Insect DSC1 channels have sequences that are intermediate between voltage-gated Na+ and Ca2+ channels but have hitherto been classified as the former. Gosselin-Badaroudine et al. clone and characterize honeybee DSC1, revealing high selectivity for Ca2+ and suggesting reclassification of DSC1 homologues as Ca2+ channels.
Abstract
Bilaterian voltage-gated Na+ channels (NaV) evolved from voltage-gated Ca2+ channels (CaV). The Drosophila melanogaster Na+ channel 1 (DSC1), which features a D-E-E-A selectivity filter sequence that is intermediate between CaV and NaV channels, is evidence of this evolution. Phylogenetic analysis has classified DSC1 as a Ca2+-permeable Na+ channel belonging to the NaV2 family because of its sequence similarity with NaV channels. This is despite insect NaV2 channels (DSC1 and its orthologue in Blatella germanica, BSC1) being more permeable to Ca2+ than Na+. In this study, we report the cloning and molecular characterization of the honeybee (Apis mellifera) DSC1 orthologue. We reveal several sequence variations caused by alternative splicing, RNA editing, and genomic variations. Using the Xenopus oocyte heterologous expression system and the two-microelectrode voltage-clamp technique, we find that the channel exhibits slow activation and inactivation kinetics, insensitivity to tetrodotoxin, and block by Cd2+ and Zn2+. These characteristics are reminiscent of CaV channels. We also show a strong selectivity for Ca2+ and Ba2+ ions, marginal permeability to Li+, and impermeability to Mg2+ and Na+ ions. Based on current ion channel nomenclature, the D-E-E-A selectivity filter, and the properties we have uncovered, we propose that DSC1 homologues should be classified as CaV4 rather than NaV2. Indeed, channels that contain the D-E-E-A selectivity sequence are likely to feature the same properties as the honeybee’s channel, namely slow activation and inactivation kinetics and strong selectivity for Ca2+ ions.
INTRODUCTION
Voltage-gated Na+ and Ca2+ channels (NaV and CaV) belong to the large family of ion channels and feature four homologous domains, each containing six transmembrane (TM) segments. NaV channels initiate the action potentials of many excitable cells, thus regulating their electrical signals. CaV channels have been attributed more diverse roles, which is expected, because they would have emerged earlier in evolution and are permeable to Ca2+, a major second messenger. They are involved in pacemaker cell action potentials, muscle contraction, and the Ca2+-dependent exocytosis of vesicles, which regulates hormone and neurotransmitter secretion (Catterall et al., 2005b).
NaV channels would have evolved from CaV channels (Hille, 2001). This possibility is supported by the existence of proteins such as the Drosophila melanogaster Na+ channel 1 (DSC1) and its orthologues. Indeed, those channels feature selectivity filter sequences that would be representative of an intermediate sequence between that of canonical NaV and CaV channels (Zhou et al., 2004; Liebeskind et al., 2011).
DSC1 was first identified in Drosophila using probes corresponding to the eel Na+ channel (Salkoff et al., 1987). Like other NaV and CaV channels, DSC1 is a 24-TM protein divided into four homologous domains, each featuring a voltage-sensitive domain resulting from the assembly of the first four TMs (S1–S4). The S5–S6 TMs of each domain assemble to form the pore domain, which is responsible for ion permeation and selectivity. A highly conserved motif located at the aperture of the pore is the main contributor to the selectivity of the 24-TM channels and is composed of one amino acid from each domain located between the helixes of the reentrant loop between S5 and S6 (Heinemann et al., 1992; Catterall et al., 2005b). Usually, only negatively charged amino acids (E-E-D-D or E-E-E-E in domains DI–DII–DIII–DIV) form this selectivity filter in CaV channels, whereas neutral and positively charged amino acids are involved in Na+ selective channels (D-E-K-A for NaV1 channels; Heinemann et al., 1992; Catterall et al., 2005a; Stephens et al., 2015). The selectivity filter sequence for most DSC1 homologues (D-E-E-A) appears to be a hybrid of the sequences found in NaV1 and CaV channels. Because DSC1 homologues and NaV1 channels would have evolved from a common ancestor, and based on phylogenic studies, DSC1 and its homologues have also been named NaV2 channels (Liebeskind et al., 2011).
Although the basic characterization of the Blatella germanica DSC1 orthologue (BSC1; Zhou et al., 2004) and the functional expression of DSC1 (Zhang et al., 2011) have been reported, no thorough biophysical characterizations of these channels is available. Because the low expression levels of both channels hampered their full characterization, the authors of both studies based their characterizations on tail current amplitudes or endogenous Xenopus oocyte Ca2+-activated Cl− channels as a readout of channel activity.
The complete biophysical characterization of a DSC1 homologue channel would provide a solid basis for the definitive classification of this 24-TM channel subfamily according to the current nomenclature (Catterall et al., 2005a,b). Moreover, precise knowledge of the channels’ properties may clarify the role of DSC1 orthologues. Indeed, a knockout of the DSC1 gene in Drosophila causes a “jumpy” phenotype and impairs olfaction (Kulkarni et al., 2002; Zhang et al., 2013). However, as shown by Liebeskind et al. (2011), DSC1 homologues exist also in animal species that do not have olfaction. Therefore, DSC1 orthologues may be implicated in other functions yet to be uncovered.
Here, we report the cloning, functional expression, and biophysical and pharmacological characterization of the honeybee (Apis mellifera) DSC1 homologue, which we have named AmCaV4, based on the properties uncovered and the current ion channels nomenclature (Catterall et al., 2005a,b). The work presented here provides a unique perspective on 24-TM channel family and its subfamilies.
MATERIALS AND METHODS
Cloning and sequencing
Total RNA from honeybee heads was extracted using TRIzol kits (Sigma-Aldrich). The cDNA was produced using Transcriptor first-strand cDNA synthesis kits (Roche). DNA and cDNA (produced from extracted RNA) were amplified with the Pfu enzyme. The cDNA corresponding to AmCaV4 was obtained by PCR amplification and was inserted into the pPol_Not1 vector (a gift from P. Isenring, Université Laval, Quebec City, Quebec, Canada), an oocyte expression vector containing the T7 promoter (5′ to 3′), the Xenopus laevis β-globin 5′-untranslated region, a multiple cloning site, the Xenopus β-globin 3′-untranslated region, a linearizing site, and polyA and polyC tracts. The vector containing the TipE regulatory subunit was amplified in Escherichia coli XL2 blue (Agilent Technologies) and was purified using GenElute HP Plasmid Maxiprep kits (Sigma-Aldrich). The constructs were linearized with Not1, and T7 RNA polymerase was used to make sense RNA using mMESSAGE mMACHINE T7 kits (Ambion). Genomic DNA from whole bees was extracted and sequenced using QIAamp DNA mini kits (QIAGEN) in order to identify sequence variations.
Xenopus oocytes
All experimental procedures involving Xenopus oocytes were approved by the Université Laval Institutional Animal Care Committee in line with the principles and guidelines of the Canadian Council on Animal Care (approval 2011155–1). They were prepared as described previously (Chen et al., 1992). In brief, oocytes surgically removed from frogs anesthetized with MS-222 (tricaine) were treated with 2 mg/ml collagenase (collagenase type 1A from Clostridium histolyticum; Sigma-Aldrich) in OR2 solution for 1 h. They were then incubated for at least 1 h at 18°C in OR3 solution. Stage IV and V oocytes were microinjected with mRNA corresponding to the AmCaV4 channel and the TipE, TEH4, or CaVβ regulatory subunit when specified (1 µg/µl for AmCaV4 and 1 µg/µl for the regulatory subunits, 50 nl per oocyte). The injected oocytes were incubated at 18°C in OR3 solution for at least 12 h before recordings.
Electrophysiology
Macroscopic currents from mRNA-injected oocytes were recorded in the solutions listed in Table 1 using the two-microelectrode voltage-clamp technique. Endogenous Ca2+-activated chloride currents were suppressed by injecting 50 nl of Ca2+ chelating solution before the experiments (either 10 mM EGTA, 10 mM EDTA, or 10 mM BAPTA with 10 mM HEPES adjusted to pH 7.4). Experiments in which Ca2+ currents were recorded were also performed using chloride-free extracellular solutions. The membrane potential for the two-microelectrode voltage-clamp technique was controlled using a Warner oocyte clamp (Warner Instrument Corp.). The currents were filtered at 5 kHz (–3 dB; four-pole Bessel filter). To compensate for the lack of chloride in the extracellular solution, the headstage of the Warner amplifier was attached to a plastic pool containing a 3 M NaCl solution through a silver chloride wire connected to the bath solution using an agar bridge. The bridge contained 3% agar (wt/vol), 500 mM NMDG, and 10 mM HEPES, pH 7.4, and was threaded with a thin platinum/iridium wire to decrease high frequency impedance.
Table 1. AmCaV4 ionic selectivity solutions.
| Divalent solutions | Monovalent solutions | |
|---|---|---|
| mM | mM | |
| NMDG | 120 | 22 |
| HEPES | 20 | 20 |
| Permeant ion | 2 | 100 |
For divalent solutions, the permeant ions were Mg2+, Ca2+, or Ba2+. For monovalent solutions, the permeant ions were Li+, Na+, or K+. The pH was adjusted to 7.4 using methane-thiosulfonic acid. The osmolarity of all solutions was 245–260 mosmol.
For the cell-attached patch experiments, the vitelline membrane was removed with forceps after briefly placing an oocyte in the hyperosmotic solution (see Solutions and reagents section). Command pulses were generated and currents were recorded using pCLAMP software v10.3 and an Axopatch 200B amplifier (Molecular Devices). Patch electrodes (3–7 MΩ) fashioned from borosilicate glass (Corning 8161) were coated with HIPEC (Corning) to reduce the capacitance and noise emissions. Single-channel currents were filtered at 2 KHz and were sampled at 100 kHz. Traces were low-pass filtered at 1.5 kHz using a digital filter integrated in the Clampfit software. Capacitance transients were eliminated by averaging recordings without openings and subtracting this average from all recordings.
Data analysis and statistics
The electrophysiological data were analyzed using Clampfit (pCLAMP v10.0; Molecular Devices) and custom scripts written using MATLAB (The MathWorks Inc.). The results are expressed as means ± SEM. Statistical comparisons were performed using a one-way ANOVA with Bonferroni’s post-hoc test in SigmaPlot (Systat Software). Differences were deemed significant at P < 0.05. The p-values and number of measurements are indicated in the text or figure legends.
Sequence analysis
AmCaV4 sequence was scanned with HMMER3 using hidden Markov model (HMM) profiles generated with the seed alignments available on Pfam and NCBI’s conserved domains database for the h gate motif (Finn et al., 2014; Marchler-Bauer et al., 2015). The sequence was also scanned using a custom HMM profile generated from an alignment of the α-interacting domain motif conserved in many CaV (Pragnell et al., 1994).
A sequence alignment using the previously characterized AmNav1 sequence (Gosselin-Badaroudine et al., 2015), the AmCav4 sequence, and the GenBank sequences used by Gur Barzilai et al. (2012) was performed using Clustal omega (Sievers et al., 2011). Low-quality alignment regions were removed with the TrimAl program (Capella-Gutiérrez et al., 2009). The maximum-likelihood phylogenetic tree was built using the ProML, SeqBoot, and Consense programs in the PHYLIP package.
Solutions and reagents
The ionic composition of the solutions used for electrophysiological recordings on AmCaV4 and AmCaV4/E1797K are shown in Table 1 and Table 2, respectively.
Table 2. AmCaV4/E1797K ionic selectivity solutions.
| Ringer | 0 Na+ | 0 K+ | 0 Ca2+ | 0 Mg2+ | |
|---|---|---|---|---|---|
| mM | mM | mM | mM | mM | |
| NaCl | 116 | 0 | 116 | 116 | 116 |
| KCl | 2 | 2 | 0 | 2 | 2 |
| CaCl2 | 1.8 | 1.8 | 1.8 | 0 | 1.8 |
| MgCl2 | 2 | 2 | 2 | 2 | 0 |
| HEPES | 5 | 5 | 5 | 5 | 5 |
| Choline-Cl− | 0 | 116 | 2 | 1.8 | 2 |
The pH was adjusted to 7.4 using CsOH. The osmolarity of all solutions was 245–260 mosmol.
The hyperosmotic solution used for the single-channel recordings was composed of 200 mM K-aspartate, 20 mM KCl, 1 MgC12, 10 mM EGTA, and 10 mM HEPES, pH 7.5. For the cell-attached experiments, the bath solution was composed of 100 mM K-aspartate, 50 mM KCl, 1.5 mM CaCl2, 1 mM MgCl2, 10 mM glucose, and 10 mM HEPES, pH 7.4. The pipette solution was composed of 107 mM CaCl2 and 10 mM HEPES, pH 7.4.
The OR3 solution was composed of a 1:2 dilution of Leibovitz’s L-15 medium supplemented with 15 mM HEPES and 50 mg/ml gentamycin. The pH was adjusted to 7.6 at 22°C using 1 M NaOH.
The OR2 solution was composed of 82.5 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, and 5 mM HEPES. The pH was adjusted to 7.6 at 22°C using 1 M NaOH.
All the chemicals and drugs were from Sigma-Aldrich, except for tetrodotoxin (TTX), which was purchased from Latoxan, and Leibovitz’s L-15 medium, which was purchased from Invitrogen in powder form. All the experiments were performed at 22°C.
Online supplemental material
Fig. S1 shows that AmCaV4 is most likely a member of the channel family identified as NaV2 by Liebeskind et al. (2011). Fig. S2 shows that different chelating solutions similarly abolished contamination of the Ca2+ current by endogenous Ca2+-activated chloride currents. Fig. S3 shows that the honeybee’s TipE, TEH4, and CaVβ subunits have no effect on the expression levels, voltage dependence of activation and inactivation, activation and inactivation kinetics, or recovery from inactivation of AmCaV4. Tables S1 and S2 feature the fit parameters for the curves displayed in Fig. S3. Table S3 describes the ionic composition of the solutions used to measure the anomalous mole fraction effect. Online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201611614/DC1.
RESULTS
Cloning and sequence analysis of AmCaV4
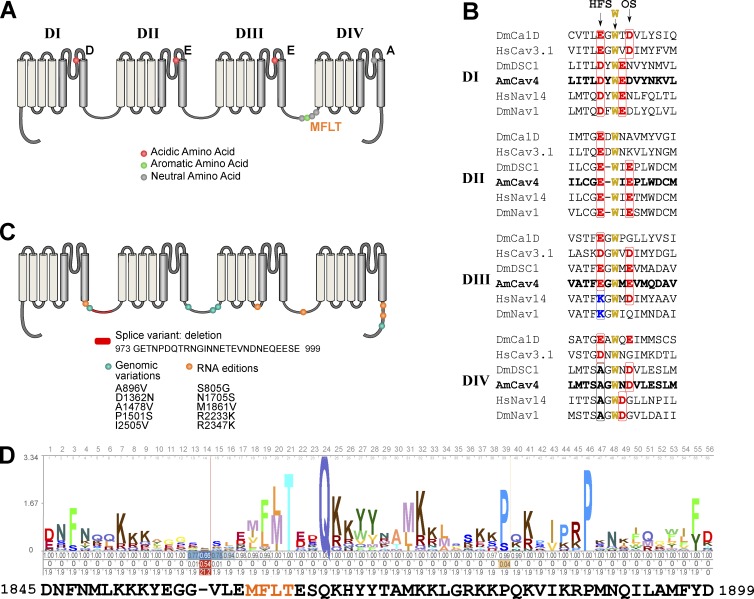
AmCaV4 was first identified based on a blast query comparing the Drosophila DSC1 sequence to the putative proteome identified by the honeybee sequencing consortium (Honeybee Genome Sequencing Consortium, 2006). The AmCaV4 gene was isolated and sequenced using genomic DNA extracted for whole honeybees and RNA extracted from honeybee heads. This led to the identification of a 2,120–amino acid protein with sequence homology to other 24-TM channels. The sequencing also revealed the presence of a splice variant, five amino acid changes caused by RNA editing, and five amino acid changes caused by genomic variations (Fig. 1). RNA editing sites were identified based on cDNA sequence variations that were not detected in the genomic DNA. Moreover, each RNA editing site was validated using at least three independent sequencing essays that were performed using at least two different oligonucleotide primer sets.
Figure 1.
Sequence analysis of AmCaV4. (A) 2D representation of the motifs in the AmCaV4 channel, including the inactivation gate, the amino acid required for voltage-sensitive domains, and the selectivity filter. (B) Alignment of the sequences contributing to the selectivity of CaV and NaV channels. The primary selectivity sequence, also often named high field site (HFS), and the outer site of the selectivity filter (OS) is shown in bold. The highly conserved tryptophan and the residues of the outer selectivity filter (which contribute to TTX binding and pore selectivity) are also shown in bold. (C) Sequence variations for AmCaV4. (D) The HMM profile calculated based on the alignment of the 72 sequences available in the National Center for Biotechnology Information’s conserved domain database for the inactivation gate of the voltage-gated sodium channel α subunits domain (cd13433) compared with the sequence identified in our AmCaV4 amino acid sequence using hmmscan.
Based on conserved domain searches and multiple sequence alignments, AmCaV4 displays the main features of traditional 24-TM channels. It is composed of four homologous domains, each containing six TM segments (Fig. 1). The fourth segment (S4) of each domain contains several positively charged residues that are conserved in numerous ion channels of different organisms. AmCaV4 also features a 56–amino acid motif with MFLT (Met-Phe-Leu-Thr) as the inactivation particle in the cytoplasmic loop between the third and fourth domain. Such a motif is usually found in NaV channels and corresponds to the hydrophobic inactivation gate. Most calcium channels feature a calcium channel β subunit interaction site. This α-interacting domain was not present in AmCaV4. AmCaV4 possesses the D-E-E-A selectivity sequence found in several bilaterian organisms, but not in mammals. Although different from the Na+-selective D-E-K-A and D-K-E-A sequences, this D-E-E-A selectivity sequence would have been a feature of the proposed ancestral bilaterian Na+ channel when it branched from Ca2+ channels (Liebeskind et al., 2011). The AmCaV4 sequence yielded a better bit-score when compared with an HMM model built from an alignment of known DSC1 homologues than it did for HMM models based on sequence alignments of NaV1 or CaV channels (bit scores of 2,759.5, 1,547.7, and 836, respectively). Thus, it is not surprising that according to the phylogenetic tree, AmCaV4 was member of the NaV2 subfamily proposed by Liebeskind et al. (2011); Fig. S1). Therefore, AmCaV4 most likely belongs to the DSC1 homologues family.
Tissue expression
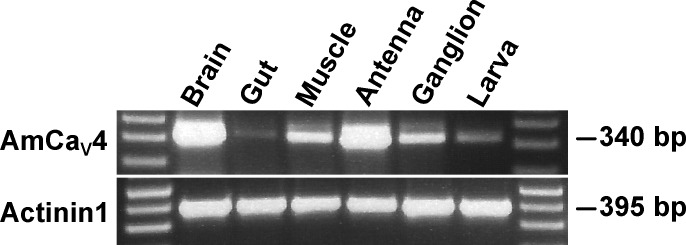
RT-PCR experiments showed that the RNA transcripts corresponding to the AmCaV4 channel are found in several tissues (Fig. 2). RNA expression levels seem higher in the brain and antennas, whereas RNA expression levels appeared much lower in the gut (Fig. 2). Actinin1 was used as positive control.
Figure 2.
Tissue expression of CaV4 in the honeybee. The tissue-specific expression of AmCaV4 was assessed by RT-PCR. All the honeybee tissue samples were processed using the same preparation steps, from dissection to gel electrophoresis. The expected weights of the amplicons are given in base pairs. Actinin1 was used as a positive control.
Functional expression of AmCaV4
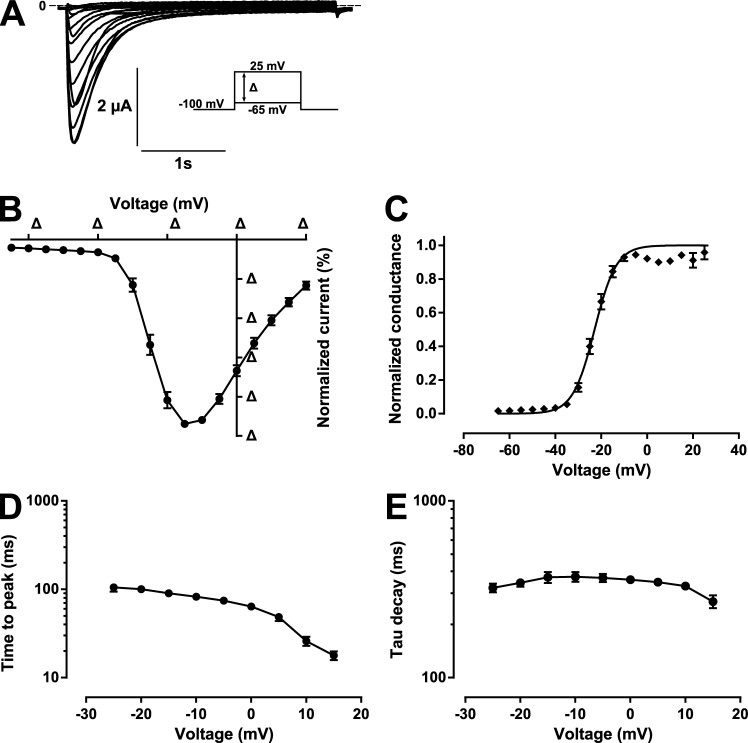
When injected with sense mRNA corresponding to the full-length AmCaV4, oocytes elicited robust inward currents upon depolarizations. The nature of the selectivity filter sequence prompted us to assess the biophysical properties of the channel in solutions where the only permeant ion was Ca2+. The oocytes expressing AmCaV4 displayed robust inward Ca2+ currents characterized by their slow kinetics (Fig. 3 A).
Figure 3.
Kinetics of the Ca2+ currents generated by AmCaV4 in response to depolarizing pulses. (A) Representative current traces from the pulse protocols applied to oocytes expressing AmCaV4. The pulse protocols consisted of imposing –65 mV to 25 mV voltage steps in 5-mV increments. The oocytes were kept at the holding potential (−100 mV) for 30 s between the voltage steps. (B) Normalized current–voltage (I-V) curve recorded in response to the protocol described in A. The curve recorded for each oocyte was normalized to its own peak current (n = 24). (C) The conductance–voltage curve calculated from the I-V curve in B fits a Boltzmann equation with the following parameters: V1/2 = −22 ± 1 mV and k = −4.5 ± 0.7 mV. The peak current measured at each potential for each oocyte was divided by (V − Vrev), where V is the test potential and Vrev is the reversal potential of the oocyte. (D) Time in milliseconds between the start of the stimulation and the peak current recorded for each test pulse. (E) Time constants of current decay. The protocol in A was used to generate transient sodium currents at different voltages. The decay of the transient current was fitted with a single exponential. The time constant of the fitted function was plotted as a function of the voltage imposed. Data are expressed as means ± SEM.
The nature of the current investigated here prompted us to confirm that our measurements were not contaminated by endogenous Ca2+-activated Cl− currents. Indeed, in recordings where no chelating solution was used, two distinct kinetics could be recorded (Fig. S2 A). Furthermore, we tested intracellular injection of 50 nl of chelating agents (either 10 mM EGTA, 10 mM EDTA, or 1 mM BAPTA) before the experiment to inhibit Ca2+ activation of Cl− channels. When chelating solutions were used, only one kinetic could be recorded (Fig. S2 B). Moreover, the voltage dependence of activation fit parameters was not dependent on the chelator used (either EGTA, EDTA, or BAPTA; Fig. S2, C and D). This indicates that endogenous Cl− current contamination was abolished by the use of any of these chelators. Because the use of EDTA and BAPTA seemed to yield slightly more leaky oocytes, we choose to perform all subsequent experiments using EGTA as a Ca2+ chelating agent.
A current–voltage (I-V) curve was generated by plotting the maximal Ca2+ current recorded as a function of the voltage imposed (Fig. 3 B) from which a G–V plot could be constructed by correcting for the ionic driving force (Fig. 3 C). The voltage dependence of AmCaV4 could be fitted to a Boltzmann equation with a V1/2 of −22 mV ± 1 mV (n = 24) and a k of −4.5 mV ± 0.7 mV (n = 24). The voltage dependence of activation parameters was compatible with both NaV1 and CaV1–2 subfamilies. However, the kinetics of the currents were very slow (Fig. 3, D and E; and Table 3).
Table 3. AmCaV4 biophysical parameters.
| Parameter | Value |
|---|---|
| Activation | |
| V1/2 (mV) | −22 ± 1 (24) |
| k (mV) | −4.5 ± 0.7 (24) |
| Inactivation | |
| V1/2 (mV) | −62.3 ± 0.6 (12) |
| k (mV) | 6.2 ± 0.2 (12) |
| Recovery from inactivation | |
| τfast (ms) | 50 ± 5 (5) |
| τslow (ms) | 4,700 ± 500 (5) |
| Afast (%) | 67 ± 2 (5) |
| Aslow (%) | 33 ± 2 (5) |
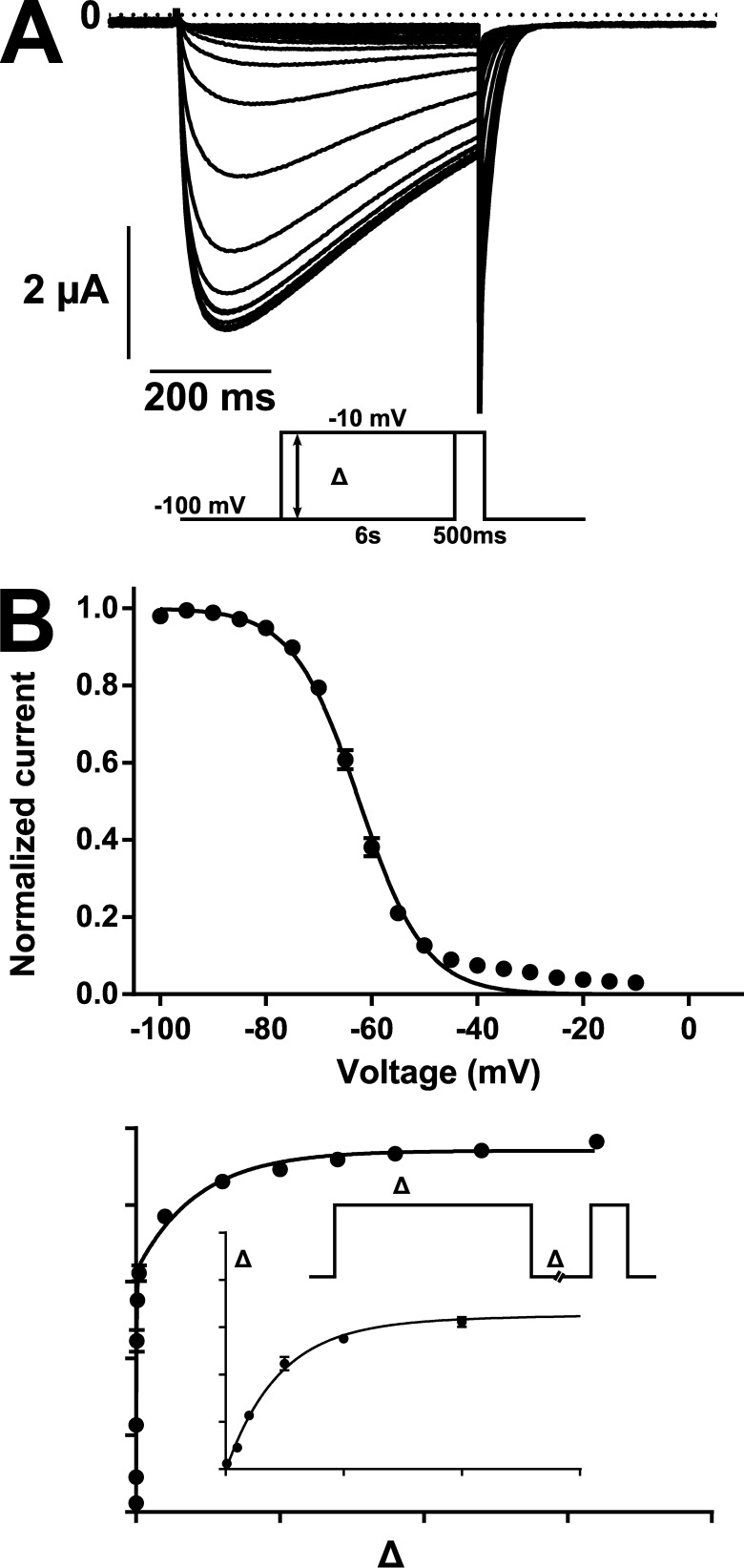
The voltage dependence of inactivation curve obtained by plotting the maximal current measured after a 6-s conditioning pulse could be fitted to a Boltzmann equation with a V1/2 of –62.3 ± 0.6 mV (n = 12) and a k of 6.2 ± 0.2 mV (n = 12; Fig. 4, A and B). Recovery from inactivation also followed slow kinetics. The peak current recorded as a function of time between depolarizations could be fitted to a two-exponential function in which the fast component represented the majority of the fit (τfast = 50 ± 5 ms and Afast = 67 ± 2%). The slow component made up the remainder of the fit (Aslow = 33 ± 2%) and had a very slow kinetic (τslow = 4.7 ± 0.5 s; Table 3).
Figure 4.
Voltage dependence of the inactivation and recovery from inactivation of AmCaV4. (A) Representative current trace recorded in response to the test pulse of the inactivation protocol given in the inset. (B, top) Voltage dependence of inactivation recorded from plotting the peak current measured with the test pulse as a function of the voltage imposed with the conditioning pulse. The current was normalized to the maximum peak current measured for each oocyte (n = 12). The voltage dependence of the inactivation of AmCaV4 fits a Boltzmann equation with the following parameters: V1/2 = −62.3 ± 0.6 mV and k = 6.2 ± 0.2 mV. The relatively poor fit between −40 and −20 mV was caused by a leak recorded in the absence of transient current was not subtracted. (Bottom) Kinetics of the recovery from inactivation of AmCaV4. The peak current measured with the test pulse normalized to the peak current measured with the conditioning pulse was plotted as a function of the time between the pulses. The data points were fitted to a two-exponential function with the following parameters: relative weight of the fast exponential = 67 ± 2%, time constant of the fast exponential = 50 ± 5 ms, and time constant of the slow exponential = 4.7 ± 00.5 s. Data are expressed as means ± SEM.
The known auxiliary subunits do not modulate AmCaV4
The TipE (temperature-induced paralysis locus E) subunit was the first NaV1 regulatory subunit to be identified in insects (Feng et al., 1995). Since then, four homologues of that protein (TEH1–4) have been reported to modulate the NaV1 channel in Drosophila and A. mellifera (Derst et al., 2006; Gosselin-Badaroudine et al., 2015). We assessed the impact of the honeybee’s TipE and TEH4 regulatory subunits on the biophysical properties of AmCaV4. We have also assessed the impact of the recently cloned honeybee CaVβb (Cens et al., 2015). Co-injection of AmCaV4 with any of those subunits had no effect on the expression levels, voltage dependence of activation and inactivation, activation and inactivation kinetics, or recovery from inactivation of AmCaV4 (Fig. S3 and Tables S1, S2, and S3).
AmCaV4 ionic selectivity
It has been postulated that the D-E-E-A motif lies midway between that of canonical NaV and CaV channels (Liebeskind et al., 2011). To investigate this possibility, we used extracellular solutions where only one ion was permeant. Normal osmolarity was maintained using NMDG.
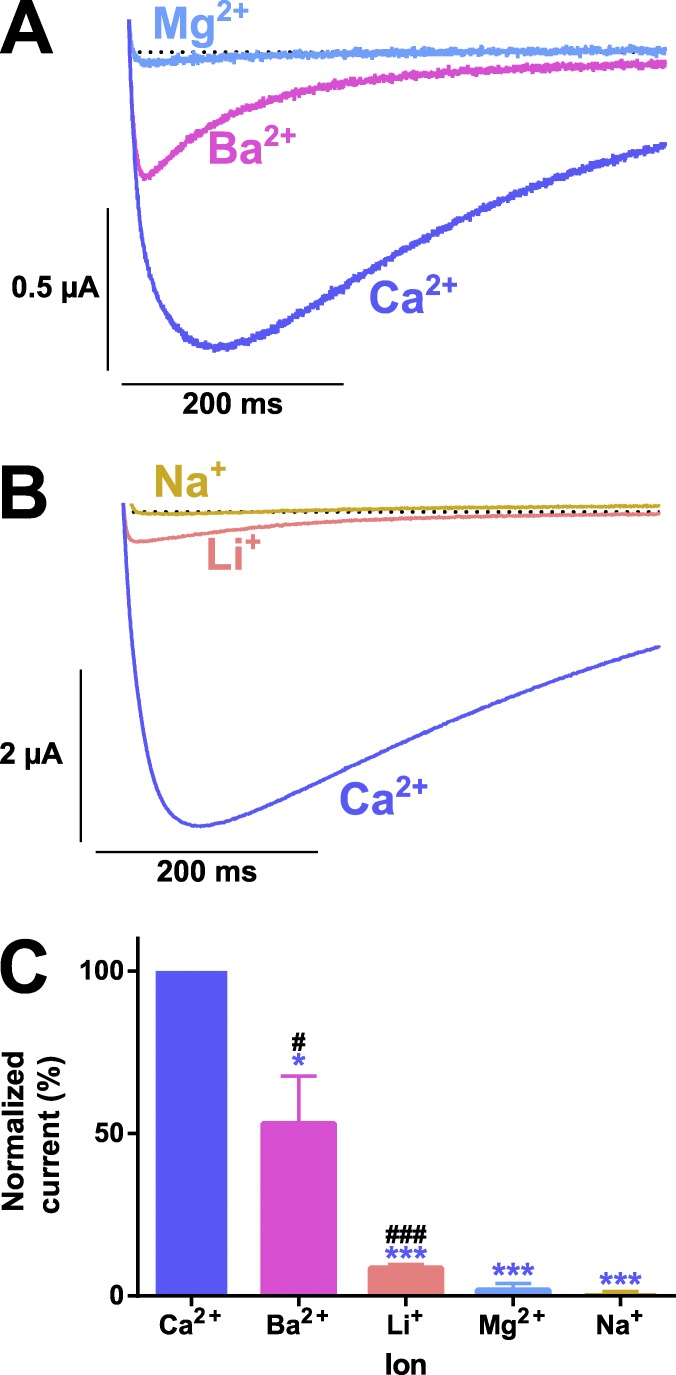
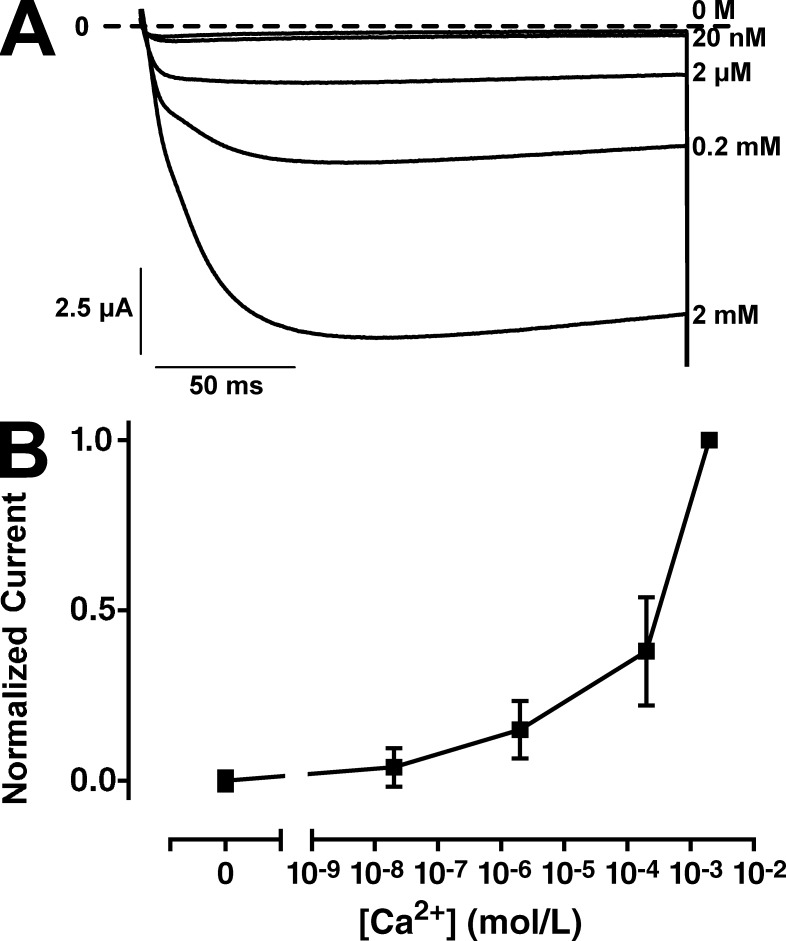
Oocytes expressing AmCaV4 displayed robust currents in the presence of Ca2+ and smaller currents in the presence of Ba2+. No currents were detected in the presence of Mg2+ (Fig. 5 A). Currents in the presence of monovalent ions displayed very small amplitudes, although they were measured in high concentration solutions (Table 1). Even at these higher concentrations, Li+ generated barely detectable currents (Fig. 5 B), and no currents were detected in Na+-containing solutions, indicating that AmCaV4 is highly selective for Ca2+ ions. This indicated that AmCaV4 would not display the anomalous mole fraction effect characteristic of other CaV channels. This effect would result in less current in the presence of both Ca2+ and Na+ than with each ion separately. Here, in the background of 100 mM Na+, reducing and removing Ca2+ from the extracellular solution resulted in decreased current amplitude until no current was detectable (Fig. 6). This confirmed that in the total absence of Ca2+, Na+ did not permeate through AmCaV4.
Figure 5.
Ionic selectivity of the wild-type AmCaV4 channel. (A) Representative current traces recorded in response to a −10-mV test pulse in extracellular solutions containing divalent ions. (B) Representative current traces recorded in response to a −10 mV test pulse in the extracellular solutions containing monovalent ions. (C) The relative permeability of the ions was calculated by dividing the peak current in a given solution by the peak current in the Ca2+ solution (n = 3). The asterisks designate values that were deemed statistically different from the relative permeation of Ca2+ determined using a one-way ANOVA (*, P < 0.05; ***, P < 0.001). The number signs designate means that were deemed statistically different from 0 (no measurable current) determined using a t test (#, P < 0.05; ###, P < 0.001). All oocytes included in these series of experiments were injected with 50 nl EGTA chelating solution prior recordings. Error bars represent SEM.
Figure 6.
The AmCaV4 channel does not display the anomalous mole fraction effect. (A) Representative current traces recorded with oocytes expressing AmCaV4 in solutions with varying concentrations of free Ca2+. (B) Normalized peak current recorded as a function of free Ca2+ in the extracellular solution. For each measurement, the peak current in a given solution was divided by the peak current measured in the 2 mM Ca2+ (n = 8–10). All solutions contained 100 mM Na+. The free Ca2+ in each solution was calculated using the MaxChelator program. The composition of the solutions is available in Table S3. Error bars represent SEM.
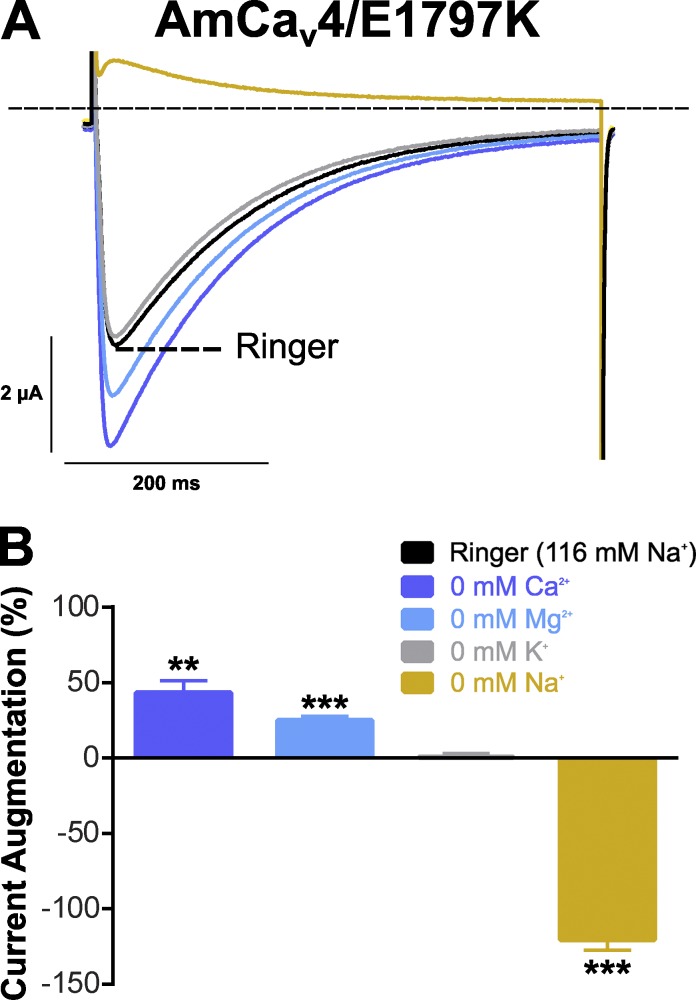
However, an AmCaV4 mutant channel with the D-E-K-A motif obtained by substituting the E1797 for a K was highly selective for Na+ ions (Fig. 7). Currents recorded in Ringer’s and modified Ringer’s solutions (Table 2) were the result of Na+ permeation, because decreasing the extracellular concentrations of other ions did not cause a decrease in current amplitude. Removing Ca2+ and Mg2+ ions only resulted in a slight increase in current amplitudes, indicating that the E1797K mutated channel can be blocked by divalent cations despite their very low affinity (in the millimolar range). Currents recorded in the absence of K+ were equivalent to those measured in normal Ringer’s solution, indicating that K+ neither blocks nor permeates the pore of the mutated channel. Removing extracellular Na+ led to outward currents, indicating that Na+ is the main component determining the current amplitude in the AmCaV4/E1797K mutant channel.
Figure 7.
Ionic selectivity of the mutated AmCaV4/E1797K channel (DEKA selectivity filter). (A) Representative current traces recorded with oocytes expressing AmCaV4/E1797K in Ringer’s and modified Ringer’s solutions. (B) Mean change in the normalized peak current after removal of a permeable ion. For each oocyte, the peak current in a given solution was divided by the peak current measured in the Ringer’s solution and subtracted by 100% (n = 5). Data are expressed as means ± SEM. Statistical differences with peak currents measured in Ringer’s solution are identified with asterisks (**, P < 0.01; ***, P < 0.001). Outward currents were measured in the absence of sodium, whereas inward currents were measured in Ringer’s solution.
Accelerated kinetics in the absence of Ca2+ permeation
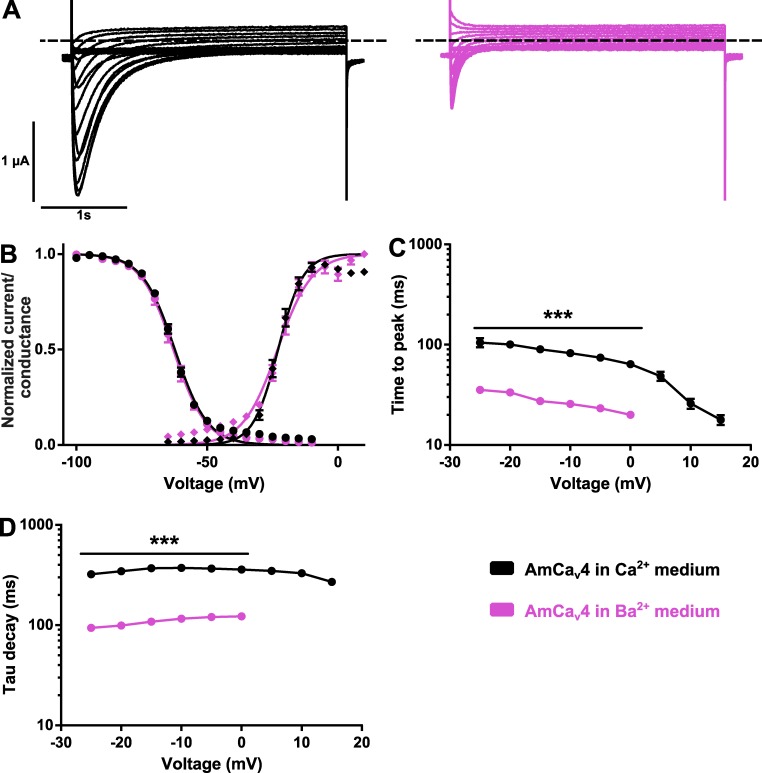
The current kinetics were accelerated in two configurations: (1) in the AmCaV4/E1797K mutation (Fig. 7 A) and (2) when Ba2+ was the permeant ion (Fig. 5 A). Currents elicited in response to depolarizing pulses with either Ca2+ or Ba2+ as permeant ion had different kinetics of activation and inactivation (Fig. 8 A). Although the voltage dependence of activation and voltage dependence of inactivation were unchanged when Ca2+ ions were removed from the extracellular medium (Fig. 8 B), the time constants characterizing the time to full activation and the delay required for inactivation of the channel were markedly altered (Fig. 8, C and D).
Figure 8.
AmCaV4 kinetics are modified in the Ba2+ solution. (A) Representative current traces recorded with oocytes expressing AmCaV4 in the Ca2+ (left) and Ba2+ (right) solutions in response to the activation protocol. (B) Voltage dependence of the activation and inactivation of AmCaV4 in the Ca2+ and Ba2+ solutions (n = 7–24 for activation, n = 7–12 for inactivation). All voltage dependence experiments were fitted with Boltzmann equations. The parameters of the fits are given in Tables S1 and S2. (C) The time to peak was significantly accelerated in the barium solution. No measurements are available for voltage steps from 5 to 15 mV because of the small current amplitudes. (D) The current decay was significantly accelerated in the barium solution. Data are expressed as means ± SEM. All oocytes included in these series of experiments were injected with 50 nl EGTA chelating solution before recordings. ***, P < 0.001.
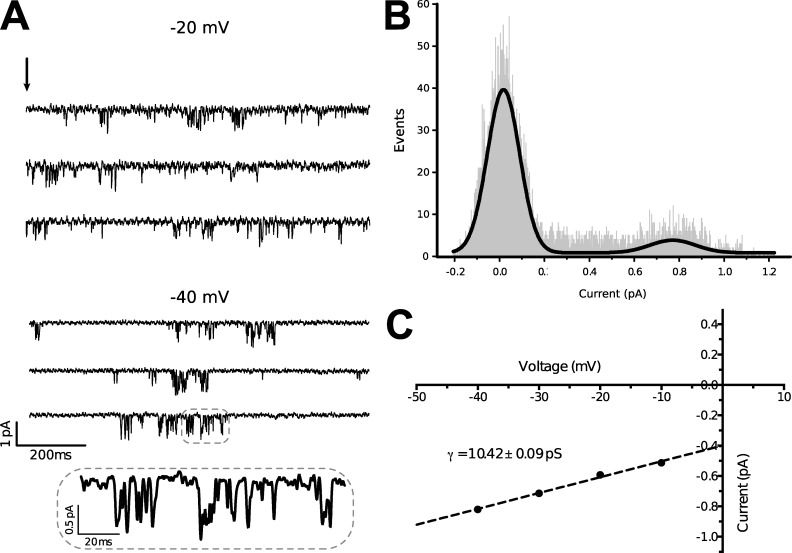
Single-channel conductance of AmCaV4
Single-channel currents were recorded for AmCaV4 from oocytes in cell-attached patches. The openings occurred in bursts (Fig. 9 A). Two-Gaussian fits on all-points histograms derived from current traces used for the amplitude measurements at −40 mV were coherent with the mean current amplitudes recorded for this voltage (Fig. 9 B). The mean current amplitudes and the standard error on these measures were then used to plot the current–voltage curve. Linear fit of those data points then yielded a single-channel conductance of 10.42 ± 0.09 pS (n = 4; Fig. 9 C).
Figure 9.
AmCaV4 single-channel conductance. (A) Single-channel traces from the cell-attached oocyte patch are illustrated for −20 and −40 mV. The arrow indicatea the onset of patch depolarization from less than −100 mV to the indicated voltage for 1 s. (B) All-points histogram of the single-channel current amplitudes at −40 mV. The smooth curve is a Gaussian fit with amplitude peaks at 0.77 pA. (C) Plot of the current–voltage relationship (n = 10–16). The straight line is a linear regression yielding a single-channel conductance of 10.42 ± 0.09 pS. Data are presented as mean ± SEM.
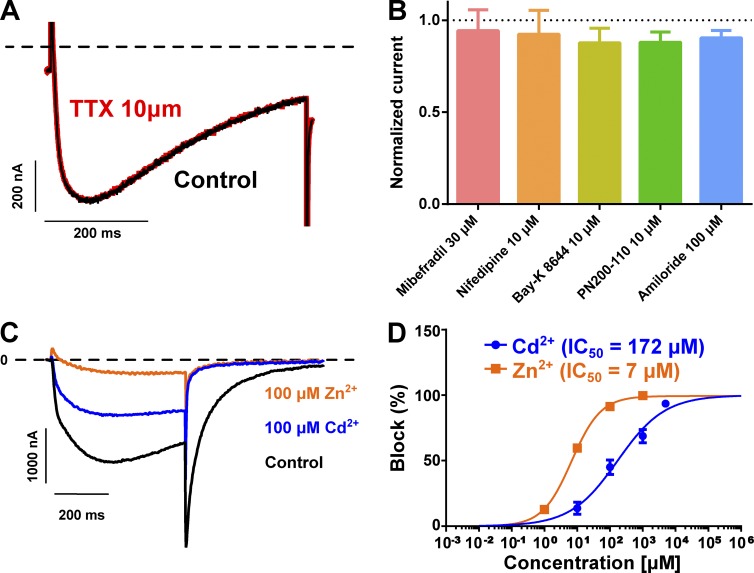
Pharmacological properties of the AmCaV4 channel
TTX block is a characteristic shared by several NaV channels. The application of TTX at concentrations as high as 10 µM did not block AmCaV4 (block = 0.3 ± 0.3%; n = 3; Fig. 10 A). Pharmacological agents known to block CaV1–3, such as mibefradil and nifedipine, did not block AmCaV4 (Fig. 10 B). Furthermore, AmCaV4 was not affected by amiloride, Bay K8644, or PN 200–110 (Fig. 10 B). However, it was blocked by micromolar concentrations of Cd2+, a divalent ion known to block both CaV and TTX-resistant NaV channels (Dascal et al., 1986; Carbonneau et al., 2002). AmCaV4 was also blocked by micromolar concentrations of Zn2+, a divalent cation known to block CaV channels (Fig. 10, C and D; Büsselberg et al., 1994).
Figure 10.
Pharmacology of AmCaV4. (A) Representative current traces recorded with an oocyte expressing AmCaV4 in response to a shortened activation protocol. AmCaV4 current was not inhibited by 10 µM TTX. (B) Known blockers and agonists of CaV channels had no effect on AmCaV4. (C) Representative current traces recorded with an oocyte expressing AmCaV4 in response to a shortened activation protocol in the Ca2+ solution. After recording the original current level, the oocyte chamber was perfused with the Ca2+ solution supplemented with 100 µM cadmium and then with the Ca2+ solution supplemented with 100 µM zinc. (D) Average block of AmCaV4 after the addition of different concentrations of cadmium and zinc to the extracellular solution (n = 3). The cumulative dose–response experiments were conducted separately for cadmium and zinc. Data are expressed as means ± SEM.
DISCUSSION
The aim of the present study was to investigate the biophysical and pharmacological properties of the AmCaV4 channel. Like other 24-TM channels, AmCaV4 has four domains, each containing six TM segments. It features a D-E-E-A selectivity filter and an MFLT inactivation particle in the DIII–DIV linker. AmCaV4 does not feature the α-interacting domain conserved in most CaV channels. This domain is known to interact with the CaV β subunits. Indeed, co-injection of AmCaV4 and AmCaVβ, an auxiliary subunit that we have previously cloned and shown to regulate A. mellifera calcium currents (Cens et al., 2015), had no effect on the expression levels, voltage dependence of activation, or voltage-dependent inactivation (Fig. S3). This indicates that AmCaV4 may not interact with the same regulatory subunits as the other CaV channels. The biophysical properties of AmCaV4 in response to activation, inactivation, and recovery from inactivation protocols were determined. No modulations were observed when either TipE or TEH4 were coexpressed with AmCaV4 (Fig. S3). The application of 10 µM TTX or 10 µM permethrin (unpublished data), which are known modulators of the NaV1 channel family, did not affect the function of the AmCaV4 channel (Fig. 10).
In addition, we determined that Ca2+ is the main permeant ion in physiological conditions. Similar results were obtained during investigations on the DSC1 channel and its homologues found in B. germanica and Nematostella vectensis (Zhou et al., 2004; Zhang et al., 2011; Gur Barzilai et al., 2012). However, unlike channels such as DSC1 and homologue channels found in B. germanica and N. vectensis, AmCaV4 is not permeable to Na+.
Our investigation also revealed that Ba2+ and Li+ permeated AmCaV4, whereas Mg2+ did not. The mutation of the selectivity sequence from D-E-E-A to D-E-K-A had a dramatic effect on channel selectivity. As expected, Na+ was the main permeant ion of the AmCaV4/E1797K mutated channel, whereas K+ had little effect on channel currents. Ca2+ and Mg2+ blocked the channel in the millimolar range, indicating that a single amino acid residue confers Ca2+ selectivity in AmCaV4.
Interestingly, the current decay kinetics of AmCaV4 appeared to be modulated by Ca2+ because they were much slower in the presence of Ca2+ (τ−10 mV = 372 ± 24 ms; n = 24) than in the presence of Ba2+ (τ−10 mV = 116 ± 2 ms; n = 7) or Li+ (τ−10 mV = 154 ± 9 ms; n = 3; Figs. 5 and 8). Similar observations on enhanced inactivation kinetics in the presence of Ba2+ have been reported in cockroach neuron (Grolleau and Lapied, 1996). Thus, the effect of Ca2+ on AmCaV4 appeared to be opposite to the effect on other CaV channels, because Ca2+ typically enhances the inactivation process via a Ca2+-dependent inactivation mechanism (Budde et al., 2002). Furthermore, the AmCaV4/E1797K mutated channel also displayed fast kinetics (τ−10 mV = 185 ± 12 ms; n = 5) of inactivation (Fig. 7). This indicates that slow inactivation kinetics may be coupled to Ca2+ permeation, as inactivation kinetics for the AmCaV4/E1797K mutated channel were recorded in the presence of extracellular Ca2+.
Here, we report higher permeability of Ca2+ than Ba2+. Previous studies on DSC1 and its B. germanica orthologue report the opposite (Zhou et al., 2004; Zhang et al., 2011). Differences in the kinetics recorded with Ba2+ and Ca2+ solutions may explain this discrepancy. Indeed, both Zhou et al. (2004) and Zhang et al. (2011) reported higher permeability for Ba2+ than Ca2+ using 15–40-ms pulses to study the properties of the channel. Because we report faster kinetics in the presence of Ba2+, it is possible that Zhou et al. (2004) and Zhang et al. (2011) underestimated the maximum amplitude of the Ca2+ current. In our hands, time to full activation at −10 mV in the presence of Ba2+ was 26 ± 1 ms (n = 7), whereas it was 82 ± 4 ms (n = 24) in Ca2+. The Ba2+ permeability represented 53 ± 15% (n = 3) of calcium permeability.
Furthermore, the single-channel conductance of AmCaV4 was determined. A single-channel conductance measured of 10.4 pS is close to the values reported for the members of the CaV3 subfamily, which range from 7.3 to 11 pS (Catterall et al., 2005b). On the other hand, the single-channel conductance reported for NaV family members is in the 20–25 pS range (Catterall et al., 2005a).
AmCaV4 was not responsive to TTX, a selective blocker of NaV channels. It was also not responsive to blockers of CaV (mibefradil and nifedipine) or to agonists of CaV (Bay K8644 and PN 200–110). AmCaV4 was, however, blocked by micromolar concentrations of Zn2+ and Cd2+. Those divalent ions are known to block both CaV and NaV channels.
Taken together, our results provide a clear basis for classifying AmCaV4 as a member of the CaV family. Therefore, it would also support renaming the family comprising DSC1 and its homologue to CaV4. Indeed, based on the consensus regarding ion channel nomenclature conventions, the name of an individual channel (a) consists of the chemical symbol of the principal permeating ion; (b) the principal physiological regulator should be indicated as a subscript; and (c) the number after the subscript indicates the gene subfamily (Catterall et al., 2005a,b). According to these conventions, the present study, and other published results, we propose that the subfamily of 24-TM channels identified as DSC1 homologues or NaV2 should be renamed CaV4 (Zhou et al., 2004; Zhang et al., 2011; Gur Barzilai et al., 2012). CaV4 channels that feature the DKEA selectivity sequence would be a special case, as they are mainly Na+ selective (Gur Barzilai et al., 2012). However, one should keep in mind that such channels are a subgroup of the much larger protein subfamily that does converge toward Ca2+ selectivity. This first thorough characterization of a CaV4 may help in determining their physiological role.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Karine Drapeau, Valérie Pouliot, and Hugo Poulin for their technical assistance.
This study was funded by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada. This work was also supported by Agence Nationale de la Recherche (grant bee-channel N° ANR-13-BSV7-0010-03). P. Gosselin-Badaroudine is the recipient of a Natural Sciences and Engineering Research Council Postgraduate Scholarship.
The authors declare no competing financial interests.
Richard W. Aldrich served as editor.
Footnotes
Abbreviations used:
- HMM
- hidden Markov model
- TM
- transmembrane
- TTX
- tetrodotoxin
References
- Budde T., Meuth S., and Pape H.-C.. 2002. Calcium-dependent inactivation of neuronal calcium channels. Nat. Rev. Neurosci. 3:873–883. 10.1038/nrn959 [DOI] [PubMed] [Google Scholar]
- Büsselberg D., Pekel M., Michael D., and Platt B.. 1994. Mercury (Hg2+) and zinc (Zn2+): two divalent cations with different actions on voltage-activated calcium channel currents. Cell. Mol. Neurobiol. 14:675–687. 10.1007/BF02088676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S., Silla-Martínez J.M., and Gabaldón T.. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25:1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonneau E., Vijayaragavan K., and Chahine M.. 2002. A tryptophan residue (W736) in the amino-terminus of the P-segment of domain II is involved in pore formation in Na(v)1.4 voltage-gated sodium channels. Pflugers Arch. 445:18–24. 10.1007/s00424-002-0887-9 [DOI] [PubMed] [Google Scholar]
- Catterall W.A., Goldin A.L., and Waxman S.G.. 2005a International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol. Rev. 57:397–409. 10.1124/pr.57.4.4 [DOI] [PubMed] [Google Scholar]
- Catterall W.A., Perez-Reyes E., Snutch T.P., and Striessnig J.. 2005b International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol. Rev. 57:411–425. 10.1124/pr.57.4.5 [DOI] [PubMed] [Google Scholar]
- Cens T., Rousset M., Collet C., Charreton M., Garnery L., Le Conte Y., Chahine M., Sandoz J.C., and Charnet P.. 2015. Molecular characterization and functional expression of the Apis mellifera voltage-dependent Ca2+ channels. Insect Biochem. Mol. Biol. 58:12–27. 10.1016/j.ibmb.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Chen L.-Q., Chahine M., Kallen R.G., Barchi R.L., and Horn R.. 1992. Chimeric study of sodium channels from rat skeletal and cardiac muscle. FEBS Lett. 309:253–257. 10.1016/0014-5793(92)80783-D [DOI] [PubMed] [Google Scholar]
- Dascal N., Snutch T., Lubbert H., Davidson N., and Lester H.. 1986. Expression and modulation of voltage-gated calcium channels after RNA injection in Xenopus oocytes. Science. 231:1147–1150. 10.1126/science.2418503 [DOI] [PubMed] [Google Scholar]
- Derst C., Walther C., Veh R.W., Wicher D., and Heinemann S.H.. 2006. Four novel sequences in Drosophila melanogaster homologous to the auxiliary Para sodium channel subunit TipE. Biochem. Biophys. Res. Commun. 339:939–948. 10.1016/j.bbrc.2005.11.096 [DOI] [PubMed] [Google Scholar]
- Feng G., Deák P., Chopra M., and Hall L.M.. 1995. Cloning and functional analysis of TipE, a novel membrane protein that enhances Drosophila para sodium channel function. Cell. 82:1001–1011. 10.1016/0092-8674(95)90279-1 [DOI] [PubMed] [Google Scholar]
- Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42(D1):D222–D230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin-Badaroudine P., Moreau A., Delemotte L., Cens T., Collet C., Rousset M., Charnet P., Klein M.L., and Chahine M.. 2015. Characterization of the honeybee AmNaV1 channel and tools to assess the toxicity of insecticides. Sci. Rep. 5:12475 10.1038/srep12475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grolleau F., and Lapied B.. 1996. Two distinct low-voltage-activated Ca2+ currents contribute to the pacemaker mechanism in cockroach dorsal unpaired median neurons. J. Neurophysiol. 76:963–976. [DOI] [PubMed] [Google Scholar]
- Gur Barzilai M., Reitzel A.M., Kraus J.E.M., Gordon D., Technau U., Gurevitz M., and Moran Y.. 2012. Convergent evolution of sodium ion selectivity in metazoan neuronal signaling. Cell Reports. 2:242–248. 10.1016/j.celrep.2012.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann S.H., Terlau H., Stühmer W., Imoto K., and Numa S.. 1992. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 356:441–443. 10.1038/356441a0 [DOI] [PubMed] [Google Scholar]
- Hille B. 2001. Ion Channels of Excitable Membranes. 3rd edition. Sinauer Associates, Sunderland. 814 pp. [Google Scholar]
- Honeybee Genome Sequencing Consortium 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 443:931–949. 10.1038/nature05260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni N.H., Yamamoto A.H., Robinson K.O., Mackay T.F.C., and Anholt R.R.H.. 2002. The DSC1 channel, encoded by the smi60E locus, contributes to odor-guided behavior in Drosophila melanogaster. Genetics. 161:1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebeskind B.J., Hillis D.M., and Zakon H.H.. 2011. Evolution of sodium channels predates the origin of nervous systems in animals. Proc. Natl. Acad. Sci. USA. 108:9154–9159. 10.1073/pnas.1106363108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I., et al. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43(D1):D222–D226. 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pragnell M., De Waard M., Mori Y., Tanabe T., Snutch T.P., and Campbell K.P.. 1994. Calcium channel beta-subunit binds to a conserved motif in the I-II cytoplasmic linker of the alpha 1-subunit. Nature. 368:67–70. 10.1038/368067a0 [DOI] [PubMed] [Google Scholar]
- Salkoff L., Butler A., Wei A., Scavarda N., Giffen K., Ifune C., Goodman R., and Mandel G.. 1987. Genomic organization and deduced amino acid sequence of a putative sodium channel gene in Drosophila. Science. 237:744–749. 10.1126/science.2441469 [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens R.F., Mehta A., Zhorov B.S., and Spafford J.D.. 2015. Alternative forms of selectivity filters and turrets, and the patterning of cysteines in extended loops provide clues to a unique extracellular domain within eukaryotic voltage-gated sodium, calcium, and NALCN channels. Front. Physiol. 6 10.3389/fphys.2015.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Liu Z., Song W., Du Y., and Dong K.. 2011. Molecular characterization and functional expression of the DSC1 channel. Insect Biochem. Mol. Biol. 41:451–458. 10.1016/j.ibmb.2011.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wang Z., Wang L., Luo N., Jiang L., Liu Z., Wu C.-F., and Dong K.. 2013. Role of the DSC1 channel in regulating neuronal excitability in Drosophila melanogaster: extending nervous system stability under stress. PLoS Genet. 9:e1003327 10.1371/journal.pgen.1003327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Chung I., Liu Z., Goldin A.L., and Dong K.. 2004. A voltage-gated calcium-selective channel encoded by a sodium channel-like gene. Neuron. 42:101–112. 10.1016/S0896-6273(04)00148-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.