Abstract
The voltage-gated proton channel (HV1) is a widely distributed, proton-specific ion channel with unique properties. Since 2006, when genes for HV1 were identified, a vast array of mutations have been generated and characterized. Accessing this potentially useful resource is hindered, however, by the sheer number of mutations and interspecies differences in amino acid numbering. This review organizes all existing information in a logical manner to allow swift identification of studies that have characterized any particular mutation. Although much can be gained from this meta-analysis, important questions about the inner workings of HV1 await future revelation.
Introduction
Voltage-gated proton channels are found in highly diverse species, from unicellular marine creatures such as dinoflagellates, diatoms, and coccolithophores (Smith et al., 2011; Taylor et al., 2011, 2012) to insects (Chaves et al., 2016), snails (Thomas and Meech, 1982; Byerly et al., 1984; Doroshenko et al., 1986), and human beings, where they are found in a variety of cells and perform many disparate functions (DeCoursey, 2013). Their unique properties (perfect H+ selectivity, ΔpH-dependent gating, extreme temperature dependence, and the ability to shift into a strikingly enhanced gating mode) are paralleled by a unique structure, the reconciliation of which is a goal of this review. Most voltage-gated ion channels are tetramers or quasi-tetramers of monomers comprising a voltage-sensing domain (VSD) S1–S4 (transmembrane [TM] segments 1–4) and a pore domain S5–S6, four of which combine to produce a single central conduction pathway. In contrast, HV1 consists of S1–S4 alone, a VSD without an explicit pore domain (Ramsey et al., 2006; Sasaki et al., 2006). In mammals and many other species (Koch et al., 2008; Lee et al., 2008; Tombola et al., 2008; Smith and DeCoursey, 2013), HV1 forms a dimer in cell membranes. However, each monomer, or protomer, has its own pore and other necessary parts and can function as a monomer (Koch et al., 2008; Tombola et al., 2008). The properties of monomeric constructs are similar in most respects to those of the dimer, but monomeric constructs open faster (Koch et al., 2008; Tombola et al., 2008; Musset et al., 2010b,c; Fujiwara et al., 2012). Several lines of evidence indicate that the two protomers comprising the HV1 dimer do not function independently, but gate cooperatively in the sense that each must undergo a voltage-dependent conformational change before either can conduct current (Gonzalez et al., 2010; Musset et al., 2010b; Tombola et al., 2010; Smith and DeCoursey, 2013).
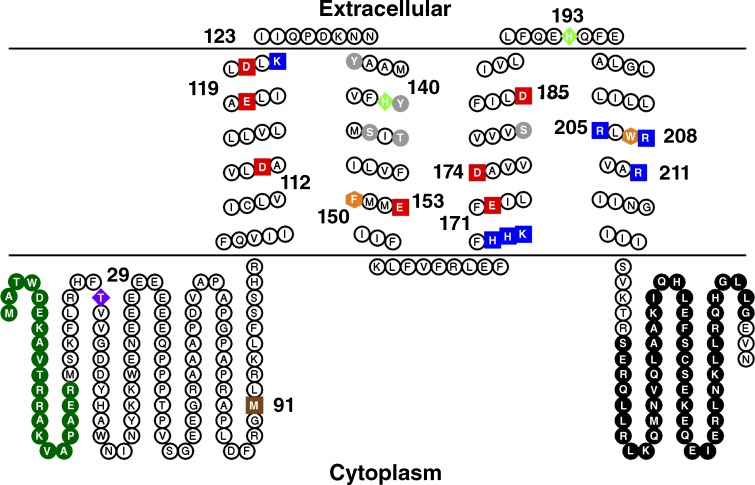
Fig. 1 illustrates schematically the entire 273–amino acid sequence of hHV1. The signature sequence that has been used successfully to identify HV1 in new species is RxWRxxR in the S4 helix (Smith et al., 2011; Rodriguez et al., 2015; Chaves et al., 2016). This sequence also identifies c15orf27 proteins, of unknown function, but these all lack Asp112 in S1, which is required for proton selectivity (Table 1). Another reported conserved motif in S2, [F,Y,W]xx[E,D]xxx[R,K], identifies some HV1 channels but is not specific to HV1, and instead identifies VSDs in general (Kang and Baker, 2016). This motif is not present in all HV1; for example, it is not present in several unicellular marine species (Taylor et al., 2011), including kHV1, which was identified by using the S4 motif (Smith et al., 2011).
Figure 1.
The amino acid sequence and schematic topology of the human voltage-gated proton channel, hHV1. Within the TM domain, acidic residues are red, basic residues are blue, aromatic residues are orange, and polar residues are gray. Specific amino acids of note, beginning at the N terminus: deletion of 1–20 (green) produces a “short” isoform common in malignant B cells (Hondares et al., 2014); Thr29 is a PKC phosphorylation site responsible for enhanced gating (Musset et al., 2010a); M91T is the first identified hHV1 mutation (Iovannisci et al., 2010); Asp112 is crucial to H+ selectivity (Musset et al., 2011); His140 and His193 coordinate Zn2+ binding (Ramsey et al., 2006); the three Arg in S4 are thought to open the conductance pathway in response to voltage (Ramsey et al., 2006; Sasaki et al., 2006; Gonzalez et al., 2013); and the C terminus has an extensive coiled-coil region (black) that holds the dimer together (Koch et al., 2008; Lee et al., 2008; Tombola et al., 2008; Fujiwara et al., 2014). The image was drawn with TOPO2 (Johns, 2016).
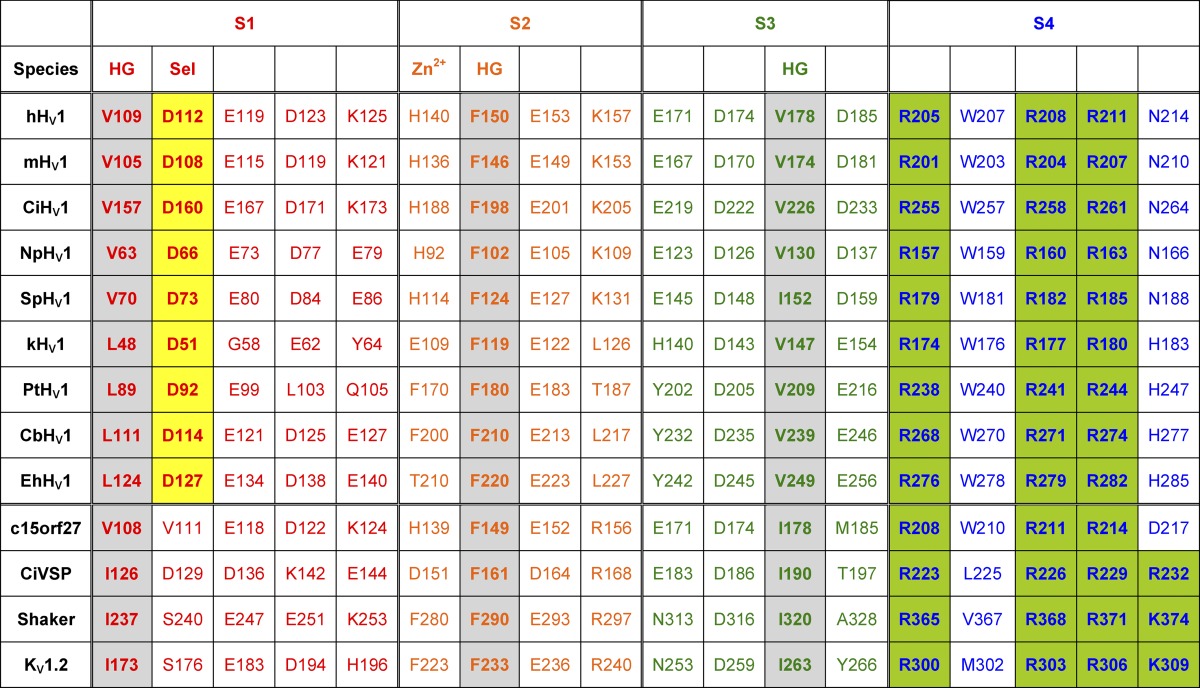
Table 1.
Numerical key to HV1 in species verified by heterologous expression
Species are indicated by one- or two-letter abbreviations: h, human; m, mouse; Ci, Ciona intestinalis; Np, Nicoletia phytophila; Sp, Strongylocentrotus purpuratus; k, Karlodinium veneficum; Pt, Phaeodactylum tricornutum; Cb, Coccolithus braarudii; Eh, Emiliania huxleyi. HG, hydrophobic gasket residues (gray); Sel, selectivity filter (yellow). Zn2+, one of the two Zn2+-binding His (in some species). Green-shaded residues sense voltage. Okamura et al. (2015) propose a slightly different hydrophobic plug based on the mHV1 crystal structure: F146, M147, L150, and F178. Several alignments of the S4 helix have been produced, which result in shifts in the register of the basic residues. Kv1.2 is human, NCBI Reference Sequence accession no. NP_004965.1; Shaker is UniProt/Swiss-Prot accession no. P08510.3.
Many reviews describe in detail the biological functions proposed for HV1 (Eder and DeCoursey, 2001; DeCoursey, 2010, 2012, 2013; Capasso et al., 2011; Demaurex, 2012; Fischer, 2012; Lishko et al., 2012; Taylor et al., 2012; Smith and DeCoursey, 2013; DeCoursey and Hosler, 2014; Seredenina et al., 2015). Here we will summarize a few aspects of functions that are relevant to the analysis that follows. The main function of HV1 in most cells is acid extrusion, although the specific consequences in each cell vary drastically. For example, HV1-mediated acid extrusion triggers capacitation in human sperm (Lishko et al., 2010), enables histamine release by basophils (Musset et al., 2008b), and exacerbates several cancers (Wang et al., 2012, 2013; Hondares et al., 2014). Acid extrusion requires extreme proton selectivity because the concentration of H+ in biological solutions is a million-fold lower than that of other major ions. HV1 are brilliantly designed and extremely efficient acid extrusion devices, changing pHi at least an order of magnitude faster than other H+ exporters (DeCoursey and Cherny, 1994), mainly because of their unique ΔpH-dependent gating mechanism (Cherny et al., 1995), which is discussed in detail below in the section Table entries defined. A second type of function of HV1 in many cells reflects the electrical consequences of the charge movement that occurs during H+ extrusion. For example, in phagocytes and certain other cells, H+ efflux serves to compensate electrically for the electron extrusion that occurs as a direct consequence of the electrogenic activity of nicotinamide adenine dinucleotide phosphate (NADPH; reduced form) oxidase and related NOX (cytochrome subunit of NADPH oxidase) isoforms (Henderson et al., 1987, 1988; DeCoursey et al., 2003). In dinoflagellates, HV1 are thought to mediate the action potential that triggers the flash in bioluminescent species (Smith et al., 2011; Taylor et al., 2012).
The respiratory burst of phagocytes, which can be elicited by pathogenic microbes, chemotactic peptides, or the phorbol ester PMA, is the manifestation of NADPH oxidase activation. During the respiratory burst, HV1 properties undergo a drastic transformation (Bánfi et al., 1999; DeCoursey et al., 2000; Murphy and DeCoursey, 2006; DeCoursey, 2016), resulting in a much higher level of activity in what has been called the enhanced gating mode (DeCoursey, 2003b). The bulk of evidence indicates that enhanced gating results from phosphorylation of HV1 (Morgan et al., 2007, Musset et al., 2010a; DeCoursey, 2016); mutations to putative phosphorylation sites are listed in Table 2.
Table 2. Changes in HV1 properties in N-terminal (1–100) mutants versus WT channels .
| Mutant | Species | Expr. system | I? | τ act | τ tail | ΔV thr | ΔpH slope | Selectivity | Other | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| hHV1S | LK35.2 wc | Yes | 2.5 | 0.75 | 5.4 | 43.2 | H+ | More profound enhanced gating | Hondares et al., 2014 | |
| T9A | hHV1S | LK35.2 pp | Yes | 0.28 | 0.9 | −18.2 | Enhanced gating lost | Hondares et al., 2014 | ||
| S77A | hHV1S | LK35.2 pp | Yes | 1.75 | 1.1 | −3.6 | nc | Hondares et al., 2014 | ||
| T9A/S77A | hHV1S | LK35.2 pp | Yes | 0.63 | 0.8 | −5.8 | Enhanced gating lost | Hondares et al., 2014 | ||
| T29A | hHV1 | LK35.2 pp | Yes | 0.23 | 1.8 | −27.6 | Enhanced gating lost | Musset et al., 2010a | ||
| T29D | hHV1 | LK35.2 pp | Yes | 0.41 | 1.0 | −12.5 | Enhanced gating lost | Musset et al., 2010a | ||
| M91T | hHV1 | COS wc | Yes | 20 | 47 | First identified naturally occurring hHV1 mutation | Iovannisci et al., 2010 | |||
| S97A | hHV1 | LK35.2 pp | Yes | 0.96 | 1.2 | 3.9 | nc | Musset et al., 2010a | ||
| S97D | hHV1 | LK35.2 pp | Yes | 0.34 | 0.9 | −3.0 | nc | Musset et al., 2010a | ||
| T29A/S97A | hHV1 | LK35.2 pp | Yes | 0.25 | 1.7 | −19.4 | Enhanced gating lost | Musset et al., 2010a | ||
| S98A | hHV1 | HEK wc | Yes | −7 | Ramsey et al., 2010 | |||||
| H99A | hHV1 | HEK wc | Yes | 13 | Ramsey et al., 2010 | |||||
| R100A | hHV1 | HEK wc | Yes | −7 | Ramsey et al., 2010 | |||||
| ΔN (1–96 deleted) | hHV1 | HEK wc | Yes | 15 | 40 | Ramsey et al., 2010 | ||||
| ΔN/ΔC | mHV1 (1–77 deleted, V216stop) | HEK wc | Yes | 0.20 | nc | nc | Loss of dimer formation | Koch et al., 2008 | ||
| ΔN/ΔC | mHV1 (1–77 deleted, V216stop) | HEK i-o | Yes | Weaker Zn2+ effects | Musset et al., 2010b |
That numerical entries are shown does not imply that any given change was significant. The entries for hHV1S are in a short isoform and are compared with full-length hHV1. HEK, HEK-293, HEK-293T, tsA, or HM1; COS, COS-7; pp, perforated patch; wc, whole cell; i-o, inside-out patch configuration. Blank entries indicate that the parameter was not examined. nc, measured, but no change. Parameters are given relative to WT in each study. For I?, yes means currents are detectable. Time constants are ratios of mutant/WT. The ΔVthreshold value is the change in absolute position of the gH–V relationship versus WT. The ΔpH slope is the slope in millivolts of the relationship between Vthreshold (or other parameters reflecting the absolute position of the gH–V relationship) and Vrev or EH (which are not identical; see section Table entries defined). When C-terminal truncations (ΔC) are indicated as XNNNstop, this means STOP replaces X at position NNN; hence, position NNN and all subsequent residues are truncated, and the last position remaining is NNN-1. The mouse N-terminal deletions (ΔN) were done by replacing P78M to initiate translation at that position.
Table 1 presents the position numbers of several key amino acids in HV1 from the nine species in which the channel has been verified by electrophysiological studies in heterologous expression systems, and the corresponding positions in two closely related molecules (c15orf27 and CiVSP) as well as in two exhaustively studied K+ channels. HV1 contains two highly conserved Asp that other voltage-gated ion channels lack, Asp112 and Asp185, as well as an anomalously located Trp207. However, HV1 lack the equivalent of Glu283 of Shaker and Na+ channels, having Ser143 instead.
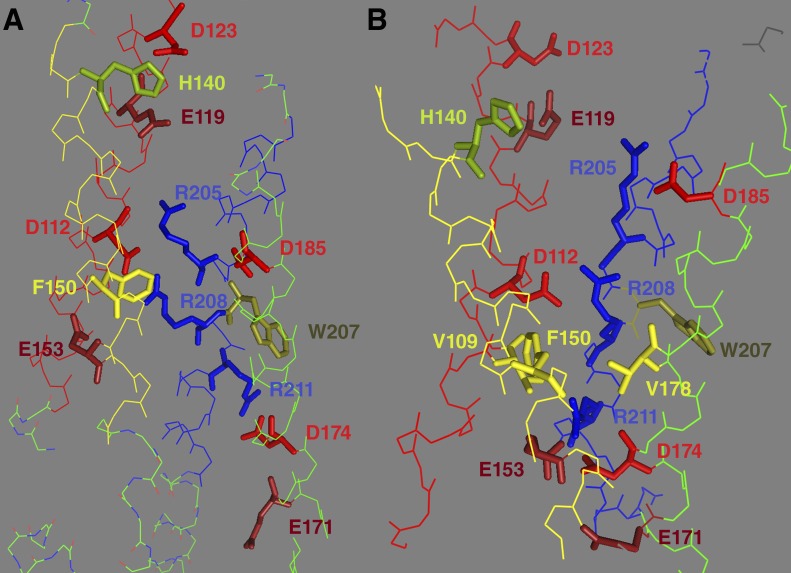
Fig. 2 shows where several key amino acids are located in the closed crystal structure of a mouse HV1 (mHV1) chimera (Fig. 2 A; Takeshita et al., 2014) and an open state model of hHV1 (Fig. 2 B; Kulleperuma et al., 2013). Numbering for both corresponds to hHV1, although the closed structure is of mHV1.
Figure 2.
Location of key amino acids. Location of some key amino acids in the crystal structure of the mHV1 chimera (Takeshita et al., 2014), labeled with hHV1 numbers (A), and in an open state model of hHV1, R2D (B; Kulleperuma et al., 2013). The channels are viewed from the side with the extracellular end at the top. An EPR study of hHV1 generally agreed with the structure of the mHV1 chimera, except in the EPR study the S2 helix (with F150 and E153) was one turn of the helix lower and S3 (with D174 and D185) was one turn higher relative to S1 and S4 (DeCoursey, 2015a; Li et al., 2015). E153 is the first amino acid replaced by the spliced-in CiVSP segment, and is actually D in the crystal. The images were produced with the PyMOL Molecular Graphics System (version 1.8; Schrödinger, LLC).
We hope that this assembly of information will in itself allow some general conclusions about structure–function relationships. Most existing data appear reasonably consistent, but in some instances, there appear to be species differences. It is unclear whether these are real or simply examples of laboratory to laboratory variation; such observations are inevitably somewhat anecdotal. We also identify examples of qualitatively different outcomes for the same mutation studied in different expression systems. In addition to listing the outcomes of mutations, the motivation for several strategies for generating mutants is discussed. The mutation studies have resulted in rapid progress in understanding how HV1 works, but many important questions remain. One word of caution must be stated: mutations are designed to test the effect of changing one or more amino acids, usually with the assumption that the rest of the protein will assemble and function exactly as the WT does. This assumption can be tested rigorously only by determining the structure of every mutant. Although this procedure became almost routine for the bacterial reaction center (Xu et al., 2004), it is not remotely possible for HV1. Nevertheless, many mutants function with little overt change beyond what might be imagined. On the other hand, mutations that alter charge may have powerful effects on structure and hence function at least locally if not globally.
Data organization and exclusions
The information in this review is organized according to the amino acid numbering of the human voltage-gated proton channel, hHV1. To make the tables more manageable, we present the mutants in numerical order (with a few exceptions whose logic may or may not become apparent) and include separate tables for the N terminus, for each TM helix (S1–S4), and for the C terminus. The boundaries of the TM helices are defined according to the electron paramagnetic resonance (EPR) study of Li et al. (2015). Double or triple mutants are listed according to the first (i.e., nearest the N terminus) position mutated (again with a few exceptions whose rationale may become apparent). The intention is to make these tables exhaustive as far as is practical, but our bias is toward electrophysiological descriptions of individual point mutants. Thus, we do not list all 109 mutants studied by Cys scanning and assessed for accessibility by PEGylation protection (Sakata et al., 2010; Kurokawa and Okamura, 2014), for example. Nor do we list all 149 positions at which Cys was introduced for EPR measurements (Li et al., 2015). These blanket mutations are interpretable mainly within the context of the entire study. We exclude most mutations examining the link between S4 and the C terminus, which involve a large variety of deletions and insertions (Fujiwara et al., 2012, 2014). Mutations to the coiled-coil region of the C terminus resulting in trimeric and tetrameric channels (Fujiwara et al., 2013a) are not discussed here. Only a fraction of a large series of Trp scanning mutants in both monomeric and dimeric constructs is listed (Okuda et al., 2016). A series of mutants and tandem constructs in which one or both of the His that bind Zn2+ (His140 and His193) were replaced (Musset et al., 2010b) is not included in the tables. A series of Cys cross-linking mutations aimed at identifying the dimer interface (Lee et al., 2008) is also omitted. Finally, we do not include domain-swap mutants, such as those of Alabi et al. (2007), in part because chimerae do not logically fit into the format of the tables.
Table entries defined
The first column lists the mutations as per the usual convention (single-letter amino acid abbreviations: WT, position counting from the N terminus, and replacement). When a study used a nonhuman species, the hHV1 equivalent is given in italics in the first column, and the actual mutation is listed in the second column. The third column gives the expression system and the voltage-clamp method used. To our knowledge, no studies exist in which different properties were observed in HEK versus COS cells (Musset et al., 2008a). However, mammalian versus amphibian studies sometimes differ. For example, D112S from three different species all expressed well and exhibited anion permeation in mammalian cells (Musset et al., 2011; Smith et al., 2011; Chaves et al., 2016), whereas currents were not observed in Xenopus laevis oocytes (Berger and Isacoff, 2011). Some proteins function better in certain expression systems: CiHV1 works well in Xenopus oocytes, whereas mHV1 does not and prefers mammalian (HEK) cells (Okuda et al., 2016). Among mammalian cells, hHV1 expressed in the B cell–related LK35.2 cell line exhibits an enhanced gating response to stimulation with PMA (Musset et al., 2010a; Hondares et al., 2014), whereas hHV1 expressed in HEK or COS cells did not respond (Musset et al., 2008a).
The fourth column (I?) simply reports whether interpretable currents were observed. A positive answer means the mutant protein is produced, reaches the plasma membrane, and functions. A negative result may have various undetermined explanations (protein misfolding, failure to traffic to the plasma membrane, disruption of gating or permeation) but is nevertheless potentially important because amino acids that play crucial roles in function may be difficult to replace without disrupting molecular function. For example, Asp112 and Arg208 form a crucial nexus that in general cannot be meddled with without altering or eliminating function. However, different laboratories may have different criteria for deciding whether small currents are “real,” small enough to be negligible, or nonexistent. This evaluation is complicated by the fact that all common mammalian expression systems (HEK, Chinese hamster ovary [CHO], and COS cells) frequently display (typically small) native voltage-gated proton currents (Cherny et al., 1997; Musset et al., 2011). To address this concern, we often introduce mutations into hHV1 in a Zn2+-insensitive background, meaning the two His primarily responsible for Zn2+ inhibition are mutated (H140A/H193A; Ramsey et al., 2006; Musset et al., 2010b). Then, if we see small currents that might be due either to native currents or to expression of a poorly conducting mutant, we add 10 µM Zn2+, which profoundly inhibits WT hHV1, but will have negligible effects on a Zn2+-insensitive mutant. This approach cannot be applied to HV1 from species lacking these His (Table 1).
The gating kinetics columns are self-evident: τact is the activation (channel opening) time constant, and τtail is the tail current or deactivation (channel closing) time constant. These are expressed as the ratio τmutant/τWT so that 1 means no change, a ratio <1 means faster than WT, and a ratio >1 means slower than WT. Because HV1 gating kinetics depends very strongly on temperature, with Q10 6–9 (DeCoursey and Cherny, 1998; Kuno et al., 2009), and is also influenced profoundly by experimental artifacts including proton depletion-induced current decay (“droop”) and pH changes, anything less than a twofold change should be viewed with skepticism.
The columns labeled ΔVthreshold (ΔVthr) and ΔpH slope embody one of the crucial and unique properties of this channel, namely its ΔpH-dependent gating. Decreasing pHi or increasing pHo shifts the gH–V relationship negatively by roughly 40 mV/U of change in pH, as originally described by Cherny et al. (1995):
| (1) |
or, generalized:
| (2) |
where Vthreshold is the most negative voltage at which detectable current can be elicited, V0 is Vthreshold at symmetrical pH (roughly 20 mV), Vslope is the steepness (in millivolts/unit pH) of the relationship (nominally 40 mV), and ΔpH = pHo − pHi. The precise value for Vslope depends on whether the abscissae are EH (the Nernst potential for H+ based on the nominal pH of the solutions) or the measured Vrev. Because Vrev often changes by less than a Nernstian amount in real experiments (almost certainly as a result of our inability to perfectly control pH, in combination with pH changes due to the measurement itself), plotting Vthreshold against the measured Vrev usually produces a larger slope. Thus, the shift of Vthreshold (vs. Vrev) for native proton currents measured in rat alveolar epithelial cells was 44 mV/U (Cherny et al., 1995). The mean shift reported in 15 types of cells was 46 mV/U (DeCoursey, 2003b). The slope for hHV1 transfected into HEK or COS cells was 39–43 mV/U (Musset et al., 2008a). There are essentially no reports of mutants in which Vslope departs convincingly from this range. Two nominal deviations from this rule are R211A (53 mV) and ΔN (28 mV; Ramsey et al., 2010), but their P-values versus WT are 0.04 and 0.02, and 2/31 values in this study could easily fall just under the arbitrary P = 0.05 cutoff by chance.
Rather than give an absolute value for the position of the gH–V relationship, such as using the parameter V0 in Eq. 2 above (which is not in common parlance), we list the change of Vthreshold (Vthr) or an equivalent parameter from control values in each study. Actual numbers will depend on conventions in each laboratory. When data were reported for asymmetrical pH, the Vthreshold value was “corrected” by shifting it by 40 mV/U change in ΔpH (Cherny et al., 1995). For reasons discussed at length elsewhere (Musset et al., 2008a), we consider it a highly questionable practice to fit whole-cell gH–V data with a Boltzmann function, as is routinely done for other voltage-gated channels. Because the distortion of current amplitudes and kinetics resulting from proton depletion are ubiquitous and profound, we prefer to quantify absolute voltage dependence by Vthreshold (the most negative voltage at which discernable time-dependent H+ currents are detected) or VgH,max/10 (the voltage at which the gH is 10% of its maximal value), both measured during small depolarizations and thus minimizing depletion. The absolute position of the gH–V relationship appears quite mutable with mutation; the extensive study of Ramsey et al. (2010) produced examples spanning >200 mV for various mutants. Given the technical difficulty and intrinsic variability of Vthreshold estimation, it would be dangerous to draw conclusions about shifts of less than ∼20 mV. There is a 30-mV range of values reported for WT hHV1 (see Table 3 in DeCoursey, 2013).
Selectivity is given only when it was explicitly evaluated. The WT channel is perfectly selective for protons, so H+ is entered. The entry Cl– means that the channel is permeable to Cl– and likely to other anions as well; Na+ means the channel is permeable to Na+ and likely other cations besides H+. The column Other simply provides concise information that does not fit elsewhere in the tables.
Table 2: The N terminus (positions 1–100 in hHV1)
N terminus
The N terminus of hHV1 comprises 100 amino acids and is intracellular. The effects of truncating the entire N terminus (ΔN) are not dramatic. Deleting both the N and C termini (ΔN/ΔC) simultaneously results in five- to sixfold faster activation, presumably because these truncations result in monomeric constructs. Although coiled-coil interactions in the C terminus are generally considered to be the main interaction that stabilizes the dimer (Koch et al., 2008; Lee et al., 2008; Tombola et al., 2008; Fujiwara et al., 2013b, 2014; Smith and DeCoursey, 2013), deleting both N and C termini (ΔN/ΔC) appeared to produce monomers more reliably (Koch et al., 2008). Nevertheless, even when both N and C termini are deleted, the VSD-only construct spontaneously dimerizes with a Kd of ∼3 µM (Li et al., 2015).
The short isoform, hHV1S
Some B cells, especially B lymphocytes from chronic lymphocytic leukemia patients or malignant B cell lines (Hondares et al., 2014), express a short isoform of hHV1 (hHV1S) that lacks the first 20 amino acids (Capasso et al., 2010). Fig. 1 shows that the 21st amino acid is Met (ATG), which acts as an alternative start site (Hondares et al., 2014). Compared with the full-length protein, hHV1S opens more slowly, and its enhanced gating response to PMA is more profound. Furthermore, hHV1S interacts less with the B cell receptor, resulting in less internalization. Together, its properties suggest that its expression may contribute to the pathogenesis of B cell malignancies (Hondares et al., 2014).
The phosphorylation site responsible for enhanced gating
When phagocytes are stimulated to undergo the respiratory burst (i.e., activation of NADPH oxidase, or NOX2), the properties of proton channels change so dramatically (Bánfi et al., 1999; DeCoursey et al., 2000; Musset et al., 2009) that at first, the appearance of a second, distinct type of proton channel (proposed to be a component of the active NADPH oxidase complex) was hypothesized (Henderson et al., 1995; Henderson and Chappell, 1996; Bánfi et al., 1999). A decade of controversy ensued (Henderson et al., 1997; Henderson, 1998; Henderson and Meech, 1999, 2002; DeCoursey et al., 2001b, 2002, 2003; Maturana et al., 2002; Touret and Grinstein, 2002; DeCoursey, 2003a,b, 2016). The idea that the gp91phox component of NADPH oxidase could function as a proton channel was not dispelled completely until well after the HVCN1 gene was identified (Ramsey et al., 2006; Sasaki et al., 2006) when the HVCN1 knockout mouse was developed, which provided the final nail in the coffin (Morgan et al., 2009; El Chemaly et al., 2010). In activated phagocytes, four characteristics of H+ currents change, all in the direction of increasing proton flux: the maximum gH increases two- to fourfold, the gH–V relationship shifts negatively by 30–40 mV, τact becomes two to five times faster, and τtail slows two- to sixfold (DeCoursey et al., 2000, 2001a,b; Cherny et al., 2001; DeCoursey, 2003a; Musset et al., 2009). This is referred to as the enhanced gating mode to emphasize that the properties of the HV1 channel change as a result of phosphorylation, as opposed to a second type of channel appearing.
The original mechanism proposed for enhanced gating of HV1 was not phosphorylation, but rather modulation of the channel by arachidonic acid generated by cPLA2α (Henderson and Chappell, 1992). Although arachidonic acid does enhance HV1 gating by a direct pharmacological effect (DeCoursey and Cherny, 1993; Kapus et al., 1994; Suszták et al., 1997; Kawanabe and Okamura, 2016), neither specific cPLA2α inhibition nor genetic knockout of cPLA2α affects the activation of NADPH oxidase or the enhanced gating of HV1 channels during the respiratory burst (Morgan et al., 2007).
Two predicted PKC phosphorylation sites, Thr29 and Ser97, were studied in hHV1 expressed in the B cell–related LK35.2 cell line (Musset et al., 2010a). Although both were detectably phosphorylated, mutation of Thr29 but not Ser97 abolished the enhanced gating response, implicating Thr29 as the key PKC phosphorylation site in hHV1 (Musset et al., 2010a). Analogous studies of the short isoform identified the same residue, Thr9, as the main phosphorylation site (Hondares et al., 2014).
The first identified human hHV1 mutation
The first naturally occurring hHV1 mutation from a human subject, M91T (Table 2), was identified by Iovannisci et al. (2010), who cloned HVCN1 genes from primary human airway tissue cultures. Unfortunately, the mutation was discovered only after the death of the donor, who consequently had no opportunity to mourn the defective nature of his/her proton channels, and we lack the opportunity to evaluate the effects of the mutation on his/her quality of life. The main effect of the mutation on HV1 expressed in COS cells is to decrease the likelihood of channel opening. It requires ∼20 mV more depolarization or ∼0.5 U of greater ΔpH (for airway epithelia, this likely means a higher pHo) to open mutant M91T channels (Iovannisci et al., 2010).
Table 3: The S1 helix (positions 101–125) and the S1–S2 linker (126–133)
Table 3. Changes in HV1 properties in S1 (101–125) and S1–S2 linker (126–133) mutants versus WT channels .
| Mutant | Species | Expr. system | I? | τ act | τ tail | ΔV thr | ΔpH slope | Selectivity | Other | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Q102C | CiHV1 [H150C] | Xenopus i-o | Yes | MTSi access | Mony et al., 2015 | |||||
| V103C | CiHV1 [V151C] | Xenopus i-o | Yes | MTSi open> closed | Mony et al., 2015 | |||||
| I105C | CiHV1 [I153C] | Xenopus i-o | Yes | MTSi open> closed | Mony et al., 2015 | |||||
| I106C | CiHV1 [I154C] | Xenopus i-o | Yes | MTSi open> closed | Mony et al., 2015 | |||||
| C107A | hHV1 | HEK wc | Yes | −17 | Ramsey et al., 2010 | |||||
| C107S | hHV1 | 90% dimer | Li et al., 2015 | |||||||
| V109C | CiHV1 [V157C] | Xenopus i-o | Yes | MTSi open> closed | Mony et al., 2015 | |||||
| V109A | hHV1 | Xenopus i-o | Yes | −11 | Hong et al., 2014 | |||||
| D112A | hHV1 | HEK wc | Yes | 59 | 38 | Ramsey et al., 2010 | ||||
| D112Aa | hHV1 | COS/HEK wc | Yes | 2.2 | 3.0 | 41b | 43 | Cl– | Musset et al., 2011 | |
| D112A | hHV1 | Vesicle flux | Slows H+ flux | Letts, 2014 | ||||||
| D112A | hHV1 | Xenopus i-o | No | Hong et al., 2014 | ||||||
| D112A | CiHV1 [D160A] | Xenopus TEVC | No | Chamberlin et al., 2015 | ||||||
| D112A | kHV1 [D51A] | COS/HEK wc | Yes | Cl– | Smith et al., 2011 | |||||
| D112A | NpHv1 [D66A] | HEK wc | Yes | Cl– | Chaves et al., 2016 | |||||
| D112C | CiHV1 [D160C] | Xenopus TEVC | No | Chamberlin et al., 2015 | ||||||
| D112C | NpHv1 [D66C] | HEK wc | No | Chaves et al., 2016 | ||||||
| D112C/R211C | CiHV1 [D160C/ R261C] | Xenopus TEVC | Yes | nc | Na+ | Chamberlin et al., 2015 | ||||
| D112N | hHV1 | HEK wc | Yes | 31 | 42 | Ramsey et al., 2010 | ||||
| D112Na | hHV1 | COS/HEK wc | Yes | 2.4 | 3.0 | 23b | 35 | Cl– | Musset et al., 2011 | |
| D112N/D185A | hHV1 | HEK wc | Yes | 103 | Ramsey et al., 2010 | |||||
| D112A | hHV1 | Xenopus i-o | No | Hong et al., 2014 | ||||||
| D112E | hHV1 | Xenopus i-o | Yes | −13 | Hong et al., 2014 | |||||
| D112E | hHV1 | COS/HEK wc | Yes | 0.18 | 7.4/.085 | −11b | 34 | H+ | Biexponential tails | Musset et al., 2011 |
| D112E | hHV1 | Xenopus i-o | Yes | −15 | Berger and Isacoff, 2011 | |||||
| D112E | kHV1 [D51E] | COS/HEK wc | Yes | H+ | Smith et al., 2011 | |||||
| D112E | NpHv1 [D66E] | HEK wc | Yes | H+ | Chaves et al., 2016 | |||||
| D112E/I127C | CiHV1 [D160A/I175C] | Xenopus i-o | Yes | nc | 4.0 | Mony et al., 2015 | ||||
| D112H | hHV1 | COS/HEK wc | Yes | 2.0 | 0.85 | 13b | 38 | Cl– | Musset et al., 2011 | |
| D112H | kHV1 [D51H] | COS/HEK wc | Yes | Cl– | Smith et al., 2011 | |||||
| D112H | NpHv1 [D66H] | HEK wc | Yes | Cl– | Chaves et al., 2016 | |||||
| D112Ka | hHV1 | COS/HEK wc | Yes | 0.8 | 0.22 | 46b | 40 | Cl– | Musset et al., 2011 | |
| D112S | hHV1 | COS/HEK wc | Yes | 2.0 | 4.1 | 25b | 38 | Cl– | Musset et al., 2011 | |
| D112S | hHV1 | Xenopus i-o | No | Berger and Isacoff, 2011 | ||||||
| D112S | hHV1 | Vesicle flux | Slows H+ flux | Letts, 2014 | ||||||
| D112S | kHV1 [D51S] | COS/HEK wc | Yes | Cl– | Smith et al., 2011 | |||||
| D112S | NpHv1 [D66S] | HEK wc | Yes | Cl– | Chaves et al., 2016 | |||||
| D112S/R211S | hHV1 | Xenopus i-o | Yes | 24 | 44 | Gu+ | At pH 8//8 | Berger and Isacoff, 2011 | ||
| D112Fa | hHV1 | COS/HEK wc | Yes | 1.6 | 0.03 | 44b | 38 | Cl– | Musset et al., 2011 | |
| D112R/R211D | hHV1 | Xenopus i-o | Yes | H+ | Berger and Isacoff, 2011 | |||||
| D112V | hHV1 | COS/HEK wc | No | Musset et al., 2011 | ||||||
| D112L | hHV1 | Vesicle flux | Slows H+ flux | Letts, 2014 | ||||||
| D112Ia | hHV1 | COS/HEK wc | No | DeCoursey, 2015b | ||||||
| D112Q | hHV1 | Xenopus i-o | No | Hong et al., 2014 | ||||||
| D112A/L108Da | hHV1 | COS/HEK wc | No | Morgan et al., 2013 | ||||||
| D112V/V109Da | hHV1 | COS/HEK wc | No | Morgan et al., 2013 | ||||||
| D112A/V109Da | hHV1 | COS/HEK wc | Yes | Cl– | Morgan et al., 2013 | |||||
| D112A/V110Da | hHV1 | COS/HEK wc | No | Morgan et al., 2013 | ||||||
| D112A/L111Da | hHV1 | COS/HEK wc | No | Morgan et al., 2013 | ||||||
| D112A/A113Da | hHV1 | COS/HEK wc | No | Morgan et al., 2013 | ||||||
| D112A/L114Da | hHV1 | COS/HEK wc | No | Morgan et al., 2013 | ||||||
| D112A/L115Da | hHV1 | COS/HEK wc | No | Morgan et al., 2013 | ||||||
| D112A/V116Da | hHV1 | COS/HEK wc | Yes | H+ | Morgan et al., 2013 | |||||
| D112V/V116Da | hHV1 | COS/HEK wc | Yes | H+ | Morgan et al., 2013 | |||||
| D112V/V116Ea | hHV1 | COS/HEK wc | Yes | H+ | Morgan et al., 2013 | |||||
| D112V/V116Sa | hHV1 | COS/HEK wc | Yes | Cl– | Morgan et al., 2013 | |||||
| D112V/V116Na | hHV1 | COS/HEK wc | Yes | Cl– | Morgan et al., 2013 | |||||
| D112A/L117Da | hHV1 | COS/HEK wc | No | Morgan et al., 2013 | ||||||
| D112A/A118Da | hHV1 | COS/HEK wc | No | Morgan et al., 2013 | ||||||
| D112N/I127C | CiHV1 [D160N/I175C] | Xenopus i-o | No | Mony et al., 2015 | ||||||
| D112N/R211S/I127C | CiHV1 [D160N/R261S/I175C] | Xenopus i-o | Yes | 15 | Mony et al., 2015 | |||||
| D112N/R211S/G199C | CiHV1 [D160N/R261S/G249C] | Xenopus i-o | Yes | 44 | Mony et al., 2015 | |||||
| D112N/G199C | CiHV1 [D160N/G249C] | Xenopus i-o | No | Mony et al., 2015 | ||||||
| A113Da | hHV1 | COS/HEK wc | Yes | H+ | Morgan et al., 2013 | |||||
| E119A | hHV1 | HEK wc | Yes | 20 | 47 | Ramsey et al., 2010 | ||||
| E119L | hHV1 | Vesicle flux | nc H+ flux | Letts, 2014 | ||||||
| E119S | mHV1 [E115S] | HEK wc | Yes | nc Zn2+ inhibition | Takeshita et al., 2014 | |||||
| E119S/D123S | mHV1 [E115S/D119S] | HEK wc | Yes | Weaker Zn2+ inhibition | Takeshita et al., 2014 | |||||
| E119A | CiHV1 [E167A] | Xenopus TEVC | Yes | 4 | Chamberlin et al., 2014 | |||||
| E119C/R205C | CiHV1 [E167C/R255C] | Xenopus TEVC | Yes | −2 | Chamberlin et al., 2014 | |||||
| E119C/R208 | CiHV1 [E167C/R258C] | Xenopus TEVC | Yes | −52 | Chamberlin et al., 2014 | |||||
| D123A | hHV1 | HEK wc | Yes | 20 | 48 | Ramsey et al., 2010 | ||||
| D123S | mHV1 [D119S] | HEK wc | Yes | nc Zn2+ inhibition | Takeshita et al., 2014 | |||||
| D123C | CiHV1 [D171C] | Xenopus i-o | Yes | MTSo open> closed | Mony et al., 2015 | |||||
| D123A | CiHV1 [D171A] | Xenopus TEVC | Yes | 72 | Chamberlin et al., 2014 | |||||
| D123A/R205N | CiHV1 [D171A/R255N] | Xenopus TEVC | Yes | 11 | Chamberlin et al., 2014 | |||||
| K125A | hHV1 | HEK wc | Yes | 19 | 47 | Ramsey et al., 2010 | ||||
| K125C | CiHV1 [K173C] | Xenopus i-o | Yes | MTSo access | Mony et al., 2015 | |||||
| I127C | CiHV1 [I175C] | Xenopus i-o | Yes | 0 | Mony et al., 2015 | |||||
| D130A | hHV1 | HEK wc | Yes | 13 | Ramsey et al., 2010 | |||||
| K131A | hHV1 | HEK wc | Yes | 33 | Ramsey et al., 2010 |
That numerical entries are shown does not imply that any given change was significant. Italicized mutant entries from nonhuman species show the hHV1 equivalent. HEK, HEK-293, HEK-293T, tsA, or HM1; COS, COS-7; Xenopus, Xenopus laevis oocyte; wc, whole cell; i-o, inside-out patch configuration; TEVC, two-electrode voltage clamp. Blank entries indicate that the parameter was not examined. nc, measured, but no change. Parameters are given relative to WT in each study. For I?, yes means currents are detectable. Time constants are ratios of mutant/WT. The ΔVthreshold value is the change in absolute position of the gH–V relationship versus WT. The ΔpH slope is the slope in millivolts of the relationship between Vthreshold (or other parameters reflecting the absolute position of the gH–V relationship) and Vrev or EH (which are not identical; see section Table entries defined). For column Other, MTS access from inside or outside (MTSi or MTSo, respectively) is listed as open>closed if the open channel was more accessible.
In an H140A/H193A (Zn2+ insensitive) background.
Previously unpublished, analyzed from data for Musset et al. (2011).
The selectivity filter, Asp112
The most intensively studied position in hHV1 is Asp112, which was implicated in proton flux (Letts, 2014) and was identified as a crucial part of the selectivity filter (Musset et al., 2011). An indication of the importance of this position is that most mutants malfunction or fail to function altogether, although with some unexplained apparent variability between species or expression systems. Mutation of Asp112 to a neutral residue in most cases results in anion conduction. Specifically, replacing the large anion methanesulfonate– in the external solution with the smaller Cl– shifts Vrev negatively, demonstrating permeability to Cl– (Musset et al., 2011). Lowering the ionic strength by 90% shifts Vrev positively, confirming anion over cation permeation (Musset et al., 2011). Quite similar phenomenology supports an identical role for the analogous Asp in the middle of the S1 helix in two evolutionarily distant species, Karlodinium veneficum (Smith et al., 2011) and Nicoletia phytophila (Chaves et al., 2016), which respectively are only 15% and 33% identical to hHV1. The conductance of Asp112 mutants appears to vary inversely with the hydrophobicity of the substituent at position 112, with two of the most hydrophobic amino acids tested, Val and Ile, eliminating current flow altogether (Musset et al., 2011; DeCoursey, 2015b). The conservative Asp→Glu mutant retains proton selectivity.
Clearly, Asp112 is crucial to the proton selectivity of hHV1. However, other Asp are present in the presumed conduction pathway, such as Asp185 (Fig. 2), but Table 5 shows that its mutation does not impair H+ flux (Letts, 2014) or H+ selectivity (Musset et al., 2011). In an attempt to determine what other requirements exist for selectivity, Asp was moved along the S1 helix to each position from 108 to 118 (Morgan et al., 2013). At most positions where Asp faced away from the pore, no current was observed. Asp produced proton selectivity at just one other position, 116. Molecular dynamics (MD) simulations suggest that the D112V/V116D construct is proton selective only when Asp116 interacts with one or more S4 Arg residues (Morgan et al., 2013). This result shows that not every near-neighbor interaction of Asp112 in its native position is necessary, but there are clearly strong constraints. The requirements for proton selectivity in hHV1 deduced from these and many other mutations include the following: (a) a carboxyl group (Asp or Glu) is required; (b) it must face the pore; (c) it must be located at a narrow point in the channel; and (d) it must be able to interact with a basic group (Arg or Lys).
Table 5. Changes in HV1 properties in S3 (166–188) and S3–S4 linker (189–196) mutants versus WT channels .
| Mutant | Species | Expr. system | I? | τ act | τ tail | ΔV thr | ΔpH slope | Selectivity | Other | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| E164A/E171A | hHV1 | HEK wc | Yes | −7 | Ramsey et al., 2010 | |||||
| H167N/H168V/K169N | hHV1 | HEK wc | Yes | −13 | 44 | Ramsey et al., 2010 | ||||
| E171A/D174A | hHV1 | HEK wc | Yes | −116 | 39 | Ramsey et al., 2010 | ||||
| E171A | hHV1 | Vesicle flux | Δ H+ fluxa | Letts, 2014 | ||||||
| D174A | hHV1 | HEK wc | Yes | −111 | 38 | Ramsey et al., 2010 | ||||
| D174A | hHV1 | Vesicle flux | Δ H+ fluxa | Letts, 2014 | ||||||
| D174N | hHV1 | HEK wc | Yes | −142 | 36 | Ramsey et al., 2010 | ||||
| D174H | hHV1 | HEK wc | Yes | −136 | 37 | Ramsey et al., 2010 | ||||
| D174E | hHV1 | HEK wc | Yes | −52 | 46 | Ramsey et al., 2010 | ||||
| D174A | CiHV1 [D222A] | Xenopus TEVC | Yes | −111 | Chamberlin et al., 2014 | |||||
| D174C/R205C | CiHV1 [D222C/R255C] | Xenopus TEVC | Yes | −95 | Chamberlin et al., 2014 | |||||
| D174C/R208C | CiHV1 [D222C/R258C] | Xenopus TEVC | Yes | 48 | Chamberlin et al., 2014 | |||||
| V178A | hHV1 | Xenopus i-o | Yes | −27 | Hong et al., 2014 | |||||
| D112V/V178D | hHV1 | COS/HEK wc | No | Morgan et al., 2013 | ||||||
| S181A | hHV1 | HEK wc | Yes | 18 | 46 | Ramsey et al., 2010 | ||||
| S181A | hHV1 | Xenopus i-o | Yes | 0 | Hong et al., 2014 | |||||
| D112V/S181D | hHV1 | COS/HEK wc | No | Morgan et al., 2013 | ||||||
| F182A | hHV1 | Xenopus i-o | Yes | −9 | Hong et al., 2014 | |||||
| D185A | hHV1 | HEK wc | Yes | 58 | 47 | Ramsey et al., 2010 | ||||
| D185M | hHV1 | COS/HEK wc | Yes | H+ | Musset et al., 2011 | |||||
| D185V | hHV1 | COS/HEK wc | Yes | 20b | 43b | H+ | Musset et al., 2011 | |||
| D185A | hHV1 | COS/HEK wc | Yes | 42b | 40b | H+ | Musset et al., 2011 | |||
| D185A | hHV1 | Vesicle flux | nc H+ flux | Letts, 2014 | ||||||
| D185N | hHV1 | COS/HEK wc | Yes | 36b | 47b | H+ | Musset et al., 2011 | |||
| D185C | CiHV1 [D233C] | Xenopus TEVC | Yes | 76 | Chamberlin et al., 2014 | |||||
| E185C/R208C | CiHV1 [D233C/R258C] | Xenopus TEVC | Yes | 4 | Chamberlin et al., 2014 | |||||
| E192A/E196A | hHV1 | HEK wc | Yes | 13 | Ramsey et al., 2010 | |||||
| H193A | hHV1 | HEK wc | Yes | H+ | 39 × Kd Zn2+ | Ramsey et al., 2006 | ||||
| H193A | hHV1 | Vesicle flux | nc H+ flux | Letts, 2014 | ||||||
| H140A/H193A | hHV1 | HEK wc | Yes | −12 | 46 | H+ | 2,000 × Kd Zn2+ | Ramsey et al., 2006 |
That numerical entries are shown does not imply that any given change was significant. Italicized mutant entries from nonhuman species show the hHV1 equivalent. In the Expression system column: HEK, HEK-293, HEK-293T, tsA, or HM1; COS, COS-7; Xenopus, Xenopus laevis oocyte; wc, whole cell; i-o, inside-out patch configuration; TEVC, two-electrode voltage clamp. Blank entries indicate that the parameter was not examined. nc, measured, but no change. Parameters are given relative to WT in each study. For I?, yes means currents are detectable. Time constants are ratios of mutant/WT. The ΔVthreshold value is the change in absolute position of the gH–V relationship versus WT. The ΔpH slope is the slope in millivolts of the relationship between Vthreshold (or other parameters reflecting the absolute position of the gH–V relationship) versus Vrev or EH (which are not identical; see section Table entries defined).
Normal initial H+ flux followed by recovery ascribed to leak induced in vesicles.
Previously unpublished, analyzed from data for Musset et al. (2011).
These conditions apparently exist only in the outer vestibule of hHV1. Furthermore, several attempts to reposition Asp into S2 or S3 failed to produce a proton-selective conductance, suggesting that, for reasons that are not at all clear, the carboxyl group must be on S1. A quantum model of the selectivity filter of hHV1 illustrates how interacting Asp and Arg side chains can selectively conduct protons while excluding other ions (Dudev et al., 2015).
Intriguingly, proton-selective conduction is preserved when Asp112 is replaced by Glu112 (Musset et al., 2011) or when Arg208 is replaced by Lys208 (Dudev et al., 2015). Clearly, this critical interaction has leeway with respect to chain length. The F1Fo ATP synthase (H+ translocating ATPase) remarkably parallels HV1 in that the proton pathway in its c subunit has an essential Asp61–Arg210 pair and Asp61 can be moved to a different location or replaced by Glu with only partial loss of function (Miller et al., 1990). It is noteworthy that in several other molecules with critical proton transport pathways, analogous substitutions impair function: Asp→Glu (Chen et al., 2000; Ruivo et al., 2012; Luoto et al., 2013), Glu→Asp (Thorndycroft et al., 2007; Cornish et al., 2011), and Lys→Arg (Balashov et al., 2013). Li et al. (2015) found that hHV1 is more mobile and dynamic than VSDs of other voltage-gated ion channels.
The series of mutations to Asp112 nicely illustrates the difficulty of interpreting mutations. For example, D112A, D112S, and D112N (Table 3) all open and close more slowly than WT. So is the function of Asp112 to speed gating? No, because D112K produced faster kinetics, and D112F changed activation and deactivation in opposite directions. So does Asp112 regulate gating kinetics? Not really, because practically every mutation for which kinetics were reported alters kinetics, by up to 100-fold. Considering that gating reflects large conformational changes and perhaps other subtler changes that involve a large fraction of the amino acids in the protein, and HV1 is a compact molecule, it is not surprising that most mutations affect gating kinetics. Their interpretation requires semantic judiciousness. Not every position whose mutation affects gating can reasonably be said to regulate gating kinetics. Nevertheless, the effects of mutations are real and suggest involvement in the process, whose mechanism may, however, be difficult to disentangle.
Countercharges in S1
A fundamental principle in the conception of how voltage gating works is that the periodically spaced cationic residues in S4 (Arg and sometimes Lys) that sense voltage interact electrostatically with anionic amino acids elsewhere in the channel protein to stabilize both closed and open states (Papazian et al., 1995; Tiwari-Woodruff et al., 1997; Lecar et al., 2003). In the General conclusions section below, we discuss alternative interpretations. Within the charge/countercharge conceptual framework, externally accessible acidic residues stabilize the open state, and internally accessible acidic groups stabilize the closed state, presumably by interacting with the cationic groups in S4. If this is the case, a neutralizing mutation to such an externally accessible amino acid should shift the gH–V relationship positively because the mutant will lose open state stabilization. Conversely, an internal acidic residue would normally stabilize the closed state, and its mutation should promote channel opening, thus shifting the gH–V relationship negatively. By these criteria, three acidic amino acids in S1 in the outer vestibule, Asp112, Glu119, and Asp123, and possibly Lys125 may be considered weak stabilizers of the open state because their neutralization by mutation produces modest positive shifts. Both Asp112 and Glu119 interact with S4 Arg residues in MD simulations of open state homology models of hHV1 (Wood et al., 2012; Kulleperuma et al., 2013) and CiHV1 (Chamberlin et al., 2014).
Table 4: The S2 helix (positions 134–156) and the S2–S3 linker (157–165)
Table 4. Changes in HV1 properties in S2 (134–156) and S2–S3 linker (157–165) mutants versus WT channels .
| Mutant | Species | Expr. system | I? | τ act | τ tail | ΔV thr | ΔpH slope | Selectivity | Other | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| Y134A | hHV1 | HEK wc | Yes | 3 | Ramsey et al., 2010 | |||||
| H140A | hHV1 | HEK wc | Yes | H+ | 9 × Kda Zn2+ | Ramsey et al., 2006 | ||||
| H140A | hHV1 | Vesicle flux | nc H+ flux | Letts, 2014 | ||||||
| H193A | hHV1 | HEK wc | Yes | H+ | 39 × Kda Zn2+ | Ramsey et al., 2006 | ||||
| H140A/ H193A | hHV1 | HEK wc | Yes | −12 | 46 | H+ | 2,000 × Kda Zn2+ | Ramsey et al., 2006 | ||
| H140A/ H193A | hHV1 | COS/HEK wc | Yes | H+ | Musset et al., 2011 | |||||
| Y141A | hHV1 | HEK wc | Yes | −27 | Ramsey et al., 2010 | |||||
| S143A | hHV1 | HEK wc | Yes | 11 | 41 | Ramsey et al., 2010 | ||||
| S143A | hHV1 | Vesicle flux | nc H+ flux | Letts, 2014 | ||||||
| D112V/S143D | hHV1 | COS/HEK wc | Yes | Cl– | Morgan et al., 2013 | |||||
| D112V/I146D | hHV1 | COS/HEK wc | No | Morgan et al., 2013 | ||||||
| D112V/L147D | hHV1 | COS/HEK wc | No | Morgan et al., 2013 | ||||||
| F150A | hHV1 | Xenopus i-o | Yes | −24 | Hong et al., 2013 | |||||
| F150C | hHV1 | Xenopus i-o | Yes | −22 | Hong et al., 2013 | |||||
| F150W | hHV1 | Xenopus i-o | Yes | −55 | Hong et al., 2013 | |||||
| E153A | hHV1 | HEK wc | Yes | −55 | 42 | Ramsey et al., 2010 | ||||
| E153A | CiHV1 [E201A] | Xenopus TEVC | No | Chamberlin et al., 2014 | ||||||
| E153G | CiHV1 [E201G] | Xenopus TEVC | No | Chamberlin et al., 2014 | ||||||
| E153N | hHV1 | HEK wc | Yes | −1.17 | 45 | Ramsey et al., 2010 | ||||
| E153A | hHV1 | Vesicle flux | nc H+ flux | Letts, 2014 | ||||||
| E153D | hHV1 | HEK wc | Yes | −23 | 37 | Ramsey et al., 2010 | ||||
| E153D/D174E | hHV1 | HEK wc | Yes | −102 | 40 | Ramsey et al., 2010 | ||||
| E153C | hHV1 | Xenopus i-o | Yes | −55 | Tombola et al., 2010 | |||||
| E153C | CiHV1 [E201C] | Xenopus TEVC | Yes | −101 | Chamberlin et al., 2014 | |||||
| E153C/R205C | CiHV1 [E201C/R255C] | Xenopus TEVC | Yes | −43 | Chamberlin et al., 2014 | |||||
| E153C/R208 | CiHV1 [E201C/R258C] | Xenopus TEVC | Yes | 37 | Chamberlin et al., 2014 | |||||
| E153C/R211C | CiHV1 [E201CR261C] | Xenopus TEVC | Yes | −60 | Chamberlin et al., 2014 | |||||
| K157A | hHV1 | HEK wc | Yes | 1 | 39 | Ramsey et al., 2010 | ||||
| K157A | hHV1 | Vesicle flux | nc H+ flux | Letts, 2014 | ||||||
| R162A | hHV1 | HEK wc | Yes | 13 | Ramsey et al., 2010 |
That numerical entries are shown does not imply that any given change was significant. Italicized mutant entries from nonhuman species show the hHV1 equivalent. HEK, HEK-293, HEK-293T, tsA, or HM1; COS, COS-7; Xenopus, Xenopus laevis oocyte; wc, whole cell; i-o, inside-out patch configuration; TEVC, two-electrode voltage clamp. Blank entries indicate that the parameter was not examined. nc, measured, but no change. Parameters are given relative to WT in each study. For I?, yes means currents are detectable. The ΔVthreshold value is the change in absolute position of the gH–V relationship versus WT. The ΔpH slope is the slope in millivolts of the relationship between Vthreshold (or other parameters reflecting the absolute position of the gH–V relationship) and Vrev or EH (which are not identical; see section Table entries defined).
Nominal Kd values are nearly meaningless for Zn2+ inhibition of HV1 because its main effects are slowing activation and shifting the gH–V relationship positively (Cherny and DeCoursey, 1999). As demonstrated in the Appendix of DeCoursey et al. (2001a), the apparent Kd derived from the ratio IH(Zn2+)/IH can vary more than three orders of magnitude depending on the test potential selected. If all measurements are done the same way, relative Kd values have meaning.
The Zn2+-binding site
The most potent inhibitor of voltage-gated proton currents is Zn2+ (Mahaut-Smith, 1989; DeCoursey, 2003b). Unlike traditional channel blockers that occlude the pore, Zn2+ shifts the gH–V relationship positively and slows activation (Cherny and DeCoursey, 1999). These effects were strongly inhibited at low pHo, indicating competition between Zn2+ and H+ for a binding site. To model the competition between H+ and Zn2+ quantitatively required assuming that Zn2+ prevents channel opening by binding to an externally accessible site on the closed channel comprising at least two titratable groups with a pKa of 6.2–6.6 (near that of His; Cherny and DeCoursey, 1999). Seven years later, the identification of the hHV1 gene confirmed this deduction because two His residues, His140 in S2 and His193 in the S3–S4 linker, were found to comprise the main sites at which Zn2+ binds to inhibit proton currents (Ramsey et al., 2006). The single mutants H140A and H193A each have diminished sensitivity to Zn2+, and the double mutant is nearly impervious. The Zn2+ sensitivity of a series of mutants in which one or both of these His were mutated to Ala, including various tandem dimers, is described elsewhere (Musset et al., 2010b) and is not included in the tables. Remarkably, the crystal structure of the closed mHV1 channel contained a Zn2+ atom tetrahedrally coordinated by the corresponding two His in the mouse (His136 and His189 in mHV1), with weaker binding to Glu115 and Asp119 (Glu119 and Asp123 in hHV1).
Countercharges in S2
S2 contains an important countercharge, Glu153, which, as seen in Table 1, is highly conserved among VSD-containing molecules (Smith et al., 2011). In neutral mutants, Vthreshold is shifted consistently negatively, in some cases by >100 mV, suggesting that this internal acidic residue stabilizes the closed state.
Charge transfer center or hydrophobic gasket
The S2 helix contains Phe150, another highly conserved residue among VSD-containing molecules (Tao et al., 2010; Smith et al., 2011) whose K+ channel correlate was described as the outer limit of the charge transfer center (Tao et al., 2010). As the positively charged Arg residues in S4 move outwards during a depolarization that opens the channel, they move past Phe150, which serves as a delimiter of internal and external accessibility. Bezanilla and colleagues include Phe150 along with two other hydrophobic residues, Val109 and Val178, in a hydrophobic gasket (or “plug”) that functions similarly (labeled HG in Table 1; Lacroix et al., 2014; Li et al., 2014, 2015; DeCoursey, 2015a). The global purpose of having a narrow isthmus of protein between the two aqueous vestibules is to focus the electric field (Yang et al., 1996, 1997; Starace and Bezanilla, 2001, 2004). This means that each gating charge (e.g., Arg) needs to move only a small distance to effectively cross the entire membrane electrical field. Mutations to Phe150 in hHV1, like those of the corresponding Phe in K+ channels, shift the gH–V relationship (Tao et al., 2010; Hong et al., 2013).
Table 5: The S3 helix (positions 166–188) and the S3–S4 linker (189–196)
Countercharges in S3
S3 contains two important countercharges. Asp174 is internally accessible and stabilizes the closed state, and neutral mutants shift the gH–V relationship strongly negatively. In the closed structure of mHV1, the Asp174 equivalent appears to interact with Arg211 in an internal pocket (Takeshita et al., 2014; Cherny et al., 2015). Conversely, Asp185 (which Table 1 shows is unique to the HV1 family) is externally accessible and stabilizes the open state, and neutral mutants shift the gH–V relationship positively. The milder effect of the Asp185 mutation mirrors its moderate interaction with Arg205 observed in MD simulations of an open state model of hHV1 (Kulleperuma et al., 2013) and with all three Arg in a model of CiHV1 (Chamberlin et al., 2014).
Table 6: The S4 helix (positions 197–218)
Table 6. Changes in HV1 properties in S4 (197–218) mutants versus WT channels .
| Mutant | Species | Expr. system | I? | τ act | τ tail | ΔV thr | ΔpH slope | Selectivity | Other | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| E196C | CiHV1 [A246C] | Xenopus TEVC | Yes | MTSo open>closed | Gonzalez et al., 2010 | |||||
| L198C | CiHV1 [I248C] | Xenopus TEVC | Yes | MTSo open>closed | Gonzalez et al., 2010 | |||||
| G199C | CiHV1 [G249C] | Xenopus i-o | Yes | 2 | Mony et al., 2015 | |||||
| L200W | CiHV1 [L250W] | Xenopus TEVC | Yes | 0.27 | 0.67 | Okuda et al., 2016 | ||||
| L200W/ΔC | CiHV1 [L250W/ΔC] | Xenopus TEVC | Yes | 0.20 | 0.019 | Okuda et al., 2016 | ||||
| I202C | CiHV1 [V252C] | Xenopus TEVC | Yes | MTSo open>closed | Gonzalez et al., 2010 | |||||
| R205A | hHV1 | HEK wc | Yes | 0.0053 | 0.086 | −1 | 48 | 0.63 × WT gating chargea | Ramsey et al., 2006, 2010 | |
| R205A | hHV1 | Vesicle flux | Δ H+ fluxb | Letts, 2014 | ||||||
| R205Hc/T222stop | hHV1 | COS wc | Yes | H+ | Accessible to external Zn2+ | Kulleperuma et al., 2013 | ||||
| R205Q | Mouse [R201Q] | HEK wc | Yes | Faster | −50 | 1.36 × WT gating chargea | Sasaki et al., 2006 | |||
| R205N | CiHV1 [R255N] | Xenopus i-o | Yes | 0.33 × WT gating charged | Gonzalez et al., 2013 | |||||
| R205N | CiHV1 [R255N] | Xenopus TEVC | Yes | −36 | 0.78 × WT gating chargea | Chamberlin et al., 2014 | ||||
| R205C | CiHV1 [R255C] | Xenopus TEVC | Yes | −37 | 1.3 × WT gating chargea | Chamberlin et al., 2014 | ||||
| R205A/R208A | hHV1 | HEK wc | Yes | 128 | 51 | Ramsey et al., 2010 | ||||
| R205A/R211A | hHV1 | HEK wc | Yes | 96 | 45 | Ramsey et al., 2010 | ||||
| L206C | CiHV1 [L256C] | Xenopus TEVC | Yes | No MTSo/i access | Gonzalez et al., 2010 | |||||
| W207A,c W207S,c or W207Fc | hHV1 | HEK/COS wc | Yes | 0.01 | 0.034 | −17.9 | 40 | H+ | Loss of selectivity at pHo > 8 | Cherny et al., 2015 |
| W207I | Mouse [W203I] | HEK wc | Yes | 0.019 | 0.059 | Tandem dimer | Okuda et al., 2016 | |||
| W207A, W207S, or W207F | kHV1 [W176A, W176S, W176F] | HEK/COS wc | Yes | 0.025 | 40 | H+ | Cherny et al., 2015 | |||
| W207A, W207S, or W207F | EhHV1 [W278A, W278S, W278F] | HEK/COS wc | Yes | 0.2 | −28.2 | 50 | H+ | Cherny et al., 2015 | ||
| W207I | CiHV1 [W257I] | Xenopus TEVC | Yes | 0.29 | 0.030 | Okuda et al., 2016 | ||||
| W207I/A210A | CiHV1 [W257I/F260A] | Xenopus TEVC | Yes | 0.23 | 0.026 | Okuda et al., 2016 | ||||
| R208A | hHV1 | HEK wc | Yes | 0.0965 | 0.075 | 7 | 45 | Ramsey et al., 2006, 2010 | ||
| R208A | hHV1 | Vesicle flux | Δ H+ fluxb | Letts, 2014 | ||||||
| R208A | hHV1 | Xenopus i-o | No | Hong et al., 2014 | ||||||
| R208K | hHV1 | Xenopus i-o | Yes | −40 | 1.7 × WT gating chargea | Hong et al., 2014 | ||||
| R208K | hHV1 | HEK/COS wc | Yes | H+ | Dudev et al., 2015 | |||||
| R208Hc/T222stop | hHV1 | COS wc | Yes | H+ | Accessible to external & maybe internal Zn2+ | Kulleperuma et al., 2013 | ||||
| R208Q | hHV1 | Xenopus i-o | No | Hong et al., 2014 | ||||||
| R208Q | Mouse [R204Q] | HEK wc | No | Sasaki et al., 2006 | ||||||
| R208N | hHV1 | Xenopus i-o | No | Hong et al., 2014 | ||||||
| R208N | CiHV1 [R258N] | Xenopus i-o | Yes | 0.50 × WT gating charged | Gonzalez et al., 2013 | |||||
| R208C | CiHV1 [R258C] | Xenopus TEVC | Yes | −10 | 0.87 × WT gating chargea | Chamberlin et al., 2014 | ||||
| V209C | CiHV1 [V259C] | Xenopus TEVC | Yes | No MTSo/i access | Gonzalez et al., 2010 | |||||
| R211A | hHV1 | HEK wc | Yes | 2.24 | 0.092 | 70 | 53 | Ramsey et al., 2006, 2010 | ||
| R211A | hHV1 | Vesicle flux | Δ H+ fluxb | Letts, 2014 | ||||||
| R211S | hHV1 | Xenopus i-o | Yes | 35 | 0.72 × WT gating chargea | Hong et al., 2014 | ||||
| R211S | hHV1 | Xenopus i-o | Yes | 87 | 49 | Gu+ | at pH 8//8 | Berger and Isacoff, 2011 | ||
| R211S/I127C | CiHV1 [R261S/I175C] | Xenopus i-o | Yes | 15 | Mony et al., 2015 | |||||
| R211Hc/T222stop | hHV1 | COS wc | Yes | H+ | Accessible to internal Zn2+ when open | Kulleperuma et al., 2013 | ||||
| R211H/D112V/V116Dc | hHV1 | COS/HEK wc | Yes | Accessible to internal Zn2+ when open | Morgan et al., 2013 | |||||
| R211Q | mHV1 [R207Q] | HEK wc | Yes | nc | Sasaki et al., 2006 | |||||
| R211N | CiHV1 [R261N] | Xenopus i-o | Yes | 0.38 × WT gating charged | Gonzalez et al., 2013 | |||||
| R211C | CiHV1 [R261C] | Xenopus TEVC | Yes | 54 | H+ | 0.78 × WT gating chargea | Chamberlin et al., 2014 | |||
| R211C | NpHv1 [R163C] | HEK wc | Yes | H+ | Chaves et al., 2016 | |||||
| I212C | CiHV1 [I262C] | Xenopus TEVC | Yes | MTSi closed>open | Gonzalez et al., 2010 | |||||
| N214K | hHV1 | HEK wc | Yes | −3 | 43 | Inward rectification | Ramsey et al., 2010 | |||
| N214R | hHV1 | HEK wc | Yes | 10 | 40 | Inward rectification | Ramsey et al., 2010 | |||
| N214R | hHV1 | Xenopus i-o | No | Tombola et al., 2008 | ||||||
| N214A | hHV1 | HEK wc | Yes | −3 | 42 | Ramsey et al., 2010 | ||||
| N214A | hHV1 | Vesicle flux | nc H+ flux | Letts, 2014 | ||||||
| N214R | mHV1 [N210R] | HEK wc | Yes | Slow | Very slow | −V | H+ | Sakata et al., 2010 | ||
| N214D | hHV1 | COS/HEK wc | Yes | H+ | Musset et al., 2011 | |||||
| N214C | CiHV1 [N264C] | Xenopus TEVC | Yes | MTSi closed>open | Gonzalez et al., 2010 | |||||
| G215A | hHV1 | COS/HEK wc | Yes | Fast | H+ | Musset et al., 2011 | ||||
| I217stop | mHV1 [I213stop] | HEK wc | Yes | Sakata et al., 2010 | ||||||
| G215stop | mHV1 [G211stop] | HEK wc | Yes | Sakata et al., 2010 | ||||||
| I213stop | mHV1 [I209stop] | HEK wc | Yes | Sakata et al., 2010 | ||||||
| A210stop | mHV1 [A206stop] | HEK wc | Yes | +V | H+ | τact has weak V dependence | Sakata et al., 2010 | |||
| L204stop | mHV1 [L200stop] | HEK wc | No | Sakata et al., 2010 |
That numerical entries are shown does not imply that any given change was significant. Italicized mutant entries from nonhuman species show the hHV1 equivalent. HEK, HEK-293, HEK-293T, tsA, or HM1; COS, COS-7; Xenopus, Xenopus laevis oocyte; wc, whole cell; i-o, inside-out patch configuration; TEVC, two-electrode voltage clamp. Blank entries indicate that the parameter was not examined. nc, measured, but no change. Parameters are given relative to WT in each study. For I?, yes means currents are detectable. Time constants are ratios of mutant/WT. The ΔVthreshold value is the change in absolute position of the gH–V relationship versus WT. The ΔpH slope is the slope in millivolts of the relationship between Vthreshold (or other parameters reflecting the absolute position of the gH–V relationship) and Vrev or EH (which are not identical; see section Table entries defined). When C-terminal truncations are indicated as XNNNstop, this means STOP replaces X at position NNN; hence, position NNN and all subsequent residues are truncated, and the last position remaining is NNN-1. For column Other, MTS access from inside or outside (MTSi or MTSo, respectively) is listed as open>closed if the open channel was more accessible.
Slope factor of gH–V relationship.
Normal initial H+ flux followed by recovery ascribed to leak induced in vesicles.
In an H140A/H193A (Zn2+ insensitive) background.
By limiting slope method.
Cys scanning reveals aqueous accessibility
A now standard approach to demonstrate aqueous accessibility of amino acids in a protein is to convert the target to Cys and then challenge the mutant with MTS reagents that react with Cys sulfhydryl groups. Whatever functional effect this reaction produces can be examined as a function of the time of exposure to determine accessibility of the Cys. Gonzalez et al. (2010) identified E196C, L198C, and I202C (all external to R1) that were accessible externally preferentially in the open state, suggesting that S4 moves outward and/or rotates. However, a smaller probe for accessibility, n-ethylmaleimide (NEM) in a PEGylation protection assay, revealed that all S4 residues external to position 203 (including the three mentioned above) are accessible, presumably in the closed state (Kurokawa and Okamura, 2014). It should be noted that the former study examined kinetics of MTS effects under voltage clamp, so the gating state was well defined. In the latter study, voltage clamp was not involved, and although at 0 mV most WT channels are closed, many mutations shift the gH–V relationship, and thus mutant channels could be open at 0 mV.
Accessibility assays are limited by the size of the probe but will also be influenced by charge. For example, aqueous accessibility determined by Cys scanning with NEM as a probe revealed greater accessibility than when using the larger AMS (4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid) as a probe (Kurokawa and Okamura, 2014). Furthermore, Ag+ as a probe revealed much greater accessibility than the larger NEM (Fillingame et al., 2002). Fillingame et al. (2002) point out that because Ag+ has an ionic radius like H3O+, it is ideal for probing proton pathways. Zn2+ has a smaller ionic radius than Ag+ (Robinson and Stokes, 1959) but is divalent. As a probe of HV1, it reveals greater accessibility than the bulkier MTS reagents (Kulleperuma et al., 2013; Morgan et al., 2013). In bulk solution, the proton diffuses almost exclusively as protonated buffer (DeCoursey, 1991; DeCoursey and Cherny, 1994, 1996). One expects that the proton permeates the aqueous vestibules of HV1 as H3O+ (DeCoursey, 2003b) and the selectivity filter as H+ (Dudev et al., 2015).
Internal accessibility assessed rigorously by Cys scanning and MTS reaction kinetics under voltage clamp indicated that I212C and N214C were both more accessible at negative voltages, indicating greater accessibility in closed channels (Gonzalez et al., 2010). The residues with state-dependent accessibility in S4 thus span two positions internally and four externally. These results strongly support the idea that S4 moves outwardly during opening, but the extent of movement could be one turn of the helix, consistent with the “one-click” model (Li et al., 2015). Also consistent with a small excursion of S4 during opening are studies using Zn2+ to probe for accessibility of Arg→His mutants. In hHV1, R205H was externally accessible and R208H was accessible externally and possibly also internally (currents were tiny), whereas R211H was accessible only internally even in the open state (Kulleperuma et al., 2013; Morgan et al., 2013).
Accessibility of S1 was explored by Cys scanning, and five residues were found to be more accessible at positive (open) voltages (Mony et al., 2015). One was external, and the rest were internal. Although the internal residues span seven positions, the fact that both the external and internal residues were more exposed in open channels suggests a widening of the vestibules rather than a large inward translational movement.
Gating charge
Numerous mutations have been performed in the S4 helix with the goal of determining the extent to which channel opening involves outward movement of positively charged groups in S4 during depolarization, as is thought to occur in most other voltage-gated ion channels. Each of the three Arg in S4 has been mutated, and the effect on gating charge was evaluated in various ways. Unfortunately, the methods for estimating gating charge are challenging, and several problems unique to HV1 make the task even more difficult. A direct approach is to measure the integral of the gating current and divide by the number of channels. However, it is nearly impossible to measure gating current in HV1 because the permeant ion cannot be removed, simple blockers have not been identified, and gating is extremely slow. The most potent inhibitor, Zn2+, does not occlude the pore, as would be required to reveal gating charge, but rather shifts the gH–V relationship positively and slows activation (Cherny and DeCoursey, 1999). Block by guanidine derivatives is also state dependent (Hong et al., 2013). Another procedure that is technically straightforward but of limited information value is to determine the slope factor of a Boltzmann function fit to the gH–V relationship. The slope factor does include the effective gating charge, but in a highly model-dependent manner; nevertheless, the steepness of the voltage dependence should diminish if gating charges are removed by mutation. Such estimates are indicated in Table 6 by footnote b. There are numerous pitfalls in this measurement, a major one being proton depletion, which produces artificial saturation of current (DeCoursey, 1991; DeCoursey and Cherny, 1994; Musset et al., 2008a), and at best, the slope provides a model-dependent estimate that almost always underestimates the true gating charge (Bezanilla and Villalba-Galea, 2013). Early studies reported slope factors corresponding to a gating charge of 1.4 e0 (Sasaki et al., 2006) for WT mHV1 or 0.9 e0 for WT hHV1 (Ramsey et al., 2006). A more meaningful approach for HV1 has been the limiting slope method, devised by Wolf Almers (Almers, 1978). This method works for a wide range of gating models, but not all (Sigg and Bezanilla, 1997), and provides gating charge estimates of ∼6 e0 for native rat proton currents (DeCoursey and Cherny, 1996, 1997), for CiHV1 (Gonzalez et al., 2010), and for hHV1 (Musset et al., 2008a) and 4 e0 for mHV1 (Fujiwara et al., 2012). The difficulty is mainly that measurements need to be extended to large negative voltages to achieve sufficiently low gH values to determine the limiting slope. When critical amino acids such as the Arg in S4 are mutated, the resulting currents are often quite small, which leads to underestimates of the gating charge. Another vexing source of error is that mutation, for example, of Arg205 (R255N in CiHV1) appears to reduce the extent of S4 movement during gating (Gonzalez et al., 2013), which in retrospect is not a very surprising result of removing one of the charges in the protein thought to move in response to voltage changes!
Three studies reported lower gating charge (Ramsey et al., 2006; Gonzalez et al., 2013; Chamberlain et al., 2014) and two reported higher gating charge (Sasaki et al, 2006; Chamberlain et al, 2014) when Arg205 was neutralized (Table 6). Two studies reported lower gating charge when Arg208 was neutralized (Gonzalez et al., 2013; Chamberlain et al., 2014). The conservative Arg→Lys mutation increased gating charge by 70% (Hong et al., 2014). Finally, both studies of neutralized Arg211 reported lower gating charge (Gonzalez et al., 2013; Chamberlain et al., 2014). It might be noted that all studies reporting higher gating charge were based on the slope of Boltzmann functions. The tables do not list a number of studies of monomeric constructs, which consistently exhibit only half the gating charge of native dimers, a manifestation of cooperative gating (Gonzalez et al., 2010, 2013; Fujiwara et al., 2012; Okuda et al., 2016).
Why is there a Trp in the middle of S4?
Tryptophan prefers the interfacial environment near membrane lipid head groups (Landolt-Marticorena et al., 1993; Killian and von Heijne, 2000), but in HV1, a perfectly conserved Trp residue (Trp207 in hHV1) is located right in the middle of the S4 TM helix. Two studies have been conducted to determine why it is there, exploring mutations in multiple species (Table 1). The most prominent effect of Trp mutation was drastic acceleration of channel gating. In hHV1, activation and deactivation were 100 times and 30 times faster, respectively (Cherny et al., 2015), whereas in CiHV1, deactivation was more profoundly accelerated (Okuda et al., 2016). Effects of Trp mutation differ dramatically among species. Table 6 shows that the acceleration of channel opening was 100-fold in hHV1, 5-fold in EhHV1, 40-fold in kHV1 (Cherny et al., 2015), 3.4-fold in CiHV1, and 53-fold in mHV1 (Okuda et al., 2016). It is noteworthy that the properties of W207A, W207S, and W207F (three substituents with quite different properties) were all modified identically in hHV1 and in kHV1, which strongly implicates a unique property of Trp at this location (Cherny et al., 2015). Two mechanisms have been proposed to explain how Trp slows gating. Based on the proximity of Trp207 and Arg211 in the closed crystal structure (Takeshita et al., 2014), cation–π interaction between Trp207 and Arg211 was postulated to stabilize closed hHV1 channels, with this interaction broken during channel opening (Cherny et al., 2015). In CiHV1, π stacking of Trp from each protomer at the dimer interface was proposed to slow deactivation (Okuda et al., 2016). This proposal was supported by Trp slowing deactivation preferentially in dimeric versus monomeric constructs (Okuda et al., 2016). Mutant cycle analysis supported the idea that π stacking of Trp at the dimer interface contributed to slow deactivation, but the slowing of activation was independent of channel dimerization. Consistent with this latter conclusion, kHV1 lacks predicted coiled coil in its C terminus and thus is likely a monomer (Smith and DeCoursey, 2013), and τact was two orders of magnitude faster in Trp mutants of kHV1.
Trp207 mutants not only opened and closed faster, but the Q10 of their gating kinetics dropped from the astronomical 6–9 of WT HV1 (DeCoursey and Cherny, 1998; Ramsey et al., 2006; Kuno et al., 2009) into the realm of ordinary voltage-gated ion channels, 3.5–4.0 (Cherny et al., 2015). Trp207 is a key component in several of the unique properties of HV1 (Cherny et al., 2015).
Does Arg211 contribute to proton selectivity?
One study concluded that Arg211 is part of the selectivity filter because 15 R211x mutants were permeable to guanidinium, Gu+, at symmetrical pH 8, whereas outward current in WT hHV1 expressed in Xenopus oocytes was blocked (Berger and Isacoff, 2011). However, this result could not be reproduced in hHV1 expressed in HEK cells, suggesting that the expression system alters this property. Large outward currents were seen in both WT and R211A channels in HEK cells at high pH (DeCoursey, 2013). Neither WT nor R211A was detectably permeant to smaller cations (unpublished data), so the Gu+ result appears anomalous, perhaps related to the ability of this chaotropic ion to disrupt hydrogen bonds, interact with hydrophobic regions of proteins, bind to sites normally occupied by water, and denature proteins (Makhatadze and Privalov, 1992; Courtenay et al., 2001; England and Haran, 2011). Although molar Gu+ is typically required for wholesale denaturation, 50 mM Gu+ sufficed to perturb the permeation pathway of voltage-gated K+ channels (Kalia and Swartz, 2011). It is likely that ions are highly concentrated in the pores of channels just as they are in the active sites of enzymes (Jimenez-Morales et al., 2012) because of the high charge density in the protein. We imagine that Gu+ tunnels through the pore in a manner that no physiological ion can reproduce, perhaps by breaking the hydrogen bonds between Asp112 and Arg208 that prevent ions other than H3O+ from entering the selectivity filter (Dudev et al., 2015).
It is difficult to imagine a role for Arg211 in selectivity because the C terminus can be truncated along with the inner part of S4 (between Arg208 and Arg211) without loss of selectivity (Sakata et al., 2010). One might argue that when Arg211 is removed, Arg208 may take over its function. However, R211H in hHV1 (Kulleperuma et al., 2013) and R211C in NpHV1 (Chaves et al., 2016) and CiHV1 (Chamberlin et al., 2015) are all proton selective.
What about Asn214?
In S4, where many VSD-containing proteins have a fourth Arg or Lys (Table 1), HV1 has Asn214. Noting that the N214R mutant did not conduct, an early suggestion was that, assuming that S4 in HV1 moves outward as it does in other channels, Asn214 might occupy a narrow constriction where it would allow protons to pass (Tombola et al., 2008). Two other groups reported that N214R did conduct (Ramsey et al., 2010; Sakata et al., 2010), but both used mammalian expression systems, as opposed to Xenopus. This may be another example in which the expression system alters the outcome. However, the currents in N214R mutants were small, so this may be a case of different laboratories having different definitions of what comprises a detectable current. Given evidence that Arg211 remains internally accessible in open hHV1 (Kulleperuma et al., 2013; Morgan et al., 2013; Li et al., 2015), Asn214 most likely remains well inside the internal vestibule.
Table 7: The C terminus (positions 219–273)
Table 7. Changes in HV1 properties in C-terminal (219–273) mutants versus WT channels .
| Mutant | Species | Expr. system | I? | τ act | τ tail | ΔV thr | ΔpH slope | Selectivity | Other | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| S219P | hHV1 | COS/HEK wc | Yes | H+ | Musset et al., 2011 | |||||
| C249S | hHV1 | Loss of dimer formation | Li et al., 2015 | |||||||
| ΔC (T222stop) | hHV1 | HEK wc | Yes | 1 | 28 | Ramsey et al., 2010 | ||||
| ΔC (T222stop) | hHV1 | Yes | 0.15 | Weaker Zn2+ effects | Musset et al., 2010b | |||||
| ΔC | mHV1 (V216stop) | HEK wc | Yes | 0.34 | nc | nc | Loss of dimer formation | Koch et al., 2008 | ||
| ΔC | mHV1 (V216stop) | HEK wc | Yes | 0.15 | 0.16 | Okuda et al., 2016 | ||||
| ΔC | CiHV1 (D275stop) | Xenopus TEVC | Yes | 0.14 | 0.075 | Okuda et al., 2016 | ||||
| V220G/ K221G/T222G | mHV1 V216G/K217G/T218G | HEK wc | Loss of cooperative gating | Fujiwara et al., 2012 |
That numerical entries are shown does not imply that any given change was significant. HEK, HEK-293, HEK-293T, tsA, or HM1; COS, COS-7; Xenopus, Xenopus laevis oocyte; wc, whole cell; i-o, inside-out patch configuration; o-o, outside-out patch configuration; TEVC, two-electrode voltage clamp. Blank entries indicate that the parameter was not examined. nc, measured, but no change. Parameters are given relative to WT in each study. For I?, yes means currents are detectable. Time constants are ratios of mutant/WT. The ΔVthreshold value is the change in absolute position of the gH–V relationship versus WT. The ΔpH slope is the slope in millivolts of the relationship between Vthreshold (or other parameters reflecting the absolute position of the gH–V relationship) and Vrev or EH (which are not identical; see section Table entries defined). When C-terminal truncations are indicated as XNNNstop, this means STOP replaces X at position NNN; hence, position NNN and all subsequent residues are truncated, and the last position remaining is NNN-1. The mouse N-terminal deletions were done by replacing P78M to initiate translation at that position. Note that a large number of mutations to the linker between S4 and the C terminus have been studied (Fujiwara et al., 2012, 2014), but their results are beyond the scope of this table and thus are not included.
The coiled-coil region holds the dimer together
The C terminus of hHV1 contains extensive typical coiled-coil sequences. When it was learned that mammalian and some other HV1 assemble into dimers in cell membranes, the coiled-coil regions were implicated in dimerization (Koch et al., 2008; Lee et al., 2008; Tombola et al., 2008; Li et al., 2010). A cross-linking study with 15 strategically located Cys mutants confirmed that the C terminus was a major point of attachment of the dimer (Lee et al., 2008). By modifying the C terminus, Okamura’s group was able to generate functioning trimers and tetramers (Fujiwara et al., 2013a). A crystal structure of the C terminus revealed that the lone Cys (Cys249 in hHV1) in the C terminus forms a disulfide bond, increasing the stability of the dimer (Fujiwara et al., 2013b). This result was confirmed by mutation of the two native Cys in hHV1; C107S was still 90% dimer, whereas C249S was <5% dimer (Li et al., 2015).
C terminus truncation was determined to produce mainly monomeric constructs (Koch et al., 2008; Tombola et al., 2008). The monomers behave differently electrophysiologically because the dimer exhibits cooperative gating (Gonzalez et al., 2010; Tombola et al., 2010). The dimer opens with sigmoid kinetics, like a classical Hodgkin-Huxley n2 mechanism (Hodgkin and Huxley, 1952). The monomer opens approximately five times faster and with exponential kinetics (Koch et al., 2008; Musset et al., 2010b,c; Tombola et al., 2010; Fujiwara et al., 2012). Because both monomers must move in response to voltage before either can open, the gating charge is twice as large in the dimer as in the monomer (Gonzalez et al., 2010, 2013; Fujiwara et al., 2012; Okuda et al., 2016).
Does the C terminus modulate gating?
The Okamura group has studied the C terminus extensively using creative approaches. They conclude that the C terminus and the S4 helix form a single rigid monolithic rod, which moves during cooperative gating (Fujiwara et al., 2012, 2014). Two types of evidence support this conclusion. First, when a rigid (AAA) or flexible (GGG) triplet was inserted between S4 and the C terminus (at 220–222, replacing VKT; Table 7), the rigid linker behaved like a WT dimer (sigmoid kinetics, slow activation, and full gating charge), whereas the floppy linker produced monomer-like behavior (exponential kinetics, fast activation, and half the gating charge). This showed that simply being a dimer was insufficient to produce cooperative gating; a continuous α helix from the C terminus through S4 was required (Fujiwara et al., 2012). In another intriguing study, 1–10 amino acids were inserted or deleted from the connection between the S4 TM segment and the C terminus. Gating kinetics exhibited a periodic dependence on the linker length, presenting slower WT-like kinetics when the whole S4-C domain was in register (Fujiwara et al., 2014). In this context, even though the VSD-only construct (lacking both N and C termini) spontaneously associates into dimers in liposomes (Li et al., 2015), it likely still functions as two independent monomers.
There is also evidence that interactions at the extracellular end of the S1 segment contribute to the dimer interface (Lee et al., 2008; Qiu et al., 2013; Hong et al., 2015). To enable this to occur, the outer ends of S4 were proposed to relax or unwind (Hong et al., 2015).
General conclusions and remaining questions
A large number of mutations result in functional proton channels. This might be inferred from the fact that there are sequence differences at most positions in HV1 among different species (Smith et al., 2011). Perhaps not surprisingly, mutations that produce nonfunctional protein tend to occur at highly conserved positions and often involve changes in charge. Both Arg208 mutation (Table 6) and Asp112 mutation (Table 3) are severely detrimental to HV1 function, which we ascribe to both their central location and essential roles in proton permeation (Kulleperuma et al., 2013; Morgan et al., 2013; Dudev et al., 2015).
Several examples exist in which HV1 channels apparently behave differently in mammalian and amphibian expression systems. Whether these discrepancies reflect differences in as yet unknown posttranslational modification can only be speculated.
Charge, countercharge, and contra-countercharge
Electrostatics is clearly of central importance in ion channel function. Voltage sensing almost certainly involves charged groups within the membrane electrical field. The idea that interaction of charge pairs (oppositely charged) helps stabilize open or closed states is well entrenched among enthusiasts of HV1 as well as other voltage-gated ion channels (Papazian et al., 1995; Tiwari-Woodruff et al., 1997). It seems reasonable to conclude that mutations that neutralize a charge will preclude this element from performing this function. The extensive study by Ramsey et al. (2010) together with other table entries is consistent with the interpretation that Asp112, Glu119, Asp123, and Asp185 in the outer vestibule stabilize the open state; internally accessible acidic groups (Glu153 and Asp174) more strongly stabilize the closed state. Interacting external and internal charge clusters have been observed consistently in MD simulations of HV1 (Ramsey et al., 2010; Kulleperuma et al., 2013; Morgan et al., 2013; Chamberlin et al., 2014).
However, large shifts of the gH–V relationship can also be observed upon replacing one uncharged residue with another one (e.g., F150W in Table 4). In fact, almost every mutation that has been examined changes Vthreshold one way or the other. Pless et al. (2011) showed convincingly that two highly conserved acids in the Shaker K+ channel VSD (equivalent to Glu153 and Asp174 in hHV1; Table 1) could be neutralized with little effect on the gK–V relationship. Four external and two internal acidic residues were identified here as counterchanges based on the observation that in neutral mutants the gH–V relationship shifted in the predicted direction; thus, by this definition, the mechanism appears to involve charge. Alternatively, these charged groups might function primarily to create an aqueous vestibule and consequently a highly focused electrical field (Pless et al., 2011). We cannot resolve the mechanism without strategically designed experiments. If the function of the acidic residues is to create aqueous vestibules, one might predict that polar substituents that replace charges should produce less drastic effects than nonpolar ones. Mining the tables, we find that existing data are neither extensive nor self-consistent enough to provide a clear answer. The reported gH–V relationship shifts (polar vs. nonpolar) are as follows: Asp112 (31, 23, 13, and 25 vs. 59, 41, and 44); Asp185 (36 vs. 58, 20, 42, and 76); Glu153 (−117 vs. −55, −55, and −101), and Asp174 (−142 and −136 vs. −111 and −111). Another point is that Lys157 is predicted by MD to be involved in an internal salt bridge in closed HV1 (Chamberlin et al., 2014), yet its neutralization has little effect on the gH–V relationship (Table 4). This question clearly requires future study.
Outstanding remaining problems
It seems surprising that the N terminus contains several sites that influence gating, despite being topologically remote from S4 (at least in the image in Fig. 1). It also seems paradoxical that truncation of the entire N terminus has less overt effect than point mutations within the N terminus. However, the N terminus is disordered, and its tertiary structure is unknown. How it interacts with the rest of the molecule is completely unknown. For example, how does phosphorylation of Thr29 alter gating so profoundly? Why does the innocuous-appearing M91T mutation impede hHV1 activation?
Voltage gating is a difficult structure–function problem because it is dynamic, not to mention three-dimensional. Despite Cys scanning and other studies designed to examine state-dependent accessibility and molecular movement, it remains unclear which parts of the HV1 molecule move during channel opening (S4, S1?), in which direction, and how far. For example, accessibility may change as a result of movement “up” or “down,” or into or out of a hydrophobic region, but it can also change simply by expansion or contraction of an aqueous vestibule, or by the helix rotating toward or away from an aqueous space.
Although much has been learned about selectivity and a model has been proposed (Dudev et al., 2015), questions still remain. Is the model correct? Do other parts of the channel contribute to selectivity? Where is the rate-limiting point of the permeation pathway?
Without a doubt, the most important question remains completely unsolved; namely, the mechanism of ΔpH-dependent gating. This unique ΔpH dependence is crucial to all of the known biological functions of HV1.
ACKNOWLEDGMENTS
We appreciate helpful corrections and comments provided by I. Scott Ramsey and Robert H. Fillingame.
This work was supported by National Institutes of Health grant GM102336 and National Science Foundation grant MCB-1242985.
The authors declare no competing financial interests.
Author contributions: T.E. DeCoursey wrote the paper. D. Morgan, B. Musset, and V.V. Cherny analyzed existing data for Tables 3 and 5. All authors read and approved the manuscript.
Lesley C. Anson served as editor.
Footnotes
Abbreviations used:
- EPR
- electron paramagnetic resonance
- MD
- molecular dynamics
- NADPH
- nicotinamide adenine dinucleotide phosphate
- NEM
- n-ethylmaleimide
- TM
- transmembrane
- VSD
- voltage-sensing domain
References
- Alabi, A.A., Bahamonde M.I., Jung H.J., Kim J.I., and Swartz K.J.. 2007. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 450:370–375. 10.1038/nature06266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almers, W. 1978. Gating currents and charge movements in excitable membranes. Rev. Physiol. Biochem. Pharmacol. 82:96–190. [DOI] [PubMed] [Google Scholar]
- Balashov, S.P., Petrovskaya L.E., Imasheva E.S., Lukashev E.P., Dioumaev A.K., Wang J.M., Sychev S.V., Dolgikh D.A., Rubin A.B., Kirpichnikov M.P., and Lanyi J.K.. 2013. Breaking the carboxyl rule: lysine 96 facilitates reprotonation of the Schiff base in the photocycle of a retinal protein from Exiguobacterium sibiricum. J. Biol. Chem. 288:21254–21265. 10.1074/jbc.M113.465138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bánfi, B., Schrenzel J., Nüsse O., Lew D.P., Ligeti E., Krause K.H., and Demaurex N.. 1999. A novel H+ conductance in eosinophils: Unique characteristics and absence in chronic granulomatous disease. J. Exp. Med. 190:183–194. 10.1084/jem.190.2.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, T.K., and Isacoff E.Y.. 2011. The pore of the voltage-gated proton channel. Neuron. 72:991–1000. 10.1016/j.neuron.2011.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezanilla, F., and Villalba-Galea C.A.. 2013. The gating charge should not be estimated by fitting a two-state model to a Q-V curve. J. Gen. Physiol. 142:575–578. 10.1085/jgp.201311056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byerly, L., Meech R., and Moody W. Jr. 1984. Rapidly activating hydrogen ion currents in perfused neurones of the snail, Lymnaea stagnalis. J. Physiol. 351:199–216. 10.1113/jphysiol.1984.sp015241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capasso, M., Bhamrah M.K., Henley T., Boyd R.S., Langlais C., Cain K., Dinsdale D., Pulford K., Khan M., Musset B., et al. 2010. HVCN1 modulates BCR signal strength via regulation of BCR-dependent generation of reactive oxygen species. Nat. Immunol. 11:265–272. 10.1038/ni.1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capasso, M., DeCoursey T.E., and Dyer M.J.S.. 2011. pH regulation and beyond: unanticipated functions for the voltage-gated proton channel, HVCN1. Trends Cell Biol. 21:20–28. 10.1016/j.tcb.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin, A., Qiu F., Rebolledo S., Wang Y., Noskov S.Y., and Larsson H.P.. 2014. Hydrophobic plug functions as a gate in voltage-gated proton channels. Proc. Natl. Acad. Sci. USA. 111:E273–E282. 10.1073/pnas.1318018111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin, A., Qiu F., Wang Y., Noskov S.Y., and Larsson H.P.. 2015. Mapping the gating and permeation pathways in the voltage-gated proton channel Hv1. J. Mol. Biol. 427:131–145. 10.1016/j.jmb.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves, G., Derst C., Franzen A., Mashimo Y., Machida R., and Musset B.. 2016. Identification of an HV1 voltage-gated proton channel in insects. FEBS J. 283:1453–1464. 10.1111/febs.13680 [DOI] [PubMed] [Google Scholar]
- Chen, K., Hirst J., Camba R., Bonagura C.A., Stout C.D., Burgess B.K., and Armstrong F.A.. 2000. Atomically defined mechanism for proton transfer to a buried redox centre in a protein. Nature. 405:814–817. 10.1038/35015610 [DOI] [PubMed] [Google Scholar]
- Cherny, V.V., and DeCoursey T.E.. 1999. pH-dependent inhibition of voltage-gated H+ currents in rat alveolar epithelial cells by Zn2+ and other divalent cations. J. Gen. Physiol. 114:819–838. 10.1085/jgp.114.6.819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny, V.V., Markin V.S., and DeCoursey T.E.. 1995. The voltage-activated hydrogen ion conductance in rat alveolar epithelial cells is determined by the pH gradient. J. Gen. Physiol. 105:861–896. 10.1085/jgp.105.6.861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny, V.V., Henderson L.M., and DeCoursey T.E.. 1997. Proton and chloride currents in Chinese hamster ovary cells. Membr. Cell Biol. 11:337–347. [PubMed] [Google Scholar]
- Cherny, V.V., Henderson L.M., Xu W., Thomas L.L., and DeCoursey T.E.. 2001. Activation of NADPH oxidase-related proton and electron currents in human eosinophils by arachidonic acid. J. Physiol. 535:783–794. 10.1111/j.1469-7793.2001.00783.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherny, V.V., Morgan D., Musset B., Chaves G., Smith S.M.E., and DeCoursey T.E.. 2015. Tryptophan 207 is crucial to the unique properties of the human voltage-gated proton channel, hHV1. J. Gen. Physiol. 146:343–356. 10.1085/jgp.201511456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish, A.J., Gärtner K., Yang H., Peters J.W., and Hegg E.L.. 2011. Mechanism of proton transfer in [FeFe]-hydrogenase from Clostridium pasteurianum. J. Biol. Chem. 286:38341–38347. 10.1074/jbc.M111.254664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtenay, E.S., Capp M.W., and Record M.T. Jr. 2001. Thermodynamics of interactions of urea and guanidinium salts with protein surface: relationship between solute effects on protein processes and changes in water-accessible surface area. Protein Sci. 10:2485–2497. 10.1110/ps.ps.20801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E. 1991. Hydrogen ion currents in rat alveolar epithelial cells. Biophys. J. 60:1243–1253. 10.1016/S0006-3495(91)82158-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E. 2003a. Interactions between NADPH oxidase and voltage-gated proton channels: why electron transport depends on proton transport. FEBS Lett. 555:57–61. 10.1016/S0014-5793(03)01103-7 [DOI] [PubMed] [Google Scholar]
- DeCoursey, T.E. 2003b. Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 83:475–579. 10.1152/physrev.00028.2002 [DOI] [PubMed] [Google Scholar]
- DeCoursey, T.E. 2010. Voltage-gated proton channels find their dream job managing the respiratory burst in phagocytes. Physiology (Bethesda). 25:27–40. 10.1152/physiol.00039.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E. 2012. Voltage-gated proton channels. Compr. Physiol. 2:1355–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E. 2013. Voltage-gated proton channels: molecular biology, physiology, and pathophysiology of the HV family. Physiol. Rev. 93:599–652. 10.1152/physrev.00011.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E. 2015a. Structural revelations of the human proton channel. Proc. Natl. Acad. Sci. USA. 112:13430–13431. 10.1073/pnas.1518486112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E. 2015b. The voltage-gated proton channel: a riddle, wrapped in a mystery, inside an enigma. Biochemistry. 54:3250–3268. 10.1021/acs.biochem.5b00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E. 2016. The intimate and controversial relationship between voltage gated proton channels and the phagocyte NADPH oxidase. Immunol. Rev. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E., and Cherny V.V.. 1993. Potential, pH, and arachidonate gate hydrogen ion currents in human neutrophils. Biophys. J. 65:1590–1598. 10.1016/S0006-3495(93)81198-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E., and Cherny V.V.. 1994. Voltage-activated hydrogen ion currents. J. Membr. Biol. 141:203–223. 10.1007/BF00235130 [DOI] [PubMed] [Google Scholar]
- DeCoursey, T.E., and Cherny V.V.. 1996. Effects of buffer concentration on voltage-gated H+ currents: does diffusion limit the conductance? Biophys. J. 71:182–193. 10.1016/S0006-3495(96)79215-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E., and Cherny V.V.. 1997. Deuterium isotope effects on permeation and gating of proton channels in rat alveolar epithelium. J. Gen. Physiol. 109:415–434. 10.1085/jgp.109.4.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E., and Cherny V.V.. 1998. Temperature dependence of voltage-gated H+ currents in human neutrophils, rat alveolar epithelial cells, and mammalian phagocytes. J. Gen. Physiol. 112:503–522. 10.1085/jgp.112.4.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E., and Hosler J.. 2014. Philosophy of voltage-gated proton channels. J. R. Soc. Interface. 11:20130799. 10.1098/rsif.2013.0799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E., Cherny V.V., Zhou W., and Thomas L.L.. 2000. Simultaneous activation of NADPH oxidase-related proton and electron currents in human neutrophils. Proc. Natl. Acad. Sci. USA. 97:6885–6889. 10.1073/pnas.100047297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E., Cherny V.V., DeCoursey A.G., Xu W., and Thomas L.L.. 2001a. Interactions between NADPH oxidase-related proton and electron currents in human eosinophils. J. Physiol. 535:767–781. 10.1111/j.1469-7793.2001.00767.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E., Cherny V.V., Morgan D., Katz B.Z., and Dinauer M.C.. 2001b. The gp91phox component of NADPH oxidase is not the voltage-gated proton channel in phagocytes, but it helps. J. Biol. Chem. 276:36063–36066. 10.1074/jbc.C100352200 [DOI] [PubMed] [Google Scholar]
- DeCoursey, T.E., Morgan D., and Cherny V.V.. 2002. The gp91phox component of NADPH oxidase is not a voltage-gated proton channel. J. Gen. Physiol. 120:773–779. 10.1085/jgp.20028704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCoursey, T.E., Morgan D., and Cherny V.V.. 2003. The voltage dependence of NADPH oxidase reveals why phagocytes need proton channels. Nature. 422:531–534. 10.1038/nature01523 [DOI] [PubMed] [Google Scholar]
- Demaurex, N. 2012. Functions of proton channels in phagocytes. Wiley Interdiscip. Rev. Membr. Transp. Signal. 1:3–15. 10.1002/wmts.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroshenko, P.A., Kostyuk P.G., and Martynyuk A.E.. 1986. Transmembrane outward hydrogen current in intracellularly perfused neurones of the snail Helix pomatia. Gen. Physiol. Biophys. 5:337–350. [PubMed] [Google Scholar]
- Dudev, T., Musset B., Morgan D., Cherny V.V., Smith S.M.E., Mazmanian K., DeCoursey T.E., and Lim C.. 2015. Selectivity mechanism of the voltage-gated proton channel, HV1. Sci. Rep. 5:10320. 10.1038/srep10320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder, C., and DeCoursey T.E.. 2001. Voltage-gated proton channels in microglia. Prog. Neurobiol. 64:277–305. 10.1016/S0301-0082(00)00062-9 [DOI] [PubMed] [Google Scholar]
- El Chemaly, A., Okochi Y., Sasaki M., Arnaudeau S., Okamura Y., and Demaurex N.. 2010. VSOP/Hv1 proton channels sustain calcium entry, neutrophil migration, and superoxide production by limiting cell depolarization and acidification. J. Exp. Med. 207:129–139. 10.1084/jem.20091837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- England, J.L., and Haran G.. 2011. Role of solvation effects in protein denaturation: from thermodynamics to single molecules and back. Annu. Rev. Phys. Chem. 62:257–277. 10.1146/annurev-physchem-032210-103531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingame, R.H., Angevine C.M., and Dmitriev O.Y.. 2002. Coupling proton movements to c-ring rotation in F1Fo ATP synthase: aqueous access channels and helix rotations at the a-c interface. Biochim. Biophys. Acta. 1555:29–36. 10.1016/S0005-2728(02)00250-5 [DOI] [PubMed] [Google Scholar]
- Fischer, H. 2012. Function of proton channels in lung epithelia. Wiley Interdiscip. Rev. Membr. Transp. Signal. 1:247–258. 10.1002/wmts.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, Y., Kurokawa T., Takeshita K., Kobayashi M., Okochi Y., Nakagawa A., and Okamura Y.. 2012. The cytoplasmic coiled-coil mediates cooperative gating temperature sensitivity in the voltage-gated H+ channel Hv1. Nat. Commun. 3:816. 10.1038/ncomms1823 [DOI] [PubMed] [Google Scholar]
- Fujiwara, Y., Kurokawa T., Takeshita K., Nakagawa A., Larsson H.P., and Okamura Y.. 2013a. Gating of the designed trimeric/tetrameric voltage-gated H+ channel. J. Physiol. 591:627–640. 10.1113/jphysiol.2012.243006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, Y., Takeshita K., Nakagawa A., and Okamura Y.. 2013b. Structural characteristics of the redox-sensing coiled coil in the voltage-gated H+ channel. J. Biol. Chem. 288:17968–17975. 10.1074/jbc.M113.459024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara, Y., Kurokawa T., and Okamura Y.. 2014. Long α helices projecting from the membrane as the dimer interface in the voltage-gated H+ channel. J. Gen. Physiol. 143:377–386. 10.1085/jgp.201311082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, C., Koch H.P., Drum B.M., and Larsson H.P.. 2010. Strong cooperativity between subunits in voltage-gated proton channels. Nat. Struct. Mol. Biol. 17:51–56. 10.1038/nsmb.1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez, C., Rebolledo S., Perez M.E., and Larsson H.P.. 2013. Molecular mechanism of voltage sensing in voltage-gated proton channels. J. Gen. Physiol. 141:275–285. 10.1085/jgp.201210857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, L.M. 1998. Role of histidines identified by mutagenesis in the NADPH oxidase-associated H+ channel. J. Biol. Chem. 273:33216–33223. 10.1074/jbc.273.50.33216 [DOI] [PubMed] [Google Scholar]
- Henderson, L.M., and Chappell J.B.. 1992. The NADPH-oxidase-associated H+ channel is opened by arachidonate. Biochem. J. 283:171–175. 10.1042/bj2830171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, L.M., and Chappell J.B.. 1996. NADPH oxidase of neutrophils. Biochim. Biophys. Acta. 1273:87–107. 10.1016/0005-2728(95)00140-9 [DOI] [PubMed] [Google Scholar]
- Henderson, L.M., and Meech R.W.. 1999. Evidence that the product of the human X-linked CGD gene, gp91-phox, is a voltage-gated H+ pathway. J. Gen. Physiol. 114:771–786. 10.1085/jgp.114.6.771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, L.M., and Meech R.W.. 2002. Proton conduction through gp91phox. J. Gen. Physiol. 120:759–765. 10.1085/jgp.20028708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, L.M., Chappell J.B., and Jones O.T.G.. 1987. The superoxide-generating NADPH oxidase of human neutrophils is electrogenic and associated with an H+ channel. Biochem. J. 246:325–329. 10.1042/bj2460325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, L.M., Chappell J.B., and Jones O.T.G.. 1988. Superoxide generation by the electrogenic NADPH oxidase of human neutrophils is limited by the movement of a compensating charge. Biochem. J. 255:285–290. [PMC free article] [PubMed] [Google Scholar]
- Henderson, L.M., Banting G., and Chappell J.B.. 1995. The arachidonate-activable, NADPH oxidase-associated H+ channel. Evidence that gp91-phox functions as an essential part of the channel. J. Biol. Chem. 270:5909–5916. 10.1074/jbc.270.11.5909 [DOI] [PubMed] [Google Scholar]
- Henderson, L.M., Thomas S., Banting G., and Chappell J.B.. 1997. The arachidonate-activatable, NADPH oxidase-associated H+ channel is contained within the multi-membrane-spanning N-terminal region of gp91-phox. Biochem. J. 325:701–705. 10.1042/bj3250701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, A.L., and Huxley A.F.. 1952. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 117:500–544. 10.1113/jphysiol.1952.sp004764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondares, E., Brown M.A., Musset B., Morgan D., Cherny V.V., Taubert C., Bhamrah M.K., Coe D., Marelli-Berg F., Gribben J.G., et al. 2014. Enhanced activation of an amino-terminally truncated isoform of the voltage-gated proton channel HVCN1 enriched in malignant B cells. Proc. Natl. Acad. Sci. USA. 111:18078–18083. 10.1073/pnas.1411390111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, L., Pathak M.M., Kim I.H., Ta D., and Tombola F.. 2013. Voltage-sensing domain of voltage-gated proton channel Hv1 shares mechanism of block with pore domains. Neuron. 77:274–287. 10.1016/j.neuron.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, L., Kim I.H., and Tombola F.. 2014. Molecular determinants of Hv1 proton channel inhibition by guanidine derivatives. Proc. Natl. Acad. Sci. USA. 111:9971–9976. 10.1073/pnas.1324012111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, L., Singh V., Wulff H., and Tombola F.. 2015. Interrogation of the intersubunit interface of the open Hv1 proton channel with a probe of allosteric coupling. Sci. Rep. 5:14077. 10.1038/srep14077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovannisci, D., Illek B., and Fischer H.. 2010. Function of the HVCN1 proton channel in airway epithelia and a naturally occurring mutation, M91T. J. Gen. Physiol. 136:35–46. 10.1085/jgp.200910379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Morales, D., Liang J., and Eisenberg B.. 2012. Ionizable side chains at catalytic active sites of enzymes. Eur. Biophys. J. 41:449–460. 10.1007/s00249-012-0798-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns, S.J. 2016. TOPO2. Transmembrane protein display software. http://www.sacs.ucsf.edu/TOPO2 (accessed April 21, 2016).
- Kalia, J., and Swartz K.J.. 2011. Elucidating the molecular basis of action of a classic drug: guanidine compounds as inhibitors of voltage-gated potassium channels. Mol. Pharmacol. 80:1085–1095. 10.1124/mol.111.074989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B.E., and Baker B.J.. 2016. Pado, a fluorescent protein with proton channel activity can optically monitor membrane potential, intracellular pH, and map gap junctions. Sci. Rep. 6:23865. 10.1038/srep23865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapus, A., Romanek R., and Grinstein S.. 1994. Arachidonic acid stimulates the plasma membrane H+ conductance of macrophages. J. Biol. Chem. 269:4736–4745. [PubMed] [Google Scholar]
- Kawanabe, A., and Okamura Y.. 2016. Effects of unsaturated fatty acids on the kinetics of voltage-gated proton channels heterologously expressed in cultured cells. J. Physiol. 594:595–610. 10.1113/JP271274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian, J.A., and von Heijne G.. 2000. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 25:429–434. 10.1016/S0968-0004(00)01626-1 [DOI] [PubMed] [Google Scholar]
- Koch, H.P., Kurokawa T., Okochi Y., Sasaki M., Okamura Y., and Larsson H.P.. 2008. Multimeric nature of voltage-gated proton channels. Proc. Natl. Acad. Sci. USA. 105:9111–9116. 10.1073/pnas.0801553105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulleperuma, K., Smith S.M.E., Morgan D., Musset B., Holyoake J., Chakrabarti N., Cherny V.V., DeCoursey T.E., and Pomès R.. 2013. Construction and validation of a homology model of the human voltage-gated proton channel hHV1. J. Gen. Physiol. 141:445–465. 10.1085/jgp.201210856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno, M., Ando H., Morihata H., Sakai H., Mori H., Sawada M., and Oiki S.. 2009. Temperature dependence of proton permeation through a voltage-gated proton channel. J. Gen. Physiol. 134:191–205. 10.1085/jgp.200910213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa, T., and Okamura Y.. 2014. Mapping of sites facing aqueous environment of voltage-gated proton channel at resting state: a study with PEGylation protection. Biochim. Biophys. Acta. 1838:382–387. 10.1016/j.bbamem.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Lacroix, J.J., Hyde H.C., Campos F.V., and Bezanilla F.. 2014. Moving gating charges through the gating pore in a Kv channel voltage sensor. Proc. Natl. Acad. Sci. USA. 111:E1950–E1959. 10.1073/pnas.1406161111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landolt-Marticorena, C., Williams K.A., Deber C.M., and Reithmeier R.A.. 1993. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J. Mol. Biol. 229:602–608. 10.1006/jmbi.1993.1066 [DOI] [PubMed] [Google Scholar]
- Lecar, H., Larsson H.P., and Grabe M.. 2003. Electrostatic model of S4 motion in voltage-gated ion channels. Biophys. J. 85:2854–2864. 10.1016/S0006-3495(03)74708-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.Y., Letts J.A., and Mackinnon R.. 2008. Dimeric subunit stoichiometry of the human voltage-dependent proton channel Hv1. Proc. Natl. Acad. Sci. USA. 105:7692–7695. 10.1073/pnas.0803277105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts, J.A. 2014. Functional and structural studies of the human voltage-gated proton channel. PhD thesis. The Rockefeller University, New York. 209 pp. [Google Scholar]
- Li, Q., Wanderling S., Paduch M., Medovoy D., Singharoy A., McGreevy R., Villalba-Galea C.A., Hulse R.E., Roux B., Schulten K., et al. 2014. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat. Struct. Mol. Biol. 21:244–252. 10.1038/nsmb.2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Shen R., Treger J.S., Wanderling S.S., Milewski W., Siwowska K., Bezanilla F., and Perozo E.. 2015. Resting state of the human proton channel dimer in a lipid bilayer. Proc. Natl. Acad. Sci. USA. 112:E5926–E5935. 10.1073/pnas.1515043112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.J., Zhao Q., Zhou Q., Unno H., Zhai Y., and Sun F.. 2010. The role and structure of the carboxyl-terminal domain of the human voltage-gated proton channel Hv1. J. Biol. Chem. 285:12047–12054. 10.1074/jbc.M109.040360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lishko, P.V., Botchkina I.L., Fedorenko A., and Kirichok Y.. 2010. Acid extrusion from human spermatozoa is mediated by flagellar voltage-gated proton channel. Cell. 140:327–337. 10.1016/j.cell.2009.12.053 [DOI] [PubMed] [Google Scholar]
- Lishko, P.V., Kirichok Y., Ren D., Navarro B., Chung J.J., and Clapham D.E.. 2012. The control of male fertility by spermatozoan ion channels. Annu. Rev. Physiol. 74:453–475. 10.1146/annurev-physiol-020911-153258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoto, H.H., Nordbo E., Baykov A.A., Lahti R., and Malinen A.M.. 2013. Membrane Na+-pyrophosphatases can transport protons at low sodium concentrations. J. Biol. Chem. 288:35489–35499. 10.1074/jbc.M113.510909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaut-Smith, M.P. 1989. The effect of zinc on calcium and hydrogen ion currents in intact snail neurones. J. Exp. Biol. 145:455–464. [DOI] [PubMed] [Google Scholar]
- Makhatadze, G.I., and Privalov P.L.. 1992. Protein interactions with urea and guanidinium chloride: A calorimetric study. J. Mol. Biol. 226:491–505. 10.1016/0022-2836(92)90963-K [DOI] [PubMed] [Google Scholar]
- Maturana, A., Krause K.H., and Demaurex N.. 2002. NOX family NADPH oxidases: Do they have built-in proton channels? J. Gen. Physiol. 120:781–786. 10.1085/jgp.20028713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M.J., Oldenburg M., and Fillingame R.H.. 1990. The essential carboxyl group in subunit c of the F1F0 ATP synthase can be moved and H+-translocating function retained. Proc. Natl. Acad. Sci. USA. 87:4900–4904. 10.1073/pnas.87.13.4900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mony, L., Berger T.K., and Isacoff E.Y.. 2015. A specialized molecular motion opens the Hv1 voltage-gated proton channel. Nat. Struct. Mol. Biol. 22:283–290. 10.1038/nsmb.2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, D., Cherny V.V., Finnegan A., Bollinger J., Gelb M.H., and DeCoursey T.E.. 2007. Sustained activation of proton channels and NADPH oxidase in human eosinophils and murine granulocytes requires PKC but not cPLA2 α activity. J. Physiol. 579:327–344. 10.1113/jphysiol.2006.124248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, D., Capasso M., Musset B., Cherny V.V., Ríos E., Dyer M.J.S., and DeCoursey T.E.. 2009. Voltage-gated proton channels maintain pH in human neutrophils during phagocytosis. Proc. Natl. Acad. Sci. USA. 106:18022–18027. 10.1073/pnas.0905565106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, D., Musset B., Kulleperuma K., Smith S.M.E., Rajan S., Cherny V.V., Pomès R., and DeCoursey T.E.. 2013. Peregrination of the selectivity filter delineates the pore of the human voltage-gated proton channel hHV1. J. Gen. Physiol. 142:625–640. 10.1085/jgp.201311045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, R., and DeCoursey T.E.. 2006. Charge compensation during the phagocyte respiratory burst. Biochim. Biophys. Acta. 1757:996–1011. 10.1016/j.bbabio.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Musset, B., Cherny V.V., Morgan D., Okamura Y., Ramsey I.S., Clapham D.E., and DeCoursey T.E.. 2008a. Detailed comparison of expressed and native voltage-gated proton channel currents. J. Physiol. 586:2477–2486. 10.1113/jphysiol.2007.149427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musset, B., Morgan D., Cherny V.V., MacGlashan D.W. Jr., Thomas L.L., Ríos E., and DeCoursey T.E.. 2008b. A pH-stabilizing role of voltage-gated proton channels in IgE-mediated activation of human basophils. Proc. Natl. Acad. Sci. USA. 105:11020–11025. 10.1073/pnas.0800886105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musset, B., Cherny V.V., Morgan D., and DeCoursey T.E.. 2009. The intimate and mysterious relationship between proton channels and NADPH oxidase. FEBS Lett. 583:7–12. 10.1016/j.febslet.2008.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musset, B., Capasso M., Cherny V.V., Morgan D., Bhamrah M., Dyer M.J.S., and DeCoursey T.E.. 2010a. Identification of Thr29 as a critical phosphorylation site that activates the human proton channel Hvcn1 in leukocytes. J. Biol. Chem. 285:5117–5121. 10.1074/jbc.C109.082727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musset, B., Smith S.M.E., Rajan S., Cherny V.V., Sujai S., Morgan D., and DeCoursey T.E.. 2010b. Zinc inhibition of monomeric and dimeric proton channels suggests cooperative gating. J. Physiol. 588:1435–1449. 10.1113/jphysiol.2010.188318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musset, B., Smith S.M.E., Rajan S., Cherny V.V., Morgan D., and DeCoursey T.E.. 2010c. Oligomerization of the voltage-gated proton channel. Channels (Austin). 4:260–265. 10.4161/chan.4.4.12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musset, B., Smith S.M.E., Rajan S., Morgan D., Cherny V.V., and DeCoursey T.E.. 2011. Aspartate 112 is the selectivity filter of the human voltage-gated proton channel. Nature. 480:273–277. 10.1038/nature10557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura, Y., Fujiwara Y., and Sakata S.. 2015. Gating mechanisms of voltage-gated proton channels. Annu. Rev. Biochem. 84:685–709. 10.1146/annurev-biochem-060614-034307 [DOI] [PubMed] [Google Scholar]
- Okuda, H., Yonezawa Y., Takano Y., Okamura Y., and Fujiwara Y.. 2016. Direct interaction between the voltage sensors produces cooperative sustained deactivation in voltage-gated H+ channel dimers. J. Biol. Chem. 291:5935–5947. 10.1074/jbc.M115.666834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazian, D.M., Shao X.M., Seoh S.A., Mock A.F., Huang Y., and Wainstock D.H.. 1995. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 14:1293–1301. 10.1016/0896-6273(95)90276-7 [DOI] [PubMed] [Google Scholar]
- Pless, S.A., Galpin J.D., Niciforovic A.P., and Ahern C.A.. 2011. Contributions of counter-charge in a potassium channel voltage-sensor domain. Nat. Chem. Biol. 7:617–623. 10.1038/nchembio.622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, F., Rebolledo S., Gonzalez C., and Larsson H.P.. 2013. Subunit interactions during cooperative opening of voltage-gated proton channels. Neuron. 77:288–298. 10.1016/j.neuron.2012.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, I.S., Moran M.M., Chong J.A., and Clapham D.E.. 2006. A voltage-gated proton-selective channel lacking the pore domain. Nature. 440:1213–1216. 10.1038/nature04700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, I.S., Mokrab Y., Carvacho I., Sands Z.A., Sansom M.S.P., and Clapham D.E.. 2010. An aqueous H+ permeation pathway in the voltage-gated proton channel Hv1. Nat. Struct. Mol. Biol. 17:869–875. 10.1038/nsmb.1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, R.A., and Stokes R.H.. 1959. Electrolyte Solutions, the Measurement and Interpretation of Conductance, Chemical Potential, and Diffusion in Solutions of Simple Electrolytes. Second edition. Butterworths Scientific Publications, London. 559 pp. [Google Scholar]
- Rodriguez, J.D., Haq S., Nowak K.F., Morgan D., Cherny V.V., Bernstein S., Sapp M.S., Curcuru J.R., Antchouey C., Nowak S.J., et al. 2015. Characterization and subcellular localization of HV1 in Lingulodinium polyedrum confirms its role in bioluminescence. Biophys. J. 108:425a. 10.1016/j.bpj.2014.11.2325 [DOI] [Google Scholar]
- Ruivo, R., Bellenchi G.C., Chen X., Zifarelli G., Sagné C., Debacker C., Pusch M., Supplisson S., and Gasnier B.. 2012. Mechanism of proton/substrate coupling in the heptahelical lysosomal transporter cystinosin. Proc. Natl. Acad. Sci. USA. 109:E210–E217. 10.1073/pnas.1115581109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata, S., Kurokawa T., Nørholm M.H., Takagi M., Okochi Y., von Heijne G., and Okamura Y.. 2010. Functionality of the voltage-gated proton channel truncated in S4. Proc. Natl. Acad. Sci. USA. 107:2313–2318. 10.1073/pnas.0911868107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, M., Takagi M., and Okamura Y.. 2006. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 312:589–592. 10.1126/science.1122352 [DOI] [PubMed] [Google Scholar]
- Seredenina, T., Demaurex N., and Krause K.H.. 2015. Voltage-gated proton channels as novel drug targets: From NADPH oxidase regulation to sperm biology. Antioxid. Redox Signal. 23:490–513. 10.1089/ars.2013.5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigg, D., and Bezanilla F.. 1997. Total charge movement per channel. The relation between gating charge displacement and the voltage sensitivity of activation. J. Gen. Physiol. 109:27–39. 10.1085/jgp.109.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S.M.E., and DeCoursey T.E.. 2013. Consequences of dimerization of the voltage-gated proton channel. Prog. Mol. Biol. Transl. Sci. 117:335–360. 10.1016/B978-0-12-386931-9.00012-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S.M.E., Morgan D., Musset B., Cherny V.V., Place A.R., Hastings J.W., and DeCoursey T.E.. 2011. Voltage-gated proton channel in a dinoflagellate. Proc. Natl. Acad. Sci. USA. 108:18162–18167. 10.1073/pnas.1115405108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starace, D.M., and Bezanilla F.. 2001. Histidine scanning mutagenesis of basic residues of the S4 segment of the Shaker K+ channel. J. Gen. Physiol. 117:469–490. 10.1085/jgp.117.5.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starace, D.M., and Bezanilla F.. 2004. A proton pore in a potassium channel voltage sensor reveals a focused electric field. Nature. 427:548–553. 10.1038/nature02270 [DOI] [PubMed] [Google Scholar]
- Suszták, K., Mócsai A., Ligeti E., and Kapus A.. 1997. Electrogenic H+ pathway contributes to stimulus-induced changes of internal pH and membrane potential in intact neutrophils: role of cytoplasmic phospholipase A2. Biochem. J. 325:501–510. 10.1042/bj3250501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita, K., Sakata S., Yamashita E., Fujiwara Y., Kawanabe A., Kurokawa T., Okochi Y., Matsuda M., Narita H., Okamura Y., and Nakagawa A.. 2014. X-ray crystal structure of voltage-gated proton channel. Nat. Struct. Mol. Biol. 21:352–357. 10.1038/nsmb.2783 [DOI] [PubMed] [Google Scholar]
- Tao, X., Lee A., Limapichat W., Dougherty D.A., and MacKinnon R.. 2010. A gating charge transfer center in voltage sensors. Science. 328:67–73. 10.1126/science.1185954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, A.R., Chrachri A., Wheeler G., Goddard H., and Brownlee C.. 2011. A voltage-gated H+ channel underlying pH homeostasis in calcifying coccolithophores. PLoS Biol. 9:e1001085. 10.1371/journal.pbio.1001085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, A.R., Brownlee C., and Wheeler G.L.. 2012. Proton channels in algae: reasons to be excited. Trends Plant Sci. 17:675–684. 10.1016/j.tplants.2012.06.009 [DOI] [PubMed] [Google Scholar]
- Thomas, R.C., and Meech R.W.. 1982. Hydrogen ion currents and intracellular pH in depolarized voltage-clamped snail neurones. Nature. 299:826–828. 10.1038/299826a0 [DOI] [PubMed] [Google Scholar]
- Thorndycroft, F.H., Butland G., Richardson D.J., and Watmough N.J.. 2007. A new assay for nitric oxide reductase reveals two conserved glutamate residues form the entrance to a proton-conducting channel in the bacterial enzyme. Biochem. J. 401:111–119. 10.1042/BJ20060856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari-Woodruff, S.K., Schulteis C.T., Mock A.F., and Papazian D.M.. 1997. Electrostatic interactions between transmembrane segments mediate folding of Shaker K+ channel subunits. Biophys. J. 72:1489–1500. 10.1016/S0006-3495(97)78797-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola, F., Ulbrich M.H., and Isacoff E.Y.. 2008. The voltage-gated proton channel Hv1 has two pores, each controlled by one voltage sensor. Neuron. 58:546–556. 10.1016/j.neuron.2008.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombola, F., Ulbrich M.H., Kohout S.C., and Isacoff E.Y.. 2010. The opening of the two pores of the Hv1 voltage-gated proton channel is tuned by cooperativity. Nat. Struct. Mol. Biol. 17:44–50. 10.1038/nsmb.1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touret, N., and Grinstein S.. 2002. Voltage-gated proton “channels”: a spectator’s viewpoint. J. Gen. Physiol. 120:767–771. 10.1085/jgp.20028706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Li S.J., Wu X., Che Y., and Li Q.. 2012. Clinicopathological and biological significance of human voltage-gated proton channel Hv1 protein overexpression in breast cancer. J. Biol. Chem. 287:13877–13888. 10.1074/jbc.M112.345280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Wu X., Li Q., Zhang S., and Li S.J.. 2013. Human voltage-gated proton channel hv1: a new potential biomarker for diagnosis and prognosis of colorectal cancer. PLoS One. 8:e70550. 10.1371/journal.pone.0070550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, M.L., Schow E.V., Freites J.A., White S.H., Tombola F., and Tobias D.J.. 2012. Water wires in atomistic models of the Hv1 proton channel. Biochim. Biophys. Acta. 1818:286–293. 10.1016/j.bbamem.2011.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Q., Axelrod H.L., Abresch E.C., Paddock M.L., Okamura M.Y., and Feher G.. 2004. X-Ray structure determination of three mutants of the bacterial photosynthetic reaction centers from Rb. sphaeroides; altered proton transfer pathways. Structure. 12:703–715. 10.1016/j.str.2004.03.001 [DOI] [PubMed] [Google Scholar]
- Yang, N., George A.L. Jr., and Horn R.. 1996. Molecular basis of charge movement in voltage-gated sodium channels. Neuron. 16:113–122. 10.1016/S0896-6273(00)80028-8 [DOI] [PubMed] [Google Scholar]
- Yang, N., George A.L. Jr., and Horn R.. 1997. Probing the outer vestibule of a sodium channel voltage sensor. Biophys. J. 73:2260–2268. 10.1016/S0006-3495(97)78258-4 [DOI] [PMC free article] [PubMed] [Google Scholar]