Abstract
Early adversity increases risk for developing psychopathology. Epigenetic modification of stress reactivity genes is a likely mechanism contributing to this risk. The glucocorticoid receptor (GR) gene is of particular interest because of the regulatory role of the GR in hypothalamic–pituitary–adrenal (HPA) axis function. Mounting evidence suggests that early adversity is associated with GR promoter methylation and gene expression. Few studies have examined links between GR promoter methylation and psychopathology, and findings to date have been mixed. Healthy adult participants (N=340) who were free of psychotropic medications reported on their childhood experiences of maltreatment and parental death and desertion. Lifetime depressive and anxiety disorders and past substance-use disorders were assessed using the Structured Clinical Interview for the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition. Methylation of exon 1F of the GR gene (NR3C1) was examined in leukocyte DNA via pyrosequencing. On a separate day, a subset of the participants (n=231) completed the dexamethasone/corticotropin-releasing hormone (Dex/CRH) test. Childhood adversity and a history of past substance-use disorder and current or past depressive or anxiety disorders were associated with lower levels of NR3C1 promoter methylation across the region as a whole and at individual CpG sites (P<0.05). The number of adversities was negatively associated with NR3C1 methylation in participants with no lifetime disorder (P=0.018), but not in those with a lifetime disorder. GR promoter methylation was linked to altered cortisol responses to the Dex/CRH test (P<0.05). This study presents evidence of reduced methylation of NR3C1 in association with childhood maltreatment and depressive, anxiety and substance-use disorders in adults. This finding stands in contrast to our prior work, but is consistent with emerging findings, suggesting complexity in the regulation of this gene.
Introduction
Substantial evidence from animal models and human studies implicates impairments in glucocorticoid signaling in stress-related psychiatric disorders, such as major depressive disorder (MDD) and post-traumatic stress disorder (PTSD). Changes in glucocorticoid receptor (GR) number and function in the brain and in peripheral cells such as leukocytes have been shown with PTSD, MDD and early stress exposure,1, 2, 3, 4, 5, 6, 7, 8 and alterations of diurnal and reactive cortisol concentrations are seen in adults and children with these conditions.9, 10, 11 GR-mediated negative feedback has a critical role in dampening activity of the hypothalamic–pituitary–adrenal (HPA) axis so that changes in GR number or function can influence activity of this system and, consequently, the biological adaptation to stressful or traumatic experiences. Because childhood maltreatment and other adverse experiences are major risk factors for the development of psychopathology, including depressive, anxiety and substance-use disorders,12, 13, 14, 15 stress-induced alterations in glucocorticoid signaling may be a mechanism of these associations.
Expression of the human GR gene, NR3C1, is regulated by a variety of transcriptional and translational mechanisms.16 A growing area of research is aimed at determining whether early-life stress is associated with epigenetic changes to the promoter region of NR3C1 that alter GR expression and potentially alter HPA-axis homeostasis and responses to stress. Epigenetic modifications to the genome allow for altered gene expression but do not change the DNA sequence and thus permit elaboration of the genome beyond what is determined by DNA base coding.17, 18 Methylation, involving the addition of a methyl group to DNA, is thought to be the most stable form of epigenetic alteration.18, 19 Typically, this occurs at sites where a cytosine nucleotide occurs next to a guanine nucleotide (CpG dinucleotides), although non-CpG methylation has been discovered in embryonic stem cells and in some adult brain, skeletal muscle, and hematopoietic cells.20 Methylation at promoter CpG sites can lead to alterations in chromatin architecture and inhibit transcription factor (TF) binding, often resulting in reduced gene expression. Consistent with this, genes such as NR3C1 that are highly expressed typically have low levels of promoter methylation.17, 21, 22
Differential methylation has been shown to be associated with stress exposure early in life. Early studies in rodents with naturally occurring differences in maternal care showed that low levels of care were associated with greater methylation of the GR gene, specifically in the promoter region homologous to the human exon 1F.23, 24 The epigenetic effects of differential maternal care were associated with long-lasting effects on stress responses.23, 24 Subsequently, a number of studies in humans across the developmental spectrum have documented a relationship between early environmental exposures and increased methylation of the alternate first exons in the NR3C1 promoter.25, 26
Beginning with prenatal exposure, several studies have demonstrated that exposure to maternal depression, anxiety or trauma during gestation is associated with greater levels of methylation of the 1F region in DNA from placental tissue and umbilical cord blood.27, 28, 29, 30, 31 Childhood maltreatment was associated with increased NR3C1 methylation in leukocyte DNA from children aged 11–14 years32 and in saliva DNA from preschool-aged children.33 Two studies of adolescents have also shown positive associations of methylation of the NR3C1 promoter in peripheral blood DNA with prenatal exposure to intimate partner violence34 or traumatic experiences in childhood.35 The enduring effects of early stress are appreciated from evidence that alterations of NR3C1 promoter methylation were seen in adults with a history of childhood adversity. In a preliminary study of healthy adults with no lifetime psychopathology, our group found positive associations of 1F methylation with childhood parental loss and maltreatment that were not accounted for by recent perceived stress.36 Taken together, these findings suggest that altered NR3C1 methylation is linked to early stress exposure, and thus might predispose to the development of psychopathology.
Early-life adversity is a known risk factor for psychopathology later in life,12, 13, 14, 15 and differential methylation of genes important to the regulation of stress, such as the GR, may be a mechanism of this effect. However, few studies have examined the relationship between NR3C1 methylation, childhood adversity and psychopathology. We recently reported that methylation of saliva DNA NR3C1 in association with maltreatment and other adversities in maltreated preschool-aged children mediated the link between early stress exposure and internalizing behaviors (that is, withdrawn, somatic, anxious and depressed).37 In a study of post-mortem brain from suicides, subjects with a history of childhood abuse had differential NR3C1 promoter methylation compared with suicide completers without a history of childhood abuse and controls without a history of childhood abuse or suicide.38 Several investigations found altered methylation of sites in the NR3C1 promoter region in those with psychopathology.39, 40, 41, 42, 43, 44, 45, 46 A few studies have examined effects of childhood adversity within the sample as a whole, with some finding no effect39, 43, 44, 45 and others finding an effect of early-life stress on increased NR3C1 methylation.41, 47, 48 However, these have not specifically examined the differential contributions of early stress and psychopathology to NR3C1 methylation. Moreover, most prior work in individuals with psychiatric disorders included participants on psychotropic medication, which can alter methylation levels.48, 49, 50
Another issue requiring further study is the direction of methylation effects in association with early stress and psychopathology. For early stress, most work has identified hypermethylation of NR3C1 in the 1F region or its rat homolog.25 However, a number of these studies reported lower levels of methylation in some of the 1F CpG sites,28, 32, 35, 38 and examination of methylation at specific CpG sites may be important to understand the regulation of this gene.51, 52 For psychopathology, findings as to the direction of methylation effects have been mixed. Increased levels of NR3C1 promoter methylation have been shown among children and adolescents with depression or internalizing problems;46, 53 however, a study of depressed outpatients documented decreased levels of methylation in comparison with controls.42 Increased methylation has been documented in borderline personality disorder39, 41 and in women with comorbid borderline personality disorder and bulimia nervosa.43 In contrast, several studies have demonstrated lower levels of NR3C1 methylation in patients with PTSD as compared with healthy controls45, 54 and PTSD symptoms in men.44 Finally, recent research has shown that externalizing disorders and problems are also characterized by hypomethylation of NR3C1.40, 55
In order to examine these issues, the present study investigated methylation of the NR3C1 1F promoter region in adults with and without a history of childhood adversity, and with and without a lifetime psychiatric diagnosis of depressive, anxiety or substance-use disorder.
Materials and methods
Participants
Three-hundred and forty adults, 213 women and 127 men, aged 18–65 (32.9±11.5) years, were recruited in a series of related studies using separate local, newspaper and internet advertisements directed toward healthy adults, adults with depression, individuals with a history of early parental loss and adults with a history of early-life stress. This sample did not include the participants from our prior study of childhood adversity and NR3C1 methylation.36 The study was approved by the Butler Hospital Institutional Review Board, and, after complete description of the study to the participants, voluntary written informed consent was obtained.
Exclusions included prescription medication use other than oral contraceptives, reported acute or chronic medical illness, pregnancy, a history of brain injury or seizure disorder, night-shift work and current alcohol- or substance-use disorders because of possible effects on activity of the HPA axis. In addition, excluded were those with a lifetime history of bipolar disorder, psychotic disorder and obsessive–compulsive disorder. The subset of participants who completed the dexamethasone/corticotropin-releasing hormone (Dex/CRH) test (n=231) completed a physical examination, electrocardiogram and standard laboratory studies to further rule out acute or unstable medical illness. Oral contraceptives were allowed, with usage accounted for in analyses of cortisol concentrations.
Assessment of diagnoses and symptoms
Axis I psychiatric diagnoses were assessed using the Structured Clinical Interview for the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition.56 The 30-item Inventory of Depressive Symptomatology, Self-Report version,57 the State-Trait Anxiety Inventory58 and the Perceived Stress Scale59 were included to assess symptoms of depression and anxiety and recent perceived stress.
Childhood adversity
Participant interview was used to elicit the history of loss of a parent before the age of 18 (N=88). This category included death (n=38) of a parent and/or prolonged separation/desertion (that is, parent deserted for at least 6 months with no attempts at contact or responses to child's attempts, n=51).
The 28-item Childhood Trauma Questionnaire60 is a self-report measure that generates a total score summarizing five types of childhood maltreatment (physical, sexual and emotional abuse, physical and emotional neglect), with high internal consistency, test–retest reliability and convergent validity.60, 61, 62, 63, 64, 65 Threshold scores corresponding with 'moderate' or 'severe' exposure on each maltreatment subscale were used to define the presence of each adversity type on this scale.63 Adversity was considered present if the score was greater than or equal to 13 for emotional abuse, 10 for physical abuse, 8 for sexual abuse, 10 for physical neglect and 15 for emotional neglect.
Participants who experienced parental death or desertion or at least one of the maltreatment types assessed by the Childhood Trauma Questionnaire were considered to have experienced childhood adversity. In addition, an index of early stress exposure was created by summing the number of adversities (parental death, parental desertion and the five types of childhood maltreatment on the Childhood Trauma Questionnaire).
NR3C1 promoter methylation sequencing
Whole blood was frozen within 20 min of blood draw at −80 °C until processing. Samples were labeled with a numerical code only, and laboratory personnel were blind to subject characteristics. DNA was extracted from frozen whole blood using PAXgene reagents (Qiagen, Valencia, CA, USA) according to the manufacturer's directions. Methylation at the NR3C1 promoter region was examined with a quantitative pyrosequencing approach following the method of Oberlander and colleagues as previously described.31, 36 The region analyzed contains 13 CpGs and encompasses exon 1F, the human homolog of the rat exon 17. Sodium bisulfite modification of 500 ng of DNA was performed using the EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA) following the manufacturer's protocol. For quality control, PCR products were visualized and sized on an agarose gel (FlashGel, Lonza, Basel, Switzerland); samples were run in triplicate for pyrosequencing and data points were rejected if they were greater or less than two times the s.d. of the mean of the triplicate group. Peripheral blood lymphocyte DNA that was not sodium bisulfite-modified was included in each pyrosequencing run and served as a control for nonspecific amplification. Methylation quantification was performed using the Pyromark Software (Qiagen). The percent of alleles that were methylated in the cell population examined was used in statistical analyses.
Dex/CRH test
On a subsequent visit, 231 of the participants completed the Dex/CRH test. The night before the test, an oral dose of dexamethasone 1.5 mg was self-administered at 2300 hours. The following day, participants arrived at 1200 hours and were given lunch. A topical anesthetic (lidocaine 2.5% and prilocaine 2.5%) was applied to the participant's forearm between 1230 and 1245 hours. At 1300 hours an indwelling intravenous catheter was inserted in the forearm by a highly experienced research nurse. Participants then remained in a semi-recumbent position throughout the procedure except to use the bathroom. They were permitted to read or watch pre-selected materials that did not contain emotionally charged material. Vital signs were monitored throughout the test. At 1500 hours, CRH 100 μg (corticorelin ovine triflutate, Acthrel, Ferring Pharmaceuticals, Parsippany, NJ, USA) reconstituted in 2 ml of 0.9% NaCl solution was infused intravenously over 30 s. Blood samples were drawn at 1459, 1530, 1545, 1600, 1615 and 1700 hours and assayed for cortisol. The 1459 hours sample represents the response to dexamethasone (post-Dex) before administration of CRH infusion.
Hormone assays
Plasma cortisol concentrations were assayed in duplicate using the following assays and following the manufacturers' instructions. Samples were labeled with a numerical code only and laboratory personnel were blind to subject characteristics. The Gamma Coat cortisol I-125 coated-tube radioimmunoassay kit (INCSTAR, Stillwater, MN, USA) was used for 106 participants. Intra- and interassay CVs observed for quality assessment samples (5 and 20 μg /dl) were less than 5% and 10%, respectively. The double antibody DSL-2000 Cortisol Radioimmunoassay Kit (Diagnostic Systems Laboratories, Webster, TX, USA) was used for 81 participants. The intra- and interassay coefficients of variation are 5.3% and 7.0%, respectively. The cortisol Chemiluminescence Immunoassay kit (Beckman-Coulter, Brea, CA, USA) was used for 44 participants. The intra- and interassay coefficients of variation are both 4.3%. Variation between these three assay types was assessed in n=19 samples that were analyzed using more than one assay type, and results showed very high correlations (r=0.92–0.98). Area under the curve (AUC) over time for cortisol response to the Dex/CRH test was calculated using the trapezoidal rule. Post-Dex and AUC values were log-transformed because of positive skew.
Statistical analysis
In order to test hypothesized effects of early stress and psychopathology, four groups were examined: participants with no early adversity and no lifetime psychiatric diagnosis (n=96, No Adversity/No Disorder group), participants with early adversity but no lifetime diagnosis (n=60, Adversity/No Disorder group), participants with a lifetime diagnosis but no history of early adversity (n=51, No Adversity/Disorder group) and those with both early adversity and a lifetime diagnosis (n=133, Adversity/Disorder group). Subsequent analyses examined effects of specific disorder categories (lifetime MDD, lifetime depressive disorder (including MDD), lifetime PTSD, lifetime anxiety disorder (including PTSD) and past substance-use disorder) and childhood adversity types (physical abuse, sexual abuse, emotional abuse, physical neglect, emotional neglect, parental death and parental desertion). Effects of the number of adversities on methylation were also examined.
Our primary analyses examined effects of the adversity/disorder grouping on mean percent methylation of CpG sites across the 1F region. Methylation was correlated across CpG sites in this region. As with our prior work, CpG 7–13 were most highly intercorrelated (r=0.72-0.91) and CpG 1–6 were more modestly intercorrelated (r=0.42–0.66). Correlations across CpG 1–6 and CpG 7–13 were r=0.33–0.78. Given this intercorrelation, we examined mean methylation across the entire region, and, in the case of a significant overall effect on mean methylation, we then conducted follow-up analyses to examine individual CpG sites. We did not adjust for multiple comparisons because individual sites were only examined as post hoc comparisons in the event of a significant omnibus test. Outliers, defined as values more than three s.d.'s from the mean, were Winsorized by setting them to the next highest value within three s.d.'s. The number of outliers at each individual CpG site ranged from 1 (CpG 5) to 9 (CpG 1), with a mean of 5.4 outliers at each site (1.6% of the sample).
Analyses were conducted with SPSS version 20 (IBM, Armonk, NY, USA). All analyses were two-tailed with alpha=0.05 and data conformed to the assumptions of the statistical tests. Levene's test of the homogeneity of variance demonstrated that for mean methylation across the 13 CpG sites, the variances were equal across the four Adversity/Disorder groups (F(3, 336)=1.27, P=0.286). With N=340, we have power ≥0.80 to detect effect sizes as small as f=0.18, a small-medium-sized effect.66 Demographic characteristics, as well as recent perceived stress levels and subsyndromal symptoms of depression and anxiety, were examined and controlled where appropriate. General linear models, controlling for relevant covariates, were used to test for effects of adversity and psychopathology. Partial correlations, controlling for age, sex and oral contraceptive use, were used to examine associations of NR3C1 methylation with post-Dex and AUC cortisol in the Dex/CRH test. Error bars in figures represent s.e.m.
Results
Preliminary analyses
Participant characteristics are displayed in Table 1. One hundred and ninety-three participants reported a history of early adversity; 184 had a past substance-use disorder or a current or lifetime depressive or anxiety disorder (see table for types of adversity and disorders). Participants in the Adversity/Disorder group were significantly older than those in the other groups (Table 1), and age was associated with methylation at CpG 1 (r=0.13, P=0.020) and CpG 2 (r=0.13, P=0.014). Gender did not differ according to group; however, women had greater methylation at CpG 3 than men (t=−2.01, P=0.046). To control for these associations, age and gender were included as covariates in models testing associations of the Adversity/Disorder groupings and NR3C1 methylation. The Adversity/Disorder groups differed with respect to the proportion of participants who identified their race as white versus those who identified another race (Table 1). However, there were no differences in NR3C1 methylation at any of the CpG sites based on race, and associations of the Adversity/Disorder groupings and the number of adversities with NR3C1 did not differ when race was included in the models; therefore, it was not further considered. There was no difference in mean methylation across the 13 CpG sites between women with and without use of oral contraceptives. For analyses testing associations of the Adversity/Disorder grouping and the Dex/CRH test response, use of oral contraceptives was associated with greater post-dexamethasone cortisol (t=−6.19, P<0.001) and greater cortisol at 120 min following CRH administration (t=−2.03, P=0.043); this variable was controlled in addition to age and gender.
Table 1. Adversity/Disorder grouping variable descriptives.
| No Adversity/No Disorder (n=96) | Adversity/No Disorder (n=60) | No Adversity/Disorder (n=51) | Adversity/Disorder (n=133) | P | |
|---|---|---|---|---|---|
| Age M (s.d.) | 29.60 (10.04)a | 31.90 (10.19)a | 32.08 (11.29)a | 35.95 (12.37)b | 0.00 |
| Sex N (%) male | 38 (39.6) | 22 (36.7) | 21 (41.2) | 46 (34.6) | 0.81 |
| Race N (%) white | 74 (77.1)a | 37 (61.7)b | 46 (90.2)a | 108 (81.2)a | 0.00 |
| Education N (%), ≥ college degree | 54 (56.3) | 24 (40.0) | 23 (45.1) | 44 (33.1) | 0.35 |
| Emotional abuse N (%) | — | 15 (25.0)a | -— | 68 (51.1)b | 0.00 |
| Physical abuse N (%) | — | 10 (16.7)a | — | 48 (36.1)b | 0.01 |
| Sexual abuse N (%) | — | 13 (21.7)a | — | 50 (37.6)b | 0.03 |
| Emotional neglect N (%) | — | 16 (26.7)a | — | 72 (54.1)b | 0.00 |
| Physical neglect N (%) | — | 10 (16.7)a | — | 45 (33.8)b | 0.02 |
| Parental death N (%) | — | 15 (25.0) | — | 23 (17.3) | 0.26 |
| Parental desertion N (%) | — | 21 (35.0) | — | 30 (22.6) | 0.13 |
| Number of adversities M (s.d.) | — | 1.73 (1.16)a | — | 2.62 (1.51)b | 0.00 |
| Current MDD N (%) | — | — | 9 (17.6)a | 44 (33.1)b | 0.04 |
| Past MDD N (%) | — | — | 18 (35.3) | 43 (32.3) | 0.70 |
| Current dysthymia/Dep NOS N (%) | — | — | 4 (7.8) | 16 (12.0) | 0.41 |
| Past dysthymia/Dep NOS N (%) | — | — | 4 (7.8) | 20 (15.0) | 0.20 |
| Current PTSD N (%) | — | — | 0 (0.0) | 4 (3.0) | 0.21 |
| Past PTSD N (%) | — | — | 2 (3.9) | 11 (8.3) | 0.30 |
| Current social phobia N (%) | — | — | 2 (3.9) | 7 (5.3) | 0.71 |
| Past social phobia N (%) | — | — | 1 (2.0) | 5 (3.8) | 0.54 |
| Current other anxiety disorder N (%) | — | — | 4 (7.8) | 4 (3.0) | 0.15 |
| Past other anxiety disorder N (%) | — | — | 2 (3.9) | 11 (8.3) | 0.30 |
| Past substance disorder N (%) | — | — | 27 (52.9)a | 48 (36.1)b | 0.04 |
| IDSSR M (s.d.) | 6.39 (5.90)a | 9.29 (6.69)a | 16.43 (12.87)b | 21.81 (13.57)c | 0.00 |
| STAI—Trait M (s.d.) | 27.48 (7.28)a | 31.43 (9.79)b | 38.14 (12.55)c | 39.41 (11.27)c | 0.00 |
| STAI—State M (s.d.) | 26.56 (6.94)a | 27.95 (7.13)a | 35.32 (12.98)b | 34.43 (10.10)b | 0.00 |
| PSS M (s.d.) | 16.33 (5.67)a | 18.89 (6.81)b | 23.88 (9.64)c | 23.83 (8.47)c | 0.00 |
Abbreviations: ANOVA, analysis of variance; Dep, depression; IDSSR, Inventory of Depressive Symptomatology, Self-Report; MDD, major depressive disorder; NOS, not otherwise specified; PTSD, post-traumatic stress disorder; STAI, State-Trait Anxiety Inventory.
Note: In four group analyses, P-values indicate ANOVA significance level. In two group analyses, P-values indicate t-test and ?2-significance level. Different superscripts within the same line indicate groups are significantly different from one another at P<0.05. Other anxiety disorders include generalized anxiety disorder, panic disorder, agoraphobia and anxiety disorder NOS.
Associations of Adversity/Disorder grouping variable with NR3C1 methylation
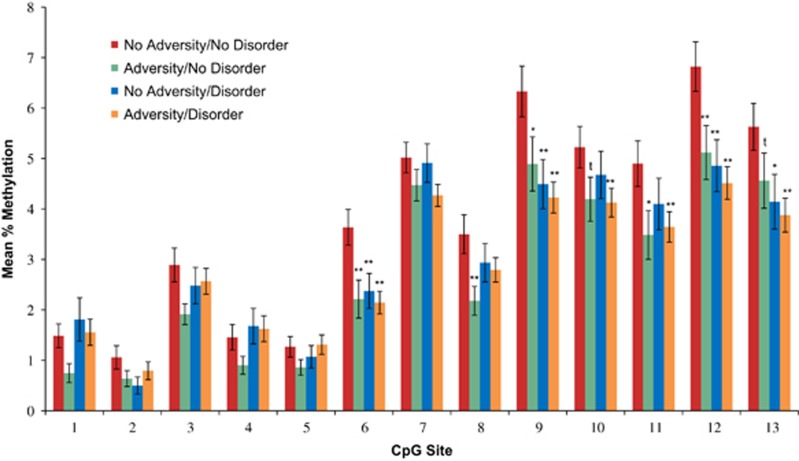
The Adversity/Disorder grouping variable was associated with mean methylation across the 13 CpG sites (F(3, 334)=3.68, P=0.012). The No Adversity/No Disorder group had significantly higher levels of mean methylation than the Adversity/No Disorder group (P=0.013) and the Adversity/Disorder group (P=0.002), and this difference was at the trend level for the No Adversity/Disorder group (P=0.06). Post hoc examination of individual CpG sites (Figure 1) revealed that there were significant differences between the four groups at CpG 6 (F=6.65, P<0.001), CpG 9 (F=6.28, P<0.001), CpG 10 (F=2.70, P=0.046), CpG 11 (F=2.82, P=0.039), CpG 12 (F=7.35, P<0.001) and CpG 13 (F=4.18, P=0.006), and a trend-level effect at CpG 8 (F=2.39, P=0.069). The No Adversity/No Disorder group had the highest levels of methylation, and this was the only group to differ significantly from any other group. The two Adversity groups showed the most consistent differences across this region, and, in addition, the No Adversity/Disorder group also had significantly lower levels of methylation at CpGs 6, 9, 12 and 13.
Figure 1.
Group differences in methylation at individual NR3C1 CpG sites Note. tP<0.10; *P<0.05; **P<0.01.
Analysis of diagnostic categories
Participants with any lifetime depressive disorder (F=7.54, P=0.007, n=115), any lifetime anxiety disorder (F=5.89, P=0.017, n=40) and any past substance-use disorder (F=3.89, P=0.050, n=75) had lower levels of methylation than those in the No Adversity/No Disorder group (n=96) across the 13 CpG sites. Table 2 shows results of post hoc analyses of associations of individual CpG sites with these disorders. There was no significant difference in mean methylation between those with No Adversity/No Disorder and those with lifetime MDD (P=0.132, n=97) or those with lifetime PTSD (P=0.422, n=16).
Table 2. CpG site-specific methylation for significant disorder and adversity types in comparison with No Adversity/No Disorder group.
| CpG site | Depressive Disorder (n=115) | Anxiety Disorder (n=40) | Past Substance Disorder (n=75) | Emotional Abuse (n=83) | Emotional Neglect (n=88) | Physical Neglect (n=55) | Parental Desertion (n=51) |
|---|---|---|---|---|---|---|---|
| Mean | 7.54** | 5.89* | 3.89* | 6.45* | 6.63* | 7.19** | 6.99** |
| 1 | ns | ns | ns | ns | ns | ns | ns |
| 2 | ns | ns | ns | ns | ns | ns | ns |
| 3 | ns | ns | ns | ns | ns | ns | ns |
| 4 | ns | ns | ns | ns | ns | ns | ns |
| 5 | ns | ns | ns | ns | ns | ns | ns |
| 6 | 14.56*** | 11.75*** | 7.40** | 8.02** | 11.60*** | 8.65** | 12.67*** |
| 7 | 3.44t | 5.08* | ns | 3.59t | 3.38t | 4.37* | ns |
| 8 | ns | ns | ns | 3.49t | ns | 3.15t | 3.34t |
| 9 | 19.56*** | 14.05*** | 8.25** | 11.02*** | 12.35*** | 12.71*** | 8.77** |
| 10 | 6.09* | 5.28* | ns | 6.06* | 5.33* | 9.47** | 5.18* |
| 11 | 6.07* | 5.59* | 3.15t | 5.76* | 4.84* | 6.96** | 5.79* |
| 12 | 21.02*** | 13.01*** | 8.64** | 14.03*** | 15.94*** | 17.66*** | 11.24*** |
| 13 | 12.48*** | 12.47*** | 7.03** | 7.76** | 8.07** | 10.33** | 5.68* |
Abbreviation: ns, not significant.
Note: All table values represent the F statistic. tP<0.10, *P<0.05, **P<0.01, ***P<0.001.
Analysis of adversity types
Participants who experienced emotional abuse (F=6.45, P=0.012, n=83), emotional neglect (F=6.63, P=0.011, n=88), physical neglect (F=7.19, P=0.008, n=55) and parental desertion (F=6.99, P=0.009, n=51) had lower levels of methylation than those in the No Adversity/No Disorder group across the 13 CpG sites. Table 2 shows results of post hoc analyses of associations of individual CpG sites with these adversity types. The reduced mean methylation in those with physical abuse (n=58), sexual abuse (n=63) and parental death (n=38) in comparison with those in the No Adversity/No Disorder group (n=96) did not reach significance (P=0.15, 0.17 and 0.11, respectively).
Associations of the number of adversities with NR3C1 methylation
The number of adversities was negatively associated with mean methylation across the 13 CpG sites (r=−0.11, P=0.046), and post hoc analyses of individual sites showed significant negative associations with CpGs, 6, 7, 9, 10, 11, 12 and 13 (P-values<0.05). Participants with disorders (lifetime depressive and anxiety disorders and past substance-use disorders) had a greater number of total adversities and were more likely to have each type of maltreatment (Table 1). There was a significant interaction of the number of adversities and lifetime psychiatric disorder (F(1, 334)=5.84, P=0.016), such that number of adversities was negatively associated with mean methylation across the region in participants with no lifetime disorder (r=−0.19, P=0.017), but not in those with a lifetime disorder (r=−0.03, not significant). Post hoc examination of the individual CpG sites revealed that this effect was significant for CpG 2 (F=4.50, P=0.035), CpG 3 (F=6.92, P=0.009), CpG 6 (F=4.00, P=0.046), CpG 8 (F=5.12, P=0.024), CpG 10 (F=4.30, P=0.039), CpG 11 (F=4.60, P=0.033) and CpG 12 (F=4.22, P=0.041). There were trend-level effects of the interaction on methylation at CpGs 1, 9 and 13 (P=0.085, 0.057 and 0.092, respectively). Consistent with the effect for mean methylation across the 13 CpG sites, the number of adversities was negatively associated with individual CpG site methylation in those participants without psychopathology (r's range from −0.14 to −0.19, P's range from 0.02 to 0.09), but was not associated with methylation at the individual CpG sites in participants with lifetime psychiatric disorders (r's range from −0.06 to 0.07, P's range from 0.35 to 0.89).
Sensitivity analyses
As shown in Table 1, there were significant differences between the four groups in perceived stress (F=20.13, P<0.001), depressive symptoms (F=44.10, P<0.001), state-related anxiety symptoms (F=17.27, P<0.001) and trait-related anxiety symptoms (F=26.40, P<0.001). Mean methylation across the 13 CpG sites was negatively associated with trait-related anxiety symptoms and with perceived stress at the trend level (r=−0.11, P=0.057; r=−0.11, P=0.066, respectively), but not with depressive symptoms or state-related anxiety symptoms. When added to the model assessing effects of the Adversity/Disorder grouping, none of these variables was significantly associated with mean methylation across the 13 CpG sites, and the Adversity/Disorder grouping variable remained a significant predictor of NR3C1 methylation.
Cortisol response to the Dex/CRH test
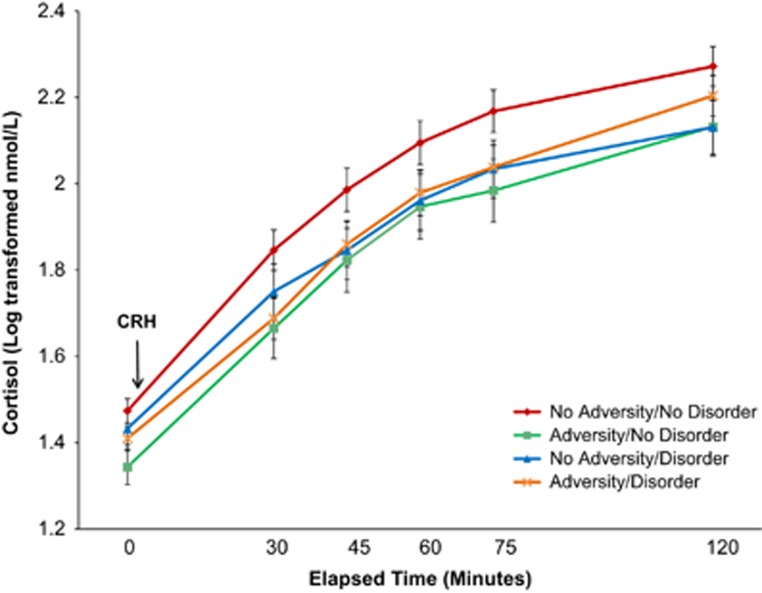
Cortisol response to the Dex/CRH test according to group is shown in Figure 2. Although the No Adversity/No Disorder group had the highest cortisol concentrations in the test, this did not reach significance (F(3, 220)=2.08, P=0.103). Similarly, effects of the Adversity/Disorder grouping did not reach significance for post-Dex cortisol (F(3, 222)=2.48, P=0.062) or cortisol AUC (F(3, 221)=2.23, P=0.086).
Figure 2.
Cortisol response to dexamethasone/corticotropin-releasing hormone (Dex/CRH) test in relation to adversity/disorder grouping. Note: the general linear model testing the effect of group on cortisol response over time did not reach significance (F(3, 220)=2.08, P=0.103).
Associations of NR3C1 methylation and cortisol response to the Dex/CRH test are displayed in Table 3. Mean methylation across the 13 sites was positively associated with post-Dex cortisol (r=0.148, P<0.05). Post hoc analyses of individual CpG sites showed that post-Dex cortisol had significant positive associations with methylation at CpG 2, CpG 5, CpG 7, CpG 9 and CpG 12 (P<0.05) and CpG 1 and CpG 3 (P<0.01), and there was a trend-level association with CpG 6, CpG 8, CpG 11 and CpG 13 (P<0.10). Cortisol AUC was associated with mean methylation across the 13 CpG sites at the trend level (P<0.10), and exploratory analyses at individual CpG sites showed significant effects at CpG 8 and CpG 11 (P<0.05), and at trend level for CpG 6 and 7 (P<0.10).
Table 3. Associations of NR3C1 methylation at individual CpG sites and cortisol concentrations in the Dex/CRH test.
| Mean CpG 1 - 13 | CpG 1 | CpG 2 | CpG 3 | CpG 4 | CpG 5 | CpG 6 | CpG 7 | CpG 8 | CpG 9 | CpG 10 | CpG 11 | CpG 12 | CpG 13 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Post-Dex | 0.148* | 0.209** | 0.136* | 0.206** | 0.106 | 0.147* | 0.113t | 0.162* | 0.118t | 0.155* | 0.098 | 0.122t | 0.143* | 0.126t |
| AUC | 0.117t | 0.056 | 0.028 | 0.083 | 0.026 | 0.058 | 0.122t | 0.114t | 0.166* | 0.098 | 0.095 | 0.139* | 0.093 | 0.099 |
Abbreviations: AUC, area under the curve; Dex/CRH, dexamethasone/corticotropin-releasing hormone.
Note: values indicate Pearson's Coefficient; tP<0.10, *P<0.05, **P<0.01.
Discussion
In this study we found that adults with a history of childhood adversity had reduced levels of methylation in exon 1F of the promoter of the GR gene. In addition to lower levels of methylation across the region, the groups with early adversity showed lower methylation at several individual CpG sites. The group with lifetime depressive, anxiety and substance-use disorders who did not have a history of childhood adversity had trend-level reductions in mean methylation across this region, with a few of the individual CpG sites showing significantly lower methylation in this group. Consistent with the expectation that lower methylation would be associated with greater GR numbers and enhanced glucocorticoid negative feedback, lower methylation at several sites in this region was associated with lower post-Dex cortisol responses to the Dex/CRH test. Those with early adversity and/or psychopathology tended to have lower cortisol responses to the test, although this was not statistically significant (in the subset of the sample that participated in the Dex/CRH test).
These findings stand in contrast to prior work that has documented higher levels of methylation of this region of NR3C1 with early stress.27, 31, 35, 36, 38, 48 However, some studies have found reduced methylation at individual CpG sites in NR3C1 1F in association with early stress.28, 35, 38 In addition, although two studies of subjects with internalizing symptoms have shown increased NR3C1 methylation as compared with controls,46, 53 there is evidence of decreased methylation at two CpG sites within the 1F region among patients with depression.42 Reduced levels of methylation in this region have also been reported in studies of patients with PTSD,44, 45, 54 and externalizing disorders.40 Individuals using psychotropic medication were excluded from the present study. It is important to note that most of the prior work on NR3C1 methylation in patients with psychiatric conditions did not exclude patients on psychotropic medication that can alter methylation levels.48, 49, 50 Our finding of reduced levels of methylation could be due to a loss of methyl groups over time, or inhibition of methylation in response to stress and trauma. In rats, the region of NR3C1 homologous to the 1F promoter is epigenetically regulated in response to chronic stress exposure in adulthood.67 Thus, it is possible that some of the group differences we observed may have occurred in response to experiences occurring over time in adulthood; however, it is important to note that effects of current symptoms or perceived stress did not account for our group findings.
We believe this is the first study to examine NR3C1 methylation in adults with and without a history of childhood adversity and with and without depressive, anxiety or substance-use disorders. The groups with early adversity, with or without psychopathology, had significantly lower methylation across the 13 sites and at several individual CpG sites. Participants with lifetime depressive, anxiety or substance-use disorders but no childhood adversity had numerically lower mean methylation across this region; however, this contrast did not reach statistical significance. Post hoc examination of individual CpG sites suggested that the group with psychopathology alone had lower methylation of some, but not all, of the sites that were significant for early adversity. The group with both adversity and lifetime psychopathology tended to show the greatest difference from controls; however, it should be noted that this group had more adverse early experiences, and that methylation in this group did not differ significantly from those with adversity but without lifetime psychopathology. The number of adversities was associated with lower methylation across the region and at several individual CpG sites in those without lifetime disorders, suggesting a dose effect. Taken together, these findings suggest that childhood adversity influences NR3C1 methylation and may represent a risk factor for the development of psychopathology. Psychiatric disorders may contribute to alterations in methylation of NR3C1 to a lesser degree, possibly via effects of these disorders on behavioral and physiological factors involved in stress responses.
We also examined the contributions of specific types of adversity and disorders. All adversity types were associated with lower methylation, and adversity types that tend to be chronic, including emotional abuse, emotional neglect, physical neglect and parental desertion were statistically significant. In contrast, physical abuse, sexual abuse and parental death did not reach statistical significance; these forms of adversity may be less pervasive and chronic than the other types, and children with such experiences may also experience buffering effects of positive emotional influences. Turning to individual disorder types, lifetime depressive disorder, lifetime anxiety disorder and past substance abuse showed significant effects, whereas lifetime PTSD did not, and lifetime MDD did not reach significance. However, it is important to note that lifetime MDD (n=97) was included in the significant effect for the lifetime depressive disorder (N=115) group. The number of participants with lifetime PTSD was small (n=16), limiting our power to detect an effect.
In our prior preliminary work with a sample of 99 healthy adults with no lifetime psychopathology, we found that early adversity was associated with greater methylation in this region of NR3C1. Although the same technique was used to measure methylation in both samples, methylation levels in the current sample were lower overall in comparison with the prior sample. The previous sample was younger, with a mean age of 27, and there may be loss of methylation with age;68, 69 however, we did control for age in the analyses. In our prior study, effects of early adversity were seen at CpGs 1–4, a region where methylation was particularly low in the current sample; therefore, there may be a 'floor effect'. In the present study, hypomethylation with adversity and psychopathology occurred in regions with overall higher levels of methylation. With respect to cortisol responses to the Dex/CRH test, in our prior study we found attenuated cortisol responses in association with higher methylation at CpG 2 and in exploratory analyses of sites CpG 5 and 7–13. We noted that this contrasted with prior work and seemed unlikely to reflect a direct effect of methylation-induced decreases in GR expression. In this much larger study of individuals with and without early adversity and with and without certain lifetime disorders, we find small but significant positive associations of post-dexamethasone cortisol with mean methylation and at several CpG sites across this region, as well as a trend for cortisol AUC. The direction of this effect is in keeping with findings of most, but not all, other studies,26 and with the expectation that lower methylation would be associated with increased gene expression and thus with enhanced HPA-axis negative feedback and reduced cortisol responses. These results from our study and others suggest that methylation of NR3C1 and its relationship to HPA-axis function is complex. It is possible that other factors such as participant age at the time of exposure to critical stressors, unmeasured adverse experiences, chronicity or severity of stressors, methylation of other HPA-regulatory genes or other factors inherent to depressive, anxiety and substance-use disorders also influenced NR3C1 methylation.26
Differential methylation of key TF-binding sites within the promoter regions is one mechanism by which adversity could exert influence on NR3C1 expression. The 1F region of NR3C1 contains binding sites for the nerve growth factor-inducible protein A (NGFI-A) that increases expression of GR, an effect that can be inhibited by methylation of these sites in both humans and the homologous 17 region in rodents.23, 38 Several,23, 24, 25, 26, 31, 32, 36, 70 but not all27, 28, 29, 33 studies have found increased methylation at the canonical NGFI-A-binding site (CpG 3 and CpG 4 in the present study), in humans and rodents with early adverse environmental exposures; this may have a functional effect on HPA-axis activity.28, 31, 71 Other studies have also reported a positive association between adversity and methylation of other canonical and non-canonical NGFI-A-binding sites within the 1F region.29, 38, 53 Decreased methylation within the NGFI-A-binding site has been documented in MDD42 and PTSD.42, 45 In the present study we did not find an effect of the adversity/disorder grouping on methylation at this binding site; however, the number of adversities was negatively linked to methylation at CpG 3 in those with no psychopathology, and there was a significant positive correlation between CpG 3 methylation and post-dexamethasone cortisol.
Some evidence indicates that interference with NGFI-A binding is not sufficient to reduce GR transcription and that methylation of other TF-binding sites across the 1F region may be involved.67 Armstrong et al.51 ran a sequence-based in silico query to identify other possible transcription factor-binding sites within NR3C1 1F. Numerous potential binding sites were identified, including sites for the alcohol dehydrogenase regulator 1, specificity protein 1 and heat shock factor.51 Some of these factors, such as heat shock factor that lies in proximity to CpG 9, are known to be involved in immune and stress responses.72 Consistent with this, CpG 9 showed differential methylation for all adversity types and diagnostic categories in this study. Putative binding sites for alcohol dehydrogenase regulator 1 reside in proximity to CpGs 4, 10, 11 and 13, which showed differential methylation with many adversity and diagnostic categories in our analyses. Putative sites for specificity protein 1 lie in proximity to CpGs 7 and 12, units that also showed differential methylation with several of our adversity and diagnostic categories. Differential methylation of CpGs within these binding sites may interfere with TF binding and regulate GR expression in response to adversity.
However, it is important to note that, consistent with findings from numerous research laboratories,8, 31, 40, 42, 45, 47, 53, 73, 74 levels of NR3C1 exon 1F methylation in this study were quite low, and may not be high enough to interfere with TF binding. Low levels of methylation typically characterize regions of the genome that are open to regulation by methylation.22, 75, 76 In contrast, high methylation is seen in areas where CpG density is low and transcription is not usually initiated.75 Our investigation was limited by the lack of gene expression data, but our finding linking lower levels of methylation at several CpG sites to lower Dex/CRH cortisol responses is consistent with prior work45, 77 and suggests that reduced methylation could have a functional effect on expression of GR.
This study is limited by the use of whole blood containing DNA from multiple different cell types. It is possible that our results were affected by differences in methylation across leukocyte types that might co-vary with adversity and psychopathology. We attempted to minimize such effects by excluding participants with acute or chronic illness or pregnancy. Our use of peripheral blood limits our ability to draw conclusions about central processes. However, peripheral and central DNA methylation and expression is likely to be more highly co-regulated for systems that have widespread effects, such as the glucocorticoid and immune systems.73, 74 We did not assess lifetime externalizing disorders; recent work suggests that externalizing disorders are associated with a decrease in NR3C1 methylation levels;40 therefore, such a history may contribute to the methylation patterns observed in this study. Other limitations include the cross-sectional and retrospective nature of the study and adversity data, the inclusion of participants taking oral contraceptives and the lack of control for menstrual phase in females. However, in contrast to most studies of NR3C1 methylation in psychiatric conditions, a major strength of this study is that we excluded individuals taking other medications as well as individuals with acute or chronic medical conditions. Finally, it should be noted that we did not include examination of additional genes or markers of global methylation. Epigenetic changes to other genes that are involved in the response to adversity and risk for psychopathology and other poor health outcomes78, 79, 80 should be considered in future work.
In conclusion, our findings of lower levels of methylation of the 1F promoter region of NR3C1 in relation to early stress and psychiatric conditions that are often associated with stress exposure suggest that these relationships are more complex than previously understood. Future work is needed to replicate these findings and clarify the mechanisms and sequelae of differential methylation of NR3C1 in association with early stress and psychopathology.
Acknowledgments
This research was supported by National Institute of Mental Health grants R21 MH091508 (ART), R01 MH101107 (ART) and R01 MH068767-08S1 (LLC). NSP is supported by Veterans Administration grant 1IK2CX000724. KKR is supported by National Institute of Mental Health grant 1R25 MH101076. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NIMH or the Department of Veterans Affairs.
The authors declare no conflict of interest.
References
- Anacker C, Zunszain PA, Carvalho LA, Pariante CM. The glucocorticoid receptor: pivot of depression and of antidepressant treatment? Psychoneuroendocrinology 2011; 36: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barden N. Implication of the hypothalamic-pituitary-adrenal axis in the physiopathology of depression. J Psychiatry Neurosci 2004; 29: 185–193. [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Binder EB. Allele-specific epigenetic modification: a molecular mechanism for gene-environment interactions in stress-related psychiatric disorders? Epigenomics 2013; 5: 109–112. [DOI] [PubMed] [Google Scholar]
- van Zuiden M, Geuze E, Willemen HL, Vermetten E, Maas M, Heijnen CJ et al. Pre-existing high glucocorticoid receptor number predicting development of posttraumatic stress symptoms after military deployment. Am J Psychiatry 2011; 168: 89–96. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Flory JD, Pratchett LC, Buxbaum J, Ising M, Holsboer F. Putative biological mechanisms for the association between early life adversity and the subsequent development of PTSD. Psychopharmacology 2010; 212: 405–417. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Seckl J. Minireview: stress-related psychiatric disorders with low cortisol levels: a metabolic hypothesis. Endocrinology 2011; 152: 4496–4503. [DOI] [PubMed] [Google Scholar]
- van Zuiden M, Kavelaars A, Vermetten E, Olff M, Geuze E, Heijnen C. Pre-deployment differences in glucocorticoid sensitivity of leukocytes in soldiers developing symptoms of PTSD, depression or fatigue persist after return from military deployment. Psychoneuroendocrinology 2015; 51: 513–524. [DOI] [PubMed] [Google Scholar]
- Yehuda R, Lowy MT, Southwick SM, Shaffer D, Giller EL Jr.. Lymphocyte glucocorticoid receptor number in posttraumatic stress disorder. Am J Psychiatry 1991; 148: 499–504. [DOI] [PubMed] [Google Scholar]
- Morris MC, Compas BE, Garber J. Relations among posttraumatic stress disorder, comorbid major depression, and HPA function: a systematic review and meta-analysis. Clin Psychol Rev 2012; 32: 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetler C, Miller GE. Depression and hypothalamic-pituitary-adrenal activation: a quantitative summary of four decades of research. Psychosom Med 2011; 73: 114–126. [DOI] [PubMed] [Google Scholar]
- Tyrka AR, Burgers DE, Philip NS, Price LH, Carpenter LL. The neurobiological correlates of childhood adversity and implications for treatment. Acta Psychiatr Scand 2013; 128: 434–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enns MW, Cox BJ, Afifi TO, De Graaf R, Ten Have M, Sareen J. Childhood adversities and risk for suicidal ideation and attempts: a longitudinal population-based study. Psychol Med 2006; 36: 1769–1778. [DOI] [PubMed] [Google Scholar]
- Hill J, Davis R, Byatt M, Burnside E, Rollinson L, Fear S. Childhood sexual abuse and affective symptoms in women: a general population study. Psychol Med 2000; 30: 1283–1291. [DOI] [PubMed] [Google Scholar]
- Hill J, Pickles A, Burnside E, Byatt M, Rollinson L, Davis R et al. Child sexual abuse, poor parental care and adult depression: evidence for different mechanisms. Br J Psychiatry 2001; 179: 104–109. [DOI] [PubMed] [Google Scholar]
- Carr CP, Martins CM, Stingel AM, Lemgruber VB, Juruena MF. The role of early life stress in adult psychiatric disorders: a systematic review according to childhood trauma subtypes. J Nerv Ment Dis 2013; 201: 1007–1020. [DOI] [PubMed] [Google Scholar]
- Vandevyver S, Dejager L, Libert C. Comprehensive overview of the structure and regulation of the glucocorticoid receptor. Endocr Rev 2014; 35: 671–693. [DOI] [PubMed] [Google Scholar]
- Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology 2013; 38: 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. The dynamic epigenome and its implications in toxicology. Toxicol Sci 2007; 100: 7–23. [DOI] [PubMed] [Google Scholar]
- Byun HM, Nordio F, Coull BA, Tarantini L, Hou L, Bonzini M et al. Temporal stability of epigenetic markers: sequence characteristics and predictors of short-term DNA methylation variations. PLoS One 2012; 7: e39220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinney SE. Mammalian non-cpg methylation: stem cells and beyond. Biology 2014; 3: 739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One 2011; 6: e14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 2007; 39: 457–466. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR et al. Epigenetic programming by maternal behavior. Nat Neurosci 2004; 7: 847–854. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Szyf M, Meaney MJ. From maternal care to gene expression: DNA methylation and the maternal programming of stress responses. Endocr Res 2002; 28: 699. [DOI] [PubMed] [Google Scholar]
- Turecki G, Meaney MJ. Effects of the social environment and stress on glucocorticoid receptor gene methylation: a systematic review. Biol Psychiatry 2014; 79: 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma-Gudiel H, Cordova-Palomera A, Leza JC, Fananas L. Glucocorticoid receptor gene (NR3C1) methylation processes as mediators of early adversity in stress-related disorders causality: a critical review. Neurosci Biobehav Rev 2015; 55: 520–535. [DOI] [PubMed] [Google Scholar]
- Conradt E, Lester BM, Appleton AA, Armstrong DA, Marsit CJ. The role of DNA methylation of NR3C1 and 11beta-HSD2 and exposure to maternal mood disorder in utero on newborn neurobehavior. Epigenetics 2013; 8: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hompes T, Izzi B, Gellens E, Morreels M, Fieuws S, Pexsters A et al. Investigating the influence of maternal cortisol and emotional state during pregnancy on the DNA methylation status of the glucocorticoid receptor gene (NR3C1) promoter region in cord blood. J Psychiatric Res 2013; 47: 880–891. [DOI] [PubMed] [Google Scholar]
- Mulligan CJ, D'Errico NC, Stees J, Hughes DA. Methylation changes at NR3C1 in newborns associate with maternal prenatal stress exposure and newborn birth weight. Epigenetics 2012; 7: 853–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgatroyd C, Quinn JP, Sharp HM, Pickles A, Hill J. Effects of prenatal and postnatal depression, and maternal stroking, at the glucocorticoid receptor gene. Transl Psychiatry 2015; 5: e560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics 2008; 3: 97–106. [DOI] [PubMed] [Google Scholar]
- Romens SE, McDonald J, Svaren J, Pollak SD. Associations between early life stress and gene methylation in children. Child Dev 2015; 86: 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Parade SH, Eslinger NM, Marsit CJ, Lesseur C, Armstrong DA et al. Methylation of exons 1D, 1 F, and 1H of the glucocorticoid receptor gene promoter and exposure to adversity in preschool-aged children. Dev Psychopathol 2015; 27: 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radtke KM, Ruf M, Gunter HM, Dohrmann K, Schauer M, Meyer A et al. Transgenerational impact of intimate partner violence on methylation in the promoter of the glucocorticoid receptor. Transl Psychiatry 2011; 1: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap LJ, Riese H, Hudziak JJ, Verbiest MM, Verhulst FC, Oldehinkel AJ et al. Glucocorticoid receptor gene (NR3C1) methylation following stressful events between birth and adolescence. The TRAILS study. Transl Psychiatry 2014; 4: e381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Price LH, Marsit C, Walters OC, Carpenter LL. Childhood adversity and epigenetic modulation of the leukocyte glucocorticoid receptor: preliminary findings in healthy adults. PLoS One 2012; 7: e30148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parade SH, Ridout KK, Seifer R, Armstrong DA, Marsit CJ, McWilliams MA et al. Methylation of the glucocorticoid receptor gene promoter in preschoolers: links with internalizing behavior problems. Child Dev 2016; 87: 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonte B, Szyf M et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci 2009; 12: 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammann G, Teschler S, Haag T, Altmuller F, Tuczek F, Dammann RH. Increased DNA methylation of neuropsychiatric genes occurs in borderline personality disorder. Epigenetics 2011; 6: 1454–1462. [DOI] [PubMed] [Google Scholar]
- Heinrich A, Buchmann AF, Zohsel K, Dukal H, Frank J, Treutlein J et al. Alterations of glucocorticoid receptor gene methylation in externalizing disorders during childhood and adolescence. Behav Genet 2015; 45: 529–536. [DOI] [PubMed] [Google Scholar]
- Martin-Blanco A, Ferrer M, Soler J, Salazar J, Vega D, Andion O et al. Association between methylation of the glucocorticoid receptor gene, childhood maltreatment, and clinical severity in borderline personality disorder. J Psychiatric Res 2014; 57: 34–40. [DOI] [PubMed] [Google Scholar]
- Na KS, Chang HS, Won E, Han KM, Choi S, Tae WS et al. Association between glucocorticoid receptor methylation and hippocampal subfields in major depressive disorder. PLoS One 2014; 9: e85425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger H, Labonte B, Groleau P, Turecki G, Israel M. Methylation of the glucocorticoid receptor gene promoter in bulimic women: associations with borderline personality disorder, suicidality, and exposure to childhood abuse. Int J Eat Disord 2013; 46: 246–255. [DOI] [PubMed] [Google Scholar]
- Vukojevic V, Kolassa IT, Fastenrath M, Gschwind L, Spalek K, Milnik A et al. Epigenetic modification of the glucocorticoid receptor gene is linked to traumatic memory and post-traumatic stress disorder risk in genocide survivors. J Neurosci 2014; 34: 10274–10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Flory JD, Bierer LM, Henn-Haase C, Lehrner A, Desarnaud F et al. Lower methylation of glucocorticoid receptor gene promoter 1 F in peripheral blood of veterans with posttraumatic stress disorder. Biol Psychiatry 2015; 77: 356–364. [DOI] [PubMed] [Google Scholar]
- Dadds MR, Moul C, Hawes DJ, Mendoza Diaz A, Brennan J. Individual differences in childhood behavior disorders associated with epigenetic modulation of the cortisol receptor gene. Child Dev 2015; 86: 1311–1320. [DOI] [PubMed] [Google Scholar]
- Melas PA, Wei Y, Wong CC, Sjoholm LK, Aberg E, Mill J et al. Genetic and epigenetic associations of MAOA and NR3C1 with depression and childhood adversities. Int J Neuropsychopharmacol 2013; 16: 1513–1528. [DOI] [PubMed] [Google Scholar]
- Perroud N, Paoloni-Giacobino A, Prada P, Olie E, Salzmann A, Nicastro R et al. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Transl Psychiatry 2011; 1: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott E, Ezra-Nevo G, Regev L, Neufeld-Cohen A, Chen A. Resilience to social stress coincides with functional DNA methylation of the Crf gene in adult mice. Nat Neurosci 2010; 13: 1351–1353. [DOI] [PubMed] [Google Scholar]
- Lotsch J, Schneider G, Reker D, Parnham MJ, Schneider P, Geisslinger G et al. Common non-epigenetic drugs as epigenetic modulators. Trends Mol Med 2013; 19: 742–753. [DOI] [PubMed] [Google Scholar]
- Armstrong DA, Lesseur C, Conradt E, Lester BM, Marsit CJ. Global and gene-specific DNA methylation across multiple tissues in early infancy: implications for children's health research. FASEB J 2014; 28: 2088–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskalakis NP, Yehuda R. Site-specific methylation changes in the glucocorticoid receptor exon 1 F promoter in relation to life adversity: systematic review of contributing factors. Front Neurosci 2014; 8: 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap LJ, van Oort FV, Verhulst FC, Oldehinkel AJ, Riese H. Methylation of NR3C1 and SLC6A4 and internalizing problems. The TRAILS study. J Affect Disord 2015; 180: 97–103. [DOI] [PubMed] [Google Scholar]
- Labonte B, Azoulay N, Yerko V, Turecki G, Brunet A. Epigenetic modulation of glucocorticoid receptors in posttraumatic stress disorder. Transl Psychiatry 2014; 4: e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemin C, Provencal N, Suderman M, Cote SM, Vitaro F, Hallett M et al. DNA methylation signature of childhood chronic physical aggression in T cells of both men and women. PLoS One 2014; 9: e86822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Gibbon M, Spitzer RL, Williams JB, Benjamin L (1997). Structured Clinical Interview for DSM-IV Axis I (SCID-I), Clinical Version.
- Rush AJ, Gullion CM, Basco MR, Jarrett RB, Trivedi MH. The Inventory of Depressive Symptomatology (IDS): psychometric properties. Psychol Med 1996; 26: 477–486. [DOI] [PubMed] [Google Scholar]
- Spielberger CD, Gorsuch RL, Lushene RE STAI State-Trait Anxiety Inventory Forma Y. Edizione Italiana a Cura di L Pedrabissi e M Santinello. Firenze, Organizzazioni Speciali 1989.
- Cohen S, Kamarck T, Mermelstein R. A global measure of perceived stress. J Health Soc Behav 1983; 24: 385–396. [PubMed] [Google Scholar]
- Bernstein DP, Stein JA, Newcomb MD, Walker E, Pogge D, Ahluvalia T et al. Development and validation of a brief screening version of the Childhood Trauma Questionnaire. Child Abuse Negl 2003; 27: 169–190. [DOI] [PubMed] [Google Scholar]
- Paivio SC. Stability of retrospective self-reports of child abuse and neglect before and after therapy for child abuse issues. Child Abuse Negl 2001; 25: 1053–1068. [DOI] [PubMed] [Google Scholar]
- Paivio SC, Cramer KM. Factor structure and reliability of the Childhood Trauma Questionnaire in a Canadian undergraduate student sample. Child Abuse Negl 2004; 28: 889–904. [DOI] [PubMed] [Google Scholar]
- Bernstein D, Fink L. Childhood Trauma Questionnaire: a Retrospective Self-Report Manual. The Psychological Corporation: San Antonio, TX, USA, 1998. [Google Scholar]
- Bernstein DP, Ahluvalia T, Pogge D, Handelsman L. Validity of the Childhood Trauma Questionnaire in an adolescent psychiatric population. J Am Acad Child Adolesc Psychiatry 1997; 36: 340–348. [DOI] [PubMed] [Google Scholar]
- Fink LA, Bernstein D, Handelsman L, Foote J, Lovejoy M. Initial reliability and validity of the childhood trauma interview: a new multidimensional measure of childhood interpersonal trauma. Am J Psychiatry 1995; 152: 1329–1335. [DOI] [PubMed] [Google Scholar]
- Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2nd edn, Lawrence Erlbaum: Hillsdale, NJ, USA, 1988. [Google Scholar]
- Witzmann SR, Turner JD, Meriaux SB, Meijer O.C, Muller CP. Epigenetic regulation of the glucocorticoid receptor promoter 1(7) in adult rats. Epigenetics 2012; 7: 1290–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Avraham D. Epigenetics of aging. Adv Exp Med Biol 2015; 847: 179–191. [DOI] [PubMed] [Google Scholar]
- Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H et al. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev 2009; 130: 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, D'Alessio AC, Brown SE, Hellstrom IC, Dymov S, Sharma S et al. The transcription factor nerve growth factor-inducible protein a mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J Neurosci 2007; 27: 1756–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud LR, Papandonatos GD, Rodriguez D, McCallum M, Salisbury AL, Phipps MG et al. Maternal smoking during pregnancy and infant stress response: test of a prenatal programming hypothesis. Psychoneuroendocrinology 2014; 48: 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerfelt M, Morimoto RI, Sistonen L. Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol 2010; 11: 545–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M. The early-life social environment and DNA methylation. Clin Genet 2012; 81: 341–349. [DOI] [PubMed] [Google Scholar]
- Szyf M. The genome- and system-wide response of DNA methylation to early life adversity and its implication on mental health. Can J Psychiatry 2013; 58: 697–704. [DOI] [PubMed] [Google Scholar]
- Lokk K, Modhukur V, Rajashekar B, Martens K, Magi R, Kolde R et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol 2014; 15: r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage VR, Jarmasz JS, Murugeshan N, Del Bigio MR, Rastegar M, Davie JR. DNA modifications: function and applications in normal and disease States. Biology 2014; 3: 670–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij SR, Costello PM, Veenendaal MV, Lillycrop KA, Gluckman PD, Hanson MA et al. Associations between DNA methylation of a glucocorticoid receptor promoter and acute stress responses in a large healthy adult population are largely explained by lifestyle and educational differences. Psychoneuroendocrinology 2012; 37: 782–788. [DOI] [PubMed] [Google Scholar]
- Klengel T, Pape J, Binder EB, Mehta D. The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology 2014; 80: 115–132. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Zhang H, Ge W, Weder N, Douglas-Palumberi H, Perepletchikova F et al. Child abuse and epigenetic mechanisms of disease risk. Am J Prev Med 2013; 44: 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weder N, Zhang H, Jensen K, Yang BZ, Simen A, Jackowski A et al. Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. J Am Acad Child Adolesc Psychiatry 2014; 53: 417–424 e415. [DOI] [PMC free article] [PubMed] [Google Scholar]