Abstract
Pigmentation processes occur from invertebrates to mammals. Owing to the complexity of the pigmentary system, in vivo animal models for pigmentation study are limited. Planarians are capable of regenerating any missing part including the dark-brown pigments, providing a promising model for pigmentation study. However, the molecular mechanism of planarian body pigmentation is poorly understood. We found in an RNA interference screen that a forkhead containing transcription factor, Albino, was required for pigmentation without affecting survival or other regeneration processes. In addition, the body color recovered after termination of Albino double stranded RNA feeding owing to the robust stem cell system. Further expression analysis revealed a spatial and temporal correlation between Albino and pigmentation process. Gene expression arrays revealed that the expression of three tetrapyrrole biosynthesis enzymes, ALAD, ALAS and PBGD, was impaired upon Albino RNA interference. RNA interference of PBGD led to a similar albinism phenotype caused by Albino RNA interference. Moreover, PBGD was specifically expressed in pigment cells and can serve as a pigment cell molecular marker. Our results revealed that Albino controls planarian body color pigmentation dominantly via regulating tetrapyrrole biogenesis. These results identified Albino as the key regulator of the tetrapyrrole-based planarian body pigmentation, suggesting a role of Albino during stem cell-pigment cell fate decision and provided new insights into porphyria pathogenesis.
Keywords: body color, FoxP, PBGD , pigmentation, planarian, tetrapyrrole
Introduction
Biological pigments provide basic granules to form overall body color of an organism as well as critical compartments for cytochrome within a single cell. A comprehensive understanding of the pigmentation process offers us a theoretical foundation for treatments of pigment-related disorders and especially for stem cell-based regenerative medicine. In this case, an in vivo model for pigmentation study is of great importance. Owing to possessing powerful regenerative abilities, planarian serves as a distinctive model for stem cell and regeneration studies [1, 2]. Neoblasts, the planarian adult stem cells, respond to injury and then proliferate and differentiate into corresponding cells required for complete regeneration including pigment cells [3–6]. A cell subpopulation among the neoblasts called cNeoblasts is pluripotent and single cell transplantation of cNeoblasts is able to rescue lethally irradiated planarian [7]. Pigmentary system derived from adult stem cell in vivo is a unique model for pigmentation study. These features confer planarian to be an advantageous model for pigmentation study. However, regulation of the pigmentation process, especially body pigmentation in planarian, is poorly understood.
Transcription factors have critical roles in guiding the regeneration processes [8, 9]. Vital transcription factors have been identified in planarian polarity establishment [10–14], nerve system [15–17], eye [18–20], pharynx [21, 22], germline [23–25], gut [7, 26] and nephridia [27] development and regeneration. Among these, forkhead containing transcription factors FoxD and FoxA independently regulate anterior pole establishment and pharynx regeneration, indicating that Fox family genes show significant roles during planarian development and regeneration relevant to their functions in mammals [11, 12, 21, 22, 28]. Still, the role that transcription factors have in planarian body pigmentation remains unknown.
Planarian eyes contain pigment cells that utilize melanin as photosensitive pigments and Gtso, Djsix-1,Djeya,Smed-sp6–9,Smed-dlx and Smed-ovo are reported to be critical factors for the generation of eye pigment cup cells in which melanin is synthesized [18–20, 29]. However, the pigments that form planarian body color require further investigations. Krugelis-Macrae [30] suggested the occurrence of porphyrin, a cyclic form of tetrapyrroles in planarian Dugesia dorotocephala, by chemical and spectral experiments and demonstrated that planarians are able to convert delta-aminolevuliwic acid to porphobilinogen [31, 32]. This porphyrin-like pigment was further localized in epidermal rhabdites [33], suggesting that the tetrapyrrole-like pigment is responsible for planarian body color. Recently, body pigment cells have been observed by using electron microscopy in D. gonocephala, D. ryukyuensis and Schmidtea mediterranea, whereas chromatograph results suggested that a tryptophan-based pigment ommochrome is responsible for the body color [34–37]. However, molecular or functional validations shall be carried out to confirm these findings and, more importantly, how this process is regulated remains unclear. Tetrapyrroles, referred to as ‘the pigment of life’, are a family of fundamental compounds generated from four porphobilinogen molecules under the catalysis of porphobilinogen deaminase (PBGD) [38–41]. As the intermediate products for heme, chlorophyll and cobalamin, tetrapyrroles typically serve as the colored chromophores. Tetrapyrrole-based biomolecules modulate cellular response to light, oxygen and other environmental factors, whereby tetrapyrrole dysregulation results in serious diseases in human [42, 43]. Whether tetrapyrroles are responsible for planarian body pigments needs further molecular validations.
In this work, we screened for transcription factors that are specifically required for planarian S. mediterranea body pigmentation. Our result revealed that a forkhead domain transcription factor is required for body pigmentation without affecting regeneration. We further examined its downstream targets via microarray. Interestingly, this forkhead domain transcription factor controls the expression of enzymes for tetrapyrrole synthesis, and loss of one of these enzymes, PBGD, resulted in the same albinism phenotype. Thus, we report a critical transcription factor that controls body color via regulating tetrapyrrole biogenesis in planarian.
Results
Planarian body pigment accumulates during juvenile development and regeneration
Newly hatched worms were born colorless and the pigmentation process took place after the hatching, and it took 12 days for a newborn planarian to get pigmented but the eye spots were pigmented at birth (Supplementary Figure S1A). Moreover, the newly generated pigments emerged randomly without a particular pattern, suggesting an irregular growth status and shape of pigment cells (Supplementary Figure S1A). The difference between body color pigmentation and eyespot pigmentation indicates the presence of two separate pigment systems for eyespot and body pigments. Applying the modified transmission electron microscopy (TEM) protocol [35], we confirmed that planarian pigment granules lay between circular and longitudinal muscle cells just beneath the basal lamina (Supplementary Figure S1B), in line with the previous publications [34–37]. Unlike the vital organs of planarian, the body color took more than a fortnight to recover post amputation (Supplementary Figure S1C). The worms regenerated visible eye spots within 6 days, whereas the blastema remained unpigmented. The first visible pigment within the newly regenerated blastema appeared ~8 days post amputation (dpa), and at least 6 more days were indispensable for a full recovery of body color (Supplementary Figure S1C). A schematic diagram showed the pigment cell location: pigment cells locate between the circular and longitudinal muscles beneath the basal lamina (Supplementary Figure S1D). Thus, planarians displayed a different dynamics from the eyespot to generate and regenerate pigment cells and pigments, and the pigmented cells are located between the muscle cells beneath the basal lamina without an orderly pattern.
Albino is required for planarian body pigmentation
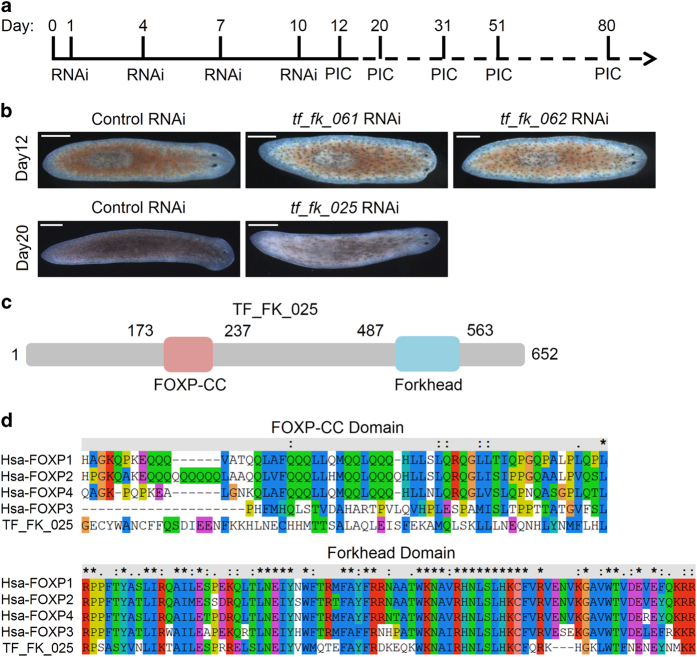
In order to explore the key factors during the pigmentation process of planarian, we carried out an RNA interference (RNAi) screen against ~600 unreported transcription factors. In the RNAi experiments, we fed the worm with in vitro synthesized double stranded RNA (dsRNA) four times within 10 days and observed the phenotype caused by dsRNA feeding (Figure 1a). In the screen, three genes were identified to be required for planarian body color maintenance: RNAi either of tf_fk_061 or tf_fk_062 led to pigment clumping and head regression resulting in worm lysis eventually, whereas RNAi of tf_fk_025 caused albinism without affecting the homeostasis or regeneration (Figure 1b). Using 5ʹ and 3ʹ rapid amplification of complementary DNA (cDNA) ends, a full length of 652 amino acids for tf_fk_025 was acquired and, using PFAM (Database of protein families. http://pfam.xfam.org/) alignments, a FOXP-CC domain and a Forkhead domain from its amino acids 173 to 237 and from 487 to 563 were identified, respectively (Figure 1c). The FOXP-CC and Forkhead domains were found to be conserved with human FOXP homologs analyzed using ClustalX2 (Figure 1d). As expected, TF_FK_025 was clustered with the human FOXP subfamily (Supplementary Figure S1E). Interestingly, both TF_FK_061 and TF_FK_062 also contain a Forkhead domain (Supplementary Figure S1F). As tf_fk_025 RNAi leads to a gradual loss of color, we name this planarian FoxP gene smed-Albino (henceforth referred to as Albino).
Figure 1.
Planarian pigmentation process and transcription factors involved. (a) Experimental design of RNAi screen; worms were fed dsRNA-mixed liver on days 1, 4, 7 and 10 for four times and pictures were collected from the twelfth day post first RNAi. (b) Genes identified from the RNAi screen that affect the pigmentary system during homeostasis. Scale bar: 200 μm. (c) Conserved domain analysis revealed a FOXP-CC domain and a Forkhead domain within protein TF_FK_025. (d) ClustX2 analysis of TF_FK_025 with human FOXP family proteins (Star indicates same amino acid).
Albino is specifically required for pigmentation without affecting worm survival or regeneration
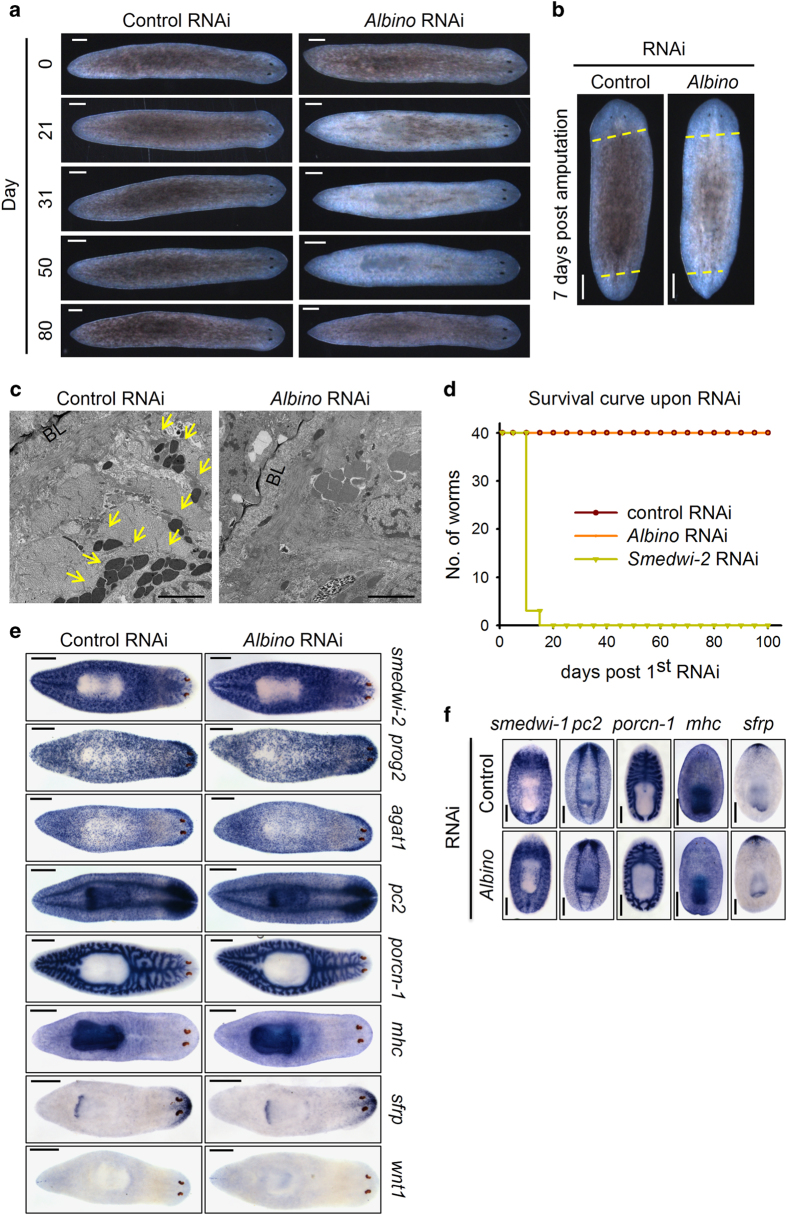
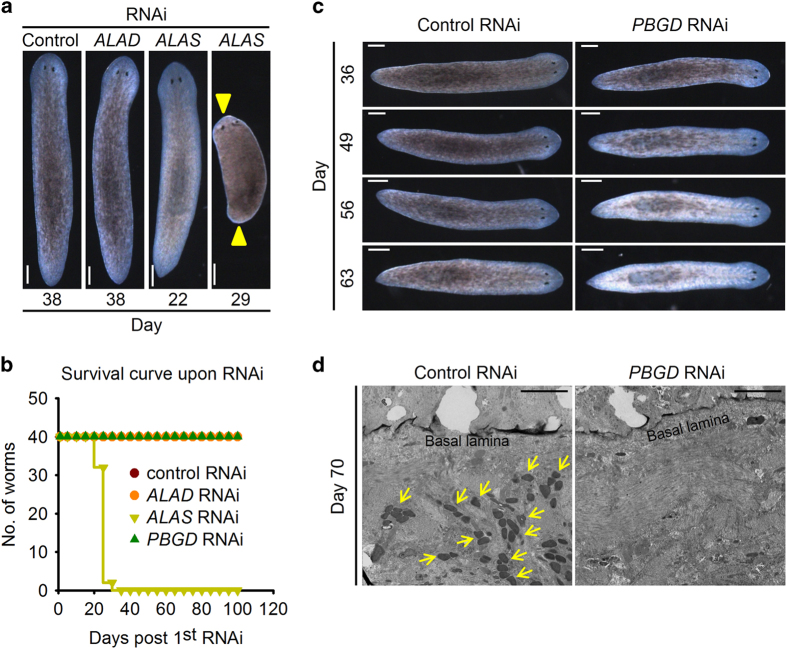
To gain insight into Albino function, we observed the albinism resulted from Albino RNAi in detail. Albino RNAi worms lost body color gradually from Day 21 post RNAi and became completely albino on around Day 50 post RNAi (Figure 2a and Supplementary Figure S2A). Amputated worms upon Albino RNAi successfully regenerated lost head and tail just as the control RNAi worms, but the newly regenerated parts were unpigmented and the original parts became albino as the intact worms (Figure 2b). Transmission electron microscopy confirmed that the pigment granules lost upon Albino RNAi, as we observed no pigment granules within the basal lamina where the pigment cells reside (Figure 2c and Supplementary Figure S2B). To ascertain whether Albino RNAi affects the worm survival, we fed the worms food containing Albino dsRNA for over 100 days and the worms survived with albinism while smedwi-2 RNAi worms died [44] within 20 days (Figure 2d). In situ hybridization (ISH) of specimen 30 days post control or Albino RNAi revealed that neither stem cells, nerve system, gut, muscle nor polarity markers were affected (Figure 2e), although the regenerating worms also showed no differences in the expression pattern for makers of stem cells, nerve system, gut, muscle and anterior polarity (Figure 2f). As the stem cell system was unaffected upon Albino RNAi, we hypothesized that albino worms would recover as the RNAi effects decrease. As expected, planarian subjected to four rounds of dsRNA feeding and reverted with normal food first became albino and then became re-pigmented around 80 days post first RNAi (Figure 2a). These results indicate that the Forkhead domain containing gene Albino is required for the pigmentation process of planarian without affecting the worm survival or regeneration, and the robust stem cell system allows the worms to become re-pigmented once the RNAi is terminated.
Figure 2.
Albino is required for pigmentation without affecting survival or regeneration. (a) Albino RNAi worms lost body color. Scale bar: 200 μm. (b) Regenerated Albino RNAi trunk fragment with albino phenotype. Scale bar: 200 μm. (c) Transmission electron micrographs showing the pigment loss in Albino RNAi worms. Scale bar: 20 μm. Yellow arrows indicate pigment granules. (d) Survival curve of RNAi worms. (e) ISH of stem cell progeny and tissue-specific markers of control and Albino RNAi intact planarian. Scale bar: 500 μm. (f) ISH of stem cell progeny and tissue-specific markers of control and Albino RNAi-amputated trunks. Scale bar: 500 μm.
Albino enriches in epidermal region and newly regenerated blastema
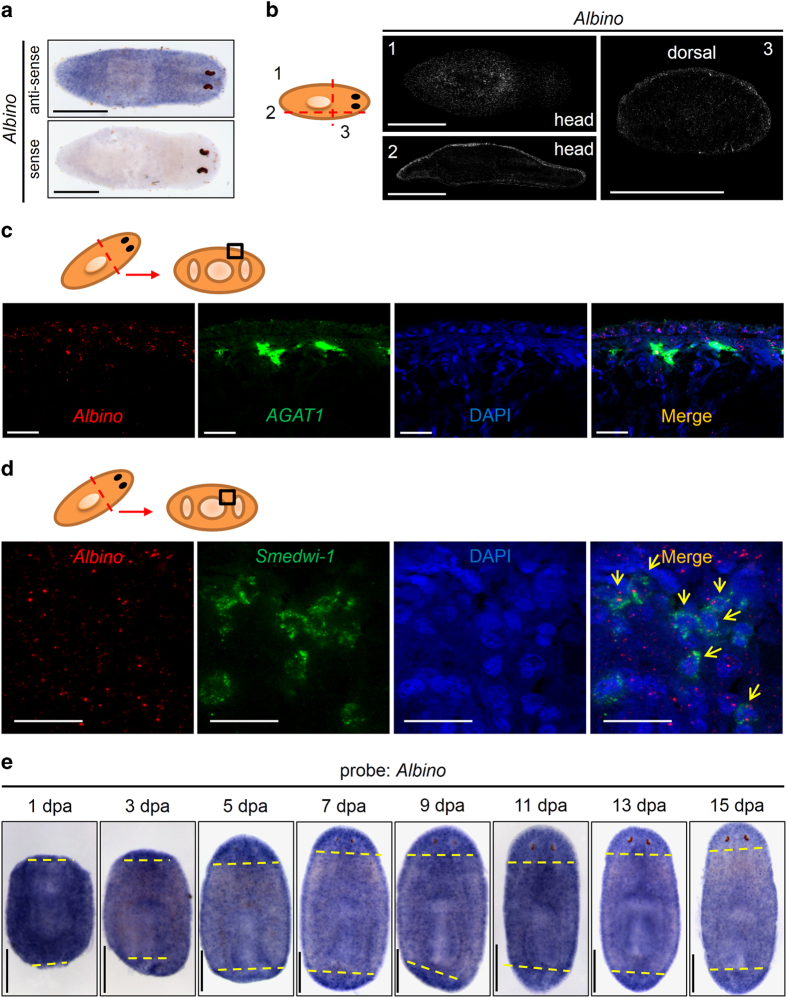
To understand how Albino controls pigmentation, systematic expression analyses were carried out (Figure 3). We first synthesized an antisense probe for Albino detection and a sense probe to validate the signals. Whole-mount ISH confirmed the specificity of the signals (Figure 3a). As shown in whole-mount- and section fluorescence in situ hybridization (FISH), Albino is ubiquitously expressed throughout the body of a whole worm. Strong signals were observed at the surface and edges, indicating superficial expression enrichment (Figure 3b). Vibration sections were performed to gain insight into the detailed expression patterns. We found a clear enrichment of Albino around the worms in the epidermal region in both transverse and vertical sections, and Albino was also observed to express in mesenchymal tissues (Figure 3b). To further elucidate Albino expression patterns, we applied double FISH (DFISH) for Albino with prog2 and AGAT1, two stem cell progeny markers known to express at the superficial region of the worms. It was clear in the highly magnified panels that Albino expressed in cells beneath epidermal cells as well as in epidermal cells, and colocalized with both prog2- and AGAT1-positive cells (Figure 3c and Supplementary Figure S3A).
Figure 3.
Albino enriches at epidermal region and during regenerating blastema. (a) WISH in intact animals. Scale bar: 500 μm. (b) Vibration sections of FISH animals. Section thickness: 80 μm and scale bar: 500 μm. Images are z-stacks of 10 μm. (c) Double FISH with AGAT1 and Albino in wild-type animals showing the dorsal body wall. Scale bar: 20 μm. Cartoon indicates region of interest. Images are single confocal sections. (d) Double FISH with smedwi-1 and Albino in wild-type animals showing a transect section. Scale bar: 25 μm. Yellow arrows indicate smedwi-1 and Albino double-positive cells. Images are single confocal sections. (e) ISH in regenerating animals detecting Albino. Yellow dashes indicate amputation sites. Scale bar: 500 μm.
As we observed a mesenchymal-like expression, we next explored the possibility of whether Albino expresses in smedwi-1-positive adult stem cells. We observed an expression of Albino in fluorescence-activated cell-sorting sorted X1 cells with quantitative polymerase chain reaction (qPCR) and a weak reduction of Albino signals in γ-ray-irradiated worms using ISH (Supplementary Figure S3B and C). DFISH showed the colocalization of smedwi-1 with Albino, suggesting a role that Albino may have in pigment cell fate commitment (Figure 3d). In addition, no defects in food uptake were noted in Albino RNAi worms as the worms were fed with food with Albino dsRNA for over 100 days (Figure 2d).
Because it took more than 2 weeks to accomplish pigmentation during regeneration, we hypothesized that Albino enriches at the unpigmented blastema during regeneration. We observed the expression pattern of Albino in wild-type regenerating specimen from the first to fifteenth day of regeneration. As expected, we found an enrichment of Albino signals in the newly regenerated areas, suggesting a requirement of this putative transcription factor (Figure 3e). In addition, we found in 2 days' post-hatching worms that Albino is not as ubiquitous as in mature planarian according to the fact that the newly hatched worms are not fully pigmented (Figure 1b and Supplementary Figure S3D). These data demonstrated that the Albino expression pattern meets the criteria for a pigment cell regulator both spatially and temporally, suggesting a role of Albino as a molecular ‘switch’ for pigment generation in planarian.
Tetrapyrrole pathway enzyme expressions depend on Albino
We then used planarian-customized mRNA expression array to elucidate the underlying mechanism. We applied two biological replicas of worms subjected to four rounds of either Albino or control RNAi and collected RNA at 7 days post the last RNAi even though the worms were not complete albino. Genes with more than twofold decrease in both replicas are listed in Table 1. Interestingly, the most significantly reduced gene is PBGD, which is a critical enzyme in biogenesis of tetrapyrrole. We further noticed in Table 1 that two additional enzymes required fortetrapyrrole biosynthesis, delta-aminolevulinic acid dehydratase (ALAD) and 5-aminolevulinic acid synthase (ALAS), were also downregulated upon Albino RNAi, implying that Albino governs planarian pigmentation through tetrapyrrole pigment pathway. Tetrapyrrole comprises four pyrrole rings and is synthesized from glycine and succinyl CoA under the catalysis of ALAS, ALAD and PBGD. If further catalyzed, tetrapyrrole forms, cyclic tetrapyrrole, the cores of natural pigment hemoglobin or chlorophyll [38, 43]. Because of the high degree of conjugation in tetrapyrrole, it is commonly colored as chromophores. Thus, we aligned the planarian amino-acid sequences of ALAD, ALAS and PBGD with the corresponding human and mouse genes, and found that these genes were clustered with their own family with identity of ~50% (Supplementary Figure S4A and B). ClustalX2-aligned results revealed that the amino-acid sequence of PBGD shares a high similarity with human and mouse homologs (Supplementary Figure S4C). We thus named these genes smed-ALAD, smed-ALAS and smed-PBGD (henceforth abbreviated to ALAD, ALAS and PBGD, respectively).
Table 1. List of downregulated genes upon Albino RNAi in array analysis.
| Probe | Fold change | BLAST | GI number |
|---|---|---|---|
| 09827 | 0.01 | Porphobilinogen deaminase isoform 1 | 84609767 |
| 00920 | 0.04 | Threonine dehydratase catabolic | 113467172 |
| 07123 | 0.05 | Saposin | 113466959 |
| 07980 | 0.11 | 5-Aminolevulinic acid synthase | 84613222 |
| 04488 | 0.13 | Synergin gamma | 84612706 |
| 01820 | 0.13 | Zinc-finger protein 474 | 84600384 |
| 03955 | 0.14 | Carbonic anhydrase | 84614305 |
| 01039 | 0.17 | Fucolectin | 84599334 |
| 05195 | 0.18 | Ferritin | 84598371 |
| 06678 | 0.19 | Lipase | 84592609 |
| 06806 | 0.20 | Elav1 (HUR) | 84613280 |
| 02989 | 0.21 | Y box protein 4-like protein | 84613514 |
| 02568 | 0.22 | Hexokinase | 84591284 |
| 00559 | 0.23 | C3H-zinc-finger-containing protein 1 | 84600517 |
| 06162 | 0.23 | Granulin-like protein | 84613008 |
| 02604 | 0.24 | Ferritin | 84596222 |
| 00736 | 0.25 | Lipase | 84610165 |
| 01255 | 0.29 | Serine protease inhibitor-1 | 84601442 |
| 01819 | 0.31 | Trans-1,2-dihydrobenzene-1,2-diol dehydrogenase | 84597330 |
| 03500 | 0.37 | Sodium-dependent glucose transporter 1 | 84613285 |
| 09907 | 0.37 | Delta-aminolevulinic acid dehydratase | 116034941 |
| 03741 | 0.41 | Lectin 1 | 84597836 |
| 00821 | 0.42 | Lipase | 84598548 |
Abbreviation: BLAST, The Basic Local Alignment Search Tool; GI, GenInfo Identifier; RNAi, RNA interference.
Fold changes were average from two replicas.
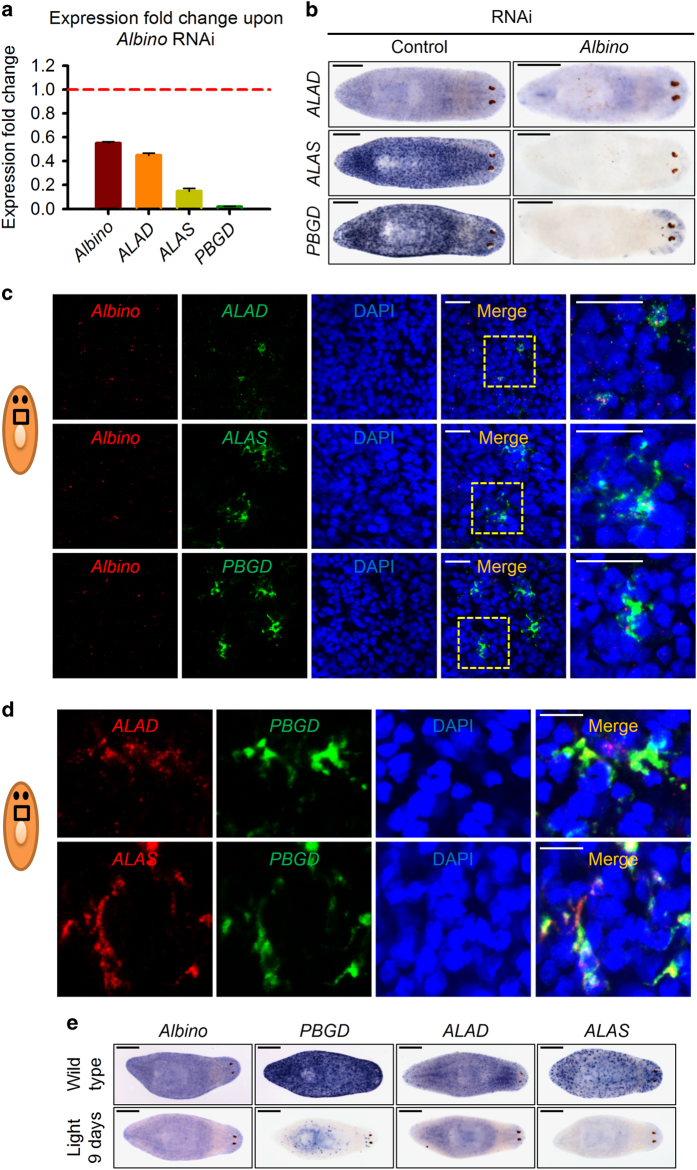
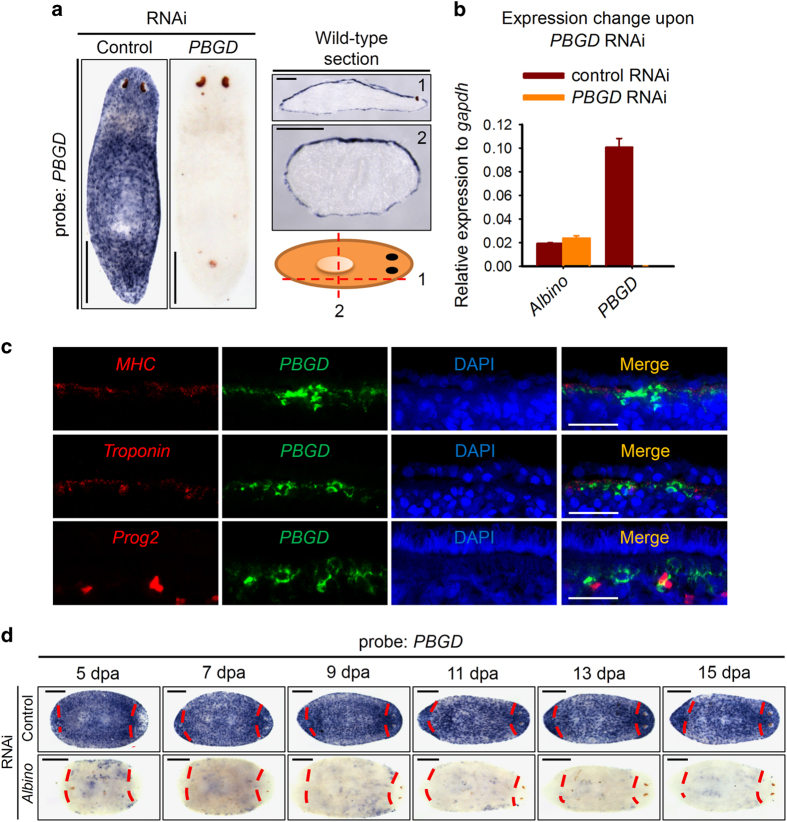
The confirming qPCR results showed that the fold changes of ALAD, ALAS and PBGD were identical to those in the mRNA array. Moreover, PBGD is notably reduced by more than 50-folds, suggesting that PBGD expression is largely dependent on Albino (Figure 4a). ISH confirmed the decrease in tetrapyrrole synthesis enzymes and, although with different expression levels, we noticed a similar expression pattern of all three enzymes at superficial layers of worms (Figure 4b). As expected, we found that Albino colocalized with these enzymes at superficial regions (Figure 4c). In addition, these enzymes also colocalized with each other as revealed by DFISH at the same region that Albino is expressed (Figure 4d).
Figure 4.
Albino regulates expression of tetrapyrrole biosynthetic enzymes in planarian. (a) Expression fold changes of tetrapyrrole biosynthetic enzymes upon Albino RNAi. Shown are averages of three independent experiments; error bars=s.d. (b) WISH for tetrapyrrole biosynthetic enzymes in worms upon control or Albino RNAi, indicating the reduction of tetrapyrrole biosynthetic enzyme expressions. WISH samples were collected 7 days post fourth RNAi. Scale bar: 500 μm. (c) Representative double FISH results of ALAD, ALAS and PBGD with Albino in wild-type animals. Scale bar: 20 μm. Images are single confocal sections. (d) Representative double FISH results of ALAD and ALAS with PBGD in wild-type animals. Scale bar: 20 μm. Images are single confocal sections. (e) ISH of either wild-type worms or worms received 9 days of continuous direct light for Albino, PBGD, ALAD and ALAS.
Tetrapyrrole could be further catalyzed into heme, which is a critical biological pigment required for many cell activities such as electron transfer, catalysis and molecule transport [45]. We next examined the remaining enzymes (Uroporphyrinogen Decarboxylase, UROD; Uroporphyrinogen III Synthase, URO3S; Coproporphyrinogen Oxidase, CPOX; Protoporphyrinogen Oxidase, PPOX) required for heme biosynthesis. However, no significant changes of these enzymes at the mRNA expression level were observed (Supplementary Figure S5A). Besides, these enzymes exhibited different expression patterns from the tetrapyrrole enzymes (Supplementary Figure S5B), indicating that heme is not involved in the pigmentation process in planarian. We also tested the expression-level changes of stem cell progenies and critical enzymes in melanin and ommochrome pathways and found that no changes over twofold were observed upon Albino RNAi in either stem cell progenies or critical enzymes in the melanin pathway (Supplementary Figure S5C). A significant reduction in kynurenine-3-monooxygenase (Kmo), an important enzyme in the ommochrome pathway, was detected using qPCR (Supplementary Figure S5C) and was subsequently confirmed by ISH (Supplementary Figure S5D). Interestingly, Albino colocalized with Kmo2 (Supplementary Figure S5E), suggesting that this enzyme is partially involved during planarian pigmentation.
Our laboratory noted a phenomenon of light bleaching several years ago: planarian lost body color by excreting the pigmented mass from the pharynx, whereby the worms became unpigmented after 9 days' exposure to continuous light (Supplementary Figure S5F). This result was supported by a recent report [46]. It is significant to understand whether the Albino-mediated pigmentary system was affected by this progress. As expected, the expression levels of both Albino and tetrapyrrole-related enzymes were decreased in light-bleached worms, further indicating that Albino-mediated tetrapyrroles are responsible for the body color of planarian (Figure 4e).
Taken together, our data demonstrate that the expression of tetrapyrrole biosynthesis enzymes depends on the expression of Albino. The colocalization of Albino and these enzymes provides the basis for a regulatory network between Albino and these enzymes. In addition, it is noteworthy that Albino is required for the expression of Kmo2, an enzyme in ommochrome biosynthesis.
PBGD RNAi resulted in albinism during regeneration and homeostasis
RNAi experiments of these enzymes were carried out in order to elucidate their functions in planarian pigmentation. RNAi of ALAD caused no significant change in pigmentation, homeostasis or regeneration (Figure 5a and b and Supplementary Figure S6A). Surprisingly, ALAS RNAi resulted in a slight body color loss at ~22 days post first RNAi, but a strong defect in homeostasis 7 days later starting with a regression from both head and tail (Figure 5a and b). It is rather unusual that Albino RNAi did not lead to homeostasis defects even when the worms were continuously interfered with Albino dsRNA for more than 100 days. However, we did observe a mesenchymal and gut-like expression pattern of ALAS (Supplementary Figure S6B), and Albino RNAi did not result in a complete loss of the ALAS mRNA level (Supplementary Figure S4A). Thus, it is possible that ALAS is involved in additional processes other than Albino-mediated pigmentation and is required for the homeostasis of planarian independently of Albino. Although PBGD-interfered worms displayed no survival or regeneration abnormalities, they exhibited Albino RNAi-like albino phenotype at ~60 days post first RNAi (Figure 5b and c and Supplementary Figure S6A). Although PBGD RNAi caused a delayed phenotype emergence compared with Albino RNAi, worms that lost PBGD eventually became completely albino (Figure 5c). We thus used TEM to examine subcellular changes upon PBGD RNAi and detected similar pigment granules lost in PBGD RNAi worms (Figure 5d and Supplementary Figure S6C).
Figure 5.
PBGD RNAi results in albinism. (a) RNAi of tetrapyrrole biosynthetic enzymes. Yellow arrows indicate regressions. Scale bar: 200 μm. (b) Survival curve of RNAi worms. (c) PBGD RNAi worms lost body color gradually and got totally albino ~60 days post first RNAi. (d) Transmission electron micrographs showing the pigment lost in PBGD RNAi worms. Scale bar: 20 μm. Yellow arrows indicate pigment granules.
Gene functions of the remaining enzymes (for heme biosynthesis: CPOX, PPOX, UROD and URO3S; for ommochrome biosynthesis: Afmid1 (arylformamidase), Afmid2, Kmo1 and Kmo2) were detected (Supplementary Figure S6D and E). However, loss of any of these enzymes did not lead to pigmentation defects like Albino RNAi. Nonetheless, a slight body color alteration was observed under Kmo2 RNAi, suggesting that ommochrome derived from tryptophan may partially be involved in the body color of planarian. Conversely, Tyrosinase (tyr), an enzyme critical for melanin biosynthesis, RNAi led to eyespot pigmentation failure only, rather than body pigment loss (Supplementary Figure S6F). These results revealed that, among the Albino-regulated tetrapyrrole biosynthesis enzymes, PBGD in particular is required for planarian body color maintenance, suggesting that PBGD serves as the downstream target of Albino in regulating the body pigment of planarian. Further validation with chromatin immunoprecipitation was required to demonstrate a direct regulation between Albino and PBGD. Moreover, we provide evidence that tetrapyrrole is involved as body pigment across evolution, demonstrating an exceptional evolutionary position of planarians.
PBGD labels planarian pigment cells
We next confirmed the superficial expression of PBGD by whole-mount and cross-section ISH (Figure 6a). The expression of PBGD is efficiently knocked down without affecting Albino expression, suggesting that Albino functions upstream of PBGD (Figure 6a and b). We noticed that, unlike Albino or ALAS, PBGD expressed only at the superficial level, prompting us to investigate whether PBGD labels planarian pigment cells specifically. To gain insight into the accurate location at which PBGD-positive cells reside, we investigated the expression relationship between PBGD and stem cell progeny prog2 and muscle cell markers mhc and troponin. We observed that PBGD-positive cells were situated between muscle cells just beneath the basal lamina (Figure 6c), a unique location for pigment cells (Figure 1d). Unlike Albino, PBGD is restricted within pigment cells without expressing in epidermal cells. Moreover, PBGD also enriched at newly regenerated blastema since 7 dpa (Figure 6d). The enrichment lasted for another 7 days during which planarians became pigmented (Figure 1a). Thus, the function and expression pattern of PBGD indicated that PBGD serves as a perfect molecular maker for labeling planarian pigment cells.
Figure 6.
PBGD labels pigment cells in planarian. (a) WISH and frozen section of PBGD in wild-type animals showing an epidermal-specific expression pattern. Scale bar: 200 μm. (b) Relative expression level to gapdh. Shown are averages of three independent experiments; error bars=s.d. (c) Double FISH for PBGD with mhc, troponin and prog2 in wild-type animals at dorsal body wall showing PBGD-positive cells lying in between muscle cells. Scale bar: 20 μm. Images are single confocal sections. (d) WISH showing expression patterns of PBGD in regenerating control or Albino RNAi worms. Red dashes indicate amputation sites. Scale bar: 200 μm.
Albino bridges between adult stem cells and PBGD-positive pigment cells
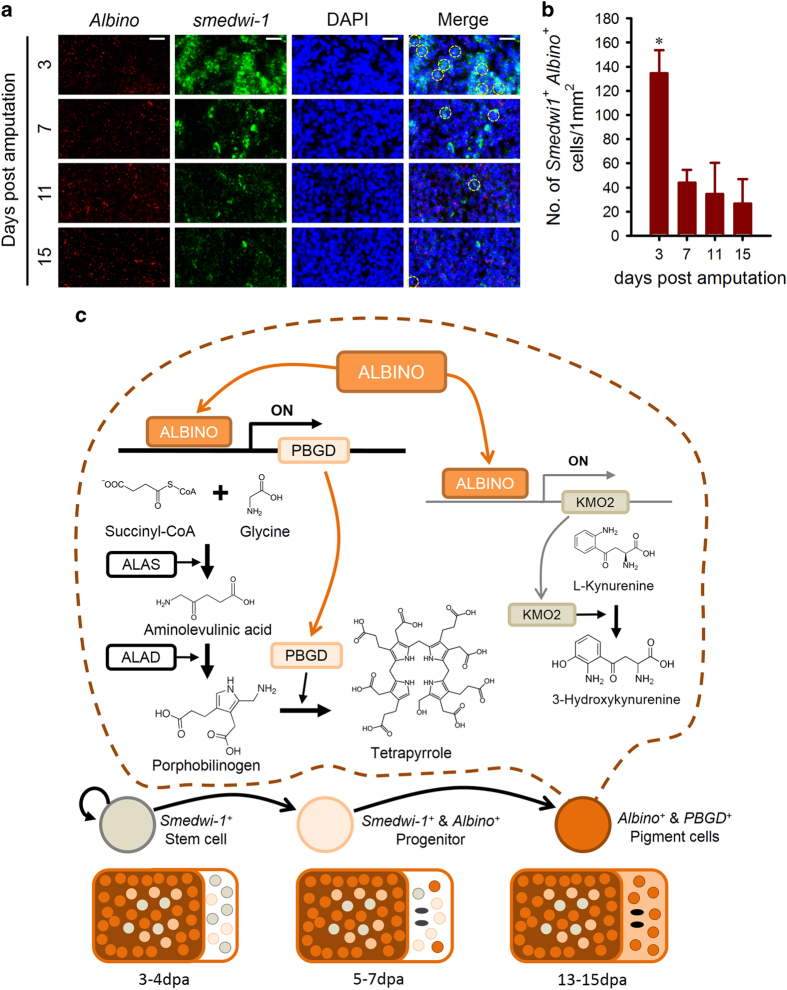
RNAi of Albino not only sabotaged the pre-existing Albino expression but also blocked the enrichment of PBGD at blastema during regeneration (Figure 6d). We next studied the expression dynamics of Albino together with smedwi-1 and PBGD during regeneration. DFISH showed at 3 dpa that a significant number of smedwi-1 and Albino double-positive cells emerged at the blastema and, as regeneration proceeded, the number of smedwi-1 and Albino double-positive cells declined to a relatively stable level (Figure 7a and b and Supplementary Figure S7A), suggesting that the fate of most pigment cells was determined right after the local neoblast proliferation [47]. However, the Albino and PBGD double-positive cells did not begin to concentrate at blastema until 7 dpa and enrichments of Albino and PBGD double-positive cells last until the pigmentation of blastema is finished (Supplementary Figure S7B), suggesting that Albino has a regulatory role during the fate decision of the stem cell subpopulation that differentiate into pigment cells.
Figure 7.
Albino mediates neoblast-pigment cell differentiation. (a) Double FISH for smedwi-1 with Albino at different times of regeneration showing that smedwi-1 and Albino colocalize at the blastema during regeneration. Scale bar: 20 μm. Yellow circles indicate double-positive cells, whereas red and green circles indicate Albino and smedwi-1 single-positive cells, respectively. Images are single confocal sections. (b) Quantification of smedwi-1 and Albino double-positive cells. Cells in 0.1 mm2 were counted in three independent experiments. Error bar=s.d.; *P<0.0001; significance determined with Student’s t-test. (c) Cartoon illustrates that Albino controls the expression of PBGD, and smedwi-1-positive neoblasts initially specialize into smedwi-1 and Albino double-positive cells and then fully differentiate into pigment cells expressing Albino and PBGD. During regeneration, Albino-expressing neoblasts accumulate in the blastema at 3 dpa and initiate the expression of tetrapyrrole biogenesis enzymes. The Albino and PBGD double-positive cells accumulate within blastema since 7 dpa, and finally these cells generate tetrapyrroles that get the planarians pigmented.
Our data thus suggest that, during the pigmentation of newly regenerated blastema, a subpopulation of smedwi-1-positive stem cells express Albino and, under the control of Albino, tetrapyrrole biosynthesis enzymes begin to express and synthesize tetrapyrrole as the body pigment of planarians. Meanwhile, the expression of Kmo2, a key enzyme in ommochrome biosynthesis, also depends on Albino (Figure 7c).
Discussion
Dysregulation of pigmentation process leads to serious disorders in humans [48]. We set up a planarian pigmentation model to search for de novo mechanisms during the differentiation from stem cells to pigment cells. In this work, we identified a forkhead domain containing transcription factor Albino required for planarian body color pigmentation. We elucidated that transcription factor Albino is required for the expression of ALAS, ALAD and PBGD expression, thereby controling the tetrapyrrole biosynthesis. Interestingly, apart from the tetrapyrrole biosynthesis pathway, the expression of Kmo2, a key enzyme in the ommochrome pathway, also depends on Albino, suggesting that Albino controls two pigment biosynthesis pathways within planarian body pigment cells (Figure 7c). Meanwhile, our results revealed that PBGD is specifically expressed within planarian body pigment cells, thus providing the best marker that labels planarian body pigment cells. Besides, the regulatory relationship between Albino and tetrapyrrole enzymes suggests new prospects in porphyria pathogenesis study.
The role of Albino in adult stem cell and other non-pigment-related cells
Our result demonstrated that Albino has a critical role in regulating tetrapyrrole biosynthetic enzyme expression that takes place in pigment cells. However, the role of Albino in adult stem cell and other non-pigment-related cells remains unclear. The neoblasts that participated in regeneration in planarian are heterogeneous, consisting of pluripotent stem cells and lineage-committed progenitors [49]. Transcription factors have critical roles in guiding the specification processes [8, 9]. According to our ISH results, Albino is broadly expressed, but Albino RNAi showed no other defects but in pigmentation. We assume that Albino functions in a modest manner in non-pigment cells without affecting the cell survival or other biological processes. However, as Albino is not completely knocked out, it is possible that the RNAi efficiency is critical for Albino functions. An Albino knockout strain will be of great help in understanding the function of this transcription factor. Nonetheless, we provided planarian as an alternative model for pigmentation study, as markers labeling early and late differentiated pigment cells are available. By monitoring Albino expression, we could observe the early specialization from neoblasts to pigment cells under various stimuli, such as light bleaching.
Albino simultaneously controls two pigment biosynthesis pathways
Our study presents the link between forkhead genes and pigmentary system development by demonstrating that Albino controls the expressions of tetrapyrrole biosynthetic enzymes that in turn generate tetrapyrrole. Albino RNAi resulted in over 100-fold decrease in PBGD expression, suggesting a direct regulatory relationship between Albino and PBGD. We provided evidence that tetrapyrroles are used as body pigment in planarian across evolution, suggesting a unique evolutionary position of planarians. However, as we failed in generating a planarian-specific Albino antibody, we could not elucidate the direct regulatory mechanism. A functional Albino antibody is urgently in need for further analyses. We also noticed a significant expression reduction in Kmo2, a critical enzyme in ommochrome generation, upon Albino RNAi. Although only a very weak depigmentation was observed upon Kmo2 RNAi, we revealed a similar expression pattern between Kmo2 and tetrapyrrole enzymes, suggesting that Albino simultaneously controls two pigment biosynthesis pathways.
Identification of planarian pigment cell markers provides a new model for pigmentation study
Expression analysis of PBGD showed a specific expression within the muscle cells around the surface of planarians. In addition, the shape of PBGD-positive cells resembles the shape of pigment cells under the observation of TEM. Meanwhile, RNAi of PBGD led to complete albinism, although it took more time than Albino RNAi. We conclude from the expression pattern and RNAi phenotype that PBGD-positive cells represent planarian pigment cells and thus provide a molecular marker for pigment cells of planarians. Meanwhile, as Kmo2 RNAi showed a rather weak body color reduction (Supplementary Figure S6E), which is supported by the recently published paper [46], thus PBGD is a better marker for labeling planarian pigment cells.
Several in vivo and in vitro models have been established for pigmentation study, especially for melanocyte studies [50, 51]. However, thus far, the complexity of the vertebrate pigmentary system made it difficult to observe pigmentation and depigmentation dynamically. Stubenhaus et al. [46] recently suggested that depigmentation induced by intense light in planarian models the pathophysiology of acute porphyrias. Our results elaborated Albino as the upstream regulator of two evolutionarily conserved planarian body pigment synthesis pathways, and this warrants further investigation on whether Albino has a role in the pathophysiology of acute porphyrias. We have shown that planarian lost body color upon Albino or PBGD dsRNA feeding. This depigmentation process provides a new in vivo animal model to study the biological alteration upon pigment loss. Meanwhile, when dsRNA foods were substituted with normal foods, planarians became re-pigmented under the effects of unaffected and pluripotent cNeoblasts system [7]. The re-pigmentation process provides a controllable pigmentation model from specialized stem cells. Our model provides the basis for elucidating the essential role of Albino in the stem cell-pigment cell fate decision.
Materials and Methods
Planarian culture
Clonal lines of hermaphroditic and asexual (CIW4) S. mediterranea were maintained as previously described [52] and supplied with 0.21 g l−1 Instant Ocean salts. Animals were fed weekly with homogenized calf liver. Animals were starved for 1 week before any experiments. For irradiation, planarians were exposed to 100 Gray of gamma irradiation using a sealed source of Cesium 137 (Gammacell3000, MDS Dordion, Chalk River, ON, Canada). The animals were kindly provided by P Newmark (University of Illinois at Urbana-Champaign/Howard Hughes Medical Institute, Urbana, IL, USA), P Reddien (Massachusetts Institute of Technology/Howard Hughes Medical Institute, Cambridge, MA, USA) and N Oviedo (University of California, Merced, CA, USA).
RNAi experiments
We use in vitro synthesized dsRNA for RNAi experiments. Each time, we prepared 400 ng dsRNA mixed with 5 µl liver for each worm and the total volume depends on the number of worms. The worms were fed four times in screening, Albino, ALAD and PBGD RNAi, two times for smedwi-2, ALAS and UROD RNAi, eight times for CPOX, PPOX, URO3S, Afmid1 and/or Afmid2Kmo1 and/or Kmo2 RNAi. We fed worms for 12 times in survival experiments of Albino, ALAD and PBGD RNAi. We fed the worms on days 1, 4, 7 and 10 for the first four times and fed the worms every 10 days with liver mixed with corresponding dsRNA. We observed the successful food uptake for every single experiment. At least 10 worms were used in each RNAi experiment and at least three independent experiments were carried out for each gene. Phenotypes shown in all replicates are presented; otherwise the specific number was labeled.
Gene cloning
In the transcription factor screen, we used a BLAST-based reciprocal best-hit method, in combination with protein sequence alignment and phylogenetic analysis as described previously. We searched for forkhead containing proteins in both planarian genome database SmedGD (http://smedgd.neuro.utah.edu), the Plan Mine (http://planmine.mpi-cbg.de) and the hermaphroditic strain (Expressed Sequence Tag) database [53–55]. We obtained the full-length sequences with the RNA ligase-mediated rapid amplification of cDNA ends kit (Ambion, Austin, TX, USA) and aligned with online PFAM (http://pfam.xfam.org/) and local ClustalX2. Phylogenetic trees were constructed with ClustalX2 using the neighbor-joining algorithm with 1000 trials of bootstrap and 120 random seeds. All sequences were deposited in GenBank.
Transmission electron microscopy
TEM was performed as previously described [35]. In brief, worms were first fixed with primary fixative. After being washed with EM buffer, secondary fixation was performed with osmium tetroxide. Ultrathin sections were stained with uranyl acetate and lead citrate, and were observed with a TEM (H-7650; Hitachi High Technologies America, Inc., Pleasanton, CA, USA).
Whole mount in situ hybridization and Fluorescence in situ hybridization
Whole mount in situ hybridization (WISH) and FISH were performed as previously described [56, 57]. In brief, worms were killed in 5% n-acetyl cysteine (Sigma-Aldrich, St Louis, MO, USA), fixed in 4% paraformaldehyde (PFA), permeabilized using reduction buffer and dehydrated in a graded series of methanol in PBSTx before bleaching. After rehydration, hybridizations were performed with 0.1~0.5 ng per ulriboprobes. For WISH, we use anti-digoxigenin-alkaline phosphatase, 1:4 000 (Roche, Pleasanton, CA, USA). Signal was developed using nitro-blue tetrazolium chloride (NBT)/5-bromo-4-chloro-3'-indolyphosphate p-toluidine salt (BCIP) substrate (1:50; Roche). For FISH or double FISH, we first generated fluorescein isothiocyanate (FITC)-tyramide and rhodamine-tyramide as previously described [58] by using fluorescein mono-n-hydroxysuccinimide-ester (46410; Pierce, Waltham, MA, USA), rhodamine mono-n-hydroxysuccinimide-ester (46406; Pierce) and tyramide (T-2879; Sigma-Aldrich). For double FISH, peroxidase (POD) antibodies (11207733910, 11426346910, Roche) were used in 1:500 and inactivation was performed with 4% PFA for 60 min. Within a given experiment, all samples were developed in the fluorescent substrate for the same length of time and imaged using identical exposure conditions. All sections were performed post WISH or FISH. Frozen sections was performed as previously described [59] with modifications. In brief, FISH-stained animals were transferred to a graded series of sucrose in PBS before embedding in optimum cutting temperature compound (Sakura, Torrance, CA, USA). Specimens were frozen in liquid nitrogen and were sectioned in a cryostat (Microm HM550; Thermo Fisher Scientific, Waltham, MA, USA) at 16 µm at −20 °C. Vibration sections were made to obtain thicker sections. Specimens were embedded with low-melting-point agarose and then the sections were performed with Automated Vibratome (VT1200 S, Leica, Wetzlar, Germany) at 80 µm. The sections were placed on charged slides (Premiere, Shanghai, China) and mounted with Mowiol mounting medium before imaging.
Image acquisition, processing and quantification
Live animals, WISH samples mounted with Mowiol mounting medium, were photographed using a microscope (SteREODiscovery.V20; Carl Zeiss, Jena, Germany) equipped with a Plan Apochromat ×1.0 objective and a digital microscope camera (AxioCamHRc; Carl Zeiss) automated using the AxioVision Rel.4.8 software (Carl Zeiss). FISH specimens were mounted with fluorescence mounting medium (Dako, Glostrup, Denmark) or Mowiol mounting medium, and the images were captured with a laser-scanning confocal microscope (True Confocal Scanner SP5; Leica; HCX Plan Apochromat confocal scanning ×10/0.4 NA, ×20/0.7 NA, ×40/0.85 NA or ×63/1.40 NA oil immersion objective lens) using the LAS AF software (Leica). Images were processed with the LAS AF Lite software and Photoshop software (Adobe, San Jose, CA, USA) and were quantified using the QWin software (Leica) or the ImageJ software (National Institutes of Health, Bethesda, MD, USA). All ISH experiments were performed, imaged and processed identically (at room temperature, 22 °C) to allow direct comparison between experimental animals and controls.
RNA extraction, qPCR and gene expression profiling
qRT-PCR was performed as previously described [60]. In brief, total RNA was isolated using TRIZOL (Invitrogen, Waltham, MA, USA). cDNAs were generated from 300 to 500 ng of total RNA with the FastQuant RT Kit with gDNAse (Tiangen, Beijing, China). Gene-specific primers were designed with Oligo Perfect designer (Invitrogen). qPCRs were performed with the Ace Q qPCR SYBR Green Master Mix Kit (Vazyme, Nanjing, China). At least three biological replicates were performed, and each experiment was performed with triplicate or quadruplicate PCR reactions. Data are expressed using the comparative cycle threshold method. Relative expression levels were normalized to the levels of GAPDH (AY068133) mRNA and plotted with SigmaPlot 11.0 (Systat Software Inc., San Jose, CA, USA) Gene arrays applied the Agilent Custom array described previously [6]. The RNAi worms were fed with dsRNA for four times and RNA samples were collected using TRIZOL (Invitrogen) 7 days post the last feeding. Those genes with a decrease in expression of upto twofolds in both Albino RNAi groups were selected (Table 1). Genes with significant changes upon Albino RNAi were attached in Supplementary Table S1 ‘Differential expressed genes in microarray and gene information’.
Light-bleaching experiments
Worms were placed under direct light for 9 days. An 11-watt fluorescent lamp was placed 15 cm above the 10 cm dishes. The illuminance was 3 000 lux detected using digital light meter (TES Electrical Electronic Corp., Taipei, Taiwan). Each dish contains 10 or less worms and the culture water was replaced every day.
Flow cytometry
Sorting by flow cytometry was performed as previously described [61, 62]. In brief, planarians were diced into small pieces on a cold plate and incubated in 1 mg ml−1 collagenase (diluted in calcium- and magnesium-free medium plus 1% bovine serum albumin) as previously described. Dissociated cells were filtered with a cell strainer (BD, Franklin Lakes, NJ, USA) and stained with 0.2 μg ml−1 calcein acetoxymethylester and 18 μg ml−1 Hoechst 33342 for an appropriate time. After incubating with 5 μg ml−1 propidium iodide, analyses and sorts were performed using the FACSAria II (BD) or MoFlo XDP (Beckman Coulter, Brea, CA, USA). Data were processed using FlowJo V7.6.5 (Tree Star Inc., Ashland, OR, USA).
Statistical analysis
Results are presented as means±s.d., and statistical analyses were performed in SigmaPlot 11.0 using the Student’s t-test for two groups or one-way analysis of variance for three or more groups. P<0.05 was considered significant.
Acknowledgments
We thank P Newmark, P Reddien and N Oviedo for kindly providing worms. We thank XX Qiu and J Chen for critically reading the manuscript, FH Guo for assistance in amputation experiments and all the members of Jing laboratory for comments. We thank A Zeng and staff in the core facility (Institute of Health Sciences) for technical assistance. This work was supported in part by the National Natural Science Foundation of China (81130005, 91339205, 31229002 and 31401238), and the Chinese Academy of Sciences (XDA01040306).
The authors declare no conflict of interest.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Discovery website.)
References
- Reddien PW , Sanchez Alvarado A . Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol 2004; 20: 725–757. [DOI] [PubMed] [Google Scholar]
- Rink JC . Stem cell systems and regeneration in planaria. Dev Genes Evol 2013; 223: 67–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo E , Baguna J . Regeneration and pattern formation in planarians. I. The pattern of mitosis in anterior and posterior regeneration in Dugesia (G) tigrina, and a new proposal for blastema formation. J Embryol Exp Morphol 1984; 83: 63–80. [PubMed] [Google Scholar]
- Guedelhoefer OCT , Sanchez Alvarado A . Amputation induces stem cell mobilization to sites of injury during planarian regeneration. Development 2012; 139: 3510–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D , Lapan SW , Wilkinson AW et al. A molecular wound response program associated with regeneration initiation in planarians. Genes Dev 2012; 26: 988–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng A , Li YQ , Wang C et al. Heterochromatin protein 1 promotes self-renewal and triggers regenerative proliferation in adult stem cells. J Cell Biol 2013; 201: 409–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE , Wang IE , Reddien PW . Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 2011; 332: 811–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML , Kravarik KM , Lapan SW et al. Neoblast specialization in regeneration of the planarian Schmidtea mediterranea. Stem Cell Rep 2014; 3: 339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH , Newmark PA . On the organ trail: insights into organ regeneration in the planarian. Curr Opin Genet Dev 2015; 32C: 37–46. [DOI] [PubMed] [Google Scholar]
- Koinuma S , Umesono Y , Watanabe K et al. The expression of planarian brain factor homologs, DjFoxG and DjFoxD. Gene Express Patterns 2003; 3: 21–27. [DOI] [PubMed] [Google Scholar]
- Scimone ML , Lapan SW , Reddien PW . A forkhead transcription factor is wound-induced at the planarian midline and required for anterior pole regeneration. PLoS Genet 2014; 10: e1003999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogg MC , Owlarn S , Perez Rico YA et al. Stem cell-dependent formation of a functional anterior regeneration pole in planarians requires Zic and Forkhead transcription factors. Dev Biol 2014; 390: 136–148. [DOI] [PubMed] [Google Scholar]
- Blassberg RA , Felix DA , Tejada-Romero B et al. PBX/extradenticle is required to re-establish axial structures and polarity during planarian regeneration. Development 2013; 140: 730–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC , Wang IE , Reddien PW . pbx is required for pole and eye regeneration in planarians. Development 2013; 140: 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowles MW , Brown DD , Nisperos SV et al. Genome-wide analysis of the bHLH gene family in planarians identifies factors required for adult neurogenesis and neuronal regeneration. Development 2013; 140: 4691–4702. [DOI] [PubMed] [Google Scholar]
- Currie KW , Pearson BJ . Transcription factors lhx1/5-1 and pitx are required for the maintenance and regeneration of serotonergic neurons in planarians. Development 2013; 140: 3577–3588. [DOI] [PubMed] [Google Scholar]
- Marz M , Seebeck F , Bartscherer K . A Pitx transcription factor controls the establishment and maintenance of the serotonergic lineage in planarians. Development 2013; 140: 4499–4509. [DOI] [PubMed] [Google Scholar]
- Mannini L , Rossi L , Deri P et al. Djeyes absent (Djeya) controls prototypic planarian eye regeneration by cooperating with the transcription factor Djsix-1. Dev Biol 2004; 269: 346–359. [DOI] [PubMed] [Google Scholar]
- Lapan SW , Reddien PW . dlx and sp6-9 control optic cup regeneration in a prototypic eye. PLoS Genet 2011; 7: e1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapan SW , Reddien PW . Transcriptome analysis of the planarian eye identifies ovo as a specific regulator of eye regeneration. Cell Rep 2012; 2: 294–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler CE , Seidel CW , McKinney SA et al. Selective amputation of the pharynx identifies a FoxA-dependent regeneration program in planaria. eLife 2014; 3: e02238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinuma S , Umesono Y , Watanabe K et al. Planaria FoxA (HNF3) homologue is specifically expressed in the pharynx-forming cells. Gene 2000; 259: 171–176. [DOI] [PubMed] [Google Scholar]
- Wang Y , Zayas RM , Guo T et al. Nanos function is essential for development and regeneration of planarian germ cells. Proc Natl Acad Sci USA 2007; 104: 5901–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y , Stary JM , Wilhelm JE et al. A functional genomic screen in planarians identifies novel regulators of germ cell development. Genes Dev 2010; 24: 2081–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong T , Stary JM , Wang Y et al. Molecular markers to characterize the hermaphroditic reproductive system of the planarian Schmidtea mediterranea. BMC Dev Biol 2011; 11: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsthoefel DJ , James NP , Escobar DJ et al. An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Dev Cell 2012; 23: 691–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML , Srivastava M , Bell GW et al. A regulatory program for excretory system regeneration in planarians. Development 2011; 138: 4387–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA , Barton MC . Many forkheads in the road to regulation. Symposium on forkhead transcription factor networks in development, signalling and disease. EMBO Rep 2008; 9: 721–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda D , Gonzalez J , Callaerts P et al. Searching for the prototypic eye genetic network: Sine oculis is essential for eye regeneration in planarians. Proc Natl Acad Sci USA 2000; 97: 4525–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krugelis-Macrae E . The occurrence of porphyrin in the planarian. Biol Bull 1956; 110: 69–76. [Google Scholar]
- Macrae EK . Porphyrin synthesis in the pianarian. I. Conversion of delta-aminolevulinic acid to porphobilinogen. J Exp Zool 1959; 142: 667–674. [DOI] [PubMed] [Google Scholar]
- Macrae EK . Porphyrin synthesis in the planarian. Ii. Incorporation of delta-aminolevulinic acid-4-C14 into uroporphyrin. J Exp Zool 1963; 153: 255–262. [DOI] [PubMed] [Google Scholar]
- Macrae EK . Localization of porphyrin fluorescence in planarians. Science 1961; 134: 331–332. [DOI] [PubMed] [Google Scholar]
- Hase S , Wakamatsu K , Fujimoto K et al. Characterization of the pigment produced by the planarian, Dugesia ryukyuensis. Pigment Cell Res 2006; 19: 248–249. [DOI] [PubMed] [Google Scholar]
- Brubacher JL , Vieira AP , Newmark PA . Preparation of the planarian Schmidtea mediterranea for high-resolution histology and transmission electron microscopy. Nat Protoc 2014; 9: 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladini G , Medolago-Albani L , Margotta V et al. The pigmentary system of planaria. I. Morphology. Cell Tissue Res 1979; 199: 197–202. [DOI] [PubMed] [Google Scholar]
- Palladini G , Medolago-Albani L , Margotta V et al. The pigmentary system of planaria. II. Physiology and functional morphology. Cell Tissue Res 1979; 199: 203–211. [DOI] [PubMed] [Google Scholar]
- Battersby AR , Fookes CJ , Matcham GW et al. Biosynthesis of the pigments of life: formation of the macrocycle. Nature 1980; 285: 17–21. [DOI] [PubMed] [Google Scholar]
- Mochizuki N , Tanaka R , Grimm B et al. The cell biology of tetrapyrroles: a life and death struggle. Trends Plant Sci 2010; 15: 488–498. [DOI] [PubMed] [Google Scholar]
- Vavilin DV , Vermaas WF . Regulation of the tetrapyrrole biosynthetic pathway leading to heme and chlorophyll in plants and cyanobacteria. Physiol Plant 2002; 115: 9–24. [DOI] [PubMed] [Google Scholar]
- Breinig S , Kervinen J , Stith L et al. Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase. Nat Struct Biol 2003; 10: 757–763. [DOI] [PubMed] [Google Scholar]
- Kauppinen R . Porphyrias. Lancet 2005; 365: 241–252. [DOI] [PubMed] [Google Scholar]
- Yin L , Bauer CE . Controlling the delicate balance of tetrapyrrole biosynthesis. Philos Trans R Soc Lond B Biol Sci 2013; 368: 20120262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW , Oviedo NJ , Jennings JR et al. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science 2005; 310: 1327–1330. [DOI] [PubMed] [Google Scholar]
- Smith AT , Pazicni S , Marvin KA et al. Functional divergence of heme-thiolate proteins: a classification based on spectroscopic attributes. Chem Rev 2015; 115: 2532–2558. [DOI] [PubMed] [Google Scholar]
- Stubenhaus BM , Dustin JP , Neverett ER et al. Light-induced depigmentation in planarians models the pathophysiology of acute porphyrias. eLife 2016; 5: e14175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenemoser D , Reddien PW . Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol 2010; 344: 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaidou E , Katsambas AD . Pigmentation disorders: hyperpigmentation and hypopigmentation. Clin Dermatol 2014; 32: 66–72. [DOI] [PubMed] [Google Scholar]
- Reddien PW . Specialized progenitors and regeneration. Development 2013; 140: 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendreau I , Angers L , Jean J , Pouliot R . In: Andrades JA (ed.), Pigmented Skin Models: Understand the Mechanisms of Melanocytes, Regenerative Medicine and Tissue Engineering. InTech: Croatia, 2013.
- Hoekstra HE . Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity 2006; 97: 222–234. [DOI] [PubMed] [Google Scholar]
- Newmark PA , Sanchez Alvarado A . Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Dev Biol 2000; 220: 142–153. [DOI] [PubMed] [Google Scholar]
- Robb SM , Gotting K , Ross E et al. SmedGD 2.0: the Schmidtea mediterranea genome database. Genesis 2015; 53: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb SM , Ross E , Sanchez Alvarado A . SmedGD: the Schmidtea mediterranea genome database. Nucleic Acids Res 2008; 36: D599–D606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayas RM , Hernandez A , Habermann B et al. The planarian Schmidtea mediterranea as a model for epigenetic germ cell specification: analysis of ESTs from the hermaphroditic strain. Proc Natl Acad Sci USA 2005; 102: 18491–18496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson BJ , Eisenhoffer GT , Gurley KA et al. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn 2009; 238: 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RS , Newmark PA . In situ hybridization protocol for enhanced detection of gene expression in the planarian Schmidtea mediterranea. BMC Dev Biol 2013; 13: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopman AH , Ramaekers FC , Speel EJ . Rapid synthesis of biotin-, digoxigenin-, trinitrophenyl-, and fluorochrome-labeled tyramides and their application for in situ hybridization using CARD amplification. J Histochem Cytochem 1998; 46: 771–777. [DOI] [PubMed] [Google Scholar]
- Tu KC , Pearson BJ , Sanchez Alvarado A . TORC1 is required to balance cell proliferation and cell death in planarians. Dev Biol 2012; 365: 458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YQ , Zeng A , Han XS et al. Argonaute-2 regulates the proliferation of adult stem cells in planarian. Cell Res 2011; 21: 1750–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T , Asami M , Higuchi S et al. Isolation of planarian X-ray-sensitive stem cells by fluorescence-activated cell sorting. Dev Growth Differ 2006; 48: 371–380. [DOI] [PubMed] [Google Scholar]
- Peiris TH , Weckerle F , Ozamoto E et al. TOR signaling regulates planarian stem cells and controls localized and organismal growth. J Cell Sci 2012; 125: 1657–1665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.