Abstract
The endoplasmic reticulum (ER) plays a central role in the biogenesis of most membrane proteins. Among these are proteins localized to the surface of lipid droplets (LDs), fat storage organelles delimited by a phospholipid monolayer. The LD monolayer is often continuous with the membrane of the ER allowing certain membrane proteins to diffuse between the two organelles. In these connected organelles, how some proteins concentrate specifically at the surface of LDs is not known. Here, we show that the ERAD ubiquitin ligase Doa10 controls the levels of some LD proteins. Their degradation is dependent on the localization to the ER and appears independent of the folding state. Moreover, we show that by degrading the ER pool of these LD proteins, ERAD contributes to restrict their localization to LDs. The signals for LD targeting and Doa10‐mediated degradation overlap, indicating that these are competing events. This spatial control of protein localization is a novel function of ERAD that might contribute to generate functional diversity in a continuous membrane system.
Keywords: Doa10, endoplasmic reticulum, ERAD, lipid droplets, protein degradation
Subject Categories: Membrane & Intracellular Transport, Protein Biosynthesis & Quality Control
Introduction
The endoplasmic reticulum (ER) plays a central role in the biogenesis of membrane and secretory proteins, facilitating the folding and the post‐translational modifications necessary for their function (Braakman & Hebert, 2013). Protein folding in the ER is under the surveillance of stringent quality control and polypeptides failing to acquire the native structure are eliminated by ER‐associated degradation (or ERAD) (Smith et al, 2011; Christianson & Ye, 2014; Ruggiano et al, 2014). This process involves the recognition of a substrate, its ubiquitination by an ER ubiquitin ligase, membrane extraction facilitated by the cytoplasmic Cdc48 ATPase, and delivery to the proteasome for degradation. These events are coordinated by ER membrane‐embedded protein complexes that have at their core an ubiquitin ligase. While highly conserved across eukaryotes, the mechanisms of ERAD are better characterized in the yeast S. cerevisiae. In yeast, genetic and biochemical studies identified three ubiquitin ligase complexes involved in ERAD, the Hrd1, Doa10, and Asi complexes, showing different specificity for misfolded proteins (Hampton et al, 1996; Swanson et al, 2001; Carvalho et al, 2006; Denic et al, 2006; Gauss et al, 2006; Foresti et al, 2014; Khmelinskii et al, 2014). Besides misfolded proteins, Hrd1 and Doa10 complexes were shown to degrade some folded, functional proteins but in a regulated manner, only upon a specific signal. Regulated degradation is important to control certain ER functions, such as sterol biosynthesis (Ruggiano et al, 2014). The Asi complex localizes specifically to the inner nuclear membrane (INM), preventing the accumulation of misfolded proteins in this highly specialized ER domain (Boban et al, 2006; Foresti et al, 2014; Khmelinskii et al, 2014). Moreover, the Asi complex degrades ER proteins mistargeted to the INM, suggesting that ERAD also integrates spatial cues.
The ER also has a direct role in the biogenesis of other organelles, such as lipid droplets (LDs) (Thiam et al, 2013; Pol et al, 2014). These storage organelles consist of a core of neutral lipids, mainly triacylglycerides (TAG) and sterol esters, enclosed by a phospholipid monolayer and a set of LD‐specific proteins, primarily enzymes promoting the synthesis, remodeling, and consumption of lipids in LDs. Therefore, the metabolic status of individual LDs is largely determined by the proteins at their surface. Typical membrane proteins with hydrophilic domains on both sides of the bilayer are not favorably accommodated in the monolayer of LDs; the association of proteins with the surface of these organelles is instead mediated by either amphipathic helices or hydrophobic hairpins (Thiam et al, 2013). Proteins with the former motif are recruited to LDs directly from the cytoplasm. In contrast, proteins of the latter type are initially targeted and membrane‐inserted in the ER, and subsequently targeted to the LD monolayer, which in yeast and in a large fraction of mammalian LDs is continuous with the outer leaflet of the ER membrane (Jacquier et al, 2011; Wilfling et al, 2013). How, among all the ER membrane proteins containing hydrophobic hairpins, some concentrate specifically at the LD monolayer is not entirely clear. In a few cases, positively charged amino acids flanking the hairpin favor their retention in LDs; however, a consensus signal or sequence has not been identified (Ingelmo‐Torres et al, 2009). Moreover, it is unclear why some proteins concentrate in LDs soon after their integration at the ER, while others accumulate slower and, in some cases, depending on the metabolic state of the cells. For example, in quiescent yeast cells the enzyme Dga1 localizes to LDs where it synthesizes TAG, while in dividing cells a prominent fraction of Dga1 localizes to the ER where it is less active (Oelkers et al, 2002; Sorger & Daum, 2002; Jacquier et al, 2011; Markgraf et al, 2014).
Here, we identified a subset of LD proteins as substrates of the ERAD ubiquitin ligase Doa10. We show that ERAD targets specifically the ER pool of these proteins. The common feature among these Doa10 clients is the presence of a hydrophobic hairpin involved in LD targeting. We show that the signals for Doa10‐mediated degradation and for LD targeting overlap, indicating that these are competing events and a potential target for regulation. Altogether, our data indicate that ERAD‐mediated degradation of a subset of LD proteins in the ER restricts their localization to the LD surface. We propose a novel function of ERAD in protein spatial control that contributes to organelle identity by limiting the accumulation of LD‐specific proteins in the ER.
Results
ERAD degrades LD proteins
Quantitative proteomic screenings recently performed in our laboratory generated a long list of potential endogenous ERAD substrates (Foresti et al, 2013, 2014). Among these, the LD proteins Pgc1, Dga1, and Yeh1 were over‐represented in the doa10Δ mutant in comparison with wt cells, suggesting that their abundance might be controlled by ERAD. Curiously, doa10Δ cells also have defects in LD morphology strengthening a potential connection between ERAD and LD regulation (Fei et al, 2008, 2009). The levels of other LD proteins, such as Erg6, Pet10, Osw5, or Hfd1, were unaffected in doa10Δ cells, as detected by SILAC and cycloheximide chase experiments. Here, we focused on Pgc1 to characterize the Doa10‐mediated degradation of certain LD proteins. To directly assess the role of Doa10 in controlling Pgc1 levels, we performed cycloheximide chase experiments. In wt cells, endogenously expressed Pgc1 was short‐lived (half‐life of ~45 min). In agreement with the proteomics data, its degradation was significantly delayed in cells lacking the ubiquitin ligase Doa10 or its binding partners Ubc6 and Ubc7, but was not affected in hrd1Δ cells, lacking another ERAD ubiquitin ligase (Fig 1A). To further characterize Pgc1 degradation, we analyzed its ubiquitination. While in wt cells ubiquitin‐conjugated 3HA‐Pgc1 was readily detected, Pgc1 ubiquitination was reduced in cells lacking Doa10 (Fig 1B). Although decreased, neither the degradation nor the ubiquitination of Pgc1 was blocked in doa10Δ cells, prompting us to search for additional ubiquitin ligases involved in its degradation. While deletion of all three ERAD ubiquitin ligases in doa10Δ hrd1Δ asi1Δ cells did not further stabilize Pgc1 (Fig EV1A), the soluble ubiquitin ligase Ubr1 was redundant with Doa10 as Pgc1 was more stable in doa10Δ ubr1Δ mutant (Fig EV1B). In fact, in ubr1Δ cells Pgc1 degradation was slowed down, even if not to the same extent as in doa10Δ mutant. Similar results were obtained for the LD proteins Dga1 and Yeh1, also identified in the SILAC dataset (Fig EV2). In agreement with these findings, Ubr1 was shown to be redundant with Doa10 in the degradation of other membrane‐bound substrates (Stolz et al, 2013), suggesting a broad role of this soluble ubiquitin ligase in ERAD.
Figure 1. The LD protein Pgc1 is an ERAD substrate.
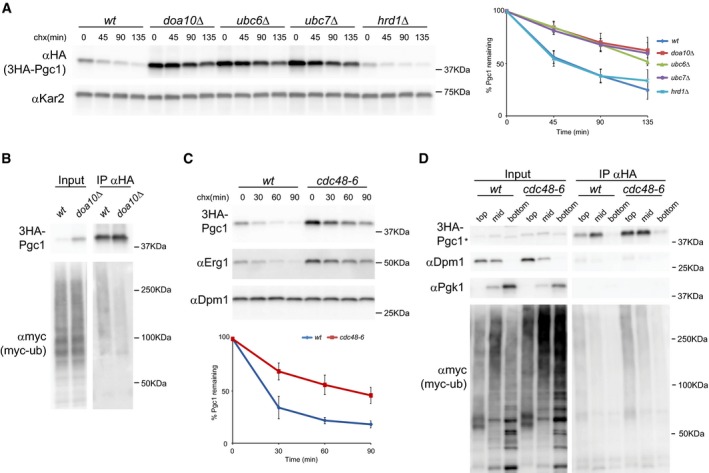
- The degradation of 3HA‐Pgc1 was analyzed in cells with the indicated genotype upon inhibition of protein synthesis with cycloheximide (chx). A plasmid‐borne 3HA‐Pgc1 expressed from the endogenous promoter was used. 3HA‐Pgc1 was detected with anti‐HA antibodies. Kar2 was used as a loading control and detected with anti‐Kar2 antibodies. The graph on the right shows the average of four independent experiments; error bars represent the standard deviation.
- The ubiquitination of 3HA‐Pgc1 was analyzed in cells with the indicated genotype and expressing myc‐tagged ubiquitin. 3HA‐Pgc1 was immunoprecipitated using anti‐HA and polyubiquitin‐conjugated 3HA‐Pgc1 was detected using anti‐myc antibody. Input fraction corresponds to 2% of the total amount used for IP.
- The degradation of 3HA‐Pgc1 was analyzed as in (A) in cells bearing the CDC48 temperature‐sensitive allele cdc48‐6. Cells were grown 2.5 h at 37°C prior to addition of cycloheximide. Inactivation of Cdc48 mutant protein was confirmed by stabilization of the ERAD substrate Erg1 in the same cells. The graph shows the average of four independent experiments; error bars represent the standard deviation.
- Membrane association of ubiquitinated 3HA‐Pgc1 was analyzed by density gradient centrifugation followed by immunoprecipitation. Cells of the indicated genotype and expressing myc‐tagged ubiquitin were grown as in (C). Lysates were subjected to centrifugation on an Optiprep density gradient. Collected fractions were analyzed by Western blotting before and after immunoprecipitation with anti‐HA antibodies. Dpm1 and Pgk1 were used as markers for membrane‐bound and soluble fractions, respectively. Input fraction corresponds to 10% of the total amount used for IP. “Top”, “mid”, and “bottom” indicate the fractions from the gradient. Asterisk marks a faster migrating unspecific band in the input bottom fraction.
Source data are available online for this figure.
Figure EV1. Pgc1 degradation is independent of the vacuolar protease Pep4 but requires Doa10 and the cytoplasmic ubiquitin ligase Ubr1.
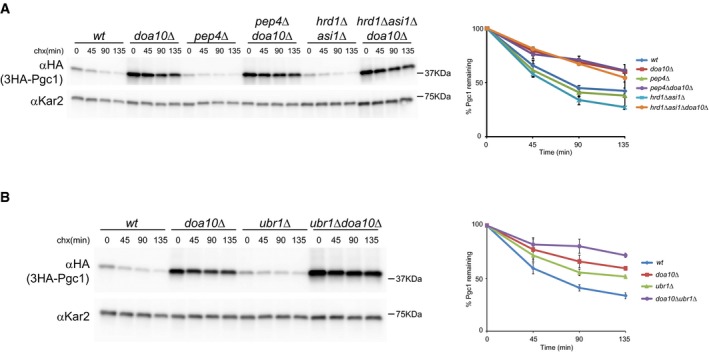
-
A, BThe degradation of 3HA‐Pgc1 was analyzed in cells with the indicated genotype as described in Fig 1A. The graphs show the average of two independent experiments; error bars represent the standard deviation.
Figure EV2. The LD proteins Dga1 and Yeh1 are degraded by the ERAD ubiquitin ligase Doa10.
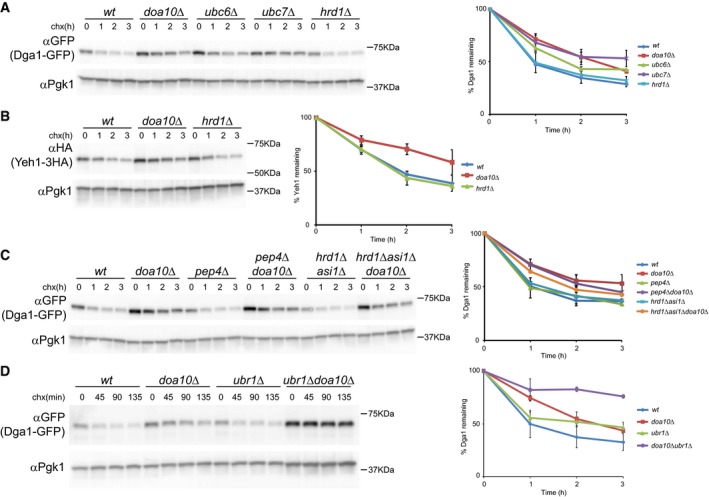
-
AThe degradation of Dga1‐GFP was analyzed in cells with the indicated genotype as described in Fig 1A. A plasmid‐borne GFP‐Dga1 expressed from the constitutive ADH1 promoter was used. The graph shows the average of two independent experiments; error bars represent the standard deviation.
-
BThe degradation of Yeh1‐3HA was analyzed in cells with the indicated genotype as described in Fig 1A. A plasmid‐borne Yeh1‐3HA expressed from the endogenous promoter was used. The graph shows the average of two independent experiments; error bars represent the standard deviation.
-
C, DThe degradation of Dga1‐GFP was analyzed in cells with the indicated genotype as described in Fig 1A. The graphs show the average of two independent experiments; error bars represent the standard deviation.
Next, we tested the involvement of Cdc48 in the degradation of LD proteins. Inactivation of Cdc48 in cells expressing the temperature‐sensitive allele cdc48‐6 (Schuberth & Buchberger, 2005) strongly delayed the degradation of Pgc1 (Fig 1C) and Dga1 (Fig EV3A). Moreover, cdc48‐6 cells accumulated large amounts of membrane‐bound, ubiquitinated Pgc1 (Fig 1D). These findings are consistent with the well‐characterized role of Cdc48 in releasing ubiquitinated ERAD substrates from the ER membrane into the cytoplasm for degradation by the proteasome (Bays et al, 2001; Ye et al, 2001; Braun et al, 2002; Jarosch et al, 2002; Rabinovich et al, 2002; Stein et al, 2014). However, it should be noted that a less stringent Cdc48 allele, cdc48‐3 (Latterich et al, 1995), while stabilizing other ERAD substrates including the Doa10 substrate Erg1, had a negligible effect on the degradation of Pgc1 and Dga1 (Fig EV3B and C). To resolve the inconsistency between the two CDC48 mutant alleles on the degradation of Pgc1, we sequenced them. Interestingly, we found that cdc48‐3 is mutated in the D1 ATPase domain (P257L and R387K), whereas, in agreement with its tighter phenotype, cdc48‐6 contains mutations in both D1 (P257L) and D2 (A540T) ATPase domains. Thus, while membrane extraction of ER luminal and polytopic proteins requires wt Cdc48, ERAD of LD proteins needs only residual Cdc48 activity. This decreased requirement for Cdc48 suggests that membrane extraction of hairpin‐containing proteins like Pgc1 and Dga1 may need only a single Cdc48 ATPase cycle while extraction of polytopic ERAD substrates might require processive and/or multiple rounds of Cdc48 activity.
Figure EV3. Degradation of Pgc1 and Dga1 in cdc48 mutant cells.
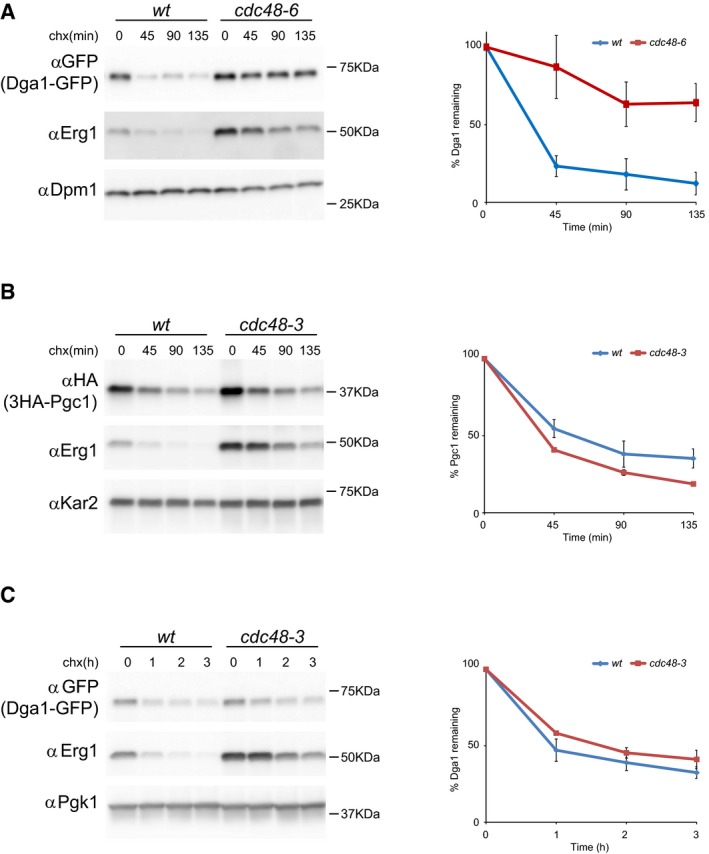
-
AThe degradation of Dga1‐GFP in temperature‐sensitive cdc48‐6 mutant cells was analyzed as in Fig 1C. The graph shows the average of three independent experiments; error bars represent the standard deviation.
-
B, CThe degradation of 3HA‐Pgc1 and Dga1‐GFP in temperature‐sensitive cdc48‐3 mutant cells was analyzed as described in Fig 1C. The graphs show the average of two independent experiments; error bars represent the standard deviation.
Mutants with impaired proteasomal function, such as pre2, showed delayed elimination of Pgc1 and Dga1, indicating that their degradation is proteasome‐dependent (Fig EV4; Heinemeyer et al, 1993). Consistent with the proteasome involvement, impairment of vacuolar proteolytic activity by PEP4 mutation did not affect Pgc1 or Dga1 turnover (Figs EV1A and EV2C; Ammerer et al, 1986). Altogether, these data show that Pgc1, Dga1, and Yeh1 are bona fide ERAD substrates of the Doa10 complex.
Figure EV4. Pgc1 and Dga1 are degraded by the proteasome.
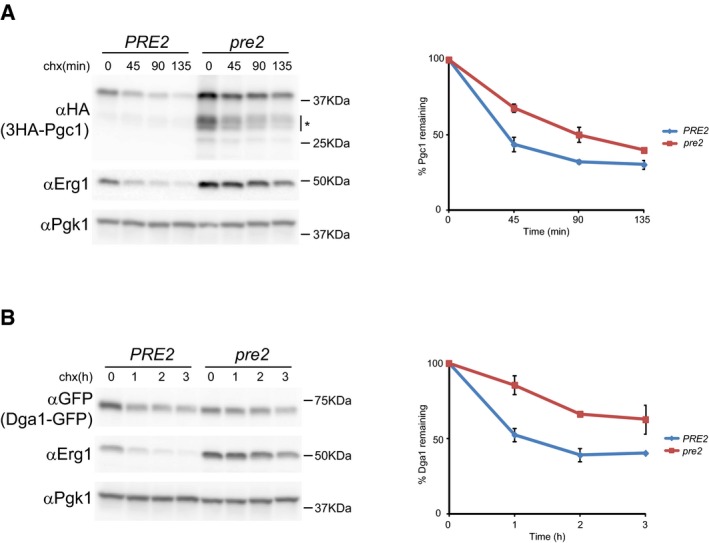
- The degradation of 3HA‐Pgc1 was analyzed in proteasome‐deficient (pre2) and wt control (PRE2) cells as described in Fig 1A. Asterisk indicates degradation products accumulating in proteasome‐deficient cells. The graph shows the average of two independent experiments; error bars represent the standard deviation.
- The degradation of Dga1‐GFP was analyzed in proteasome‐deficient (pre2) and wt control (PRE2) cells as in Fig 1A. The graph shows the average of two independent experiments; error bars represent the standard deviation.
Pgc1 is a membrane‐anchored protein stably associated with LDs
Pgc1 is a protein of unknown function predicted to have a glycerophosphodiester phosphodiesterase motif and to associate with membranes through a C‐terminal hydrophobic segment (Beilharz et al, 2003; Fernandez‐Murray & McMaster, 2005; Fisher et al, 2005). In recent proteomic analysis, it was identified as a high‐confidence LD protein (Grillitsch et al, 2011; Currie et al, 2014). Indeed, when expressed from the constitutive alcohol dehydrogenase (ADH1) promoter as a N‐terminal GFP fusion, GFP‐Pgc1 localized to LDs in both wt and doa10Δ cells (Fig 2A). In the doa10Δ mutant, GFP‐Pgc1 was also detected at the nuclear and cortical ER (Fig 2A). In most wt cells, GFP‐Pgc1 was virtually undetectable at the ER. Next, we analyzed the role of the C‐terminal hydrophobic region in Pgc1 membrane association (Fig 2B). Upon subcellular fractionation, endogenously expressed Pgc1 bearing a N‐terminal HA epitope was found in the microsomal fraction (Fig 2C, mock), which under the isolation conditions also contains LDs. Importantly, the majority of Pgc1 maintained its microsomal association after alkaline treatment, which removes peripherally associated proteins such as Kar2, and it was only released upon detergent solubilization of membranes. A similar behavior was displayed by a truncated version encoding the last 47 amino acids of Pgc1 (3HA‐GFP‐Pgc1275–321) encompassing the predicted hydrophobic region (Fig 2D). Thus, Pgc1 is anchored to membranes through its hydrophobic C‐terminal region. In silico analysis through the server TOPCONS predicted that this hydrophobic C‐terminus adopts a hairpin configuration, which is typical of many other LD proteins (including Dga1; Tsirigos et al, 2015). A number of hairpin‐containing LD proteins have been shown to target the ER membrane before concentrating on LDs (Jacquier et al, 2011; Wilfling et al, 2013). Several lines of evidence indicate that Pgc1 displays a similar behavior. First, in cells lacking LDs, such as the are1Δ are2Δ lro1Δ dga1Δ mutant deficient in neutral lipid synthesis (Sandager et al, 2002; Sorger et al, 2004), Pgc1 co‐localized with the ER marker Sec63‐Cherry (Fig 3A). Second, experiments where LD formation was initiated upon galactose induction of DGA1 expression in are1Δ are2Δ lro1Δ mutant (Jacquier et al, 2011) showed that ER labeling of GFP‐Pgc1 decreases and the protein gradually concentrated on LDs as they formed. The kinetics of Pgc1 accumulation in LDs was comparable to the one of Erg6, a well‐characterized LD marker protein (Fig 3B) (Grillitsch et al, 2011; Jacquier et al, 2011; Currie et al, 2014). To exclude that LD‐associated Pgc1 had been recruited from a cytosolic pool, similar LD induction experiments were performed in cells expressing Pgc1 fused to photoconvertible EOS (EOS‐Pgc1) (McKinney et al, 2009). EOS‐Pgc1 photoconverted at the ER was detected at LDs as these formed (Fig 3C). These experiments demonstrate that Pgc1 traffics through the ER en route to LDs. Importantly, the pool of Pgc1 at LDs is relatively stable, not diffusing back to the ER, as revealed in photobleaching experiments (Fig 3D), suggesting that the LD monolayer provides a more favorable environment to Pgc1 hydrophobic hairpin.
Figure 2. Pgc1 behaves as an integral membrane protein.
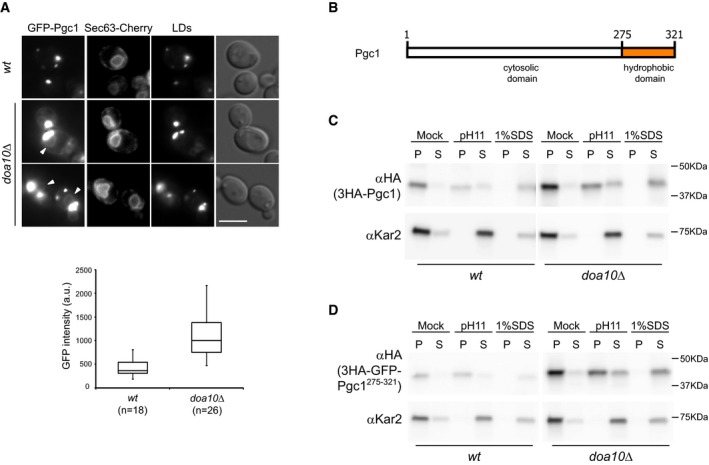
- Localization of chromosomally expressed GFP‐Pgc1 from the constitutive ADH1 promoter in wt and doa10Δ cells. Arrowheads indicate GFP‐Pgc1 labeling at the ER, which is stained by Sec63‐Cherry. LDs were visualized upon staining with the neutral lipid dye MDH. On the bottom, box plot with quantitation of GFP‐Pgc1 fluorescence intensity at the nuclear envelope in wt and doa10Δ. Horizontal lines indicate median, first and third quartiles of the distribution; whiskers indicate maximum and minimum values. GFP‐Pgc1 measurements were taken as described in the Materials and Methods section. Scale bar: 5 μm.
- Schematic representation of Pgc1. The location of the predicted hydrophobic domain is indicated.
- Crude membranes from wt and doa10Δ cells expressing 3HA‐Pgc1 were subjected to the indicated treatments and subsequently fractionated into membrane pellet (P) and supernatant (S).
- Crude membranes from wt and doa10Δ cells expressing 3HA‐GFP‐Pgc1275–321 were analyzed as in (C).
Source data are available online for this figure.
Figure 3. Pgc1 localizes to the ER before stably concentrating on LDs.
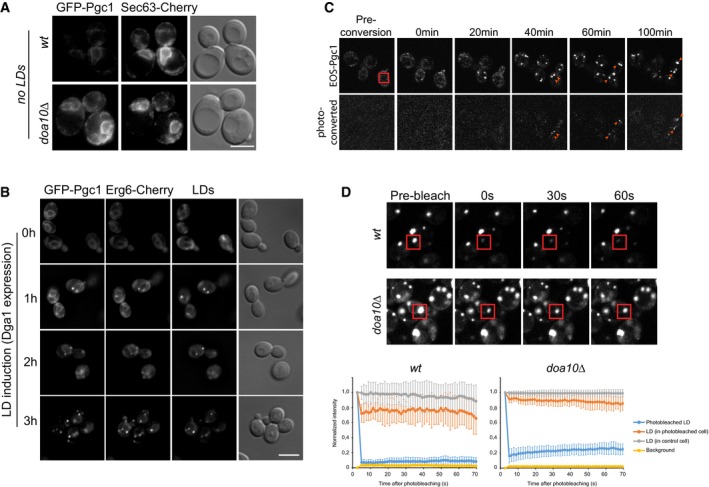
- Localization of GFP‐Pgc1 in are1Δ are2Δ lro1Δ dga1Δ cells in the presence (no LDs) or absence of DOA10 (no LDs doa10Δ). The ER was visualized by expression of Sec63‐Cherry. Scale bar: 5 μm.
- GFP‐Pgc1 was expressed under the ADH1 promoter; LD formation was stimulated by galactose‐induced expression of DGA1 in are1Δ are2Δ lro1Δ. Fluorescence microscopy was used to follow GFP‐Pgc1 localization over time. LDs were visualized upon staining with the neutral lipid dye MDH. Scale bar: 5 μm.
- Photoconversion of tdEOS‐Pgc1 expressed under the ADH1 promoter; LD formation was stimulated by galactose‐induced expression of DGA1 in are1Δ are2Δ lro1Δ doa10Δ cells. The red square marks the photoconverted region. The time (in minutes) after photoconversion is indicated. Arrowheads point to LDs containing photoconverted tdEOS‐Pgc1. Scale bar: 5 μm.
- FRAP experiment of GFP‐Pgc1 in wt and doa10Δ cells. Representative examples are shown. The bleached areas are marked by red squares and include a LD adjacent to ER. The time (in seconds) after photobleaching is indicated. Each graph shows average fluorescence intensities for 10 cells normalized to pre‐bleached plotted over time. Error bars indicate standard deviation.
Pgc1 is degraded by Doa10 at the ER
Since the Doa10 complex localizes exclusively at the ER, it would be expected that restricting Pgc1 to this organelle, as in the are1Δ are2Δ lro1Δ dga1Δ mutant, would result in its faster degradation. Indeed, Pgc1 degradation was significantly accelerated in this mutant (Fig 4A). In the absence of LDs, Pgc1 degradation was still dependent on Doa10, as the protein was stabilized by additional mutation of this ubiquitin ligase while deletion of HRD1 had no effect. On the other hand, expansion of LD surface by oleate feeding, decreased TAG lipolysis (in tgl3Δ tgl4Δ tgl5Δ cells) or both delayed Pgc1 turnover (Fig 4B). Importantly, the kinetics of degradation of Vma12‐Ndc10C', a Doa10 substrate that does not localize to LDs (Furth et al, 2011), was unaffected indicating that the treatment affects specifically LD‐localized Pgc1. These experiments show that Doa10 promotes the degradation of the ER pool of Pgc1 while LD‐localized Pgc1 is spared from degradation.
Figure 4. Pgc1 is degraded by Doa10 at the ER .
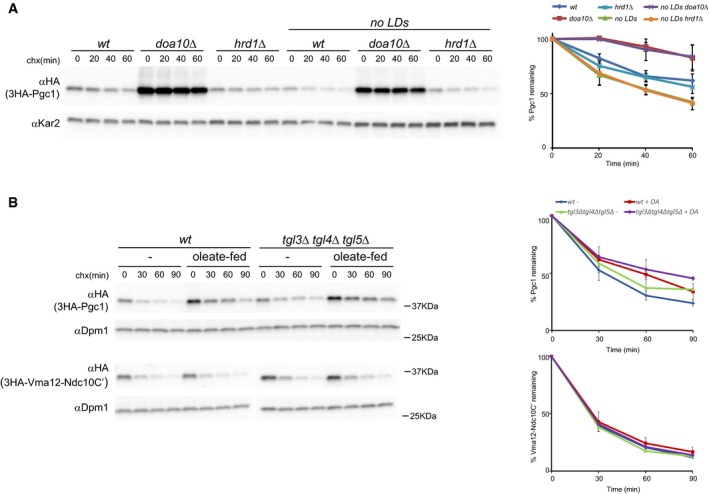
- The degradation of 3HA‐Pgc1 was analyzed in cells with the indicated genotype as in Fig 1A. The graph shows the average of three independent experiments; error bars represent the standard deviation.
- The degradation of the Doa10 substrates 3HA‐Pgc1 and 3HA‐Vma12‐Ndc10C' was analyzed in cells of the indicated genotypes treated with oleic acid. 3HA‐Pgc1 and 3HA‐Vma12‐Ndc10C' were detected with anti‐HA antibodies. Dpm1 was used as a loading control and detected with anti‐Dpm1 antibodies. The graph shows the average of three independent experiments; error bars represent the standard deviation.
Source data are available online for this figure.
Pgc1 hydrophobic hairpin is necessary and sufficient for Doa10‐dependent degradation
Next, we analyzed whether Pgc1 degradation required its hydrophobic hairpin. Derivatives of Pgc1 in which the hydrophobic hairpin (residues 275–321) was replaced by the membrane anchor (MA) of the ER proteins Scs2 (Pgc1Scs2MA; Loewen et al, 2007) or Bos1 (Pgc1Bos1MA; Lian & Ferro‐Novick, 1993) were generated and their localization analyzed by fluorescence microscopy. In both wt and doa10Δ cells, the two chimeric proteins co‐localized with the ER marker Sec63 and were excluded from LDs (Fig 5A). These data indicate that LD targeting of Pgc1 requires its MA. Despite their ER localization, the chimeric constructs Pgc1Scs2MA and Pgc1Bos1MA were stable, showing that Pgc1 hydrophobic hairpin is also required for the Doa10‐mediated ERAD (Fig 5B). Conversely, the construct 3HA‐GFP‐Pgc1275–321 encoding for Pgc1 hydrophobic hairpin was extremely short‐lived in wt cells while its turnover was strongly delayed in doa10Δ mutants (Fig 5C). Thus, Pgc1 hydrophobic hairpin is necessary and sufficient for its Doa10‐dependent degradation. The extremely short half‐life of 3HA‐GFP‐Pgc1275–321 in wt cells precluded its detection by fluorescence microscopy. In contrast, in doa10Δ cells, 3HA‐GFP‐Pgc1275–321 was readily detected and showed a dual localization, with a pool at the ER and another at LDs (Fig 5D). Altogether, these data indicate that Pgc1 hydrophobic hairpin is necessary and sufficient for its LD targeting. Moreover, they show that the same region of Pgc1 acts as degradation signal (or degron) for Doa10‐mediated ERAD. The overlap of signals promoting LD localization and ERAD targeting offers the potential for regulating these competing events, for example, depending on the metabolic status of the cells.
Figure 5. Pgc1 hydrophobic hairpin is necessary and sufficient for Doa10‐dependent degradation.
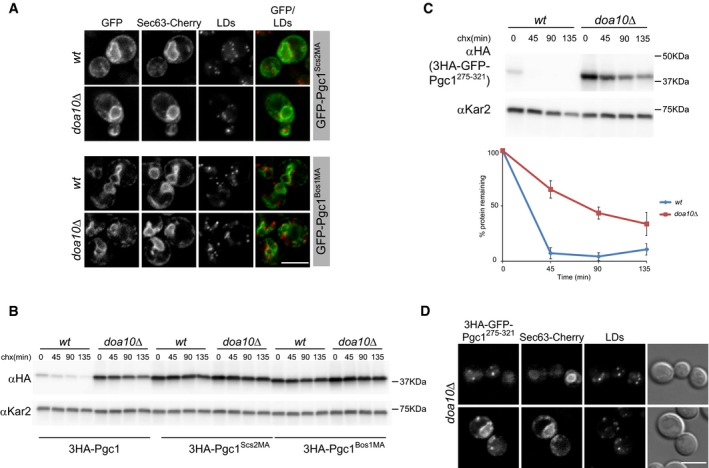
- Localization of GFP‐tagged Pgc1 derivatives in which the hydrophobic hairpin (aa275–321) was replaced by membrane anchor of Scs2 (Pgc1Scs2MA) or Bos1 (Pgc1Bos1MA). ER and LDs were visualized by expression of Sec63‐Cherry and MDH staining, respectively. Scale bar: 5 μm.
- The degradation of 3HA‐Pgc1 or the indicated chimeras in wt and doa10Δ cells was analyzed as in Fig 1A. The blot is representative of at least three independent experiments.
- The degradation of 3HA‐GFP‐Pgc1275–321 in wt and doa10Δ cells was analyzed as in Fig 1A. The graph shows the average of four independent experiments; error bars represent the standard deviation.
- Localization of 3HA‐GFP‐Pgc1275–321 in doa10Δ cells. The ER was visualized by expression of Sec63‐Cherry. Scale bar: 5 μm.
Source data are available online for this figure.
Hairpins of LD proteins can serve as degrons for Doa10 ERAD
Several proteins exchanging between ER and LDs were shown to associate with membranes through a hydrophobic hairpin (Jacquier et al, 2011; Wilfling et al, 2013). Among these are the yeast lipases Tgl1 and Yeh1, the diacylglycerol acyltransferase Dga1, its mammalian homologue DGAT2, or the Drosophila GPAT4. Our results indicate that Pgc1 might be another of such proteins. Since Pgc1, Yeh1, and Dga1 are Doa10 substrates, we wondered whether hairpins mediating LD targeting of certain proteins could function as a general degradation signal for Doa10. To directly test this possibility, we replaced the hydrophobic hairpin of Pgc1 with the heterologous hairpin of GPAT4. This domain (residues 160–216) was shown to be sufficient to target mCherry to LDs in Drosophila cultured cells (Wilfling et al, 2013). While in wt cells 3HA‐Pgc1‐GPAT4160–216 was a short‐lived protein, deletion of DOA10 strongly increased the half‐life of the chimeric protein (Fig 6A). Like Pgc1, 3HA‐Pgc1‐GPAT4160–216 was further stabilized in doa10Δ ubr1Δ. In contrast, deletion of HRD1 had no effect on 3HA‐Pgc1‐GPAT4160–216 degradation (Fig EV5). Importantly, the chimeric construct GFP‐Pgc1‐GPAT4160–216 localized to LDs in wt cells, indicating that GPAT4 hydrophobic hairpin is a functional LD targeting signal in yeast (Fig 6B). Moreover, conditions that strongly stabilized GFP‐Pgc1‐GPAT4160–216, such as doa10Δ ubr1Δ mutant, lead to its accumulation at the ER besides LDs (Fig 6B). To further characterize the behavior of 3HA‐Pgc1‐GPAT4160–216, we analyzed its degradation in the absence of LDs. Like Pgc1, the degradation of the chimeric protein was strongly accelerated in are1Δ are2Δ lro1Δ dga1Δ cells (no LDs) (half‐life < 20′ in no LDs vs. ~45′ in wt cells; Fig 6C). Importantly, degradation of 3HA‐Pgc1‐GPAT4160–216 in this background was substantially delayed by additional deletion of DOA10 (Fig 6C). The longer half‐life of Pgc1‐GPAT4160–216 in the mutant are1Δ are2Δ lro1Δ dga1Δ doa10Δ allowed us to confirm that in the absence of LDs, the GFP‐tagged chimeric protein indeed localized to the ER, like full‐length Pgc1 (Fig 6D). Thus, the chimera containing the well‐characterized GPAT4 hairpin behaves as wt Pgc1, indicating that this LD targeting motif serves as a generic Doa10 degron.
Figure 6. A heterologous hydrophobic hairpin functions as a Doa10 degron.
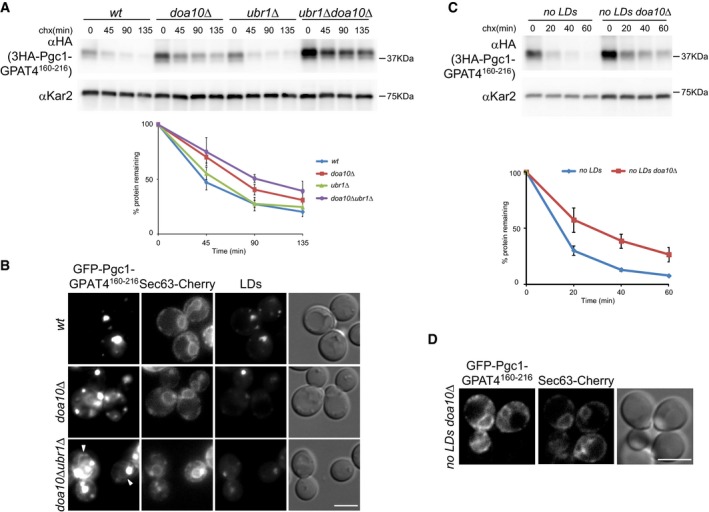
- The degradation of 3HA‐Pgc1‐GPAT160–216 containing GPAT4 hydrophobic hairpin (aa160–216) replacing the one of Pgc1 was analyzed in cells with the indicated genotype as in Fig 1A. The graph shows the average of three independent experiments; error bars represent the standard deviation.
- Localization of GFP‐Pgc1‐GPAT4160–216 in cells with the indicated genotype. Arrowheads indicate GFP‐Pgc1‐GPAT4160–216 labeling at the ER, marked with Sec63‐Cherry. LDs were visualized upon staining with MDH. Scale bar: 5 μm.
- The degradation of 3HA‐Pgc1‐GPAT160–216 in cells with the indicated genotype was analyzed as in Fig 1A. The graph shows the average of three independent experiments; error bars represent the standard deviation.
- Localization of GFP‐Pgc1‐GPAT160–216 in are1Δ are2Δ lro1Δ dga1Δ doa10Δ mutant (no LDs doa10Δ). The ER was visualized by expression of Sec63‐Cherry. Scale bar: 5 μm.
Source data are available online for this figure.
Figure EV5. The chimeric protein 3HA‐Pgc1‐GPAT160–216 is a Doa10 substrate.
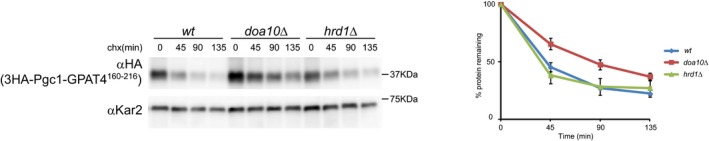
The degradation of 3HA‐Pgc1‐GPAT160–216 was analyzed in cells with the indicated genotype as in Fig 1A. The graph shows the average of three independent experiments; error bars represent the standard deviation.
Discussion
Here, we uncover a new class of substrates of the ERAD ubiquitin ligase Doa10. These are proteins that contain a hydrophobic hairpin and that localize to ER and LD membranes. By degrading specifically the ER pool, ERAD restricts their localization to LDs, thereby contributing to maintain the individual membrane identities of the ER and LDs. These findings reveal a function for the ERAD pathway that is distinct from its role in protein quality control or in lipid‐dependent degradation of sterol enzymes (Ruggiano et al, 2014). We call this novel function “protein spatial control” since it leads to the degradation of a protein based on its localization rather than its folding status.
The involvement of quality control systems in the degradation of mislocalized proteins has been described in different contexts. For example, tail‐ and GPI‐anchored proteins failing to insert in the ER membrane are selectively targeted for degradation (Hessa et al, 2011; Ast et al, 2014; Rodrigo‐Brenni et al, 2014). Similarly, ER and peroxisomal proteins erroneously inserted in mitochondria outer membrane are degraded by an ill‐defined process involving the ATPase Msp1/Atad1 (Chen et al, 2014; Okreglak & Walter, 2014). Thus, sequential, non‐redundant quality control processes prevent proteins to accumulate in the inappropriate cellular compartment. The process described here expands this concept to proteins in continuous but distinct membrane regions, such as ER and LDs. Spatial control of LD proteins resembles the degradation of certain proteins in the INM by the Asi complex, a recently identified ERAD branch (Foresti et al, 2014; Khmelinskii et al, 2014). In this case, Asi‐mediated ERAD excludes mislocalized proteins from the INM, therefore contributing to maintain the identity of this ER domain. We speculate that protein spatial control by ERAD might be a general mechanism to generate heterogeneity and/or functional domains, such as INM and LDs, in the continuous membrane of the ER.
How ERAD substrates are recognized based on their localization is not clear yet. In the case of LD proteins, the simplest possibility is that the hydrophobic hairpin, while in a bilayer membrane, may display some conformation instability or features typical of a misfolded protein, which target them to ERAD as “quality control” substrates. At the LD monolayer, the hairpins might adopt a more favorable conformation, decreasing the mobility of proteins back into the ER and as such keeping them away from the ERAD machinery and sparing them from degradation. Indeed, photobleaching experiments indicate that Pgc1 has a long dwell time at the LD monolayer. Similarly, GPAT4 is quite stable once at the LD monolayer (Wilfling et al, 2013). Given the overlap of the signals for LD targeting and Doa10 degradation, there is the potential for modulation of the two distinct outcomes by other factors. Such a mechanism might explain the changes of Dga1 localization under different metabolic states (Sorger & Daum, 2002; Markgraf et al, 2014). However, the identification of factors involved in the recognition of the hydrophobic hairpins is necessary to understand the process.
The regulation of LD proteins by ERAD is likely a conserved process in eukaryotes. A recent report showed that DGAT2 is also degraded by ERAD (Choi et al, 2014). Although in this case the ubiquitin ligase involved was Gp78, the homologue of the yeast Hrd1, the general process has remarkable similarities with our findings. The degradation of DGAT2 required its hydrophobic hairpin while a block in DGAT2 degradation led to its accumulation in the ER and increased TAG levels. Similarly, yeast cells lacking DOA10 display higher levels of TAG, in agreement with increased levels of Dga1 in this mutant (Fei et al, 2009; our unpublished data). Other hairpin‐containing LD proteins are also involved in synthesis, modification, and turnover of lipids at the core and monolayer of LDs. Therefore, deregulation of their levels and distribution might account for the LD defects in doa10Δ cells. The ubiquitin ligase Ubr1 can to some extent compensate for the loss of DOA10, both in the degradation of LD proteins (our data) and of some ER membrane proteins (Stolz et al, 2013). These findings point to a more prominent role of a cytosolic ubiquitin ligase in ERAD, and future studies should address how Ubr1 is recruited to its ER membrane‐bound substrates.
Additional links between ERAD and LDs have been previously reported, in particular the dynamic localization of some ERAD components to ER and LD membranes (Klemm et al, 2011; Spandl et al, 2011; Wang & Lee, 2012; Jo et al, 2013; Olzmann et al, 2013). If in most cases this has not been functionally dissected, for UBXD8, an adaptor for the Cdc48/p97 ATPase, its LD localization is important to regulate cellular TAG levels. Under conditions disfavoring lipolysis, UBXD8 relocates from the ER to LDs where it inhibits the activity of a major TAG lipase ATGL (Olzmann et al, 2013). Interestingly, this function of UBXD8 did not appear to involve proteolysis suggesting multiple and complex connections between ERAD and LD regulation that should be the focus of future studies.
Materials and Methods
Reagents
Rat monoclonal anti‐hemagglutinin (HA) antibody (clone 3F10) was purchased from Roche and used at 1:2,000 dilution for immunoblot and 1:1,000 for immunoprecipitation; mouse anti‐myc antibody was purchased from Roche and used at 1:1,000 dilution; rabbit polyclonal anti‐GFP and anti‐Kar2 antibodies were purchased from Santa Cruz Biotechnology and used at 1:1,000 dilution; anti‐Pgk1 antibody was purchased from Invitrogen and used at 1:10,000 dilution; rat monoclonal (3H9) anti‐GFP antibody was purchased from Chromotek and used at 1:2,000 dilution; anti‐Dpm1 antibody was purchased from Life Technologies and used at 1:3,000 dilution; rabbit polyclonal anti‐Erg1 antibody was raised against the full‐length protein as described in Foresti et al (2013). Cycloheximide (Sigma‐Aldrich) was used at 250 μg/ml. Monodansyl pentane (MDH) was purchased from Abgent and used at 0.1 mM. All other reagents and chemicals were purchased from Sigma‐Aldrich.
Yeast strains and growth
Yeast strains were isogenic to wild‐type BY4741 (Mata ura3∆0 his3∆1 leu2∆0 met15∆0), BY4742 (Matα ura3∆0 his3∆1 leu2∆0 lys2∆0), FY251 (Mata ura3‐52 his3∆200 leu2∆1 trp1∆63), or DF5 (Mata his3∆200 leu2‐3, 2‐112 lys2‐801 ura3‐52 trp1‐1). Single or multiple deletion mutants were obtained by transformation using PCR‐based homologous recombination (Longtine et al, 1998) or by crossing haploid cells of opposite mating types. The list of strains is available in Table EV1. Cells were grown in minimal medium supplemented with the appropriate amino acids. For galactose induction, cells were pre‐cultured in 2% raffinose medium for 24 h and induced by 2% galactose in early logarithmic phase.
For the oleic acid treatment, logarithmic YPD cultures were grown in the presence of 0.1% oleic acid and 1% Brij‐58 or 1% Brij‐58 alone for at least 10 generations before cycloheximide addition.
Plasmids
A complete list of the plasmids used in this study is available in Table EV2. To generate pPC882, SEC63‐mCHERRY sequence was amplified from yPC4314 using primers 185‐721 and cloned into pRS416 between XhoI and XbaI sites.
To generate pPC1040, PGC1 promoter (550 bp) was amplified from BY4741 genomic DNA with primers 1515‐1516; 3HA‐PGC1 was amplified with its own terminator from yPC6800 genomic DNA with primers 1517‐1518. The fusion PCR product obtained with primers 1515‐1518 (introducing SacI and PstI restriction sites, respectively) was cloned into pRS315 between SacI and PstI sites. To generate pPC1051, PGC1 promoter was amplified from BY4741 genomic DNA with primers 1515‐2091; GFP‐PGC1 was amplified with its own terminator from yPC6834 genomic DNA with primers 2092‐1518. The fusion PCR product obtained with primers 1515‐1518 was cloned into pRS315 between SacI and PstI sites.
To generate pPC1145, ADH1 promoter‐GFP‐PGC1 was amplified from yPC6834 genomic DNA with primers 1779‐1518. The PCR product was cloned into pRS315 between SacI and PstI sites.
The DNA sequence encoding aa160–216 from GPAT4 was amplified from Drosophila S2 cells cDNA using primers 2066‐2067. Other plasmids encoding PGC1 are derived from pPC1040, pPC1051, and pPC1145 via sub‐cloning, fusion PCR, or site‐directed mutagenesis.
To generate pPC1196, ADH1 promoter‐DGA1‐GFP was amplified from yPC7249 genomic DNA with primers 1779‐185. PCR product was cloned into pRS415 between SacI and XhoI sites.
To generate pPC1299, YEH1‐3HA was amplified with its own promoter from yPC9214 genomic DNA with primers 185‐2148. The PCR product was cloned into pRS316 between XhoI and NotI sites.
The primers used are listed in Table EV3.
Cycloheximide shut‐off experiments
Cycloheximide shut‐off experiments in exponentially growing cells (OD600 ≤ 1) were performed at 30°C, unless differently specified. Whole‐cell extracts for each time point were prepared as in Kushnirov (2000) and analyzed by SDS–PAGE and Western blot.
Microsome preparation and alkaline extraction
Microsomes were prepared from exponentially growing cells (OD600 = 1) essentially as described in Liu et al (2011) and resuspended in 10 mM Hepes pH 7.4. For extraction of membrane proteins, equal amounts of microsomes were treated with 10 mM Hepes pH 7.4, or 0.2 M Na2CO3 pH 11 in water for 1 h at 4°C or 1% SDS in 10 mM Hepes for 1 h at room temperature. After incubation, samples were separated into pellet and supernatant by centrifugation at 100,000 g. Supernatant fractions were TCA‐precipitated. Pellets were resuspended in Laemmli buffer and analyzed by SDS–PAGE and Western blot.
Detection of ubiquitinated Pgc1
Logarithmically growing cells were harvested and washed with 10 mM sodium azide. Cell lysis was performed with glass beads in 0.8% SDS, 50 mM Tris–HCl pH 7.4 buffer containing 20 mM NEM and 0.1 mM PMSF. Pgc1 was immunoprecipitated with anti‐HA antibodies in 0.2% SDS, 1% NP‐40, 50 mM Tris–HCl pH 7.4. Immunoprecipitated material was analyzed by SDS–PAGE and Western blotting.
Detection of membrane‐associated ubiquitinated Pgc1
Membrane floatation was performed essentially as described (Bagnat et al, 2000). Fractions were collected and proteins precipitated with TCA. The pellets were resuspended in buffer containing 2 M urea, 50 mM Tris–HCl pH 7.4, 1 mM EDTA, 1% SDS and heated at 65°C for 10 min. After dilution with a buffer containing 50 mM Tris–HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X‐100, Pgc1 was immunoprecipitated using anti‐HA antibodies. Immunoprecipitated material was analyzed by SDS–PAGE and Western blotting.
Microscopy
Fluorescence microscopy was performed at room temperature in a Zeiss Cell Observer HS equipped with a CMOS camera (Hamamatsu ORCA‐Flash 4.0) controlled by 3i Slidebook 6.0 software. A 100× 1.40 oil immersion objective was used. GFP, mCherry, and MDH signals were detected using GFP, RFP, and DAPI filters, respectively, with standard settings.
Cells were imaged in logarithmic growth phase. Images were acquired using the same settings, and brightness and contrast were processed in a similar manner. For GFP‐Pgc1‐expressing cells, GFP pixel intensities at the NE were measured using the line tool in ImageJ on a single‐plane image. Intensity values were adjusted for background and were used to calculate median, first and third quartiles of the distribution for a set of images (wt or doa10Δ). Data were displayed in a box plot; whiskers extend to the highest and lowest value of the distribution.
Photobleaching experiments were performed on cells grown in YPD medium. Early stationary cells were diluted into the same medium to OD600 0.2 and grown up to OD600 1.2 before being transferred to a concanavalin A‐pre‐treated chamber. Live imaging was performed on a confocal Leica TCS SP5 microscope using a HCX PL APO CS100 × 1.40 oil objective and controlled by the LAS AF software.
Bleaching experiments were performed using the point bleach option of the FRAP module. Photobleaching was applied to LD in contact with the nuclear envelope. Three pre‐bleach images were acquired followed by 800 ms of photobleaching at 80% laser power. Images were acquired every 1.32 s.
For analysis, the fluorescence intensity of four regions of interest was measured: the photobleached LD, a non‐photobleached LD in the bleached cell to check for diffusion‐dependent changes in fluorescence, a LD in a cell that was not photobleached to check for overall fluorescence variation, and region outside of the irradiated cell to check for overall background fluorescence. The fluorescence recovery values of the bleached region were background‐subtracted and normalized to the average of the pre‐bleaching values. The normalized recovery values were plotted after adjusting for the slow decay of fluorescence caused by imaging using areas of the image distant from the bleached region as described (Shibata et al, 2008). For each genotype, average and standard deviation were calculated from 10 photobleaching events.
For photoconversion experiments, cells were imaged using a laser scanning confocal microscope Leica TCS SP5 AOBS (inverted) with a 63×/1.4 oil immersion objective, using 13% of argon laser intensity (488 nm line) and 10% of DPPSS 561 laser intensity (561 nm line) at 30% output. Photoconversion was applied on a ROI as indicated in the figure with the laser 405 diode (405 nm line) at 10% laser intensity during 10 s. After conversion, a single image was taken every 20 min. Images were analyzed using ImageJ.
Author contributions
PC conceived and supervised the project. PC, AR, and GM designed the experiments and analyzed the data. AR and GM performed most of the experiments. LB performed the FRAP and photoconversion experiments. All authors discussed the results. PC and AR wrote the manuscript with input from GM.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Review Process File
Source Data for Figure 1
Source Data for Figure 1D
Source Data for Figure 2
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
We thank E. Sabidó and C. Chiva for help with the mass spectrometry, Y. Barral, B. Crosas, and S. Jentsch for yeast strains and plasmids, and O. Foresti for discussion and critical reading of the manuscript. A. Ruggiano was supported by a “La Caixa” graduate fellowship; P. Carvalho is supported by CRG, an International Early Career Award from the HHMI, the EMBO Young Investigator Program, and grants from the Spanish MCCIN and ERC (FP7/2007‐2013 ERC grant agreement no. 309477 DROPFAT). We acknowledge support of the Spanish Ministry of Economy and Competitiveness, “Centro de Excelencia Severo Ochoa 2013–2017”, SEV‐2012‐0208.
The EMBO Journal (2016) 35: 1644–1655
References
- Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH (1986) PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol Cell Biol 6: 2490–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ast T, Aviram N, Chuartzman SG, Schuldiner M (2014) A cytosolic degradation pathway, prERAD, monitors pre‐inserted secretory pathway proteins. J Cell Sci 127: 3017–3023 [DOI] [PubMed] [Google Scholar]
- Bagnat M, Keranen S, Shevchenko A, Simons K (2000) Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl Acad Sci USA 97: 3254–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays NW, Wilhovsky SK, Goradia A, Hodgkiss‐Harlow K, Hampton RY (2001) HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell 12: 4114–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilharz T, Egan B, Silver PA, Hofmann K, Lithgow T (2003) Bipartite signals mediate subcellular targeting of tail‐anchored membrane proteins in Saccharomyces cerevisiae. J Biol Chem 278: 8219–8223 [DOI] [PubMed] [Google Scholar]
- Boban M, Zargari A, Andreasson C, Heessen S, Thyberg J, Ljungdahl PO (2006) Asi1 is an inner nuclear membrane protein that restricts promoter access of two latent transcription factors. J Cell Biol 173: 695–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman I, Hebert DN (2013) Protein folding in the endoplasmic reticulum. Cold Spring Harb Perspect Biol 5: a013201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S (2002) Role of the ubiquitin‐selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J 21: 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P, Goder V, Rapoport TA (2006) Distinct ubiquitin‐ligase complexes define convergent pathways for the degradation of ER proteins. Cell 126: 361–373 [DOI] [PubMed] [Google Scholar]
- Chen YC, Umanah GK, Dephoure N, Andrabi SA, Gygi SP, Dawson TM, Dawson VL, Rutter J (2014) Msp1/ATAD1 maintains mitochondrial function by facilitating the degradation of mislocalized tail‐anchored proteins. EMBO J 33: 1548–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Kim H, Kang H, Lee SY, Lee SJ, Back SH, Lee SH, Kim MS, Lee JE, Park JY, Kim J, Kim S, Song JH, Choi Y, Lee S, Lee HJ, Kim JH, Cho S (2014) Regulation of diacylglycerol acyltransferase 2 protein stability by gp78‐associated endoplasmic‐reticulum‐associated degradation. FEBS J 281: 3048–3060 [DOI] [PubMed] [Google Scholar]
- Christianson JC, Ye Y (2014) Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat Struct Mol Biol 21: 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie E, Guo X, Christiano R, Chitraju C, Kory N, Harrison K, Haas J, Walther TC, Farese RV Jr (2014) High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J Lipid Res 55: 1465–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS (2006) A luminal surveillance complex that selects misfolded glycoproteins for ER‐associated degradation. Cell 126: 349–359 [DOI] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, Wenk MR, Parton RG, Yang H (2008) Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol 180: 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Wang H, Fu X, Bielby C, Yang H (2009) Conditions of endoplasmic reticulum stress stimulate lipid droplet formation in Saccharomyces cerevisiae . Biochem J 424: 61–67 [DOI] [PubMed] [Google Scholar]
- Fernandez‐Murray JP, McMaster CR (2005) Glycerophosphocholine catabolism as a new route for choline formation for phosphatidylcholine synthesis by the Kennedy pathway. J Biol Chem 280: 38290–38296 [DOI] [PubMed] [Google Scholar]
- Fisher E, Almaguer C, Holic R, Griac P, Patton‐Vogt J (2005) Glycerophosphocholine‐dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae . J Biol Chem 280: 36110–36117 [DOI] [PubMed] [Google Scholar]
- Foresti O, Rodriguez‐Vaello V, Funaya C, Carvalho P (2014) Quality control of inner nuclear membrane proteins by the Asi complex. Science 346: 751–755 [DOI] [PubMed] [Google Scholar]
- Foresti O, Ruggiano A, Hannibal‐Bach HK, Ejsing CS, Carvalho P (2013) Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. eLife 2: e00953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth N, Gertman O, Shiber A, Alfassy OS, Cohen I, Rosenberg MM, Doron NK, Friedler A, Ravid T (2011) Exposure of bipartite hydrophobic signal triggers nuclear quality control of Ndc10 at the endoplasmic reticulum/nuclear envelope. Mol Biol Cell 22: 4726–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Jarosch E, Sommer T, Hirsch C (2006) A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol 8: 849–854 [DOI] [PubMed] [Google Scholar]
- Grillitsch K, Connerth M, Kofeler H, Arrey TN, Rietschel B, Wagner B, Karas M, Daum G (2011) Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim Biophys Acta 1811: 1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton RY, Gardner RG, Rine J (1996) Role of 26S proteasome and HRD genes in the degradation of 3‐hydroxy‐3‐methylglutaryl‐CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell 7: 2029–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer W, Gruhler A, Mohrle V, Mahe Y, Wolf DH (1993) PRE2, highly homologous to the human major histocompatibility complex‐linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem 268: 5115–5120 [PubMed] [Google Scholar]
- Hessa T, Sharma A, Mariappan M, Eshleman HD, Gutierrez E, Hegde RS (2011) Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature 475: 394–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelmo‐Torres M, Gonzalez‐Moreno E, Kassan A, Hanzal‐Bayer M, Tebar F, Herms A, Grewal T, Hancock JF, Enrich C, Bosch M, Gross SP, Parton RG, Pol A (2009) Hydrophobic and basic domains target proteins to lipid droplets. Traffic 10: 1785–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R (2011) Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae . J Cell Sci 124: 2424–2437 [DOI] [PubMed] [Google Scholar]
- Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T (2002) Protein dislocation from the ER requires polyubiquitination and the AAA‐ATPase Cdc48. Nat Cell Biol 4: 134–139 [DOI] [PubMed] [Google Scholar]
- Jo Y, Hartman IZ, DeBose‐Boyd RA (2013) Ancient ubiquitous protein‐1 mediates sterol‐induced ubiquitination of 3‐hydroxy‐3‐methylglutaryl CoA reductase in lipid droplet‐associated endoplasmic reticulum membranes. Mol Biol Cell 24: 169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A, Blaszczak E, Pantazopoulou M, Fischer B, Omnus DJ, Le Dez G, Brossard A, Gunnarsson A, Barry JD, Meurer M, Kirrmaier D, Boone C, Huber W, Rabut G, Ljungdahl PO, Knop M (2014) Protein quality control at the inner nuclear membrane. Nature 516: 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm EJ, Spooner E, Ploegh HL (2011) Dual role of ancient ubiquitous protein 1 (AUP1) in lipid droplet accumulation and endoplasmic reticulum (ER) protein quality control. J Biol Chem 286: 37602–37614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV (2000) Rapid and reliable protein extraction from yeast. Yeast 16: 857–860 [DOI] [PubMed] [Google Scholar]
- Latterich M, Frohlich KU, Schekman R (1995) Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell 82: 885–893 [DOI] [PubMed] [Google Scholar]
- Lian JP, Ferro‐Novick S (1993) Bos1p, an integral membrane protein of the endoplasmic reticulum to Golgi transport vesicles, is required for their fusion competence. Cell 73: 735–745 [DOI] [PubMed] [Google Scholar]
- Liu Q, Siloto RM, Snyder CL, Weselake RJ (2011) Functional and topological analysis of yeast acyl‐CoA:diacylglycerol acyltransferase 2, an endoplasmic reticulum enzyme essential for triacylglycerol biosynthesis. J Biol Chem 286: 13115–13126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen CJ, Young BP, Tavassoli S, Levine TP (2007) Inheritance of cortical ER in yeast is required for normal septin organization. J Cell Biol 179: 467–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR‐based gene deletion and modification in Saccharomyces cerevisiae . Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Markgraf DF, Klemm RW, Junker M, Hannibal‐Bach HK, Ejsing CS, Rapoport TA (2014) An ER protein functionally couples neutral lipid metabolism on lipid droplets to membrane lipid synthesis in the ER. Cell Rep 6: 44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL (2009) A bright and photostable photoconvertible fluorescent protein. Nat Methods 6: 131–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL (2002) The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem 277: 8877–8881 [DOI] [PubMed] [Google Scholar]
- Okreglak V, Walter P (2014) The conserved AAA‐ATPase Msp1 confers organelle specificity to tail‐anchored proteins. Proc Natl Acad Sci USA 111: 8019–8024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Richter CM, Kopito RR (2013) Spatial regulation of UBXD8 and p97/VCP controls ATGL‐mediated lipid droplet turnover. Proc Natl Acad Sci USA 110: 1345–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol A, Gross SP, Parton RG (2014) Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J Cell Biol 204: 635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar‐Nun S (2002) AAA‐ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum‐associated protein degradation. Mol Cell Biol 22: 626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo‐Brenni MC, Gutierrez E, Hegde RS (2014) Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol Cell 55: 227–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiano A, Foresti O, Carvalho P (2014) Quality control: ER‐associated degradation: protein quality control and beyond. J Cell Biol 204: 869–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandager L, Gustavsson MH, Stahl U, Dahlqvist A, Wiberg E, Banas A, Lenman M, Ronne H, Stymne S (2002) Storage lipid synthesis is non‐essential in yeast. J Biol Chem 277: 6478–6482 [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A (2005) Membrane‐bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER‐associated protein degradation. Nat Cell Biol 7: 999–1006 [DOI] [PubMed] [Google Scholar]
- Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK (2008) The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem 283: 18892–18904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS (2011) Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334: 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger D, Athenstaedt K, Hrastnik C, Daum G (2004) A yeast strain lacking lipid particles bears a defect in ergosterol formation. J Biol Chem 279: 31190–31196 [DOI] [PubMed] [Google Scholar]
- Sorger D, Daum G (2002) Synthesis of triacylglycerols by the acyl‐coenzyme A:diacyl‐glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae . J Bacteriol 184: 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandl J, Lohmann D, Kuerschner L, Moessinger C, Thiele C (2011) Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J Biol Chem 286: 5599–5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, Ruggiano A, Carvalho P, Rapoport TA (2014) Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell 158: 1375–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A, Besser S, Hottmann H, Wolf DH (2013) Previously unknown role for the ubiquitin ligase Ubr1 in endoplasmic reticulum‐associated protein degradation. Proc Natl Acad Sci USA 110: 15271–15276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R, Locher M, Hochstrasser M (2001) A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER‐associated and Matalpha2 repressor degradation. Genes Dev 15: 2660–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam AR, Farese RV Jr, Walther TC (2013) The biophysics and cell biology of lipid droplets. Nat Rev Mol Cell Biol 14: 775–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsirigos KD, Peters C, Shu N, Kall L, Elofsson A (2015) The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43: W401–W407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CW, Lee SC (2012) The ubiquitin‐like (UBX)‐domain‐containing protein Ubx2/Ubxd8 regulates lipid droplet homeostasis. J Cell Sci 125: 2930–2939 [DOI] [PubMed] [Google Scholar]
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, Liu X, Buhman KK, Coleman RA, Bewersdorf J, Farese RV Jr, Walther TC (2013) Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell 24: 384–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414: 652–656 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Review Process File
Source Data for Figure 1
Source Data for Figure 1D
Source Data for Figure 2
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6


