Summary
Emerging evidence shows that hydrocarbonoclastic bacteria (HCB) may be commonly found associated with phytoplankton in the ocean, but the ecology of these bacteria and how they respond to crude oil remains poorly understood. Here, we used a natural diatom-bacterial assemblage to investigate the diversity and response of HCB associated with a cosmopolitan marine diatom, S. costatum, to crude oil. Pyrosequencing analysis revealed a dramatic transition in the diatom-associated bacterial community, defined initially by a short-lived bloom of Methylophaga (putative oil-degraders) that was subsequently succeeded by a distinct groups of HCB (Marinobacter, Polycyclovorans, Arenibacter, Parvibaculum, Roseobacter clade), including putative novel phyla, as well as other groups with previously unqualified oil-degrading potential. Interestingly, these oil-enriched organisms contributed to the apparent and exclusive biodegradation of substituted and non-substituted polycyclic aromatic hydrocarbons (PAHs), thereby suggesting that the HCB community associated with the diatom is tuned to specializing in the degradation of PAHs. Furthermore, the formation of marine oil snow (MOS) in oil-amended incubations was consistent with its formation during the Deepwater Horizon oil spill. This work highlights the phycosphere of phytoplankton as an underexplored biotope in the ocean where HCB may contribute importantly to the biodegradation of hydrocarbon contaminants in marine surface waters.
Keywords: eukaryotic phytoplankton, hydrocarbon-degrading bacteria, biodegradation, crude oil, Skeletonema, marine environment
Introduction
Marine eukaryotic phytoplankton, or micro-algae, are primary producers that contribute significantly to the earth's total produced oxygen (accounting for approx. 50% in the atmosphere) (Field et al., 1998) and are pivotal in controlling atmospheric carbon dioxide levels through their capacity to fix carbon photoautotrophically (Raven, 1994). Over the years it has become apparent that eukaryotic phytoplankton and bacteria share an intimate co-existence. Eukaryotic phytoplankton harbour on their cell surface, or phycosphere, communities of bacteria whose presence is non-coincidental. Several studies investigating the relationship between phytoplankton and their bacterial associates have shown evidence of some form of symbiotic relationship in this eukaryotic-prokaryotic partnership, often one that is mediated through a reciprocated sharing of a specific nutrient(s). Amin et al., (2009), for example, showed a mutual sharing of iron and fixed carbon between several species of phytoplankton and bacteria, whilst Kazamia et al., (2012) reported the supply of bacterial-produced vitamin B12 to the eukaryote partner in exchange for fixed carbon. The significance of these symbiotic relationships is highlighted by the fact that few phytoplankton cultures can be maintained in the laboratory and sub-cultured for long periods axenically (i.e. completely devoid of their bacterial community).
Emerging evidence over the past decade has revealed the occurrence of eukaryotic phytoplankton as a potentially important biotope for finding hydrocarbon-degrading bacteria (Green et al., 2004; Gutierrez et al., 2014), including obligate hydrocarbonoclastic bacteria (Gutierrez et al., 2012a,b, 2013). The ecology and life cycle of these types of bacteria in the ocean, respective to their possible interactions with and close dependence on other organisms, is an area that is in a nascent phase of understanding. A common misconception when isolating bacteria from seawater is that they are thought to have been occurring in a free-living state, and frequently no attention is given to the possibility that they may have been associated, or attached, to other planktonic surfaces. Aside from the many salts and dissolved inorganic and organic carbon (DIC, DOC) that constitute seawater, the ocean contains enormous quantities of transparent exopolymer (TEP), particulate organic matter (POM), and planktonic eukaryotic organisms (e.g. phytoplankton, zooplankton), all of which can act as a surface to which bacteria can be found attached. The occurrence of hydrocarbon-degrading bacteria living with marine eukaryotic phytoplankton is an intriguing pairing that remains to be qualified in terms of establishing the underlying reason(s) for this partnership. There is evidence that this association may stem from the capacity of eukaryotic phytoplankton cells to accumulate hydrocarbons on their cell surfaces. Some studies, for example, have shown phytoplankton cells to accumulate PAH molecules from the surrounding seawater (Binark et al., 2000; Kowalewska, 1999), which would create a PAH-enriched zone around the phycosphere of the phytoplankton cells and in turn attract PAH-degrading bacteria to colonise this zone. The ability of some eukaryotic phytoplankton to synthesize PAHs (Andelman & Suess, 1970; Gunnison & Alexander, 1975), or long-chain hydrocarbon-like compounds, such as alkenones (Marlowe et al., 1984), may also explain the occurrence of hydrocarbon-degrading bacteria found associated with these organisms.
Sea surface oil slicks can mediate the accumulation of high concentrations of hydrocarbons, particularly PAHs, within the upper layers of the water column. During the Deepwater Horizon (DwH) disaster, for example, high concentrations of n-alkanes were measured in both surface and plume waters at the spill site (Hazen et al., 2010), whereas high-molecular-weight PAHs were found to preferentially predominate in surface waters (Diercks et al., 2010; Hazen et al., 2010; Camilli et al., 2010). PAHs are recognized as high-priority pollutants in the environment (Agency for Toxic Substances and Disease Registry, 2007), and their accumulation in the upper layers of the water column, in particular the euphotic zone, where primary production is mainly confined, can have significant impacts to phytoplankton (Harrison et al., 1986). This was evidenced within the first few weeks following the onset of the DwH spill on April 15, 2010, where a dramatic decline in phytoplankton occurred at the vicinity of the spill site, even before the application of dispersant (Abbriano et al., 2011). This Gulf of Mexico phytoplankton community may have acted as a natural seed of hydrocarbon-degrading bacteria in surface oil slicks and sub-surface dispersed oil in the upper water column. However, the response of natural bacterial communities associated with eukaryotic phytoplankton to oil contamination has, to our knowledge, not been addressed. In light of the evidence showing the occurrence of hydrocarbon-degrading bacteria associated with phytoplankton (Green et al., 2006; Gutierrez et al., 2012a,b; Gutierrez et al., 2013; Gutierrez et al., 2014), this putative symbiotic relationship, including how these natural consortia respond to oil, deserves attention as it would improve our understanding of the biotopes and ecology of oil-degrading bacteria in the ocean.
Photosynthetically enhanced biodegradation of oil hydrocarbons has been demonstrated using artificial combinations of phytoplankton-bacterial consortia (Borde et al., 2003; Muñoz et al., 2003; Safanova et al., 1999; Warshawsky et al., 2007). However, little is known about this process in natural assemblages of these organisms. Furthermore, experiments to address this in the field can be quite challenging and lack the level of control required to make accurate interpretations. Here, we conducted a laboratory-based simulated oil-spill simulated experiment using a non-axenic culture of the marine diatom Skeletonema costatum to investigate its response to crude oil and that of its associated bacterial community. 16S rRNA gene pyrosequence analysis and qPCR were used to monitor the response of the total bacterial community and that of specific hydrocarbon-degrading taxa in oil-amended and non-amended incubations, as well as to infer their possible contribution to the overall biodegradation of the oil.
Results
Degradation of PAHs and ANS crude oil
Table 1 shows the degradation of each of the five PAHs as measured during incubation of non-axenic S. costatum CCAP 1077/1C with the five different PAH compounds. For enrichments utilizing ONR7a medium, significant (P < 0.05) amounts of naphthalene and phenanthrene were degraded after 28 days – respectively 86.2% ± 5.2% and 72.5% ± 3.0%. No significant degradation of anthracene, pyrene or fluorene was detected in incubations using ONR7a medium. For the enrichments utilizing ZM/100 medium, significant amounts of naphthalene, phenanthrene and fluorene were degraded after 28 days – respectively 94.6% ± 4.8%, 81.7% ± 2.2% and 17.7% ± 6.0% of the total initial amount added. No degradation of anthracene or pyrene was measured in these enrichments. Uninoculated controls showed no significant loss of PAHs. Degradation studies in our laboratory with axenic cultures of S. costatum showed an inability to utilize hydrocarbons as a carbon and energy source (unpublished results).
Table 1.
Biodegradation of various PAH compounds by the bacterial community associated with non-axenic S. costatum strain CCAP 1077/1C.
| Treatment | Avg % (± SD) of compound biodegraded a |
|---|---|
| PAH enrichments in ONR7a: | |
| Naphthalene | 86.2 ± 5.2 |
| Phenanthrene | 72.5 ± 3.0 |
| Anthracene | – |
| Pyrene | – |
| Fluorene | – |
| PAH enrichments in ZM/100: | |
| Naphthalene | 94.6 ± 4.8 |
| Phenanthrene | 81.7 ± 2.2 |
| Anthracene | – |
| Pyrene | – |
| Fluorene | 17.7 ± 6.0 |
Values are from triplicate measurements and expressed as a percentage of compound biodegraded after 28 days relative to uninoculated / acid-inhibited controls.
–, no significant degradation measured.
In the crude oil enrichment experiment, we determined the concentrations of various aromatic and aliphatic hydrocarbon species at the time-point when the cultures were amended with ANS crude oil (day 7) and at the termination of the enrichment (day 64). For this, designated cultures were sacrificed for whole-culture extraction, as described above. Compared with acid-inhibited controls, the concentrations of some hydrocarbon species were found to have significantly decreased (P < 0.1) after 64 days in the uninhibited incubations. As shown in Table 2, these hydrocarbons, and the percent they were biodegraded compared to their concentrations in acid-inhibited controls, were naphthalene (19.9 ± 1.1%), 1-methylnaphthalene (5.8 ± 0.4%), 2-methylnaphthalene (29.5 ± 0.9%), biphenyl (11.1 ± 0.2%), C1-naphthalenes (19.6 ± 1.5%) and phenanthrene (2.0 ± 0.2%). No significant biodegradation of aliphatic hydrocarbons analysed (C1-C40) was measured in the uninhibited incubations after 64 days.
Table 2.
Hydrocarbons biodegraded during enrichment of non-axenic S. costatum strain CCAP 1077/1C with ANS crude oil.
| Hydrocarbon | Avg % (± SD) of compound biodegraded a |
|---|---|
| Naphthalene | 19.9 ± 1.1 |
| 1-Methylnaphthalene | 5.8 ± 0.4 |
| 2-Methylnaphthalene | 29.5 ± 0.9 |
| Biphenyl | 11.1 ± 0.2 |
| C1-Naphthalenes | 19.6 ± 1.5 |
| Phenanthrene | 2.0 ± 0.2 |
Values are from triplicate measurements and expressed as a percentage of compound biodegraded after 64 days relative to acid-inhibited controls.
S. costatum and prokaryotic population dynamics
To assess the diatom and its associated prokaryotic (bacterial and archaeal) community response to crude oil, changes in Chla a and total DAPI counts were determined. Chl a concentrations increased from initial values of 0.06 ± 0.00 to 0.16 ± 0.01 μg ml-1 at day 7 across all incubations (Fig. 1), except in the acid-inhibited controls (K7-9) as was expected (results not shown). Amendment of flasks O1-3 with crude oil at day 7 (Fig. 1a), however, had a marked effect on Chl a concentrations compared to the untreated control incubations (Fig. 1b). Whilst growth of the diatom continued in the untreated control, with Chl a concentrations reaching 2.41 ± 0.11 μg ml-1 by the end of the experiment at day 64, in the oil-treated incubations, however, its growth was completely suppressed within 4 days after addition of the oil at day 7 (Fig. 1a). Chl a concentrations reached maximum levels at day 11 (0.25 ± 0.02 cells ml-1) and thereafter gradually decreased to 0.16 ± 0.05 cells ml-1 compared to 2.4 ± 0.11 cells ml-1 in the untreated incubations (C4-6) at day 64. Microscopic examination of the culture liquid from the oil-treated incubations a day after addition of the oil revealed the majority of the diatom cells to have taken on a dark-brown appearance and over time to progressively become bleached. Conversely, S. costatum cells in the untreated incubations (C4-6) displayed a healthy reddish-brown appearance.
Figure 1.
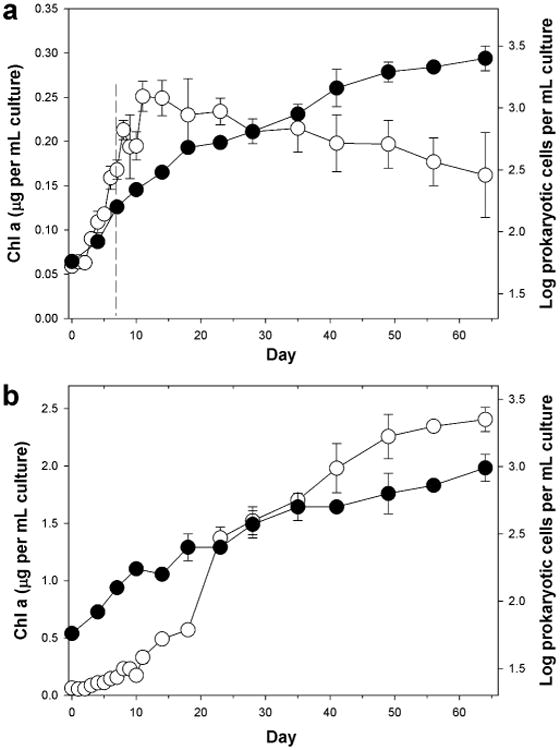
Chlorophyll a concentrations (open circles) and prokaryotic (bacterial and archaeal) cell numbers (solid circles) in incubations of non-axenic S. costatum strain CCAP1077/1C amended with (a) or without (b) ANS crude oil – respectively flasks O1-3 and C4-6. The hatched line at day 7 (top panel) indicates the time-point the oil was added. Note difference in scale of the left-hand y-axis in the two plots. Some error bars are smaller than the symbol.
Prokaryotic (bacteria and archaea) abundance in the oil-amended incubations (O1-3) increased at an average rate of 3.8 × 104 cells L-1 day-1 between days 7 to 41 and reached a maximum cell concentration of 2.6 × 106 cells L-1 at day 64 (Fig. 1a). Prokaryotic numbers in the untreated control (Fig. 1b) showed a similar trend, but their rate of increase (1.1 × 104 cells L-1 day-1) and final abundance at day 64 (9.7 × 105 cells L-1) were markedly lower compared to that in the oil-treated incubations (Fig. 1a). Visually, the oil-treated “live” incubations gradually became more turbid compared to the untreated controls after day 7, which was indicative of prokaryotic growth due to the addition of the oil. We note that whilst our DAPI counts demonstrate an expected pattern for prokaryotic dynamics in the oil-amended and untreated control incubations, these counts are markedly underestimated because we observed high numbers of DAPI-stained prokaryotic cells associated with marine oil snow (MOS), or oil flocs, that were practically impossible to count accurately due to their density and localisation within and on the surface of the MOS flocs.
Dynamics of qPCR-targeted hydrocarbon-degraders
The gene copy number of P. algicola and Marinobacter were quantified by qPCR in the oil-treated (flasks O1-3) and untreated (flasks C4-6) incubations. As shown in Figure 2, the gene copy number of Marinobacter increased by ca. 2 orders of magnitude by day 8 (1 day after addition of the oil) in both the oil-treated (Fig. 2a) and untreated (Fig. 2b) incubations. Thereafter, the dynamic of these organisms became markedly delineated between both treatments. In the oil-treated incubations, the Marinobacter gene copy number continued to increase, reaching maximal concentrations (7.4 ± 0.5 Log genes mL-1) by day 24 – ca. 5 orders of magnitude higher compared to initial concentrations (Fig. 2a). By day 8 in the untreated control incubations, however, the gene copy number for these organisms had reached maximal concentrations (4.5 ± 0.1 Log genes mL-1) and remained at these levels for the remainder of the experiment (Fig. 2b).
Figure 2.
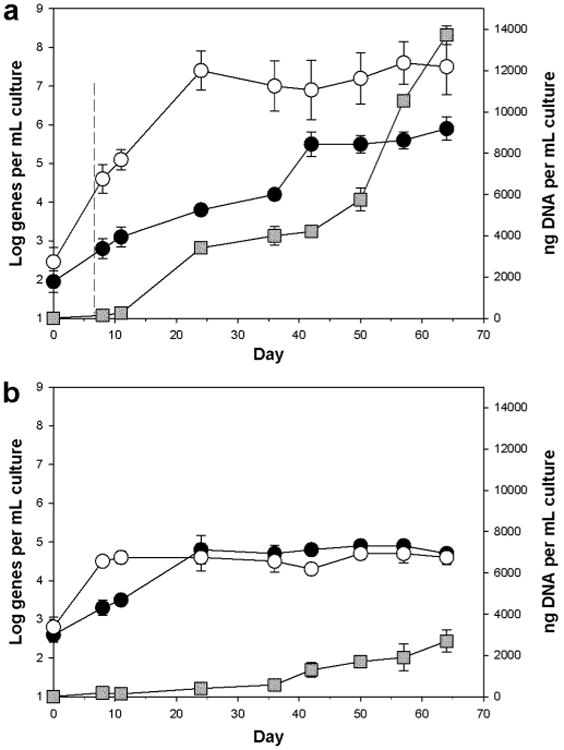
Abundance of Polycyclovorans algicola TG408 (solid circles) and Marinobacter (open circles) 16S rRNA genes in incubations of non-axenic S. costatum strain CCAP1077/1C amended with (a) or without (b) ANS crude oil – respectively flasks O1-3 and C4-6. Each circular point on the graph represents the average and standard deviation of triplicate qPCR measurements calculated as the abundance of 16S rRNA genes per mL of culture. Squares are the average and standard deviation of triplicate measurements of the total mass of DNA per sample. The hatched line at day 7 (top panel) indicates the time-point the oil was added. Some error bars are smaller than the symbol.
In the case of P. algicola, the gene copy number for these organisms increased by almost one order of magnitude by day 8 in both treatments – from initial concentrations of 1.9 ± 0.3 Log genes mL-1 to 2.8 ± 0.3 Log genes mL-1 in the oil-treated incubations (Fig. 2a), and from initial concentrations of 2.6 ± 0.2 Log genes mL-1 to 3.3 ± 0.2 Log genes mL-1 in the untreated incubations (Fig. 2b). In the untreated controls, the gene copy number of these organisms continued to increase and reached maximal levels (4.8 ± 0.3 Log genes mL-1) by day 24, thereafter remained at these levels for the remainder of the experiment. In the oil-treated incubations, however, the gene copy number continued to gradually increase, reaching 4.2 ± 0.2 Log genes mL-1 by day 36. It then increased by more than another order of magnitude to 5.5 ± 0.3 Log genes mL-1 within a period of 6 days, and remained at these concentrations for the remainder of the experiment.
For both the oil treated and untreated incubations the concentration of total DNA, as a proxy for biomass (bacterial, archaeal and S. costatum), showed a general increase throughout the experiment (Fig. 2), though it was markedly more pronounced in the oil-treated incubations (Fig. 2a). For the latter incubations, total DNA concentrations appeared to partially reflect the dynamics of Marinobacter and P. algicola gene copy number up until day 42. Thereafter, DNA concentrations dramatically increased until the termination of the experiment, reaching maximal concentrations of 13.7 ± 0.5 ng DNA L-1. On the other hand, total DNA concentrations in the untreated control incubations reached maximal levels of only 2.7 ± 0.6 ng DNA L-1 by the termination of the experiment. As expected, no increase in DNA mass was measured in the acid-inhibited incubations (results not shown).
Bacterial community analysis
Barcoded 16S rRNA gene pyrosequencing was used to analyze the bacterial communities present in two replicates of each of the oil-treated and untreated incubations – respectively, Flasks O1 and O2, and Flasks C4 and C5. This was performed at day 7 (immediately prior to oil addition), and then at days 10, 35 and 64. Initially, before addition of the oil, the 16S rRNA gene pyrosequencing results revealed a complex bacterial community associated with S. costatum that was dominated (57-70% of total 16S rRNA gene sequence reads) by cultured and uncultured members of the Bacteroidetes, mainly of the family Flavobacteriaceae and members of the Class Flavobacteria, with minority representation from Flammeovirgaceae, Sphingobacteriales and Flexibacteraceae. The community was also dominated by a complex assemblage of Gammaproteobacteria and Alphaproteobacteria that, respectively, included members of the Piscirickettsiaceae and of the Rhodobacteraceae and unclassified Rhizobiales (Fig. 3; Table S1). Minority groups included members of the Hyphomonadaceae and Rhodobacterales. By day 10, the Flavobacteriaceae, and to a lesser extent other unclassified Bacteroidetes, were the major dominating groups. By day 35 and 64, the dominant members of the community belonged to the Order Xanthomonadales, Alphaproteobacteria, Flavobacteriaceae, Rhodobacteriaceae and Rhizobiales. Lower levels of representation to total sequences in each the libraries at the different time-points were also contributed by unclassified Proteobacteria and members of the Order Pirellulales (Phylum Planctomycetes).
Figure 3.
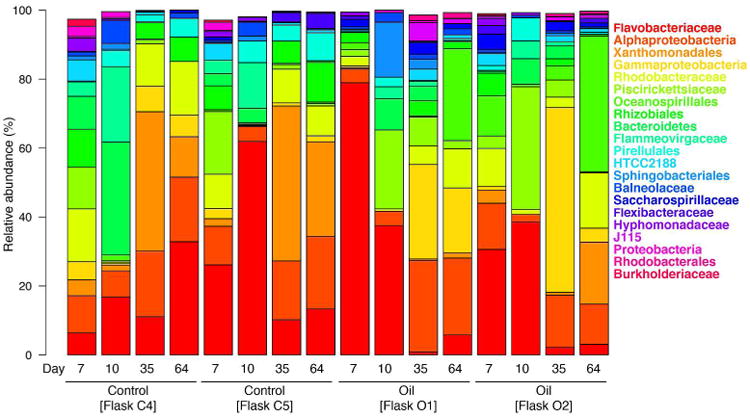
Composition of bacterial 16S rRNA gene pyrosequencing reads from incubations of Skeletonema costatum amended with or without ANS crude oil. The relative abundance of taxa present at >1% relative abundance is shown. Sequences were classified to family-level taxonomy when possible and otherwise a higher-level classification is shown.
Three days following addition of the oil at day 7 to Flasks O1 and O2, the community changed compared to the untreated (no oil) control and became dominated by members of the family Piscirickettsiacae, mainly Methylophaga. By day 35, the major dominating groups were members of the Class Gammaproteobacteria, including the Marinobacter group (Fig. 4). By day 64, members of the Order Xanthomonadales and family Rhodobacteraceae dominated the community as reflected also in the untreated control incubations. However, compared to the untreated control incubations, members of the Oceanospirillales, particularly Marinobacter, were markedly enriched. Overall, ten OTUs were identified to have become enriched by the oil: OTU-512 (Parvibaculum), OTU-282 (Saccharospirillum), OTU-142 (Arenibacter), OTU-9 (Seohaeicola), OTU-256 (Rhoseovarius), OTU-506 (Parvularcula), OTU-202 (Arenibacter), OTU-499 (Methylophaga), OTU-477 (Marinobacter) and OTU-758 (Marinobacter). As shown in Fig. 4, the latter three OTUs showed the strongest succession pattern in the oil-amended incubations, with OTU-499 (Methylophaga) peaking in relative abundance on day 10, OTU-477 (Marinobacter) on day 35, and OTU-758 (Marinobacter) on day 64.
Figure 4.
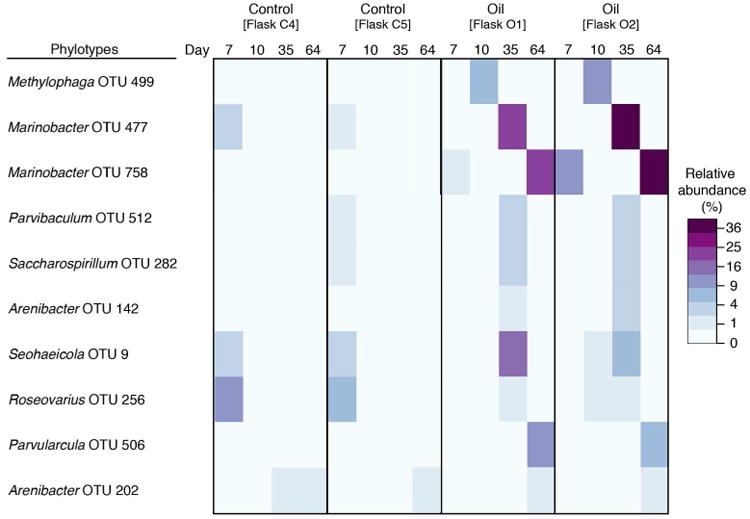
Heatmap of all OTUs enriched in both oil flasks, but not controls. OTUs were considered enriched if there was a mean increase of at least 1% relative abundance (e.g. shift from 1 to 2%) in at least one time point for replicate incubations, and if the difference was statistically significant (P < 0.05, FDR-corrected for multiple comparisons). Color key indicates square-root normalized relative abundance (%). A strong succession pattern in oil flasks was apparent for three OTUs: OTU-499 (Piscirickettsiaceae) peaked in relative abundance on day 10; OTU-477 (unclassified Gammaproteobacteria) peaked in relative abundance on day 35; and OTU-758 (unclassified Oceanospirillales) peaked in relative abundance on day 64.
Discussion
Although the properties that govern the toxicity of hydrocarbons to phytoplankton remain poorly understood, the gradual decline of the diatom following exposure to the ANS crude oil was not unexpected. Toxicological effects of hydrocarbons to phytoplankton, particularly PAHs (Harrison et al., 1986; Ozhan et al., 2014a), have been reported in several studies (Ostgaard et al., 1984a; Ostgaard et al., 1984b; Sargian et al., 2005), including to S. costatum species (Huang et al., 2011); PAHs have been reported to cause toxicity to phytoplankton at concentrations as low as 1 μg L-1 (Ozhan et al., 2014b and references therein). Drawing parallels with the DwH oil spill, a marked decline in phytoplankton abundance, lasting several weeks, was measured immediately following the Macondo blowout in the Gulf of Mexico and was likely associated with exposure of phytoplankton cells to toxic concentrations of Macondo oil hydrocarbons (Abbriano et al., 2011). Furthermore, the aggregation and formation of MOS in the oil-enrichment incubations (flasks O1-O3) following addition of the crude oil at day 7 was consistent with the formation of large quantities of MOS comprising microbial (phytoplankton and bacterial) flocs observed in surface waters within the first week of the DwH blowout in the field (Diercks et al., 2010a; Diercks et al., 2010b; Passow et al., 2012). Their formation has been demonstrated in laboratory roller table bottle incubations using fresh oil slick samples and Gulf of Mexico surface water (Ziervogel et al., 2012); MOS is suggested to originate through the production of copious quantities of extracellular polymeric substances (EPS) by eukaryotic phytoplankton (Passow & Alldredge, 1994), in particular diatoms (Passow et al., 1994), and their interaction with dissolved hydrocarbons and/or emulsified oil droplets (Passow et al., 2012). EPS produced by certain groups of oil-degrading bacteria (Alteromonas, Halomonas, Pseudoalteromonas) that were enriched during the DwH spill has also been shown to trigger the formation of MOS in the presence of crude oil (Gutierrez et al., 2013; and unpublished results). Hence, by nature of their stickiness, these macromolecules, whether produced by S. costatum or any of its associated bacterial members, may have participated in the observed aggregation and subsequent sedimentation of the diatom in our oil-treated incubations.
While the addition of the ANS crude oil at day 7 impacted the diatom negatively, pyrosequencing analysis revealed a dramatic transition in the associated bacterial community. The community was initially dominated by members of the Flavobacteriaceae and Alphaproteobacteria – groups that are typically found associated with marine phytoplankton (Buchan et al., 2014) – and included recognized oil-degrading taxa such as Marinobacter, as well as putative oil-degrading bacterial groups. After addition of the oil, the community shifted and became dominated initially by members of the Piscirickettsiaceae, including Methylophaga. A few studies have reported the enrichment of Methylophaga in oil-contaminated field samples and in laboratory experiments with oil or individual hydrocarbons (Röling et al., 2002; Yakimov et al., 2005; Coulon et al., 2007; Vila et al., 2010). Whilst members of this genus are known to exhibit an almost exclusive requirement for C1 sources (methanol, methylamine, dimethylsulfide) as sole growth substrates, with the exception of some species that are also capable of utilizing fructose (Janvier and Grimont, 1995), recent work has shown that some Methylophaga species possess the ability to utilize hydrocarbons as a sole source of carbon and energy (Mishamandani et al., 2014). Intriguingly, the Methylophaga identified in some of these previous studies were, like in the present study, found to have bloomed in abundance within 3 days of exposure to hydrocarbons and thereafter decreased to almost undetectable levels. The reason for this initial and ephemeral bloom by this group of organisms is intriguing and warrants further investigation. In the meantime, however, we posit that this short-lived bloom may be due to the ability of these Methylophaga to respond quickly upon exposure to oil hydrocarbons before succumbing, within just a few days, to being outcompeted by other members of the Gammaproteobacteria. We also note that these Methylophaga that bloomed in the oil-treated incubations, represented by OTU-499, may represent novel species since they share <98% 16S rRNA gene sequence identity to their closest relative, the type strain Methylophaga nitratireducenticrescens strain JAM1T (Auclair et al., 2010).
Following this initial bloom of Methylophaga and by other members of the Piscirickettsiaceae, our community analysis revealed that S. costatum harbors bacterial groups, such as Arenibacter (OTUs 142 and 202) and Marinobacter (OTUs 477 and 758) – genera comprising members reported to degrade hydrocarbons (Gutierrez et al., 2014; Yakimov et al., 2007) – that were able to respond positively to the crude oil. As expected, P. algicola strain TG408T, which was previously identified living associated with S. costatum and other eukaryotic phytoplankton species, and which is a specialist aromatic hydrocarbon-degrader (Gutierrez et al., 2013), had also become positively enriched by the oil. Intriguingly, a few reports have described the identification of organisms, albeit uncultivated clones, with >97% 16S rRNA gene sequence identity to strain TG408T by sequencing surveys of marine, brackish or sediment environments (Patel et al., 2014; Zeng et al., 2014) and near-coastal terrestrial oil-contaminated environments (Kim & Crowley, 2007). A possible explanation for this may lie in the fastidious and narrow substrate spectrum of these organisms, and/or their ecology, which may be somewhat confined to living associated with eukaryotic phytoplankton (Gutierrez et al., 2013). This may explain why these organisms have remained practically recalcitrant to routine cultivation efforts. Their association with eukaryotic phytoplankton may be viewed as a co-evolutionary adaptation based on the ability of phytoplankton to synthesize PAH chemicals, or to sequester them from the surrounding seawater in the marine water column. Roseovarius was also found enriched in the crude oil amended incubations. This is a taxonomic group of the Roseobacter clade which is commonly found in high abundance during algal blooms (Gonzalez et al., 2000). Their enrichment can be attributed to encoding multiple ring-cleaving pathways that participate in the degradation of monocyclic and polycyclic aromatic hydrocarbons (Moran et al., 2007). Based on a BLASTn analysis of their 16S rRNA gene, these oil-enriched organisms may represent novel species since they shared <96% identity to closest type strains.
Other members of the bacterial community associated with the diatom that had also become enriched included Parvibaculum (OTU-512), Saccharospirillum (OTU-282), Seohaeicola (OTU-9) and Parvularcula (OTU-506). Based on a BLASTn analysis of their 16S rRNA gene sequences, these organisms may also represent novel species since they each shared <97% identity to closest type strains. Of these four genera, only Parvibaculum contains members that have been reported to degrade hydrocarbons – namely Parvibaculum hydrocarboniclasticum strain EPR92T (Rosario-Passapera et al., 2012) and Parvibaculum lavamentivorans strain DS-1T (Schleheck et al., 2004). Whilst there are no reports, to our knowledge, that describe the ability of any member within the genera Saccharospirillum, Seohaeicola and Parvularcula to degrade hydrocarbons, further work would be needed to evaluate this phenotype for these organisms. It should be noted that Saccharospirillum is most closely related to Oceanospirillum and to members of the family Halomonadaceae (Labrenz et al., 2003), both groups of which contain oil-degrading representatives and, hence, suggesting the possibility that this group may have contributed to the biodegradation of the crude oil in the oil-amended incubations.
Marinobacter is recognized as a generalist oil-degrader with a preference for utilizing aliphatic/n-alkane hydrocarbons as a sole carbon and energy source (Yakimov et al., 2005). During an oil spill at sea, the microbial response often commences with the enrichment of aliphatic-degrading bacteria, which is typically succeeded by groups of bacteria with a preference for utilising aromatic hydrocarbons (Head et al., 2006 and references therein). Upon challenge with the ANS crude oil, the observed initial increase in abundance of Marinobacter by day 24, followed thereafter by the PAH-degrader TG408 by day 42, reflects this bacterial succession that is commonly observed during an oil spill at sea. However, the results of our hydrocarbon analysis did not appear to support this aliphatic-to-aromatic hydrocarbon degradation trend since the aromatic fraction of the ANS crude oil was, based on our biodegradation analysis, preferentially degraded over the aliphatic/n-alkane fraction. In fact, we found no quantifiable biodegradation of any aliphatic hydrocarbons (C1-C40) in the ANS crude oil (Table 2). Whilst members of the Marinobacter genus are commonly associated with the degradation of aliphatic/n-alkane hydrocarbons (Gauthier et al., 1992; McGowan et al., 2004), these organisms can be quite versatile and some species have been reported to degrade aromatic hydrocarbons (Gao et al., 2013; Hedlund et al., 2001). Strains of Marinobacter with the ability to degrade PAHs, such as naphthalene and phenanthrene, have been isolated by our group from various species of marine eukaryotic phytoplankton, including the S. costatum strain employed in this study (unpublished results). These results indicate that S. costatum harbours a community of hydrocarbon-degrading bacteria that are better adapted to degrading aromatic hydrocarbons. Possibly the aromatic-degradation pathway of these phycosphere-dwelling hydrocarbon-degraders is almost always in a state of up-regulation – i.e. constantly primed – for degrading aromatic hydrocarbons. Indeed, eukaryotic phytoplankton can synthesize aromatic chemicals that could support an actively-primed community of associated PAH-degrading bacteria (Andelman & Suess, 1970; Gol'man et al., 1973; Gunnison & Alexander, 1975; Pastuska, 1961; Zelibor et al., 1988).
Interestingly, the bacterial community throughout the duration of this 64-day enrichment was found to maintain a high species diversity. This is atypical to what is commonly observed in laboratory oil-enrichment experiments and in the field during a spill where, quite often, one or more oil-degrading bacterial groups become markedly dominant and representing up to 70% of total 16S rRNA gene sequencing libraries (Head et al., 2006; Yakimov et al., 2007). As noted earlier, the death and observed aggregation and sedimentation of the diatom to the bottom of the culture flasks (O1-O3) following exposure to the ANS crude oil is analogous to the observed formation of MOS during the DwH oil spill. We posit that in these oil-amended incubations, MOS formation likely supported opportunistic heterotrophs, not necessarily just oil-degraders, within the Gammaproteobacteria which had become markedly enriched. Studies exploring the impact of high-molecular-weight dissolved organic matter (DOM) on microbial community structure and activity have also shown a selective enrichment of marine heterotrophs within this Class under DOM-amendment derived from phytoplankton blooms (McCarren et al., 2010). Therefore, other groups within the diatom-associated bacterial community that are not necessarily capable of utilizing oil hydrocarbons could have been supported by the rich supply of DOM and MOS, and thereby providing a mechanism for supporting a wider diversity of species in the oil-amended incubations.
To the best of our knowledge, this study represents the first systematic investigation on the dynamics of the bacterial community associated with a marine eukaryotic phytoplankton in response to crude oil, including a paralleled analysis of the biodegradative capacity of the community to degrade various hydrocarbon species that are present in crude oil. Moreover, this work has added to our current knowledge on the diversity of hydrocarbon-degrading communities associated with phytoplankton in the ocean, and their capacity to respond to petrochemicals. Taken collectively with results from other studies that have described the isolation of novel taxa of oil-degrading bacteria from marine phytoplankton across the three major lineages (diatoms, dinoflagellates, coccolithophores) (Green et al., 2006; Gutierrez et al., 2012a,b; Gutierrez et al., 2013; Gutierrez et al., 2014;), this work highlights the phycosphere of eukaryotic phytoplankton as an underexplored biotope in the ocean where novel taxa of oil-degrading bacteria are found and that would be capable of responding to oil contamination. We hope that this work will spur other investigators to conduct their studies on oil biodegradation processes in ocean systems by taking into account the importance of eukaryotic phytoplankton as a reservoir of hydrocarbon-degrading bacterial communities. Field research on the potential role of phytoplankton-associated bacteria in oil bioremediation would be an appropriate extension of our microcosm studies. This algal-bacterial association may have potentially profound implications for the natural purging of the marine water column and help contribute to the overall health of the marine ecosystem.
Experimental Procedures
Micro-algal strain used
The algal strain used in this investigation was a non-axenic laboratory culture of the marine diatom Skeletonema costatum strain CCAP 1077/1C that was originally isolated from an unspecified location in the North Sea. The strain was purchased from the Culture Collection of Algae and Protozoa (CCAP; Oban, Scotland) and maintained in f/2 + Si algal medium (Guillard, 1975; Guillard and Ryther, 1962) in a temperature-controlled 12°C illuminated incubator.
Measurement of PAH degradation in enrichments with S. costatum
The ability of the prokaryotic community associated with S. costatum strain CCAP 1077/1C to degrade PAHs was determined in acid-washed (0.1 N HCl) steam-sterilized glass test tubes (13 ×100 mm) fitted with screw caps lined with Teflon-lined silicone septa (Chromacol). Stock solutions of naphthalene (NAP) (ca. 9,000 mg/liter), phenanthrene (PHE), anthracene (ANT), pyrene (PYR) and fluorene (FLU) (each at ca. 3,000 mg/liter) dissolved in acetone were prepared – when used, the acetone was allowed to volatilize prior to inoculation. Two types of media were used in these PAH enrichment experiments: ONR7a, which is defined synthetic seawater medium (Dyksterhouse et al., 1995), and ZM/100, which is a 100-fold dilution of Zobell's 2216 marine medium (Zobell, 1941). Enrichments were performed with individual PAH compounds rather than a PAH mixture because of the possibility that competitive inhibition might occur (Stringfellow & Aitken, 1995). For each enrichment on a PAH compound (NAP, PHE, ANT, PYR or FLU) and using one media type (ONR7a or ZM/100), one set of 3 test tubes was prepared, each containing a PAH and 2.8 ml of the respective medium. The 3 test tubes were then each inoculated with 200 μl of exponentially-growing culture of S. costatum. Uninoculated controls, acid-killed controls, and tubes that were inoculated but without any added PAH were also prepared. The initial amounts of the PAHs in the respective tubes were 0.9 ± 0.02 mg for NAP, 0.33 ± 0.01 mg for PHE, 0.35 ± 0.01 mg for ANT, 0.42 ± 0.02 mg for PYR, and 0.47 ± 0.02 mg for FLU. All test tubes were incubated in the dark with gentle shaking (100 rpm) at 21°C. A spectrophotometric method was used to quantify the amount of each PAH after 28 days incubation in the enrichments, as previously described (Gutierrez et al., 2012b). A significant decrease (P < 0.05) in the PAH concentration measured in the inoculated test tubes, relative to the uninoculated controls, was indicative of degradation.
Crude oil enrichment set-up
To examine the dynamics of the diatom and its associated bacterial community in response to crude oil exposure, nine 1-litre Erlenmeyer glass flasks were prepared, each containing 500 ml of f/2 + Si media and inoculated with a growing culture of the diatom Skeletonema costatum CCAP1077/1C to 1% (v/v). To three of the flasks (designated K7, K8, K9), 85% phosphoric acid was added to a final concentration of 3% to serve as the acid-inhibited control. All nine flasks were incubated in a temperature-controlled 12°C illuminated incubator with a 12:12 light/dark cycle and at a photon flux density of 25-50 μmol s-1 m-2, which is at the lower end of light intensity for the North Sea from where the diatom was originally isolated. On day 7, Alaska North Slope (ANS) crude oil was added to three of the “live” (uninhibited) flasks (designated O1, O2, O3) at 1% v/v – an amount that has been used to simulate laboratory-controlled oil enrichments (Piehler et al., 1999). An equal amount of the oil was also added at this time point to the acid-inhibited flasks K7, K8 and K9. Oil was not added to the other three culture flasks (designated C4, C5, C6) to serve as untreated controls. ANS crude oil was obtained from the cargo hold of the Exxon Valdez tanker in 1989 and has since been stored at -20°C. It was selected for use in this study because its composition has been well characterized. It is considered a moderately heavy crude oil consisting of 36.8% naphthenes, 27.3% alkanes, 25.3% aromatics, and 10.6% polar and other compounds (Elmendorf et al., 1994). Samples for algal and bacterial counts were taken daily in the first 10 days, then every 4-5 days until day 28, and then every 7 days until the termination of the experiment at day 64. Samples for molecular analysis were taken at days 0, 7, 10, 22, 35, 42, 50, 56 and 64.
In order to analyze for changes in the composition of the oil due to biodegradation, an additional eight flasks were prepared in the same way with f/2 + Si medium and the diatom. Of these eight flasks, four were treated with acid (as above) to act as the acid-inhibited controls, and all eight flasks were incubated in parallel together with the flasks above. Similarly to the treatment of the nine flasks above (O1-3, C4-6, K7-9), ANS crude oil was added to these eight flasks at the 7-day time point. Immediately after addition of the oil, two of the uninhibited and two of the acid-treated flasks were randomly selected and sacrificed for extraction of total petroleum hydrocarbons (TPH) and subsequent analysis for individual hydrocarbon constituents by gas chromatography/mass spectrometry (GC-MS), as detailed below. The other four flasks (two uninhibited and two acid-treated) were kept in the temperature-controlled illuminated incubator. At the termination of the experiment (day 64), these other four flasks were extracted for TPH for GC-MS analysis (described below).
Hydrocarbon analysis
Each flask was sacrificed at specified time points for extraction of TPH by placing the contents into 250-ml separatory funnels with dichloromethane (DCM) at an oil/water mix to DCM ratio of 2:1. The DCM fraction was removed and the oil/water mix re-extracted an additional two times. The extracts were combined and stored at 4°C for subsequent analysis of individual aromatic and aliphatic hydrocarbon constituents.
Quantification of up to 50 aromatic hydrocarbon constituents was measured by GC-MS at the Department of Environmental and Molecular Toxicology, North Carolina State University, as previously described (Page et al., 1995, Stout et al., 2001, Douglas et al., 1996). An Agilent 6890 gas chromatograph, with a Restek RTX-5MS and Integra-Guard column (30 m length × 0.25 mm internal diameter × 0.25 μm film thickness) that was connected to an Agilent 5973 mass selective detector, was used and operated in selected ion monitoring (SIM) mode. Helium was the carrier gas. Pressure in the inlet was held at 25 psi using a splitless injection technique. Each sample (2 μl) was injected into a 300°C single taper inlet liner with a deactivated glass wool plug. The carrier gas pressure was reduced one minute after injection and flow was kept constant at 1.2 ml/min throughout the run. The initial temperature was 40°C for one minute, increased at 25°C/min to 100°C, then ramped at 5°C/min to a final temperature of 310°C, which was held for 15 minutes. The transfer line was heated to 300°C, the mass spectrometry source was heated to 230°C, and the quadrupole was heated to 150°C.
Quantification of aliphatic hydrocarbon constituents was performed by Fugro ERT in Edinburgh using a HP 6890 Series gas chromatograph (GC) with 7673 auto-injector on a 100%-dimethylpolysiloxane bonded fused silica column (60 m length × 0.32 mm internal diameter × 0.25 μm film thickness). Samples (1 μl) were injected and run with hydrogen as the carrier gas at a constant flow rate of 3.8 ml/min. The oven temperature program was set to 40°C for 4 minutes, 40°C to 330°C at 15°C/min, and 330°C for 7 minutes. The source detector temperature was 350°C.
Concentrations of aromatic and aliphatic hydrocarbon species/groups that were biodegraded after 64 days were calculated by subtracting the respective hydrocarbon concentrations measured in the acidified controls from those of the non-acidified incubations. A Student's t-test was performed to test for significant differences (P < 0.05) in the degradation of the hydrocarbons analysed between the different treatments.
Quantification of the prokaryotic population and S. costatum by DAPI staining and Chlorophyll a measurements
Samples (2 ml) for Chlorophyll a (Chl a) determinations were extracted using a modified version of EPA Method 445 (Arar and Collins, 1997). For this, each 2-ml sample was added to 50 ml of sterile-filtered (0.1 μm) natural seawater immediately upon collection. The suspensions were filtered through a 25 mm Glass Fiber Filter (GF/F) and then placed in 50-ml Falcon centrifuge tubes (BD Biosciences) containing 10 ml of 90% acetone. The tubes were sonicated in an iced water bath for 10 minutes in the dark and then maintained at -20°C for ca. 20 hours. The samples were then centrifuged to remove particulate matter and the supernatant fractions allowed to equilibrate in the dark at room temperature prior to fluorometric analysis on a Fluoromax-4 fluorometer (Horiba Scientific) using an excitation wavelength of 430 nm with a 5 nm slit width. Emission was measured by recording the maximum reading from 500-750 nm by increments of 5 nm. Chl a concentrations were calculated from a standard curve constructed from serial dilutions of a Chl a spinach extract (Sigma-Aldrich, Saint Louis, MO) as per the method of Welshemeyer (1994).
Samples (2 ml) for prokaryotic cell counts were fixed with 2% paraformaldehyde (PFA) and stored at 4°C prior to staining with DAPI (4′6-diamidino-2-phenylindole) for fluorescence microscopy, as adapted from the method of Porter & Feig (1980). For this, each 2-ml PFA-fixed sample was diluted with a known volume of filter-sterilised seawater and then stained with DAPI to a final concentration of 0.01 μg/ml for 10 min. DAPI-stained solutions were then filtered through 0.2 um pore-size GTBP polycarbonate filters (Millipore, 25 mm diameter) using a glass vacuum filtration system (Millipore). Each filter was embedded in Citifluor mounting solution (AF1, glycerol/PBS solution) and visualized with an Olympus BX51 epifluorescense microscope (Tokyo, Japan) equipped with a Hamamatsu C8484 digital camera (Hamamatsu City, Japan). Cells were enumerated with MetaMorph image analysis software version 7.6.0.0 (Sunnyvale, CA, USA) from at least 10 microscopic fields of view (selected randomly) and expressed per ml of original sample volume as calculated per the method of Bloem and Vos (2008).
Extraction of DNA
Cell biomass from samples taken at each sampling point was collected by filtration using a glass vacuum filtration system (Millipore) with 47-mm cellulose acetate membrane filters (0.2 um pore size; Millipore) and the filters stored at -20°C. DNA was extracted from one quarter of each filter using the FastDNA spin kit (MP Biomedicals, Solon, OH) as per the manufacturer's instructions. All DNA extracts were purified using the QIAquick nucleotide removal kit (Qiagen, Valencia, CA) and eluted into a final volume of 30 μl of 1× TE buffer. Purified DNA was stored at -20°C for subsequent quantification and molecular analysis.
Real-time quantitative PCR
Preliminary results from our laboratory indicated the occurrence of Marinobacter associated with this and other species of Skeletonema (unpublished results), so we decided to quantify the abundance of Marinobacter 16S rRNA genes by quantitative PCR (qPCR) in the oil-enrichment incubations (Flasks O1-3) and non-oil treated controls (C4-6). For this, we adapted an oligonucleotide FISH (fluorescence in-situ hybridization) probe, Mrb-0625-a (5′-CAGTTCGAAATGCCGTTCCCA-3′), that was recently designed and optimized for the detection of Marinobacter in environmental samples by FISH (McKay et al., 2014), for the quantitative detection of Marinobacter in this study. The adapted primer, designated here MB1r for qPCR, targets ∼75% of the monophyletic Marinobacter group (McKay et al., 2014) and was used together with the universal eubacterial primer 533f (5′-GTGCCAGCAGCCGCGGTAA-3′) (Weisburg et al., 1991). The optimal annealing temperature for the 533f/MB1r primer set (62°C) was determined using an Eppendorf (Hauppauge, NY) Mastercycler gradient thermal cycler. The template for construction of a standard curve for qPCR was the 16S rRNA gene of Marinobacter algicola (DSM 16394) that was amplified using PCR with primers 27f (Wilmotte et al., 1993) and 1492r (Lane, 1991), and the resultant amplicon purified using the QIAquick nucleotide removal kit (Qiagen, Valencia, CA). The limit of quantification for the target strain using these primers was 114 gene copies per reaction. When threshold cycle (CT) values beyond the highest value in the linear range of the standard curve (CT range, 12.1 to 34.5) were measured, the gene was considered detected but below the quantification limit of the assay. The amplification efficiency of the primers (Pfaffl, 2001) was determined to be 1.9.
Primers for qPCR targeting Polycyclovorans algicola strain TG408, a novel PAH-degrader previously identified living associated with S. costatum 1077/1C and other species of eukaryotic phytoplankton (Gutierrez et al., 2013), were used to track the abundance of this bacterial strain in the oil enrichments and untreated controls. These TG408-specific qPCR primers are Pcy120f (5′-TACATAGGAATCTGCCCGA-3′) and Pcy223r (5′-AGACATAGGCTCCTCCAA-3′), and the qPCR method for their use is described elsewhere (Gutierrez et al., 2013).
DNA extracted from incubations with and without oil was quantified using a NanoDrop ND-3300 fluorospectrometer (Thermo, Waltham, MA) and the Quant-iT Picogreen double-stranded DNA (dsDNA) kit (Invitrogen, Carlsbad, CA). Identification of the target organisms (Marinobacter or P. algicola TG408) in these extracts was determined by qPCR as described previously (Singleton et al., 2006). Duplicate reactions were performed on each triplicate DNA extraction (from triplicate samples) taken at each time point.
Barcoded amplicon pyrosequencing
Barcoded 16S rRNA gene pyrosequencing was used to analyse the bacterial community in the purified DNA extracts from samples collected at days 7 (prior to oil amendment), 10, 35 and 64. Ten-fold dilutions of extracted DNA in water were used as a template for triplicate PCR reactions for each sample. A two-step barcoded PCR method was used, as previously described by Berry et. al (2011). The first PCR step employed non-barcoded primers 909f (5′-ACTCAAAKGAATWGACGG-3′) and 1492r (5′-NTACCTTGTTACGACT-3′). Each 20 μl of PCR master mix contained 1.25 U Taq DNA polymerase, 50 mM KCl, 30 mM Tris-HCl, 1.5 mM Mg2+, 0.2% Igepal®-CA630, and 200 μM of each dNTP, 0.3 μM of each primer, and 1 μl of DNA as template. PCR was run using an Eppendorf (Hauppauge, NY, USA) Mastercycler Gradient thermal cycler before verification of the proper amplicon size on a 1% agarose gel. An additional set of PCR reactions (in triplicate) was run in parallel using E. coli DNA as template to act as an internal control for the pyrosequencing data analysis; E. coli was chosen because it is not part of the bacterial community associated with S. costatum 1077/1C.
For the second PCR step, the products from step 1 were re-amplified using the same primers, with the exception that the reverse primer (1492r) was linked to a 2-bp spacer on the 5′-end of the primer sequence and a 8-bp barcode sequence that was unique to each sample (Hamady et al., 2008). In addition, amplification was performed for only 5 cycles in order to reduce PCR bias introduced by the adapter and barcode sequence during amplification (Berry et al., 2011). The reactions were run on a 1% agarose gel to verify the proper amplicon size. Triplicate reactions were then pooled and purified with a QIAquick PCR Purification Kit (Qiagen) and eluted in 30 μl of 10 mM Tris-HCl (pH 8.5) buffer. The DNA concentration of pooled amplicons was then quantified as described above prior to combining into a single sample at a concentration suitable for pyrosequencing. The E. coli PCR product was added at 100-fold lower concentration, as internal control, than the samples (Berry et al., 2011). The sample was submitted to the High-Throughput Sequencing Facility at the University of North Carolina at Chapel Hill for sequencing using the 454 Life Sciences Titanium platform (Roche Diagnostics, Branford, CT, USA).
Pyrosequencing reads were trimmed and filtered using the LUCY programme with a minimum PHRED score of 27.5 and minimum length of 200 nt to remove low-quality regions, short reads and plastid sequences (Kunin et al., 2010). Reads were de-multiplexed based on an 8-nt barcode identifier and the primer and barcode regions were removed using QIIME (Caporaso et al., 2010). To form OTUs, the reads were clustered at 97% sequence identity with UCLUST (Edgar, 2010) and the most abundant unique read within each cluster was used as its representative sequence. Initial phylogenetic identification was made using BLAST (Altschul et al., 1990) and chimeras were detected with Chimera Slayer (Haas et al., 2011) and removed. Reads with a significant BLAST match to S. costatum (FJ002160) were also removed prior to analysis. Sequence data were submitted to the European Nucleotide Archive Sequence Read Archive under the study accession numbers (ERS725072– ERS725089).
Phylogenetic tree
Representative 16S rRNA gene sequences of OTUs representing dominant hydrocarbon-degraders identified from the pyrosequencing analysis were aligned and manually curated in ARB (Ludwig et al., 2004). The sequences and type strains (from the Living Tree Project) (Yarza et al., 2010) with the highest similarity from the SILVA SSU NR 119 NR database (Quast et al., 2013) were also used for tree construction. A neighbour-joining tree was constructed with Jukes-Cantor correction and bootstrapped replication (n=1000), and Verrucomicrobium spinosum (X90515) was used as an outgroup.
Supplementary Material
Figure 5.
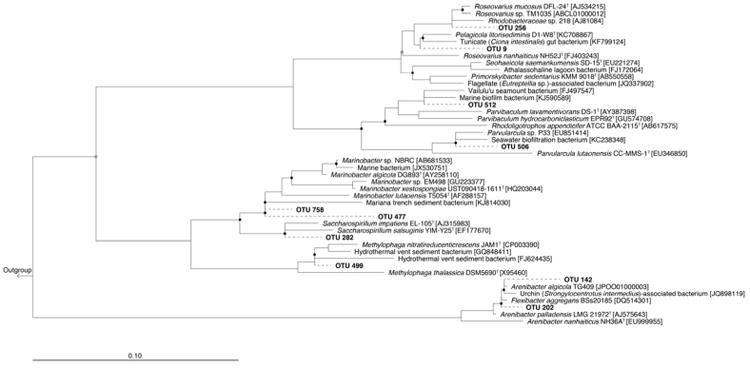
Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences (>1,200 bp), showing the 10 OTUs enriched in the oil-amended incubations and representatives of related taxa. Parvibaculum hydrocarboniclasticum and Parvibaculum lavamentivorans are included as representatives of hydrocarbon-degraders. Filled circles indicate nodes with bootstrap values (1,000 bootstrap replications) greater than 90%; open circles indicate bootstrap values greater than 60%. Enriched OTUs (shown in bold) were added using the ARB maximum parsimony quick-add feature (dashed branches). GenBank accession numbers are shown in parentheses. Bar, 5 substitutions per 100 nucleotide positions.
Acknowledgments
This work was supported by a Marie Curie International Outgoing Fellowship (PIOF-GA-2008-220129) within the 7th European Community Framework Programme. Partial support was also provided through the US National Institute of Environmental Health Sciences, grant 5 P42ES005948, and a MASTS Visiting Fellowship to David Berry. We also thank Damian Shea, of North Carolina State University, for assistance with GC-MS analysis of aromatic hydrocarbons and for providing us with the ANS crude oil, and David Singleton for assistance with qPCR.
Footnotes
The authors declare no conflict of interest.
References
- 1.Abbriano RM, Carranza MM, Hogle SL, Levin RA, Netburn AN, Seto KL, Snyder SM, Franks PJS SIO280. Deepwater Horizon oil spill: A review of the planktonic response. Oceanography. 2011;24:294–301. [Google Scholar]
- 2.Agency for Toxic Substances and Disease Registry. CERCLA priority list of hazardous substances. Agency for Toxic Substances and Disease Registry; Atlanta, GA: 2007. http://www.atsdr.cdc.gov/SPL/index.html. [Google Scholar]
- 3.Andelman JB, Suess MJ. Polynuclear aromatic hydrocarbons in the water environment. Bull World Health Organ. 1970;43:479–508. [PMC free article] [PubMed] [Google Scholar]
- 4.Borde X, Guieysse B, Delgado O, Muñoz R, Hatti-Kaul R, Nugier-Chauvin C, Patin H, Mattiasson B. Synergistic relationships in algal-bacterial microcosms for the treatment of aromatic pollutants. Bioresour Technol. 2003;86:293–300. doi: 10.1016/s0960-8524(02)00074-3. [DOI] [PubMed] [Google Scholar]
- 5.Buchan A, LeCleir GR, Gulvik CA, Gonzalez JM. Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol. 2014;12:686–698. doi: 10.1038/nrmicro3326. [DOI] [PubMed] [Google Scholar]
- 6.Camilli R, Reddy CM, Yoerger DR, Van Mooy BAS, Jakuba MV, Kinsey JC, McIntyre CP, Sylva SP, Maloney JV. Tracking hydrocarbon plume transport and bio- degradation at deepwater horizon. Sci. 2010;330:201–204. doi: 10.1126/science.1195223. [DOI] [PubMed] [Google Scholar]
- 7.Diercks AR, Highsmith RC, Asper VL, Joung DJ, Zhou Z, Guo L, Shiller AM, Joye SB, Teske AP, Guinasso N, Wade TL, Lohrenz SE. Characterization of subsurface polycyclic aromatic hydrocarbons at the deepwater horizon site. Geophys Res Lett. 2010a;37:L20602. [Google Scholar]
- 8.Diercks AR, Asper VL, Highsmith R, Woolsey M, Lohrenz S, McLetchie K, Gossett A, Lowe M, III, Joung D, McKay L, Joye S, Teske A. NIUST – Deepwater Horizon Oil Spill Response Cruise. OCEANS 2010, in OCEANS-IEEE series 2010b [Google Scholar]
- 9.Gao W, Cui ZS, Li Q, Xu GS, Jia XJ, Zheng L. Marinobacter nanhaiticus sp. nov., polycyclic aromatic hydrocarbon-degrading bacterium isolated from the sediment of the South China Sea. Antonie Van Leeuwenhoek. 2013;103:485–491. doi: 10.1007/s10482-012-9830-z. [DOI] [PubMed] [Google Scholar]
- 10.Gauthier MJ, Lafay B, Christen R, Fernandez L, Acquaviva M, Bonin P, Bertrand JC. Marinobacter hydrocarbonoclasticus gen. nov., sp. nov., a new extremely halotolerant, hydrocarbon-degrading marine bacterium. Int J Syst Bacteriol. 1992;42:568–576. doi: 10.1099/00207713-42-4-568. [DOI] [PubMed] [Google Scholar]
- 11.Gol'man LP, Mikhaseva MF, Reznikov VM. Infrared spectra of lignin preparations of pteridophytes and seaweeds. Dokl Akad Nauk BSSR. 1973;17:1031–1033. [Google Scholar]
- 12.González JM, Simo R, Massana R, Covert JS, Casamayor EO, Pedoros-Alio C, et al. Bacterial community structure associated with dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl Environ Microbiol. 2000;66:4237–4246. doi: 10.1128/aem.66.10.4237-4246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green DH, Bowman JP, Smith EA, Gutierrez T, Bolch CJS. Marinobacter algicola sp. nov., isolated from laboratory cultures of paralytic shellfish toxin-producing dinoflagellates. Int J Syst Evol Microbiol. 2006;56:523–527. doi: 10.1099/ijs.0.63447-0. [DOI] [PubMed] [Google Scholar]
- 14.Green DH, Llewellyn LE, Negri AP, Blackburn SI, Bolch CJS. Phylogenetic and functional diversity of the cutlivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum. FEMS Microbiol Ecol. 2004;47:345–357. doi: 10.1016/S0168-6496(03)00298-8. [DOI] [PubMed] [Google Scholar]
- 15.Gunnison D, Alexander M. Basis for the resistance of several algae to microbial decomposition. Appl Microbiol. 1975;29:729–738. doi: 10.1128/am.29.6.729-738.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez T, Aitken MD. Role of methylotrophs in the degradation of hydrocarbons during the Deepwater Horizon oil spill. The ISME J. 2014;8:2543–2545. doi: 10.1038/ismej.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutierrez T, Green DH, Nichols PD, Whitman WB, Semple KT, Aitken MD. Algiphilus aromaticivorans gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium isolated from a culture of the marine dinoflagellate Lingulodinium polyedrum, and proposal of Algiphilaceae fam. nov. Int J Syst Evol Microbiol. 2012a;62:2743–2749. doi: 10.1099/ijs.0.033324-0. [DOI] [PubMed] [Google Scholar]
- 18.Gutierrez T, Green DH, Whitman WB, Nichols PD, Semple KT, Aitken MD. Polycyclovorans algicola gen. nov., sp. nov., an aromatic hydrocarbon-degrading marine bacterium found associated with laboratory cultures of marine phytoplankton. Appl Environ Microbiol. 2013;79:205–214. doi: 10.1128/AEM.02833-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutierrez T, Nichols PD, Whitman WB, Aitken MD. Porticoccus hydrocarbonoclasticus sp. nov., an aromatic hydrocarbon-degrading bacterium identified in laboratory cultures of marine phytoplankton. Appl Environ Microbiol. 2012b;78:628–637. doi: 10.1128/AEM.06398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutierrez T, Rhodes G, Mishamandani S, Berry D, Whitman WB, Nichols PD, Semple KT, Aitken MD. PAH degradation of phytoplankton-associated Arenibacter and description of Arenibacter algicola sp. nov., an aromatic hydrocarbon-degrading bacterium. Appl Environ Microbiol. 2014;80:618–628. doi: 10.1128/AEM.03104-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutierrez T, Singleton DR, Aitken MD, Semple KT. Stable-isotope probing of an algal bloom identifies uncultivated members of the Rhodobacteraceae associated with low molecular-weight PAH degradation. Appl Environ Microbiol. 2011;77:7856–7860. doi: 10.1128/AEM.06200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hazen TC, Dubinsky EA, DeSantis TZ, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Sci. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 23.Head IM, Jones MD, Roling WFM. Marine microorganisms make a meal of oil. Nat Rev Microbiol. 2006;4:173–182. doi: 10.1038/nrmicro1348. [DOI] [PubMed] [Google Scholar]
- 24.Hedlund BP, Geiselbrecht AD, Staley JT. Marinobacter strain NCE312 has a Pseudomonas-like naphthalene dioxygenase. FEMS Microbiol Lett. 2001;201:47–51. doi: 10.1111/j.1574-6968.2001.tb10731.x. [DOI] [PubMed] [Google Scholar]
- 25.Huang YJ, Jiang ZB, Zeng JN, Chen QZ, Zhao YQ, Liao YB, Shou L, Xu XQ. The chronic effects of oil pollution on marine phytoplankton in a subtropical bay, China. Environ Monitoring Assess. 2011;176:517–530. doi: 10.1007/s10661-010-1601-6. [DOI] [PubMed] [Google Scholar]
- 26.Kim JS, Crowley DE. Microbial diversity in natural asphalts of the Rancho La Brea Tar Pits. Appl Environ Microbiol. 2007;73:4579–4591. doi: 10.1128/AEM.01372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labrenz M, Lawson PA, Tindall BJ, Collins MD, Hirsch P. Saccharospirillum impatiens gen. nov., sp. nov., a novel γ-Proteobacterium isolated from hypersaline Ekho Lake (East Antarctica) Int J Syst Evol Microbiol. 2003;53:653–660. doi: 10.1099/ijs.0.02406-0. [DOI] [PubMed] [Google Scholar]
- 28.Marlowe IT, Green JC, Neal AC, Brassell SC, Eglinton G, Course PA. Long chain (n-C37-C39) alkenones in the Prymnesiophyceae. Distribution of alkenones and other lipids and their taxonomic significance. British Phycol J. 1984;19:203–216. [Google Scholar]
- 29.McGowan L, Herbert R, Muyzer G. A comparative study of hydrocarbon degradation by Marinobacter sp., Rhodococcus sp. and Corynebacterium sp. isolated from different mat systems. Ophelia. 2004;58:271–281. [Google Scholar]
- 30.Moran MA, Belas R, Schell MA, Gonzalez JM, Sun F, Sun S, et al. Ecological genomics of marine roseobacters. Appl Environ Microbiol. 2007;73:4559–4569. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muñoz R, Guieysse B, Mattiasson B. Phenanthrene biodegradation by an algal-bacterial consortium in two-phase partitioning bioreactors. Appl Microbiol Biotechnol. 2003;61:261–267. doi: 10.1007/s00253-003-1231-9. [DOI] [PubMed] [Google Scholar]
- 32.Myklestad SM. Release of extracellular products by phytoplankton with special emphasis on polysaccharides. Sci Total Environ. 1995;165:155–164. [Google Scholar]
- 33.Ostgaard K, Eide I, Jensen A. Exposure of phytoplankton to Ekofisk crude oil. Mar Environ Res. 1984a;11:183–200. [Google Scholar]
- 34.Ostgaard K, Hegseth EN, Jensen A. Species-dependent sensitivity of marin planktonic algae to Ekofisk crude oil under different light conditions. Botanica Marina. 1984b;27:309–318. [Google Scholar]
- 35.Ozhan K, Miles MS, Gao H, Bargu S. Relative phytoplankton growth responses to physically- and chemically-dispersed South Louisiana sweet crude oil. Environ Monitoring Assess. 2014a;186:3941–3956. doi: 10.1007/s10661-014-3670-4. [DOI] [PubMed] [Google Scholar]
- 36.Ozhan K, Parsons ML, Bargu S. How were phytoplankton affected by the Deepwater Horizon oil spill? BioScience. 2014b;64:829–836. [Google Scholar]
- 37.Passow U, Alldredge AL. Distribution, size and bacterial colonization of transparent exopolymer particles (TEP) in the ocean. Mar Ecol Progr Ser. 1994;113:185–198. [Google Scholar]
- 38.Passow U, Alldredge AL, Logan BE. The role of particulate carbohydrate exudates in the flocculation of diatom blooms. Deep Sea Res. I. 1994;4:335–357. [Google Scholar]
- 39.Passow U, Ziervogel K, Asper V, Diercks A. Marine snow formation in the aftermath of the Deepwater Horizon oil spill. Environ Res Lett. 2012;7 doi: 10.1088/1748-9326/7/3/035301. [DOI] [Google Scholar]
- 40.Pastuska G. Die Kieselgelschicht-Chromatographie von Phenolen und Phenolcarbensiuren. I Z Anal Chem. 1961;179:355–358. [Google Scholar]
- 41.Patel V, Munot H, Shouche YS, Madamwar D. Response of bacterial community structure to seasonal fluctuation and anthropogenic pollution on coastal water of Alang-Sosiya ship breaking yard, Bhavnagar, India. Bioresour Technol. 2014;161:362–370. doi: 10.1016/j.biortech.2014.03.033. [DOI] [PubMed] [Google Scholar]
- 42.Rosario-Passapera R, Keddis R, Wong R, Lutz RA, Starovoytov V, Vetriani C. Parvibaculum hydrocarboniclasticum sp. nov., a mesophilic, alkane-oxidizing alphaproteobacterium isolated from a deep-sea hydrothermal vent on the East Pacific Rise. Int J Syst Evol Microbiol. 2012;62:2921–2926. doi: 10.1099/ijs.0.039594-0. [DOI] [PubMed] [Google Scholar]
- 43.Safanova ET, Dmitrieva IA, Kvitko KV. The interaction of algae with alcanotrophic bacteria in black oil decomposition. Res Conserv Recycling. 1999;27:193–201. [Google Scholar]
- 44.Sargian P, Mostajir B, Chatila K, Ferreyra GA, Pelletier E, Demers S. Non-synergistic effects of water-soluble crude oil and enhanced ultraviolet-B radiation on a natural plankton assemblage. Mar Ecol Progr Ser. 2005;294:63–77. [Google Scholar]
- 45.Schleheck D, Tindall BJ, Rosselló-Mora R, Cook AM. Parvibaculum lavamentivorans gen. nov., sp. nov., a novel heterotroph that initiates catabolism of linear alkylbenzenesulfonate. Int J Syst Evol Microbiol. 2004;54:1489–1497. doi: 10.1099/ijs.0.03020-0. [DOI] [PubMed] [Google Scholar]
- 46.Singleton DR, Sangaiah R, Gold A, Ball LM, Aitken MD. Identification and quantification of uncultivated proteobacteria associated with pyrene degradation in a bioreactor treating PAH-contaminated soil. Environ Microbiol. 2006;8:1736–1745. doi: 10.1111/j.1462-2920.2006.01112.x. [DOI] [PubMed] [Google Scholar]
- 47.Stringfellow WT, Aitken MD. Competitive metabolism of naphthalene, methylnaphthalenes, and fluorene by phenanthrene-degrading pseudomonads. Appl Environ Microbiol. 1995;61:357–362. doi: 10.1128/aem.61.1.357-362.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teira E, Lekunberri I, Gasol JM, Nieto-Cid M, Alvarez-Salgado XA, Figueiras FG. Dynamics of the hydrocarbon-degrading Cycloclasticus bacteria during mesocosm-simulated oil spills. Environ Microbiol. 2007;9:2551–2562. doi: 10.1111/j.1462-2920.2007.01373.x. [DOI] [PubMed] [Google Scholar]
- 49.Warshawsky D, LaDow K, Schneider J. Enhanced degradation of benzo[a]pyrene by Mycobacterium sp. in conjunction with green alga. Chemosphere. 2007;69:500–506. doi: 10.1016/j.chemosphere.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 50.Yakimov MM, Timmis KN, Golyshin PN. Obligate oil-degrading marine bacteria. Curr Opin Biotechnol. 2007;18:257–266. doi: 10.1016/j.copbio.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 51.Zelibor JL, Romankiw L, Hatcher PG, Colwell RR. Comparative analysis of the chemical composition of mixed and pure cultures of green algae and their decomposed residues by 13C nuclear magnetic resonance spectroscopy. Appl Environ Microbiol. 1988;54:1051–1060. doi: 10.1128/aem.54.4.1051-1060.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeng J, Deng LJ, Lou K, Zhang T, Yang HM, Shi YW, Lin Q. Molecular characterization of the planktonic microorganisms in water of two mountain brackish lakes. J Basic Microbiol. 2014;54:509–520. doi: 10.1002/jobm.201300187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


