Abstract
Homologous to the E6-associated protein carboxyl terminus domain containing 3 (HECTD3) is an E3 ubiquitin ligase which ubiquitinates caspase-8, caspase-9 and promotes cancer cell survival. Aberrant HECTD3 expression is frequently involved in various types of cancer progression. However, to date, the regulation of HECTD3 remains unclear. Here, we demonstrated that miR-153 functions as a negative regulator of HECTD3 and sensitizes cisplatin-induced apoptosis in triple-negative breast cancer cells MDA-MB-231 and BT-549. Luciferase reporter assay demonstrated that miR-153 suppresses HECTD3 expression through directly targeting its mRNA within the 3’-Untranslated Region (3’UTR). Additionally, the expression levels of miR-153 and HECTD3 are inversely correlated in breast cancer cell lines. Furthermore, ectopic expression of miR-153 promotes apoptosis in MDA-MB-231 and BT-549 cells treated with cisplatin or TNF-α, and miR-153 inhibitor treatment inhibits cisplatin induced apoptosis in MDA-MB-231 and BT-549 cells. Moreover, stable overexpression of HECTD3 abrogates the sensitization effect of miR-153 to cisplatin treatment in MDA-MB-231 cells, and miR-153 inhibitor protects cells against cisplatin cytotoxicity in control cells, but not in the stable knockdown HECTD3 MDA-MB-231 cells. More importantly, breast cancer patients with higher expression levels of miR-153 had significant higher 5-year survival rate in PROGmiR database (P<0.05). Taken together, our study indicated that miR-153 inhibits TNBC survival by targeting HECTD3 and functions as a potent tumor suppressor.
Keywords: HECTD3, miR-153, apoptosis
Introduction
Breast cancer is the most frequently diagnosed malignancy and the second leading cause of cancer-related death among females worldwide [1]. Triple negative breast cancer (TNBC) is the most aggressive subtype of breast cancer, known as being negative for human epidermal growth factor receptor 2 (HER2), estrogen receptor (ER), and progesterone receptor (PR), which results in shorting of effective targets in clinical therapies with a poor prognosis [2]. During the past few decades, a large amount of study focused on uncovering the molecular mechanisms underlying the pathogenesis of TNBC, in which apoptosis-associated genes contribute to this process by evasion of apoptosis. However, the pathogenesis is still not completely understood. Therefore, it is urgent to clarify the TNBC aggressiveness and identify novel, efficient targets for therapeutic intervention.
Ubiquitination as one of the crucial protein modifications in eukaryotes is involved in regulating an enormous range of biological processes, such as apoptosis that plays an important role in tumorigenesis and development [3,4]. Therefore, dysfunction of ubiquitination may lead to the occurrence of diseases including breast cancer [5,6]. Many E3s potentially act as oncogenes or tumor suppressors owing to their genetic and expression alterations in cancer. HECTD3 is an E3 ubiquitin ligase belonging to the HECT (homologous to E6-AP COOH terminus) domain-containing E3s. Previous studies have revealed that HECTD3 could promote cell survival through targeting MALT1 and caspase8 in a nondegradative approach [7,8]. Moreover, it is frequently overexpressed in breast carcinomas [8]. As a potential oncogene, there is not yet any relevant report about the regulation of HECTD3 at present. Therefore, we determine to study the post-transcriptional regulation of HECTD3.
MicroRNAs (miRNAs) are a class of endogenous small noncoding single-stranded RNAs of approximately 20-22 nucleotides (nt) long. miRNAs regulate the expression of protein-coding genes by binding to the complementary sequence of the mRNA 3’UTR [9]. To date, there are over 2,500 miRNAs that have been identified in human genome [10]. miRNAs participate in regulating almost all vital cellular processes, including development, differentiation, proliferation and apoptosis. And numerous studies have uncovered that dysregulation of miRNAs is associated with the initiation, progression and metastasis of human cancers, including mammary carcinoma [11]. The TNBC subtype exhibits the greatest resistance to chemotherapy. Growing evidences have indicated a strong association between miRNAs and chemosensitivity. For instance, miRNA-26b, miR-497 and other miRNAs enhance the chemosensitivity of cancer cells [12,13]. Since the important roles of miRNAs in cancers and the limitations of nowadays available diagnostic methods having exposed, miRNAs are emerging as diagnostic, prognostic and predictive biomarkers.
In this study, we identified that miR-153 can directly target and suppress HECTD3 expression. Moreover, miR-153 sensitizes TNBC cells (MDA-MB-231) to apoptosis by down-regulating HECTD3. As above, miR-153 seems to be a tumor suppressor miRNA (tsmiR), which is compatible with the information in PROGmiR database that miR-153 positively correlates with the 5-year survival rate of breast cancer. These findings establish a new mechanism of apoptosis regulation pathway, and provide a novel potential prognostic biomarker and therapeutic target.
Materials and methods
Cell culture
The MDA-MB-231, BT-549, MDA-MB-468, SKBR3, MCF7 and SW527 cell lines used in this study were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in DMEM containing 10% fetal bovine serum (FBS). HEK293T cells were cultured in DMEM containing 10% FBS.
Plasmid construction and Transient Transfection
The full-length 3’UTR of HECTD3 was cloned from HEK-293T genomic DNA and then inserted into the pIS0 luciferase vector to generate pIS0-HECTD3-3’UTR. Mutant construct of HECTD3 3’UTR, named pIS0-HECTD3-3’UTR-MUT, which carried a substitution of six nucleotides within the core binding sites of HECTD3-3’UTR, was carried out using Site-Directed Mutagenesis Kit (SBS Genetech, Beijing, China). HECTD3-pLenti6 was described in a previous study [8].
Transient transfection for plasmids and miRNAs was performed using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions.
Antibodies and other reagents
The anti-HECTD3 rabbit polyclonal antibody was described in a previous study [7]. The anti-β-actin mouse monoclonal Ab, anti-cleaved-caspase-7, anti-cleaved-caspase-3 and anti-caspase-3 antibodys were acquired from Cell Signaling (Danvers, MA). The anti-PARP and anti-HA antibodys were from Santa Cruz Biotechnology (Santa Cruz, CA). All miRNAs and inhibitor of miR-153 were purchased from GenePharma (Shanghai, China). Cisplatin was purchased from Hospira (Austrilia).
Lentivirus production and transduction
Virus particles were harvested 48 hours after pLenti6 and HECTD3-pLenti6 constructs transfection with the PLP1, PLP2 and PLP/VSVG constructs into HEK293T cells by using Lipofectamine 2000 reagent (Invitrogen). MDA-MB-231 cells were infected with lentivirus plus 5 mg/ml polybrene (Sigma, St Louis, MO, USA).
Real-time quantitative PCR (RT-qPCR)
Total RNA was extracted with TRIzol (Invitrogen, USA) and reverse-transcribed into cDNA using Quantscript RT kit produced by Tiangen (Beijing, China) in accordance with the manufacturer’s protocol. RT-qPCR was carried out using SYBR Premix Ex TaqTM II (TaKaRa, Japan) on Step-one plus real-time PCR system (Applied Biosystems, Foster City, CA, USA) with miR-153 and HECTD3 specific primers. The primer sequences were as followed: hsa-miR-153-R: CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGATCACTTT; hsa-miR-153-F: ACACTCCAGCTGGGTTGCATAGTCACAAAAGT; HECTD3-F: GGGCTACGAGACCACAGACG; HECTD3-R: GCAAGGCCAAGAGGGACAC.
Cell viability analysis (SRB assay)
Cells in 24-well plates were fixed in situ by gentle addition of 0.4 ml cold TCA (10%) per well and incubated for 1 hour at 4°C. Supernatant was discarded and plates were washed 5 times with water and air dried. 400 µl of sulforhodamine B (SRB) solution 0.4% (w/v) in 1% acetic acid was added to each well, and plates incubated for 10 minutes at RT. After staining, unbound dye was removed by washing 5 times with 1% acetic acid and plates were air dried. Bound stain was solubilized with 10 mM Tris base. Absorbance was read on an automated plate reader at 515 nm. The cell viability experiments were conducted in triplicate. The data were pooled to generate means ± standard deviation and analyzed by t-test.
Dual luciferase assay
HEK293T cells were seeded in 24-well plates. The next day, the cells were transfected with the HECTD3 3’UTR luciferase reporter constructs for 24 hours, and an internal control pRL-β-actin combined with miR-153 and control in triplicate. The cells were continually cultured for 24 hours, and luciferase activities were measured by using the dual luciferase reporter assay system (Promega, Madison, WI, USA).
Statistical analysis
Data presented are mean ± SD of a minimum of three independent experiments. The statistical significance was determined by Student’s unpaired t-test for all experiments. P values less than 0.05 were considered to be of statistical significance. Data and statistical analysis were performed using GraphPad Prism version 5.01 for Windows, GraphPad Software (La Jolla, CA, USA).
Results
MiR-153 directly targets HECTD3
We have previously demonstrated that HECTD3 is an oncogene and promotes cancer cell survival. However, the regulation of HECTD3 is yet unknown. Here, we identified eight putative miRNA-binding sites in the 3’UTR of HECTD3 mRNA by three algorithms: TargetScan, miRanda, and Pictar (Figure 1A). To narrow down the scope, we respectively transfected these eight miRNAs and control into MDA-MB-231 cells. The HECTD3 expression was determined using Western blot and five miRNAs (miR-26b, miR-1297, miR-153, miR-939 and miR-875-5p) were found to significantly down-regulate HECTD3 expression level (Figure 1B and 1C). To further test the specific regulation through the predicted binding sites, we cloned the 3’UTR of HECTD3 into the pISO reporter plasmid downstream of luciferase gene and transfected miRNA mimics or control and reporter plasmids into HEK293T cells. The luciferase reporter assays showed that miR-153 prominently suppressed the luciferase activity for over 80% (P<0.001) and other four miRNAs suppressed only for 40-60% (Figure 1D). Furthermore, this suppressive effect of miR-153 was rescued by several nucleotide substitutions in the core binding sites (Figure 1E, 1F). These results suggested that miR-153 can directly target and suppress the HECTD3 expression in MDA-MB-231 cells.
Figure 1.
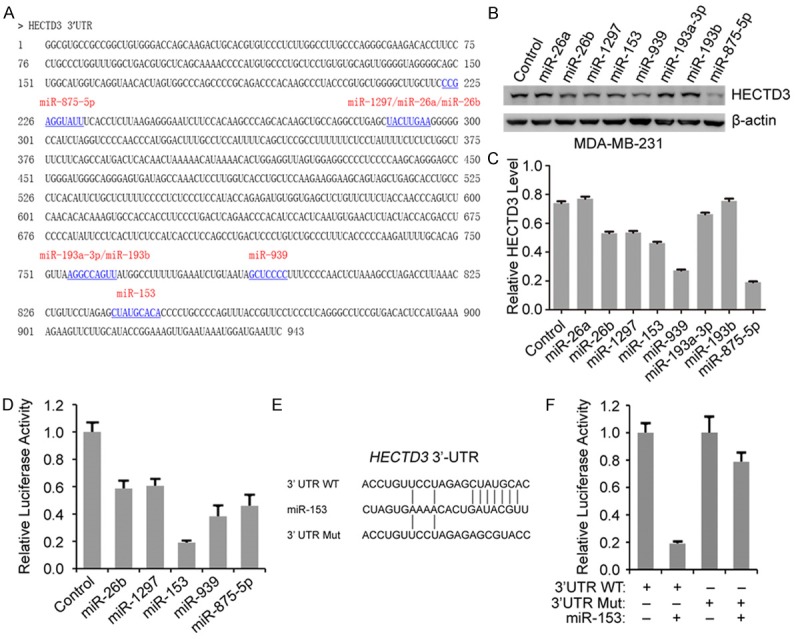
MiR-153 directly targets HECTD3 mRNA 3’UTR. A. Sequence of the 3’UTR region of HECTD3 with putative miRNA-binding sites (underlined) predicted by three algorithms: TargetScan, miRanda, and Pictar. B. Western blot analysis for HECTD3 protein expression level in MDA-MB-231 cells after transfection with eight miRNAs as indicated for 2 days. C. HECTD3 levels were quantified in panel B by Image J software. D. HEK293T cells were transfected with the HECTD3 3’UTR luciferase reporter and an internal control pRL-β-actin constructs combined with indicated miRNAs or control for 24 hours. Then the luciferase activities were determined with Dual-Luciferase Reporter System. E. Sequences of wild type and mutant miR-153-binding sites in the 3’UTR of HECTD3 mRNA. F. HEK293T cells were transfected with the HECTD3 3’UTR or HECTD3 3’UTR-MUT luciferase reporter and an internal control pRL-β-actin combined with miR-153 and control for 24 hours, and luciferase activities were measured.
To further investigate the relationship between HECTD3 and miR-153, we measured the HECTD3 protein level and mature miR-153 level in five breast cancer cell lines (MDA-MB-468, SKBR3, MCF7, SW527 and MDA-MB-231) and an immortalized human breast epithelial cell line 184A1. Compared with 184A1, HECTD3 is over-expressed in the breast cancer cell lines (Figure 2A, 2B) and miR-153 is down-regulated in the five breast cancer cell lines (Figure 2C). The results revealed that there is an inverse relationship between miR-153 and HECTD3, which further confirmed that miR-153 negatively regulates HECTD3.
Figure 2.
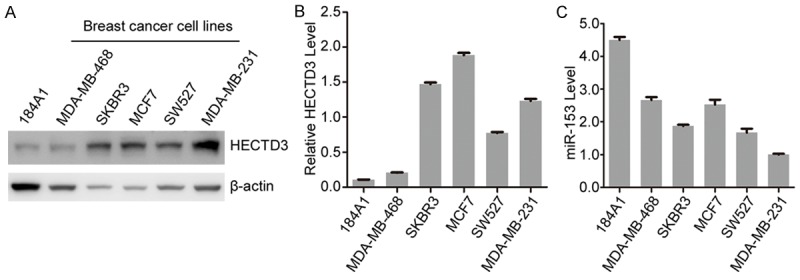
MiR-153 is negatively correlated to HECTD3 in breast cancer cell lines. (A) The expression of the HECTD3 protein in all the above breast cell lines was determined by Western blot. (B) HECTD3 levels were quantified as indicated in panel (A) by Image J software. (C) The miR-153 expression levels of the six breast cell lines were detected by RT-qPCR.
MiR-153 promotes apoptosis of breast cancer cells in the presence of TRAIL and cisplatin through targeting HECTD3
Based on miR-153 down-regulating HECTD3 and being involved in apoptosis regulation, we postulated that re-expression of miR-153 could sensitize breast cancer cells to apoptosis inducers (such as TNF-α and cisplatin). Hence, we transfected miR-153 mimics or control (NC oligonucleotides) into MDA-MB-231 cells treated with or without TNF-α and cisplatin. The miR-153 and HECTD3 mRNA expression were determined by qRT-PCR and HECTD3 protein level, apoptosis were determined by Western blot. As expected, the endogenous HECTD3 mRNA level was significantly repressed by overexpressing miR-153 (Figure 3A). Meanwhile, apoptosis indicated by cleaved caspase-3/7 was increased in MDA-MB-231 cells (Figure 3B) and BT-549 cells (Figure 3C) with miR-153 transfection in the presence of TNF-α or cisplatin. Subsequently, cell viability was measured by SRB assay to characterize the function of miR-153 in MDA-MB-231 cells. As shown in Figure 3D, cell viability was significantly decreased in the miR-153 transfected cells treated with cisplatin. These data are consistent with our previous report that HECTD3 depletion could sensitize cancer cells to apoptotic stimuli. On the other hand, the inhibitor of miR-153 significantly up-regulates the endogenous HECTD3 mRNA and protein level through decreasing miR-153 level in MDA-MB-231 cells (Figure 4A, 4B). Moreover, apoptosis indicated by cleaved caspase-3/7 was decreased in MDA-MB-231 cells (Figure 4B) and BT-549 cells (Figure 4C) with miR-153 transfection in the presence of cisplatin, and cell viability was significantly increased in MDA-MB-231 cells transfected the miR-153 inhibitor in the presence of cisplatin (Figure 4D).
Figure 3.
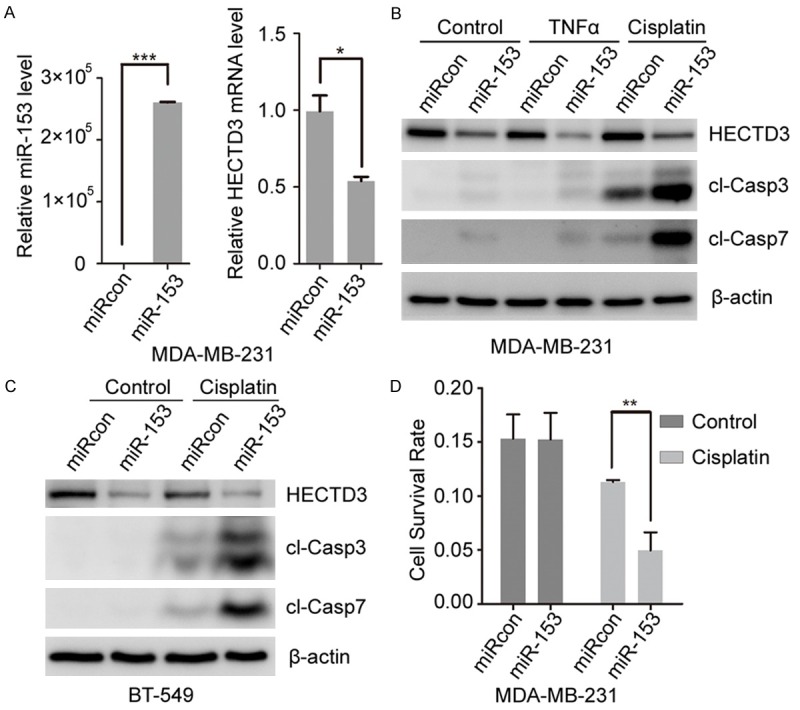
MiR-153 sensitized TRAIL and cisplatin induced cell apoptosis in MDA-MB-231 cells. (A) MDA-MB-231 cells were transfected with miR-153 for 2 days, the expression of miR-153 and HECTD3 mRNA were measured by RT-qPCR. (B, C) MDA-MB-231 (B) and BT-549 (C) cells were transfected with miR-153 for one day and treated with cisplatin or TNF-α overnight, and then apoptosis was measured by cleaved caspase3 (cl-casp3) and cleaved caspase7 (cl-casp7). (D) MDA-MB-231 cells were transfected with miR-153 for one day and treated with cisplatin overnight, and cell viability was determined by the SRB assay.
Figure 4.
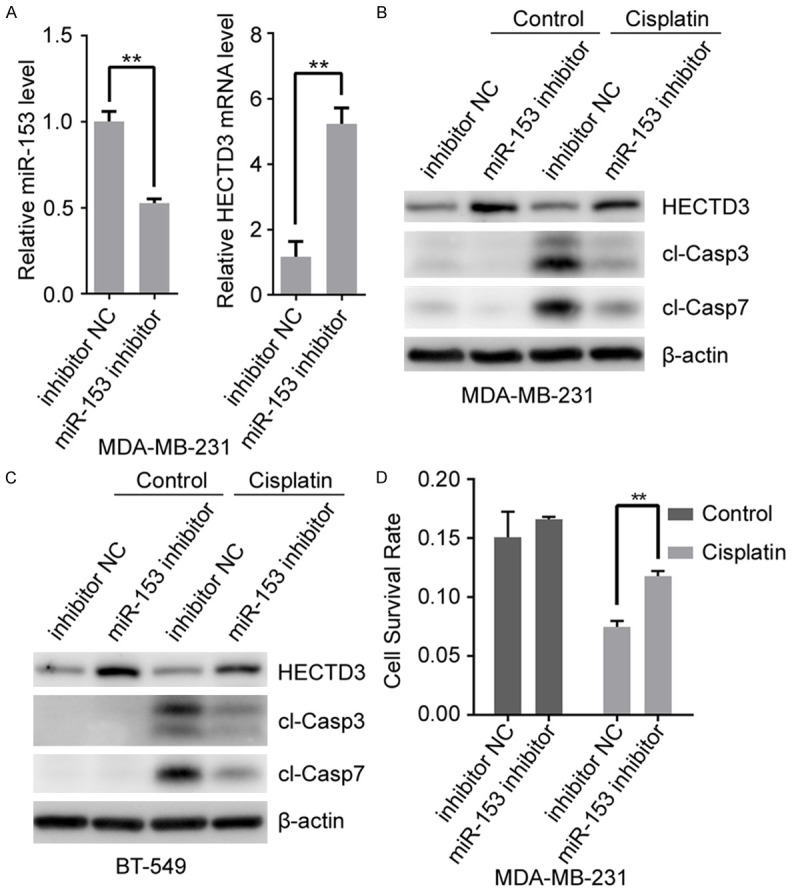
MiR-153 inhibitor protects cell against cisplatin induced apoptosis. (A) MDA-MB-231 cells were transfected with miR-153 inhibitor for 2 days, the expression of miR-153 and HECTD3 mRNA were measured by RT-qPCR. (B, C)MDA-MB-231 (B) and BT-549 (C) cells were transfected with miR-153 inhibitor for one day and treated with cisplatin overnight, and then apoptosis was measured by cleaved caspase3 (cl-casp3) and cleaved caspase7 (cl-casp7). (D) MDA-MB-231 cells were transfected with miR-153 inhibitor for one day and treated with cisplatin overnight, and cell viability was determined by the SRB assay.
To confirm that miR-153 sensitized cisplatin-induced cytotoxicity through down-regulating HECTD3, we transfected miR-153 into stable overexpressing HECTD3 and control MDA-MB-231 cells by lentivirus system (Figure 5A) and treated the cells with cisplatin, and apoptosis was determined by Western blot. As shown in Figure 5B, overexpression of HECTD3 inhibited the cytotoxicity of miR-153 combining with cisplatin. On the other hand, we transfected miR-153 inhibitor into stable knockdown HECTD3 and control MDA-MB-231 cells by lentivirus system (Figure 5C) and treated the cells with cisplatin. The results demonstrated that miR-153 inhibitor inhibits cisplatin induced apoptosis in control cells, but not in the stable knockdown HECTD3 MDA-MB-231 cells (Figure 5D).
Figure 5.
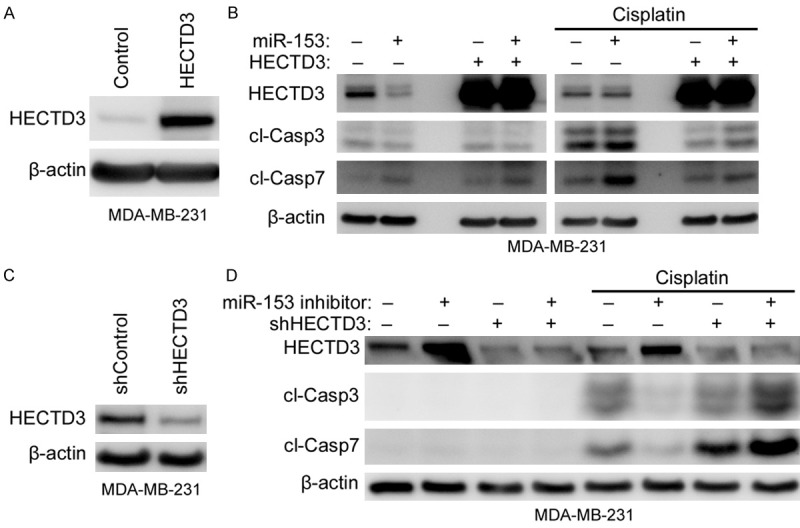
MiR-153 sensitized cancer cells to cisplatin induced apoptosis through targeting HECTD3. (A, B) Stable overexpressing HECTD3 or control MDA-MB-231 cells (A) were transfected with miR-153 for one day and treated with cisplatin overnight, and then apoptosis was measured by cl-casp3 and cl-casp7 (B). (C, D) Stable knockdown HECTD3 or control MDA-MB-231 cells (C) were transfected with miR-153 inhibitor for one day and treated with cisplatin overnight, and then apoptosis was measured by cl-casp3 and cl-casp7 (D).
Collectively, these results suggested that HECTD3 is a direct target of miR-153, and the regulatory effect of miR-153 on breast cancer cell apoptosis is at least in part through the down-regulation of HECTD3.
To further investigate the impact of miR-153 on the breast cancer outcome, Kaplan-Meier survival analysis of patients with breast cancer in PROGmiR database was performed. The result demonstrated that the 5-year survival rate was remarkably elevated in patients with higher expression of miR-153 (Figure 6). Down-regulation of miR-153 predicted poor prognosis, which suggested that miR-153 might play an important role in breast cancer progression.
Figure 6.
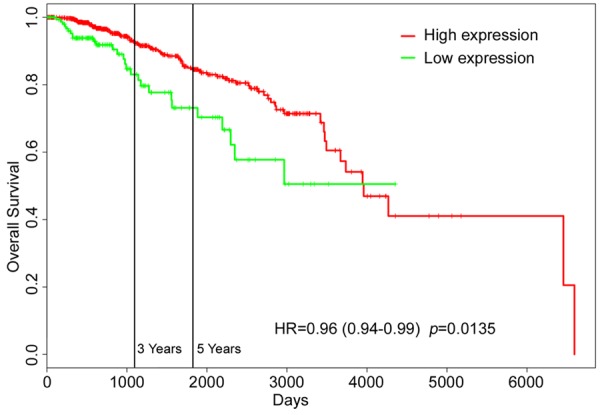
MiR-153 is associated with 5-year survival rate of breast cancer patients. Kaplan-Meier survival analysis of patients with breast cancer, stratified by expression of miR-153. Data was obtained from the PROGmiR database.
Discussion
Although TNBC represents less than 20% of all breast cancer types, it accounts for a disproportionate and large number of breast cancer deaths with poor outcomes [14,15]. TNBC exhibiting resistance to chemotherapeutic drugs has become a priority outstanding issue we are being confronted with, therefore there is an urgent need to develop new therapies to prevent chemoresistance. Meanwhile increasing evidence revealed that multiple genes and signalling pathways are involved in this process [16]. As previously reported, HECTD3 decreases chemosensitivity of TNBC and may become a prospective target for TNBC therapy.
miRNAs have become an accepted important post-transcriptional regulation since its discovery. Previous study showed that more than half of the human miRNA genes are located in cancer-associated genomic regions or in fragile sites, which suggested that miRNAs play a key role in oncogenesis [17]. It was reported that miR-153 could negatively regulate metastases of gastric cancer, inhibit epithelial-to-mesenchymal transition (EMT) in hepatocellular carcinoma and breast cancer, and suppress migration and invasion of human non-small-cell lung cancer [18-21]. Together, these results revealed that miR-153 probably acts as a tumor suppressor in human cancers although there was a report demonstrated that miR-153 silencing induces apoptosis in TNBC cell line MDA-MB-231 [22]. Our data demonstrated that miR-153 promotes TNBC cell MDA-MB-231 and BT-549 TNF-α- and cisplatin-mediated apoptosis by suppressing HECTD3 expression. This regulatory relationship was confirmed by the negative correlation of miR-153 and HECTD3 in breast cancer cell lines. Down-regulation of HECTD3 mediated by miR-153 significantly sensitizes cisplatin-induced apoptosis and decreases viability in cancer cells, and exogenous HECTD3 could efficiently eliminate the sensitization of miR-153 to cisplatin-induced cytotoxicity.
As we have discussed above, miR-153 targets several critical genes regulating metastasis, apoptosis and EMT in human cancers. Hence, the treatment effect of miR-153 is much better than therapies aiming at one single oncogene. Our data implicated combined treatment with miR-153 and chemotherapeutic drugs might effectively enhance chemotherapy-induced cytotoxicity in TNBC cells, which had tremendous clinical significance. Additionally, miR-153 is positively associated with the 5-year survival rate of breast cancer and high level of miR-153 predicates good prognosis in PROGmiR database. We speculated that miR-153 mainly plays a key role in apoptosis of TNBC cells. Taken together, our results indicated that miR-153 functions as a tumor suppressor in TNBC, which suggested that the miR-153-HECTD3 axis might be a potential therapeutic target for TNBC.
Acknowledgements
This project was funded by National Basic Research Program of China (2013CB911004) (to Y. L.).
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 3.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 4.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 5.Chaugule VK, Walden H. Specificity and disease in the ubiquitin system. Biochem Soc Trans. 2016;44:212–227. doi: 10.1042/BST20150209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohta T, Wu W, Koike A, Asakawa H, Koizumi H, Fukuda M. Contemplating chemosensitivity of basal-like breast cancer based on BRCA1 dysfunction. Breast Cancer. 2009;16:268–274. doi: 10.1007/s12282-009-0115-y. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Chen X, Wang Z, Zhao D, Chen H, Chen W, Zhou Z, Zhang J, Li H, Chen C. The HECTD3 E3 ubiquitin ligase suppresses cisplatin-induced apoptosis via stabilizing MALT1. Neoplasia. 2013;15:39–48. doi: 10.1593/neo.121362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Kong Y, Zhou Z, Chen H, Wang Z, Hsieh YC, Zhao D, Zhi X, Huang J, Zhang J, Li H, Chen C. The HECTD3 E3 ubiquitin ligase facilitates cancer cell survival by promoting K63-linked polyubiquitination of caspase-8. Cell Death Dis. 2013;4:e935. doi: 10.1038/cddis.2013.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762–776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 12.Wang L, Jiang CF, Li DM, Ge X, Shi ZM, Li CY, Liu X, Yin Y, Zhen L, Liu LZ, Jiang BH. MicroRNA-497 inhibits tumor growth and increases chemosensitivity to 5-fluorouracil treatment by targeting KSR1. Oncotarget. 2016;7:2660–2671. doi: 10.18632/oncotarget.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao N, Wang R, Zhou L, Zhu Y, Gong J, Zhuang SM. MicroRNA-26b suppresses the NF-kappaB signaling and enhances the chemosensitivity of hepatocellular carcinoma cells by targeting TAK1 and TAB3. Mol Cancer. 2014;13:35. doi: 10.1186/1476-4598-13-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnedos M, Bihan C, Delaloge S, Andre F. Triple-negative breast cancer: are we making headway at least? Ther Adv Med Oncol. 2012;4:195–210. doi: 10.1177/1758834012444711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng M, Wu Z, Wu A, Huang Z, He N, Xie X. MiR-145 promotes TNF-alpha-induced apoptosis by facilitating the formation of RIP1-FADDcaspase-8 complex in triple-negative breast cancer. Tumour Biol. 2016 doi: 10.1007/s13277-015-4631-4. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 16.O’Reilly EA, Gubbins L, Sharma S, Tully R, Guang MH, Weiner-Gorzel K, McCaffrey J, Harrison M, Furlong F, Kell M, McCann A. The fate of chemoresistance in triple negative breast cancer (TNBC) BBA Clin. 2015;3:257–275. doi: 10.1016/j.bbacli.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song YX, Yue ZY, Wang ZN, Xu YY, Luo Y, Xu HM, Zhang X, Jiang L, Xing CZ, Zhang Y. MicroRNA-148b is frequently down-regulated in gastric cancer and acts as a tumor suppressor by inhibiting cell proliferation. Mol Cancer. 2011;10:1. doi: 10.1186/1476-4598-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Zhai L, Zhao C, Lv S. MiR-153 inhibits epithelial-mesenchymal transition by targeting metadherin in human breast cancer. Breast Cancer Res Treat. 2015;150:501–509. doi: 10.1007/s10549-015-3346-y. [DOI] [PubMed] [Google Scholar]
- 19.Shan N, Shen L, Wang J, He D, Duan C. MiR-153 inhibits migration and invasion of human non-small-cell lung cancer by targeting ADAM19. Biochem Biophys Res Commun. 2015;456:385–391. doi: 10.1016/j.bbrc.2014.11.093. [DOI] [PubMed] [Google Scholar]
- 20.Wang Z, Liu C. MiR-153 regulates metastases of gastric cancer through Snail. Tumour Biol. 2015 doi: 10.1007/s13277-015-3846-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 21.Xia W, Ma X, Li X, Dong H, Yi J, Zeng W, Yang Z. miR-153 inhibits epithelial-to-mesenchymal transition in hepatocellular carcinoma by targeting Snail. Oncol Rep. 2015;34:655–662. doi: 10.3892/or.2015.4008. [DOI] [PubMed] [Google Scholar]
- 22.Anaya-Ruiz M, Cebada J, Delgado-Lopez G, Sanchez-Vazquez ML, Perez-Santos JL. miR-153 silencing induces apoptosis in the MDA-MB-231 breast cancer cell line. Asian Pac J Cancer Prev. 2013;14:2983–2986. [PubMed] [Google Scholar]


