Abstract
SMAD7 is a key inhibitor of transforming growth factor β (TGFβ) receptor signaling, which regulates the alteration of cancer cell invasiveness through epithelial-mesenchymal cell conversion. Since microRNAs (miRNAs) play a potential role in the tumorigenesis, cancer cell growth and metastases of oral squamous cell carcinoma (OSCC), determination of the involved miRNAs that may regulate SMAD7-mediated OSCC cell invasion appears to be one important question. Here, we found that the levels of miR-497 were significantly increased and the levels of SMAD7 were significantly decreased in OSCC specimens, compared to the paired adjacent non-tumor tissue. Moreover, miR-497 and SMAD7 inversely correlated in OSCC specimens. The 5-year survival of the patients with higher miR-497 levels in the resected OSCC was worse than those high miR-497 levels. Bioinformatics analyses showed that miR-497 targeted the 3’-UTR of SMAD7 mRNA to inhibit its translation, which was proved by luciferase reporter assay. Furthermore, miR-497 overexpression increased SMAD7-suppressed cell invasion, while miR-497 depletion decreased SMAD7-suppressed cell invasion in OSCC cells, in both a transwell cell invasion assay and a scratch would healing assay. Together, our data suggest that suppression of miR-497 in OSCC cells may promote cancer cell invasion via suppression of SMAD7, and highlight miR-497 as an intriguing therapeutic target to prevent OSCC metastases.
Keywords: Aral squamous cell carcinoma (OSCC), SMAD7, miR-497, cancer cell invasion
Introduction
Oral squamous cell carcinoma (OSCC) appear to be malignant due to its aggressive characteristics [1-5]. Hence, studies on the molecular regulation of OSCC invasion are critical for generating novel therapeutic strategies. The epithelial-mesenchymal transition (EMT) is characterized by the feature of loss of cell polarity and cell-cell adhesion of epithelial cells to allow them to gain migratory and invasive properties [6-8]. EMT is essential for numerous biological processes during development, and in wound healing, organ fibrotic remodeling and in cancer initiation and progression [6-8]. Among all factors that induce EMT, transforming growth factor β1 (TGFβ1) is the most important and potential one [6-14]. TGFβ receptor signaling initiates by the binding of a ligand to a type II TGFβ receptor, which catalyzes the phosphorylation of a type I TGFβ receptor, and subsequently the phosphorylation of two intracellular proteins SMAD2 and SMAD3 to form heteromeric complexes with SMAD4. The activated SMAD complexes are then translocated to the nucleus, where they modulate gene transcription [9,15,16]. SMAD7 is a general antagonist against TGFβ receptor signaling. Activation of TGFβ receptor signaling is essential for EMT to occur in cancer [17,18]. Thus, SMAD7 appeared to be a tumor suppressor.
MicroRNAs (miRNAs) are non-coding small RNAs that have established functions in regulation of the protein translation through their base-pairing with the 3’-untranslated region (3’-UTR) of the target mRNAs [19,20]. It is well-known that miRNAs regulate cancinogenesis [21-23]. Among all miRNAs, miR-497 has been only recently recognized as a tumor-associated microRNA in breast cancer [24-27], colorectal carcinoma [28], and lung cancer [29]. However, a role of miR-497 in CRC has not been ill-defined before.
Here, we studied the regulation of SMAD7-regulated OSCC cell invasion by miRNAs. We found that the levels of miR-497 were significantly increased and the levels of SMAD7 were significantly decreased in OSCC specimens, compared to the paired adjacent non-tumor tissue. Moreover, miR-497 and SMAD7 inversely correlated in OSCC specimens. The 5-year survival of the patients with higher miR-497 levels in the resected OSCC was worse than those high miR-497 levels. Bioinformatics analyses showed that miR-497 targeted the 3’-UTR of SMAD7 mRNA to inhibit its translation, which was proved by luciferase reporter assay. Furthermore, miR-497 overexpression increased SMAD7-suppressed cell invasion, while miR-497 depletion decreased SMAD7-suppressed cell invasion in OSCC cells, in both a transwell cell invasion assay and a scratch would healing assay.
Materials and methods
Patient tissue specimens
A total of 30 resected specimens from OSCC patients were collected from 2010 to 2014 in at Hospital of Stomatology affiliated to Zhejiang University, and were included in this study. OSCC specimens were compared with the paired adjacent non-tumor bone tissue (NT). All specimens had been histologically and clinically diagnosed at Hospital of Stomatology affiliated to Zhejiang University. All specimens had been histologically and clinically diagnosed independently by two experienced pathologists. For the use of these clinical materials for research purposes, prior patient’s consents and approval from the Institutional Research Ethics Committee were obtained.
Culture of a human OSCC cell line
SCC-15 is a human OSCC line purchased from American Type Culture Collection (ATCC, Rockville, MD, USA), and was cultured in RPMI 1640 medium supplemented with 15% heat-inactivated fetal bovine serum (FBS; Sigma-Aldrich, St Louis, MO, USA), 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Carlsbad, CA, USA) in a humidified atmosphere of 5% CO2 at 37°C.
Plasmid transfection
MiR-497-modulating and SMAD7-modulating plasmids were prepared from a backbone plasmid containing a GFP reporter under CMV promoter (pcDNA3.1-CMV-GFP, Clontech, Mountain View, CA, USA). The miR-497 mimic, or antisense, or control null, or short-hairpin interfering RNA for SMAD7 (shSMAD7) was all purchased from Sigma-Aldrich, and digested with Xhol and BamHI and then subcloned with a 2A into the pcDNA3.1-CMV-GFP plasmid. Sequencing was performed to confirm the correct orientation of the new plasmid. Transfection was done using Lipofectamine 2000 reagent (Invitrogen), according to the instructions of the manufacturer. One day after transfection, the transfected cells were purified by flow cytometry based on GFP expression.
Western blot
The protein was extracted from the patients’ specimens, or from the cultured cells, and then subjected to regular procedure for Western blot. The membrane blots were first probed with a primary antibody. After incubation with horseradish peroxidase-conjugated second antibody, autoradiograms were prepared using the enhanced chemiluminescent system to visualize the protein antigen. The signals were recorded using X-ray film. Primary antibodies were rabbit anti-SMAD7 and anti-α-tubulin (Cell Signaling, San Jose, CA, USA). Secondary antibody is HRP-conjugated anti-rabbit (Jackson ImmunoResearch Labs, West Grove, PA, USA). Blotting images were representatives from 5 repeats. α-tubulin was used as a protein loading control.
RT-qPCR
Total RNA was extracted from resected specimens or from cultured cells using a miRNeasy mini kit (Qiagen, Hilden, Germany). Complementary DNA (cDNA) was randomly primed from total RNA using the Omniscript reverse transcription kit (Qiagen). Quantitative PCR (RT-qPCR) were performed in duplicates with QuantiTect SYBR Green PCR Kit (Qiagen). All primers were purchased from Qiagen. Data were collected and analyzed using 2-ΔΔCt method for quantification of the relative mRNA expression levels. Values of genes were first normalized against α-tubulin, and then compared to controls.
MicroRNA target prediction and 3’-UTR luciferase-reporter assay
MiRNAs targets were predicted with the algorithms TargetSan (https://www.targetscan.org) [30]. Luciferase-reporters were successfully constructed using molecular cloning technology. The SMAD7 3’-UTR reporter plasmid (SMAD7 3’-UTR) and SMAD7 3’-UTR reporter plasmid with a mutant at the miR-497 binding site (SMAD7 3’-UTR mut) were purchased from Creative Biogene (Shirley, NY, USA). OSCC cells were co-transfected with SMAD7 3’-UTR/SMAD7 3’-UTR mut and miR-497/as-miR-497/null by Lipofectamine 2000 (5×104 cells per well). Cells were collected 24 hours after transfection for assay using the dual-luciferase reporter assay system gene assay kit (Promega, Beijing, China), according to the manufacturer’s instructions. The normalized control was null-transfected OSCC cells with 3’-UTR of SMAD7 mRNA (wild type).
Transwell cell invasion assay
Cells (104) were plated into the top side of polycarbonate transwell filter coated with Matrigel in the upper chamber of the BioCoatTM Invasion Chambers (Becton-Dickinson Biosciences, Bedford, MA, USA) and incubated at 37°C for 22 hours. The cells inside the upper chamber with cotton swabs were then removed. Invasive cells on the lower membrane surface were fixed, stained with hematoxylin, and counted for 10 random 100X fields per well. Cell counts are expressed as the mean number of cells per field of view. Five independent experiments were performed and the data are presented as mean ± standard deviation (SD).
Scratch wound healing assay
Cells were seeded in 24-well plates at a density of 104 cells/well in complete media and cultured to confluence. The cell monolayer was serum starved overnight before experiment. Confluent cell monolayer were then scraped with a pipette tip to generate scratch wounds followed by cell debris removal. Cells were then incubated at 37°C for 24 hours. Time lapse images were captured after 12 hours, after which the migration areas are determined by subtracting the wound area at the indicated time periods from the initial wound area, using by NIH ImageJ software (Bethesda, MA, USA).
Statistical analysis
All statistical analyses were carried out using the SPSS 17.0 statistical software package. Bivariate correlations were calculated by Spearman’s Rank Correlation Coefficients. Kaplan-Meier curves were sued to analyze the patient survival by miR-497 levels. All values are depicted as mean ± SD and are considered significant if p < 0.05. All data were statistically analyzed using one-way ANOVA with a Bonferroni correction, followed by Fisher’s Exact Test for comparison of two groups.
Results
Increased miR-497 correlates with decreased SMAD7 in OSCC specimens
In 30 OSCC specimens, we detected significantly lower levels of SMAD7 by Western blot, compared to paired adjacent non-tumor tissue (NT; Figure 1A). Moreover, we detected significantly higher levels of miR-497 in OSCC specimens, compared to NT (Figure 1B). Then we examined the relationship between miR-497 and SMAD7 in OSCC specimens. Thus, we performed a correlation test using the 30 OSCC specimens. A strong inverse correlation was detected between miR-497 and SMAD7 (Figure 1C, ɤ=-0.72, p<0.0001, n=30), suggesting that a regulatory relationship between miR-497 and SMAD7 may be present in OSCC. Next, we investigated whether the levels of miR-497 in OSCC tissue may correlate with 5-year survival of the patients. The median value for miR-497 in all 30 cases was chosen as the cutoff point for separating miR-497-high cases (n=15) from miR-497-low cases (n=15). Kaplan-Meier curves were performed, showing that miR-497-high OSCC patients had a significantly shorter 5-year survival, compared to miR-497-low OSCC patients (Figure 1D).
Figure 1.
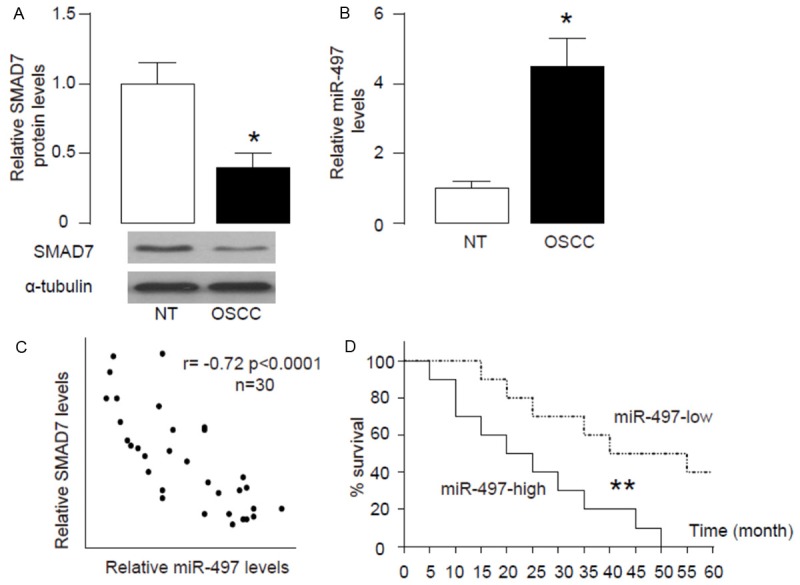
Increased miR-497 correlates with decreased SMAD7 in OSCC specimens. (A, B) Western blot for SMAD7 (A) and RT-qPCR on miR-497 (B) were performed on paired OSCC and the adjacent non-tumor bone tissue (NT) from 30 patients. (C) A correlation test between SMAD7 and miR-497 (ɤ=-0.72, p<0.0001, N=30). (D) The median value for miR-497 in all 30 cases was chosen as the cutoff point for separating miR-497-high cases (n=15) from miR-497-low cases (n=15). Kaplan-Meier curves were performed, showing that miR-497-high OSCC patients had a significantly worse 5-year survival, compared to miR-497-low OSCC patients. *p<0.05. **p<0.01. N=30.
MiR-497 targets 3’-UTR of SMAD7 mRNA to inhibit its protein translation
In order to prove presence of a regulatory relationship between miR-497 and SMAD7 in OSCC cells, we used bioinformatics analyses to predict binding of miR-497 on SMAD7 mRNA. We detected a specific miR-497 binding site on the 3’-UTR (from 55th to 62th base site) of the SMAD7 mRNA (Figure 2A). Next, we analyzed whether this binding of miR-497 to SMAD7 mRNA may alter the SMAD7 levels. Thus, we either overexpressed miR-497, or inhibited miR-497 in a human OSCC cell line SCC-15, by a miR-497-expressing plasmid, or a plasmid carrying miR-497 antisense (as-miR-497), respectively. The SCC-15 cells were also transfected with a null plasmid, to be used as a control for miR-497 modification (Nul). First of all, the alteration of miR-497 levels in SCC-15 cells was confirmed by RT-qPCR (Figure 2B). SCC-15 cells were then transfected with 1 μg plasmids of miR-497-modification plasmids, with plasmids carrying a luciferase reporter for 3’-UTR of SMAD7 mRNA or a luciferase reporter for 3’-UTR of SMAD7 mRNA with mutate at the miR-497 binding site (mut). The luciferase activities were determined in these cells, and our data showed that SMAD7 3’-UTR plus miR-497 had the most repression for SMAD7, and the 3’-UTR SMAD7 mutant plus miR-497 had much lower repression. Moreover, the 3’-UTR in the presence of as-miR-454 restored expression of SMAD7 (Figure 2C). These data demonstrate that miR-497 may target 3’-UTR of SMAD7 mRNA to inhibit its translation.
Figure 2.
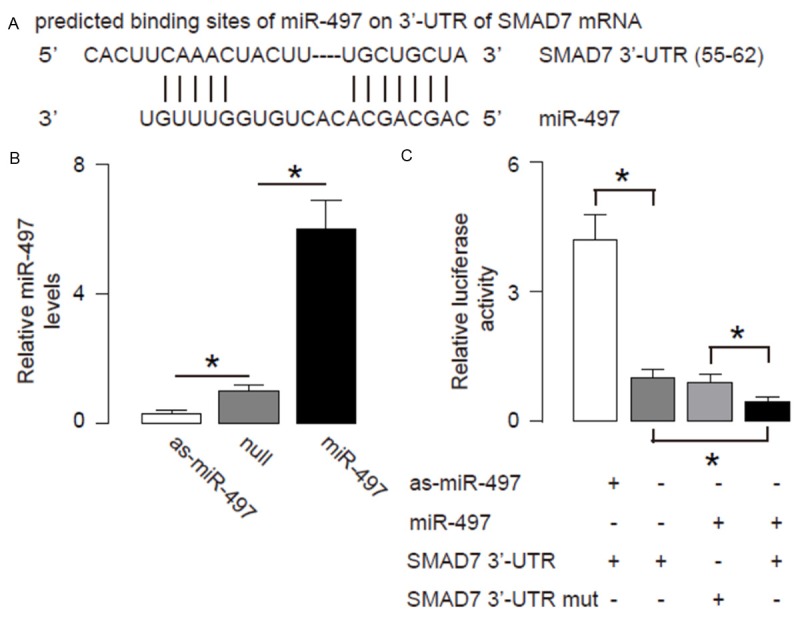
MiR-497 targets 3’-UTR of SMAD7 mRNA to inhibit its protein translation. A. Bioinformatics analyses of binding of miR-497 to the 3’-UTR of SMAD7 mRNA. B-C. We either overexpressed miR-497, or inhibited miR-497 in OSCC cells by transfecting the cells with a miR-497-expressing plasmid, or with a plasmid carrying miR-497 antisense (as-miR-497). The OSCC cells were also transfected with a null plasmid as a control (null). B. RT-qPCR for miR-497 in miR-497 modified OSCC cells. C. MiR-497-modified OSCC cells were then transfected with 1 μg of SMAD7 3’-UTR luciferase-reporter/3’-UTR of SMAD7 mRNA with one mutate at the miR-497 binding site (mut) and miR-497-modified plasmids. The luciferase activities were quantified. *p<0.05. N=5.
MiR-497 reduces SMAD7 protein in OSCC cells
Next, we analyzed the effects of modification of miR-497 levels on SMAD7 in OSCC cells. In order to confirm that the effects of miR-497 depletion on SMAD7, we further knocked down SMAD7 by shRNA in OSCC cells that had expressed antisense of miR-497 (as-miR-497-shSMAD7). We found that alteration of miR-497 in OSCC cells did not change SMAD7 mRNA (Figure 3A). However, overexpression of miR-497 significantly decreased SMAD7 protein (Figure 3B). On the other hand, inhibition of miR-497 significantly increased SMAD7 protein (Figure 3B). These data suggest that MiR-497 inhibits SMAD7 protein translation in OSCC cells.
Figure 3.
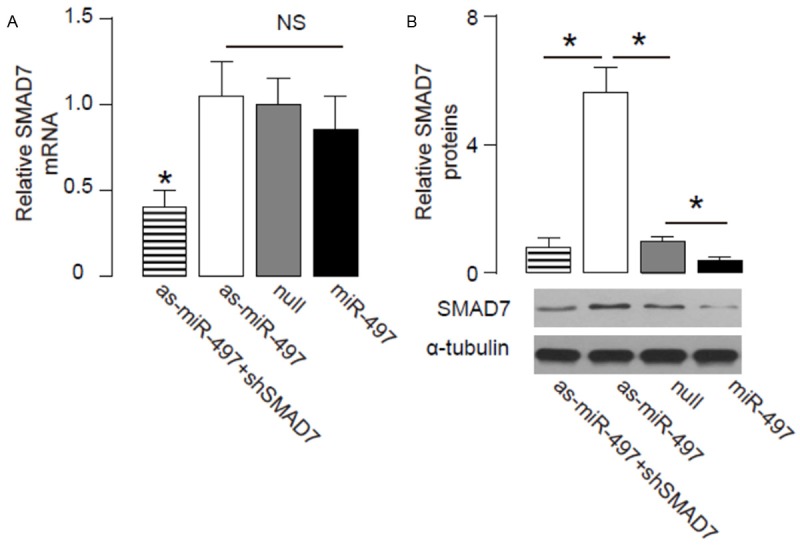
MiR-497 reduces SMAD7 protein in OSCC cells. (A-B) RT-qPCR (A) and Western blot (B) for SMAD7 in miR-497 (plus SMAD7)-modified cells. *p<0.05. NS: non-significant. N=5.
MiR-497 suppresses OSCC cell invasion
We found that overexpression of miR-497 resulted in decreases in cell invasion of OSCC cells in a transwell cell invasion assay, shown by quantification (Figure 4A), and by representative images (Figure 4B). Similarly, depletion of miR-497 resulted in increases in cell invasion of OSCC cells, shown by quantification (Figure 4A), and by representative images (Figure 4B). SMAD7 depletion abolished the effects of as-miR-497 expression on cell invasion (Figure 4A, 4B). We also used scratch wound healing assay to confirm our finding in transwell cell invasion assay. We found that overexpression of miR-497 resulted in decreases in cell migration in vitro (Figure 5A, 5B). Similarly, depletion of miR-497 resulted in increases in cell migration in vitro (Figure 5A, 5B). SMAD7 depletion abolished the effects of as-miR-497 expression on cell migration (Figure 5A, 5B). Together, our data suggest that miR-497 inhibits OSCC cell invasion through SMAD7 suppression (Figure 6).
Figure 4.
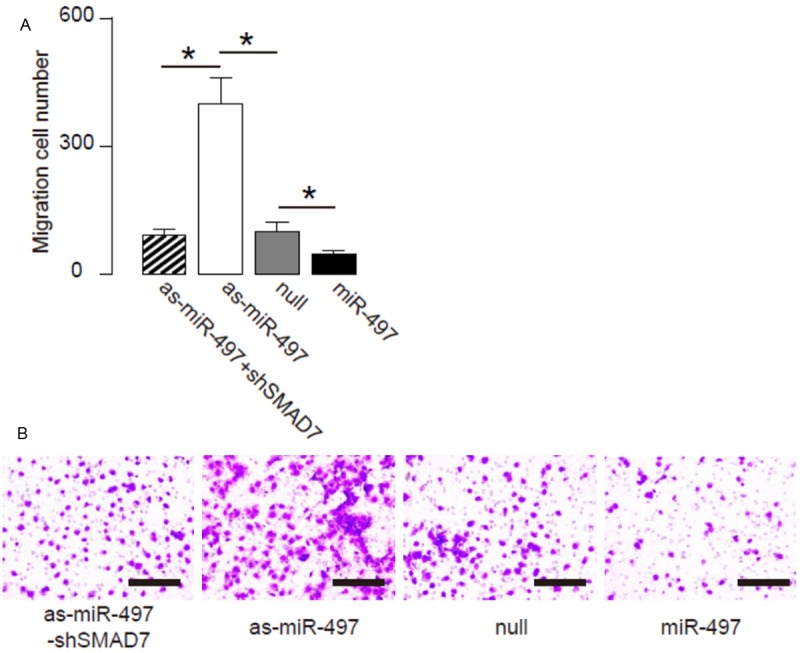
MiR-497 suppresses OSCC cell invasion in a transwell cell invasion assay. (A, B) Cell invasiveness of miR-497 (plus SMAD7)-modified OSCC cells in a transwell cell invasion assay, shown by quantification (A), and by representative images (B). *p<0.05. N=5.
Figure 5.
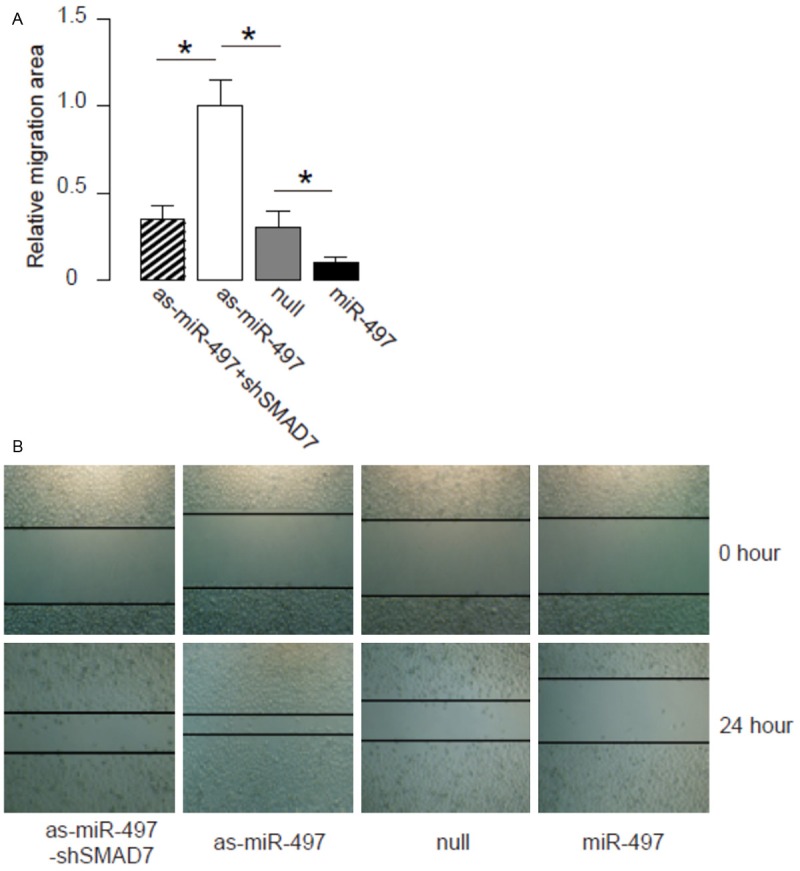
MiR-497 suppresses OSCC cell migration in a scratch wound healing assay. (A, B) Cell invasiveness of miR-497 (plus SMAD7)-modified OSCC cells in a scratch wound healing assay, shown by quantification (A), and by representative images (B). *p<0.05. N=5.
Figure 6.
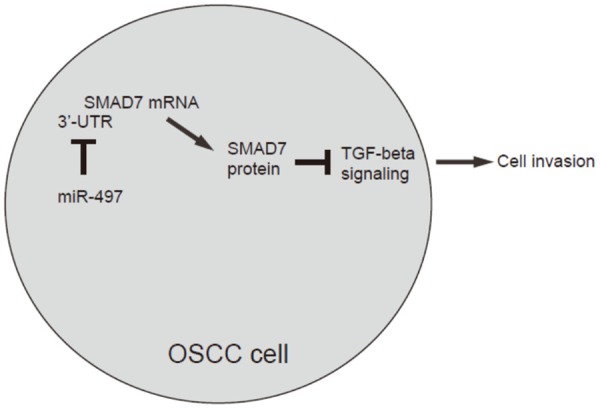
Schematic of the model MiR-497 inhibits OSCC cell invasion and migration through SMAD7 suppression.
Discussion
MiRNAs have demonstrated roles in the invasion and metastases of malignant tumor cells. However, miR-497 has been only recently recognized as a tumor-associated microRNA in breast cancer [24-27], colorectal carcinoma [28], and lung cancer [29]. For example, Shen et al. showed that miR-497 may serve as a tumor suppressor gene in breast cancer. They found that up-regulation of miR-497 expression caused cellular growth inhibition and apoptotic enhancement, as well as G0/G1 phase arrest, suggesting miR-497 as a potential therapeutic target for the treatment of breast cancer [24]. Moreover, Jiang et al. showed that the regulation of MMP7 by Nrdp1 in colorectal carcinoma cells could be inhibited by miR-497 through suppressing Nrdp1 translation, which highlights a novel molecular regulatory machinery that regulates metastasis of colorectal carcinoma [28]. Most recently, Gu et al. reported that miR-497 played a role in suppression of VEGF-A-mediated non-small cell lung cancer cell growth and invasion [29]. However, none of the above studies showed that SMAD7 could be target for miR-497.
To the best of our knowledge, we are the first to report miR-497 as a suppressor of SMAD7 in OSCC. Since we aimed to study the regulation of SMAD7 in OSCC, we performed sequence matching, and only found several miRNAs that target SMAD7. However, most of these candidate miRNAs did not alter their expression level in OSCC, compared to NT. On the other hand, we specifically detected a significant increase in miR-497 in OSCC specimens, compared to NT. Most importantly, the levels of miR-497 and SMAD7 inversely correlated. Thus, we hypothesize that miR-497 may target and regulate SMAD7 in OSCC cells. Moreover, high miR-497 was found to be associated with an overall poor prognosis, and may be useful for predicting 5-year survival.
Using in vitro assay, we further demonstrate that alteration in miR-497 levels does not affect SMAD7 mRNA, but regulated SMAD7 protein level. Using promoter luciferase assay, we found that miR-497 inhibited SMAD7 through translation suppression. Moreover, miR-497-induced SMAD7 in OSCC cells directly regulated cell invasion, independently in transwell cell invasion assay and in a scratch wound healing assay. These data suggest that miR-497/SMAD7 regulatory axis may play a critical role in regulation of OSCC cell invasion. When a miRNA molecule is attached as a perfect match to a target mRNA, it causes the mRNA degradation therefore the mRNA levels would be diminished by a RT-qPCR, which was not the case in the current study. These data suggest that a partial interaction between the miR-497 and the 3’-UTR of the SMAD7 gene. In case that the miRNA does not form a perfect match with the mRNA from the target gene, the translation process stops at that point and hence the protein production is reduced, as described here.
To summarize, here we propose a model that miR-497 enhances OSCC metastases through SMAD7 inhibition. UP-regulation of miR-497 appears to directly contribute to the distal metastases of primary OSCC and subsequently poor prognosis. Together, our study highlights miR-497 as a promising novel target for treating OSCC via preventing OSCC metastases.
Disclosure of conflict of interest
None.
References
- 1.Bodner L, Manor E, Friger MD, van der Waal I. Oral squamous cell carcinoma in patients twenty years of age or younger--review and analysis of 186 reported cases. Oral Oncol. 2014;50:84–89. doi: 10.1016/j.oraloncology.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Zafereo ME. Evaluation and staging of squamous cell carcinoma of the oral cavity and oropharynx: limitations despite technological breakthroughs. Otolaryngol Clin North Am. 2013;46:599–613. doi: 10.1016/j.otc.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Andisheh-Tadbir A, Khademi B, Kamali F, Fattahi MJ, Malekzadeh M, Taghva M. Upregulation of serum vascular endothelial growth factor and matrix metalloproteinase-3 in patients with oral squamous cell carcinoma. Tumour Biol. 2014;35:5689–5693. doi: 10.1007/s13277-014-1753-z. [DOI] [PubMed] [Google Scholar]
- 4.Zhang E, Liu S, Xu Z, Huang S, Tan X, Sun C, Lu L. Pituitary tumor-transforming gene 1 (PTTG1) is overexpressed in oral squamous cell carcinoma (OSCC) and promotes migration, invasion and epithelial-mesenchymal transition (EMT) in SCC15 cells. Tumour Biol. 2014;35:8801–8811. doi: 10.1007/s13277-014-2143-2. [DOI] [PubMed] [Google Scholar]
- 5.Bu J, Bu X, Liu B, Chen F, Chen P. Inhibition of metastasis of oral squamous cell carcinoma by anti-PLGF treatment. Tumour Biol. 2015;36:2695–2701. doi: 10.1007/s13277-014-2892-y. [DOI] [PubMed] [Google Scholar]
- 6.Willis BC, Borok Z. TGF-beta-induced EMT: mechanisms and implications for fibrotic lung disease. Am J Physiol Lung Cell Mol Physiol. 2007;293:L525–534. doi: 10.1152/ajplung.00163.2007. [DOI] [PubMed] [Google Scholar]
- 7.Morrison CD, Parvani JG, Schiemann WP. The relevance of the TGF-beta Paradox to EMT-MET programs. Cancer Lett. 2013;341:30–40. doi: 10.1016/j.canlet.2013.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heldin CH, Vanlandewijck M, Moustakas A. Regulation of EMT by TGFbeta in cancer. FEBS Lett. 2012;586:1959–1970. doi: 10.1016/j.febslet.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 9.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao X, Gaffar I, Guo P, Wiersch J, Fischbach S, Peirish L, Song Z, El-Gohary Y, Prasadan K, Shiota C, Gittes GK. M2 macrophages promote beta-cell proliferation by up-regulation of SMAD7. Proc Natl Acad Sci U S A. 2014;111:E1211–1220. doi: 10.1073/pnas.1321347111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yi JJ, Barnes AP, Hand R, Polleux F, Ehlers MD. TGF-beta signaling specifies axons during brain development. Cell. 2010;142:144–157. doi: 10.1016/j.cell.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewen ME, Sluss HK, Whitehouse LL, Livingston DM. TGF beta inhibition of Cdk4 synthesis is linked to cell cycle arrest. Cell. 1993;74:1009–1020. doi: 10.1016/0092-8674(93)90723-4. [DOI] [PubMed] [Google Scholar]
- 13.Naka K, Hoshii T, Muraguchi T, Tadokoro Y, Ooshio T, Kondo Y, Nakao S, Motoyama N, Hirao A. TGF-beta-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature. 2010;463:676–680. doi: 10.1038/nature08734. [DOI] [PubMed] [Google Scholar]
- 14.Xiao X, Wiersch J, El-Gohary Y, Guo P, Prasadan K, Paredes J, Welsh C, Shiota C, Gittes GK. TGFbeta Receptor Signaling Is Essential for Inflammation-Induced but Not beta-Cell Workload-Induced beta-Cell Proliferation. Diabetes. 2013;62:1217–1226. doi: 10.2337/db12-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Fan Q, He S, Tang T, Liao Y, Xie J. MicroRNA-21 negatively regulates Treg cells through a TGF-beta1/Smad-independent pathway in patients with coronary heart disease. Cell Physiol Biochem. 2015;37:866–878. doi: 10.1159/000430214. [DOI] [PubMed] [Google Scholar]
- 16.Ji Y, Gao F, Sun B, Hao J, Liu Z. Angiotensin-Converting Enzyme 2 Inhibits Apoptosis of Pulmonary Endothelial Cells During Acute Lung Injury Through Suppressing SMAD2 Phosphorylation. Cell Physiol Biochem. 2015;35:2203–2212. doi: 10.1159/000374025. [DOI] [PubMed] [Google Scholar]
- 17.Smith AL, Iwanaga R, Drasin DJ, Micalizzi DS, Vartuli RL, Tan AC, Ford HL. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;31:5162–5171. doi: 10.1038/onc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Li Z, He W, Xu L, Wang J, Shi J, Sheng M. Effects of Astragaloside IV Against the TGF-beta1-Induced Epithelial-to-Mesenchymal Transition in Peritoneal Mesothelial Cells by Promoting Smad 7 Expression. Cell Physiol Biochem. 2015;37:43–54. doi: 10.1159/000430332. [DOI] [PubMed] [Google Scholar]
- 19.Di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23:3–11. doi: 10.1016/j.gde.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereira DM, Rodrigues PM, Borralho PM, Rodrigues CM. Delivering the promise of miRNA cancer therapeutics. Drug Discov Today. 2013;18:282–289. doi: 10.1016/j.drudis.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Mei Q, Li F, Quan H, Liu Y, Xu H. Busulfan inhibits growth of human osteosarcoma through miR-200 family microRNAs in vitro and in vivo. Cancer Sci. 2014;105:755–762. doi: 10.1111/cas.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang F, Xiao W, Sun J, Han D, Zhu Y. MiRNA-181c inhibits EGFR-signaling-dependent MMP9 activation via suppressing Akt phosphorylation in glioblastoma. Tumour Biol. 2014;35:8653–8658. doi: 10.1007/s13277-014-2131-6. [DOI] [PubMed] [Google Scholar]
- 23.Liu G, Jiang C, Li D, Wang R, Wang W. MiRNA-34a inhibits EGFR-signaling-dependent MMP7 activation in gastric cancer. Tumour Biol. 2014;35:9801–9806. doi: 10.1007/s13277-014-2273-6. [DOI] [PubMed] [Google Scholar]
- 24.Shen L, Li J, Xu L, Ma J, Li H, Xiao X, Zhao J, Fang L. miR-497 induces apoptosis of breast cancer cells by targeting Bcl-w. Exp Ther Med. 2012;3:475–480. doi: 10.3892/etm.2011.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang Y, Zou C, Zhang X, Liu S, Wang X, Zhao D, Sun Q, Zeng Z, Dress A, Lin MC, Kung HF, Rui H, Liu LZ, Mao F, Jiang BH, Lai L. Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. Clin Cancer Res. 2011;17:1722–1730. doi: 10.1158/1078-0432.CCR-10-1800. [DOI] [PubMed] [Google Scholar]
- 26.Wang S, Li H, Wang J, Wang D. Expression of microRNA-497 and its prognostic significance in human breast cancer. Diagn Pathol. 2013;8:172. doi: 10.1186/1746-1596-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Luo Q, Li X, Gao Y, Long Y, Chen L, Huang Y, Fang L. MiRNA-497 regulates cell growth and invasion by targeting cyclin E1 in breast cancer. Cancer Cell Int. 2013;13:95. doi: 10.1186/1475-2867-13-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Y, Meng Q, Qi J, Shen H, Sun S. MiR-497 promotes metastasis of colorectal cancer cells through Nrdp1 inhibition. Tumour Biol. 2015;36:7641–7647. doi: 10.1007/s13277-015-3489-9. [DOI] [PubMed] [Google Scholar]
- 29.Gu A, Lu J, Wang W, Shi C, Han B, Yao M. Role of miR-497 in VEGF-A-mediated cancer cell growth and invasion in non-small cell lung cancer. Int J Biochem Cell Biol. 2016;70:118–125. doi: 10.1016/j.biocel.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 30.Coronnello C, Benos PV. ComiR: Combinatorial microRNA target prediction tool. Nucleic Acids Res. 2013;41:W159–164. doi: 10.1093/nar/gkt379. [DOI] [PMC free article] [PubMed] [Google Scholar]


