJ, Steinhoff MM, Zhang C, Lawrence WD. Maspin expression in CIN 3, microinvasive squamous cell carcinoma, and invasive squamous cell carcinoma of the uterine cervix. Mod Pathol. 2005;18:1102–1106. doi: 10.1038/modpathol.3800393. [DOI] [PubMed] [Google Scholar]
- 10.Wang F, Li Y, Zhou J, Xu J, Peng C, Ye F, Shen Y, Lu W, Wan X, Xie X. miR-375 is down-regulated in squamous cervical cancer and inhibits cell migration and invasion via targeting transcription factor SP1. Am J Pathol. 2011;179:2580–2588. doi: 10.1016/j.ajpath.2011.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, Khan SA. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575–2582. doi: 10.1038/sj.onc.1210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Wang HK, McCoy JP, Banerjee NS, Rader JS, Broker TR, Meyers C, Chow LT, Zheng ZM. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA. 2009;15:637–647. doi: 10.1261/rna.1442309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao GD, Sellors JW, Sun HK, Zhang X, Bao YP, Jeronimo J, Chen W, Zhao FH, Song Y, Cao Z, Zhang SK, Xi MR, Qiao YL. p16INK4A immunohistochemical staining and predictive value for progression of cervical intraepithelial neoplasia grade 1: a prospective study in China. Int J Cancer. 2014;134:1715–1724. doi: 10.1002/ijc.28485. [DOI] [PubMed] [Google Scholar]
- 14.Wang HL, Lu DW. Detection of human papillomavirus DNA and expression of p16, Rb, and p53 proteins in small cell carcinomas of the uterine cervix. Am J Surg Pathol. 2004;28:901–908. doi: 10.1097/00000478-200407000-00009. [DOI] [PubMed] [Google Scholar]
- 15.Negri G, Vittadello F, Romano F, Kasal A, Rivasi F, Girlando S, Mian C, Egarter-Vigl E. p16INK4a expression and progression risk of low-grade intraepithelial neoplasia of the cervix uteri. Virchows Arch. 2004;445:616–620. doi: 10.1007/s00428-004-1127-9. [DOI] [PubMed] [Google Scholar]
- 16.del Pino M, Garcia S, Fuste V, Alonso I, Fuste P, Torne A, Ordi J. Value of p16(INK4a) as a marker of progression/regression in cervical intraepithelial neoplasia grade 1. Am J Obstet Gynecol. 2009;201:488, e481–487. doi: 10.1016/j.ajog.2009.05.046. [DOI] [PubMed] [Google Scholar]
- 17.Ozaki S, Zen Y, Inoue M. Biomarker expression in cervical intraepithelial neoplasia: potential progression predictive factors for low-grade lesions. Hum Pathol. 2011;42:1007–1012. doi: 10.1016/j.humpath.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 18.Carozzi F, Gillio-Tos A, Confortini M, Del Mistro A, Sani C, De Marco L, Girlando S, Rosso S, Naldoni C, Dalla Palma P, Zorzi M, Giorgi-Rossi P, Segnan N, Cuzick J, Ronco G. Risk of high-grade cervical intraepithelial neoplasia during follow-up in HPV-positive women according to baseline p16-INK4A results: a prospective analysis of a nested substudy of the NTCC randomised controlled trial. Lancet Oncol. 2013;14:168–176. doi: 10.1016/S1470-2045(12)70529-6. [DOI] [PubMed] [Google Scholar]
- 19.Kjaer S, Hogdall E, Frederiksen K, Munk C, van den Brule A, Svare E, Meijer C, Lorincz A, Iftner T. The absolute risk of cervical abnormalities in high-risk human papillomavirus-positive, cytologically normal women over a 10-year period. Cancer Res. 2006;66:10630–10636. doi: 10.1158/0008-5472.CAN-06-1057. [DOI] [PubMed] [Google Scholar]
- 20.Bjerregaard B, Larsen OB. The Danish Pathology Register. Scand J Public Health. 2011;39:72–74. doi: 10.1177/1403494810393563. [DOI] [PubMed] [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 23.Cleveland WS. Robust Locally Weighted Regression and Smoothing Scatterplots. Journal of the American Statistical Association. 1979:829–836. [Google Scholar]
- 24.Hochberg Y, Benjamini Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 25.Benita Y, Kikuchi H, Smith AD, Zhang MQ, Chung DC, Xavier RJ. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009;37:4587–4602. doi: 10.1093/nar/gkp425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin-Rendon E, Hale SJ, Ryan D, Baban D, Forde SP, Roubelakis M, Sweeney D, Moukayed M, Harris AL, Davies K, Watt SM. Transcriptional profiling of human cord blood CD133+ and cultured bone marrow mesenchymal stem cells in response to hypoxia. Stem Cells. 2007;25:1003–1012. doi: 10.1634/stemcells.2006-0398. [DOI] [PubMed] [Google Scholar]
- 27.Flamant L, Roegiers E, Pierre M, Hayez A, Sterpin C, De Backer O, Arnould T, Poumay Y, Michiels C. TMEM45A is essential for hypoxia-induced chemoresistance in breast and liver cancer cells. BMC Cancer. 2012;12:391. doi: 10.1186/1471-2407-12-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S, Stewart S, Nagtegaal I, Luo J, Wu Y, Colditz G, Medina D, Allred DC. Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer. Cancer Res. 2012;72:4574–4586. doi: 10.1158/0008-5472.CAN-12-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayez A, Malaisse J, Roegiers E, Reynier M, Renard C, Haftek M, Geenen V, Serre G, Simon M, de Rouvroit CL, Michiels C, Poumay Y. High TMEM45A expression is correlated to epidermal keratinization. Exp Dermatol. 2014;23:339–344. doi: 10.1111/exd.12403. [DOI] [PubMed] [Google Scholar]
- 30.Mattiuzzo NR, Toulza E, Jonca N, Serre G, Guerrin M. A large-scale multi-technique approach identifies forty-nine new players of keratinocyte terminal differentiation in human epidermis. Exp Dermatol. 2011;20:113–118. doi: 10.1111/j.1600-0625.2010.01188.x. [DOI] [PubMed] [Google Scholar]
- 31.Gerber PA, Hevezi P, Buhren BA, Martinez C, Schrumpf H, Gasis M, Grether-Beck S, Krutmann J, Homey B, Zlotnik A. Systematic identification and characterization of novel human skin-associated genes encoding membrane and secreted proteins. PLoS One. 2013;8:e63949. doi: 10.1371/journal.pone.0063949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hudson LG, Gale JM, Padilla RS, Pickett G, Alexander BE, Wang J, Kusewitt DF. Microarray analysis of cutaneous squamous cell carcinomas reveals enhanced expression of epidermal differentiation complex genes. Mol Carcinog. 2010;49:619–629. doi: 10.1002/mc.20636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heselmeyer-Haddad K, Sommerfeld K, White NM, Chaudhri N, Morrison LE, Palanisamy N, Wang ZY, Auer G, Steinberg W, Ried T. Genomic amplification of the human telomerase gene (TERC) in pap smears predicts the development of cervical cancer. Am J Pathol. 2005;166:1229–1238. doi: 10.1016/S0002-9440(10)62341-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, Rafidi K, Seftor E, Sager R. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;263:526–529. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]
- 35.Yang Y, Shi H, Li X, Yi Y. Effects of shRNA targeting maspin on invasion of gastric carcinoma SGC7901 cell line. Oncol Rep. 2011;25:259–265. [PubMed] [Google Scholar]
- 36.Lin Z, Liu Y, Sun Y, He X. Expression of Ets-1, Ang-2 and maspin in ovarian cancer and their role in tumor angiogenesis. J Exp Clin Cancer Res. 2011;30:31. doi: 10.1186/1756-9966-30-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Umekita Y, Yoshida H. Expression of maspin is up-regulated during the progression of mammary ductal carcinoma. Histopathology. 2003;42:541–545. doi: 10.1046/j.1365-2559.2003.01620.x. [DOI] [PubMed] [Google Scholar]
- 38.Song SY, Son HJ, Kim MH, Nam ES, Rhee JC, Park C. Prognostic significance of maspin expression in human gastric adenocarcinoma. Hepatogastroenterology. 2007;54:973–976. [PubMed] [Google Scholar]
- 39.Bettstetter M, Woenckhaus M, Wild PJ, Rummele P, Blaszyk H, Hartmann A, Hofstadter F, Dietmaier W. Elevated nuclear maspin expression is associated with microsatellite instability and high tumour grade in colorectal cancer. J Pathol. 2005;205:606–614. doi: 10.1002/path.1732. [DOI] [PubMed] [Google Scholar]
- 40.Payne CM, Holubec H, Crowley-Skillicorn C, Nguyen H, Bernstein H, Wilcox G, Bernstein C. Maspin is a deoxycholate-inducible, anti-apoptotic stress-response protein differentially expressed during colon carcinogenesis. Clin Exp Gastroenterol. 2011;4:239–253. doi: 10.2147/CEG.S24093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao D, Zhang Q, Wu LS, Salaria SN, Winter JW, Hruban RH, Goggins MS, Abbruzzese JL, Maitra A, Ho L. Prognostic significance of maspin in pancreatic ductal adenocarcinoma: tissue microarray analysis of 223 surgically resected cases. Mod Pathol. 2007;20:570–578. doi: 10.1038/modpathol.3800772. [DOI] [PubMed] [Google Scholar]
- 42.Kim J, Jang KT, Kim KH, Park JW, Chang BJ, Lee KH, Lee JK, Heo JS, Choi SH, Choi DW, Rhee JC, Lee KT. Aberrant maspin expression is involved in early carcinogenesis of gallbladder cancer. Tumour Biol. 2010;31:471–476. doi: 10.1007/s13277-010-0056-2. [DOI] [PubMed] [Google Scholar]
- 43.Roelens J, Reuschenbach M, von Knebel Doeberitz M, Wentzensen N, Bergeron C, Arbyn M. p16INK4a immunocytochemistry versus human papillomavirus testing for triage of women with minor cytologic abnormalities: a systematic review and meta-analysis. Cancer Cytopathol. 2012;120:294–307. doi: 10.1002/cncy.21205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLaughlin-Drubin ME, Crum CP, Munger K. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci U S A. 2011;108:2130–2135. doi: 10.1073/pnas.1009933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiffman M, Khan MJ, Solomon D, Herrero R, Wacholder S, Hildesheim A, Rodriguez AC, Bratti MC, Wheeler CM, Burk RD. A study of the impact of adding HPV types to cervical cancer screening and triage tests. J Natl Cancer Inst. 2005;97:147–150. doi: 10.1093/jnci/dji014. [DOI] [PubMed] [Google Scholar]