Abstract
Background
The opportunistic bacterial pathogen Legionella pneumophila uses substrate effectors of Dot/Icm type IVB secretion system (T4BSS) to accomplish survival and replication in amoebae cells and mammalian alveolar macrophages. During the conversion between its highly resistant, infectious dormant form and vigorously growing, uninfectious replicative form, L. pneumophila utilizes a complicated regulatory network in which proteolysis may play a significant role. As a highly conserved core protease, ClpP is involved in various cellular processes as well as virulence in bacteria, and has been proved to be required for the expression of transmission traits and cell division of L. pneumophila.
Results
The clpP-deficient L. pneumophila strain failed to replicate and was digested in the first 3 h post-infection in mammalian cells J774A.1. Further investigation demonstrates that the clpP deficient mutant strain was unable to escape the endosome-lysosomal pathway in host cells. We also found that the clpP deficient mutant strain still expresses T4BSS components, induces contact-dependent cytotoxicity and translocate effector proteins RalF and LegK2, indicating that its T4BSS was overall functional. Interestingly, we further found that the translocation of several effector proteins is significantly reduced without ClpP.
Conclusions
The data indicate that ClpP plays an important role in regulating the virulence and effector translocation of Legionella pneumophila.
Electronic supplementary material
The online version of this article (doi:10.1186/s12866-016-0790-8) contains supplementary material, which is available to authorized users.
Keywords: Legionella pneumophila, ClpP, Virulence, T4BSS, Effectors, Substrate, Translocation
Background
First isolated in 1977, Legionella pneumophila, a Gram-negative, intracellular bacterial pathogen is the agent causing the severe form of pneumonia named Legionnaires’ disease, as well as the less severe flu-like Pontiac fever [1]. It has drawn much attention for its capability of intracellular replication in both protozoa and human beings. After the endocytosis by protozoan hosts like amoebae or human alveolar macrophages, the Legionella-containing vacuole (LCV) inhibits phagolysosomal fusion and recruits mitochondria followed by the association of ribosome-studded membranes that later disguise LCV as endoplasmic reticulum (ER). Within this ER-like compartment, the bacterium replicates to high numbers and eventually is released through lysing the host cell for the next invasion [2].
During this process, L. pneumophila requires most protein products of 27 dot/icm (defect in organelle trafficking/intracellular multiplication) genes to constitute a type IVB secretion system (T4BSS) [2]. Although neither the composition nor the function of T4BSS has been fully understood in L. pneumophila, progress has been achieved in identifying and characterizing the Dot/Icm proteins. DotC, DotD, DotF, DotG and DotH comprise the core of the secretion complex which spans across the bacterial membrane. DotC and DotD are outer-membrane lipoproteins and required for DotH to target the outer membrane [3]. DotH may be the out-membrane channel through which substrates get delivered following the transit from the DotF-DotG inner-membrane proteins with the assistance of the DotL–DotM ATPase [2]. DotB, also an ATPase, interacts with DotL and may play a role in various functions such as the assembly of T4BSS, retraction of pili and/or export of substrates [4, 5]. IcmQ participates in the membrane pore formation [6], and IcmT is crucial for pore formation-mediated escape of L. pneumophila from protozoan or mammalian cells [7]. DotL is proposed to be a type IV coupling protein (T4CP) of T4BSS and interacts with other inner-membrane proteins including DotN, DotM and IcmS/W, a heterodimer complex functions as T4BSS adaptor, to constitute the T4CP subcomplex, a very important complex for T4BSS to facilitate substrate secretion [8, 9].
Through the T4BSS, L. pneumophila secretes a large number of substrate proteins called effectors that interfere with the host pathways to help bacteria evade the endosome-lysosomal pathway and replicate in host cells [2]. The effector RalF, which has guanine nucleotide exchange activity and mediates the exchange of GDP for GTP, disturbs vesicle traffic between the ER and Golgi and further promotes the biogenesis of LCV by modulating the activity and localization of the key intracellular regulator Arf1 [10]. The effector AnkB is important for the moorage of K48-linked polyubiquitinated proteins when it is anchored into the phagosome membrane by host-mediated farnesylation and interacts with the SCF1 E3 ubiquitin ligase complex. Then the K48-linked proteins are degraded and the amino acids are utilized for bacterial intracellular proliferation [11, 12]. LegK2, whose deletion causes reduced cytotoxicity, and adversely affect the intracellular survival and replication of L. pneumophila, acts as a protein kinase [13]. So far more than 300 effectors have been identified but many of them are considered functionally redundant, only a few are indispensable for the intracellular proliferation of L. pneumophila, such as MavN and SdhA [14–16].
During the shift between extracellular and intracellular environments, L. pneumophila encounters different growth conditions and has to respond accordingly to survive. To make the appropriate responses, L. pneumophila has developed a complex network to modulate the transition at different phases. Proteolysis has been regarded as an important and precise regulatory mechanism for both eukaryote and prokaryote to adapt to a variety of growth conditions by removing short-lived regulatory proteins, as well as misfolded and damaged proteins [17]. It is now clear that cellular proteolysis is carried out by the energy-dependent proteases such as the Lon and Clp proteases and the eukaryotic 26S proteasome [17]. To date, Clp protease is the most characterized protease in prokaryotes. It consists of two functional subunits: a cylinder-like proteolytic core named ClpP which is widely distributed and highly conserved, and two chaperone rings with ATPase activity such as ClpA, ClpC, ClpE or ClpX [17, 18]. The protease core consists of 14 ClpP serine peptidase subunits stacked in two heptameric rings, forming an internal chamber in which the active sites are sequestered from the cytoplasm [19]. The Clp ATPases are responsible for the recognition, unfolding and translocation of substrates into the degradation chamber [20].
It is widely accepted that Clp proteases are involved in many physiological processes of bacteria. In a range of low GC Gram-positive bacteria including Bacillus subtilis, Listeria monocytogenes and Lactococcus lactis, ClpP-deficient mutants suffer restricted growth at high temperatures [21–24]. ClpP is also considered as the major determinant for the turnover of bulk proteins in B. subtilis at the transition from exponential stage to competence and further sporulation stages [24, 25]. Moreover, both ClpP and its ATPase chaperones play significant roles in virulence expression and regulation in various bacterial pathogens. For instance, S. aureus cells lacking the ClpB chaperone are unable to replicate intracellularly in bovine cells [26]. The absence of ClpP in L. monocytogenes results in the lack of listeriolysin O, a major virulence factor implicated in phagosome lysis [23, 27]. Our recent research has shown that ClpP is required for the transmission traits of Legionella pneumophila during the transition in its biphasic life cycle, including some traits associated with virulence such as cytotoxicity and intracellular proliferation in the amoebae host Acanthamoeba castellanii [28]. In this report, studies were focused on the function of Legionella pneumophila ClpP in the mammalian cell J774A.1 and results revealed that the deletion of clpP severely impaired the bacterial virulence and the translocation of several T4BSS effectors though the functional integrity of T4BSS was not fully neutralize.
Results
ClpP is essential for intracellular proliferation of L. pneumophila in macrophages
We have shown previously that ClpP is essential for intracellular multiplication of L. pneumophila in amoebae A. castellanii [28]. To investigate whether ClpP also affects intracellular proliferation of L. pneumophila in macrophages, the wild type, the clpP-deficient mutant, the constitutive complementation, and the Dot/Icm-deficient dotA mutant strains were grown to the stationary phase and used to infect Mus musculus macrophages. The infection was allowed to proceed for 5 days and the intracellular proliferation was evaluated by plating the cell lysate onto BCYE plates per 24 h. As shown in Fig. 1, the wild type Lp02pj and the complemented strain Xp02c exhibited essentially identical proliferation rate in J774A.1 cells. In contrast, both the clpP-deficient mutant Xp02pj and the dotA mutant strain Lp03pj showed significantly impaired multiplication. These results indicate that ClpP is essential for the intracellular growth of L. pneumophila in macrophage.
Fig. 1.
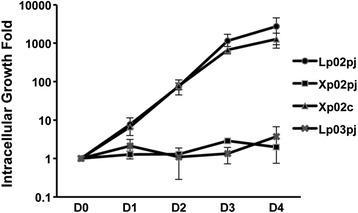
Intracellular growth of clpP-deficient L. pneumophila strain Xp02 in J774A.1 was impaired. J774A.1 cells were seeded in 24-well plates and infected with L. pneumophila at an MOI of 1. At each time point, cells were lysed and the CFU was determined by plating dilutions onto BCYE plates. The intracellular growth kinetics of Lp02 (●), clpP mutant (■), clpP complemented strain (▲), and dotA mutant Lp03 (X) were shown. Points indicate mean values and error bars indicate standard deviations of three independent experiments
clpP mutant is degraded rapidly upon entry into the host cells
Avirulent L. pneumophila strains decrease rapidly in the first hours of phagocytosis [29]. Because the clpP-deficient strain showed severely reduced cytotoxicity and intracellular replication (Fig. 1 and [28]), it is of interest to explore whether ClpP is essential for preventing L. pneumophila from degradation after uptake into host cells. To this end, J774A.1 were exposed to the stationary-phase L. pneumophila strains and co-cultures were maintained for 3 h before the cells were lysed. The significantly lower percentage (p < 0.01) of bacteria residing within host cells 3 h post infection (Fig. 2) demonstrated the impaired survival capability of clpP-deficient mutant after phagocytosis. As a negative control, the dotA-deficient mutant Lp03 similarly exhibited impaired survival capability compared with the wild type strain Lp02 (Fig. 2). These data indicate the rapid degradation of the clpP-deficient bacteria post infection and suggest that clpP mutant cannot escape the host defense systems.
Fig. 2.
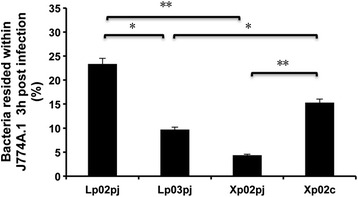
The L. pneumophila clpP mutant Xp02 was degraded within 3 h of phagocytosis. J774A.1 cells were seeded in 24-well plates and infected with L. pneumophila at an MOI of 1. After 3 h, the survival of bacteria was determined by plating dilutions onto BCYE plates and calculating the numbers of CFU of L. pneumophila lysed from infected J774A.1. *, p < 0.05; **, p < 0.01. The phagocytosis assay was carried out in triplicate. Shown are the averages and standard deviations of three independent counts
The clpP mutant fails to evade the late endosome-lysosomal pathway
To assess whether the clpP-deficient mutant could survive the endosome- lysosomal pathway, the molecular morphology of phagosomes containing L. pneumophila was investigated in J774A.1. Lysosomal-associated membrane protein 1 (LAMP-1) and the Texas-red conjugated ovalbumin (TroV) were utilized as markers of the late endosome and lysosome, respectively. The wild type strain Lp02pj that was grown in broth until it reached exponential phase was used as the negative control (labeled as EpLp02pj in Fig. 3c and d) for the lysosome fusion assay because the phagosomes containing Dot/Icm-deficient dotA mutant strain do not acquire lysosomal TroV in the first several hours post-infection [30]. Predictably, the wild type strain Lp02pj that can replicate in host cells was excluded from LAMP-1 stained compartments, while the clpP-deficient mutant Xp02pj was frequently colocalized with LAMP-1 staining, just like the negative control Lp03pj (Fig. 3a). Staining results showed that 31 % of the Lp02pj-containing phagosomes exhibited detectable LAMP-1 accumulation, whereas 65 % of the Xp02pj-containing phagosomes were LAMP-1 positive. Although higher than that of Lp02pj-containing phagosomes (p < 0.01), the percentage of LAMP-1 positive Xp02pj-containing phagosomes was still significantly lower than that of Lp03pj-containing phagosomes (p < 0.01) (Fig. 3b). Similar results were obtained in the lysosome fusion assay in which the percentages of TroV positive phagosomes containing Lp02pj, Xp02pj or EpLp02pj were about 27, 63 or 95 %, respectively (Fig. 3c and d). These means were significantly different from each other (p < 0.01). Based on these findings, we conclude that ClpP is important for L. pneumophila to escape the late endosome-lysosomal pathway of mammalian host cells.
Fig. 3.
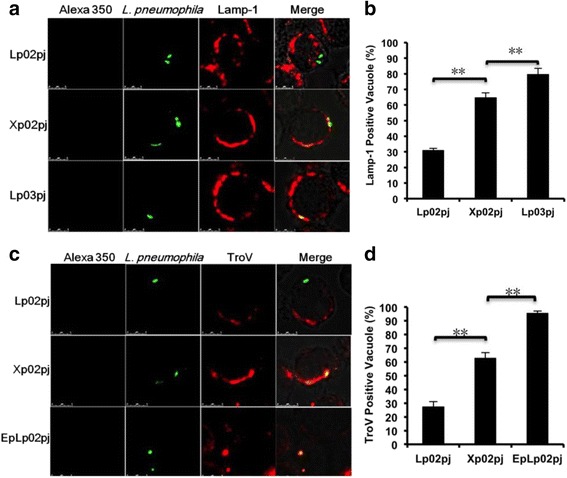
Immunofluorescence analysis of the late endosome and lysosome in macrophages infected with L. pneumophila. J774A.1 cells were incubated with the wild-type L. pneumophila or mutant strains for 2 h, then fixed and stained with a monoclonal antibody specific to LAMP-1 or TroV to identify the macrophage late endosomes or lysosomes. a Phagosomes containing wild-type strain Lp02pj were not co-localized with late endosomes, whereas phagosomes containing Xp02pj or Lp03pj were stained and colocalized with LAMP-1. b Meanwhile the percentages of L. pneumophila-containing phagosomes fused with the late endosomes were determined. **, p < 0.01. c The fusion of phagosomes with lysosomes was also examined using confocal microscopy and d quantified as above. **, p < 0.01. The immunofluorescence assay was carried out in triplicate. Shown are the averages and standard deviations of three independent counts. The number of J774A.1 cells for each count is about 100
Loss of ClpP does not affect the expression of dot/icm components
To survive and multiply within phagocytic host cells, L. pneumophila manipulates host cellular processes, alters the host endocytic pathway, thereby inhibits rapid phagosome-lysosome fusion and creates a niche for its replication. These are achieved with the help of Dot/Icm T4BSS and effectors [2, 14]. In view of this, we presumed it might be the abnormal function of T4BSS or interrupted secretion of effectors that caused the impaired survival and intracellular proliferation of the clpP-deficient strain in host cells. To test this hypothesis, we measured the transcription activity of dot/icm genes in 9 operon areas [31] by examining the promoter activities of these genes. Promoters were fused with gfp reporter gene and transformed into both Xp02 and Lp02. The fluorescence intensities, as well as the expression of DotH (IcmK), DotI (IcmL) and DotG (IcmE), which have been proved to be components of the Dot/Icm system [3], were measured. Plate patching results showed that there were no significant differences in fluorescence intensities between Lp02 and Xp02 harboring the icm-gfp fusion plasmids (Fig. 4a-c). The fluorescence in the liquid culture confirmed the results of patching and only a slight reduction of fluorescence intensity (p > 0.05) was found in the icmR-gfp-harboring Xp02pG2, compared to that in Lp02pG2 (Fig. 4d). Furthermore, Western blot analysis indicated that there were no differences between the expression levels of DotH, DotI, DotG, IcmS and IcmW in Lp02 and Xp02 (Fig. 4e). Taken together, these results indicate that loss of ClpP might not significantly influence the expression of the T4BSS components of L. pneumophila.
Fig. 4.

Analysis of the promoter activitiy and the expression of the dot/icm components. a-c Promoter activities of 9 dot/icm operons were examined through streaking and comparing the bacterial strains harboring the promoter-gfp fusion plasmids on plates. d Quantified fluorescence intensities of the wild-type Lp02 and the clpP mutant Xp02 harboring icmR:gfp fusion plasmid. There is no statistical difference between the two samples. e Western blot analysis of the expression of DotH, DotI, DotG, IcmS and IcmW in the wild type and the clpP deficient mutant strain. A western blot using antibody to isocitrate dehydrogenase (ICDH) was used to confirm even protein levels of the samples. Shown are the averages and standard deviations of 2 to 3 independent experiments, each performed in triplicate
The T4BSS secretion apparatus is still functional in the L. pneumophila clpP deficient mutant
The Dot/Icm T4BSS system of L. pneumophila consists of 27 proteins [2]. Because not all antibodies to these components are available, we cannot determine whether the T4BSS of the clpP deficient mutant still functions normally through Western blot analysis. To investigate the integrity of T4BSS complex in the clpP mutant, we utilized contact-dependent cytotoxicity assay. It is well comprehended that L. pneumophila triggers contact-dependent cytotoxicity, which is actually caspase-1-mediated or caspase-3-mediated cell death, through delivering flagellin protein into host cells and this process requires a functional Dot/Icm T4BSS complex [32, 33]. L. pneumophila strains harboring pJB908 were grown to post-exponential phase in broth and co-cultured with J774A.1 at an MOI of 50 and 100. After incubation for 2 h, the permeabilization of cells was detected by the release of the intracellular enzyme lactate dehydrogenase (LDH). At an MOI of 50, both the wild type and the complemented strains showed high contact-dependent cytotoxicity and the LDH release was 50 and 48 %, respectively. In contrast, the LDH release of the dotA-deficient strain Lp03 was only 9 %. Intriguingly, the LDH release of the clpP mutant was 34 % and remained cytotoxic, although it was lower than that of the wild type and the complemented strains (p > 0.05) (Fig. 5). Similar results were observed when the MOI was raised to 100 (Fig. 5). These findings imply that the T4BSS complex in the clpP mutant may be still functional and overall integrated.
Fig. 5.
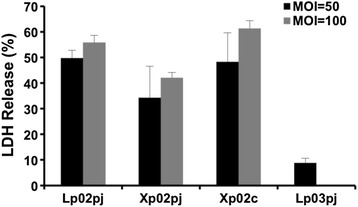
The clpP-deficient L. pneumophila still has a functional T4BSS. Contact-dependent cytotoxicity of the wild-type strain Lp02, the clpP mutant Xp02, the complemented strain Xp02c and the dotA mutant Lp03. Infection was performed at an MOI of 50 and 100, and cell permeabilization was detected by the release of the intracellular enzyme lactate dehydrogenase (LDH). Shown are the averages and standard deviations of three independent experiments, each performed in triplicate (duplicate for cytotoxicity assays). And there is no statistical difference between the strains
ClpP controls the translocation efficiency of some T4BSS effectors
Since the T4BSS apparatus is still functional in the clpP mutant, we therefore examined if the translocation frequency of effectors is impaired. A well-established reporter gene for translocation assay, cyaA [34], was fused with seven effector coding genes, respectively. The resulting plasmids were transformed into the wild type Lp02, the clpP deficient strain Xp02 and the dotA mutant strain Lp03, respectively. The recombined strains were then infected J774A.1 cells and the fused genes were expressed under IPTG-induction for translocation frequency assay. As shown in Fig. 6, the cAMP levels of RalF and LegK2 showed no difference between Lp02 and Xp02, respectively, whereas the cAMP levels of SidA, SidB, SidD, SidF and LegU1 in Xp02 were significantly lower than that in Lp02, which decreased to 3.27, 0.74, 7.64, 0.25 and 6.06 % of that in Lp02, respectively. Immunoblotting assay showed that all Cya fusion proteins were expressed abundantly (Additional file 1: Figure S1). These results indicate that the translocation of some T4BSS effectors is partly controlled by ClpP.
Fig. 6.
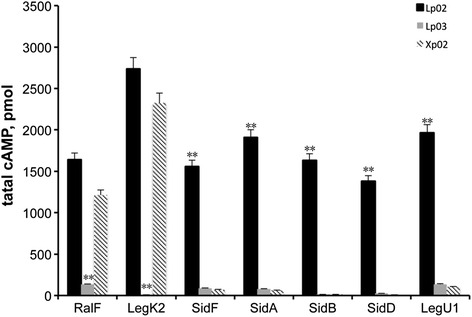
clpP-deficiency impaired the translocation of some effectors. J774A.1 cells were infected with the wild type (Lp02), the dotA mutant (Lp03) and the clpP mutant (Xp02) L. pneumophila strains expressing the indicated Cya hybrid proteins. After 1 h of infection, tissue culture cells were lysed and cAMP was extracted from the sample. Total cAMP production induced by translocation of the hybrid was quantified using an enzyme-immunoassay system, indicated as pmol. Results represent average values of experiments performed in triplicate. **, p < 0.01
Discussion
In the current study, we found that the L. pneumophila clpP deficient mutant exhibited poor survival and intracellular multiplication in J774A.1 cells (Figs. 1 and 2). Furthermore, the mutant strain could not escape the late endosome-lysosomal pathway (Fig. 3). Thus, consistent with our previous results obtained in the amoebae host A. castellanii [28], ClpP may be required for the expression of virulence in L. pneumophila. To investigate how ClpP regulates the virulence, we tested whether the deficiency of clpP could affect the component expression and the function of T4BSS complex. The 27 proteins of dot/icm components may not be significantly affected based on the results of transcriptional activity assay together with immunoblotting analysis (Fig. 4). Although we did not examine the expression of all 27 proteins, the findings that the clpP mutant could still induce contact-dependent cytotoxicity against host cells (Fig. 5), together with that both RalF and LegK2 could be translocated into host cells, indicated that the T4BSS function was not compromised seriously in the absence of ClpP. However, the fact that the secretion of some effectors was impaired in the clpP-deficient strain suggested that one of the strategies for ClpP to affect the virulence of Legionella pneumophila is via regulating the translocation of effectors.
The phenotype of clpP-deficient L. pneumophila resembles that of the IcmS/W mutants, the T4BSS chaperone. They all exhibit significantly impaired intracellular replication, but still maintain fair contact-dependent cytotoxicity against host cells (Figs. 1 and 5) [35, 36]. Moreover, the translocation of three IcmS/W-mediated effectors (SidA, SidB and SidD) and LegU1 was impaired obviously in the absence of ClpP, and the translocation of non-IcmS/W-mediated RalF was unchanged without ClpP (Fig. 6) [37]. More interestingly, the non-IcmS/W-mediated effector SidF showed reduced translocation efficiency in clpP-deletion mutant (Fig. 6), which was similar in icmS/icmW double mutant, and the translocation of SidA, SidB and SidD in clpP-deletion mutant was more neutralized than that in icmS or icmW single mutant [37]. Taken together, a hypothesis that clpP-deletion mutant might resemble an absolutely IcmSW-abolishing mutant could be proposed. Currently, how ClpP affects IcmS/W subcomplex is difficult to be clarified because the expression level of each protein in clpP-deletion mutant is unchanged (Fig. 4e). Previous studies have shown that ClpP affects the virulence expression in some gram-positive pathogens such as Staphylococcus aureus, Streptococcus pneumoniae and L. monocytogenes [23, 26, 38, 39]. More details about the virulence regulation by ClpP have been revealed in Salmonella enterica serovar Typhimurium and Yersinia pestis, where ClpP governs the protein levels of important virulence-regulating factors for type III secretory systems (T3SS), RpoS and YomA [40, 41]. For pathogens containing T4BSS, little information about the association between Clp proteases and other virulence-related factors is available. Recently studies on the DotL-IcmSW coupling subcomplex of L. pneumophila T4BSS have revealed that ClpAP protease is responsible for the degradation of DotL in the absence of IcmS. Briefly, in the absence of IcmS/W mediation, abundant effector proteins could be trapped within DotL, and subsequently the jamming complex is subjected to specific degradation by ClpAP [8, 42]. Interestingly, although DotL degradation by ClpP requires specific recognition by ClpA, the single deletion of clpA does not affect the intracellular replication or the protein level of DotL [8]. Thus, it is possible that ClpP may affect DotL-IcmSW-mediated effector translocation through other processes bypassing ClpA. On the one hand, ClpP may influence the optimal assembly of T4BSS rather than protein expression, especially in DotL-IcmSW coupling subcomplex. On the other hand, successful effector translocation mediated by IcmS/W might need essential cleavage or modification by ClpP protease. In the second situation, translocation signal peptides may be involved. Up to now, a C-terminal signal peptide has been proven to be essential for the translocation of nearly all T4BSS effectors and an internal signal sequence has been found to be important in IcmS/W-mediated translocation [43, 44]. Thus, the latter signal peptide may be the possible target of ClpP modification or regulation. In our future proposal, the association of ClpP with the internal signal peptide in IcmS/W-mediated effectors needs to be explored, and we also hope to examine the assembly status of the DotL-IcmSW subcomplex in clpP-deletion mutant.
It is also possible that the neutralized intracellular replication is partially due to the impaired stress tolerance of clpP-deficient L. pneumophila. In both natural aquatic and intracellular environment, L. pneumophilla would encounter various stresses [45, 46]. Legitimately, a rapid responding system involving proteolytic procedures would be beneficial to the stress tolerance of L. pneumophilla. Studies have revealed that ClpP protease plays significant roles in DNA repair and other stress responses and helps bacteria adapt to many harsh conditions [38, 47–49]. Likewise, L. pneumophila ClpP is required for the stationary-phase resistance against various stresses such as oxidation, acidity, osmotic stress and inappropriate temperatures [28]. Among these stresses, acid resistance might be the most critical in Legionella’s intravacuolar survival. L. pneumophila could neutralize the acidic environment of phagosome and subsequently maintain a nearly neutral-pH vacuole during the first hours of uptake [50]. However, about 18 h later L. pneumophila would still encounter the acidic environment when the LCV mature into low-pH and endocytic compartments [51]. Considering our previous finding that the clpP mutant strain exhibited reduced acid resistance and vulnerable cell surface compared to the wild type strain [28], we wonder whether and how much the “weakness” the clpP mutant displays under unfavorable environments contributes to the disability of virulence expression, this assumption needs deeper exploration.
Conclusion
In this study, our data show that the loss of clpP prevents L. pneumophila from evading the endocytic pathway and replicating in host cells. Our results also revealed that the clpP-deficiency affects the translocation of some T4BSS effectors without impairing the integrity of T4BSS. Taken together, L. pneumophila ClpP is an indispensable factor for the virulence and the translocation of some T4BSS effectors.
Methods
Bacterial strains, cells and reagents
The bacterial strains, plasmids and primers used in this work are listed in Tables 1, 2 and 3, respectively. L. pneumophila strains were cultured on buffered charcoal yeast extract (BCYE) plates, or in N-(2-acetamido)-2-aminoethanesul-fonic acid (ACES)-buffered yeast extract (AYE) medium [52], supplemented with 5 μg/ml chloramphenicol when necessary. Escherichia coli strains were cultured on Luria-Bertani (LB) agar plates, or in LB broth, supplemented with 30 μg/ml chloramphenicol or 100 μg/ ml ampicillin when necessary. A. castellanii (ATCC 30234) was grown in proteose yeast extract glucose medium (PYG) at 30 °C [53]. J774A.1 (ATCC TIB-67) cells were maintained in Roswell Park Memorial Institute 1640 (RPMI1640) medium supplemented with 10 % fetal bovine serum (FBS) (Gibco), at 37 °C and in 5 % (v/v) CO2. Bacto yeast extract and proteose peptone were obtained from Becton Dickinson Biosciences. All other reagents were purchased from Sigma Co., unless specified otherwise.
Table 1.
Bacterial strains used in this study
| Strain | Characteristics | Reference or source |
|---|---|---|
| E.coli DH5α | host strain used for cloning | Lab collection |
| Lp02 | Virulent L. pneumophila serogroup 1, strain Philadelphia, rpsL, HsdR − , Thy − | Lab collection |
| Lp03 | Virulent L. pneumophila serogroup 1, strain Philadelphia, dot03, rpsL, HsdR − , Thy − | [58] |
| Xp02 | Lp02 with clpP deletion | This study |
| Xp02c | Xp02 containing pXL901 for complementation | This study |
| Lp02pj | Lp02 containing pJB908 | This study |
| Lp03pj | Lp03 containing pJB908 | This study |
| Xp02pj | Xp02 containing pJB908 | This study |
| Lp02pGX* | Lp02 containing plasmids pGB908X*
with icm promoter-gfp fusions |
This study |
| Xp02pGX* | Xp02 containing plasmids pGB908X*
with icm promoter-gfp fusions |
This study |
| Lp03pG9 | Lp03 containing plasmid pGB9089 with icmV promoter-gfp fusion |
This study |
The X* represents the numbers of the plasmids or bacterial strains containing the icm promoter-gfp fusions: 1. icmTS, 2. icmR, 3. icmQ, 4. icmPO, 5. icmMLKEGCD, 6. icmJB, 7. icmHF, 8. icmWX, 9. icmV
Table 2.
Plasmids used in this study
| Plasmids | Characteristics | Reference |
|---|---|---|
| pBRDX | Suicide delivery vector, rdxA sacB Cm | [54] |
| pBRΔclpP | pBRDX::clpP for clpP deletion | This study |
| pJB908 | Insert thy gene, mutate mob into pkB5 | [4] |
| pBC(gfp)Pmip | ColE1 ori Cm Pmip gfpmut2 | [59] |
| pZL01 | Insert SacI/PstI fragment encoding GFP, Pmip into pJB908 | This study |
| pXL901 | Complementation plasmid, derived from pZL01, with gfpmut2 changed for clpP | This study |
| pTLpflaG | Insert BamHI/SphI fragment encoding GFP, flaA promoter into pTLP6 | [57] |
| pGB908 | Insert XbaI/SphI fragment of gfpmut3 into pJB908 | This study |
| pGB908X* | Insert 9 icm promoters of operon areas into pGB908 | This study |
| pCyaSidJ | Insert EcoRI/SalI fragment of CyaA catalytic domain and SidJ into pKB5 | [34, 50] |
The X* represents the numbers of the plasmids or bacterial strains containing the icm promoter-gfp fusions: 1. icmTS, 2. icmR, 3. icmQ, 4. icmPO, 5. icmMLKEGCD, 6. icmJB, 7. icmHF, 8. icmWX, 9. icmV
Table 3.
Primers used in this study
| Primer | 5’ → 3’ sequence | Source |
|---|---|---|
| XP-F1 | TGGTCCGGATCCCTGCCAGTAGGTCCTATAAG | This study |
| XP-P1 | TATGACATACAAGTTGCTGGACATTCTAC | This study |
| XP-F2 | CAACTTGTATGTCATAGGAACGCTCACC | This study |
| XP-P2 | TGGTCCAGATCTTGGGAAAATTGACAAACCGT | This study |
| XL091F | TGGTGGTCTAGATTTAAGAAGGAGATATACATATGCCAGGCTATTCAGATAAT | This study |
| XL091R | TGGTGGGCATGCTTATAAGTCTGTAGAATGTCCAGC | This study |
| Rac-F | CGGGATCCATGCATCCAGAAATTGAAAAGGC | This study |
| Rac-R | ACGCGTCGACTTATTTCTTATAACTCGATCTACTTTC | This study |
| LK2c-F | CGGGATCCATGGTTTATTACATAAATTTGAAGGAACAA | This study |
| LK2c-R | ACGCGTCGACTTAGCTTGGGCCTCGCATCA | This study |
| SDAc-F | CGGGATCCTTGGTATATTATGAGATCATTAAGGATAT | This study |
| SDAc-R | ACGCGTCGACTTAAATAGTAAGACTCGAGTTAGTTG | This study |
| SDBc-F | CGGGATCCATGGCTAAAATTTATAATGCACCAAAAC | This study |
| SDBc-R | ACGCGTCGACCTAATTTATTTCTGGTATACTTTTTACG | This study |
| SDDc-F | CGGGATCCTTGGTATATTATGAGATCATTAAGGATAT | This study |
| SDDc-R | ACGCGTCGACTTAAATAGTAAGACTCGAGTTAGTTG | This study |
| SDFc-F | CGGGATCCATGCCACGAATCACTGAAAATAT | This study |
| SDFc-R | ACGCGTCGACTTAGAAGTTTACTGGCGTGG | This study |
| LU1c-F | CGGGATCCATGAAAGCAAAATACGAC | This study |
| LU1c-R | ACGCGTCGACCTACAATGGCTCACATTGGC | This study |
DNA manipulation, chromosomal in-frame deletion, complementation assay
All DNA manipulations were performed according to standard protocols and the in-frame deletion was carried out as described before [28] except that the suicide vector was substituted for pBRDX and the L. pneumophila strain for Lp02. Briefly, the upstream and downstream flanking sequences of clpP were amplified by PCR using PXP-F1/PXP-R1 and PXP-F2/PXP-R2 primer pairs, respectively. The PCR products were mixed as templates for the subsequent fusion PCR using PXC-F1/PXC-R2 as the primer pair. The Fusion PCR product was digested with BamHI and BglII, and sub-cloned into the pBRDX vector [54], yielding pBRΔclpP. Then, pBRΔclpP was introduced into the wild-type Lp02 strain by electroporation and chloramphenicol-resistant colonies were selected on BCYE-Cm plates. Transformants were patched and inoculated into AYE broth and then spread on BCYE plates containing 20 μg/ ml metronidazole. Bacteria were cultured for about 3 days at 37 °C to screen for strains without the suicide vector. Positive colonies were verified by PCR and sequencing.
In the complementation assay, a RSF1010 pKB5-derived vector pJB908 was utilized as the cloning backbone [4]. The ColE1-type plasmid pBC(gfp)Pmip, which had been used as a complementation vector previously [28], was digested with SacI and PstI. The fragment was ligated with pJB908, yielding pZL01. Then, the sequence of clpP was amplified using PXL091F/PXL091R and digested with XbaI and SphI. Finally, the digestion product was ligated with pZL01 to construct pXL091, in which Pmip controlled the transcription of clpP gene constitutively. Because the resulting plasmid pXL091 contains the thymidylate synthetase gene originating from pJB908 backbone, transformants harboring pXL091 could be easily selected on BCYE plates devoid of thymidine, without addition of any antibiotics.
Phagocytosis assay and intracellular growth assay
For phagocytosis, J774A.1 cells were seeded into 24-well tissue culture plates (2.5 × 105 cells per well) and allowed to adhere overnight. L. pneumophila strains harboring pJB908 were grown to stationary phase, which were predominantly motile (OD600 3.7–4.5), and used for infection at a multiplicity of infection (MOI) of 1 after suspension in culture medium. The infection was synchronized by centrifugation at 500 g for 10 min and incubating for 30 min. The extracellular bacteria were removed by washing 3 times. After another 3 h of incubation, cells were lysed with 0.05 % saponin for plating dilutions onto BCYE plates and colony forming unit (CFU) counting. The percentages of bacteria resided within host cells were calculated as below: Phagocytosis percentage (%) = 100 x (CFU of cell lysate after incubating for 3 h)/(CFU of bacterial suspensions added at the initiation of infection).
The intracellular growth assay in J774A.1 cells was performed using a similar protocol as the assay in A. castellanii [28], except that the cell culture medium and the washing buffer were replaced with pre-warmed RPMI1640, the MOI was reduced from 10 to 1, and the lysing reagent was changed to 0.05 % saponin.
Contact-dependent cytotoxicity assay
J774A.1 cells were seeded into 96-well plates (1.0 × 105 cells/well) 24 h prior to infection. L. pneumophila strains harboring pJB908 were grown to post-exponential phase (OD600 3.0–3.7) in AYE broth, then harvested and used to infect J774A.1 cells at an MOI of 50 and 100. The culture plates were centrifuged at 400 g for 10 min and incubated for 2 h at 37 °C. After incubation, the cytosolic enzyme lactate dehydrogenase (LDH) release of supernatants was determined using the CytoTox-ONE 96 cytotoxicity assay kit (Promega), according to the instructions provided by the manufacturer. The level of Legionella-induced contact-dependent cytotoxicity was calculated as below: LDH release (%) = 100 x (experimental – culture medium background)/(maximum LDH release – culture medium background). Maximum LDH release was the LDH release from cells treated with 0.9 % Triton X-100, used as a positive control.
Cya translocation assays
To construct the CyaA fusions, the sequences of ralF, legK2, sidA, sidB, sidD, sidF and legU1 were amplified using the primers PRAc-F/PRAc-R, PLK2c-F/PLK2c-R, PSDAc-F/PSDAc-R, PSDBc-F/PSDBc-R, PSDDc-F/PSDDc-R, PSDFc-F/PSDFc-R and PLU1c-F/PLU1c-R respectively. Then the DNA products were digested with BamHI and SalI and ligated into the BamHI/SalI site of a pJB2581-derived plasmid with cyaA-sidJ fusion [34, 50], yielding pCA1-pCA7.
Cya translocation assay was carried out as described before [55]. Briefly, J774A.1 cells were seeded into 96-well plates (2.5 × 105 cells/well) and infected with L. pneumophila strains expressing CyaA fusion proteins at an MOI of 30. After incubation for 1 h at 37 °C, cells were washed and lysed, and total cAMP was extracted and determined using cAMP Enzyme Immunoassay Kit (Sigma-Aldrich). Creation of the fusion proteins was assessed by Western blotting using a monoclonal antibody to CyaA (CyaA (3D1), Santa Cruz Biotechnology).
Immunoblotting
L. pneumophila bacteria were harvested by centrifugation and washed in pre-cooled sterile water for 3 times. Then the samples were resuspended in 1 × Laemmli Sample Buffer, boiled for 10 min and centrifuged at 13,000 rpm for 10 min at 4 °C. Supernatants were then collected and loaded on SDS-polyacrylamide gels for analysis. The primary antibodies used and their dilutions were as follows: rabbit Anti-DotH, DotG and DotI (1:750; gifts from Dr. Luo ZQ), rabbit Anti-IcmS, IcmW and ICDH (1:200, 1:1000 and 1:2000; gifts from Dr. Vogel JP). The secondary antibodies were horseradish peroxidase (HRP)-conjugated goat anti-mouse or goat anti-rabbit (1:10000; Sigma). SuperSignal West Pico (Pierce) was used for signal detection.
Interaction of phagosomes with the endocytic pathway
The fusion of Legionella-containing phagosomes with the late endosome was assessed by detecting the co-localization of phagosomes with the lysosome-associated membrane protein 1 (LAMP-1) [52]. J774A.1 cells were seeded into 24-well tissue culture plates with 12-mm coverslips (1 × 105 cells/well) and incubated overnight at 37 °C. Then the cells were infected with L. pneumophila harboring pJB908 (OD600 3.7–4.5) at an MOI = 5. After incubation for 1 h, extracellular bacteria were eliminated by treating with 100 μg/ml gentamicin sulfate for 0.5 h. Then the coverslips were washed, transferred to clean wells, and fixed with 4 % paraformaldehyde solution for 10 min. To label extracellular bacteria, cells blocked with PBSSG (PBS containing 5 % sucrose and 2 % goat serum) buffer for 1 h were loaded with mouse anti-Legionella primary antibody (1:150; Santa Cruz Biotechnology), followed by Alex Fluo 350 conjugated goat anti-mouse IgG secondary antibody (1:2000; Molecular Probes). After being permeabilized with cold methanol for 20 s, intracellular bacteria were also blocked and labeled as above, with secondary antibody changed to the Alex Fluo 488 conjugated goat anti-mouse IgG. Finally, cells were loaded with rat anti-mouse Lamp-1 primary antibody (1:150; Santa Cruz Biotechnology) and Alex Fluo 594 conjugated goat anti-rat IgG secondary antibody (1:2000; Molecular Probes).
To assess the fusion with lysosomes, Texas-red conjugated ovalbumin (TroV) was used to label the lysosomes as described previously [51, 52]. Before the infection, 500 μl of pre-warmed TroV (100 μg/ml; Molecular Probes) was added to each well and incubated with cells for 0.5 h at 37 °C. Then the cells were washed 3 times for later use. The following procedures including infection, fixation, blocking, permeabilization and bacteria labeling were performed as in the Lamp-1 co-localization assay.
Construction of icm:gfp fusion plasmids and fluorescence analysis
To analyze the activities of icm promoters, a GFP coding gene gfpmut3, the product of which emits intense fluorescence [56], was employed. The coding sequence of gfpmut3 was cut out from pTLpflaG [57] with XbaI and SphI, and ligated with pJB908, yielding pGB908. Then the DNA sequences containing icm promoters of the 9 operon areas were amplified respectively and cloned into the multiple cloning site (MCS) upstream of gfpmut3, yielding pGB908X (from pGB9081 to pGB9089). The resulting plasmids were transformed into L. pneumophila strains by electroporation and the positive transformants were screened on thymidine-free BCYE plates.
For fluorescence analysis, L. pneumophila strains harboring pGB908X were inoculated into 5 ml AYE broth, and grown at 37 °C, 220 rpm for 20 h. Then the cultures were expanded into 25 ml AYE in flasks with initial optical densities (OD600) at approximate 0.2–0.3, and incubated to a stationary phase (OD600 3.7–4.5). Subsequently 100 μl of bacteria culture was transferred into 96-well fluorometer plates for fluorescence quantification on a microplate reader (TECAN INFINITE M200) with excitation at 488 nm and emission at 540 nm. Bacteria with pGB908X were also streak inoculated on thymidine-free BCYE plates and the GFP fluorescence intensity was estimated by direct observation.
Statistical analysis
Basic statistical analyses were performed using Excel, and one-way ANOVA was performed using SPSS followed by a post hoc Student-Newman-Keul’ s test.
Abbreviations
CFU, Colony-forming units; dot, defect in organelle trafficking; GFP, green fluorescent protein; icm, intracellular multiplication; Lamp-1, lysosome associated membrane protein 1; LCV, Legionella containing vacuole; MOI, multiplicity of infection; T4BSS, type IVB secretion system; TroV, texas-red conjugated ovalbumin.
Acknowledgements
We thank Dr. Zhao-Qing Luo, Dr. Paul Hoffman and Dr. Joseph Vogel for their kindly gifts of strains, antibodies and vectors.
Funding
This work was supported by the National Natural Science Foundation of China (No. 30970123) and the Natural Science Foundation of Guangdong province (No. 2016A030311036) to Yong-jun Lu.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BBZ, LXH and YJL designed the experiments and drafted the manuscript. BBZ and LXH performed the experiments. YLZ participated in the study design and performed the phagocytosis analysis. All authors read and approved the final manuscript.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Additional file
Immunoblots of whole-cell bacterial extracts expressing indicated Cya hybrid proteins from the wild type (Lp02) and the clpP mutant (Xp02) probed with monoclonal antibody specific to the CyaA epitope (Santa Cruz Biotechnology sc-13582) and ICDH with anti-ICDH antibody (a kind gift with Dr. Vogel JP). (TIF 3783 kb)
Contributor Information
Bei-bei Zhao, Email: bbsq2332@163.com.
Xiang-hui Li, Email: millerlee@126.com.
Yong-lun Zeng, Email: 506365841@qq.com.
Yong-jun Lu, Phone: +86-020-84110778, Email: luyj@mail.sysu.edu.cn.
References
- 1.Cunha BA, Burillo A, Bouza E. Legionnaires’ disease. Lancet. 2016;387(10016):376-85. [DOI] [PubMed]
- 2.Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7(1):13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent CD, Friedman JR, Jeong KC, Buford EC, Miller JL, Vogel JP. Identification of the core transmembrane complex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol. 2006;62(5):1278–1291. doi: 10.1111/j.1365-2958.2006.05446.x. [DOI] [PubMed] [Google Scholar]
- 4.Sexton JA, Pinkner JS, Roth R, Heuser JE, Hultgren SJ, Vogel JP. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J Bacteriol. 2004;186(6):1658–1666. doi: 10.1128/JB.186.6.1658-1666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sexton JA, Yeo HJ, Vogel JP. Genetic analysis of the Legionella pneumophila DotB ATPase reveals a role in type IV secretion system protein export. Mol Microbiol. 2005;57(1):70–84. doi: 10.1111/j.1365-2958.2005.04667.x. [DOI] [PubMed] [Google Scholar]
- 6.Dumenil G, Montminy TP, Tang M, Isberg RR. IcmR-regulated membrane insertion and efflux by the Legionella pneumophila IcmQ protein. J Biol Chem. 2004;279(6):4686–4695. doi: 10.1074/jbc.M309908200. [DOI] [PubMed] [Google Scholar]
- 7.Bitar DM, Molmeret ML, Abu Kwalk Y. Structure-function analysis of the C-terminus of IcmT of Legionella pneumophila in pore formation-mediated egress from macrophages. FEMS Microbiol Lett. 2005;242(1):177–184. doi: 10.1016/j.femsle.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Vincent CD, Friedman JR, Jeong KC, Sutherland MC, Vogel JP. Identification of the DotL coupling protein subcomplex of the Legionella Dot/Icm type IV secretion system. Mol Microbiol. 2012;85(2):378–391. doi: 10.1111/j.1365-2958.2012.08118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buscher BA, Conover GM, Miller JL, Vogel SA, Meyers SN, Isberg RR, Vogel JP. The DotL protein, a member of the TraG-coupling protein family, is essential for Viability of Legionella pneumophila strain Lp02. J Bacteriol. 2005;187(9):2927–2938. doi: 10.1128/JB.187.9.2927-2938.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donaldson JG, Jackson CL. Regulators and effectors of the ARF GTPases. Curr Opin Cell Biol. 2000;12(4):475–482. doi: 10.1016/S0955-0674(00)00119-8. [DOI] [PubMed] [Google Scholar]
- 11.Price CT, Al-Quadan T, Santic M, Rosenshine I, Abu Kwaik Y. Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science. 2011;334(6062):1553–1557. doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
- 12.Al-Quadan T, Price CT, Abu Kwaik Y. Exploitation of evolutionarily conserved amoeba and mammalian processes by Legionella. Trends Microbiol. 2012;20(6):299–306. doi: 10.1016/j.tim.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hervet E, Charpentier X, Vianney A, Lazzaroni JC, Gilbert C, Atlan D, Doublet P. Protein kinase LegK2 is a type IV secretion system effector involved in endoplasmic reticulum recruitment and intracellular replication of Legionella pneumophila. Infect Immun. 2011;79(5):1936–1950. doi: 10.1128/IAI.00805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 15.Isaac DT, Laguna RK, Valtz N, Isberg RR. MavN is a Legionella pneumophila vacuole-associated protein required for efficient iron acquisition during intracellular growth. Proc Natl Acad Sci U S A. 2015;112(37):E5208–E5217. doi: 10.1073/pnas.1511389112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harding CR, Stoneham CA, Schuelein R, Newton H, Oates CV, Hartland EL, Schroeder GN, Frankel G. The Dot/Icm effector SdhA is necessary for virulence of Legionella pneumophila in Galleria mellonella and A/J mice. Infect Immun. 2013;81(7):2598–2605. doi: 10.1128/IAI.00296-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- 18.Kress W, Mutschler H, Weber-Ban E. Both ATPase domains of ClpA are critical for processing of stable protein structures. J Biol Chem. 2009;284(45):31441–31452. doi: 10.1074/jbc.M109.022319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Hartling JA, Flanagan JM. The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91(4):447–456. doi: 10.1016/S0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- 20.Zolkiewski M. A camel passes through the eye of a needle: protein unfolding activity of Clp ATPases. Mol Microbiol. 2006;61(5):1094–1100. doi: 10.1111/j.1365-2958.2006.05309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Msadek T, Dartois V, Kunst F, Herbaud ML, Denizot F, Rapoport G. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol Microbiol. 1998;27(5):899–914. doi: 10.1046/j.1365-2958.1998.00735.x. [DOI] [PubMed] [Google Scholar]
- 22.Frees D, Ingmer H. ClpP participates in the degradation of misfolded protein in Lactococcus lactis. Mol Microbiol. 1999;31(1):79–87. doi: 10.1046/j.1365-2958.1999.01149.x. [DOI] [PubMed] [Google Scholar]
- 23.Gaillot O, Pellegrini E, Bregenholt S, Nair S, Berche P. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol Microbiol. 2000;35(6):1286–1294. doi: 10.1046/j.1365-2958.2000.01773.x. [DOI] [PubMed] [Google Scholar]
- 24.Kock H, Gerth U, Hecker M. The ClpP peptidase is the major determinant of bulk protein turnover in Bacillus subtilis. J Bacteriol. 2004;186(17):5856–5864. doi: 10.1128/JB.186.17.5856-5864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamoen LW, Venema G, Kuipers OP. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology. 2003;149(Pt 1):9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
- 26.Frees D, Chastanet A, Qazi S, Sorensen K, Hill P, Msadek T, Ingmer H. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol Microbiol. 2004;54(5):1445–1462. doi: 10.1111/j.1365-2958.2004.04368.x. [DOI] [PubMed] [Google Scholar]
- 27.Gaillot O, Bregenholt S, Jaubert F, Di Santo JP, Berche P. Stress-induced ClpP serine protease of Listeria monocytogenes is essential for induction of listeriolysin O-dependent protective immunity. Infect Immun. 2001;69(8):4938–4943. doi: 10.1128/IAI.69.8.4938-4943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li XH, Zeng YL, Gao Y, Zheng XC, Zhang QF, Zhou SN, Lu YJ. The ClpP protease homologue is required for the transmission traits and cell division of the pathogen Legionella pneumophila. BMC Microbiol. 2010;10:54. doi: 10.1186/1471-2180-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hilbi H, Segal G, Shuman HA. Icm/dot-dependent upregulation of phagocytosis by Legionella pneumophila. Mol Microbiol. 2001;42(3):603–617. doi: 10.1046/j.1365-2958.2001.02645.x. [DOI] [PubMed] [Google Scholar]
- 30.Joshi AD, Sturgill-Koszycki S, Swanson MS. Evidence that Dot-dependent and -independent factors isolate the Legionella pneumophila phagosome from the endocytic network in mouse macrophages. Cell Microbiol. 2001;3(2):99–114. doi: 10.1046/j.1462-5822.2001.00093.x. [DOI] [PubMed] [Google Scholar]
- 31.Gal-Mor O, Zusman T, Segal G. Analysis of DNA regulatory elements required for expression of the Legionella pneumophila icm and dot virulence genes. J Bacteriol. 2002;184(14):3823–3833. doi: 10.1128/JB.184.14.3823-3833.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2(3):e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santic M, Asare R, Doric M, Abu Kwaik Y. Host-dependent trigger of caspases and apoptosis by Legionella pneumophila. Infect Immun. 2007;75(6):2903–2913. doi: 10.1128/IAI.00147-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bardill JP, Miller JL, Vogel JP. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol. 2005;56(1):90–103. doi: 10.1111/j.1365-2958.2005.04539.x. [DOI] [PubMed] [Google Scholar]
- 35.Coers J, Kagan JC, Matthews M, Nagai H, Zuckman DM, Roy CR. Identification of Icm protein complexes that play distinct roles in the biogenesis of an organelle permissive for Legionella pneumophila intracellular growth. Mol Microbiol. 2000;38(4):719–736. doi: 10.1046/j.1365-2958.2000.02176.x. [DOI] [PubMed] [Google Scholar]
- 36.Vincent CD, Vogel JP. The Legionella pneumophila IcmS-LvgA protein complex is important for Dot/Icm-dependent intracellular growth. Mol Microbiol. 2006;61(3):596–613. doi: 10.1111/j.1365-2958.2006.05243.x. [DOI] [PubMed] [Google Scholar]
- 37.Cambronne ED, Roy CR. The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 2007;3(12):e188. doi: 10.1371/journal.ppat.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frees D, Qazi SN, Hill PJ, Ingmer H. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol Microbiol. 2003;48(6):1565–1578. doi: 10.1046/j.1365-2958.2003.03524.x. [DOI] [PubMed] [Google Scholar]
- 39.Ibrahim YM, Kerr AR, Silva NA, Mitchell TJ. Contribution of the ATP-dependent protease ClpCP to the autolysis and virulence of Streptococcus pneumoniae. Infect Immun. 2005;73(2):730–740. doi: 10.1128/IAI.73.2.730-740.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackson MW, Silva-Herzog E, Plano GV. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol Microbiol. 2004;54(5):1364–1378. doi: 10.1111/j.1365-2958.2004.04353.x. [DOI] [PubMed] [Google Scholar]
- 41.Knudsen GM, Olsen JE, Aabo S, Barrow P, Rychlik I, Thomsen LE. ClpP deletion causes attenuation of Salmonella Typhimurium virulence through mis-regulation of RpoS and indirect control of CsrA and the SPI genes. Microbiology. 2013;159(Pt 7):1497–1509. doi: 10.1099/mic.0.065797-0. [DOI] [PubMed] [Google Scholar]
- 42.Sutherland MC, Nguyen TL, Tseng V, Vogel JP. The Legionella IcmSW complex directly interacts with DotL to mediate translocation of adaptor-dependent substrates. PLoS Pathog. 2012;8(9):e1002910. doi: 10.1371/journal.ppat.1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci U S A. 2005;102(3):826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeong KC, Sutherland MC, Vogel JP. Novel export control of a Legionella Dot/Icm substrate is mediated by dual, independent signal sequences. Mol Microbiol. 2015;96(1):175–188. doi: 10.1111/mmi.12928. [DOI] [PubMed] [Google Scholar]
- 45.Hales LM, Shuman HA. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J Bacteriol. 1999;181(16):4879–4889. doi: 10.1128/jb.181.16.4879-4889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Molofsky AB, Swanson MS. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol Microbiol. 2004;53(1):29–40. doi: 10.1111/j.1365-2958.2004.04129.x. [DOI] [PubMed] [Google Scholar]
- 47.Gonzalez M, Rasulova F, Maurizi MR, Woodgate R. Subunit-specific degradation of the UmuD/D’ heterodimer by the ClpXP protease: the role of trans recognition in UmuD’ stability. EMBO J. 2000;19(19):5251–5258. doi: 10.1093/emboj/19.19.5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michel A, Agerer F, Hauck CR, Herrmann M, Ullrich J, Hacker J, Ohlsen K. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J Bacteriol. 2006;188(16):5783–5796. doi: 10.1128/JB.00074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savijoki K, Ingmer H, Frees D, Vogensen FK, Palva A, Varmanen P. Heat and DNA damage induction of the LexA-like regulator HdiR from Lactococcus lactis is mediated by RecA and ClpP. Mol Microbiol. 2003;50(2):609–621. doi: 10.1046/j.1365-2958.2003.03713.x. [DOI] [PubMed] [Google Scholar]
- 50.Xu L, Shen X, Bryan A, Banga S, Swanson MS, Luo ZQ. Inhibition of host vacuolar H + −ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 2010;6(3):e1000822. doi: 10.1371/journal.ppat.1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sturgill-Koszycki S, Swanson MS. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J Exp Med. 2000;192(9):1261–1272. doi: 10.1084/jem.192.9.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bachman MA, Swanson MS. RpoS co-operates with other factors to induce Legionella pneumophila virulence in the stationary phase. Mol Microbiol. 2001;40(5):1201–1214. doi: 10.1046/j.1365-2958.2001.02465.x. [DOI] [PubMed] [Google Scholar]
- 53.Moffat JF, Tompkins LS. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect Immun. 1992;60(1):296–301. doi: 10.1128/iai.60.1.296-301.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LeBlanc JJ, Davidson RJ, Hoffman PS. Compensatory functions of two alkyl hydroperoxide reductases in the oxidative defense system of Legionella pneumophila. J Bacteriol. 2006;188(17):6235–6244. doi: 10.1128/JB.00635-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ninio S, Zuckman-Cholon DM, Cambronne ED, Roy CR. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol Microbiol. 2005;55:912–926. [DOI] [PubMed]
- 56.Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173(1 Spec No):33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 57.Hammer BK, Swanson MS. Co-ordination of legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol Microbiol. 1999;33(4):721–731. doi: 10.1046/j.1365-2958.1999.01519.x. [DOI] [PubMed] [Google Scholar]
- 58.Swanson MS, Isberg RR. Identification of Legionella pneumophila mutants that have aberrant intracellular fates. Infect Immun. 1996;64(7):2585–2594. doi: 10.1128/iai.64.7.2585-2594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kohler R, Bubert A, Goebel W, Steinert M, Hacker J, Bubert B. Expression and use of the green fluorescent protein as a reporter system in Legionella pneumophila. Mol Gen Genet. 2000;262(6):1060–1069. doi: 10.1007/PL00008649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.


