Summary
Broadly neutralizing antibodies (bNAbs) against the HIV-1 envelope glycoprotein (Env) suppress viremia in animal models of HIV-1 and humans. To achieve potent activity without the emergence of viral escape mutants, co-administration of different bNAbs is necessary to target distinct epitopes essential for viral fitness. Here we report the development of bispecific anti-Env neutralizing antibodies (biNAbs) with potent activity. Synergistic activity of biNAbs was achieved by combining an engineered hinge domain of IgG3 to increase Fab domain flexibility necessary for hetero-bivalent binding to the Env trimer, while retaining the functional properties of the IgG1-Fc. Compared to unmodified biNAbs, hinge domain variants exhibited substantially improved neutralization activity, with particular combinations showing evidence of synergistic neutralization potency in vitro and enhanced in vivo therapeutic activity in HIV-1-infected humanized mice. These findings suggest innovative strategies for generating biNAbs with enhanced neutralization breadth and potency, representing ideal candidate molecules for the control of HIV-1 infection.
Introduction
Recent development and characterization of antibodies against the envelope glycoprotein of HIV-1 (Env) with broad and potent neutralizing activity suggested their passive administration as an effective strategy for the prevention or treatment of HIV-1 infection in humans (Burton and Mascola, 2015; Klein et al., 2013). Indeed, a number of recent studies in murine and non-human primate models of HIV-1 infection (Barouch et al., 2013; Bournazos et al., 2014; Halper-Stromberg et al., 2014; Horwitz et al., 2013; Klein et al., 2012b; Shingai et al., 2013), as well as in HIV-1-infected humans showed that broadly neutralizing anti-Env antibodies (bNAbs) have the capacity to confer both effective pre-exposure prophylaxis and therapeutic control of viremia (Caskey et al., 2015; Lynch et al., 2015). In contrast to conventional antiretroviral pharmacologic strategies, HIV-1 control by anti-Env bNAbs is mediated by pleiotropic effector functions, including viral neutralization and clearance, elimination of HIV-1 infected cells, as well as stimulation of host immune responses and induction of long-term immunity (Barouch et al., 2013; Bournazos et al., 2015; Bournazos et al., 2014; Bournazos and Ravetch, 2015; Haigwood et al., 2004; Halper-Stromberg et al., 2014; Ng et al., 2010; Pietzsch et al., 2012).
Despite the profound benefits of anti-Env bNAb therapy for the control of HIV-1 infection, two main limitations exist: (i) no single bNAb has the capacity to efficiently neutralize all virus strains and (ii) bNAb administration is often associated with the emergence of virus escape mutants in the respective bNAb-targeting epitope (Diskin et al., 2013; Horwitz et al., 2013; Klein et al., 2012b). Co-administration of two or more bNAbs with non-overlapping epitope specificities is therefore necessary to overcome these limitations and confer robust control of HIV-1 replication (Klein et al., 2012b). Indeed, previous studies in murine models of HIV-1 infection indicated that the use of single bNAbs is inadequate for effective control of virus replication, as the administered bNAb often exerts significant selection pressure on the virus, leading to the rapid generation of escape mutants (Diskin et al., 2013; Horwitz et al., 2013; Klein et al., 2012b). These limitations suggest the development of a new class of antibody-based molecules that would combine the breadth, potency, and antigenic specificity of two bNAbs, required for the effective control of HIV-1 infection. Indeed, bispecific anti-Env bNAbs (biNAbs) represent an attractive strategy for the prevention and treatment of HIV-1 infection, as they offer unique advantages over conventional, monospecific antibodies, providing a compelling platform for the development of single therapeutic molecules with improved in vivo protective activity.
Recent attempts to generate anti-HIV-1 bispecific antibody-based molecules mainly focused on targeting HIV-1-infected cells through the use of anti-Env specificities combined with anti-CD3 (Pegu et al., 2015; Sung et al., 2015); a concept originally developed for anti-tumor bispecific molecules (Chames and Baty, 2009). This approach induces clearance of HIV-1-infected cells, by increasing the recruitment of cytotoxic T cells to Env-expressing infected cells, promoting their lysis. Since these molecules specifically target infected cells without any direct effects on virus neutralization, their use in prophylactic regiments to block HIV-1 infection is rather limited. Additionally, despite the reported low toxicity of these molecules, targeting host receptors, like CD3 still poses concerns over the long-term safety of this approach, especially in HIV-1 infected, immunocompromised individuals. In addition, such molecules exhibit relatively short half-life, as their interaction with host cells enhances their clearance from circulation. Likewise, limitations associated with targeting host receptors (toxicity, enhanced clearance) are also expected for anti-CD4/anti-Env bispecific antibodies (Pace et al., 2013).
We therefore aimed to develop and characterize biNAbs targeting exclusively the HIV-1 Env trimer, focusing particularly on the optimization of these biNAbs to exhibit significantly improved, neutralization activity, compared to unmodified, monospecific bNAbs. Although several strategies have been previously described for the generation of antibodies or antibody-based molecules with dual specificities (Spiess et al., 2015), in the present study, a major prerequisite for the development of anti-HIV-1 Env biNAbs was to maintain the physiological IgG architecture, preserving thereby the favorable pharmacokinetic properties of IgG (half-life of 3 weeks for human IgG1) and its Fc effector function, which has been previously shown to contribute to the in vivo protective activity of anti-HIV-1 bNAbs (Bournazos et al., 2014; Halper-Stromberg et al., 2014; Hessell et al., 2007). Previous attempts to generate anti-HIV-1 Env biNAbs based on the IgG1 structure yielded biNAbs with high breadth and potency, as well as sufficient in vivo stability and half-life (Asokan et al., 2015). However, none of these anti-Env biNAbs exhibited improved neutralization potency compared to their respective parental bNAbs, suggesting that IgG1 biNAbs offer no advantages over conventional, monospecific bNAbs (Asokan et al., 2015).
We therefore aimed to generate anti-Env biNAbs with improved neutralization potency by exploiting the unusual structure of the IgG3 hinge domain engineered to increase Fab domain flexibility, favoring thereby hetero-bivalent interactions with the Env trimer. Using this strategy and a panel of bNAbs targeting distinct epitopes on the HIV-1 Env, we have generated and characterized the in vitro neutralization activity of several biNAb combinations, identifying particular biNAbs with evidence for synergistic activity i.e. exhibiting increased neutralization potency compared to their respective parental, monospecific bNAbs. These hinge domain-engineered biNAbs exhibited improved neutralization potency and enhanced in vivo protective activity in HIV-1-infected humanized mice, suggesting their potential clinical use for the control of HIV-1 infection in humans.
Results
Generation and characterization of anti-HIV-1 biNAbs
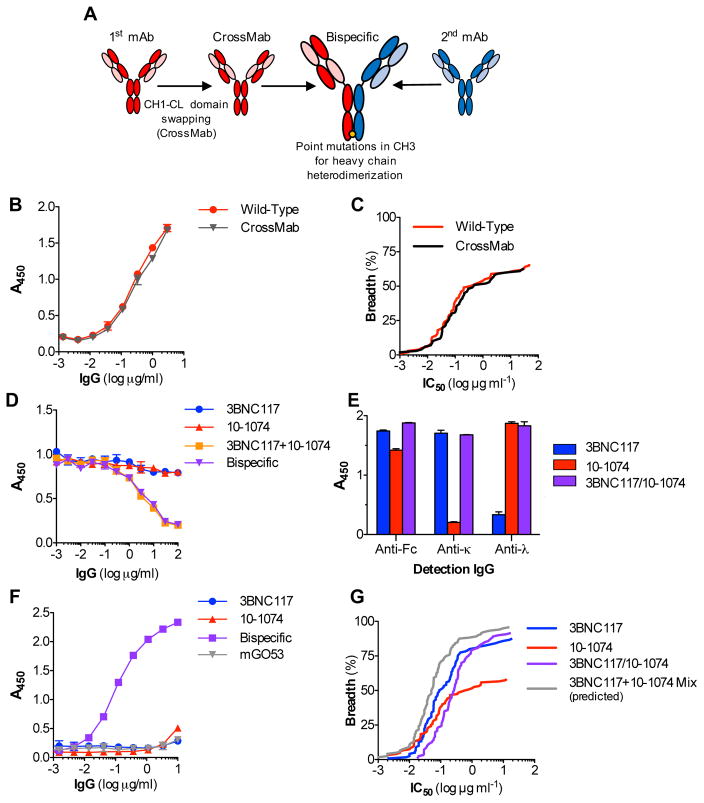
To study the activity of anti-Env biNAbs, we initially selected two mAbs with broad and potent in vitro and in vivo activity: 3BNC117 (Scheid et al., 2011) and 10-1074 (Mouquet et al., 2012), which target the CD4 binding site (CD4bs) and the V3 sites of Env, respectively. For the generation of 3BNC117/10-1074 biNAbs, we employed a combination of previously described strategies (Merchant et al., 1998; Ridgway et al., 1996; Schaefer et al., 2011) to ensure proper pairing of the heavy and light chains of the two parental mAbs and generation of heavy chain heterodimers (Fig. 1A). Pairing of the correct heavy chain with the respective light chain was achieved by swapping the CH1 domain with the constant domain of the light chain (CL) for one mAb (10-1074), while preserving the wild-type domain organization for the other mAb (3BNC117)(Schaefer et al., 2011). This approach (CrossMab (Schaefer et al., 2011)) had no measurable effect on the antigenic specificity and neutralization activity of the 10-1074 mAb, as determined by antigen (gp140)-specific ELISA and TZMbl-based in vitro neutralization assay (Montefiori, 2005) against an extended multiclade virus panel, respectively (Fig. 1B–C). Heterodimerization of the heavy chain was achieved by introducing mutations in the CH3 domain that alter the physical and chemical properties of the two mAb heavy chains, favoring heterodimer formation (Merchant et al., 1998; Ridgway et al., 1996). Consistent with previous reports (Asokan et al., 2015; Merchant et al., 1998), this approach resulted in efficient expression of biNAbs, typically yielding >90% heterodimers. By taking advantage of the differences in the light chain isotype (κ for 3BNC117 and λ for 10-1074) and the Env epitope specificity (CD4bs and V3 for 3BNC117 and 10-1074, respectively), competition and epitope-specific ELISA assays confirmed the dual specificity of the resulting 3BCN117/10-1074 biNAb (Fig. 1D–F).
Figure 1. Overview of biNAb generation and neutralization activity of 3BNC117/10-1074 IgG1 biNAbs.
(A) Overview of the strategy for generating biNAbs to ensure proper heavy-light chain pairing and heterodimerization. For correct heavy-light chain pairing, one of the parental mAbs was expressed in the CrossMab format (CH1-CL swapping), while for the other mAb, the wild-type domain architecture was maintained. Heavy chain heterodimerization was achieved by introduction of point mutations in the CH3 domain of the two mAbs. (B) Binding specificity for gp140 and (C) in vitro neutralization activity of wild-type and CrossMab variant of 10-1074 was assessed by ELISA using recombinant gp140 and standardized TZMbl neutralization assay, respectively. No difference in the antigen specificity and in vitro neutralization activity was noted between the different 10-1074 mAb variants. (D–F) ELISA assays to determine dual specificity of the 3BNC117/10-1074 biNAb. (D) Competition ELISA with increasing concentrations of each of the parental bNAbs or a mixture (1:1) of the two bNAbs to determine dual specificity of the biNAb. (E) 3BNC117/10-1074 biNAb, 3BNC117 and 10-1074 bNAbs were immobilized to gp140-coated microtiter plates and detected using Fc domain- or light chain (κ or λ) subclass-specific secondary IgG. BiNAb was detected with both the anti-kappa and anti-lambda secondary antibodies, whereas 3BNC117 and 10-1074 only with anti-kappa or anti-lambda, respectively. (F) Epitope-specific ELISA using a CD4bs antigen (2-CC Core) for capture and an anti-lambda detection antibody to confirm bispecific activity. Data are represented as mean ± SEM. (G) In vitro neutralization breadth and potency plot of 3BNC117/10-1074 IgG1 biNAb against an extended (120 strain) multiclade virus panel. Neutralization activity of their respective parental IgG1 bNAbs was included for comparison. 3BNC117/10-1074 IgG1 biNAb exhibited marked reduction in neutralization potency compared to the activity of a mix of their parental mAbs (determined based on the activity of the most potent parental mAb for a given virus strain) See also related Table S1.
IgG1 biNAbs exhibit reduced neutralization potency compared to their respective parental bNAbs
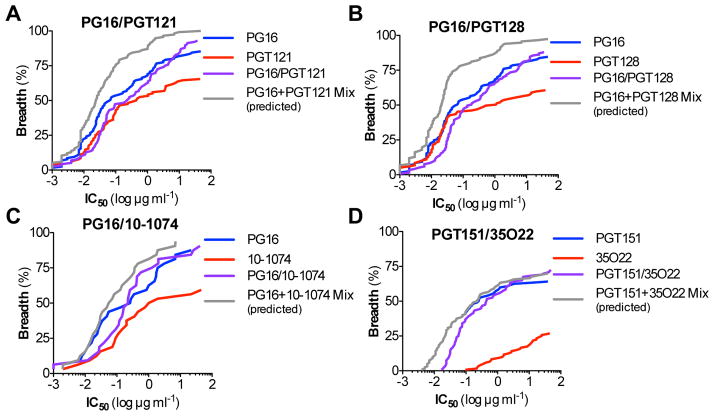
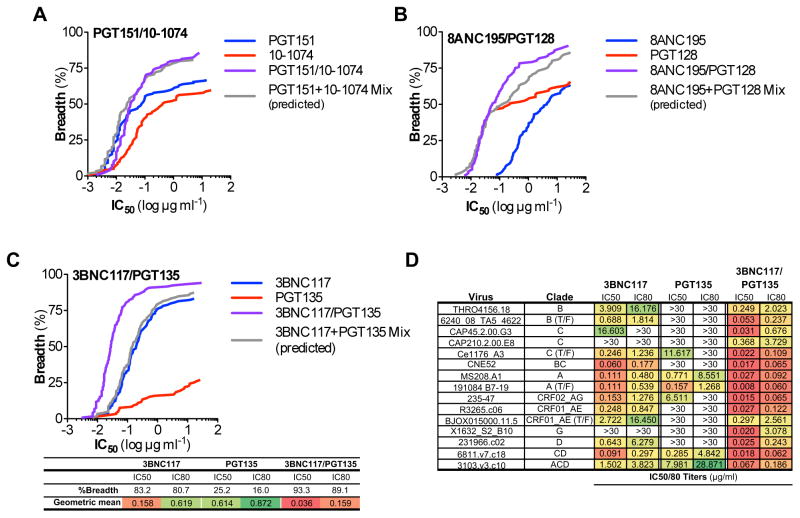
Using a standardized TZMbl assay (Montefiori, 2005), evaluation of the in vitro neutralization activity of the 3BNC117/10-1074 biNAb against an extended multiclade virus panel revealed a marginal increase in the neutralization breadth over 3BNC117 and 10-1074 bNAbs (Fig. 1G, Table S1). However, neutralization potency was markedly reduced when compared to the activity of the parental bNAbs (either alone or as a mix, calculated based on the lowest IC50 titer of the two bNAbs for a given virus). For the majority of the viruses tested, the observed IC50 titers of the biNAb were the average from the two parental bNAbs (Table S1). To determine whether the observed reduction in the neutralization potency was related to the epitope specificity of the selected bNAbs, we extended our study by focusing on different regions of Env, including the V1/2 epitope (targeted by the PG16 bNAb (Walker et al., 2009)), the V3 epitope (targeted by the 10-1074 (Mouquet et al., 2012), PGT121 and PGT128 bNAbs (Pejchal et al., 2011; Walker et al., 2011)), as well as antibodies against the gp120/41 interface (PGT151 (Blattner et al., 2014) and 35O22 (Huang et al., 2014)). Similar to the 3BNC117/10-1074 combination, V1/2-V3 biNAbs (PG16/10-1074, PG16/PGT121, PG16/PGT128) as well as the gp120/41 interface biNAb, PGT151/35O22 exhibited compromised neutralization potency and none of the combinations achieved the activity of their respective parental bNAbs (Fig. 2A–D, Tables S2–S5). These findings clearly suggest that biNAbs, irrespective of their epitope specificities, fail to recapitulate the neutralization activity of their respective parental bNAbs, offering no advantages over conventional, monospecific bNAbs.
Figure 2. In vitro neutralization activity of IgG1 biNAbs targeting different HIV-1 Env epitopes.
In vitro neutralization activity against an extended multiclade virus panel was assessed for IgG biNAbs targeting different, non-overlapping epitopes on HIV-1 Env, using standardized TZMbl neutralization assays. Breadth (% viruses neutralized) vs. potency (IC50 titer (μg/ml)) plots of (A) PG16/PGT121, (B) PG16/PGT128, (C) PG16/10-1074, and (D) PGT151/35O22 IgG1 biNAbs are presented. Neutralization activity of their respective parental IgG1 bNAbs and the predicted activity of the mix of the two parental bNAbs (determined based on the activity of the most potent parental mAb for a given virus strain) was included for comparison. Neutralization data are presented in Tables S2–S5.
Generation and evaluation of IgG hinge domain variants based on the IgG3 hinge structure
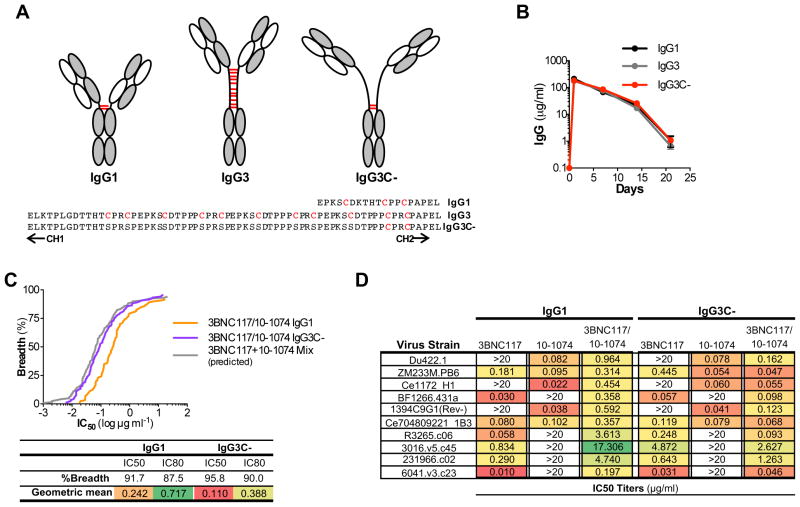
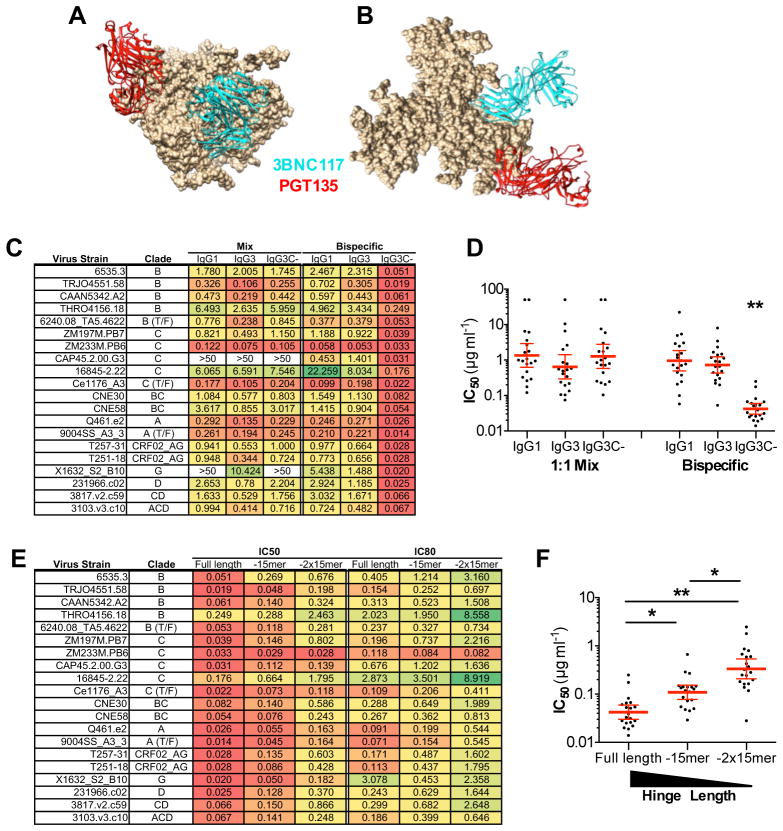
A key immune evasion mechanism of HIV-1 against host antibody responses is the remarkably low density of Env molecules on the viral surface (Zhu et al., 2006), as well as the unique Env trimeric architecture, which preclude high-avidity bivalent interactions of IgG (Klein and Bjorkman, 2010). It is therefore likely that the tested biNAbs would exhibit predominantly monovalent binding to their respective epitopes, possibly accounting for the lack of synergistic activity. Indeed, given the relative rigidity and the short length of the hinge domain of IgG1, concurrent binding of the two Fab arms within the same Env trimer is largely restricted. Overcoming this limitation and favoring intra-trimeric, bivalent interactions of biNAbs should augment their neutralization activity through enhanced avidity (Galimidi et al., 2015; Klein and Bjorkman, 2010; Klein et al., 2009). Among the human IgG subclasses, IgG3 encompasses an exceptionally long and flexible hinge domain, with distinct structural and functional characteristics (Roux et al., 1998; Roux et al., 1997) (Fig. 3A). It is comprised of a 17mer amino acid sequence followed by three 15mer repeats that are highly homologous to the IgG1 hinge structure and represent genomic duplication events of the ancestral hinge-encoding exon, conserved among all other IgG subclasses (Roux et al., 1998). Previous biophysical studies on the IgG3 hinge indicated that the hinge domain spans an over 110Å distance and the unique primary amino acid composition of the CH1 proximal 17mer confers increased flexibility of the Fab arms, allowing for greater degree of rotation compared to IgG1 (117° vs. 136°)(Roux et al., 1998).
Figure 3. Generation and characterization of biNAb hinge variants.
(A) Schematic representation and primary amino acid sequence of the hinge domain of IgG1, IgG3 and the generated variant, IgG3C-. Cysteine residues participating in inter-chain disulfide bonds are depicted in red. IgG1 hinge variants of 10-1074 bNAb were generated by switching the IgG1 hinge region with either the IgG3 or IgG3C- hinge. (B) In vivo stability and half-life was compared between the various 10-1074 hinge-domain variants (mean±SEM, n=4 mice/group). No differences among the hinge domain variants were noted in terms of in vivo pharmacokinetics (B) and protein stability (see also related Figure S1). (C) Comparison of the in vitro neutralization activity against an extended (120 strains) multiclade virus panel of 3BNC117/10-1074 biNAbs expressed as IgG1 or IgG3C- hinge variants (IC50/80 titers are presented in Table S6). As control, the predicted neutralization of the mix of the two parental bNAbs (3BNC117 and 10-1074) was included. (D) Representative IC50 titers (μg/ml) of IgG1 and IgG3C- hinge domain variants of 3BNC117/10-1074 biNAbs and the parental bNAbs (3BNC117 and 10-1074) showing the improved neutralization activity of the IgG3C- hinge variant compared to the wild-type, unmodified IgG1 biNAb.
Based on these unique properties of the IgG3 hinge domain structure, we aimed to develop biNAbs with increased Fab domain flexibility, using the IgG3 hinge as template. To further increase the inherent IgG3 hinge flexibility, we generated an “open” IgG3-based hinge variant (IgG3C-), in which all the cysteine residues have been replaced with serines with the exception of the last two, CH2 proximal residues that are necessary to maintain the structural integrity of the Fc domain through inter-heavy chain disulfide bonding (Fig. 3A). Hinge domain variants of 10-1074 IgG1 were expressed as chimeric molecules, in which the wild-type hinge domain (IgG1) was replaced with that of either IgG3 or the IgG3C- variant. With the exception of the hinge domain, all other domains of the constant region (CH1, CH2, CH3) were of the IgG1 subclass to preserve the effector function and half-life of wild-type IgG1. Indeed, hinge domain variants of 10-1074 demonstrated comparable binding affinity to the different classes of human and mouse Fcγ receptors (FcγRs), suggesting a minimal role for the hinge region in Fc-FcγR interactions (Fig. S1C). Likewise, no differences among the hinge domain variants were noted in terms of protein stability and in vivo pharmacokinetics (Fig. 3B, Fig. S1A–B).
BiNAbs with engineered hinge domain structures exhibit improved neutralization activity
Having established that modifications in the hinge domain had no impact on FcγR binding and IgG half-life and stability, we assessed the neutralization activity of 3BNC117/10-1074 biNAb hinge variants in TZMbl assays (Montefiori, 2005) against an extended multiclade virus panel. Compared to the wild-type, unmodified IgG1 biNAb, IgG3C- hinge variant of the 3BNC117/10-1074 biNAb presented improved activity (Fig. 3C–D). For the vast majority of the tested virus strains (120 stains), a consistent decrease in both the IC50 and IC80 titers was evident for the IgG3C- biNAb, indicating improved neutralization activity compared to IgG1 biNAb (IC50 (μg/ml): 0.242 vs. 0.110; IC80 (μg/ml): 0.717 vs. 0.388 for IgG1 vs. IgG3C-, respectively). In addition, the observed neutralization breadth and potency of the IgG3C- biNAb was comparable to that expected from the mix of the two parental antibodies (Fig. 3C, Table S6). Indeed, whereas IgG1 biNAb exhibited the average neutralization potency of the two parental bNAbs (3BNC117 and 10-1074), IgG3C- biNAb demonstrated activity equal or in some cases better than that of the parental bNAbs (Fig. 3D). In contrast, no difference in the neutralization activity was noted among hinge domain variants of the parental, monospecific antibodies (3BNC117 and 10-1074), demonstrating activity similar to that of their wild-type IgG1 counterparts (Fig. S2A–B). These findings indicate that alterations in the Fab arm flexibility through modifications in the hinge domain structure lead to improved neutralization activity of biNAbs, suggesting that hinge structures engineered for increased length and flexibility could lead to the development of biNAbs with enhanced breadth and potency.
Evaluation of the neutralization activity of a panel of hinge-modified biNAbs reveals particular combinations with synergistic activity
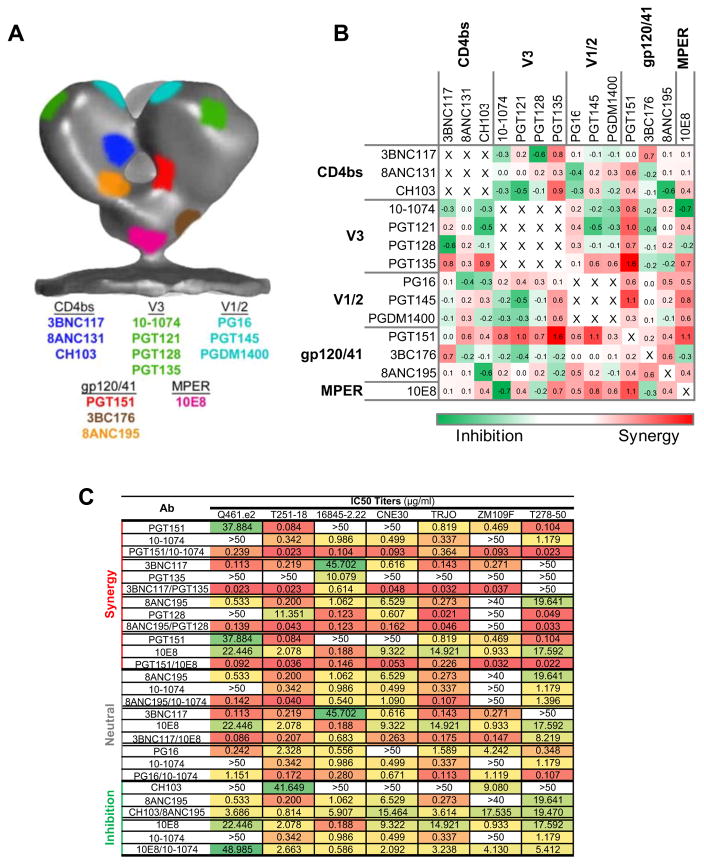
Given the intrinsic flexibility of the Env trimer (Ward and Wilson, 2015), as well as the differences in the angle and orientation by which the Fab domains of antibodies target their respective epitopes on Env, it is difficult to accurately predict the distance required for two Fab arms of a biNAb combination to preferentially exhibit bivalent, intra-trimeric binding and thus confer synergistic activity. To add to this complexity, there are multiple examples of bNAbs that recognize conformational epitopes on Env, or inhibit recognition by other bNAbs upon binding, either by inducing structural changes to the Env trimer or by steric inhibition mechanisms (Burton and Mascola, 2015; Derking et al., 2015; Julien et al., 2013; Ward and Wilson, 2015). In an attempt to identify particular biNAb combinations that would exhibit synergistic activity, a panel of bNAbs targeting various distinct epitopes on the Env trimer with different angle and orientation (Burton and Mascola, 2015) was selected (3BNC117 (Scheid et al., 2011), CH103 (Liao et al., 2013), and 8ANC131 (Zhou et al., 2015) for CD4bs, PG16 (Walker et al., 2009), PGT145 (Walker et al., 2011), and PGDM1400 (Sok et al., 2014) for V1/2, 10-1074 (Mouquet et al., 2012), PGT121, PGT128 (Pejchal et al., 2011; Walker et al., 2011), and PGT135 (Kong et al., 2013) for V3, PGT151 (Blattner et al., 2014), 3BC176 (Klein et al., 2012a; Lee et al., 2015) and 8ANC195 (Scharf et al., 2015) for the gp120/gp41 interface and 10E8 (Huang et al., 2012) for MPER)(Fig. 4A). These bNAbs were cloned as IgG3C- hinge domain variants and biNAbs were generated with non-overlapping epitope specificities. As we have previously described for 3BNC117 and 10-1074 bNAbs, no difference in the neutralization activity was observed between IgG1 and IgG3C- hinge variants for all the parental, monospecific antibodies (Fig. S2, data not shown). Analysis of the neutralization activity of the various biNAb combinations against a panel of 7 cross-clade, tier 2/3 viruses identified particular combinations exhibiting higher (synergistic), equal (neutral) or lower (inhibitory) activity compared to their respective parental bNAb counterparts (also in the IgG3C- hinge variant format)(Fig. 4B–C, Table S7).
Figure 4. Neutralization activity of a panel of hinge domain engineered (IgG3C-) biNAbs.
A panel of bNAbs were selected targeting distinct epitopes on Env and biNAb combinations encompassing the IgG3C- hinge variant were generated with non-overlapping epitope specificities. (A) Epitope mapping on the surface of the HIV-1 Env trimer showing the binding sites of the selected bNAbs (color-coded to match their respective HIV-1 Env epitope). In vitro neutralization activity against a cross-clade, tier 2/3 virus panel (7 strains) was assessed and combinations with variable degree of synergy were identified (B: grid showing the fold change (log) in activity of biNAb over the respective parental bNAb with the most potent (lowest IC50) activity for a given virus strain; C: IC50 titers (μg/ml) of example biNAb combinations exhibiting synergistic, neutral or inhibitory activity). Complete neutralization data are presented in Table S7.
Three particular biNAb combinations that showed evidence for synergistic activity were selected for further analysis, based on their epitope specificities, evidence for efficient expression, sufficient in vivo half-life, and long-term protein stability. These combinations included: PGT151/10-1074, 8ANC195/PGT128, and 3BNC117/PGT135 biNAbs. Evaluation of their neutralization activity against an extended multiclade virus panel revealed that the 8ANC195/PGT128 biNAb combination presented a modest improvement in activity, while no differences were noted for the PGT151/10-1074 biNAb (Fig. 5A–B, Tables S8–9). In contrast, the 3BNC117/PGT135 biNAb exhibited augmented neutralization breadth and potency, surpassing the activity of the parental bNAbs (3BNC117 and PGT135)(Fig. 5C–D, Table S10). For the vast majority of the tested virus strains (119 strains), 3BNC117/PGT135 biNAb demonstrated lower IC50 and IC80 titers compared to the most potent of the parental bNAbs (3BNC117 or PGT135) for each tested virus strain. Overall, the 3BNC117/PGT135 IgG3C- biNAb was capable of neutralizing over 93% of the tested viruses, with an average (geometric mean) IC50 of 0.036 μg/ml, representing one of the most potent anti-HIV-1 Env antibodies characterized so far.
Figure 5. In vitro neutralization breadth and potency of selected IgG3C- hinge biNAbs.
In vitro neutralization activity against an extended multiclade virus panel (119 strains) was assessed for IgG3C- hinge variants of PGT151/10-1074 (A), 8ANC195/PGT128 (B), 3BNC117/PGT135 (C) biNAbs and their respective parental bNAbs (also as IgG3C- hinge variants). Complete neutralization data are presented in Tables S8–S10. Predicted neutralization activity of the mix of the two parental bNAbs (determined based on the activity of the most potent parental mAb for a given virus strain) was included for comparison. (D) Representative IC50/80 titers (μg/ml) of 3BNC117, PGT135, and 3BNC117/PGT135 biNAb showing substantially improved neutralization activity of the biNAb over the parental bNAbs.
The synergistic neutralization activity of the 3BNC117/PGT135 biNAb is dependent on hinge length and flexibility
The synergistic activity observed for the 3BNC117/PGT135 IgG3C- biNAb might actually reflect the capacity of this biNAb combination for bivalent binding of the two Fab arms accomplished by the flexible IgG3C- hinge structure. Indeed, comparison of the gp120-bound PGT135 Fab crystal structure to other anti-V3 bNAbs (PGT122, or PGT128) revealed a unique orientation and angle for PGT135 (Kong et al., 2013) that when combined with the 3BNC117 bNAb, as in the case of 3BNC117/PGT135 IgG3C- biNAb, would facilitate intra-trimeric bivalent interactions (Fig. 6A–B), as the length and flexibility of the IgG3C- hinge could sufficiently span the distance between the two Fab arms bound to the Env trimer. To test this assumption and provide additional mechanistic insights into the requirements for the enhanced synergistic neutralization activity of the 3BNC117/PGT135 IgG3C- biNAb, we assessed the in vitro neutralization activity of different hinge domain variants with variable lengths and properties.
Figure 6. Hinge length and flexibility requirements for the enhanced neutralization activity of the 3BNC117/PGT135 biNAb.
(A) Side and (B) top view of 3BNC117 (cyan; PDB ID: 4JPV) and PGT135 (red; PDB ID: 4JM2) Fabs bound to the Env trimer (PDB ID: 4NCO). The crystal structures of PGT135 and 3BNC117 Fab complexed with gp120 (JRFL and 93TH057, respectively) were aligned onto the crystal structure of the BG505.SOSIP.664 trimer (PDB ID: 4NCO). The distance between the ends of the two Fabs was calculated to be 67Å, suggesting that the IgG3C- hinge variant might facilitate bivalent, intra-trimeric interactions of the 3BNC117/PGT135 biNAb. (C–D) In vitro neutralization activity (IC50 titers (μg/ml)) of 3BNC117 + PGT135 bNAb mix or 3BNC117/PGT135 biNAb expressed with the following hinge domain structures: IgG1, IgG3 and IgG3C- (see also related Fig. 3A for sequence). 3BNC117/PGT135 IgG3C- biNAb exhibited significantly improved neutralization activity (**p<0.001) compared to IgG1 or IgG3 biNAb hinge variants. IC50 titer (μg/ml) results are presented as geometric mean±95%CI. (E–F) IC50 and IC80 titers (μg/ml) of 3BNC117/PGT135 IgG3C- biNAbs with variable hinge domain length. Shorter variants of IgG3C- (“Full length”) were generated by deleting either one (“-15mer”) or two (“-2x15mer”) of the three 15-mer repeats (EPKSSDTPPPSPRSP). (F) Comparison of the neutralization activity between 3BNC117/PGT135 IgG3C- biNAb (full length) and shortened hinge domain variants revealed that enhanced neutralization activity is correlated with hinge domain length. IC50 titer (μg/ml) results are presented as geometric mean±95%CI. *p<0.05; **p<0.001.
3BNC117/PGT135 biNAbs and their respective parental bNAbs (3BNC117 and PGT135) were expressed as variants encompassing the hinge domain structure of wild-type IgG1, IgG3, or IgG3C- (open hinge structure based on wild-type IgG3 sequence) and their neutralization activity was assessed against a multiclade, 20-strain panel. When the activity of a 1:1 mix of the two parental bNAbs was compared among the different hinge domain variants, no significant differences were noted, with all the hinge variants exhibiting comparable neutralization activity (Fig. 6C–D). In contrast, 3BNC117/PGT135 biNAb demonstrated significantly augmented activity only in the IgG3C-, but not in the IgG1 or IgG3 hinge domain format, indicating that the synergistic activity of the 3BNC117/PGT135 biNAb is dependent upon the unique structure and flexibility of the IgG3C- hinge variant. To provide further evidence on the role of the hinge length in the improved neutralization activity of the 3BNC117/PGT135 biNAb, two different variants of the IgG3C- hinge structure were generated, which lacked either one (-15mer) or two (-2x15mer) of the three 15 amino acid repeats (EPKSSDTPPPSPRSP) that comprise part of the IgG3C- hinge sequence (Fig. 3A). 3BNC117/PGT135 biNAbs encompassing the shortened hinge domain variants were generated and their activity was compared to that of IgG3C-. Assessment of the in vitro neutralization activity revealed that hinge length is correlated with the neutralization potency of the 3BNC117/PGT135 biNAb, with shorter hinge variants (“-15mer” or “-2x15mer”) exhibiting impaired neutralization activity (Fig. 6E–F). These findings suggest that the observed enhancement in the in vitro neutralization activity of the 3BNC117/PGT135 biNAb is dependent upon hinge length and flexibility, that likely favors intra-trimeric, bivalent Env interactions.
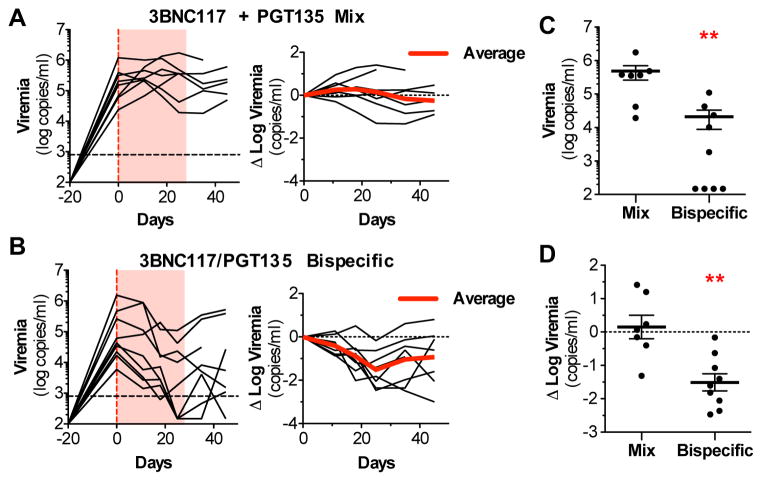
3BNC117/PGT135 IgG3C- biNAb confer enhanced in vivo protective activity in HIV-1 infected humanized mice
In order to determine whether the observed increase in neutralization potency of the 3BNC117/PGT135 IgG3C- biNAb also translates to improved therapeutic efficacy in vivo, we evaluated its capacity to suppress viremia in humanized mice (human CD34+-reconstituted NRG mice (Klein et al., 2012b)) with established HIV-1 infection. HIV-1-infected humanized mice were treated either with a mix (1:1) of 3BNC117 and PGT135 bNAbs or with 3BNC117/PGT135 biNAb (all in IgG3C- hinge variant format). Among the two experimental groups, no differences in the in vivo half-life or the serum antibody levels were noted (Fig. S3A–B). Quantitation of plasma viremia revealed that 3BNC117/PGT135 biNAb treatment decreased viremia by an average of 1.5 log10 during the treatment period (Fig. 7B–D) with the majority (7/9) of treated animals exhibiting substantially reduced plasma viremia levels (Δ log10 viremia <−1.0). Mice that failed to demonstrate robust response to biNAb therapy were often associated with mutations in the biNAb-targeting epitopes (V3 or CD4bs; Fig. S3C–D). In contrast, administration of the 3BNC117 + PGT135 bNAb mix had minimal effect on plasma viremia (mean Δ log10 viremia: 0.15; Fig. 7A, C–D), These findings suggest that the 3BNC117/PGT135 IgG3C- biNAb represents a unique biNAb molecule with in vivo therapeutic activity, with potential for future pre-clinical development.
Figure 7. In vivo evaluation of the therapeutic activity of the 3BNC117/PGT135 IgG3C-biNAb in HIV-1-infected humanized mice.
(A–B) The in vivo therapeutic activity of 3BNC117/PGT135 IgG3C- biNAb was compared to that of a mix of 3BNC117 + PGT135 IgG3C- bNAbs in HIV-1-infected humanized mice. Mice with established viremia were treated for 4 weeks (red shaded area) with bNAb mix (A) or biNAb (B) and plasma viremia was monitored to assess the in vivo protective activity of antibodies. (C) Comparison of viremia levels or (D) change from baseline at the end of the treatment period revealed significant differences between the mix- and biNAb-treated mice (**p<0.005; mean±SEM, n=7 or 9). See also related Figure S3.
Discussion
Several recent studies using anti-Env antibodies with broad and potent neutralizing activity revealed their capacity to confer both effective pre-exposure prophylaxis and therapeutic control of viremia in murine and non-human primate HIV-1 disease models, as well as in HIV-1 infected humans (Barouch et al., 2013; Bournazos et al., 2014; Caskey et al., 2015; Halper-Stromberg et al., 2014; Horwitz et al., 2013; Klein et al., 2012b; Lynch et al., 2015; Shingai et al., 2013). Studies on animal models for HIV-1 infection suggested that sustained therapeutic viremia suppression is accomplished by co-administration of a cocktail of bNAbs targeting key epitopes on the HIV-1 Env that are essential for viral fitness, overcoming thereby the emergence of virus escape mutants. Bispecific anti-HIV-1 Env antibodies represent an ideal therapeutic approach that would combine the breadth, antigenic specificity and neutralization potency of two bNAbs, into a single molecule, facilitating preclinical evaluation and development. In addition to the additive effect on the neutralization breadth, favoring hetero-bivalent interactions of the two Fab arms could confer synergistic activity (Klein et al., 2009), yielding biNAbs with substantially improved neutralization potency compared to conventional bNAbs. Indeed, the use of DNA- and protein-based structures to link two Fab arms at optimal distances necessary to achieve hetero-bivalent, intra-trimeric interactions resulted in enhanced in vitro neutralization potency (Galimidi et al., 2015).
Based on these observations, we developed and characterized biNAbs with non-overlapping epitope specificities to identify particular biNAb combinations that would demonstrate potent and synergistic neutralization activity. Given the significance of the FcγR-mediated pathways in the in vivo protective activity of anti-HIV-1 bNAbs (Bournazos et al., 2014; Halper-Stromberg et al., 2014; Hessell et al., 2007), our major goal was to generate biNAbs with the wild-type IgG structure, while avoiding irregular, non-physiological architectures used in the past (Spiess et al., 2015). This approach ensured that the resulting biNAb would have the capacity to interact with FcγRs and exhibit long and stable in vivo pharmacokinetics, with minimal immunogenicity potential. Indeed, all the generated biNAb variants exhibited identical in vivo half-life and affinity for the different classes of FcγRs. Attempts to generate anti-HIV-1 Env biNAbs based on the IgG1 structure were generally characterized by lower neutralization potency compared to their corresponding parental bNAbs, irrespective of their epitope specificity (Fig. 1G, 2; Tables S1–5). Similar findings have also been reported in a recent study that generated and characterized a number of anti-Env IgG1 biNAbs (Asokan et al., 2015), suggesting that IgG1 biNAbs offer no advantages over conventional, monospecific bNAbs. Since the observed impairment in the neutralization potency possibly reflects the lack of sufficient flexibility of the two Fab arms to achieve bivalent, intra-trimeric interactions in the IgG1 format, hinge domain engineered biNAbs were generated with improved Fab domain flexibility, based on the naturally-occurring hinge domain of IgG3, which is characterized by a uniquely long and flexible structure (Roux et al., 1998; Roux et al., 1997). This approach incorporates minimal changes to the overall IgG structure, while it achieves significant enhancement in the neutralization breadth and potency.
Among all the tested hinge-engineered biNAb combinations, 3BNC117/PGT135 showed evidence for synergistic activity, surpassing the potency of both parental bNAbs (3BNC117 and PGT135). Indeed, assessment of its neutralization potency indicated that for the vast majority of the tested viruses, the 3BNC117/PGT135 biNAb exhibits lower IC50 and IC80 titers, and for over a third of the tested viruses, the improvement in the neutralization potency exceed 10-fold compared to 3BNC117 and PGT135 bNAbs. Its enhanced activity was largely attributed to the hinge length and flexibility, as hinge domain variants with shorter length or decreased flexibility also exhibited reduced neutralization breadth and potency. These findings indicate that the improved neutralization activity of the 3BNC117/PGT135 IgG3C- biNAb is likely the result of bivalent, intra-trimeric interactions, accomplished by the unique structure of the engineered hinge domain of the IgG3C- variant. As previously suggested (Galimidi et al., 2015), intra-trimeric, heterobivalent crosslinking of the two Fab arms increases the overall avidity of the Env trimer – biNAb interaction, leading thereby to augmented neutralization potency.
Although several mechanisms of bNAb binding interference have been described, even for bNAbs with non-overlapping epitope specificities like 3BNC117 and PGT135 (Derking et al., 2015), our findings suggest that the observed effect of the 3BNC117/PGT135 biNAb could not be attributed to conformational changes induced upon bNAb binding, as no change in the neutralization activity was evident for the mix of the two bNAbs (3BNC117 + PGT135)(Figure 6C–D). However, a recent study that analyzed the binding profile of a panel of bNAbs to the soluble native BG505.SOSIP.664 gp140 trimer revealed that PGT135 inhibited unidirectionally the binding of anti-CD4bs, VRC01-like antibodies to the gp140 trimer, due to reorientation of a glycan structure (predominantly Asn386 and/or Asn392 glycans), partly occluding the CD4bs epitope (Derking et al., 2015). Since all VRC01-like bNAbs exhibit substantially higher affinity for Env compared to PGT135, it is unlikely that such cross-bNAb inhibition would occur for strains sensitive to both bNAbs. Based on these findings, in the context of the 3BNC117/PGT135 biNAb, it is expected that for virus strains sensitive to 3BNC117, increased neutralization potency would be expected through initial binding of the 3BNC117 arm (due to higher affinity) followed by the PGT135. In contrast, for viruses that are resistant to 3BNC117, but sensitive to PGT135, no improvement in the neutralization activity is expected, due to PGT135-mediated occlusion of the CD4bs epitope. Indeed, assessment of the extended panel neutralization data (Table S10) indicates that for the few virus strains that are resistant to 3BNC117, but sensitive to PGT135 (like CNE20, CNE21, X2088_c9), no enhanced neutralization activity is evident for the 3BNC117/PGT135 biNAb and the observed neutralization potency is lower than that of the PGT135 bNAb, further supporting the notion that the enhanced neutralization activity of the 3BNC117/PGT135 biNAb is the result of bivalent, intra-trimeric interactions.
Despite the augmented neutralization potency of the 3BNC117/PGT135 IgG3C- biNAb, it was essential to determine whether its in vitro activity also translates to enhanced in vivo therapeutic efficacy. We therefore assessed its in vivo activity in HIV-1-infected humanized mice; a robust model that recapitulates human HIV-1 infection and has been systematically used in the past to accurately investigate the in vivo activity of bNAbs (Bournazos et al., 2014; Diskin et al., 2013; Halper-Stromberg et al., 2014; Horwitz et al., 2013; Klein et al., 2012b). Consistent with the in vitro findings, 3BNC117/PGT135 IgG3C- biNAb showed improved in vivo protective activity, probably reflecting its enhanced neutralization activity. However, a role for Fc effector functions in the in vivo protective activity of this biNAb could not be excluded. In addition to enhancing neutralization, hetero-bivalent biNAb binding to the Env trimer could also lead to more stable biNAb-Env interactions, facilitating thereby clearance of viral particles and infected cells through FcγR-mediated mechanisms.
Collectively, these findings suggest that the hinge-engineered 3BNC117/PGT135 biNAb represents one of the most potent anti-HIV-1 bNAbs developed to date, exhibiting high neutralization breadth and potency, as well as improved in vivo activity. To comparatively benchmark the activity of the 3BNC117/PGT135 biNAb with previously characterized bNAbs, its neutralization breadth and potency was compared to several bNAbs targeting distinct epitopes on gp140. As shown in Figure S4A, the 3BNC117/PGT135 biNAb displays increased neutralization potency, compared to the most potent bNAbs so far characterized, including VRC07, PGT121, and PG9 (using data reported in Kong et al., (2015)). Additionally, to gain further insights on how the activity of this biNAb compares to physical combinations of bNAbs, we directly compared the activity observed in this study for 3BNC117/PGT135 biNAb with that previously reported for several bNAb combinations (Kong et al., 2015). Compared to all two-bNAb combinations, the 3BNC117/PGT135 biNAb demonstrated higher neutralization activity, neutralizing >80% of viral strains with IC50 <0.1 μg/ml (Figure S4B). Its potency was only marginally lower compared to that observed for the three-bNAb combinations. This analysis provides a direct measure of the neutralization activity of the 3BNC117/PGT135 biNAb against other bNAbs and bNAb combinations, highlighting the improved potency of this biNAb over conventional, monospecific bNAbs.
In summary, the present study described an approach for the generation of anti-HIV-1 Env biNAbs with physiological IgG architecture, FcγR binding profile and pharmacokinetic properties. Compared to conventional, monospecific bNAbs, biNAbs with hinge domain engineered structures exhibit potent neutralization activity with improved breadth and potency and enhanced in vivo activity. These unique advantages of hinge domain-optimized biNAbs represent a platform technology that can also be extended to other viral and cellular targets.
Experimental Procedures
A detailed description of the experimental procedures is provided in the Supplemental Experimental Procedures section.
In Vitro Neutralization Assay
In vitro neutralization activity of antibodies was assessed against different HIV-1 envelope pseudoviruses, using a standardized TZMbl neutralization assay, as previously described (Montefiori, 2005). For assessing the IC50/80 titers of a mix of two bNAbs, antibodies were mixed at a ratio of 1:1 (experimental IC50/80). Predicted (theoretical IC50/80) neutralization activity of a mix of two mAbs was determined by selecting the lowest IC50/80 titer of the two mAbs for a given virus. For combinations involving 2 mAbs with non-overlapping epitope specificities, theoretical IC50/80 has been shown to be comparable to the neutralization activity of the experimental IC50/80 (Kong et al., 2015).
In Vivo Experiments
All in vivo experiments were performed in compliance with federal laws and institutional guidelines and have been approved by the Rockefeller University Institutional Animal Care and Use Committee. Humanized NRG mice were generated, as previously described (Klein et al., 2012b). Mice with a measurable human CD45+ graft (10–15 weeks, males and females) were infected following i.p. injection of HIV-1T251-18 (180 ng p24). Viral load was quantified 3 weeks post-infection and mice with viral loads >103 copies/ml were included in treatment experiments. Mice were randomly assigned to experimental groups and both groups had comparable baseline average viremia levels. Antibodies (either 1 mg of 3BNC117/PGT135 biNAb or 1 mg 1:1 mix of 3BNC117 (0.5 mg) and PGT135 (0.5mg)) were administered biweekly s.c. for 4 weeks. Each experimental group consisted of 9 mice; a group size previously determined to sufficiently detect response to antibody therapy (Bournazos et al., 2014; Klein et al., 2012b). Plasma HIV-1 viral load was determined by quantitative reverse-transcriptase PCR as previously described (Bournazos et al., 2014; Klein et al., 2012b).
Statistical Analysis
Results from multiple experiments are presented as mean ± standard error of the mean (SEM). IC50 titers are presented as geometric mean ± 95% CI. For comparison of the in vitro neutralization activity, Kruskal-Wallis test was used to test for differences in the IC50/80 titers, and where statistically significant effects were found, post hoc analysis using Dunn’s multiple comparison test was performed. For comparison of viremia between the two experimental groups, Mann-Whitney non-parametric test (two-sided) was used. Data were analyzed with Graphpad Prism software (Graphpad) and P values of <0.05 were considered to be statistically significant.
Supplementary Material
Acknowledgments
We wish to thank P. Smith, H. Lee, M. Saito, M. Ye, E. Lam, P. Bartel, T. Schoofs, J. Cetrulo-Lorenzi, and L. Nogueira (Rockefeller University) for technical help and advice, M. Louder and J. Mascola (VRC, NIAID) for providing the raw neutralization data for the bNAb combinations (Fig. S4) and the NIH AIDS Reagent Program for obtaining the T251-18 env construct. Research reported in this publication was supported in part by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers AI081677 and AI100148 (to J.V.R. and M.C.N.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Research support was also provided by the Bill & Melinda Gates Foundation grant OPP1033115 and OPP1124068 to J.V.R. and M.C.N. and OPP1032144 to M.S.S. S.B. was supported by an amfAR Mathilde Krim Fellowship in Basic Biomedical Research (108977-57-RKVA).
Footnotes
Author Contributions
S.B. designed and performed experiments, analyzed data and wrote the manuscript, A.G. assisted in protein expression, M.S.S. performed the in vitro neutralization assays, M.C.N. designed experiments, J.V.R. designed and directed the study.
All authors declare no conflict of interest.
References
- Asokan M, Rudicell RS, Louder M, McKee K, O’Dell S, Stewart-Jones G, Wang K, Xu L, Chen X, Choe M, et al. Bispecific Antibodies Targeting Different Epitopes on the HIV-1 Envelope Exhibit Broad and Potent Neutralization. J Virol. 2015;89:12501–12512. doi: 10.1128/JVI.02097-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang HW, Shekhar K, Gupta S, et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Peña AT, Cupo A, Julien JP, van Gils M, Lee PS, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014;40:669–680. doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S, DiLillo DJ, Ravetch JV. The role of Fc-FcγR interactions in IgG-mediated microbial neutralization. J Exp Med. 2015;212:1361–1369. doi: 10.1084/jem.20151267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S, Klein F, Pietzsch J, Seaman MS, Nussenzweig MC, Ravetch JV. Broadly Neutralizing Anti-HIV-1 Antibodies Require Fc Effector Functions for In Vivo Activity. Cell. 2014;158:1243–1253. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S, Ravetch JV. Fcγ receptor pathways during active and passive immunization. Immunol Rev. 2015;268:88–103. doi: 10.1111/imr.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Mascola JR. Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol. 2015;16:571–576. doi: 10.1038/ni.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caskey M, Klein F, Lorenzi JC, Seaman MS, West AP, Buckley N, Kremer G, Nogueira L, Braunschweig M, Scheid JF, et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature. 2015;522:487–491. doi: 10.1038/nature14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chames P, Baty D. Bispecific antibodies for cancer therapy: the light at the end of the tunnel? MAbs. 2009;1:539–547. doi: 10.4161/mabs.1.6.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derking R, Ozorowski G, Sliepen K, Yasmeen A, Cupo A, Torres JL, Julien JP, Lee JH, van Montfort T, de Taeye SW, et al. Comprehensive antigenic map of a cleaved soluble HIV-1 envelope trimer. PLoS Pathog. 2015;11:e1004767. doi: 10.1371/journal.ppat.1004767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Klein F, Horwitz JA, Halper-Stromberg A, Sather DN, Marcovecchio PM, Lee T, West AP, Gao H, Seaman MS, et al. Restricting HIV-1 pathways for escape using rationally designed anti-HIV-1 antibodies. J Exp Med. 2013;210:1235–1249. doi: 10.1084/jem.20130221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimidi RP, Klein JS, Politzer MS, Bai S, Seaman MS, Nussenzweig MC, West AP, Bjorkman PJ. Intra-spike crosslinking overcomes antibody evasion by HIV-1. Cell. 2015;160:433–446. doi: 10.1016/j.cell.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigwood NL, Montefiori DC, Sutton WF, McClure J, Watson AJ, Voss G, Hirsch VM, Richardson BA, Letvin NL, Hu SL, et al. Passive immunotherapy in simian immunodeficiency virus-infected macaques accelerates the development of neutralizing antibodies. J Virol. 2004;78:5983–5995. doi: 10.1128/JVI.78.11.5983-5995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halper-Stromberg A, Lu CL, Klein F, Horwitz JA, Bournazos S, Nogueira L, Eisenreich TR, Liu C, Gazumyan A, Schaefer U, et al. Broadly Neutralizing Antibodies and Viral Inducers Decrease Rebound from HIV-1 Latent Reservoirs in Humanized Mice. Cell. 2014;158:989–999. doi: 10.1016/j.cell.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell AJ, Hangartner L, Hunter M, Havenith CE, Beurskens FJ, Bakker JM, Lanigan CM, Landucci G, Forthal DN, Parren PW, et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- Horwitz JA, Halper-Stromberg A, Mouquet H, Gitlin AD, Tretiakova A, Eisenreich TR, Malbec M, Gravemann S, Billerbeck E, Dorner M, et al. HIV-1 suppression and durable control by combining single broadly neutralizing antibodies and antiretroviral drugs in humanized mice. Proc Natl Acad Sci U S A. 2013;110:16538–16543. doi: 10.1073/pnas.1315295110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Gaebler C, Mouquet H, Sather DN, Lehmann C, Scheid JF, Kraft Z, Liu Y, Pietzsch J, Hurley A, et al. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J Exp Med. 2012a;209:1469–1479. doi: 10.1084/jem.20120423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012b;492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JS, Bjorkman PJ. Few and far between: how HIV may be evading antibody avidity. PLoS Pathog. 2010;6:e1000908. doi: 10.1371/journal.ppat.1000908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JS, Gnanapragasam PN, Galimidi RP, Foglesong CP, West AP, Bjorkman PJ. Examination of the contributions of size and avidity to the neutralization mechanisms of the anti-HIV antibodies b12 and 4E10. Proc Natl Acad Sci U S A. 2009;106:7385–7390. doi: 10.1073/pnas.0811427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R, Louder MK, Wagh K, Bailer RT, deCamp A, Greene K, Gao H, Taft JD, Gazumyan A, Liu C, et al. Improving neutralization potency and breadth by combining broadly reactive HIV-1 antibodies targeting major neutralization epitopes. J Virol. 2015;89:2659–2671. doi: 10.1128/JVI.03136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Leaman DP, Kim AS, Torrents de la Peña A, Sliepen K, Yasmeen A, Derking R, Ramos A, de Taeye SW, Ozorowski G, et al. Antibodies to a conformational epitope on gp41 neutralize HIV-1 by destabilizing the Env spike. Nat Commun. 2015;6:8167. doi: 10.1038/ncomms9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch RM, Boritz E, Coates EE, DeZure A, Madden P, Costner P, Enama ME, Plummer S, Holman L, Hendel CS, et al. Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med. 2015;7:319ra206. doi: 10.1126/scitranslmed.aad5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant AM, Zhu Z, Yuan JQ, Goddard A, Adams CW, Presta LG, Carter P. An efficient route to human bispecific IgG. Nat Biotechnol. 1998;16:677–681. doi: 10.1038/nbt0798-677. [DOI] [PubMed] [Google Scholar]
- Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol. 2005;12:12.11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109:E3268–3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CT, Jaworski JP, Jayaraman P, Sutton WF, Delio P, Kuller L, Anderson D, Landucci G, Richardson BA, Burton DR, et al. Passive neutralizing antibody controls SHIV viremia and enhances B cell responses in infant macaques. Nat Med. 2010;16:1117–1119. doi: 10.1038/nm.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CS, Song R, Ochsenbauer C, Andrews CD, Franco D, Yu J, Oren DA, Seaman MS, Ho DD. Bispecific antibodies directed to CD4 domain 2 and HIV envelope exhibit exceptional breadth and picomolar potency against HIV-1. Proc Natl Acad Sci U S A. 2013;110:13540–13545. doi: 10.1073/pnas.1304985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegu A, Asokan M, Wu L, Wang K, Hataye J, Casazza JP, Guo X, Shi W, Georgiev I, Zhou T, et al. Activation and lysis of human CD4 cells latently infected with HIV-1. Nat Commun. 2015;6:8447. doi: 10.1038/ncomms9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietzsch J, Gruell H, Bournazos S, Donovan BM, Klein F, Diskin R, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, et al. A mouse model for HIV-1 entry. Proc Natl Acad Sci U S A. 2012;109:15859–15864. doi: 10.1073/pnas.1213409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway JB, Presta LG, Carter P. ‘Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 1996;9:617–621. doi: 10.1093/protein/9.7.617. [DOI] [PubMed] [Google Scholar]
- Roux KH, Strelets L, Brekke OH, Sandlie I, Michaelsen TE. Comparisons of the ability of human IgG3 hinge mutants, IgM, IgE, and IgA2, to form small immune complexes: a role for flexibility and geometry. J Immunol. 1998;161:4083–4090. [PubMed] [Google Scholar]
- Roux KH, Strelets L, Michaelsen TE. Flexibility of human IgG subclasses. J Immunol. 1997;159:3372–3382. [PubMed] [Google Scholar]
- Schaefer W, Regula JT, Bähner M, Schanzer J, Croasdale R, Dürr H, Gassner C, Georges G, Kettenberger H, Imhof-Jung S, et al. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc Natl Acad Sci U S A. 2011;108:11187–11192. doi: 10.1073/pnas.1019002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf L, Wang H, Gao H, Chen S, McDowall AW, Bjorkman PJ. Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env. Cell. 2015;162:1379–1390. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M, Lifson JD, et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013;503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sok D, van Gils MJ, Pauthner M, Julien JP, Saye-Francisco KL, Hsueh J, Briney B, Lee JH, Le KM, Lee PS, et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc Natl Acad Sci U S A. 2014;111:17624–17629. doi: 10.1073/pnas.1415789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C, Zhai Q, Carter PJ. Alternative molecular formats and therapeutic applications for bispecific antibodies. Mol Immunol. 2015;67:95–106. doi: 10.1016/j.molimm.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Sung JA, Pickeral J, Liu L, Stanfield-Oakley SA, Lam CY, Garrido C, Pollara J, LaBranche C, Bonsignori M, Moody MA, et al. Dual-Affinity Re-Targeting proteins direct T cell-mediated cytolysis of latently HIV-infected cells. J Clin Invest. 2015;125:4077–4090. doi: 10.1172/JCI82314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward AB, Wilson IA. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem Sci. 2015;40:101–107. doi: 10.1016/j.tibs.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, et al. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Liu J, Bess J, Chertova E, Lifson JD, Grisé H, Ofek GA, Taylor KA, Roux KH. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.