Abstract
Dysfunction of ribosome biogenesis induces divergent ribosome-related diseases including ribosomopathy and occasionally results in carcinogenesis. Although many defects in ribosome-related genes have been investigated, little is known about contribution of ribosomal RNA (rRNA) in ribosome-related disorders. Meanwhile, microRNA (miRNA), an important regulator of gene expression, is derived from both coding and noncoding region of the genome and is implicated in various diseases. Therefore, we performed in silico analyses using M-fold, TargetScan, GeneCoDia3, and so forth to investigate RNA relationships between rRNA and miRNA against cellular stresses. We have previously shown that miRNA synergism is significantly correlated with disease and the miRNA package is implicated in memory for diseases; therefore, quantum Dynamic Nexus Score (DNS) was also calculated using MESer program. As a result, seventeen RNA sequences identical with known miRNAs were detected in the human rRNA and termed as rRNA-hosted miRNA analogs (rmiRNAs). Eleven of them were predicted to form stem-loop structures as pre-miRNAs, and especially one stem-loop was completely identical with hsa-pre-miR-3678 located in the non-rDNA region. Thus, these rmiRNAs showed significantly high DNS values, participation in regulation of cancer-related pathways, and interaction with nucleolar RNAs, suggesting that rmiRNAs may be stress-responsible resident miRNAs which transmit stress-tuning information in multiple levels.
1. Introduction
It has recently been revealed that dysregulation of ribosome biogenesis is implicated in various diseases termed ribosomopathy such as Diamond-Blackfan anemia (DBA), Shwachman-Diamond syndrome (SDS), X-linked dyskeratosis congenita (DKC), Treacher Collins syndrome (TCS), and cartilage hair hypoplasia (CHH) [1–3]. The most studied ribosomopathy, DBA, is a rare congenital hypoplastic anemia and its pathogenesis is associated with defects in various ribosomal protein (RP) genes such as RPS19, RPS24, RPL5, and RPL11. Mutation in RPS and RPL genes results in significant reduction in the amount of mature 40S and 60S subunit, respectively [4]. Other ribosomopathies, SDS, DKC, TCS, and CHH, are caused by gene defects on SBDS, DKC1, TCOF1, and RMRP, respectively, which encode proteins involved in ribosome biogenesis [2]. However, what mechanism is linked to these proteins in the pathogenesis of ribosomopathies? Whether cancer is related to them? These are still unsolved.
Ribosomal RNA (rRNA) is the most abundant noncoding RNA gene in cells and is essential for the structure and function of ribosomes. All four eukaryotic rRNAs, such as 18S, 5.8S, 28S, and 5S, are highly conserved across human and related species, and their biogenesis is strictly regulated by several mechanisms [5–8]. RNA45S, also called RN45S, the 45S gene, or rDNA, is an operon containing 18S, 5.8S, and 28S RNA genes [8–11]. On the other hand, the 5S RNA gene is coded alone. Among eukaryotes, the RNA45S and 5S RNA genes are transcribed by Pol I and Pol III, respectively [12, 13]. The first step in rRNA gene transcription in humans is the formation of the preinitiation complex (PIC) on the core promoter and the upstream control element of rDNA. PIC attracts Pol I, and a full-length rRNA precursor called 47S rRNA is transcribed. 47S rRNA is processed into 45S rRNA by cleaving fixed positions on the 3′ and 5′ external transcribed spacers (ETS) in the nucleus and is then divided into 21S and 32S rRNA by either of the two processes [8]. Finally, 18S-E, 6S, and 28S rRNAs are generated through various mechanisms and transported into the cytoplasm to construct the mature ribosomal complex.
RNA45S genes in humans are located on chromosomes (Chr) 13, 14, 15, 21, and 22 [14]. These acrocentric chromosomes have multiple copies of the 45S RNA gene on the p12 region in their short arms. This tandemly repeated rRNA gene copy is commonly called an rDNA repeat or rRNA gene cluster, and each repeating unit consists of a nontranscribed spacer (NTS) and the RNA45S gene. RNA45S also contains a 5′ ETS, an internal transcribed spacer (ITS), and a 3′ ETS in addition to the 18S, 5.8S, and 28S rRNA genes. On the other hand, the 5S rRNA gene is only located at the q42 region of chromosome 1. The copy number of rDNA is important for normal cell functions although the majority of rDNA copies are transcriptionally silent; therefore, reduced rDNA copy number after cell stress is repaired by a specific amplification system. It has also been reported that perturbation in the copy number and stability of rRNA gene caused by mutations in rRNA-related enzymes or cell senescence are linked to various cellular dysfunctions and insufficiency of genome integrity [15–18].
MicroRNA (miRNA) is an essential regulator of gene expression and a member of the small noncoding RNA family, which are RNAs approximately 22 nucleotides long [19]. Sequence complementarity-based interactions between miRNA and its target mRNA suppress and occasionally augment the translation of mRNAs into proteins [20–23]. One miRNA regulates multiple mRNAs; thus, one mRNA is targeted by multiple miRNAs [24–26]. Almost all functional genes in humans are under the control of miRNAs [27]. Therefore, alterations in the miRNA profile after injury, infection, or chemical treatment can alter various functions, such as immunoreactivity, cell proliferation and differentiation, apoptosis, and carcinogenesis [28–32]. The expression profile of miRNA genes is deeply associated with a considerable number of human diseases including cancer [28, 29]. There are several reports about miRNA dynamics after cell stresses, for instance, participation of poly(ADP-ribose) in controlling miRNA activity in the cytoplasm [33]. In the deep insight of miRNA-disease relationship, it needs huge efforts to make complete data for clinical validation of miRNA-mRNA associations in diseases. Therefore, it has been shown that computational analysis is required for miRNA research and increasing number of disease-related miRNA databases and computational analyses have recently been established [34, 35]. The miRNA genes are scattered throughout the genome, and miRNAs are created through many complexed processing pathways [36–38]. Most miRNA genes are transcribed by RNA polymerase II (Pol II) as hairpin-shaped primary miRNA (pri-miRNA), and the pri-miRNA is processed into pre-miRNA after cleavage of the 5′-cap and 3′-polyA tail by the microprocessor complex, which is composed of Drosha and DiGeorge syndrome chromosomal region 8 (DCGR8). These are the RNase III proteins and double-stranded RNA binding proteins, respectively. Subsequently, pre-miRNA is exported to the cytoplasm by exportin-5 and further processed into the miRNA: miRNA∗ duplex by cleavage of the 5′- and 3′-termini and loop domain by Dicer, which is an RNase III-like protein. This duplex is finally loaded into the RNA-induced silencing complex, and a duplex chain is selected thermodynamically to function as mature miRNA [39]. However, some noncanonical pathways are used to mature miRNA [36, 40]. For example, dme-mir-1003 is the first discovered mirtron, which is a pri-miRNA that exists as an intron of pre-mRNA and is processed into pre-miRNA without the Drosha canonical processor [41]. This means that all protein-coding, noncoding, intergenic, and intragenic regions can become miRNA hosts.
According to the RNA wave 2000 model advocated by Fujii, miRNA genes are the RNA information genes with four critical characteristics: (1) the miRNA gene is a mobile genetic element that induces transcriptional and posttranscriptional silencing via networking processes; (2) the RNA information supplied by miRNA genes expands to intracellular, intercellular, intraorgan, interorgan, intraspecies, and interspecies under a lifecycle in the global environment; (3) mobile miRNAs self-proliferate; and (4) cells contain resident and genomic miRNAs [42, 43]. miRNAs can be classified into genomic and resident miRNAs. The former are miRNAs preserved in DNA as miRNA genes, and the latter are miRNAs stored in a non-DNA form. The greatest difference between genomic and resident miRNAs is the expression regulatory mechanism. Most known miRNAs are genomic because their expression levels are controlled by a specific transcriptional factor, RNA polymerase, and so forth [44–46]. However, some miRNAs, such as mmu-miR-712, dme-miR-10404, and hsa-miR-663, are typical resident miRNAs because they do not require specific transcriptional factors or nucleases to exert their functions [47, 48].
In particular, it is anticipated that resident miRNAs and other cytoplasmic RNAs play more important roles in cells with unique cytoplasmic or genomic characteristics, such as erythrocytes, spermatozoa, and oocytes, than those of other cells. Erythrocytes contain diverse and abundant RNA species, including cytoplasmic miRNAs that contribute to regulating erythropoiesis and malarial resistance, although erythrocytes have been thought to contain no RNA because they are anucleated [49, 50]. Given that erythrocytes are the most abundant cell in blood, a large number of erythrocyte-contained miRNAs may be circulating. Spermatozoa are characterized by minimal cytoplasm and extremely condensed DNA. However, various RNAs are abundant in the cytoplasm of spermatozoa, such as rRNA, transfer RNA (tRNA), piwi-interacting RNA, and miRNA, and have important roles before and after fertilization [51–53]. Oocytes are transcriptionally silent cells; therefore, the many pooled mRNA and noncoding RNAs in the cytoplasm, such as miRNAs, are essential to complete late oogenesis and early embryogenesis without de novo transcription [53, 54]. Only the resident RNAs in these cells are considered information transmitters or memorizing devices, rather than DNA. Furthermore, as miRNA is self-reproducible, an identical miRNA could become both genomic and resident miRNA [42]. The quantities of tRNA and rRNA decrease under stress, suggesting that resident miRNAs help with biological regulation under stress [55]. Thus, we hypothesized that cytoplasmic tRNA and/or rRNA is a pool of self-reproducible resident miRNAs.
tRNA is another functional noncoding RNA that is most abundant (approximately 10% of RNAs) in cells next to rRNA (approximately 80% of RNAs). Recent studies have discovered that transfer RNA-derived RNA fragments (tRFs) are generated from tRNAs as terminal functional products. In the case of murine gammaherpesvirus 68, viral miRNAs were generated by Pol III [56]. Further, a number of endoribonucleases including Dicer and Angiogenin are implicated in the production of tRFs from tRNA transcripts [57, 58]. tRFs exist in various species, such as humans, cows, flies, and plants, and work as gene expression regulators, similar to miRNA [57, 59–63]. Some tRFs were listed as miRNAs in the miRBase (now dead entries). Other common characteristics between miRNAs and tRFs are their interactions with Argonaute (AGO) proteins, significant changes in expression levels during disease and aging, and circulation in a steady form [61–63]. Several reports have shown that tRFs are occasionally more abundant than miRNAs [64].
We considered the possibility that RNA fragments may be derived from rRNA in a manner similar to how tRFs are derived from tRNA because several tRF-related endoribonucleases have common activity of nuclease [58]. Till date, to the best of our knowledge, only a few studies have reported biogenesis and functions of rRNA-derived miRNAs or miRNA-like fragments, although many rRNA-annotated fragments of miRNA-like size have been detected in deep sequencing data from RNA studies [65]. Chak et al. revealed that the novel miRNA hairpin named mir-10404/mir-ITS1 exists in the ITS1 region of Drosophila rDNA [47]. Son et al. also discovered that mmu-miR-712 is coded in ITS2 of mouse 45S precursor RNA (Rn45s) and hsa-miR-663 is coded in the ITS1 region of human RNA45S [48]. Furthermore, Drosha-related proteins are included in rRNA processing pathways [66]. These ITS-derived miRNAs are supposed to be generated upon degradation of the ITS region, similar to the generation of mirtrons in the nucleoplasm or cytoplasm.
The effects of tRNA or rRNA degradation and processing on cell activities in response to stress are important. The small RNA molecules derived through this process play an important role in the transition from fine-tuning to stress-tuning functions. Other ncRNA species such as SINE, especially human Alu elements, have also been revealed to be contained in nucleolus and control the size of nucleoli adopting to cell circumstances [67].
Therefore, we examined whether rRNAs contain functional small RNAs and confirmed the relationship between ribosome and disease shown in previous studies. Moreover, how rRNA-hosted microRNA analogs (rmiRNAs) contribute to the stress response as nongenomic memory in the nucleolus and cytoplasm was also investigated using multiple computer-based tools and databases to find stress-tuning RNA interaction in transcriptional and posttranscriptional level. The quantum relationships among miRNAs were also calculated as Dynamic Nexus Score (DNS) by MESer program that we have previously developed and its significance in stress response was discussed.
2. Method
2.1. Sequence Data Collection
All miRNA sequence data used in this study were downloaded as miRNA.dat, hairpin.fa, and mature.fm from miRBase (http://www.mirbase.org) in release 21 (June 2014) [68]. This includes 2,588 and 1,915 mature miRNA sequences of human and mouse, respectively. Sequences of rRNAs were obtained from European Nucleotide Archive (ENA, http://www.ebi.ac.uk/ena) release 127 (April 6, 2016) and National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov) in FASTA format [69]. After comparing and merging latest rRNA sequence data, two rRNA coding sequences, RNA45S and human rDNA complete repeating unit, were selected as the source of rRNA sequence (Supplemental Table 1 in Supplementary Material available online at http://dx.doi.org/10.1155/2016/7562085). The latter source involves the sequence of the former but their sequences have some differences even in common regions, for instance, slightly polymorphisms in 18S and moderate ones in 28S, ITS, and ETS region. Sequences of tRNA and tRF were also obtained from Genomic tRNA database (GtRNAdb, http://gtrnadb.ucsc.edu) and tRFdb (http://genome.bioch.virginia.edu/trfdb), respectively [70, 71].
2.2. Definition of Passenger Strand
Passenger strands of miRNAs whose guide strands were found in the rRNA sequences were researched referring to stem-loop structure in miRBase. If a passenger strand is not recorded in miRBase, a sequence which is complement to the guide strand was defined as the passenger strand in this study.
2.3. Secondary Structure Prediction
To determine the secondary structures of found miRNA-like sequences, M-fold was used in a condition of 37 Celsius degrees and 1 M NaCl. Any other options which influence prediction results were set in default (RNA sequence is linear, percent suboptimality number is 5, upper bound on the number of computed foldings is 50, the window parameter is default, the maximum interior/bulge loop size is 30, the maximum asymmetry of an interior/bulge loop is 30, and the maximum distance between paired bases is no limit).
2.4. Chromosome Confirmation
For browsing miRNA locations on each chromosome visually, UCSC Genome Browser on Human Dec. 2013 (GRCh38/hg38) Assembly (https://genome.ucsc.edu/cgi-bin/hgTracks?db=hg38) was used.
2.5. Calculations of DNS
Dynamic Nexus Score (DNS) was prepared as a quantum-based score for evaluating quantum interactions between or among miRNAs [72]. DNS calculation of rRNA-derived miRNA and tRF was performed by using the original program, MESer (http://meser.mirna-academy.org). Computational results were statistically analyzed with Microsoft Office Excel 2013 (Microsoft Japan Co., Ltd., Tokyo, Japan).
2.6. Target Prediction and Ontology Analysis
Putative targets of rmiRNAs were predicted under the seed theory by using TargetScan (http://www.targetscan.org/) [73]. Validated targets of rmiRNAs were confirmed in miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) [74]. Selection of top 10 targets in miRTarBase was conducted by referring to the number of validation methods (primary) and the number of reports (secondary). Categorization of putative target genes in Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was accomplished by using GeneCoDis3 web service (http://genecodis.cnb.csic.es/) [75].
2.7. Alu Sequence and Target Site Prediction
Sequence data of human Alu family were downloaded from SINE Base (http://sines.eimb.ru), last update May 28, 2015 [76]. Target sites of rmiRNAs in Alu sequences were predicted using RNA22 version 2.0 (https://cm.jefferson.edu/rna22/) with default settings (sensitivity of 63%, specificity of 61%, seed size of 7, allow maximum of 1 UN-paired bases in seed, minimum number of paired-up bases in heteroduplex being 12, maximum folding energy for heteroduplex being −12 Kcal/mol, and maximum number of G : U wobbles allowed in seed region being no limit) [77].
3. Results
3.1. Pre-miRNA Sequence in Human rRNA
To investigate whether miRNAs also exist in human rRNA, we firstly collected base sequences of pre-miRNAs, mature miRNAs, and rDNA. Then the sequence of rRNA and its adjacent regions, RNA45S, and rDNA-repeating unit, respectively, were searched for 2,588 human pre-miRNA sequences by using a simple C++ based detection program we developed for this study. As a result, an identical sequence to pre-miR-3687 was detected from rDNA-repeating unit although the known location of the miR-3687 gene was distinct from rRNA coding region. However, other 2,587 pre-miRNA sequences were not found in any rRNA-related sequences. For further similar sequencing research, a detection of mature miRNA sequences instead of pre-miRNA from rRNA gene was also performed. Subsequently, seventeen RNA alignments identical to human mature miRNAs, namely, miR-663a, miR-663b-3p, miR-1268a, miR-1268b, miR-1275, miR-3648, miR-3656-3p, miR-3687-3p, miR-4417, miR-4466, miR-4488, miR-4492-3p, miR-4508, miR-4516, miR-4532, miR-6087, and miR-6724, were detected from the human rRNA sequences (Table 1). In detail, miR-1268a and miR-1268b were detected from only RNA45S, and miR-1275, miR-3687, and miR-6724 were detected from only rDNA-repeating unit.
Table 1.
Detected mature miRNAs from rRNA gene and adjacent region.
| miR name | Mature sequence | Region | Location |
|---|---|---|---|
| miR-663a | AGGCGGGGCGCCGCGGGACCGC | 5′ ETS | 2049–2071 |
| miR-663b | GGUGGCCCGGCCGUGCCUGAGG | 5′ ETS | 2113–2135 |
| miR-1268a | CGGGCGUGGUGGUGGGGG | 3′ ETS | 13102–13119 |
| miR-1268b | CGGGCGUGGUGGUGGGGGUG | 3′ ETS | 13102–13121 |
| miR-1275 | GUGGGGGAGAGGCUGUC | (NTS) | 42294–42310 |
| miR-3648 | AGCCGCGGGGAUCGCCGAGGG | 5′ ETS | 2513–2533 |
| miR-3656 | GGCGGGUGCGGGGGUGG | 28S | 8524–8540 |
| miR-3687 | CCCGGACAGGCGUUCGUGCGACGU | (5′ ETS) | 2888–2911 |
| miR-4417 | GGUGGGCUUCCCGGAGGG | 5′ ETS | 2412–2429 |
| miR-4466 | GGGUGCGGGCCGGCGGGG | (5′ ETS) | 631–648 |
| miR-4488 | AGGGGGCGGGCUCCGGCG | 28S | 8510–8527 |
| miR-4492 | GGGGCUGGGCGCGCGCC | 28S | 10851–10867 |
| miR-4508 | GCGGGGCUGGGCGCGCG | 28S | 10849–10865 |
| miR-4516 | GGGAGAAGGGUCGGGGC | 28S | 11049–11065 |
| miR-4532 | CCCCGGGGAGCCCGGCG | 28S | 11227–11243 |
| miR-6087 | UGAGGCGGGGGGGCGAGC | 28S | 12007–12024 |
| miR-6724 | CUGGGCCCGCGGCGGGCGUGGGG | (NTS) | 42320–42342 |
Seventeen sequences homologous to mature human miRNAs were detected from rRNA gene coding region. Note that miR-1268a and miR-1268b were found in only RNA45S and miR-1275, miR-3687, miR-4466, and miR-6724 were found in only rDNA-repeating unit. This data might be caused by differences in base alignment between two rRNA sequence data.
Among these detected miRNAs, miR-1268a, miR-3648, miR-3687, miR-4508, and miR-6724 were originated in rDNA containing chromosomes, Chr 15, Chr 21, Chr 21, Chr 15, and Chr 21, respectively. However, their locations were different from rRNA coding regions (data not shown). This suggests that the detected miRNAs might also have been transcribed from rRNA-related regions as well as above Chr loci or that the miRNAs may be generated through further processing of transcribed rRNA gene.
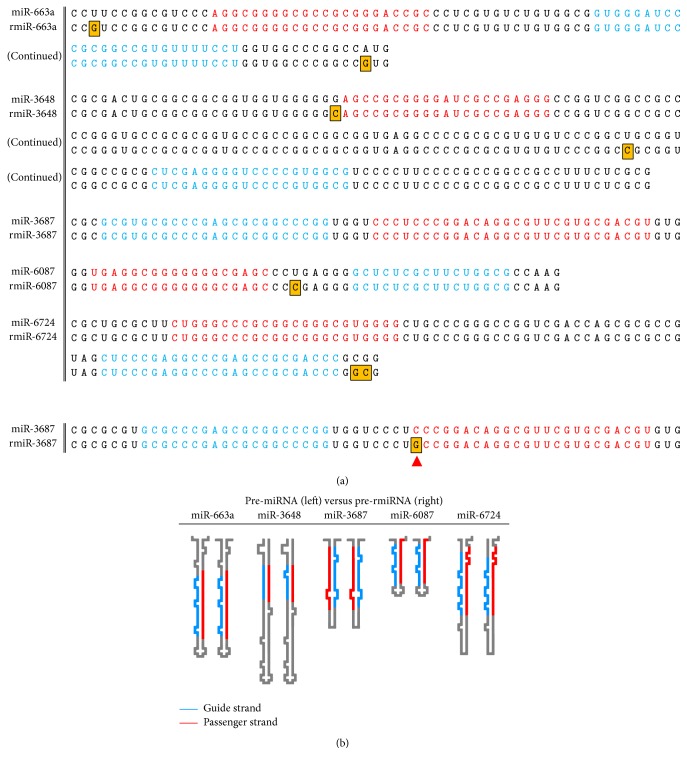
To further examine whether the detected miRNAs could form miRNA/miRNA∗ duplex and/or stem-loop structure like miR-3687, their passenger strand sequences were also searched in the rDNA repeat region. Passenger strand sequences were determined referring to their putative stem-loop structure in miRBase. As a result, passenger strands of miR-663a, miR-3648, miR-3687, miR-6087, and miR-6724 were found nearby their guide strands (Figure 1(a)). This result indicated that these rRNA-hosted miRNA-like RNAs could form stem-loop or at least miRNA/miRNA∗ duplex. Intriguingly, a passenger strand of miR-3687 was detected from RNA45S although its guide strand was not detected. Since these rRNA-hosted RNA pieces, especially miR-3687, possess high concordance rate to each known pre-miRNA sequence, therefore we termed them rRNA-hosted miRNA analog (rmiRNA).
Figure 1.
An overview of pre-rmiRNAs with high similarity to canonical pre-miRNAs. (a) Sequence comparison between detected precursor-rmiRNAs (pre-rmiRNAs) and their canonical pre-miRNAs. Mature (guide) miRNA strand is colored with red and passenger is colored with blue. Differences in base sequences between pre-rmiRNAs and pre-miRNAs are highlighted in yellow and box lines. (b) Comparison of secondary structure of pre-rmiRNAs with that of their canonical pre-miRNAs. These rmiRNAs contain the identical sequences to canonical ones in both guide and passenger. This group contains only a few polymorphisms in loop and terminal region.
For more rigorous verification, putative precursor sequences of rmiRNAs were predicted by referring to the sequences and structures of known human pre-miRNAs identical to detected rmiRNAs (Figure 1(a)). Subsequently, secondary structures of pre-rmiRNA sequences were predicted by using M-fold software. The same prediction for canonical pre-miRNA sequences were also performed and used as positive controls for comparison, and it was proven that all of five pre-rmiRNA candidates could form hairpin-loop structures which have high similarities to that original pre-miRNAs form (Figure 1(b)).
3.2. Exploration for Noncanonical Passenger Strands and Precursors
Since one pre-rmiRNA sequence is identical with pre-miR-3687 and four pre-rmiRNAs which have high similarity to known pre-miRNAs were detected in rDNA, we thought that other twelve rmiRNAs also could form stem-loop structure with different style. To examine this hypothesis, we have carefully investigated adjacent regions of detected guide strands. Primarily, some RNA sequences were clipped out as putative pre-rmiRNA. Each of them contained guide strand sequence and had the same length to its canonical pre-miRNA. Next, these putative pre-rmiRNAs were compared with its canonical pre-miRNA in base sequences and then secondary structures. Of twelve putative pre-rmiRs, pre-rmiR-663a showed the highest similarity to pre-miR-663b in both base sequence and precursor structure (Supplemental Figure 1A). Other three pre-rmiRNAs, pre-rmiR-3656, pre-rmiR-4417, pre-rmiR-4466, and pre-rmiR-4508 also showed high similarity in secondary structures to pre-miR-3656, pre-miR-4417, pre-miR-4466, and pre-miR-4508, respectively, although their precursor sequences showed low similarities to canonical ones (Supplemental Figure 2A). These results implied that these rmiRNAs have obtained new passenger strand to maintain their function as mature miRNAs. On the other hand, putative pre-rmiR-1268b sequence generated in accordance with the rules above did not form stem-loop structure according to M-fold prediction. However, we found that pre-rmiRNA-1268b could construct stem-loop structure with a slight modification such as lengthening of the 3′ terminal region (Supplemental Figure 1A).
Furthermore, it was ascertained that the left six rmiRNAs, namely, rmiR-1268a, rmiR-1275, rmiR-4488, rmiR-4492, rmiR-4516, and miR-4532, also could form stem-loop structure by further modification. We conceived an idea of “reversed pattern” of primary structure; for instance, miR-1275 usually exists as 5p sequence in pre-miR-1275 but might exist as 3p sequence in pre-rmiR-1275. To examine this idea, broader region analysis was performed and some new candidate rmiRNA sequences were predicted (Supplemental Figure 1B). As a result, it was confirmed that all of new rmiRNAs can form well-ordered stem-loop structure (Table 2 and Supplemental Figure 2B).
Table 2.
A list of all determined pre-rmiRNA sequence and its location.
| rmiR name | Sequence of pre-rmiRNAs | Region | Location |
|---|---|---|---|
| rmiR-663a | CCGUCCGGCGUCCCAGGCGGGGCGCCGCGGGACCGCCCUCGUGUCUGUGGCGGUGGGAUCCCGCGGCCGUGUUUUCCUGGUGGCCCGGCCGUG | ETS | 2028–2119 |
| rmiR-663b | GGGGCCGAGGGCCGUCCGGCGUCCCAGGCGGGGCGCCGCGGGACCGCCCUCGUGUCUGUGGCGGUGGGAUCCCGCGGCCGUGUUUUCCUGGUGGCCCGGCCGUGCCUGAGGUUUC | ETS | 2025–2140 |
| rmiR-1268a | CUUCCUCCCUCCCGGCCUCUCCCGCCGACCGCGGGCGUGGUGGUGGGGGU | 3′ ETS | 13071–13123 |
| rmiR-1268b | CCGCGGGCGUGGUGGUGGGGGUGUGGGGGGGAGGGCGCGCGACCCCGGUCGGCGCGCCCCGCUUC | 3′ ETS | 13099–13163 |
| rmiR-1275 | AGCCCGGCUGGCCCGGUGGCGCCAGAGCUGUGGCCGGUCGCUUGUGAGUCACAGCUCUGGCGUGCAGGUUUAUGUGGGGGAGAGGCUGUCGCU | (NTS) | 42221–42313 |
| rmiR-3648 | CGCGACUGCGGCGGCGGUGGUGGGGGCAGCCGCGGGGAUCGCCGAGGGCCGGUCGGCCGCCCCGGGUGCCGCGCGGUGCCGCCGGCGGCGGUGAGGCCCCGCGCGUGUGUCCCGGCCGCGGUCGGCCGCGCUCGAGGGGUCCCCGUGGCGUCCCCUUCCCCGCCGGCCGCCUUUCUCGCG | ETS | 2486–2665 |
| rmiR-3656 | CUCCCUUCCCCCGCCGCCCCUCCUCCUCCUCCCCGGAGGGGGCGGGCUCCGGCGGGUGCGGGGGUGGGC | 28S | 8474–8542 |
| rmiR-3687 | CGCGCGUGCGCCCGAGCGCGGCCCGGUGGUCCCUCCCGGACAGGCGUUCGUGCGACGUGUG | (ETS) | 2854–2914 |
| rmiR-3687∗ | CGCGCGUGCGCCCGAGCGCGGCCCGGUGGUCCCUGCCGGACAGGCGUUCGUGCGACGUGUG | ETS | 2857–2917 |
| rmiR-4417 | GCGUGGGGCCCGGUGGGCUUCCCGGAGGGUUCCGGGGGUCGGCCUGCGGCGCGU | ETS | 2400–2454 |
| rmiR-4466 | UCGCGGGUGCGGGCCGGCGGGGUCCUCUGACGCGGCAGACAGCCCUGCCUGUCG | (ETS) | 627–680 |
| rmiR-4488 | CCGCCCUCCCUUCCCCCGCCGCCCCUCCUCCUCCUCCCCGGAGGGGGCGGGCUCCGGCGGGUGCGGGGGUGGGCGG | 28S | 8468–8544 |
| rmiR-4492 | GGGGCGCGAAGCGGGGCUGGGCGCGCGCCGCGGCUGGACGAGGCGCCGCCGCCCCCCCCACGCCCGGGGCACCCCCCUCGCGGCCC | 28S | 10838–10924 |
| rmiR-4508 | GGCGCGAAGCGGGGCUGGGCGCGCGCCGCGGCUGGACGAGGCGCCGCCGCCCCCCCCACGCCCGGGGCAC | 28S | 10841–10910 |
| rmiR-4516 | CCGUCCUCCCCCCUCCCCGGGGGAGCGCCGCGUGGGGGCGGCGGCGGGGGGAGAAGGGUCGGGGCGG | 28S | 11001–11067 |
| rmiR-4532 | GACGCGAGCCGGGCCCUUCCCGUGGAUCGCCCCAGCUGCGGCGGGCGUCGCGGCCGCCCCCGGGGAGCCCGGCGGGCGCCGGCGC | 28S | 11169–11254 |
| rmiR-6087 | GGUGAGGCGGGGGGGCGAGCCCCGAGGGGCUCUCGCUUCUGGCGCCAAG | 28S | 12005–12052 |
| rmiR-6724 | CGCUGCGCUUCUGGGCCCGCGGCGGGCGUGGGGCUGCCCGGGCCGGUCGACCAGCGCGCCGUAGCUCCCGAGGCCCGAGCCGCGACCCGGCG | (NTS) | 42310–42401 |
Note that some of them are overlapping each other. ∗rmiR-3687 indicates the sequence identical with pre-miR-3687 except for a point mutation in the guide sequence (see the lower part of Figure 1(a)).
3.3. DNS Computation and Comparison
Because concordance of so many sequences must not be detected accidentally, it is natural to consider hidden mechanisms on the background. We previously developed a quantum-based score, Dynamic Nexus Score (DNS), to evaluate miRNA/miRNA interactions and demonstrated that biological activity of the miRNA synergy is positively correlated with DNS value. The average DNS value among mature rmiRNAs was calculated through MESer computer program. DNSs of 1,032 human tRFs and all of 2,588 human miRNAs were also calculated as controls. Surprisingly, the average DNS of rmiRNAs marked 130.23; it was much higher than that of tRFs (40.76) and all human miRNAs (38.31) (Supplemental Figures 3A and 3B). Additionally, DNS values between tRFs, rmiRNAs, and all miRNAs were also calculated. As a result, it was confirmed that the miRNA pairs including rmiRNAs had relatively high DNS values (Supplemental Figure 3C), and this meant that rmiRNAs might induce miRNA-miRNA synergy to accelerate their biofunctions.
3.4. Target Prediction and Ontology Analysis
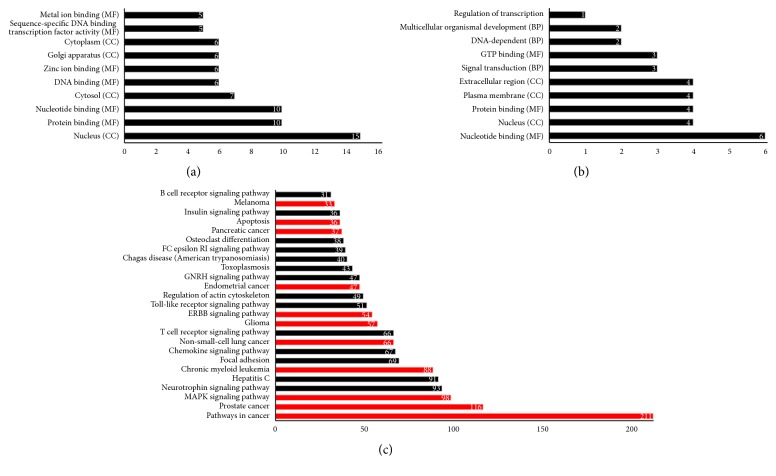
To investigate targets of rmiRNAs, we used TargetScan and miRTarBase. TargetScan was used for collecting putative target genes predicted by the seed theory-based algorithm; in contrast, miRTarBase was used for collecting experimentally validated target genes. In this experiment, we focused on top 5 high DNS of rmiRNAs, namely, rmiR-1268, rmiR-3656, rmiR-4466, rmiR-6087, and rmiR-6724, and these rmiRNAs were located at separated regions of the rDNA-repeating unit, such as 5′ ETS, 28S rRNA, 3′ ETS, and NTS. Top 10 targets of top 5 DNS rmiRNAs (total 50 targets) were extracted from both TargetScan and miRTarBase (Supplemental Table 2); subsequently, their classification in GO biological process (BP), molecular function (MF), and cellular component (CC) were performed and their results were listed through GeneCodis3 web tool (Figures 2(a) and 2(b)). Intriguingly, three gene ontology (GO) terms, namely, nucleus (CC), protein binding (MF), and nucleotide binding (MF), were commonly ranked on top 3 place between TargetScan and miRTarBase. Moreover, almost all their biofunctions were commonly related to gene regulation such as transcription and nucleotide binding although the greater parts of the GO analysis results were different in detail. Contributions of total 50 targets in biological pathway were also analyzed using GeneCodis3 with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway option. However, no pathway was presented in both cases of predicted target (TargetScan) and validated targets (miRTarBase). Therefore, to investigate in larger scale, we extracted all predicted targets in TargetScan having more than 0.1 cumulative weighted context++ score. KEGG pathway analysis of these targets was conducted and various biological pathways were successfully indicated. The results showed that the majority of putative targets were related to cancer or cancer-related pathways such as MAPK signaling and ERBB signaling (Figure 2(c)). This suggests that rmiRNAs have an inclination to target cancer-related genes and might have some important roles in anticancer or antistress pathways.
Figure 2.
GO and KEGG analysis for predicted targets of rmiRNAs. (a) GO characterization of top 10 targets of the top 5 high DNS rmiRNAs in miRTarBase. (b) GO characterization of top 10 putative targets of the top 5 high DNS rmiRNAs in TargetScan. (c) KEGG pathway annotations of putative target genes having less than −0.1 cumulative weighted context++ score in TargetScan. Cancer and cancer-related pathways were colored with red.
3.5. Prediction of rmiRNA Targets in Alu
Numerous Alu element-containing RNAs exist in the nucleolus and participate in the synthesis of rRNAs. Therefore, to seek RNA-RNA relationship between nucleolar function and rmiRNA, target sites of rmiRNAs inside Alu sequence were searched. Six branched members of Alu family, namely, Alu consensus, AluJo, AluSz, AluSc, AluSp, and AluY, were processed using RNA22 tool and then 5, 8, 7, 5, 4, and 3 putative rmiRNA target sites were detected, respectively (Supplemental Table 3). Several rmiRNA target sites were conserved among these Alu sequences. Nine of 17 rmiRNAs have potent target sites on Alu sequences and 3 of 5 high DNS rmiRNAs such as miR-4466, miR-6087, and miR-6724-5p were included.
4. Discussion
Ribosomes, large ribonucleoprotein complexes composed of various RPs and rRNAs, are a molecular machine that translate mRNAs into proteins and exists in all living cells [78, 79]. Proper function of the ribosome is essential for normal cell activities; therefore, modification and assembling of RPs and rRNAs are strictly and intricately regulated in the ribosome construction process [78–80]. Ribosomal dysfunction is associated with various diseases represented by ribosomopathies such as DBA, TCS, and SDS and is caused by mutation in ribosome-related genes, ribosomal haploinsufficiency, cellular stresses from chemical or infection, and so forth [1–3]. Several ribosomopathies increase the risk of carcinogenesis and cancer cells often have abnormality in the ribosome function due to mutation in RP and ribosome-related processor genes that cause ribosomopathies [4].
The synthesis of the ribosome itself largely contributes to malignant cell proliferation [81–83]. Ribosomal biogenesis is generally upregulated in the G1 phase dividing cells because enhanced protein synthesis is required to produce viable daughter cells [84]. Thus, inhibiting ribosomal synthesis causes G1 phase arrest and hinders cell proliferation reversibly in normal cells [85]. The upregulation of ribosomal synthesis is also observed in various cancer cells [82, 83]; therefore, inhibiting ribosomal synthesis has been recognized as a potent and novel anticancer strategy [81, 86–88]. This method particularly showed apoptosis-inducing effects in various malignant cells that synthesized ribosomes at a high rate and exhibited sufficient efficacy with only 3 h transient treatments. This inhibition was accomplished by deleting ribosomal protein genes or by cell treatments with actinomycin D, doxorubicin, 5-fluorouracil, and CX-5461, which have significant tumor suppressing effects [85, 88, 89]. However, this antitumor effect of inhibiting ribosomal biogenesis is not dependent on suppressing protein synthesis but on ribosomal biogenesis itself [81]. This anticancer mechanism presumably depends on stabilizing p53 by inhibiting a p53 degrading protein, called Murine Double Minute 2, by competitively combining with a ribosomal protein, which becomes free because it no longer participates in the construction of ribosomes [81, 88]. Additional evidences declaring associations between ribosome and cellular dysfunction have been reported. 28S rRNA has been identified as a novel fusion partner of carcinoma-related genes such as BCL6, BCL11B, IGKV3-20, and COG1 in gastric lymphoma or hematopoietic tumors [90, 91]. Mutation or inhibition of specific genes associated with ribosome construction pathway such as DKC1, AROS, and several snoRNAs have been identified to impair normal cell functions and sometimes cause carcinogenesis in various cell types [92–95]. Angiogenin-mediated rRNA transcription has been revealed to be related to squamous cell carcinoma [96]. Almost all these ribosome-related genes are considered as potent therapeutic targets for ribosomopathies.
Similar to rRNA, tRNA is the second most abundant noncoding RNA and contains many miRNA-like fragments as tRFs. Various tRFs are generated from mature or pre-tRNAs by ordered cleavage processing and they have similar characteristics to miRNAs, such as evolutionary conservation, target RNA recognition, translational regulation, circulation, and interaction with AGO proteins [57, 59–63, 97, 98]. In addition, miRNAs derived from pre-tRNA or tRNA–miRNA encoded by tRNA genes have been reported [59, 99]. Therefore, it is appropriate that miRNAs or miRNA-like functional fragments are also generated from rRNA gene-related regions.
As a sequel to computational analysis, we found that 17 pre-miRNA-like arrays, which contained identical sequences to known human mature miRNAs, were located in the rRNA gene coding region; therefore, we called them rDNA-hosted pre-miRNA analogs (rmiRNA). We also performed supplemental examination with mouse miRNAs and rDNA and found several sequences which are identical to miR-696, miR-712-5p, miR-712-3p, miR-714, miR-466i-5p, miR-5099, and miR-6538. Of these miRNAs, miR-696, miR-712, and miR-714 have already been reported as rRNA-locating ones [47]. According to Son et al., mmu-miR-712 is located at ITS2 of mouse rRNA gene (Rn45s) and hsa-miR-663 is located at ITS1 region of human rRNA gene (RNA45S) [48]. In our research, miR-712 was likewise found in ITS2 of Rn45s; however, hsa-miR-663 was not found in ITS1 but 5′ ETS of RNA45S. This discrepancy may be caused by the differences in detection tool or the version of RNA45S sequence data. For instance, they used MirEval web tool for sequence analysis and their study probably may be based on the older version of rRNA sequence data.
We also discovered that almost all of these rmiRNAs formed stem-loop structures and were located in RNA45S rather than the NTS (Figure 1, Supplemental Figure 2, and Table 2). Putative pre-rmiRNAs which have very similar sequences with their original miRNAs also showed similarity to their original miRNAs in the thermodynamic stability of stem-loop structures (Supplemental Figure 4A). Moreover, putative pre-miRNAs which have moderate substitutions in their sequences except the guide strands likewise showed similar stabilities and some showed more stable stem-loop structures than their original pre-miRNAs. This indicates that rmiRNAs have adequate potential stability to form stem-loop structures regarded as pre-miRNA. These results suggest that a large number of rmiRNAs are continually transcribed because rRNA is the most abundant noncoding RNA in eukaryotic cells. rDNA is transcribed so frequently that the rDNA region on the genome forms multicopies in the nucleolus [100]. Furthermore, rmiRNA may be noncanonically generated from rRNA in the cytoplasm because cytoplasmic RNA is a miRNA source [55].
All pooled miRNAs, such as rmiRNAs and/or tRFs, are important for immunoreactivity, transcriptional regulation, gene mobility, and cytoplasmic memory [17]. Sharma et al. indicated that paternal diet alters RNA information, such as population and composition of tRFs, in spermatozoa and influences progeny phenotype [101]. Likewise, although it was not confirmed in this study whether rDNA-hosted miRNAs would work in vivo with known identical miRNAs, rmiRNAs may function as resident miRNAs and participate in determining genotype and memorization. The number of stress-sensitive miRNAs is insufficient to exert an immediate response to cell damage if these miRNAs are generated only by transcription from DNA. Therefore, rapid generation of miRNAs from ready-made RNAs, such as rRNA and tRNA, should be considered. In addition, rmiRNAs and their identical miRNAs may work together because homologous miRNAs at different loci function together [102].
According to previous reports, rRNA-contained miRNAs such as miR-663, miR-1275, miR-3648, miR-3656, miR-3687, miR-4417, and miR-4516 are associated with tumor suppression, carcinomas, neuronal differentiation, breast cancer, breast cancer/neuronal differentiation, breast cancer, and regulation of signal transducer and activator of transcription 3, respectively [103–108]. However, the functions of residual rRNA-hosted miRNAs remain unclear. This motivated us to predict the targets of rmiRNAs and a number of putative and validated target genes which were associated with cancer-related pathways were found (Figure 2(c)). Therefore, RNA-RNA and/or RNA-protein interactions may participate in cancer-related functions. This indicates that rmiRNAs, in addition to ribosome-associated proteins and snoRNAs, might also be implicated in cell dysregulation and dysfunctions linked to ribosomopathies in multiple steps such as transcription, posttranscription, and biofunction.
In our previous study, the DNS was positively correlated with the strength of miRNA/miRNA synergies [72, 109]. As these rmiRNAs commonly have high DNS values, rmiRNA-derived miRNAs may also function as an activity booster of other miRNAs (Supplemental Figure 2). This characteristic would be effective for quick responses to cell emergencies that are not severe enough to cause changes in intracellular RNA composition [10, 16, 110, 111]. In this study, most of the top 10 rmiRNA targets were predicted to play roles in gene regulation and participate in cancer-related pathways (Figure 2(c)). This finding indicates that the rRNA copy number and expression level may be directly associated with rmiRNA generation and regulation of the biological reactions to cell stress leading to carcinogenesis.
Given the known mechanisms of pre-rRNA processing and that of miRNA generation from rRNA, maturation of rmiRNA occurs as follows: (1) rRNA genes containing rmiRNAs are transcribed as RNA45S by Pol I [7, 9]; (2a) the RNA45S ITS and ETS are degraded by XRN1 or other nucleases after the rRNA matures, and pre-rmiRNAs located in these regions are generated simultaneously [7, 9, 48]; (2b) pre-rmiRNAs located on 18S rRNA are biologically generated upon the degradation of mature rRNAs in the ribosome or degradation of pre-rRNA in response to stress [110–112]. (3) Drosha, Dicer, or its related proteins and enzymes process pre-rmiRNAs into mature rmiRNAs. The last step in which Drosha and Dicer participate in rRNA processing has been observed in several studies. RNase III enzymes including Drosha and Dicer have a miRNA-independent role in RNA processing, because the depletion of Dicer or Drosha impairs rRNA processing but does not affect the exonuclease activities required for rRNA processing [113, 114]. Fukuda et al. revealed that the DEAD-box RNA helicase p68 (Ddx5) and p72 (Ddx17), which are subunits of the Drosha complex, are required for pre-rRNA and pri-miRNA processing. Woolnough et al. reported that the human Ago2 protein binds rRNA and interferes with the transcription of nascent human rRNA via binding with Pol III and the transcription factor III complex on the gene [66, 115]. These data indicate that rRNA processing is closely related to the miRNA processing enzymes and its related proteins. In contrast, Chak et al. reported that the generation of miR-10404 and endo-siRNA from the rRNA gene is unaffected by mutations in Drosha, Pasha, Ago2, or Dcr-2 but by Dcr-1 in Drosophila [47]. Son et al. demonstrated that the generation of pre-miR-712 is dependent on XRN1 but independent of Drosha and DGCR8 in mice [48]. Pre-miR-712 processing is a mirtron-like, but it remains unknown whether Drosha and Dicer contribute to generating rRNA-derived miRNA because the details of the roles of Drosha, Dicer, and related proteins in rRNA processing are unknown. However, these findings suggest that rRNA-derived miRNAs could be generated in both Drosha-dependent and Drosha-independent pathways.
The number of repeated rDNA arrays is strongly associated with cell senescence, gene integrity, and ribosomal function, although the majority of rDNA is inactive [15–17]. Moreover, rDNA cluster size differs among species and individuals and even in individual cells when the cells are responding to DNA damage or when the rRNA repeat number is being amplified [15–17, 116]. As these differences are inherited, it is certain that rRNA and rRNA-hosted miRNAs participate in cell identity [117]. The ETS and ITS regions are not highly conserved as compared to 18S, 5.8S, and 28S RNA. All three previously reported ITS- or ETS-derived miRNAs, such as miR-663, miR-712, and miR-10404, are human-, mouse-, and fly-specific miRNAs, respectively, and they are well conserved intraspecifically [47, 48], suggesting that variations in rmiRNAs and rDNA copy number contribute to evolution, particularly the inheritance of acquired characteristics.
Nucleolus, where rRNA is transcribed and processed, is the largest structure in the nucleus formed at rRNA coding regions on chromosomes and composed of diverse specific proteins and RNAs [118]. It has been revealed that some miRNAs exist and function in nucleolus. For instance, miR-206, a highly expressed miRNA in skeletal muscle, and several other miRNAs are detected in the nucleolus as well as in the cytoplasm with in situ hybridization [119]. Subsequently, it has been shown by deep sequencing that a set of miRNAs present in the nucleus rather than in the cytoplasm and some of them tend to accumulate at the nucleolus [120]. RNA interference (RNAi) factors such as AGO protein, Dicer, and TRBP are also found in the cell nuclei, suggesting that miRNA machinery is active even in the nucleolus [121]. Moreover, it has recently been reported that Alu element-containing Pol II transcripts (aluRNA) are abundant in nucleolus [67]. Alu element is the most abundant SINE family that comprises about 10% of the genome and exists in both noncoding and coding region including introns and 3′ UTR of mRNA transcripts [122, 123]. There are growing evidences that a portion of mRNAs have Alu-derived sequence in their 3′ UTR which can be targeted by a set of miRNAs [124–126]. Since it has been reported that the transcriptional rate of Alu is upregulated upon cellular stress and strongly influences the nucleolar size and pre-rRNA transcript rate [67, 127, 128], we supposed that aluRNAs might also be regulated by miRNAs. In our investigation, several miRNAs were detected from rRNA sequence as rmiRNA, and half of these rmiRNAs have potential target sites in Alu family sequences (Supplemental Table 3). Although it was not confirmed in this study whether rmiRNAs really regulate aluRNAs, at least, the possibility that rmiRNAs might interact with aluRNA in the nucleolus and contribute to the regulation of ribosomal function and composition upon cellular stress as a ribosomal feedback machinery was implied.
It was technically difficult to distinguish the origins of the sources using ready-made technologies, because the mature rmiRNA sequences, such as rmiR-663a/b and rmiR-1268a/b, were identical between the rDNA and non-rDNA genes. Therefore, in this study, we performed in silico analyses to by-pass this problem. No rmiRNA was detected from the 5S rRNA gene but the AGO2 protein binds to 5S rRNA [115], and AGO2 has Slicer activity [129], suggesting that various rRNA-derived specific miRNAs with different mature sequences to annotated miRNAs, that is, novel miRNAs, may be generated from the rRNA coding region. Furthermore, mature rmiRNA may have been generated in another form, such as loop miRNAs [130]. Numerous undefined RNA fragments derived from well-known RNAs or other noncoding RNAs might unveil the RNA wave enigma and implication of tumorigenesis. Therefore, additional laboratory and clinical investigations are required for discovery of the nascent human miRNAs and for decipherment of precise interaction among miRNAs, noncoding RNAs, and human cancer.
5. Conclusion
Seventeen rDNA-hosted miRNA analogs (rmiRNAs) were found in rRNA coding region by in silico analyses. These rmiRNAs might be generated from rRNA upon construction or degradation of ribosomes. The majority of predicted targets of rmiRNAs were stress- or cancer-related genes and it was indicated that rmiRNAs could also target AluRNA in nucleolus, suggesting that rmiRNAs may regulate ribosomal function at multiple levels adopting to cellular stress. While rmiRNAs showed significantly high DNS values compared to those of normal miRNAs and tRFs, rmiRNAs may efficiently boost bioactivities of other miRNAs to attenuate cell stress and tumorigenesis as a quantum memory device and a member of the resident miRNA genes. Altogether, rmiRNAs would be implicated in human ribosomopathy. In future, rmiRNA mimics or anti-rmiRNA agents may be developed to cancer therapy and there is some possibility that rmiRNAs in serum could be applied for prognosis and/or diagnosis of ribosomopathy.
Supplementary Material
Supplemental Figure 1. Sequences and secondary structures of guide-only rmiRNAs. Supplemental Figure 2. Sequences and secondary structures of reversed rmiRNAs. Supplemental Figure 3. DNS distribution of rmiRNA, tRF, and all miRNAs. Supplemental Figure 4. Free energies of secondary structures of pre-miRNAs and their original pre-miRNAs. Supplemental Table 1. Differences between two sample rRNA gene sequence. Supplemental Table 2. TOP 10 predicted and validated targets of rmiRNAs. Supplemental Table 3. Predicted interactions between Alu family and rmiRNA.
Competing Interests
The authors have declared that no competing interests exist.
References
- 1.Luft F. The rise of a ribosomopathy and increased cancer risk. Journal of Molecular Medicine. 2010;88(1):1–3. doi: 10.1007/s00109-009-0570-0. [DOI] [PubMed] [Google Scholar]
- 2.Narla A., Ebert B. L. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115(16):3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yelick P. C., Trainor P. A. Ribosomopathies: global process, tissue specific defects. Rare Diseases. 2015;3(1):p. 10. doi: 10.1080/21675511.2015.1025185.e1025185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goudarzi K. M., Lindström M. S. Role of ribosomal protein mutations in tumor development (Review) International Journal of Oncology. 2016;48(4):1313–1324. doi: 10.3892/ijo.2016.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nazar R. N., Sitz T. O., Busch H. Sequence homologies in mammalian 5.8S ribosomal RNA. Biochemistry. 1976;15(3):505–508. doi: 10.1021/bi00648a008. [DOI] [PubMed] [Google Scholar]
- 6.Zentner G. E., Saiakhova A., Manaenkov P., Adams M. D., Scacheri P. C. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Research. 2011;39(12):4949–4960. doi: 10.1093/nar/gkq1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frenk S., Oxley D., Houseley J. The nuclear exosome is active and important during budding yeast meiosis. PLoS ONE. 2014;9(9) doi: 10.1371/journal.pone.0107648.e107648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henras A. K., Plisson-Chastang C., O'Donohue M.-F., Chakraborty A., Gleizes P.-E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdisciplinary Reviews: RNA. 2015;6(2):225–242. doi: 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decatur W. A., Fournier M. J. RNA-guided nucleotide modification of ribosomal and other RNAs. The Journal of Biological Chemistry. 2003;278(2):695–698. doi: 10.1074/jbc.r200023200. [DOI] [PubMed] [Google Scholar]
- 10.Maden B. E. H., Dent C. L., Farrell T. E., Garde J., McCallum F. S., Wakeman J. A. Clones of human ribosomal DNA containing the complete 18S-rRNA and 28S-rRNA genes. Characterization, a detailed map of the human ribosomal transcription unit and diversity among clones. Biochemical Journal. 1987;246(2):519–527. doi: 10.1042/bj2460519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang K.-D., Fang S.-A., Chang F.-C., Chung M.-C. Chromosomal conservation and sequence diversity of ribosomal RNA genes of two distant Oryza species. Genomics. 2010;96(3):181–190. doi: 10.1016/j.ygeno.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Goodfellow S. J., Zomerdijk J. C. B. M. Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. In: Kundu T. K., editor. Epigenetics: Development and Disease. Vol. 61. Dordrecht, The Netherlands: Springer; 2013. pp. 211–236. (Subcellular Biochemistry). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciganda M., Williams N. Eukaryotic 5S rRNA biogenesis. Wiley Interdisciplinary Reviews: RNA. 2011;2(4):523–533. doi: 10.1002/wrna.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson A. S., Warburton D., Atwood K. C. Ribosomal DNA connectives between human acrocentric chromosomes. Nature. 1973;245(5420):95–97. doi: 10.1038/245095b0. [DOI] [PubMed] [Google Scholar]
- 15.Ide S., Miyazaki T., Maki H., Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010;327(5966):693–696. doi: 10.1126/science.1179044. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi T. Ribosomal RNA gene repeats, their stability and cellular senescence. Proceedings of the Japan Academy Series B: Physical and Biological Sciences. 2014;90(4):119–129. doi: 10.2183/pjab.90.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malone J. H. Balancing copy number in ribosomal DNA. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(9):2635–2636. doi: 10.1073/pnas.1500054112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diesch J., Hannan R. D., Sanij E. Perturbations at the ribosomal genes loci are at the centre of cellular dysfunction and human disease. Cell & Bioscience. 2014;4(1, article 43) doi: 10.1186/2045-3701-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 20.Bartel D. P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q., Yao J., Smith G. W., Dong C. Identification of a novel microRNA important for melanogenesis in alpaca (Vicugna pacos) Journal of Animal Science. 2015;93(4):1622–1631. doi: 10.2527/jas.2014-8404. [DOI] [PubMed] [Google Scholar]
- 22.Rayner K. J. MicroRNA-155 in the heart: the right time at the right place in the right cell. Circulation. 2015;131(18):1533–1535. doi: 10.1161/circulationaha.115.016327. [DOI] [PubMed] [Google Scholar]
- 23.Srikantan S., Marasa B. S., Becker K. G., Gorospe M., Abdelmohsen K. Paradoxical microRNAs: individual gene repressors, global translation enhancers. Cell Cycle. 2011;10(5):751–759. doi: 10.4161/cc.10.5.14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grey F., Tirabassi R., Meyers H., et al. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5′UTRs. PLoS Pathogens. 2010;6(6) doi: 10.1371/journal.ppat.1000967.e1000967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashimoto Y., Akiyama Y., Yuasa Y. Multiple-to-multiple relationships between MicroRNAs and target genes in gastric cancer. PLoS ONE. 2013;8(5) doi: 10.1371/journal.pone.0062589.e62589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu L., Dai W.-Q., Xu X.-F., Wang F., He L., Guo C.-Y. Effects of multiple-target anti-microRNA antisense oligodeoxyribonucleotides on proliferation and migration of gastric cancer cells. Asian Pacific Journal of Cancer Prevention. 2012;13(7):3203–3207. doi: 10.7314/APJCP.2012.13.7.3203. [DOI] [PubMed] [Google Scholar]
- 27.Lewis B. P., Burge C. B., Bartel D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 28.Fujii Y. R. Oncoviruses and pathogenic microRNAs in humans. The Open Virology Journal. 2009;3(1):37–51. doi: 10.2174/1874357900903010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ardekani A. M., Naeini M. M. The role of microRNAs in human diseases. Avicenna Journal of Medical Biotechnology. 2010;2(4):161–179. [PMC free article] [PubMed] [Google Scholar]
- 30.Gama-Carvalho M., Andrade J., Brás-Rosário L. Regulation of cardiac cell fate by microRNAs: implications for heart regeneration. Cells. 2014;3(4):996–1026. doi: 10.3390/cells3040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shenoy A., Blelloch R. H. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nature Reviews Molecular Cell Biology. 2014;15(9):565–576. doi: 10.1038/nrm3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai W.-F., Siu P. M. MicroRNAs as regulators of cutaneous wound healing. Journal of Biosciences. 2014;39(3):519–524. doi: 10.1007/s12038-014-9421-4. [DOI] [PubMed] [Google Scholar]
- 33.Leung A. K. L., Vyas S., Rood J. E., Bhutkar A., Sharp P. A., Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Molecular Cell. 2011;42(4):489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X., Liu M.-X., Yan G.-Y. RWRMDA: predicting novel human microRNA–disease associations. Molecular BioSystems. 2012;8(10):2792–2798. doi: 10.1039/c2mb25180a. [DOI] [PubMed] [Google Scholar]
- 35.Chen X., Yan G.-Y. Semi-supervised learning for potential human microRNA-disease associations inference. Scientific Reports. 2014;4, article 5501 doi: 10.1038/srep05501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim V. N., Han J., Siomi M. C. Biogenesis of small RNAs in animals. Nature Reviews Molecular Cell Biology. 2009;10(2):126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 37.Hsu P. W. C., Huang H.-D., Hsu S.-D., et al. miRNAMap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Research. 2006;34:D135–D139. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez-Mercado M., Manterola L., Lawrie C. H. MicroRNAs in lymphoma: regulatory role and biomarker potential. Current Genomics. 2015;16(5):349–358. doi: 10.2174/1389202916666150707160147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khvorova A., Reynolds A., Jayasena S. D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115(2):209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 40.Melo C. A., Melo S. A. Biogenesis and physiology of MicroRNAs. In: Fabbri M., editor. Non-Coding RNAs and Cancer. New York, NY, USA: Springer; 2014. pp. 5–24. [DOI] [Google Scholar]
- 41.Ruby J. G., Jan C. H., Bartel D. P. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448(7149):83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujii Y. R. RNA genes: retroelements and virally retroposable microRNAs in human embryonic stem cells. The Open Virology Journal. 2010;4:63–75. doi: 10.2174/1874357901004010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujii Y. R. The RNA gene information: retroelement-MicroRNA entangling as the RNA quantum code. In: Ying S.-Y., editor. MicroRNA Protocols. Totowa, NJ, USA: Humana Press; 2013. pp. 47–67. [DOI] [PubMed] [Google Scholar]
- 44.Wiesen J. L., Tomasi T. B. Dicer is regulated by cellular stresses and interferons. Molecular Immunology. 2009;46(6):1222–1228. doi: 10.1016/j.molimm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Z., Wang L. Regulation of microRNA expression and function by nuclear receptor signaling. Cell & Bioscience. 2011;1(1, article 31) doi: 10.1186/2045-3701-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuchen S., Resch W., Yamane A., et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32(6):828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chak L.-L., Mohammed J., Lai E. C., Tucker-Kellogg G., Okamura K. A deeply conserved, noncanonical miRNA hosted by ribosomal DNA. RNA. 2015;21(3):375–384. doi: 10.1261/rna.049098.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Son D. J., Kumar S., Takabe W., et al. The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nature Communications. 2013;4, article 3000 doi: 10.1038/ncomms4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doss J. F., Corcoran D. L., Jima D. D., Telen M. J., Dave S. S., Chi J. A comprehensive joint analysis of the long and short RNA transcriptomes of human erythrocytes. BMC Genomics. 2015;16, article 952 doi: 10.1186/s12864-015-2156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LaMonte G., Philip N., Reardon J., et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host & Microbe. 2012;12(2):187–199. doi: 10.1016/j.chom.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward W. S., Coffey D. S. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biology of Reproduction. 1991;44(4):569–574. doi: 10.1095/biolreprod44.4.569. [DOI] [PubMed] [Google Scholar]
- 52.Jodar M., Selvaraju S., Sendler E., Diamond M. P., Krawetz S. A. The presence, role and clinical use of spermatozoal RNAs. Human Reproduction Update. 2013;19(6):604–624. doi: 10.1093/humupd/dmt031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Govindaraju A., Uzun A., Robertson L., et al. Dynamics of microRNAs in bull spermatozoa. Reproductive Biology and Endocrinology. 2012;10, article 82 doi: 10.1186/1477-7827-10-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barckmann B., Simonelig M. Control of maternal mRNA stability in germ cells and early embryos. Biochimica et Biophysica Acta (BBA)—Gene Regulatory Mechanisms. 2013;1829(6-7):714–724. doi: 10.1016/j.bbagrm.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 55.Jin Q., Xue Z., Dong C., Wang Y., Chu L., Xu Y. Identification and characterization of MicroRNAs from tree peony (Paeonia ostii) and their response to copper stress. PLoS ONE. 2015;10(2) doi: 10.1371/journal.pone.0117584.e0117584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reese T. A., Xia J., Johnson L. S., Zhou X., Zhang W., Virgin H. W. Identification of novel microRNA-like molecules generated from herpesvirus and host tRNA transcripts. Journal of Virology. 2010;84(19):10344–10353. doi: 10.1128/jvi.00707-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee Y. S., Shibata Y., Malhotra A., Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes & Development. 2009;23(22):2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Megel C., Morelle G., Lalande S., Duchêne A.-M., Small I., Maréchal-Drouard L. Surveillance and cleavage of eukaryotic tRNAs. International Journal of Molecular Sciences. 2015;16(1):1873–1893. doi: 10.3390/ijms16011873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Green D., Fraser W. D., Dalmay T. Transfer RNA-derived small RNAs in the cancer transcriptome. Pflügers Archiv-European Journal of Physiology. 2016;468(6):1041–1047. doi: 10.1007/s00424-016-1822-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diebel K. W., Zhou K., Clarke A. B., Bemis L. T. Beyond the ribosome: extra-translational functions of tRNA fragments. Biomarker Insights. 2016;11, supplement 1:1–8. doi: 10.4137/BMI.S35904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casas E., Cai G., Neill J. D. Characterization of circulating transfer RNA-derived RNA fragments in cattle. Frontiers in Genetics. 2015;6, article 271:1–7. doi: 10.3389/fgene.2015.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karaiskos S., Naqvi A. S., Swanson K. E., Grigoriev A. Age-driven modulation of tRNA-derived fragments in Drosophila and their potential targets. Biology Direct. 2015;10, article 51 doi: 10.1186/s13062-015-0081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Loss-Morais G., Waterhouse P. M., Margis R. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biology Direct. 2013;8(1, article 6) doi: 10.1186/1745-6150-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Selitsky S. R., Baran-Gale J., Honda M., et al. Small tRNA-derived RNAs are increased and more abundant than microRNAs in chronic hepatitis B and C. Scientific Reports. 2015;5, article no. 7675 doi: 10.1038/srep07675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei H., Zhou B., Zhang F., et al. Profiling and identification of small rDNA-derived RNAs and their potential biological functions. PLoS ONE. 2013;8(2) doi: 10.1371/journal.pone.0056842.e56842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fukuda T., Yamagata K., Fujiyama S., et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nature Cell Biology. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 67.Caudron-Herger M., Pankert T., Seiler J., et al. Alu element-containing RNAs maintain nucleolar structure and function. The EMBO Journal. 2015;34(22):2758–2774. doi: 10.15252/embj.201591458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kozomara A., Griffiths-Jones S. MiRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research. 2014;42(1):D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gibson R., Alako B., Amid C., et al. Biocuration of functional annotation at the European nucleotide archive. Nucleic Acids Research. 2016;44:D58–D66. doi: 10.1093/nar/gkv1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chan P. P., Lowe T. M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Research. 2016;44(1):D184–D189. doi: 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar P., Mudunuri S. B., Anaya J., Dutta A. tRFdb: a database for transfer RNA fragments. Nucleic Acids Research. 2015;43(1):D141–D145. doi: 10.1093/nar/gku1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshikawa M., Osone T., Fujii Y. MicroRNA memory I: the positive correlation between synergistic effects of microRNAs in cancer and a novel quantum scoring system. Journal of Advances in Medical and Pharmaceutical Sciences. 2016;5(4):1–16. doi: 10.9734/jamps/2016/22134. [DOI] [Google Scholar]
- 73.Agarwal V., Bell G. W., Nam J.-W., Bartel D. P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4(2015) doi: 10.7554/eLife.05005.e05005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chou C.-H., Chang N.-W., Shrestha S., et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Research. 2016;44(1):D239–D247. doi: 10.1093/nar/gkv1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tabas-Madrid D., Nogales-Cadenas R., Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Research. 2012;40(1):W478–W483. doi: 10.1093/nar/gks402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vassetzky N. S., Kramerov D. A. SINEBase: a database and tool for SINE analysis. Nucleic Acids Research. 2013;41(1):D83–D89. doi: 10.1093/nar/gks1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Miranda K. C., Huynh T., Tay Y., et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 78.Lafontaine D. L. J., Tollervey D. The function and synthesis of ribosomes. Nature Reviews Molecular Cell Biology. 2001;2(7):514–520. doi: 10.1038/35080045. [DOI] [PubMed] [Google Scholar]
- 79.Gamalinda M., Woolford J. L. Paradigms of ribosome synthesis: lessons learned from ribosomal proteins. Translation. 2015;3(1) doi: 10.4161/21690731.2014.975018.e975018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kressler D., Hurt E., Baβler J. Driving ribosome assembly. Biochimica et Biophysica Acta (BBA)—Molecular Cell Research. 2010;1803(6):673–683. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 81.Brighenti E., Treré D., Derenzini M. Targeted cancer therapy with ribosome biogenesis inhibitors: a real possibility? Oncotarget. 2015;6(36):38617–38627. doi: 10.18632/oncotarget.5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Montanaro L., Treré D., Derenzini M. Nucleolus, ribosomes, and cancer. The American Journal of Pathology. 2008;173(2):301–310. doi: 10.2353/ajpath.2008.070752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maggi L. B., Jr., Weber J. D. Nucleolar adaptation in human cancer. Cancer Investigation. 2005;23(7):599–608. doi: 10.1080/07357900500283085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thomas G. An encore for ribosome biogenesis in the control of cell proliferation. Nature Cell Biology. 2000;2(5):E71–E72. doi: 10.1038/35010581. [DOI] [PubMed] [Google Scholar]
- 85.Volarević S., Stewart M. J., Ledermann B., et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288(5473):2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 86.Negi S. S., Brown P. Transient rRNA synthesis inhibition with CX-5461 is sufficient to elicit growth arrest and cell death in acute lymphoblastic leukemia cells. Oncotarget. 2015;6(33):34846–34858. doi: 10.18632/oncotarget.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Negi S. S., Brown P. rRNA synthesis inhibitor, CX-5461, activates ATM/ATR pathway in acute lymphoblastic leukemia, arrests cells in G2 phase and induces apoptosis. Oncotarget. 2015;6(20):18094–18104. doi: 10.18632/oncotarget.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scala F., Brighenti E., Govoni M., et al. Direct relationship between the level of p53 stabilization induced by rRNA synthesis-inhibiting drugs and the cell ribosome biogenesis rate. Oncogene. 2015;35(8):977–989. doi: 10.1038/onc.2015.147. [DOI] [PubMed] [Google Scholar]
- 89.Sulic S., Panic L., Barkic M., Mercep M., Uzelac M., Volarevic S. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes & Development. 2005;19(24):3070–3082. doi: 10.1101/gad.359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen Y.-W., Hu X.-T., Liang A. C., et al. High BCL6 expression predicts better prognosis, independent of BCL6 translocation status, translocation partner, or BCL6-deregulating mutations, in gastric lymphoma. Blood. 2006;108(7):2373–2383. doi: 10.1182/blood-2006-05-022517. [DOI] [PubMed] [Google Scholar]
- 91.Kobayashi S., Taki T., Nagoshi H., et al. Identification of novel fusion genes with 28S ribosomal DNA in hematologic malignancies. International Journal of Oncology. 2014;44(4):1193–1198. doi: 10.3892/ijo.2014.2291. [DOI] [PubMed] [Google Scholar]
- 92.Bellodi C., McMahon M., Contreras A., et al. H/ACA small RNA dysfunctions in disease reveal key roles for noncoding RNA modifications in hematopoietic stem cell differentiation. Cell Reports. 2013;3(5):1493–1502. doi: 10.1016/j.celrep.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Knight J. R. P., Willis A. E., Milner J. Active regulator of SIRT1 is required for ribosome biogenesis and function. Nucleic Acids Research. 2013;41(7):4185–4197. doi: 10.1093/nar/gkt129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sondalle S. B., Baserga S. J. Human diseases of the SSU processome. Biochimica et Biophysica Acta (BBA—Molecular Basis of Disease. 2014;1842(6):758–764. doi: 10.1016/j.bbadis.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moon Y. Ribosomal alteration-derived signals for cytokine induction in mucosal and systemic inflammation: noncanonical pathways by ribosomal inactivation. Mediators of Inflammation. 2014;2014:10. doi: 10.1155/2014/708193.708193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen L., Hu G.-F. Angiogenin-mediated ribosomal RNA transcription as a molecular target for treatment of head and neck squamous cell carcinoma. Oral Oncology. 2010;46(9):648–653. doi: 10.1016/j.oraloncology.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guzman N., Agarwal K., Asthagiri D., et al. Breast cancer-specific miR signature unique to extracellular vesicles includes ‘microRNA-like’ tRNA fragments. Molecular Cancer Research. 2015;13(5):891–901. doi: 10.1158/1541-7786.mcr-14-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kumar P., Anaya J., Mudunuri S. B., Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biology. 2014;12, article 78 doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Diebel K. W., Oko L. M., Medina E. M., et al. Gammaherpesvirus small noncoding RNAs are bifunctional elements that regulate infection and contribute to virulence in vivo. mBio. 2015;6(1) doi: 10.1128/mbio.01670-14.e01670-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tsai R. Y. L., Pederson T. Connecting the nucleolus to the cell cycle and human disease. The FASEB Journal. 2014;28(8):3290–3296. doi: 10.1096/fj.14-254680. [DOI] [PubMed] [Google Scholar]
- 101.Sharma U., Conine C. C., Shea J. M., et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351(6271):391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lovat F., Fassan M., Gasparini P., et al. miR-15b/16-2 deletion promotes B-cell malignancies. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(37):11636–11641. doi: 10.1073/pnas.1514954112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zang W., Wang Y., Wang T., et al. miR-663 attenuates tumor growth and invasiveness by targeting eEF1A2 in pancreatic cancer. Molecular Cancer. 2015;14(1, article 37) doi: 10.1186/s12943-015-0315-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 104.Fawzy I. O., Hamza M. T., Hosny K. A., Esmat G., El Tayebi H. M., Abdelaziz A. I. MiR-1275: A single microRNA that targets the three IGF2-mRNA-binding proteins hindering tumor growth in hepatocellular carcinoma. FEBS Letters. 2015;589(17):2257–2265. doi: 10.1016/j.febslet.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 105.Matamala N., Vargas M. T., González-Cámpora R., et al. Tumor MicroRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clinical Chemistry. 2015;61(8):1098–1106. doi: 10.1373/clinchem.2015.238691. [DOI] [PubMed] [Google Scholar]
- 106.Shu R., Wong W., Ma Q. H., et al. APP intracellular domain acts as a transcriptional regulator of miR-663 suppressing neuronal differentiation. Cell Death and Disease. 2015;6(2) doi: 10.1038/cddis.2015.10.e1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Murria R., Palanca S., de Juan I., et al. Immunohistochemical, genetic and epigenetic profiles of hereditary and triple negative breast cancers. Relevance in personalized medicine. American Journal of Cancer Research. 2015;5(7):2330–2343. [PMC free article] [PubMed] [Google Scholar]
- 108.Chowdhari S., Saini N. hsa-miR-4516 mediated downregulation of STAT3/CDK6/UBE2N plays a role in PUVA induced apoptosis in keratinocytes. Journal of Cellular Physiology. 2014;229(11):1630–1638. doi: 10.1002/jcp.24608. [DOI] [PubMed] [Google Scholar]
- 109.Osone T., Yoshikawa M., Fujii Y. MicroRNA memory II: a novel scoring integration model for prediction of human disease by microRNA/microRNA quantum multi-interaction. Journal of Advances in Medical and Pharmaceutical Sciences. 2016;5(3):1–18. doi: 10.9734/jamps/2016/22095. [DOI] [Google Scholar]
- 110.Allmang C., Mitchell P., Petfalski E., Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Research. 2000;28(8):1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Basturea G. N., Zundel M. A., Deutscher M. P. Degradation of ribosomal RNA during starvation: comparison to quality control during steady-state growth and a role for RNase PH. RNA. 2011;17(2):338–345. doi: 10.1261/rna.2448911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang Q., Lee I., Ren J., Ajay S. S., Lee Y. S., Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Molecular Therapy. 2013;21(2):368–379. doi: 10.1038/mt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Johanson T. M., Lew A. M., Chong M. M. W. MicroRNA-independent roles of the RNase III enzymes Drosha and Dicer. Open Biology. 2013;3(10) doi: 10.1098/rsob.130144.130144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liang X.-H., Crooke S. T. Depletion of key protein components of the RISC pathway impairs pre-ribosomal RNA processing. Nucleic Acids Research. 2011;39(11):4875–4889. doi: 10.1093/nar/gkr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Woolnough J. L., Atwood B. L., Giles K. E. Argonaute 2 binds directly to tRNA genes and promotes gene repression in cis . Molecular and Cellular Biology. 2015;35(13):2278–2294. doi: 10.1128/mcb.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stults D. M., Killen M. W., Pierce H. H., Pierce A. J. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Research. 2008;18(1):13–18. doi: 10.1101/gr.6858507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Robertus Fujii Y. The xenotropic microRNA gene information for stem cell researches and clinical applications. Stem Cell Discovery. 2013;3(1):32–36. doi: 10.4236/scd.2013.31005. [DOI] [Google Scholar]
- 118.Lam Y. W., Trinkle-Mulcahy L. New insights into nucleolar structure and function. F1000Prime Reports. 2015;7, article 48 doi: 10.12703/p7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ritland Politz J. C., Hogan E. M., Pederson T. MicroRNAs with a nucleolar location. RNA. 2009;15(9):1705–1715. doi: 10.1261/rna.1470409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bai B., Liu H., Laiho M. Small RNA expression and deep sequencing analyses of the nucleolus reveal the presence of nucleolus-associated microRNAs. FEBS Open Bio. 2014;4:441–449. doi: 10.1016/j.fob.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gagnon K., Li L., Chu Y., Janowski B., Corey D. RNAi factors are present and active in human cell nuclei. Cell Reports. 2014;6(1):211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mighell A. J., Markham A. F., Robinson P. A. Alu sequences. FEBS Letters. 1997;417(1):1–5. doi: 10.1016/s0014-5793(97)01259-3. [DOI] [PubMed] [Google Scholar]
- 123.Deininger P. Genomic Disorders. Totowa, NJ, USA: Humana Press; 2006. Alu Elements; pp. 21–34. [DOI] [Google Scholar]
- 124.Daskalova E., Baev V., Rusinov V., Minkov I. 3′UTR-located ALU elements: donors of potential miRNA target sites and mediators of network miRNA-based regulatory interactions. Evolutionary Bioinformatics Online. 2006;2:103–120. [PMC free article] [PubMed] [Google Scholar]
- 125.Hoffman Y., Dahary D., Bublik D. R., Oren M., Pilpel Y. The majority of endogenous microRNA targets within Alu elements avoid the microRNA machinery. Bioinformatics. 2013;29(7):894–902. doi: 10.1093/bioinformatics/btt044. [DOI] [PubMed] [Google Scholar]
- 126.Hoffman Y., Pilpel Y., Oren M. MicroRNAs and Alu elements in the p53-Mdm2-Mdm4 regulatory network. Journal of Molecular Cell Biology. 2014;6(3):192–197. doi: 10.1093/jmcb/mju020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gu T. J., Yi X., Zhao X. W., Zhao Y., Yin J. Q. Alu-directed transcriptional regulation of some novel miRNAs. BMC Genomics. 2009;10, article 563 doi: 10.1186/1471-2164-10-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lehnert S., Van Loo P., Thilakarathne P. J., Marynen P., Verbeke G., Schuit F. C. Evidence for co-evolution between human microRNAs and Alu-repeats. PLoS ONE. 2009;4(2) doi: 10.1371/journal.pone.0004456.e4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Miyoshi K., Tsukumo H., Nagami T., Siomi H., Siomi M. C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes & Development. 2005;19(23):2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chak L.-L., Okamura K. Argonaute-dependent small RNAs derived from single-stranded, non-structured precursors. Frontiers in Genetics. 2014;5, article 172 doi: 10.3389/fgene.2014.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Sequences and secondary structures of guide-only rmiRNAs. Supplemental Figure 2. Sequences and secondary structures of reversed rmiRNAs. Supplemental Figure 3. DNS distribution of rmiRNA, tRF, and all miRNAs. Supplemental Figure 4. Free energies of secondary structures of pre-miRNAs and their original pre-miRNAs. Supplemental Table 1. Differences between two sample rRNA gene sequence. Supplemental Table 2. TOP 10 predicted and validated targets of rmiRNAs. Supplemental Table 3. Predicted interactions between Alu family and rmiRNA.