Abstract
Intramembrane proteolysis by γ‐secretases plays major roles in disease pathology and cellular signalling, yet the dynamics of these enzyme complexes and how they recognize substrates remains poorly understood. New work in The EMBO Journal utilizes photo‐affinity cross‐linking to map APP interactions to different γ‐secretase subunits, suggesting a succession of recruitment and engagement steps that lead up to substrate cleavage.
Subject Categories: Membrane & Intracellular Transport; Molecular Biology of Disease; Post-translational Modifications, Proteolysis & Proteomics
γ‐secretases are fascinating multimeric intramembrane protease complexes involved in a wide spectrum of biological activities (Jurisch‐Yaksi et al, 2013). They cleave many and very different substrates in the membrane using an intriguing multiple turnover mechanism, which releases proteolytic fragments at both sides of the cell membrane and possibly also hydrophobic peptides into the membrane. Many of this newly generated intracellular fragments exert (or are supposed to exert) signalling functions (Jurisch‐Yaksi et al, 2013), while some of the extracellular fragments, like the Aβ fragment generated from the Alzheimer's disease‐related amyloid precursor protein (APP), play a central role in disease pathology (De Strooper & Chávez Gutiérrez, 2015). γ‐secretase complexes consist of presenilin (PSEN), nicastrin (NCT), presenilin enhancer 2 (PEN2) and anterior pharynx defective 1 (APH1) subunits, and perform endoproteolytic, carboxypeptidase‐like, and even aminopeptidase activities (De Strooper & Chávez Gutiérrez, 2015). Details of the interaction of these different subunits have recently been revealed in atomic structures obtained by cryo‐electron microscopy (Bai et al, 2015a,b). However, to unravel crucial questions such as the mechanisms that underlie the different proteolytic activities, the dynamics of the complexes or the way substrates are recognized, further work is needed.
While their relaxed specificity would suggest that γ‐secretase complexes indiscriminately cut type I integral membrane proteins after removal of their ectodomain, kinetic data indicate that γ‐secretases do distinguish between substrates (Chávez‐Gutiérrez et al, 2012). Furthermore, the strength of the interaction between enzyme and the substrate transmembrane domain (TMD) may drive endo‐proteolysis, as shown for the case of Notch receptor (Bolduc et al, 2016). Finally, differential subcellular localization of γ‐secretases and their targets also adds substrate specificity to each complex (Sannerud et al, 2016).
How exactly γ‐secretases recognize their substrates at the molecular level remains a largely unaddressed question. Answering it could have huge implications for the development of safer drugs for Alzheimer's disease and certain cancers, since it could allow the development of selective inhibitors that interfere with the processing of specific (subsets of) substrates while sparing the processing of others. Obviously, this would constitute a much safer approach than the clinically tested (and failed) broad spectrum γ‐secretase inhibitors (De Strooper & Chávez Gutiérrez, 2015).
Writing in this issue of The EMBO Journal, Fukumori and Steiner (2016) describe an innovative and extremely labour‐intensive approach to investigate how one particular substrate interacts with the γ‐secretase complex at the level of the single amino acid residue. In a real tour de force, they delineated a substrate‐binding “exosite” in the complex, from which the substrate is then transferred to a “docking site” close to (or even overlapping with) the catalytic site. These translocations imply significant conformational changes in the complex.
Fukumori and Steiner incorporated the photo‐activatable amino acid analogue p‐benzoylphenylalanine (Bpa) at 68 different positions into recombinant APP‐CTF, a 99 amino acid carboxyterminal fragment of the well‐studied γ‐secretase–substrate APP. Each of the resulting 68 substrates was incubated with solubilized γ‐secretase preparations, followed by UV light‐dependent activation of the Bpa moiety that cross‐links the substrate to the complex only if Bpa was < 3 Å distant from potential interacting residues; this can be probed by the resulting shift in migration of the cross‐linked protein in SDS–PAGE. These experiments revealed a series of interactions between the APP‐CTF substrate and the amino‐terminal fragment of the catalytic presenilin subunit of γ‐secretase (PSEN‐NTF). In contrast, the carboxyterminal fragment of presenilin (PSEN‐CTF) appeared to engage in close interactions only with amino acid residues at the membrane‐cytosol border of the substrate. Interestingly, a recent atomic model of γ‐secretase in complex with an inhibitor shows how the cytosolic end of PSEN‐CTF is inserted into the PSEN‐NTF fold, bringing the catalytic Asp385 residue at hydrogen‐bonding distance of its cognate Asp257 site within PSEN‐NTF (Fig 1; Bai et al, 2015b).
Figure 1. PSEN‐NTF‐subunit interacts with substrates during catalysis.
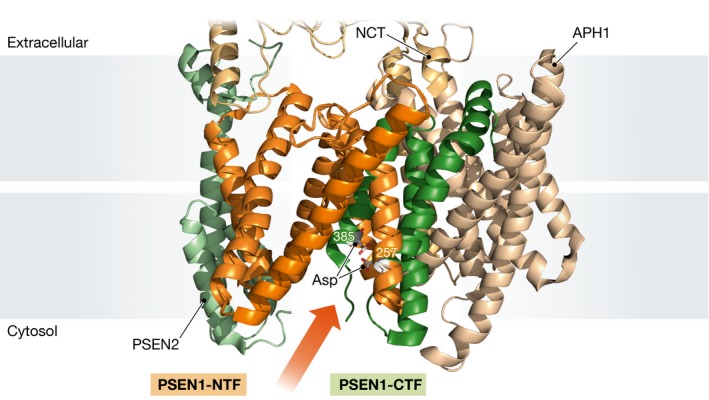
Lateral view of the membrane core of the γ‐secretase complex (PDB entry: 5FN3) with PSEN1‐NTF and ‐CTF coloured in orange and green, respectively, and catalytic Asp residues displayed as rod models. Additional subunits PSEN2, APH1 and NCT are shown in light green, pale brown and pale yellow, respectively. An arrow indicates the putative substrate entrance.
All these data taken together indicate that the substrate is embraced by the PSEN‐NTF during catalysis, except for its carboxy‐terminal end that contacts PSEN‐CTF in a hydrophilic environment. Thus, it is tempting to speculate that a hydrophobic interaction with PSEN‐NTF holds the substrate and aligns it for catalysis by exposing its cytosolic end to the hydrophilic environment where the catalytic aspartate in the PSEN‐CTF transmembrane domain (TM7) lies in waiting. However, the photo‐cross‐linking experiments not only revealed substrate interactions with the catalytic presenilin subunit, but also uncovered interactions with the PEN2 and nicastrin subunits. Nicastrin contacts mainly residues close to the substrate amino‐terminus, while PEN2 cross‐links to regions flanking the APP transmembrane domain. The nicastrin ectodomain has been suggested to bind the free amino‐terminus of substrates (Shah et al, 2005), but this model has been debated (Chavez‐Gutierrez et al, 2008; Zhao et al, 2010); more recent data point to a passive role for the nicastrin ectodomain in substrate recruitment by obstructing the entrance of substrates with bulky ectodomains (Bolduc et al, 2016). Intriguingly, both scenarios imply close proximity between the amino‐terminal region of substrates and the nicastrin ectodomain. Whether this influences substrate specificity and/or catalysis remains to be addressed.
It is highly notable that the observed contact patterns for PEN2 seem to indicate substrate binding to the unstructured, flexible N‐ and C‐termini of PEN2, rather than interactions involving the PEN2 transmembrane domains. PEN2 transmembrane domains are located at opposite from the proposed substrate entrance gate (likely PSEN TM2/TM6), while the flexible PEN2 amino‐terminus lies much closer (< 12 Å in distance) to the cytosolic end of the PSEN transmembrane domain 6 (active site) (Fig 1). Moreover, experimental elongation of the PEN2 amino‐terminus was found to affect substrate processing as observed by altered Aβ42:Aβ40 ratios (Isoo et al, 2007). Based on the new results of Fukumori and Steiner (2016), one may speculate that PEN2 amino‐terminal elongation might affect the architecture of the exosite and alter substrate translocation to the active site to favour the production of Aβ42.
Finally, Fukumori and Steiner (2016) also performed highly interesting and more dynamic pulse‐chase experiments, in which substrate binding and cross‐linking to the enzyme takes place at 4°C and is followed by incubation at 37°C to activate γ‐secretase proteolysis. This showed that substrates bound to the exosite do not get cleaved, while substrates bound to the docking/catalytic site are processed. The authors relate this to the progressive transfer of substrate from exosite to catalytic site and conclude that substrates first need to be transferred out of the exosite to subsequently be cleaved. Consistently, previous reports have shown important conformational changes in γ‐secretase upon binding of substrate (Uemura et al, 2010) or inhibitor (Li et al, 2014; Elad et al, 2015).
A better understanding of how substrates interact with γ‐secretase may help in the design of strategies to selectively inhibit, activate or modulate γ‐secretase‐mediated cleavage(s) of particular substrates. The findings of Fukumori and Steiner (2016) make a step forward into that direction. An important concept supported by the current work is that beyond the recent breakthrough studies on the atomic structure of the enzyme (Bai et al, 2015a,b), different conformations of the complex will probably have to be elucidated to obtain comprehensive understanding of the functioning of these enigmatic complexes. A new era of drug development will start once further functional–structural studies (such as those presented here by Fukumori and Steiner) combined with profound structural analysis will have brought dynamics to the structures of these complex intramembrane proteolysis machines, leading to more effective and safer drugs in the fight against Alzheimer's disease and cancer.
See also: A Fukumori & H Steiner (August 2016)
Contributor Information
Lucía Chávez‐Gutiérrez, Email: lucia.chavezgutierrez@cme.vib-kuleuven.be.
Bart De Strooper, Email: bart.destrooper@cme.vib-kuleuven.be.
References
- Bai X, Yan C, Yang G, Lu P, Sun L, Zhou R, Scheres SHW, Shi Y (2015a) An atomic structure of human γ‐secretase. Nature 525: 212–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai XC, Rajendra E, Yang G, Shi Y, Scheres SH (2015b) Sampling the conformational space of the catalytic subunit of human gamma‐secretase. Elife 4: e11182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc DM, Montagna DR, Gu Y, Selkoe DJ, Wolfe MS (2016) Nicastrin functions to sterically hinder γ‐secretase‐substrate interactions driven by substrate transmembrane domain. Proc Natl Acad Sci USA 113: E509–E518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez‐Gutiérrez L, Bammens L, Benilova I, Vandersteen A, Benurwar M, Borgers M, Lismont S, Zhou L, Van Cleynenbreugel S, Esselmann H, Wiltfang J, Serneels L, Karran E, Gijsen H, Schymkowitz J, Rousseau F, Broersen K, De Strooper B (2012) The mechanism of γ‐secretase dysfunction in familial Alzheimer disease. EMBO J 31: 2261–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez‐Gutierrez L, Tolia A, Maes E, Li T, Wong PC, de Strooper B, Chávez‐Gutiérrez L, Tolia A, Maes E, Li T, Wong PC, de Strooper B (2008) Glu(332) in the Nicastrin ectodomain is essential for gamma‐secretase complex maturation but not for its activity. J Biol Chem 283: 20096–20105 [DOI] [PubMed] [Google Scholar]
- De Strooper B, Chávez Gutiérrez L (2015) Learning by failing: ideas and concepts to tackle γ‐secretases in Alzheimer's disease and beyond. Annu Rev Pharmacol Toxicol 55: 419–437 [DOI] [PubMed] [Google Scholar]
- Elad N, De Strooper B, Lismont S, Hagen W, Veugelen S, Arimon M, Horre K, Berezovska O, Sachse C, Chavez‐Gutierrez L (2015) The dynamic conformational landscape of gamma‐secretase. J Cell Sci 128: 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumori A, Steiner H (2016) Substrate recruitment of γ‐secretase and mechanism of clinical presenilin mutations revealed by photoaffinity mapping. EMBO J 35: 1628–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoo N, Sato C, Miyashita H, Shinohara M, Takasugi N, Morohashi Y, Tsuji S, Tomita T, Iwatsubo T (2007) Aβ42 overproduction associated with structural changes in the catalytic pore of γ‐secretase: common effects of Pen‐2 N‐terminal elongation and fenofibrate. J Biol Chem 282: 12388–12396 [DOI] [PubMed] [Google Scholar]
- Jurisch‐Yaksi N, Sannerud R, Annaert W (2013) A fast growing spectrum of biological functions of gamma‐secretase in development and disease. Biochim Biophys Acta 1828: 2815–2827 [DOI] [PubMed] [Google Scholar]
- Li Y, Lu SH, Tsai CJ, Bohm C, Qamar S, Dodd RB, Meadows W, Jeon A, McLeod A, Chen F, Arimon M, Berezovska O, Hyman BT, Tomita T, Iwatsubo T, Johnson CM, Farrer LA, Schmitt‐Ulms G, Fraser PE, St George‐Hyslop PH (2014) Structural interactions between inhibitor and substrate docking sites give insight into mechanisms of human PS1 complexes. Structure 22: 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud R, Esselens C, Ejsmont P, Mattera R, Rochin L, Tharkeshwar AK, De Baets G, De Wever V, Habets R, Baert V, Vermeire W, Michiels C, Groot AJ, Wouters R, Dillen K, Vints K, Baatsen P, Munck S, Derua R, Waelkens E et al (2016) Restricted location of PSEN2/γ‐secretase determines substrate specificity and generates an intracellular Aβ pool. Cell 166: 193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, Ball H, Dann CE III, Sudhof T, Yu G (2005) Nicastrin functions as a gamma‐secretase‐substrate receptor. Cell 122: 435–447 [DOI] [PubMed] [Google Scholar]
- Uemura K, Farner KC, Hashimoto T, Nasser‐Ghodsi N, Wolfe MS, Koo EH, Hyman BT, Berezovska O (2010) Substrate docking to gamma‐secretase allows access of gamma‐secretase modulators to an allosteric site. Nat Commun 1: 130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Liu Z, Ilagan MX, Kopan R (2010) Gamma‐secretase composed of PS1/Pen2/Aph1a can cleave notch and amyloid precursor protein in the absence of nicastrin. J Neurosci 30: 1648–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]


