Abstract
In the Neurospora crassa circadian clock, a protein complex of frequency (FRQ), casein kinase 1a (CK1a), and the FRQ‐interacting RNA Helicase (FRH) rhythmically represses gene expression by the white‐collar complex (WCC). FRH crystal structures in several conformations and bound to ADP/RNA reveal differences between FRH and the yeast homolog Mtr4 that clarify the distinct role of FRH in the clock. The FRQ‐interacting region at the FRH N‐terminus has variable structure in the absence of FRQ. A known mutation that disrupts circadian rhythms (R806H) resides in a positively charged surface of the KOW domain, far removed from the helicase core. We show that changes to other similarly located residues modulate interactions with the WCC and FRQ. A V142G substitution near the N‐terminus also alters FRQ and WCC binding to FRH, but produces an unusual short clock period. These data support the assertion that FRH helicase activity does not play an essential role in the clock, but rather FRH acts to mediate contacts among FRQ, CK1a and the WCC through interactions involving its N‐terminus and KOW module.
Keywords: circadian clock, chaperone, transcriptional repressor, protein interactions, protein structure
Subject Categories: Metabolism, Structural Biology
Introduction
FRQ‐interacting RNA helicase (FRH) is an ATP‐dependent RNA helicase that functions in the central transcriptional–translational feedback loop of the Neurospora crassa circadian clock (Brunner & Kaldi, 2008; Baker et al, 2012; Cha et al, 2015; Hurley et al, 2015). The core oscillator components include the positive transcriptional activators White Collar 1 (WC‐1) and White Collar 2 (WC‐2) and the negative elements FREQUENCY (FRQ) and FRH (Brunner & Kaldi, 2008; Guo & Liu, 2010; Baker et al, 2012; Hurley et al, 2015). WC‐1 and WC‐2 form the white‐collar complex (WCC) that regulates the expression of clock‐controlled genes and responds to light (Fig 1A; Froehlich et al, 2002; He et al, 2002). FRH (1,106 residues c. 124 kDa) binds and stabilizes FRQ, the major transcriptional repressor of the oscillator (Cheng et al, 2005; Guo et al, 2009, 2010; Shi et al, 2010). FRQ is an intrinsically disordered protein (IDP) (989 residues, 108 kDa) that is largely unstructured in the absence of FRH (Hurley et al, 2013; Zhou et al, 2013, 2015; Cha et al, 2015). The WCC regulates frq expression (Froehlich et al, 2002; He et al, 2002), whereas the FRQ‐FRH complex (FFC) (Cheng et al, 2005; Shi et al, 2010) represses action of the WCC. Casein kinases 1a and 2 (CK1a and CKII) bind to the FFC and phosphorylate FRQ to regulate its activity and stability (He et al, 2006; Baker et al, 2009; Tang et al, 2009; Querfurth et al, 2011). The FFC influences the circadian cycle by mediating contact between FRQ and the WCC to promote phosphorylation of the WCC by CK1a/CKII and by targeting frq mRNA to the exosome for degradation (Fig 1A) (Brunner & Kaldi, 2008; Guo et al, 2009, 2010; Tang et al, 2009; Baker et al, 2012; Cha et al, 2015). FRH provides a stable scaffold for FRQ and thereby modulates FRQ phosphorylation, localization, and stability (Guo et al, 2010; Cha et al, 2011; Hurley et al, 2013). Furthermore, FRH interacts with the WCC in the absence of FRQ and thus serves to recruit the repressor complex to the WCC (Cheng et al, 2005; Guo et al, 2010; Shi et al, 2010). Finally, FRH binds to the light‐adaptation photoreceptor Vivid (VVD) to alter FRQ expression through repression of the WCC (Hunt et al, 2010). FRH has ATPase and helicase functions that are essential for Neurospora growth; in vitro studies have suggested that these enzymatic activities also have a role in clock function (Lauinger et al, 2014). However, FRH mutants devoid of ATPase activity maintain normal circadian cycles (Hurley et al, 2013). In contrast, a mutant FRH protein (R806H) that supports cell viability causes major clock dysfunction (Shi et al, 2010).
Figure 1. Function and structure of FRH.
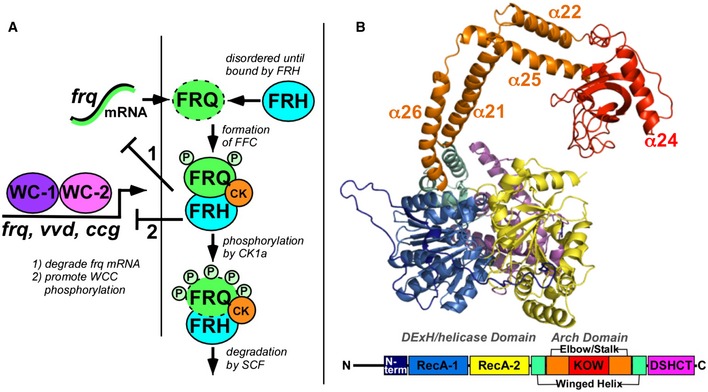
- In the Neurospora crassa circadian core clock oscillator, FRH associates with FRQ and CK1a to form a repressor complex (FFC) that inhibits the positive‐acting white‐collar transcription factors (WC‐1 and WC‐2) which form the white‐collar complex (WCC). Stoichiometries of the components are not represented in the schematic. FFC composition has been estimated as 1 FRH molecule: 2 FRQ molecules (Cheng et al, 2005; Hurley et al, 2013), or 2 FRH molecules: 2 FRQ molecules (Lauinger et al, 2014).
- Structure of FRH from the large crystallographic cell with an accordingly colored domain map below.
FRH promotes degradation of frq RNA and interacts with components of the exosome (Guo et al, 2009). The Saccharomyces cerevisiae (yeast) homolog of FRH, Mtr4p/Dob1p (Mtr4) is also a coenzyme for the nuclear exosome. Mtr4 associates with two other proteins: (i) Trf4 (or Trf5), from the Polβ superfamily of nucleotidyl transferases responsible for RNA polyadenylation; and (ii) Air2 (or Air1), a five Zn‐knuckle protein important for RNA or protein binding (LaCava et al, 2005; Jackson et al, 2010; Wang et al, 2013; Falk et al, 2014). Together Trf4‐Air2‐Mtr4p forms the TRAMP RNA surveillance complex, whose roles include 3′‐processing and polyadenylation of rRNA, snoRNAs, and snRNAs (LaCava et al, 2005). Yeasts do not possess a circadian clock and have no FRQ homolog; thus, the clock function of FRH may derive from properties not held by Mtr4. FRH and FRQ are found in both the nucleus and the cytoplasm (Cha et al, 2011); in contrast, Mtr4 is primarily present in the nucleus (Dez et al, 2006).
FRH is essential for cell viability; thus, genetic experiments to assess the role of FRH in the clock rely on the finding that either low levels of WT FRH or specific mutants (e.g., R806H) will support growth but not circadian rhythms (Cheng et al, 2005; Hurley et al, 2013). Under low levels of FRH expression, rhythmicity can be rescued by FRH mutants expressed from a separate allele that are defective in helicase/ATPase activities but still contain a unique N‐terminal region (residues 100–150) that promotes interactions with the C‐terminal regions of FRQ (residues 695–778; Guo et al, 2010; Cha et al, 2011; Hurley et al, 2013).
FRH belongs to the DEAD‐like helicase superfamily. The protein comprises several domains (Fig 1B): (i) the characteristic DExH/Dc or HELICc helicase superfamily domains (PFam:PF00270) of RecA‐like modules 1 and 2 that harbor the nucleotide and ATP‐binding sites; (ii) the DSHCT C‐terminal domain found in Dob1/Ski2/helY‐like DEAD box helicases (PF08148); (iii) the arch domain required for 5.8S rRNA processing by Mtr4 (PF13234) that itself contains the helical stalk, elbow, and KOW (Kyrpides, Ouzounis, and Woese) modules (Jackson et al, 2010; Weir et al, 2010); and (iv) a winged‐helix domain (PF00633) that connects the arch to the helicase + DSHCT core. Another FRH homolog, Ski2 (superkiller2, 36% identical to FRH), is the cytosolic homolog of Mtr4. Ski2 participates in mRNA quality control mediated by the exosome. Ski2 binds Ski3, a tetratricopeptide repeat protein, and Ski8, a WD40 protein, to form the Ski complex (Halbach et al, 2012). Despite Ski2 and FRH sharing similar cellular distributions and the tendency to interact with partners through unique N‐terminal regions (Ski2 with Ski3, FRH with FRQ), FRH is more similar to Mtr4 than Ski2 (55% vs. 35% identity). Sequence differences between FRH and Mtr4 concentrate at the N‐terminus through the first 145 residues and also in the KOW module.
Herein, we report the 3.1 Å crystal structure of FRH from Neurospora crassa, which serves as a first look at this clock‐essential RNA helicase and reveals features that distinguish FRH from other Mtr4 family members. Furthermore, we show through mutagenesis studies that single‐residue substitutions in the unique FRH N‐terminus and KOW module affect interactions between FRH and both FRQ and the WCC with impact on circadian timing.
Results
Crystal structure determination
FRH variants beginning at residue 100 or 114 (FRH‐Δ100 or FRH‐Δ114) were expressed in and purified from E. coli. Recombinant FRH was crystallized with and without ADP and a single‐stranded 13‐nucleotide RNA. FRH‐Δ114 produces two orthorhombic crystal forms, each containing a distinct conformation of the protein. The structure of FRH‐Δ114 was first determined for crystals with the larger cell dimensions to 3.1 Å resolution (Fig 1B and Appendix Table S1) using the Mtr4 structure as a molecular replacement probe (pdb 2XGJ, sequence identity 57%, Fig EV1). The small‐cell structure of FRH‐Δ114 (3.2 Å resolution) differs from that of the large cell primarily in the positioning of the long helices of the arch, which change in angle with respect to the DExH helicase core + DSHCT domain by over 20° (Fig 2). FRH‐Δ100 crystallized in the smaller cell, with and without ADP and RNA.
Figure EV1. Primary sequence comparison of FRH and Mtr4.
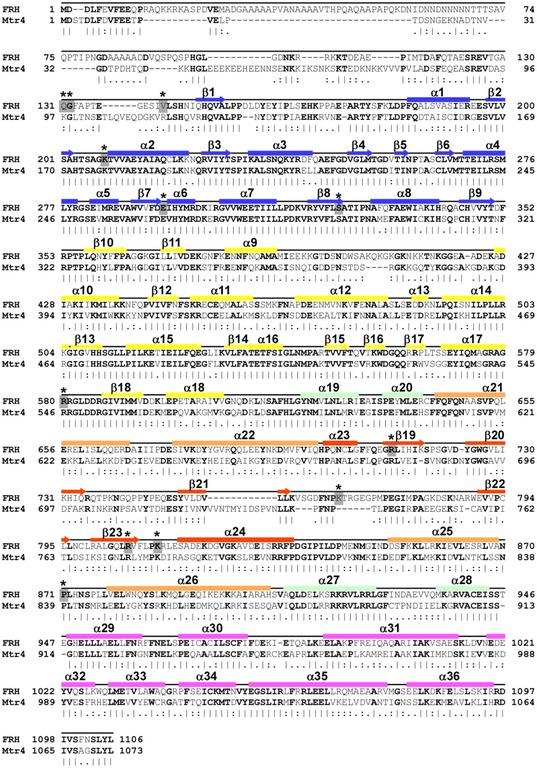
Sequence alignment of FRH and Mtr4 with secondary structure and corresponding domain identifiers colored in accordance with Fig 1. Relevant mutation sites are boxed in gray and starred.
Figure 2. Comparison of FRH and Mtr4 structures.
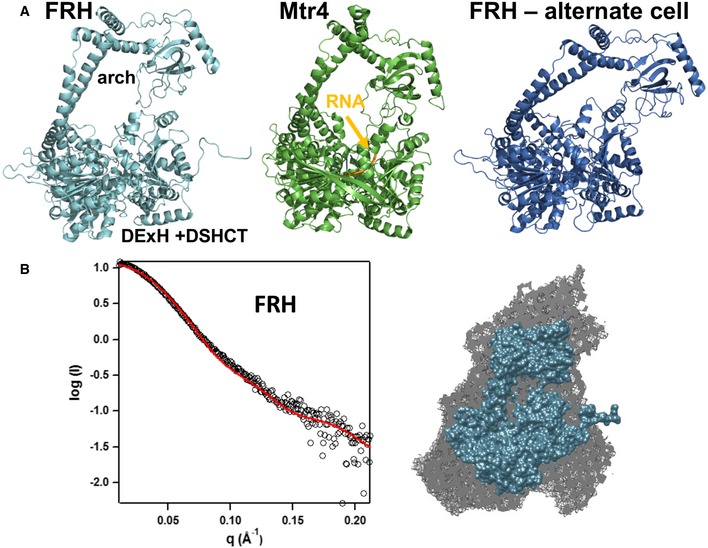
- FRH large cell (cyan), Mtr4 (pdb 2XGJ, green, RNA backbone in orange), and FRH small alternate cell (blue) differ primarily in the orientation of the arch domain. The FRH RNA‐bound structure also crystallizes in the small‐cell configuration. The two different FRH structures have an overall RMSD of 3.9 Å, and the RMSD between Mtr4 and FRH is 2.9 Å (over 734 residues, for large‐cell FRH).
- SAXS data indicate that FRH‐Δ114 is monomeric and flexible. The crystal structure of the large‐cell structure predicts the solution scattering curve (left, χ2 = 0.92) and fits well to the calculated molecular envelope (right).
Overall structure
Like Mtr4, FRH consists of five distinct domains: RecA‐1, RecA‐2, winged–helix, arch (stalk helices, elbow, and KOW) domain, and DSHCT (Fig 1B). RecA‐1 and RecA‐2 are DExH/Dc‐box helicase domains, typical of AAA‐ATPases that assume a nucleotide‐binding fold and contain the signature Walker A and Walker B motifs (Ye et al, 2004). The ATP‐binding site and RNA‐binding determinants reside at the opposite edges of the interface between the two RecA domains. The arch domain extends up into an “arm and fist” structure, with the helical stalks forming the “arm” and “elbow” (at the turn between helices) and the KOW region as the “fist”. After the arch the chain returns to the small winged‐helix and larger DSHCT domains, the latter of which juxtaposes the RecA‐2 domain to complete the RNA‐binding cleft. The N‐terminal region is highly variable between the two structures and lacks defined secondary structure until residue 148 (Fig EV2). In both FRH structures, the N‐terminal conformation is influenced by intermolecular contacts with symmetry mates that differ between the two crystal forms (Fig 3). In the absence of these contacts, the N‐terminus is likely to be highly dynamic. Overall, the FRH polypeptide is continuous with the exception of an undefined surface loop in the RecA‐2 domain between α9 and α10. The missing loop (~30 residues) contains ten lysine residues among a sequence dominated by flexible and polar side chains. Small‐angle X‐ray scattering (SAXS) confirmed the monomeric state of the protein in solution and indicated that the molecule has considerable flexibility about a core region (Fig 2B and Appendix Fig S1).
Figure EV2. Conformational variability of FRH.
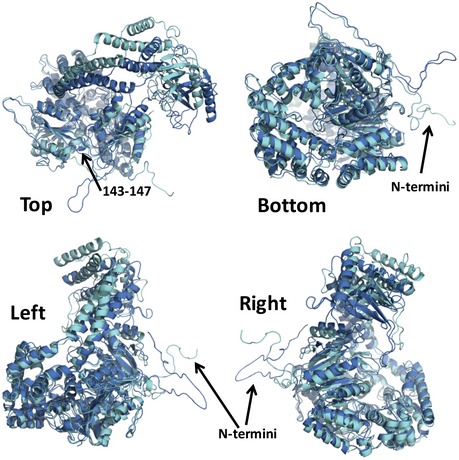
Structural overlays of large (cyan) and small (marine) cell FRH structures shown from different perspectives and emphasizing the differences in the conformations of the N‐terminus and the KOW modules.
Figure 3. The N‐terminal region of FRH is unstructured in the absence of FRQ.
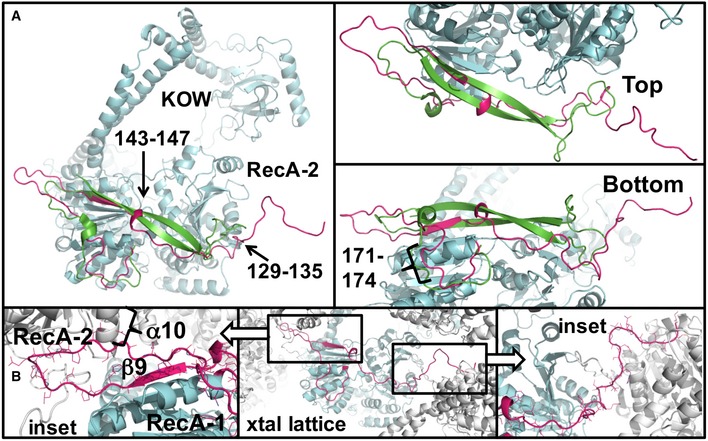
- Comparison of the FRH N‐terminus (large‐cell structure, magenta) to that of Mtr4 (green) aligned on their RecA domains. Only the additional elements of FRH are shown (cyan). The FRH N‐terminal extension has little ordered secondary structure, although regions 143–147 and 129–135 form small turns that interact with the helicase core.
- The very N‐terminus of FRH extends away from the RecA‐2 domain to interact with a symmetry‐related molecule in the crystal (below), whereas the Mtr4 N‐terminus turns back to form an outer β‐strand. Center panel shows the crystal contacts, with close‐ups for each region highlighted as insets on the left and right.
The DExH helicase core
The DExH helicase core of FRH (Fig 4 and Appendix Fig S2) is composed of two RecA domains arranged similarly to those of other RNA helicases (Erzberger & Berger, 2006; Putnam & Jankowsky, 2013; Rudolph & Klostermeier, 2015). The RecA domain contains an interior parallel β‐sheet surrounded by α‐helices, a highly conserved Walker A motif, or P‐loop, that coordinates the γ‐phosphate of ATP during hydrolysis and the less conserved Walker B motif that coordinates the magnesium ion and nucleophilic water molecule (Ye et al, 2004). The exposed edges of the interface between the two RecA domains bind ATP on one side and single‐stranded RNA on the other (Fig 5A). RecA‐1 acts as the primary site for binding ATP, whereas RecA‐2 participates in nucleotide hydrolysis. Motion between the RecA domains couples ATP hydrolysis to nucleic acid translocation (Putnam & Jankowsky, 2013).
Figure 4. The RecA and arch domains of FRH compared to Mtr4.
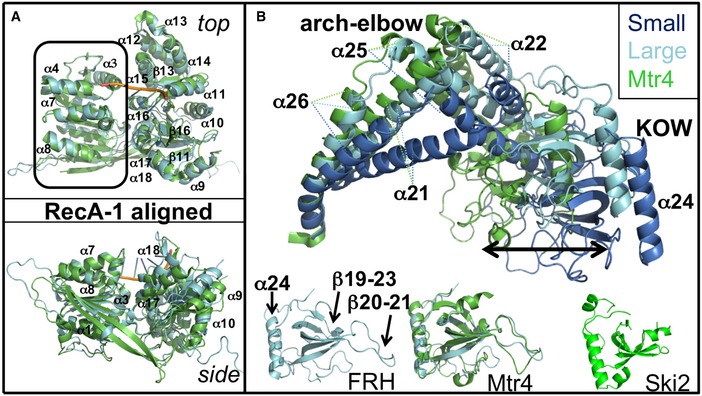
- The two RecA domains are rotated relative to one another in FRH (cyan, large cell) compared to Mtr4 (green, RNA backbone in orange). The change in positioning of RecA‐2 with respect to RecA‐1 results in an overall displacement from the Mtr4 domain juxtaposition that ranges from 1.1 to 4.0 Å throughout RecA‐2 with the largest differences at the external helices.
- Comparison of arch domains conformations from Mtr4 (green) and FRH (large cell in cyan, small cell in marine) after superposition of the most N‐terminal helical region. The isolated KOW modules of FRH, Mtr4 (with FRH superimposed), and Ski2 have considerably different loop configurations (below).
Figure 5. Interactions of FRH with RNA and ADP.
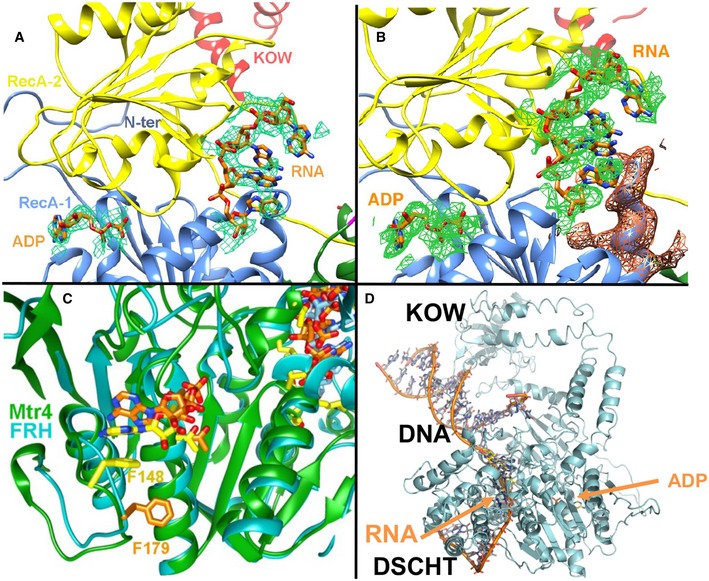
- 3.8 Å resolution mFo‐DFc omits map density (green, calculated with sharpened B‐factors and contoured at 2.0σ), reveals the presence of ADP and a short segment of RNA bound to FRH.
- 3.8 Å resolution 2mFo‐DFc calculated without ADP or RNA contributing to the model. Green electron density shown for ADP and RNA (0.6σ); orange density shown for surrounding protein (0.6σ).
- Comparison of Mtr4 and FRH ADP binding (FRH in cyan with ADP, RNA, and F179 in orange bonds; Mtr4 in green with ADP, RNA, and F148 in yellow bonds).
- Unwound DNA from Hel308 (pdb 2PR6; magenta, DNA in orange bonds) overlaid in FRH with RNA and ADP (FRH in cyan with yellow RNA and cyan ADP in stick) after superposition of the respective RecA domains. See also Appendix Fig S4. The superposition indicates that the KOW module is positioned to interact with double stranded RNA that is being unwound by the RecA and DHSCT domains.
Differences in the helicase cores of FRH and Mtr4 (67% identity, Fig EV1 and Appendix Fig S2) are primarily within the RecA interfaces (Fig 4). The individual RecA domains of the large‐cell FRH and Mtr4 structures are very similar in structure (1.3 Å RMSD over 405 residues). In RecA‐1, small differences between FRH and Mtr4 arise in α3 and the connections of β3 to α4. In RecA‐2, minor shifts are found in α‐helices 12, 13, 16, 19, and 20, and the loop conformations connecting β17 to α19 and β20 to α20. The RecA domains are rotated relative to one another when comparing the small‐ and large‐cell structures, and also in relation to their positions in Mtr4 (Fig 4 and Appendix Fig S2). Variability in RecA association reflects the well‐known importance of domain motion in helicase function (Ye et al, 2004; Buttner et al, 2007).
The arch domain
The arch domain is unique to the Mtr4/Ski2 subfamily of Ski2‐like helicases (Jackson et al, 2010; Halbach et al, 2012; Jarmoskaite & Russell, 2014), and this region shows the greatest differences in conformation between FRH and Mtr4 owing to changes in the angle of the stalk helices relative to the helicase core, conformational differences in the elbow connection and greater sequence divergence of the KOW modules (Fig 4B, 64% similar, 39% identical; Fig EV1). The stalk helices (α21, α22, α25, α26) anchor to the helicase core through winged helical domains that overlap well in both structures and hinge at a variable elbow that does not (Fig 4).
The KOW module of the arch domain is essential for RNA binding in Mtr4/Ski2 helicases (Johnson & Jackson, 2013; Taylor et al, 2014). This “fist” region is found in the bacterial transcriptional elongation factor NusG (Steiner et al, 2002), the eukaryotic chromatin elongation factor Spt5 (Meyer et al, 2015), and the higher eukaryotic protein KIN17 (Carlier et al, 2007), where it is known to mediate nucleic acid binding and protein–protein interactions. In FRH, the KOW module begins with the short helix α23 and leads into a five β‐stranded core (β19‐23) connected by flexible loops of varied lengths and ends with one 27 Å helix, α24. The KOW β‐sheets are very similar in the FRH and Mtr4 structures (RMSD 1.0 Å), but there are major differences in the long flexible loops between β20 and β21, and between β21 and β22 (Fig 4B), both of which have very different sequences in the two proteins (Fig EV1). The β20‐β21 loop that projects toward the RNA‐binding cleft is quite variable among the family members (Fig 4B), as is the connection between β21 and β22, which is not present in Ski2 (Fig 4B; Halbach et al, 2012). The KOW module also contacts the RecA‐2 domain in the region of the structurally undefined, lysine‐rich loop linking α9 and α10 (Fig 3 and Appendix Fig S3). The residue whose mutation disrupts clock, but not helicase function (R806), resides in the KOW module at an exposed position within a positively charged patch of residues that include K811, K731, R812, K766, R806, K739, R831, R832, and K824 (Fig 6).
Figure 6. Features of FRH critical for helicase and clock function.
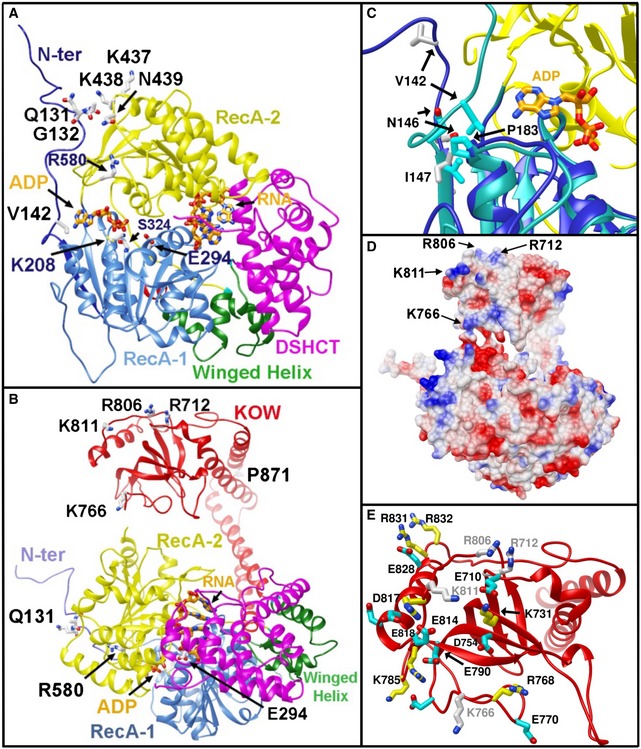
- Key residues for FRH function shown on the large‐cell structure: K208 and S324 compose the ATP‐binding site; E294 lies between the ADP and RNA pockets and participates in ATP hydrolysis; P871 resides in the elbow region of the arch domain. K811, R806, R712, and K766 contribute to a positively charged surface on the KOW domain module. R806H disrupts the clock. Q131, G132, and V142 in the N‐terminus provide interactions to the helicase RecA domains (FRH domains are colored according to Fig 1, and ADP and RNA are in orange bonds).
- Same as (A) after rotation about a horizontal axis.
- Close‐up of the ADP‐binding pocket and N‐terminal interaction for the large‐cell structure (cyan ribbons) and the small‐cell structure (blue ribbons).
- Electrostatic potential surface of FRH (blue positive, red negative) shown with the same orientation as in (B).
- Close‐up of the KOW module showing the large number of basic (aqua sticks) and acidic side chains (yellow sticks) on the surface that also includes the mutational sites R806, R712, K811, and K766 (gray sticks)
Substrate recognition
A 3.8 Å resolution structure was determined of FRHΔ100 in the presence of ADP and single‐stranded RNA (rGAUCCGCUCGAUC). Several different RNA sequences were screened for cocrystallization, with the sequence above giving the best diffracting crystals. Discernible difference electron density in the ADP‐binding pocket and in the RecA‐2/DSHCT cleft indicates that both ADP and RNA are present (Fig 5A and B and Appendix Fig S4). Because of the low resolution and the possibility for overlapping binding configurations, the RNA was modeled as poly‐A. The binding cleft is known to change conformation in response to nucleotide binding and hydrolysis (Singleton & Wigley, 2002); however, ATP‐recognition residues in FRH have conformations similar to that of the apo Mtr4 structure (Fig 5 and Appendix Fig S4). For example, an ATP‐binding loop (170–187) well conserved in AAA‐ATPases is disengaged from the nucleotide (Fig 5C) and an aromatic residue (FRH F179) does not stack with the ADP adenosine as it does in the nucleotide complexes of many other helicases (Fig 5C). The reason for these differences is unclear, but may stem from incomplete occupancy of ADP or crystal contacts made by the 170–187 loop to other FRH molecules. Several bases of RNA line the cleft formed by the RecA‐2 domain and the so‐called ratchet helix on the DSHCT domain, responsible for translocating RNA (Buttner et al, 2007). Superposition of the RecA‐1 domains of FRH with that of the Archael HEL308 SF2 DNA helicase bound to an unwrapped DNA substrate aligns the 3′‐strand of the unwound DNA with the RNA found in the FRH structure (Buttner et al, 2007). This superposition suggests that an RNA substrate bound by FRH will be in close proximity to the KOW (Fig 5D and Appendix Fig S4C). In particular, a long loop between β20 and β21 in the KOW module that varies considerably in structure and sequence among FRH, Mtr4, and Ski2 (Fig 4B) projects toward the nucleic acid in the superposition (Fig 5D and Appendix Fig S4C). Conserved positions of key residues for ATP hydrolysis and helicase activity (e.g., K208, S324, E294, R580; Fig 6) indicate that FRH maintains the substrate binding determinants and catalytic residues that are consistent with its function as an RNA helicase.
The N‐terminal FRQ interaction region
FRH requires residues 100–150 to interact with FRQ (Hurley et al, 2013); this region is mostly discernible in both structures. Conformational differences between the N‐terminal FRQ‐binding regions of FRH in the small‐ and large‐cell FRH structures underscore the intrinsic flexibility of this region when not bound to FRQ (Fig EV2). In the small‐cell structure, electron density is weak between residues 100 and 123, and there is little interaction with the RecA‐2 domain due to an excursion of residues 122–141 that wraps against the KOW fist of a neighboring molecule. The absence of secondary structure in the FRH N‐terminus (113–130) contrasts with the analogous region of Mtr4, which forms an N‐terminal β‐hairpin motif (M80‐D96). Interestingly, sequence similarity between the two proteins in this region (Fig EV1) suggests that the FRH N‐terminus may assume a similar more‐ordered structure when bound to FRQ. The FRQ‐binding residues of FRH (100–150) interact with FRQ residues Q695‐M798 (Hurley et al, 2013). The FRQ structure here is unknown, although the sequence predicts two short ordered segments (Appendix Fig S6). It follows that the FRH N‐terminal extension and the target sequence on FRQ complement each other to form a stable module upon their mutual interaction.
FRQ binding to the FRH N‐terminus may alter the properties of the ATP‐ and RNA‐binding sites on the RecA domains by removing interactions between the N‐terminus and the helicase. Residues 114–129 of the large‐cell structure do not form an outer β‐strand as is found in Mtr4 and rather have a random coil conformation that is displaced from the FRH core. Residues 130–154 then stretch across the RecA‐2 and RecA‐1 domains in an extended mode with the exception of two kink regions that each pack against one of the two RecA domains (Figs 3 and 6). Residues 129–135 form a turn that interacts with the C‐terminal end of RecA‐2 α10, whose N‐terminus connects with the Lys‐rich disordered loop proximate to the KOW module and RNA‐binding cleft (Fig 3). This contact may alter the 449–451 loop conformation and the following β‐strand, which both differ relative to Mtr4 (Fig 3). As in Mtr4, the 449–451 loop may interact with single‐stranded RNA as it is pulled through the helicase. RecA‐2 α10 also has a high degree of positive charge (from K430, K433, K437, K438, K367) and along with the 449–451 loop could be an interaction surface for a negatively charged moiety, such as phosphorylated FRQ or the WCC.
Another structured N‐terminal kink (residues 143–147) interacts with the ATP‐binding loop of RecA‐1 (residues 170–187) and may contribute to the more “open” conformation of the nucleotide binding pocket (Figs 5C and 6C). Following this segment, residues 147–154 form an irregular β‐strand (β1) that becomes the exposed edge of the RecA‐1 β‐sheet. β1 has substantial similarity in sequence and structure with the analogous segment of Mtr4. The loop that follows β1 again diverges between FRH and Mtr4 before realigning in the first RecA‐1 α‐helix (FRH residue P183).
Interactions of the FRH N‐terminus
To test how specific residues within structured regions of the FRH N‐terminus may influence clock function, we altered those that contact the core helicase and evaluated their effects on clock period and component interactions. Of these residues, Q131 stabilizes the secondary structure of the first turn and interacts with residues 437–439 at the C‐terminus of the RecA‐2 α10 helix, whereas neighboring Gly132 assumes a backbone conformation disallowed by other residue types (Fig 6). At the RecA‐2 interface, V142 buttresses the 179–187 ATP‐binding loop (Fig 6C). By conducting pull‐down assays with an epitope‐tagged FRH (Fig 7 and Appendix Fig S5), we find that FRH variants Q131A and G132V (which should disrupt the first turn conformation) have no effects on FRQ or WCC binding, or clock behavior (Fig 7). In contrast, V142G (which removes the side chain contact to the ADP‐binding region in the large‐cell structure; Fig 6C) weakens the interaction between FRH and both FRQ and the WCC (Fig 7). Thus, the relatively minor V142G substitution recapitulates previous deletion analysis that identified the 100–150 segments as an important determinant for FRQ binding (Hurley et al, 2013). Rhythmicity was monitored for cells in which the select variant FRH was expressed along with helicase competent, but clock‐deficient FRH that is necessary for viability (Appendix Fig S5) (Hurley et al, 2013). Surprisingly, FRH V142G FRH has a substantially shortened clock period of 18.7 h, compared with ~21.5 h for WT (Fig 7).
Figure 7. FRH variant interactions with the core clock components examined by race tube and immunoprecipitation assays.
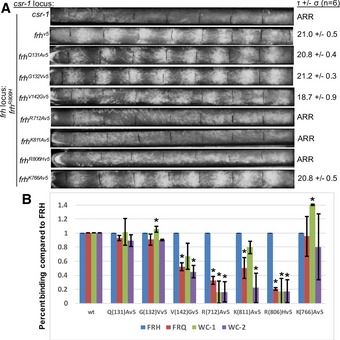
- Race tube assays with FRH mutants. Every circadian cycle N. crassa lays down aerial hyphae that appear as fluffy yellow/orange conidial bands. The distance between the bands reflects the clock period. The strains were grown in constant darkness, and growth fronts were labeled every 24 h (black vertical lines), rhythmicity on right (τ = period in hours, σ = standard deviation, n = number of race tubes; ARR, arrhythmic). The race tubes shown here are representative samples from three replicate tubes.
- Pull‐down assays evaluating interactions among FRH, FRQ, WC‐1, and WC‐2. Ratio of FRQ, WC‐1, and WC‐2 binding to the V5 tagged FRH mutants as compared to the inter‐strain variations in FRH levels, error bars represent standard deviation for the assay run in triplicate. *P < 0.05 (unpaired two‐tailed t‐test with Gaussian distribution assumption). Representative immunoblots are shown in Appendix Fig S5.
Recognition sites for partners
The structure of the ternary complex of Mtr4 with regions of the other two TRAMP complex proteins, Trf4 and Air2 (Falk et al, 2014), provides insight into how FRH may interact with its protein partners. Mtr4 binds seven residues of Trf4 on the RecA‐2 domain in the β11‐α9 region (D121‐S127), whereas Mtr4 interacts with Air2 between the RecA‐2 and DSHCT domains in α34‐35 and α15‐16 (Fig EV3). The N‐terminus of Air2 also contacts the KOW domain of a symmetry‐related molecule of Mtr4 on β19, the loop connecting β23 and α24, and the end of α24 (Fig EV3). Outside the context of the crystal, both regions of the flexible Air2 peptide may bind to the same molecule of Mtr4 (Falk et al, 2014). The Air2‐interacting residues on the KOW domain of Mtr4 are conserved in FRH as R712 and R806, the latter of which abrogates clock function when mutated to His (Hurley et al, 2013). As the FRH KOW module shares properties of the Air2‐interacting region of Mtr4, it may also mediate protein–protein interactions, if not with homologs of Air2, then possibly with other elements related to clock function. Neurospora contains homologs of Trf4 and Air2 (Fig EV3), but no binding of either to FRH was detected in proteome‐wide interaction studies (Baker, 2012).
Figure EV3. Putative TRAMP complex interactions sites.
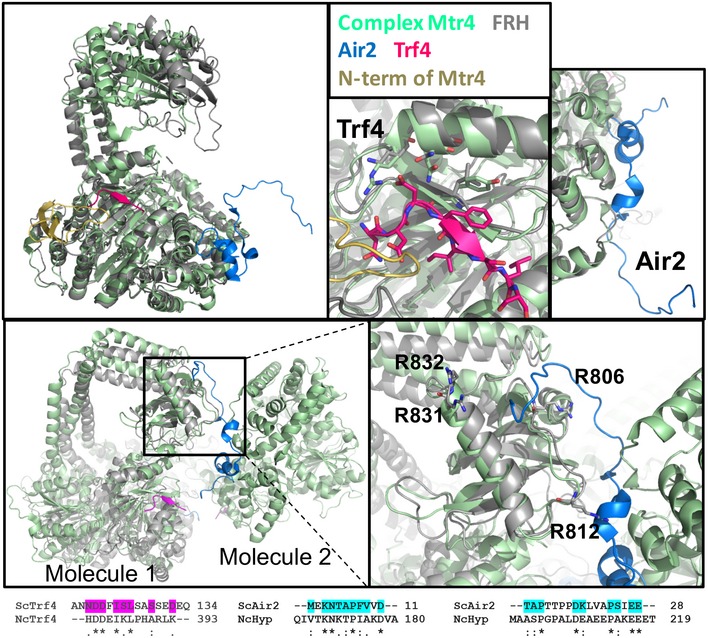
Structural comparison of the Mtr4 TRAMP complex (pdb 4U4C) with FRH (FRH in gray, Mtr4 in green with N‐terminus (added from pdb 2XGJ) in gold, Air2 in blue, Trf4 in magenta). Trf4 lies along RecA‐2, Air2 contacts the DSHCT, RecA‐2, and the KOW domain of a symmetry‐related molecule. The Trf4 and Air2 interfaces with Mtr4 are in regions well conserved by FRH. Although there is currently no data on an N.c. TRAMP complex, a Neurospora homolog of Trf4 (GenBank: EAA31314.2; NCU05588, 38% sequence identity) conserves residues that could potentially interact with α9 of FRH. A direct homolog of Air2 is even less obvious in the genome of N.c., although the hypothetical protein NcHyp (GenBank: CAD37057; NCU04617) has modest sequence similarity (33%) to yeast Air2. This putative Air2 homolog also maintains some residues that could interact with conserved residues in the FRH DSHCT domain (K1048, M1049, and E1054). Segments of Trf4 and Air2 that contact Mtr4 are aligned below with the related regions of the respective N.c. homologs; residues that make contacts are highlighted.
Interactions of the KOW module with clock components
The FRH structure places constraints on how the R806H mutant disrupts clock rhythmicity. R806 resides on an exposed surface of the KOW module, far removed from both the FRQ‐binding N‐terminus and the ATPase domains (Fig 6A, B and E). However, R806H weakens interactions between the FFC and the WCC (Shi et al, 2010). Interestingly, R806 is conserved in Mtr4, where it interacts with the Air2 N‐terminus, along with R712, which is also conserved in both proteins. R806 and R712 contribute to a positively charged surface that involves several other Arg and Lys residues (e.g., Lys811, Lys766, Lys732, Arg768, Arg812; Fig 6D and E). To test the possibility that this surface participates in protein–protein interactions important for the clock, we mutated select positively charged residues therein and evaluated their effects on binding partners as well as on clock behavior (Fig 7 and Appendix Fig S5). Like R806H, R712A weakens the interaction between FRH and FRQ and also the WCC. Not surprisingly, R712A is arrhythmic, like R806H. In contrast, K811A is arrhythmic from what primarily appears to be a defect in binding WC‐2. The differential binding of FRH to WC‐1 compared with WC‐2 in the K811A variant was unexpected and suggests independent interactions between the FRH and these two transcription factors (Fig 7 and Appendix Fig S5). Moreover, the K766A substitution increases FRH binding to WC‐1 independent of changes to WC‐2 binding, but does not affect period (Fig 7). The ability of four positively charged residues on the KOW to alter FRQ and WCC interactions strongly implicates the KOW module as a key component of the inhibitory complex formed by the FFC and the WCC.
Discussion
The structure of FRH reveals the close relationship of this DExD/H helicase to the well‐characterized yeast homolog Mtr4. FRH is essential for growth of Neurospora, but its general housekeeping functions of RNA metabolism are largely unexplored. Nevertheless, Neurospora contains within its genome distant homologs for the TRAMP components Air2 and Trp4; moreover, these putative TRAMP components conserve residues with their yeast homologs that mediate their interaction with Mtr4 (Fig EV3). The corresponding interaction residues on FRH are also somewhat conserved with Mtr4. Although FRH may then participate in a TRAMP‐like nuclear or a Ski‐like cytosolic complex, it is likely transient or of low abundance (Baker, 2012).
Unlike Mtr4, FRH is an essential component of a cellular circadian clock and it has been shown to target frq mRNA for degradation. Nevertheless, the primary role of FRH in the clock does not depend on RNA processing. Several frh mutations have been studied in attempts to associate FRH helicase activity with clock function: K208E, which eliminates ATP binding; E294Q, which prevents ATP hydrolysis; S324L, which blocks RNA unwinding; and R580P, which interferes with ATP binding and helicase activity. The structure of FRH confirms that these residues are suitably positioned to execute their assumed enzymatic activities (Fig 6A and B). In reconstitution experiments, substitutions of catalytically important FRH residues reduce CK1a phosphorylation of FRQ (Lauinger et al, 2014). Consequently, it has been proposed that global changes in FRH architecture driven by FRH ATPase activity lead to selective access of CK1a to FRQ substrate sites (Lauinger et al, 2014). The overall structure of FRH would be conducive to coupling between the helicase core and interaction regions such as the KOW module. However, in contrast to the in vitro experiments, genetic studies indicate that FRH can support clock rhythmicity even when devoid of helicase activity (Hurley et al, 2013). These experiments rely on expressing helicase‐competent FRH at low levels to support essential cellular functions independent of the clock. It has been noted that if FRH were to be active as a dimer, helicase‐deficient, FRQ‐interacting FRH subunits could recruit helicase‐competent subunits, thereby localizing both the chaperone and helicase functions in the vicinity of the FRQ substrate (Lauinger et al, 2014). However, FRH does not dimerize in solution or crystals. Furthermore, cellular pull‐down experiments of differentially tagged FRH proteins indicate that there is only one FRH in each complex with FRQ (Hurley et al, 2013).
FRH stabilizes FRQ, but it also mediates interactions between the FRQ:CK1a complex and the WCC. This latter property is altered by the R806H substitution. R806 resides on an exposed surface of the KOW module, displaced from the helicase core and RNA‐binding cleft of the RecA‐12/DSHCT module. In both Mtr4 and Ski2, similarly located arginine residues are critical for RNA binding. In Ski2, the R903E mutant impairs binding of single‐stranded RNA (Halbach et al, 2012), and this residue is conserved in Mtr4 (R678) and FRH (R712). Furthermore, in the TRAMP complex of Mtr4, the KOW binds the very N‐terminus of Air2 and may interact with Rrp6 (Jackson et al, 2010), a Ski‐type homolog for cytoplasmic rather than nuclear RNA processing. In FRH, several other charged residues, including R806, cluster with R712 on the KOW module (Fig 6E). R806 is conserved in Mtr4 (R774) as is the neighboring R831 and R832 (Mtr4 R799 and R800). FRH also contains two other positively charged residues in the connection of β25 to α26: K811 and R812. Pull‐down assays indicate that several other positively charged residues surrounding R806 (Fig 6E) also reduce interactions to the WCC (Fig 7). Interestingly, the K811A substitution affects WC‐2 binding to a larger degree than either FRQ or WC‐1 binding (Fig 7 and Appendix Fig S5). Differential binding of WC‐1 and WC‐2 by FRH K811A is unexpected because K806 and K811 are in close proximity and WC‐1 and WC‐2 are thought to be tightly associated through their PAS domains (Denault et al, 2001). Nonetheless, WC‐2 oscillates in its interaction with the cis regulatory C‐box, whereas WC‐1 largely remains associated with the C‐box throughout the circadian cycle (Belden et al, 2007). The need for a stable interaction between WC‐1 and FRH:FRQ only up to a certain threshold of affinity may be reflected in the behavior of the FRH K766A variant, which binds WC‐1 more tightly than WT, but does not affect period. Overall, the KOW module appears to interact with WC‐1 and WC‐2 independently and may even facilitate their dissociation.
Key structural differences between FRH and Mtr4 are at the N‐terminus, where FRH residues 100–150 bind to and stabilize FRQ. Variable conformations of this segment in the large‐ and small‐cell structures support the assertion that it is largely disordered in the absence of FRQ (Hurley et al, 2013). The V142G substitution, which is involved in the contact between the FRH N‐terminus and the ATP‐binding site of the RecA‐1, reduces interactions between FRH and FRQ and produces a short‐period phenotype. Thus, this FRH segment may be displaced from the FRH core domain when bound to FRQ. Single‐residue changes that cause short‐periods are uncommon and where characterized correlate with changes in FRQ stability, often through phosphorylation defects (Tang et al, 2009; Tataroglu et al, 2012). For V142G, the stability of FRQ or its sensitivity to phosphorylation may be directly modulated by its altered interaction with FRH. Conversely, the shortened period caused by V142G could derive from a weakened interaction with the WCC that effectively increases WCC activity. However, the KOW variants, which have qualitatively similar effects on FRQ and WCC binding as V142G, do not shorten period. Thus, affinity of the clock components for each other is not the only parameter important for proper function. Taken together, the structural features of FRH combined with mutational studies emphasize that FRH:FRQ and the WCC associate with a complex architecture, wherein the conformations and interactions of the components are highly co‐dependent.
Materials and Methods
Expression and purification
FRH‐Δ114 and FRH‐Δ100 in a pET28a vector with an N‐terminal 6x His tag were expressed in E. coli BL21 (DE3) cells. The gene sequence was codon optimized for expression in E. coli (Hurley et al, 2013). Full‐length recombinant FRH was not well behaved. Proteins were purified by: (i) Ni‐NTA affinity chromatography using 150 mM NaCl, 200 mM imidazole, 50 mM HEPES buffer for elution; (ii) size‐exclusion chromatography (Superdex 200, GE Healthcare) using 150 mM NaCl, 50 mM HEPES pH 7.6, and 2–5 mM DTT as the running buffer; and (iii) anion exchange chromatography with 500 mM NaCl, 50 mM HEPES pH 7.6, and 2 mM DTT as the applied gradient buffer.
Crystallization and structure determination
FRH‐Δ114 and FRH‐Δ100 were concentrated to 15–22 mg/ml following purification. Crystals were grown at 17°C using hanging drop vapor diffusion by mixing purified protein with 0.1 M sodium citrate pH 5.5–6 adjusted with HCl, and 36–44% v/v PEG600. Crystals ranged in size from ~20 to 200 microns. No additional cryoprotectant was added prior to data collection. Data were collected at Advanced Photon Source at Argonne National Laboratory beamlines (NE‐CAT) 24‐ID‐C and 24‐ID‐E. Data were processed, scaled, and analyzed in RAPD using XDS, Pointless, Aimless, Scala, Freerflag, and Mtz2various from the CCP4 Suite (Bailey, 1994). Phases were obtained by molecular replacement using Phaser (McCoy et al, 2007) with a model derived from homolog Mtr4 (PDB 2XGJ). The model was built using Coot (Emsley & Cowtan, 2004) and refined in Phenix (Adams et al, 2002) with data to 3.1 Å resolution. Both FRH‐Δ114 and FRH‐Δ100 produced crystals of varying diffraction quality. Both the large‐ and small‐cell structures reported in the absence of cofactors were obtained from FRH‐Δ114. FRH‐Δ100 produced crystals in the small cell, both with and without RNA and ADP. Data collection and refinement statistics are given in Appendix Table S1. The differences in large and small cell dimensions are > 2 Å in a, ~6 Å in b, and ~18 Å in c. The space group of both cells is P 212121, with one molecule per asymmetric unit. Estimates of solvent content vary from 46 to 56%.
For RNA and ADP structures, 1 μl of 5 mM single‐stranded RNA (5′‐rGrArUrCrCrGrCrUrCrGrArUrC‐3′) and 5 μl of 10 mM ADP (in 50 mM HEPES pH 7.5, 50 mM NaCl, 5 mM DTT buffer) were added to 20 μl of 15–22 mg/ml of FRH‐Δ100 and incubated for 30 min before distributing in crystal trays. Many different oligonucleotides were screened for cocrystallization with the one above producing the best diffracting crystals. Nonetheless, the low resolution of the RNA complex prevents sequence assignment. Thus, apparent RNA in the electron density was modeled as poly‐A4.
SAXS data were obtained at the BioSAXS beamline, G1 station, at the Cornell High Energy Synchrotron Source (CHESS). SAXS profiles were obtained of FRH‐Δ114 and FRH‐Δ100 at concentration ranging from 2 to 5 mg/ml, with the representative profiles from 3 to 2 mg/ml, respectively. Protein samples were exposed to 9.968 keV X‐ray, in an integrated capillary sample flow cell with a 2 mm path; both X‐ray path and sample cell were kept under vacuum. Samples were initially centrifuged for 30 min, and then, aliquots (30 μl) were loaded from a 96‐well tray and oscillated in the beam by an automated system. Scattering data were captured on a Pilatus 100K‐S detector (Dectris, Baden, Switzerland). The SAXS data were analyzed in BioXTAS Raw (Nielsen et al, 2009) and indicate that FRH‐Δ114 as purified is monomeric in solution. The I(0) values, the extrapolated SAXS intensities at zero angle, and Porod volumes were proportional to molecular weight (glucose isomerase (MW 173,000 Da was used as standard). For FRH‐Δ114, the radius of gyration was 40.40 Å, the D max was 135.66 Å, and Porod volume was 206,372 Å3. For FRH‐Δ100, the radius of gyration was 39.50 Å, the D max was 128.76 Å, and the Porod volume was 210,802 Å3. The protein appears mainly globular, but the Kratky plots and the molecular envelope reconstructions, computed using DAMMIF software (Franke & Svergun, 2009), indicate considerable flexibility about a core region. The envelopes were an average (DAMAVER) (Volkov & Svergun, 2003) of 20 bead models (mean normalized spatial discrepancy, NSD = 0.725 (FRH‐Δ114), 0.762 (FRH‐Δ100)), and include several protrusions from the core which likely represent conformational heterogeneity in the arch domain and the N‐terminal region (Fig 2B and Appendix Fig S1). Situs (Wriggers, 2012) was used to render the volume from the averaged bead model, and SUPCOMB (Kozin & Svergun, 2001) was used to place the protein within the envelope. FoxS was used for validation of the experimental SAXS data with the crystallographic structure (Schneidman‐Duhovny et al, 2010, 2013). The fits of both FRH‐Δ114 and FRH‐Δ100 had χ2 values of 0.92 with calculated Rg values of 34.39.
FRH protein interaction assays
Transformations were performed as described (Colot et al, 2006; Bardiya & Shiu, 2007). Race tube assays and circadian liquid culture experiments were performed as described with slight modifications (1× Vogel's, 0.1% glucose, 0.2% arginine, and 50 ng/ml biotin) (Loros et al, 1989; Garceau et al, 1997; Liu et al, 1997). For Neurospora culture, conidia were inoculated into Bird medium (Metzenberg, 2004) containing 1.8% glucose. Protein lysates were prepared on a small scale with a protease inhibitor mixture (P9599; Sigma). To perform the co‐IP assay, 1 mg of total protein extract was incubated with 30 μl of anti‐V5 antibody‐coated agarose overnight (A7345; Sigma). The agarose/beads were then washed with protein extraction buffer four times before elution with Laemmli buffer (65°C for 15 min) and loading on the gel. For Western blot analysis, equal volumes of input and immunoprecipitated protein were loaded per lane. Primary anti‐V5 antibody (Invitrogen) was diluted 1:5,000. Anti‐FRQ and anti‐WC‐1 and WC‐2 antibodies were diluted 1:250 (Garceau et al, 1997; Luo et al, 1998; Denault et al, 2001). SuperSignal West Femto ECL (Pierce) was used for signal development. Race tube assays were performed as described with slight modification (Garceau et al, 1997). The Neurospora strains used in this study are defined as 1153 (csr), 1163 (frhwt), 1634‐1 + 1634‐2 (frhQ131Av5), 1635‐1 + 1635‐2 (frhG132Vv5), 1636‐1 + 1636‐2 (frhV142Gv5), 1637‐1 + 1637‐2 (frhR712Av5), 1638‐1 + 1638‐2 (frhK811Av5), 1165 (frhR806Hv5), 1639‐1 + 1639‐2 (frhK766Av5). The Neurospora strains upon which the studied was based were obtained from the Fungal Genetics Stock Center (Kansas State University, Manhattan, KS).
Sequence analysis
Analyses of FRH and FRQ sequences for secondary structure and regions of low complexity were carried out with PONDR (Xue et al, 2010) and JPRED (Cole et al, 2008; Drozdetskiy et al, 2015).
Accession codes
Coordinates and diffraction data have been deposited in the Protein data Bank as entries 4XGT, 5DZR, and 5E02.
Author contributions
BRC, JCD, JJL, KSC, and JMH designed and interpreted the study. KSC expressed, purified, crystallized, and determined structures of FRH. JW assisted in cloning and protein expression. JMH and CSR carried out race tube and pull‐down experiments. KSC and BRC wrote the paper with input from all authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by grant P41 GM103403 from NIGMS. This research was supported by NIH grants R01‐GM079679 (to. B.R.C.) F32‐GM099391 (to K.C.), R35‐GM118022 (to JJL), R01‐GM34985 (to JCD), and R35‐GM118021 (to JCD). We thank the Fungal Genetics Stock Center (Kansas State University, Manhattan, KS) for curation and dissemination of Neurospora strains.
The EMBO Journal (2016) 35: 1707–1719
References
- Adams PD, Grosse‐Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW , Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D‐Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Bailey S (1994) THE CCP4 suite ‐ programs for protein crystallography. Acta Crystallogr D‐Biol Crystallogr 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Baker CL, Kettenbach AN, Loros JJ, Gerber SA, Dunlap JC (2009) Quantitative proteomics reveals a dynamic interactome and phase‐specific phosphorylation in the Neurospora circadian clock. Mol Cell 34: 354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL (2012) Thesis. Post‐Translational Regulation of the Neurospora crassa Circadian System. Dartmouth: Dartmouth Press; [Google Scholar]
- Baker CL, Loros JJ, Dunlap JC (2012) The circadian clock of Neurospora crassa . FEMS Microbiol Rev 36: 95–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardiya N, Shiu PKT (2007) Cyclosporin A‐resistance based gene placement system for Neurospora crassa . Fungal Genet Biol 44: 307–314 [DOI] [PubMed] [Google Scholar]
- Belden WJ, Loros JJ, Dunlap JC (2007) Execution of the circadian negative feedback loop in Neurospora requires the ATP‐dependent chromatin‐remodeling enzyme CLOCKSWITCH. Mol Cell 25: 587–600 [DOI] [PubMed] [Google Scholar]
- Brunner M, Kaldi K (2008) Interlocked feedback loops of the circadian clock of Neurospora crassa . Mol Microbiol 68: 255–262 [DOI] [PubMed] [Google Scholar]
- Buttner K, Nehring S, Hopfner KP (2007) Structural basis for DNA duplex separation by a superfamily‐2 helicase. Nat Struct Mol Biol 14: 647–652 [DOI] [PubMed] [Google Scholar]
- Carlier L, Couprie J, Le Maire A, Guilhaudis L, Milazzo‐Segalas I, Courcon M, Moutiez M, Gondry M, Davoust D, Gilquin B, Zinn‐Justin S (2007) Solution structure of the region 51‐160 of human KIN17 reveals an atypical winged helix domain. Protein Sci 16: 2750–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J, Yuan HY, Liu Y (2011) Regulation of the activity and cellular localization of the circadian clock protein FRQ. J Biol Chem 286: 11469–11478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha J, Zhou M, Liu Y (2015) Mechanism of the Neurospora circadian clock, a FREQUENCY‐centric view. Biochemistry 54: 150–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P, He Q, He QY, Wang LX, Liu Y (2005) Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev 19: 234–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36: W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC (2006) A high‐throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103: 10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denault DL, Loros JJ, Dunlap JC (2001) WC‐2 mediates WC‐1‐FRQ interaction within the PAS protein‐linked circadian feedback loop of Neurospora. EMBO J 20: 109–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Houseley J, Tollervey D (2006) Surveillance of nuclear‐restricted pre‐ribosomes within a subnucleolar region of Saccharomyces cerevisiae . EMBO J 25: 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, Barton GJ (2015) JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43: W389–W394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Crystallogr D‐Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM (2006) Evolutionary relationships and structural mechanisms of AAA plus proteins. Annu Rev Biophys Biomol Struct 35: 93–114 [DOI] [PubMed] [Google Scholar]
- Falk S, Weir JR, Hentschel J, Reichelt P, Bonneau F, Conti E (2014) The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities. Mol Cell 55: 856–867 [DOI] [PubMed] [Google Scholar]
- Franke D, Svergun DI (2009) DAMMIF, a program for rapid ab‐initio shape determination in small‐angle scattering. J Appl Crystallogr 42: 342–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich AC, Liu Y, Loros JJ, Dunlap JC (2002) White collar‐1, a circadian blue light photoreceptor, binding to the frequency promoter. Science 297: 815–819 [DOI] [PubMed] [Google Scholar]
- Garceau NY, Liu Y, Loros JJ, Dunlap JC (1997) Alternative initiation of translation and time‐specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell 89: 469–476 [DOI] [PubMed] [Google Scholar]
- Guo JH, Cheng P, Yuan HY, Liu Y (2009) The exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell 138: 1236–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Liu Y (2010) Molecular mechanism of the Neurospora circadian oscillator. Protein Cell 1: 331–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JH, Cheng P, Liu Y (2010) Functional significance of FRH in regulating the phosphorylation and stability of Neurospora circadian clock protein FRQ. J Biol Chem 285: 11508–11515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach F, Rode M, Conti E (2012) The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome. RNA 18: 124–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He QY, Cheng P, Yang YH, Wang LX, Gardner KH, Liu Y (2002) White collar‐1, a DNA binding transcription factor and a light sensor. Science 297: 840–843 [DOI] [PubMed] [Google Scholar]
- He Q, Cha JS, He QY, Lee HC, Yang YH, Liu Y (2006) CKI and CKII mediate the FREQUENCY‐dependent phosphorylation of the WHITE COLLAR complex to close the Neurospora circadian negative feedback loop. Genes Dev 20: 2552–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SM, Thompson S, Elvin M, Heintzen C (2010) VIVID interacts with the WHITE COLLAR complex and FREQUENCY‐interacting RNA helicase to alter light and clock responses in Neurospora. Proc Natl Acad Sci USA 107: 16709–16714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Larrondo LF, Loros JJ, Dunlap JC (2013) Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered neurospora clock protein FRQ. Mol Cell 52: 832–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley J, Loros JJ, Dunlap JC (2015) Chapter two ‐ dissecting the mechanisms of the clock in Neurospora In Methods in Enzymology, Amita S. (ed), Vol. 551, pp 29–52. Cambridge, MA: Academic Press; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, Johnson SJ (2010) The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J 29: 2205–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmoskaite I, Russell R (2014) RNA helicase proteins as chaperones and remodelers. Annu Rev Biochem 83: 697–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SJ, Jackson RN (2013) Ski2‐like RNA helicase structures Common themes and complex assemblies. RNA Biol 10: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozin MB, Svergun DI (2001) Automated matching of high‐ and low‐resolution structural models. J Appl Crystallogr 34: 33–41 [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D (2005) RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Lauinger L, Diernfellner A, Falk S, Brunner M (2014) The RNA helicase FRH is an ATP‐dependent regulator of CK1a in the circadian clock of Neurospora crassa . Nat Commun 5: 3598 [DOI] [PubMed] [Google Scholar]
- Liu Y, Garceau NY, Loros JJ, Dunlap JC (1997) Thermally regulated translational control of FRQ mediates aspects of temperature responses in the neurospora circadian clock. Cell 89: 477–486 [DOI] [PubMed] [Google Scholar]
- Loros JJ, Denome SA, Dunlap JC (1989) Molecular‐cloning of genes under control of the circadian clock in neurospora. Science 243: 385–388 [DOI] [PubMed] [Google Scholar]
- Luo CH, Loros JJ, Dunlap JC (1998) Nuclear localization is required for function of the essential clock protein FRQ. EMBO J 17: 1228–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg RL (2004) Bird Medium: an alternative to Vogel medium. Fungal Genet Newsl 51: 19–20 [Google Scholar]
- Meyer PA, Li S, Zhang M, Yamada K, Takagi Y, Hartzog GA, Fu J (2015) Structures and functions of the multiple KOW domains of transcription elongation factor Spt5. Mol Cell Biol 35: 3354–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SS, Toft KN, Snakenborg D, Jeppesen MG, Jacobsen JK, Vestergaard B, Kutter JP, Arleth L (2009) BioXTAS RAW, a software program for high‐throughput automated small‐angle X‐ray scattering data reduction and preliminary analysis. J Appl Crystallogr 42: 959–964 [Google Scholar]
- Putnam AA, Jankowsky E (2013) DEAD‐box helicases as integrators of RNA, nucleotide and protein binding. Biochimi Biophys Acta 1829: 884–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querfurth C, Diernfellner ACR, Gin E, Malzahn E, Hofer T, Brunner M (2011) Circadian conformational change of the Neurospora clock protein FREQUENCY triggered by clustered hyperphosphorylation of a basic domain. Mol Cell 43: 713–722 [DOI] [PubMed] [Google Scholar]
- Rudolph MG, Klostermeier D (2015) When core competence is not enough: functional interplay of the DEAD‐box helicase core with ancillary domains and auxiliary factors in RNA binding and unwinding. Biol Chem 396: 849–865 [DOI] [PubMed] [Google Scholar]
- Schneidman‐Duhovny D, Hammel M, Sali A (2010) FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res 38: W540–W544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman‐Duhovny D, Hammel M, Tainer JA, Sali A (2013) Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys J 105: 962–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Collett M, Loros JJ, Dunlap JC (2010) FRQ‐interacting RNA helicase mediates negative and positive feedback in the Neurospora circadian clock. Genetics 184: 351–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Wigley DB (2002) Modularity and specialization in superfamily 1 and 2 helicases. J Bacteriol 184: 1819–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner T, Kaiser JT, Marinkovic S, Huber R, Wahl MC (2002) Crystal structures of transcription factor NusG in light of its nucleic acid‐ and protein‐binding activities. EMBO J 21: 4641–4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CT, Li SJ, Long CZ, Cha J, Huang GC, Li LL, Chen S, Liu Y (2009) Setting the pace of the Neurospora circadian clock by multiple independent FRQ phosphorylation events. Proc Natl Acad Sci USA 106: 10722–10727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tataroglu O, Lauinger L, Sancar G, Jakob K, Brunner M, Diernfellner ACR (2012) Glycogen synthase kinase is a regulator of the circadian clock of Neurospora crassa . J Biol Chem 287: 36936–36943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LL, Jackson RN, Rexhepaj M, King AK, Lott LK, van Hoof A, Johnson SJ (2014) The Mtr4 ratchet helix and arch domain both function to promote RNA unwinding. Nucleic Acids Res 42: 13861–13872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI (2003) Uniqueness of ab initio shape determination in small‐angle scattering. J Appl Crystallogr 36: 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liang X, Chen X, Guo J (2013) Ribonucleoprotein complexes that control circadian clocks. Int J Mol Sci 14: 9018–9036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir JR, Bonneau F, Hentschel J, Conti E (2010) Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci USA 107: 12139–12144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W (2012) Conventions and workflows for using Situs. Acta Crystallogr D‐Biol Crystallogr 68: 344–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN (2010) PONDR‐FIT: a meta‐predictor of intrinsically disordered amino acids. Biochim Biophys Acta 1804: 996–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Osborne AR, Groll M, Rapoport TA (2004) RecA‐like motor ATPases—lessons from structures. Biochim Biophys Acta 1659: 1–18 [DOI] [PubMed] [Google Scholar]
- Zhou M, Guo JH, Cha J, Chae M, Chen S, Barral JM, Sachs MS, Liu Y (2013) Non‐optimal codon usage affects expression, structure and function of clock protein FRQ. Nature 495: 111–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Wang T, Fu JJ, Xiao GH, Liu Y (2015) Nonoptimal codon usage influences protein structure in intrinsically disordered regions. Mol Microbiol 97: 974–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


