Vesicular transport from mitochondria to lysosomes is an emerging mitochondrial quality control mechanism. Here, McLelland et al. identify how mitochondrial vesicles are targeted for degradation, showing that syntaxin-17 is recruited to these structures to govern their SNARE-dependent fusion with endolysosomes.
Abstract
Mitochondria are considered autonomous organelles, physically separated from endocytic and biosynthetic pathways. However, recent work uncovered a PINK1/parkin-dependent vesicle transport pathway wherein oxidized or damaged mitochondrial content are selectively delivered to the late endosome/lysosome for degradation, providing evidence that mitochondria are indeed integrated within the endomembrane system. Given that mitochondria have not been shown to use canonical soluble NSF attachment protein receptor (SNARE) machinery for fusion, the mechanism by which mitochondrial-derived vesicles (MDVs) are targeted to the endosomal compartment has remained unclear. In this study, we identify syntaxin-17 as a core mitochondrial SNARE required for the delivery of stress-induced PINK1/parkin-dependent MDVs to the late endosome/lysosome. Syntaxin-17 remains associated with mature MDVs and forms a ternary SNARE complex with SNAP29 and VAMP7 to mediate MDV–endolysosome fusion in a manner dependent on the homotypic fusion and vacuole protein sorting (HOPS) tethering complex. Syntaxin-17 can be traced to the last eukaryotic common ancestor, hinting that the removal of damaged mitochondrial content may represent one of the earliest vesicle transport routes in the cell.
Introduction
Proper mitochondrial function is paramount to neuronal survival, and deficits in mitochondrial activity may underlie neurodegenerative diseases such as Parkinson’s disease (PD). Mutations in PARKIN and PINK1 cause recessive forms of PD (Kitada et al., 1998; Valente et al., 2004), and these genes (encoding parkin, an E3 ubiquitin ligase, and PINK1, a mitochondrially targeted protein kinase) primarily function in the quality control of mitochondria; a collection of pathways regulating the removal of damaged proteins, lipids, and organelles from the mitochondrial reticulum to ensure its proper activity (Ryan et al., 2015). In one such mechanism, parkin is recruited to depolarized mitochondria by PINK1, where it initiates their autophagic turnover (termed mitophagy; Ryan et al., 2015; Yamano et al., 2016). Alternatively, in response to oxidative stress, parkin and PINK1 deliver selective, oxidized components of the mitochondrial matrix and inner membrane to the late endosome for turnover via a class of mitochondrial-derived vesicles (MDVs; Soubannier et al., 2012a,b; McLelland et al., 2014). Whereas mitophagy describes the engulfment of a fusion-incompetent mitochondrial fragment within the autophagosome, PINK1/parkin-dependent MDVs are mechanistically distinct in that vesicles containing highly selected mitochondrial cargo bud off mitochondria independently of the core mitochondrial fission GTPase Drp1, and their turnover does not require canonical autophagy machinery (Soubannier et al., 2012a; McLelland et al., 2014). Indeed, evidence from parkin- and PINK1-null flies has supported a role for these proteins in both mitophagy and selective mitochondrial protein turnover in vivo (Vincow et al., 2013). Moreover, a recent study in Drosophila melanogaster revealed a strong genetic interaction between parkin and Vps35 (Malik et al., 2015), another PD-linked gene involved in the generation of other MDV populations (Braschi et al., 2010; Wang et al., 2016), suggesting that defective MDV transport may play a role in PD pathogenesis.
Although parkin and PINK1 activity are required for the generation of oxidative stress–triggered MDVs, the mechanism by which this vesicle population reaches the lysosome remains poorly understood. As mitochondrial vesicles are membrane-bound structures, a role for membrane fusion in turnover seems apparent (Sugiura et al., 2014). SNAREs mediate most membrane fusion events in cells (mitochondrial membrane fusion being one exception) and associate via the formation of a four-helix bundle between their helical SNARE domains (designated Qa, Qb, Qc, and R based on the amino acid present at the so-called zero layer; Fasshauer et al., 1998; Sutton et al., 1998), which zipper progressively toward the membrane-bound end of the complex to bring membranes together to fuse (Hanson et al., 1997; Gao et al., 2012; Li et al., 2014). As the compartmental specificity of fusion events is encoded by the SNAREs on the vesicle and target membrane (Söllner et al., 1993), the regulation of targeting and turnover of MDVs by SNARE-dependent membrane fusion presents itself as an intriguing and logical possibility. However, given the endosymbiotic origin of mitochondria, as well as the established roles of large GTPases involved in both homotypic and heterotypic mitochondrial fusion and tethering events, a role for SNAREs in MDV targeting and fusion is not a foregone conclusion.
Here, we demonstrate that syntaxin-17 (Stx17), a Qa-SNARE, is involved in the targeting of parkin/PINK1-generated MDVs to endolysosomal compartments. We observe the loading of Stx17 onto vesicles that were budded from mitochondria in vitro and the enrichment of Stx17 on mitochondrial foci and nearby fully formed vesicles in cells. Loss of Stx17 abrogates lysosomal targeting of MDVs and prevents pathway processivity. Stx17 assembles into a ternary SNARE complex with SNAP29 and VAMP7 to mediate MDV–endolysosome fusion, which can be disrupted using structure-based mutations. Strikingly, these SNAREs are dispensable for mitophagy, implying divergent targeting mechanisms in different parkin/PINK1 mitochondrial quality control pathways. These results demonstrate that MDVs are integrated within bona fide cellular membrane trafficking pathways and identify machinery required for MDV turnover.
Results
Stx17 is biochemically enriched on mitochondrial-derived vesicles
We sought to identify the manner by which oxidative stress–triggered MDVs target to the lysosome, reasoning that this turnover pathway, in a manner similar to lysosomally directed endosomal transport, required SNARE-dependent targeting to reach the endolysosomal membrane. A portion of the Qa-SNARE Stx17 localizes to mitochondria, in addition to the ER (Hamasaki et al., 2013; Hung et al., 2014; Arasaki et al., 2015). Moreover, Stx17 can form complexes with target R-SNAREs associated with lysosomes and late endosomes (Itakura et al., 2012), and its assembly into these complexes is metabolically regulated (Guo et al., 2014), making it a prime candidate for MDV–endolysosome fusion.
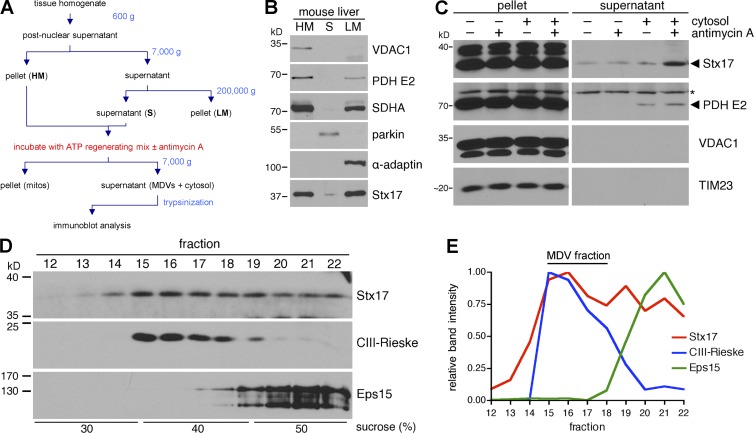
We began by testing whether Stx17, which is unique among SNAREs with its two C-terminal transmembrane domains (Kloepper et al., 2008), is biochemically enriched on MDVs, turning to in vitro MDV reconstitution assays (outlined in Fig. 1 A) that we adapted from previous work (Soubannier et al., 2012b). We fractionated mouse liver into heavy membranes (HMs; enriched in the mitochondrial proteins VDAC1, PDH E2, and SDHA), a soluble fraction (S; containing cytosol), and light membranes (LMs; containing microsomes with some mitochondrial contamination; Fig. 1 B). Endogenous Stx17 was evenly enriched in both membrane fractions (Fig. 1 B), as expected (Hamasaki et al., 2013). We then incubated the heavy membranes in the presence or absence of cytosol (fraction S) and/or antimycin A (a mitochondrial complex III inhibitor that causes oxidative stress; see Fig. 1 A or, for a detailed description of the assay, see Materials and methods), as we have previously shown that mitochondrial cargo can be extracted from the HM fraction in a manner dependent on reactive oxygen species generation and cytosolic components in vitro (Soubannier et al., 2012b). We then pelleted the mitochondria and trypsinized the MDV-containing supernatant; MDV cargoes remain trypsin-protected, as they are contained within vesicles (Soubannier et al., 2012b). Immunoblot analysis of trypsinized supernatants revealed that Stx17 was extracted from the mitochondrial fraction, along with PDH E2 (an established MDV cargo [McLelland et al., 2014]), in a cytosol- and antimycin A–dependent manner (Fig. 1 C). VDAC1 and TIM23, which do not enrich with this MDV population in vitro (Soubannier et al., 2012b) or in cells (McLelland et al., 2014), respectively, were not present in these supernatants, as expected (Fig. 1 C). Although Stx17, likely present on the outer MDV membrane, was available to trypsin digestion, its proteolytic protection in our assay was not surprising, as SNAREs are often resistant to trypsin (Poirier et al., 1998; Lawrence and Dolly, 2002a,b).
Figure 1.
Stx17 is biochemically enriched on mitochondrial-derived vesicles (MDVs). (A) Flowchart of cell-free MDV budding assay (see Materials and methods for complete details). (B) Mouse liver was fractionated (see Materials and methods), and 40 µg heavy-membrane (HM), soluble (S), and light-membrane (LM) fractions were separated by SDS-PAGE and analyzed by immunoblotting. (C) Trypsinized supernatants and pellets from in vitro MDV budding assays incubated in the presence of cytosol and/or 50 µM antimycin A (anti A) were separated, along with the reaction pellets (mitochondria), by SDS-PAGE and analyzed by immunoblotting. The asterisk indicates a nonspecific band. (D) A large-scale budding reaction (incubated with cytosol and 50 µM antimycin A) was fractionated over a discontinuous sucrose gradient, separated by SDS-PAGE and analyze by immunoblotting. (E) Intensity profiles for the proteins probed for in D, expressed as a fraction of their maximum intensity.
To ensure that Stx17 was truly enriched on membranous structures, we fractionated the supernatant from the reaction containing both antimycin A and cytosol (not treated with trypsin) on a discontinuous sucrose gradient. We have previously shown that double-membrane MDVs (the population generated by antimycin A) enrich at the 30/40% sucrose interface (Soubannier et al., 2012b). As expected, immunoblotting our fractionated sample showed an accumulation of the Rieske subunit of complex III (CIII-Rieske) around the 30/40% interface, whereas the soluble protein Eps15 remained at the bottom of the gradient, where the reaction supernatant was initially loaded (Fig. 1 D, and E, blue and green lines for CIII-Rieske and Eps15, respectively). Although a portion of Stx17 also remained at the bottom of the gradient, most Stx17 cofractionated with the MDVs (Fig. 1, D and E, red line), corroborating our earlier results with trypsinized supernatants (Fig. 1 C). Thus, Stx17 is enriched on double-membrane MDVs generated in vitro.
Stx17 is recruited to nascent vesicles prior to scission
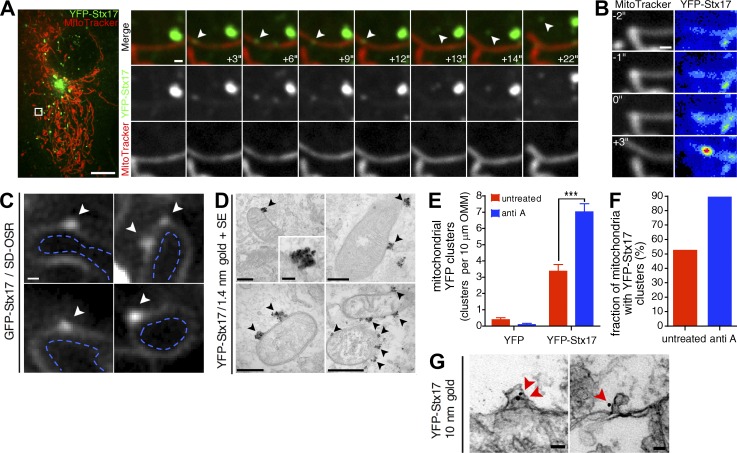
We next followed the dynamics of YFP-Stx17 by live-cell confocal microscopy in COS7 cells to determine the manner through which this SNARE was recruited to MDVs. We grew these cells in media containing galactose as a carbon source, as this increases steady-state MDV biogenesis through increased mitochondrial respiration (Soubannier et al., 2012a); unless explicitly stated, all cell-based experiments were performed under these bioenergetic conditions. In MitoTracker-labeled COS7 cells, steady-state formation and subsequent lateral disassociation of diffraction-limited YFP-Stx17 puncta on mitochondrial tubules were seen (Fig. 2 A, white arrowheads indicate an ∼0.3-µm-wide structure). We also observed much larger (>0.5 µm), stationary Stx17 structures adjacent to mitochondria (for example, the larger, brighter Stx17 structure in Fig. 2 A) that possibly corresponded to autophagosomes, as these structures form adjacent to mitochondria (Itakura et al., 2012; Hamasaki et al., 2013; Cheng et al., 2015). Focusing on the smaller Stx17 structures on mitochondria, we were able to observe a buildup of YFP-Stx17 signal (Fig. 2 B) on the mitochondrial tubule before lateral release (Fig. 2 A, rapid release occurs between frames +12 s and +13 s). We have previously observed this type of lateral release when studying the budding of MDVs off mitochondria (Neuspiel et al., 2008). Further analysis of GFP-Stx17 by superresolution microscopy (using the spinning disk Olympus superresolution [SD-OSR] system) revealed a weak, diffuse signal along the outer mitochondrial membrane (OMM), with clusters on the cytosolic side of the OMM that exceeded the resolution limit of this technique (Fig. 2 C, white arrowheads indicate GFP-Stx17 foci, whereas the dashed blue markings delineate the inside of mitochondria). We then sought to analyze these mitochondrial Stx17 foci, which were continually beneath the resolution limit of our light microscopy techniques. To this end, we labeled YFP-Stx17 with 1.4-nm gold particles in semipermeabilized cells and analyzed the samples by EM after developing the gold signal by silver enhancement (Fig. 2, D–F, SE). Similar to what we had observed by superresolution microscopy, we detected a strong enrichment of YFP-Stx17 within mitochondrial subdomains on the cytosolic face of the OMM (Fig. 2 D, black arrowheads), which increased in frequency with antimycin A treatment (Fig. 2, E and F), the trigger used to up-regulate the biogenesis of double-membrane MDVs. Importantly, we did not observe any clustering in cells transfected with YFP alone (Fig. 2 E), and these <100-nm-wide mitochondrial Stx17 foci differed morphologically from mature or preautophagosomal structures (Kishi-Itakura et al., 2014). We also observed, by immuno-EM using 10-nm gold particles (without enhancement, thus revealing membrane structure beneath the gold signal), YFP-Stx17 labeling on OMM deformations (Fig. 2 G, red arrowheads). Collectively, these confocal, superresolution and ultrastructural data indicated that Stx17 is recruited to mitochondrial subdomains during vesicle formation and remains associated with fully formed MDVs after scission, and these findings supported our biochemical studies.
Figure 2.
Dynamics and ultrastructure of mitochondrial Stx17 foci. (A) Live-cell observation of Stx17 vesicle formation and release in an untreated COS7 cell expressing YFP-Stx17 (green, 800 ms exposure time) and stained with MitoTracker Deep Red FM (red; 50-ms exposure time) by spinning disc confocal microscopy. Elapsed time is indicated in the bottom right of each merged image. The white arrowhead indicates a Stx17+ structure that forms on and subsequently dissociates from the tubule. Bars: (full image) 10 µm; (zoom-in) 0.5 µm. (B) Stx17 concentrates on the tubule before release. In the same time series from A, YFP-Stx17 signal (right) concentrates on the MitoTracker-labeled tubule (left). The YFP-Stx17 image is represented as a heatmap, and the brightness/contrast has been adjusted (from A) so that the fully formed structure is almost saturating and the concentration of signal can be clearly seen. Bar, 0.5 µm. (C) SD-OSR images of live cells transfected with GFP-Stx17. The Stx17 signal is located on the outer mitochondrial membrane, and white arrowheads indicate Stx17 foci on the OMM. Dashed blue lines delineate the mitochondrial interior. Bar, 200 nm. (D) Representative transmission electron micrographs of COS7 cells expressing YFP-Stx17, immunostained against YFP and labeled with 1.4 nm gold particles and silver enhanced (SE). Cells were treated with 25 µM antimycin A for 45 min before fixation. Black arrowheads indicate YFP-Stx17 foci on the cytosolic face of the mitochondrial outer membrane. Bars: (top left) 250 nm; (top left inset), 50 nm; (top right) 250 nm; (bottom left) 200 nm; (bottom right) 500 nm. (E) Quantification of the number of YFP-positive foci per distance of mitochondrial outer membrane, in antimycin A–treated and untreated cells expressing YFP and YFP-Stx17, in cells from D. Bars represent mean ± SEM; n = 80–112 mitochondria per condition; ***, P < 0.001. (F) Quantification of the proportion of mitochondria with YFP-Stx17 foci in cells from D. Bars represent the mean. (G) Representative transmission electron micrographs of COS7 cells expressing YFP-Stx17, immunostained against YFP and labeled with 10 nm gold particles. Red arrowheads indicate gold particles present on mitochondrial membrane deformations at the cytosolic face of the mitochondrial outer membrane. Bars, 50 nm.
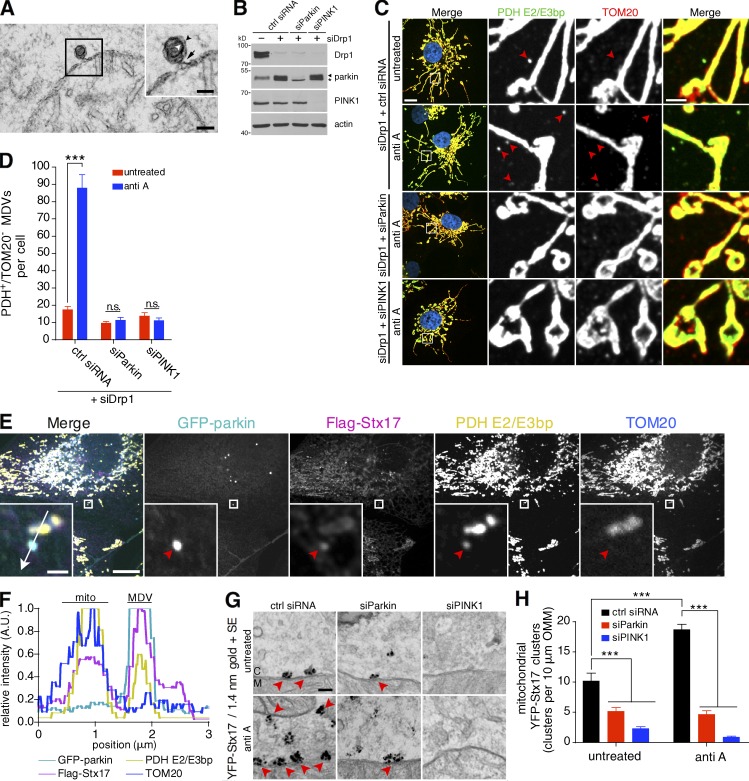
We next tested the link between these mitochondrial clusters of YFP-Stx17 and MDV biogenesis by parkin and PINK1. During biogenesis, nascent MDVs are visible under the electron microscope as 70- to 150-nm, double-membrane budding profiles (Neuspiel et al., 2008; Soubannier et al., 2012b) still attached to their parent mitochondrion via a constricted neck (Fig. 3 A). These vesicle necks are much smaller than the ∼100-nm-wide ring formed by assembled Drp1 oligomers during mitochondrial division (Rosenbloom et al., 2014), consistent with MDV formation being Drp1 independent (Neuspiel et al., 2008; Soubannier et al., 2012a; McLelland et al., 2014), and much more reminiscent of a vesicular budding process. We have previously shown that the activity of parkin and PINK1 is required to generate double-membrane MDVs in response to oxidative stress, which contain, among other cargoes, the mitochondrial matrix protein PDH E2/E3bp (detected by an antibody raised against homologous regions of the E2 and E3bp subunits, herein referred to as PDH), but lack the OMM marker TOM20 (Soubannier et al., 2012a; McLelland et al., 2014). Despite an absence of TOM20, matrix-containing MDVs have an outer membrane (Fig. 3 A). We first confirmed the involvement of endogenous parkin and PINK1 in PDH-positive/TOM20-negative MDV generation in COS7 cells by silencing both parkin and PINK1, as well as Drp1 (Fig. 3 B). In cells transfected with only siDrp1, we observed few cargo-selective puncta in the cytosol, which increased after antimycin A treatment (Fig. 3, C and D). As expected, additional silencing of parkin or PINK1 abolished MDV formation in these cells (Fig. 3, C and D). In U2OS cells stably overexpressing GFP-parkin (U2OS:GFP-parkin cells), we confirmed that Flag-Stx17 localized to PDH-positive/TOM20-negative MDVs, along with GFP-parkin (Fig. 3 E, red arrowhead), enriching on these vesicles compared with mitochondria (Fig. 3 F). Returning to ultrastructural studies, we repeated our 1.4-nm gold labeling and silver enhancement of YFP-Stx17 in cells in which parkin or PINK1 was silenced. Upon quantification of the number of clusters, we observed that YFP-Stx17 cluster formation on the OMM in the absence of parkin or PINK1 in both untreated cells and cells treated with antimycin A was greatly reduced (Fig. 3, G and H, red arrowheads). Thus, parkin and PINK1 are required for Stx17 clustering on the OMM.
Figure 3.
Stx17 localizes to parkin/PINK1-dependent MDVs. (A) Transmission electron micrograph of a COS7 cell treated with 25 µM antimycin A for 45 min before fixation. The inset (black box) shows a budding profile connected to the mitochondrial outer membrane. White and black arrowheads indicate the inner and outer vesicular membranes, respectively, and the black arrow indicates the vesicle neck. Bars: (full image) 100 nm; (inset) 50 nm. (B) COS7 cells were transfected with the indicated siRNA and 60 µg lysate from untreated cells was separated by SDS-PAGE and immunoblotted against the indicated protein. The asterisk indicates a nonspecific band. (C) Representative confocal images of antimycin A–treated cells immunostained for PDH E2/E3bp (green) and TOM20 (red; Hoescht, blue). Selective, PDH E2/E3bp+/TOM20− structures corresponding to MDVs are indicated by red arrowheads. Bars: (left) 10 µm; (right) 2 µm. (D) Quantification of the number of PDH E2/E3bp+/TOM20− MDVs per cell in cells from C treated with or without 25 µM antimycin A for 45 min. Bars represent mean ± SEM, n = 23–29 cells per condition. ***, P < 0.001; n.s., not significant. (E) Confocal image of a U2OS:GFP-parkin (cyan) cell, expressing Flag-Stx17 (magenta), treated with 25 µM antimycin A for 2 h, fixed and immunostained against PDH E2/E3bp (yellow) and TOM20 (blue). Red arrowheads in the insets indicate a PDH E2/E3bp+/TOM20− structure colocalizing with GFP-parkin and Flag-Stx17. Bars: (full image) 10 µm; (insets) 1 µm. (F) Intensity plot through the 3-µm line drawn in the merged inset in E. The MDV indicated in E, as well as a mitochondrion (mito), are indicated. A.U., arbitrary units. (G) Representative transmission electron micrographs of COS7 cells, transfected with control siRNA (ctrl siRNA) or siRNA targeting parkin (siParkin) or PINK1 (siPINK1), expressing YFP-Stx17, immunostained against YFP, and labeled with 1.4 nm gold particles and silver enhanced (SE). Cells were treated with 25 µM antimycin A for 45 min or left untreated before fixation. Red arrowheads indicate clusters of YFP-Stx17 signal. M, mitochondrion; C, cytosol. Bar, 100 nm. (H) Quantification of the number of YFP-positive foci per distance of mitochondrial outer membrane, in antimycin A–treated and untreated cells expressing YFP-Stx17, in cells from G. Bars represent mean ± SEM; n = 105–207 mitochondria per condition; ***, P < 0.001.
Stx17 mediates MDV turnover and pathway processivity
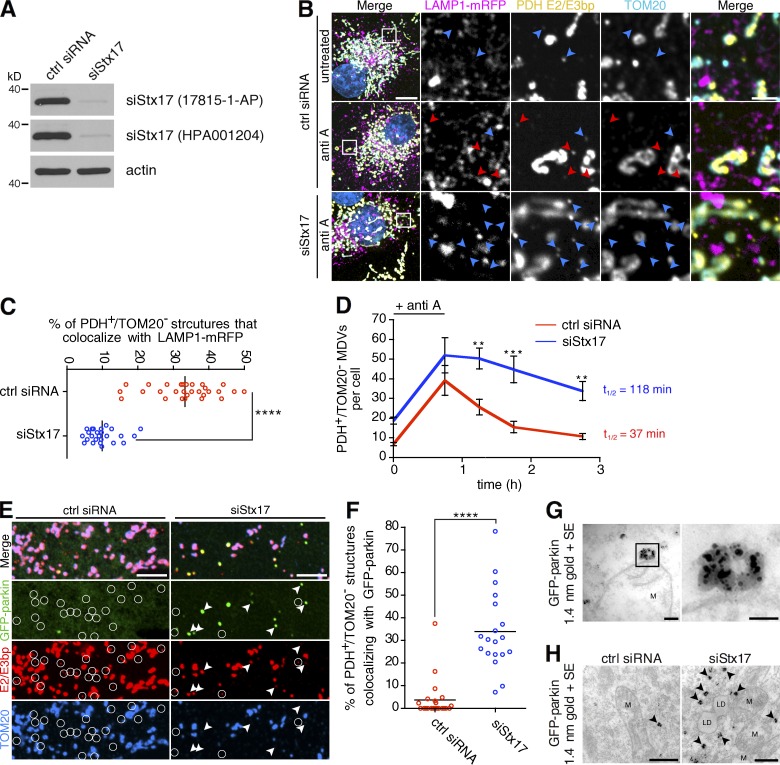
Given its previously known role in targeting starvation-induced autophagosomes to lysosomes, we next asked whether Stx17 played an analogous role in MDV trafficking. When Stx17 was silenced (siStx17) in COS7 cells (Fig. 4 A), colocalization of MDVs with LAMP1-mRFP–positive endolysosomes in antimycin A–treated cells was severely reduced in siStx17 versus control cells (Fig. 4, B and C). This suggested that loss of Stx17 may impact MDV turnover. We tested this by measuring MDV turnover kinetics in siStx17 and control cells, where cells pulsed with antimycin A were chased with media lacking the inhibitor. Here, we observed a defect in MDV degradation in cells where Stx17 was silenced (Fig. 4 D; with MDV half-lives of 37 and 118 min for control siRNA [red line] and siStx17 [blue line] conditions, respectively), supporting our lysosomal targeting data in demonstrating a role for Stx17 in MDVs turnover.
Figure 4.
Stx17 is required for the lysosomal turnover of parkin/PINK1-dependent MDVs. (A) Stx17 silencing was detected in COS7 cells by immunoblot using two different antibodies. (B) Representative confocal images of cells expressing LAMP1-mRFP (magenta) treated with antimycin A and fixed and immunostained for PDH E2/E3bp (yellow) and TOM20 (cyan; Hoescht, blue). PDH E2/E3bp+/TOM20− structures that are positive (red arrowheads) or negative (blue arrowheads) for LAMP1 are indicated. Bars: (main) 10 µm; (inset) 2 µm. (C) Quantification of the percent of PDH E2/E3bp+/TOM20− MDVs that colocalize with LAMP1-mRFP in the antimycin A–treated cells from B. Bars represent the mean; n = 29 and 25 cells for control siRNA and siStx17, respectively; ****, P < 0.0001. (D) Pulse-chase analysis of MDV turnover in siStx17 cells. Cells were pulsed with 25 µM antimycin A for 45 min in COS7 cells transfected with control siRNA (red line) or siRNA targeting Stx17 (blue line), then chased with media lacking drug for 0.5 to 2 h before fixation. The number of PDH E2/E3bp+/TOM20− structures were counted for the indicated times. Bars represent the mean ± SEM; n = 19–23 cells per condition; **, P < 0.01; ***, P < 0.001. MDV half-lives were calculated from fitted decay plots (outlined in Materials and methods; r2 = 0.98 for control siRNA and r2 = 0.91 for siStx17). (E) Stx17 silencing delays parkin dissociation from MDVs. U2OS:GFP-parkin cells were transfected with the indicated siRNA and treated with 25 µM antimycin A for 4 h, then fixed and immunostained for PDH E2/E3bp (red) and TOM20 (blue). PDH E2/E3bp+/TOM20− MDVs that are also positive (arrowheads) or negative (circles) for GFP-parkin (green) are indicated. Bars, 5 µm. (F) Quantification of the percentage of PDH E2/E3bp+/TOM20− MDVs that colocalize with GFP-parkin in cells from E. Bars represent the mean; n = 22 and 19 cells for control siRNA and siStx17, respectively; ****, P < 0.0001. (G) Transmission electron micrograph of antimycin A–treated U2OS:GFP-parkin cells, immunostained against GFP and labeled with 1.4 nm gold particles. The black box (left) magnifies a GFP-parkin cluster on the cytosolic face of the OMM (right). M, mitochondrion. Bars: (left) 100 nm; (right) 50 nm. (H) Transmission electron micrograph of antimycin A–treated U2OS:GFP-parkin cells transfected with control siRNA (left) or siRNA targeting Stx17 (siStx17, right), immunostained against GFP and labeled with 1.4 nm gold particles. Black arrowheads indicate cytosolic clusters of GFP-parkin. M, mitochondrion; LD, lipid droplet. Bars, 500 nm.
We previously suggested that parkin dissociates from matrix-containing MDVs during vesicle maturation (after scission but before fusion with the late endosome; McLelland et al., 2014); thus, a loss of Stx17-dependent MDV turnover may slow the rate of parkin dissociation from vesicles. Indeed, more parkin remained colocalized with MDVs in Stx17-silenced, antimycin A–treated U2OS:GFP-parkin cells compared with control (Fig. 4, E and F). When we immunogold-labeled GFP-parkin in antimycin A–treated, control siRNA–transfected cells, we observed few clusters of GFP-parkin, either on the cytosolic face of the OMM (Fig. 4 G, compare with YFP-Stx17 in Fig. 2 D) or in the cytosol (Fig. 4 H, left). In contrast, we observed many cytosolic GFP-parkin structures in siStx17 cells treated with antimycin A (Fig. 4 H, right). This confirmed that the accumulation of parkin seen by confocal microscopy was predominantly on ∼100-nm cytosolic structures and not on mitochondria.
Finally, these Stx17-mediated targeting defects were specific to this MDV subpopulation, as the targeting of two other classes of mitochondrial vesicles, TOM20-containing MDVs to lysosomes and MAPL-containing MDVs to peroxisomes, showed no dependence on Stx17 (Fig. S1, A and B, for TOM20 carriers; and Fig. S1, C and D, for MAPL-containing MDVs). This pathway specificity is in line with our YFP-Stx17 immunogold labeling experiment in parkin and PINK1 knockdown cells (Fig. 3, G and H), as neither of these other MDV pathways requires parkin or PINK1 (Neuspiel et al., 2008; McLelland et al., 2014). Thus, Stx17 is required for lysosomal targeting and turnover of stress-induced MDVs, and loss of Stx17-mediated fusion delays parkin dissociation from the fully formed vesicle.
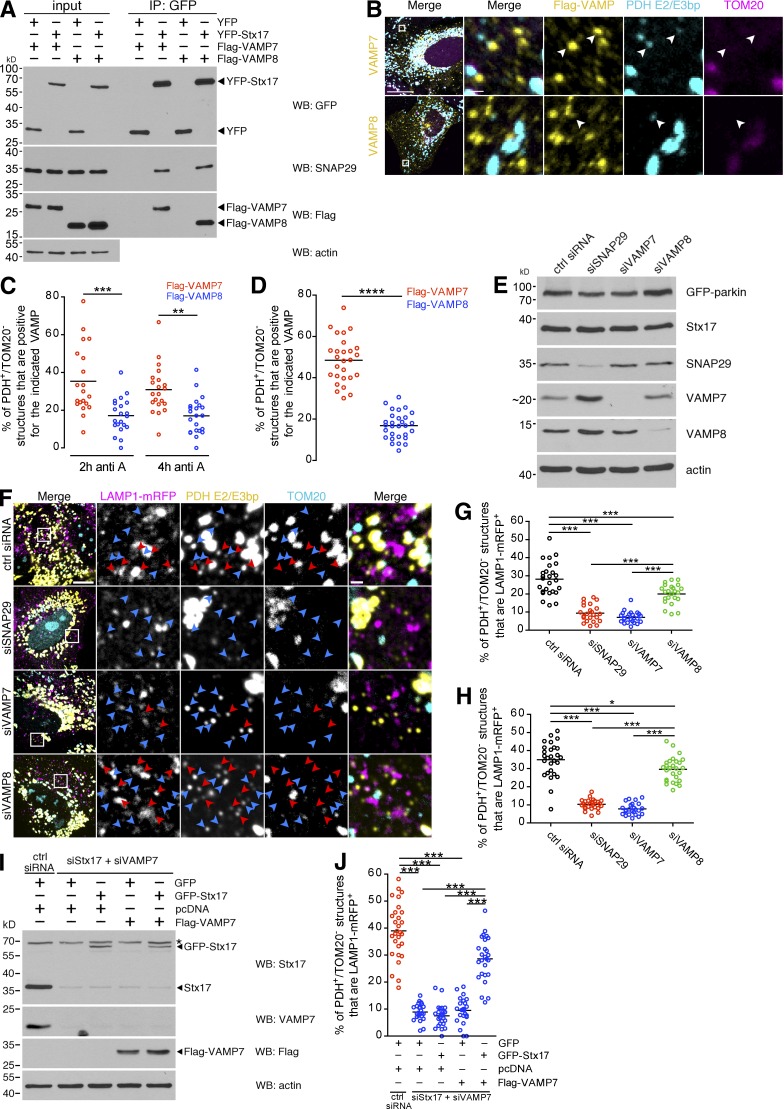
SNAP29 and VAMP7 target MDVs to endolysosomes
As the ultimate fate of parkin/PINK1-dependent MDVs is lysosomal turnover (Soubannier et al., 2012a; McLelland et al., 2014), we accordingly sought to identify the Stx17-containing SNARE complex that mediates fusion of MDVs with endolysosomes. Stx17 (a Qa-SNARE) has already been shown to complex with the Qbc-SNARE SNAP29 and the lysosomal R-SNARE VAMP8 in the context of starvation-induced autophagosome-lysosome fusion in mammals (Guo et al., 2014; Diao et al., 2015). Additionally, coimmunoprecipitation data have indicated that Stx17 also binds the R-SNARE VAMP7 in mammalian cells and in Drosophila (Itakura et al., 2012; Takáts et al., 2013). Both VAMP7 and VAMP8 are transmembrane proteins that localize to late endocytic and lysosomal compartments (Fig. S2). We confirmed this interaction data in COS7 cells, where Flag-VAMP7 and Flag-VAMP8, as well as endogenous SNAP29, coimmunoprecipitated with YFP-Stx17 (Fig. 5 A). Although PDH-positive/TOM20-negative structures colocalized with both VAMP7 and VAMP8, we observed higher colocalization with Flag-VAMP7 compared with Flag-VAMP8 in both antimycin A–treated U2OS:GFP-parkin (Fig. 5, B and C) and COS7 (Fig. 5 D) cells. To then determine whether these SNAREs, like Stx17, were involved in MDV delivery to endolysosomes, we silenced SNAP29, VAMP7, and VAMP8 in U2OS:GFP-parkin cells (Fig. 5 E). In these cells, loss of SNAP29 or VAMP7 almost completely abolished the lysosomal targeting of MDVs, with a less pronounced (but still significant) inhibition caused by VAMP8 silencing (Fig. 5, F and G), a pattern that was also observed in COS7 cells (Fig. 5 H). We reasoned that the minor targeting defect in the siVAMP8 condition may have resulted from a general impairment of lysosomal pathways, as silencing of the other SNAREs reduced endolysosomal targeting of MDVs to <10% and further supported the selectivity of this pathway for VAMP7 rather than VAMP8.
Figure 5.
The Stx17-associated SNAREs SNAP29 and VAMP7 are required for MDV turnover. (A) Coimmunoprecipitation of endogenous SNAP29 and either Flag-VAMP7 or Flag-VAMP8 with YFP-Stx17 in COS7 cells, immunoprecipitated using an anti-GFP antibody. Immunoprecipitates (IP) were separated, along with 8% input, by SDS-PAGE and immunoblotted for the indicated protein. (B) U2OS:GFP-parkin cells expressing either Flag-tagged VAMP7 or VAMP8 (yellow) were treated with 25 µM antimycin A for 2 h, then fixed and immunostained for PDH E2/E3bp (cyan) and TOM20 (magenta). Arrowheads indicate PDH E2/E3bp+/TOM20− structures that colocalize with the related VAMP. Bars: (main) 20 µm; (zoom) 1 µm. (C) Quantification of the percentage of PDH E2/E3bp+/TOM20− structures that colocalize with VAMP7 (red dots) or VAMP8 (blue dots) in U2OS cells from B treated for 2 and 4 h. Bars represent the mean; n = 20–22 cells; **, P < 0.01; ***, P < 0.001. (D) Quantification of the percent of PDH E2/E3bp+/TOM20− structures that colocalize with VAMP7 (red dots) or VAMP8 (blue dots) in COS7 cells treated with 25 µM antimycin A for 45 min. Bars represent the mean; n = 27 and 29 cells for VAMP7 and VAMP8, respectively; ****, P < 0.0001. (E) Stx17-associated SNAREs were silenced in U2OS:GFP-parkin cells, and silencing was confirmed by immunoblotting. (F) Representative confocal images of U2OS:GFP-parkin cells transfected with the indicated siRNA and LAMP1-mRFP (magenta) that were treated with 25 µM antimycin A for 2 h, then fixed and immunostained for PDH E2/E3bp (yellow) and TOM20 (cyan). PDH E2/E3bp+/TOM20− structures that are positive (red arrowheads) or negative (blue arrowheads) for LAMP1 are indicated. Bars: (main) 10 µm; (zoom) 1 µm. (G) Quantification of the percent of PDH E2/E3bp+/TOM20− structures that colocalize with LAMP1-mRFP in the U2OS cells from F. Bars represent the mean; n = 23–26 cells per condition; ***, P < 0.001. (H) Quantification of the percent of PDH E2/E3bp+/TOM20− structures that colocalize with LAMP1-mRFP in antimycin A–treated COS7 cells. Bars represent the mean; n = 24–27 cells per condition; *, P < 0.05; ***, P < 0.001. (I) GFP-Stx17 and Flag-VAMP7 were expressed in a 1:3 ratio, along with LAMP1-mRFP, in COS7 cells in which endogenous Stx17 and VAMP7 were silenced. GFP and pcDNA were used as control plasmids. Cells were lysed, and 20 µg cell lysate was separated by SDS-PAGE and immunoblotted for the indicated protein. The asterisk indicates a nonspecific band. Note that the antibody against endogenous VAMP7 does not recognize Flag-VAMP7, as this construct was created using cDNA from mouse. (J) Quantification of PDH E2/E3bp+/TOM20− structures that colocalize with LAMP1-mRFP in GFP-positive, antimycin A–treated cells from I. Bars represent the mean; n = 24–29 cells per condition; ***, P < 0.001.
To confirm that the targeting defect was not an off-target effect resulting from the siRNA, we reexpressed GFP-Stx17 and/or Flag-VAMP7 in COS7 cells in which both endogenous Stx17 and VAMP7, which induced the most severe defect, were silenced (Fig. 5 I). In these cells, only the expression of both GFP-Stx17 and Flag-VAMP7 partially rescued the targeting of MDVs to endolysosomes (Figs. 5 J and S3). Thus, Stx17, SNAP29, and VAMP7 are required for the targeting of MDVs to lysosomes, and Stx17 and VAMP7, integral membrane proteins on MDVs and endolysosomes, respectively, likely mediate fusion between their respective compartments.
Stx17, SNAP29, and VAMP7 form a SNARE complex that mediates MDV turnover
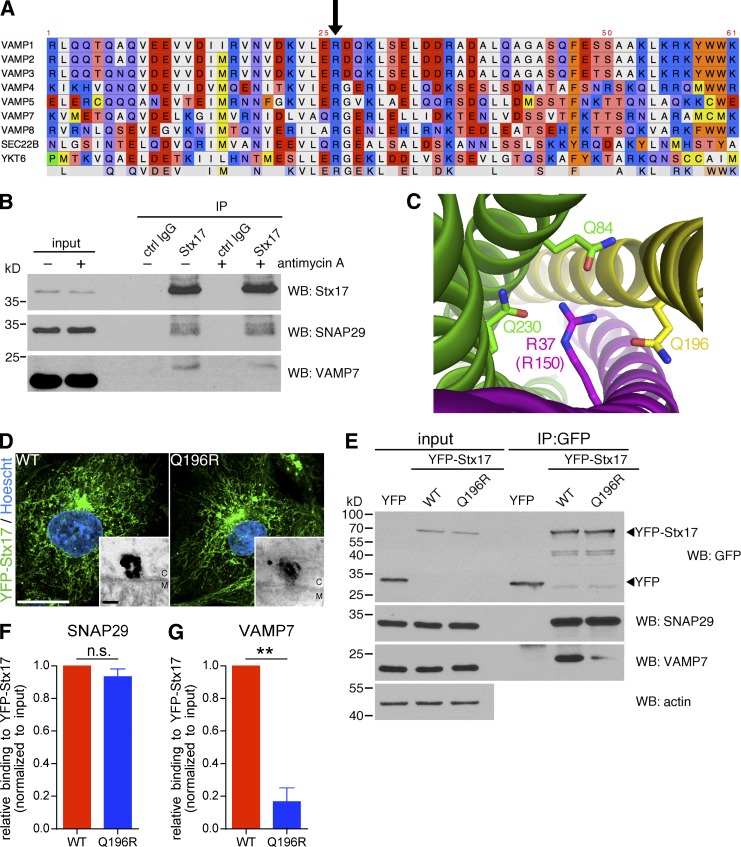
The simplest interpretation of our loss-of-function targeting experiments was that an Stx17–SNAP29–VAMP7 complex mediates fusion between MDVs and endolysosomes, analogous to the Stx17–SNAP29–VAMP8 complex involved in autophagy in mammals (Itakura et al., 2012); indeed, the SNARE domain is highly conserved across R-SNAREs (Fig. 6 A, arrow indicates the conserved arginine at the center of the domain). Interestingly, in Drosophila, an organism that lacks VAMP8, autophagosome–lysosome fusion is achieved via a Stx17–SNAP29–VAMP7 complex (Kloepper et al., 2008; Takáts et al., 2013). Although we were able to coimmunoprecipitate endogenous VAMP7 and SNAP29 with Stx17, the amounts of VAMP7 and SNAP29 binding Stx17 remained unchanged when cells were treated with antimycin A (Fig. 6 B). Thus, we turned to a structure-function approach to determine the role of an Stx17–SNAP29–VAMP7 complex in MDV turnover.
Figure 6.
Manipulation of the Stx17/VAMP7 zero layer affects complex formation and MDV turnover. (A) Primary sequence alignment of the SNARE domain of human R-SNARE proteins. The arrow indicates the conserved arginine that participates in the central ionic layer of the SNARE ternary complex. (B) Coimmunoprecipitation of endogenous SNAP29 and VAMP7 from untreated and antimycin A–treated COS7 cells with endogenous Stx17 immunoprecipitated with anti-Stx17 antibody. Anti-GFP is used as a control. Immunoprecipitates were separated, along with 5% input, by SDS-PAGE and immunoblotted for the indicated protein. Note that the VAMP7 band is shifted in the immunoprecipitation lanes because of increased glycerol content of the immunoprecipitated sample compared with the input, resulting in slower migration by SDS-PAGE. (C) Structure of the central ionic layer of the ternary SNARE complex formed by Stx17 (yellow), SNAP29 (green) and VAMP8 (magenta) solved in Diao et al. (2015) (PDB ID 4WY4). Participating residues are indicated. Note that arginine-37 in VAMP8 is equivalent to arginine-150 in VAMP7. (D) Wild-type (WT) or mutant (Q196R) YFP-Stx17 were expressed in COS7 cells, which were then fixed and prepared for immunofluorescence (YFP-Stx17, green; Hoescht, blue) or, in the case of the insets, immuno-EM. M, mitochondrion; C, cytosol. Bars: (main) 20 µm; (zoom) 50 nm. (E) Coimmunoprecipitation of endogenous SNAP29 and VAMP7 from COS7 cells expressing either an empty YFP vector, WT, or mutant (Q196R) YFP-Stx17, immunoprecipitated using an anti-GFP antibody. Immunoprecipitates were separated, along with 2.5% input, by SDS-PAGE and immunoblotted for the indicated protein. (F and G) Quantification of the amounts of SNAP29 (F) and VAMP7 (G) coimmunoprecipitated by YFP-Stx17 WT compared with Q196R, normalized to the input. Bars represent mean ± SEM; n = 3 experiments; **, P < 0.01; n.s., not significant.
To first test whether Stx17, SNAP29, and VAMP7 formed a ternary SNARE complex in vivo, we manipulated the zero layer (also known as the central ionic layer) of the potential complex (Fig. 6 C). To this end, we mutated glutamine-196 in Stx17 to arginine (Stx17Q196R) to disrupt the 3:1 ratio of glutamine to arginine at the zero layer (Ossig et al., 2000; Diao et al., 2015). In COS7 cells, we found that YFP-Stx17Q196R assembled on mitochondrial foci and localized to mitochondria in a similar manner to the wild-type (WT) construct (Fig. 6 D). Intriguingly, coimmunoprecipitation of YFP-Stx17 revealed that whereas both the WT and Q196R mutant could interact with SNAP29 (Fig. 6, E and F), the Q196R mutation drastically reduced the interaction of Stx17 with VAMP7 (Fig. 6 E, and G). Stx17 and SNAP29 have previously been shown to exist in a binary complex before ternary complex formation (Diao et al., 2015), analogous to the syntaxin-1–SNAP25 binary complex and VAMP2 on opposing membranes in the context of neuronal secretion (Rothman, 2014).
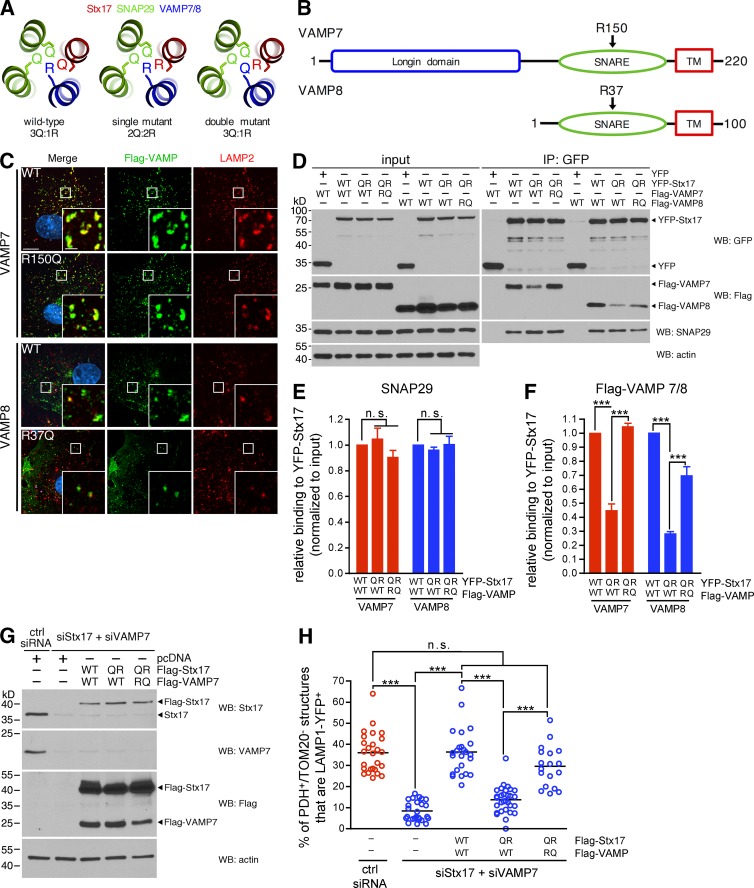
We next sough to rescue VAMP7 binding to Stx17 by restoring the 3Q:1R ratio at the zero layer (Fig. 7 A) and made the corresponding R-to-Q mutation in VAMP7 (R150Q) in parallel with VAMP8 (R37Q; Fig. 7 B), using this latter VAMP as a control for binding as it has already been shown to form a ternary SNARE complex with Stx17 and SNAP29 (Itakura et al., 2012; Diao et al., 2015). Both Flag-VAMP7R150Q and Flag-VAMP8R37Q localized to LAMP2-positive compartments, like their WT counterparts (Fig. 7 C). We then expressed combinations of zero-layer mutants of YFP-Stx17 and either Flag-VAMP7 or Flag-VAMP8 in COS7 cells. As before, the binding of SNAP29 to Stx17 was unaffected by mutation of the zero layer residues of Stx17, VAMP7, or VAMP8 (Fig. 7, D and E). Although both Flag-VAMP7WT and Flag-VAMP8WT robustly bound YFP-Stx17WT, binding to YFP-Stx17Q196R was again significantly reduced (Fig. 7, D and F). Strikingly, correcting the zero-layer ratio back to 3Q:1R from 2Q:2R with Flag-VAMP7R150Q and Flag-VAMP8R37Q rescued YFP-Stx17Q196R binding (Fig. 7, D and F). Thus, the binding of Stx17 to cognate R-SNAREs is dependent on a proper 3Q:1R ratio at the zero layer of the four-helix bundle and indicates that Stx17, SNAP29, and VAMP7 form a ternary SNARE complex in cells.
Figure 7.
SNARE complex formation and MDV turnover require a proper zero layer ratio. (A) Organization and mutation of zero layer residues in a ternary complex composed of Stx17 (red), SNAP29 (green), and VAMP7/8 (blue), based on the structure solved by Diao et al. (Diao et al., 2015; PDB ID 4WY4). (B) Domain structure of VAMP7 and VAMP8. The arrows indicate the conserved zero layer residue in the SNARE domains. TM, transmembrane domain. (C) Flag-tagged VAMP7 and VAMP8 constructs were expressed in COS7 cells, then fixed and immunostained against Flag (green) and LAMP2 (red). Bars: (main) 10 µm; (inset) 2 µm. (D) Coimmunoprecipitation of endogenous SNAP29 and Flag-tagged VAMP7 or VAMP8 zero layer mutants with YFP, YFP-Stx17 WT, or Q196R in COS7 cells, using an anti-GFP antibody. Immunoprecipitates were separated, along with 8% input, by SDS-PAGE, and immunoblotted for the indicated protein. (E and F) Quantification of the amounts of SNAP29 (E) and Flag-tagged VAMP7 and VAMP8 (F) coimmunoprecipitated by YFP-Stx17 WT compared with Q196R, normalized to the input. Bars represent mean ± SEM; n = 3 experiments; ***, P < 0.001; n.s., not significant. (G) Flag-Stx17 (WT or Q196R), Flag-VAMP7 (WT or R150Q), and LAMP1-YFP were expressed in a 3:3:1 ratio in COS7 cells in which endogenous Stx17 and VAMP7 were silenced (pcDNA3 was used as a control for the Flag vectors). Cells were lysed, and 20 µg cell lysate was separated by SDS-PAGE and immunoblotted for the indicated protein. (H) Quantification of PDH E2/E3bp+/TOM20− structures that colocalize with LAMP1-YFP in antimycin A–treated COS7 cells transfected with constructs from G. Bars represent the mean; n = 18–30 cells per condition; ***, P < 0.001; n.s., not significant.
To then determine the effect of zero layer manipulation on MDV trafficking, we reconstituted the Stx17-VAMP7 interaction in COS7 cells. We again silenced both Stx17 and VAMP7, and then reexpressed combinations of Flag-Stx17 and Flag-VAMP7 mutants (Fig. 7 G). When we looked at MDV targeting in these cells, we found that Flag-Stx17Q196R disrupted colocalization of MDVs with LAMP1-YFP compared with WT in cells coexpressing Flag-VAMP7WT and that this, like binding, was rescued by expression of Flag-VAMP7R150Q (Figs. 7 H and S4). Collectively, these data demonstrate the existence of a ternary Stx17–SNAP29–VAMP7 SNARE complex that mediates the fusion of MDVs with endolysosomes.
MDV and mitophagosome turnover use different sets of SNAREs but both require the HOPS tethering complex
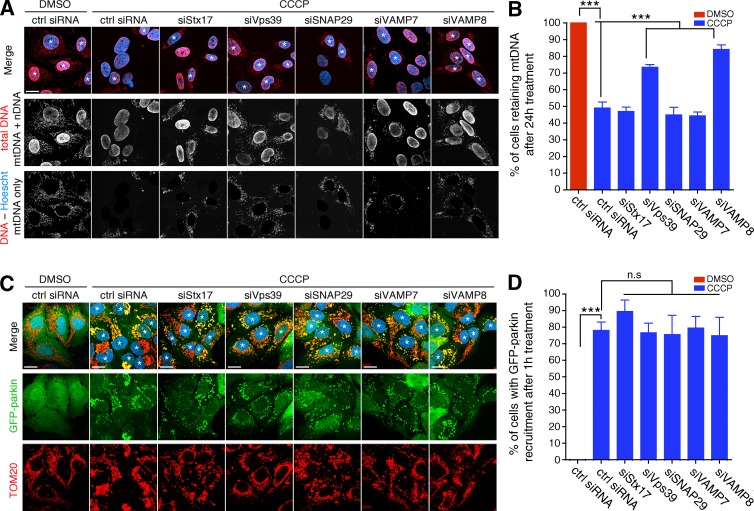
Stx17 is involved in the fusion of starvation-induced autophagosomes with lysosomes, and it has been suggested that it is the SNARE responsible for mitophagosome–lysosome fusion during parkin/PINK1-mediated mitophagy (Yoshii and Mizushima, 2015). Given our current data implicating Stx17 in MDV–endolysosome fusion, we thus sought to identify a role for this molecule in depolarization-induced mitophagy, reasoning that Stx17 may be common to both pathways. To this end, we silenced Stx17 and its associated SNAREs in U2OS:GFP-parkin cells that were grown in glucose medium (a metabolic condition under which mitophagy is permitted; Van Laar et al., 2011; McCoy et al., 2014; Lee et al., 2015) to determine which relevant membrane fusion-related genes would impair mitophagy. We additionally silenced Vps39, a component of the HOPS (homotypic fusion and vacuole protein sorting) tethering complex that regulates autophagosome–lysosome fusion and other late endosomal tethering events (Balderhaar and Ungermann, 2013; Jiang et al., 2014; Takáts et al., 2014), to serve as a positive control. After these cells were depolarized for 24 h, to our surprise, only cells in which either Vps39 or VAMP8 was silenced displayed a reduced rate of mitophagy (as visualized by a loss of either mitochondrial DNA or TOM20), with loss of any of Stx17, SNAP29, or VAMP7 having a negligible effect (Fig. 8, A and B, for mitochondrial DNA; and Fig. S5 for TOM20). To ensure that the effects seen by silencing Vps39 and VAMP8 were occurring upstream of mitophagosome turnover, we quantified the recruitment of GFP-parkin after 1 h of CCCP (a time frame in which delays in recruitment are visible [Durcan et al., 2014]) and saw no significant delay in any condition (Fig. 8, C and D).
Figure 8.
Stx17 is dispensable for depolarization-induced mitophagy. (A) Representative confocal images of U2OS:GFP-parkin (not depicted) cells (grown on glucose) treated with 20 µM CCCP or DMSO for 24 h. Mitophagy was monitored by then fixing and immunostaining for DNA (red) and counterstaining with Hoescht 33342 (blue). Cells that have retained their mitochondria are marked by asterisks. To make the presence of mitochondrial DNA (mtDNA) clearer to the reader, the Hoescht image (pertaining to nuclear DNA [nDNA]) was thresholded and subtracted from the total DNA staining to yield an image of solely mtDNA (bottom row). Bar, 20 µm. (B) Quantification of the percent of cells retaining mitochondria from cells in A. Bars represent mean ± SEM; n = 3 replicates cells per condition, with >100 cells counted per condition for each replicate. ***, P < 0.001. (C) Representative confocal images of U2OS:GFP-parkin (cells, grown on glucose, were treated with 20 µM CCCP or DMSO for 1 h to monitor GFP-parkin [green] recruitment [cells marked with asterisks] to mitochondria [TOM20, red; Hoescht, blue]). Bars, 20 µm. (D) Quantification of the percent of cells display parkin recruitment to mitochondria in cells from C. Bars represent mean ± SEM; n = 3 replicates cells per condition, with >100 cells counted per condition for each replicate; ***, P < 0.001; n.s., not significant.
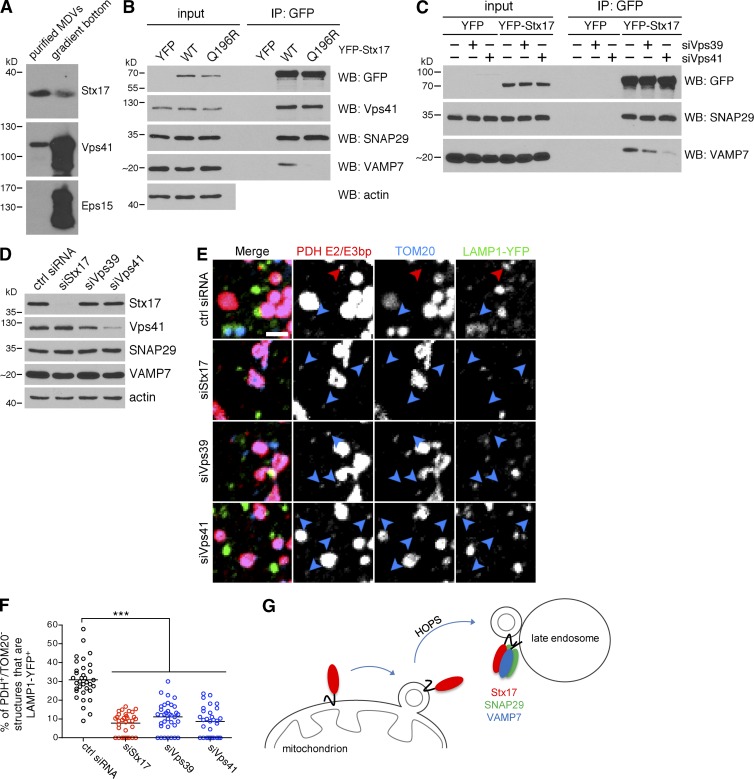
We next tested a role for the HOPS complex in MDV turnover. Similarly to Stx17, the HOPS-specific Vps41 subunit copurified with MDVs generated in cell-free reconstitution assays (Fig. 9 A), and we confirmed the association of Stx17 with the HOPS complex in cells by coimmunoprecipitating Vps41 with Stx17 (Fig. 9 B; Jiang et al., 2014; Takáts et al., 2014). Interestingly, we saw no difference in Vps41 binding between WT Stx17 and the Q196R zero layer mutant (which cannot bind cognate R-SNAREs; Fig. 6, E and G). Collectively, these data indicated that Stx17 can recruit the HOPS complex to MDVs before binding VAMP7 on the late endosome. Indeed, silencing either Vps39 or Vps41 abrogated the binding of YFP-Stx17 to VAMP7 (Fig. 9 C), as would be expected if the complex was acting as a tether. When we silenced the Vps39 and Vps41 HOPS subunits in COS7 cells (Fig. 9 D), we observed an MDV targeting defect similar to that obtained with Stx17 depletion (Fig. 9, E and F). In summary, the HOPS complex binds Stx17 on MDVs and is required for their efficient targeting to endolysosomes, likely through a conserved role in coordinating interactions at the zero layer (Starai et al., 2008; Baker et al., 2015) between Stx17 and VAMP7. Additionally, although mitophagosomes generated by depolarizing insults are degraded in a HOPS-dependent manner, this occurs largely independently of Stx17, SNAP29, and VAMP7.
Figure 9.
The HOPS complex acts as a tether during MDV turnover. (A) The HOPS subunit Vps41 is present on MDVs generated in vitro. Equal volumes of purified MDVs (equivalent to fraction 16 in Fig. 1 D) generated in a cell-free assay as in Fig. 1 D (with cytosol and 50 µM antimycin A), as well as material present at the bottom of the gradient (equivalent to fraction 22 in Fig. 1 D) were separated by SDS-PAGE and immunoblotted for the indicated protein. Eps15 is included as a control for soluble proteins. (B) Coimmunoprecipitation of endogenous Vps41 from COS7 cells expressing either an empty YFP vector (YFP; note that the blot is cropped above the ∼30-kD band corresponding to YFP), wild-type (WT), or mutant (Q196R) YFP-Stx17, immunoprecipitated using an anti-GFP antibody. Immunoprecipitates were separated, along with 2.5% input, by SDS-PAGE and immunoblotted for the indicated protein. (C) Coimmunoprecipitation of endogenous SNAP29 and VAMP7 from control, Vps39-, or Vps41-depleted COS7 cells expressing YFP or YFP-Stx17, immunoprecipitated using an anti-GFP antibody. Immunoprecipitates were separated, along with 2.5% input, by SDS-PAGE and immunoblotted for the indicated protein. (D) Immunoblot analysis of COS7 cells transfected with the indicated siRNA. (E) Representative confocal images of COS7 cells transfected with the indicated siRNA and LAMP1-YFP (green) that were treated with 25 µM antimycin A for 45 min, then fixed and immunostained for PDH E2/E3bp (red) and TOM20 (blue). PDH E2/E3bp+/TOM20− structures that are positive (red arrowheads) or negative (blue arrowheads) for LAMP1 are indicated. Bar, 2 µm. (F) Quantification of the percent of PDH E2/E3bp+/TOM20− structures that colocalize with LAMP1-YFP in the cells from E. Bars represent the mean; n = 30–35 cells per condition; ***, P < 0.001. (G) Model of MDV–late endosome fusion mediated by Stx17. Stx17 is recruited to MDVs during budding, likely mobilized from a diffuse pool present on the OMM. In a HOPS-dependent manner, Stx17 forms a ternary SNARE complex with SNAP29 and VAMP7 at the late endosome to mediate fusion.
Discussion
In this study, we have demonstrated the molecular principles that govern MDV transport to the lysosome, showing a requirement for evolutionarily conserved SNARE machinery in this process. The Qa-SNARE Stx17 localizes to the mitochondrial outer membrane, building up at foci that are then released as vesicles, and remains present on MDVs that have dissociated from the mitochondrial tubule. Stx17 also promotes the turnover of mitochondrial vesicles, and MDV–endolysosomal fusion is achieved via an Stx17–SNAP29–VAMP7 ternary SNARE complex. Endolysosomal targeting is regulated via Stx17–VAMP7 binding, as revealed by mutations that disrupt this interaction. Although the Stx17–SNAP29–VAMP7 ternary SNARE complex identified in this study is not involved in parkin-mediated mitophagy, both processes use the HOPS tethering complex for turnover (Fig. 9 G).
Manipulation of residues at the zero layer of the helical bundle identified SNARE machinery involved in the turnover of MDVs. The physiological role of residues at this position remains contentious; an all-Q zero layer can still support vacuolar fusion and exocytosis in yeast (Katz and Brennwald, 2000; Ossig et al., 2000; Fratti et al., 2007), whereas other exocytic studies have provided little coherence as to the function of disrupting the 3Q:1R ratio (Graham et al., 2001; Gil et al., 2002; Lauer et al., 2006; Watanabe et al., 2013). Here, we show that zero layer mutations in Stx17 prevent fusion of MDVs with endolysosomes through a reduction in Stx17–VAMP7 binding, and we are the first to demonstrate, to our knowledge, that defects resulting from an aberrant zero layer ratio can be functionally rescued via compensatory mutation in a mammalian system.
Accompanying our findings that SNAREs are required for heterotypic fusion between the MDV outer membrane (which is derived from the OMM) and the endolysosomal limiting membrane is the notion that this machinery is distinct from the mitochondrion’s own fuseogenic GTPases involved in homotypic fusion of the organelle (Scorrano, 2013). Indeed, although MDV transport to the lysosome is directional and destructive, the major role of mitochondrial fusion is the mixing of mitochondrial contents (Mishra and Chan, 2016), and thus, coevolution of distinct membrane fusion machineries would allow for segregated regulation of these two processes. Drawing this ancestral parallel further, the evolutionary origin of Stx17 is, like the mitochondrion, also primordial, traceable to the so-called last eukaryotic common ancestor (Koumandou et al., 2013; Arasaki et al., 2015). Moreover, Stx17 is a phylogenic “outlier” compared with its most immediate family members (Stx7, Stx13, and Stx20), all of which are much more closely interrelated (Kloepper et al., 2008), indicative of an early evolutionary origin. Together, these observations highlight the fundamental concept that, in a similar manner to other conserved trafficking pathways, vesicular trafficking from mitochondria to the lysosome may be an ancient process (Sugiura et al., 2014). Indeed, MDV release may have its origins in the ancestor of the mitochondrion itself; that is, vesicle shedding by α-proteobacteria (Deatherage and Cookson, 2012). Thus, the coevolution of mitochondria and MDV transport to the lysosome (or vacuole) remains an intriguing area of future work.
The precise mechanism by which Stx17 is recruited into clusters on the OMM is another further area of study. We provide evidence that Stx17 likely assembles at locations of parkin/PINK1-dependent MDV formation, implicating Stx17 specificity for this MDV subpopulation versus others. Although a direct interaction between Stx17 with parkin or PINK1 is unlikely, an unbiased screen for interactors of S65-phosphorylated ubiquitin (the major PINK1 enzymatic product at depolarized mitochondria; Kane et al., 2014; Kazlauskaite et al., 2014; Koyano et al., 2014) revealed SNARE-family proteins as a top hit of preferential binding partners in yeast (Swaney et al., 2015). Thus, phosphoubiquitin itself may recruit Stx17 to sites of vesicle formation. Alternatively, Stx17 may be recruited to sites of MDV formation indirectly or in parallel with other factors. For example, Stx17 may use its two transmembrane domains to detect OMM curvature (a more passive version of how atlastin, another hairpin transmembrane protein, can itself bend ER membranes; Hu et al., 2009; Daumke and Praefcke, 2011; Byrnes et al., 2013) and thus preferentially collect at sites of membrane deformation.
In the current study, the identification of a canonical SNARE pairing mechanism driving the process of MDV–endolysosomal fusion confirms that this transport route employs established, cellular trafficking machinery and highlights the integration of mitochondrion-to-lysosome vesicular transport within the cellular milieu. Additionally, in studying the mechanism of MDV delivery to lysosomes and contrasting this to mitophagy, we have further distinguished MDV transport from parkin/PINK1-dependent mitophagy, highlighting the divergence of their downstream targeting mechanisms. Studying MDV biology at the molecular level will help us understand how disruption of this mitochondrial quality control mechanism may lead to neuronal degeneration.
Materials and methods
Antibodies and other reagents
Antibodies used in this study include antiactin (MAB1501; EMD Millipore), anti–α-adaptin (sc-17771; Santa Cruz Biotechnology, Inc.), anti-CD63 (556019; BD), anti-DNA (61014; Progen), anti-Drp1 (611113; BD), anti-Eps15 (sc-534; Santa Cruz Biotechnology, Inc.), anti-Flag (anti-DDDDK, ab1257; Abcam), anti-Flag (F1804; Sigma-Aldrich), anti-GFP (ab6673; Abcam), anti-GFP (A6455; Invitrogen), anti-LAMP2 (sc-18822; Santa Cruz Biotechnology, Inc.), anti-LBPA (Z-SLBPA; Eschelon), anti-parkin (sc-32282; Santa Cruz Biotechnology, Inc.), anti-PDH E2/E3bp (ab110333; Abcam), anti-PINK1 (6946; Cell Signaling Technology), anti-PMP70 (SAB4200181; Sigma-Aldrich), anti-SDHA (ab14715; Abcam), anti-SNAP29 (ab138500; Abcam), anti-Stx17 (17815–1-AP; ProteinTech), anti-Stx17 (HPA001204; Sigma-Aldrich), anti-TIM23 (611222; BD), anti-TIP47 (Novus Biologicals), anti-TOM20 (sc-11414; Santa Cruz Biotechnology, Inc.), anti-UQCRFS1 (referred to herein as CIII-Rieske, ab14746; Abcam), anti-VAMP7 (sc-166394; Santa Cruz Biotechnology, Inc.), anti-VAMP8 (ab76021; Abcam), anti-VDAC1 (ab14734; Abcam), and anti-Vps41 (ab181078; Abcam). Unless otherwise specified, all reagents were purchased from Sigma-Aldrich. The TIP47 antibody was a gift from P. McPherson (McGill University, Montreal, Quebec, Canada).
Plasmids, subcloning, and mutagenesis
Flag-Stx17 (#45911), Flag-VAMP8 (#45912), Flag-VAMP7 (#45913), and LAMP1-mRFP (#1817) were purchased from Addgene and are described elsewhere (Sherer et al., 2003; Itakura et al., 2012). MAPL-YFP has also been previously described (Neuspiel et al., 2008). YFP-Stx17 was constructed by first performing sequential digestions of Flag-Stx17 with XbaI and then EcoRI (New England Biolabs, Inc.). YFP-C1 (mVenus-C1, plasmid 27794; Addgene) was cut in the same manner but dephosphorylated using SAP (Affymetrix) before separation on the gel. As the Stx17 cDNA contains an internal EcoRI site, XbaI-cut Flag-Stx17 was partially digested with EcoRI to yield a ∼950-bp band (representing the full cDNA) that was excised and purified. Purified components were ligated using Rapid DNA Ligation kit (Thermo Fisher Scientific) according to the manufacturer’s instructions, and positive clones were identified by sequencing. GFP-Stx17 was constructed by double-digesting YFP-Stx17 and pEGFP-C1 (Takara Bio Inc.) with XhoI and XbaI, followed by a ligation as above. Flag- and YFP-tagged Stx17Q196R were generated using a QuikChange II site-directed mutagenesis kit (Agilent Technologies) by mutagenizing Stx17WT with the primer pair of 5′-CTCCTAGTGAATTCTCGGCAGGAGAAGATTGAC-3′ and 5′-GTCAATCTTCTCCTGCCGAGAATTCACTAGGAG-3′ according to the manufacturer’s instructions. Flag-VAMP7R150Q and Flag-VAMP8R37Q were generated by the same method, using the primer pairs of 5′-CATAGATTTAGTTGCTCAACAGGGAGAAAGGTTGGAATTGC-3′/5′-GCAATTCCAACCTTTCTCCCTGTTGAGCAACTAAATCTATG-3′ and 5′-GGATCCTGGCCCAGGGGGAAAACTTG-3′/5′-CAAGTTTTCCCCCTGGGCCAGGATCC-3′ for Flag-VAMP7 and Flag-VAMP8, respectively. LAMP1-YFP was constructed by double-digesting LAMP1-mRFP with BamHI and EcoRI and ligating the 1,200-bp insert into YFP-N1 (mVenus-N1, plasmid 27793; Addgene) that had been cut in the same manner.
RNA interference
siRNA oligonucleotides targeting Drp1, parkin, and PINK1 have been described previously (McLelland et al., 2014). Nontargeting siRNA, as well as siRNA targeting Stx17 (5′-GGAAACCUUAGAAGCGGACUUAAUUdTdT-3′ Jiang et al., 2014), SNAP29 (5′-AGACAGAAAUUGAGGAGCAdTdT-3′; Guo et al., 2014), VAMP8 (5′-GCAACAAGACAGAGGAUCUdTdT-3′; Guo et al., 2014), Vps39 (5′-GGUAAAGAAGCUGAAUGACUCUGAUdTdT-3′; Jiang et al., 2014), and Vps41 (5′-GAGAAUGAAUGUAGAGAUUdTdT-3′; McEwan et al., 2015) were purchased from Thermo Fisher Scientific. siRNA targeting VAMP7 (ON-TARGETplus J-020864-05) was purchased from GE Healthcare.
Cell culture and transfection
COS7 cells were purchased from ATCC, and the U2OS:GFP and U2OS:GFP-parkin stable cell lines have been described previously (McLelland et al., 2014). Cells were maintained in DMEM supplemented with l-glutamine, penicillin/streptomycin, and 10% FBS in the presence of 10 mM galactose (Wisent). Alternatively, for the experiments depicted in Fig. 8, 25 mM glucose was used as a carbon source. Cells were typically transfected with 40 nM siRNA, or 0.5 μg/ml DNA for experiments involving a single construct, via lipofection using jetPRIME transfection reagent (Polyplus Transfection) according to the manufacturer’s instructions. Cells were treated and analyzed 3 d after transfection for siRNA and 1 d after transfection for DNA transfections. In the case of siRNA/DNA double-transfections, cells were first transfected in six-well plates with siRNA, then replated the next day in 24-well plates with coverslips (for imaging) or six-well plates (for immunoblotting). The next day, cells were transfected with the indicated DNA construct, and analysis was performed the next day. In the case of rescue experiments (Fig. 5, I and J; Fig. S3; Fig. 7, G and H; and Fig. S4), cells were transfected initially with 40 nM of each siRNA oligonucleotide. For Fig. 5 (I and J) and Fig. S3, cells were then transfected with 0.5 µg/ml LAMP1-mRFP, 0.2 µg/ml GFP/GFP-Stx17, and 0.6 pcDNA/Flag-VAMP7 (transfection was monitored by GFP fluorescence). For Fig. 7 (G and H) and Fig. S4, cells were transfected with 0.5 µg/ml LAMP1-YFP, 0.6 µg/ml pcDNA/Flag-Stx17, and 0.6 µg/ml pcDNA/Flag-VAMP7; transfection was monitored by YFP fluorescence.
Immunofluorescence, fluorescence microscopy, and microscope image acquisition
Cells on coverslips were fixed in 6% formaldehyde, washed with PBS, and permeabilized in PBS containing 0.25% Triton X-100. Coverslips were then blocked in 10% FBS in PBS. Primary antibodies were diluted in 5% FBS/PBS, and incubations were performed for 1 h at room temperature. Coverslips were then washed three times in 5% FBS/PBS, then incubated with Alexa Fluor–conjugated secondary antibodies (Thermo Fisher Scientific), diluted in 5% FBS/PBS, for 1 h at room temperature. Cells were then washed three times in PBS and mounted on glass slides using Aqua Poly/Mount (Polysciences Inc.). Cells were often counterstained with Hoescht 33342 before mounting. Confocal slices (<1 µm-thick) were acquired via a laser scanning confocal microscope (LSM710; ZEISS) through a 63× 1.4 NA or 40× 1.3 NA objective lens or a spinning disc microscope (with Andor Yokogawa system; IX81; Olympus) through a 100× 1.4 NA or 60× 1.4 NA objective lens using excitation wavelengths of 405, 488, 543, and 633 nm or 405, 488, 515, 561, and 640 nm for the laser scanning and spinning disc microscopes, respectively. Live-cell imaging was performed in a chamber heated to 37°C at 5% CO2, with images acquired every second at 800 ms (YFP) and 50 ms (MitoTracker Deep Red FM; Invitrogen) exposure times. Cells were loaded with 225 nM MitoTracker Deep Red FM for 30 min, then rinsed three times with media without dye, before imaging. Super-resolution was performed on an equivalent spinning disc set up and was achieved using the SD-OSR system (Olympus), and images were acquired using 2,000-ms exposure times. Image files were analyzed using ImageJ (National Institutes of Health). Although vesicles were counted by eye, peroxisomes were counted using the particle-counting feature of ImageJ.
Immunoprecipitation and immunoblotting
Cells were rinsed in ice-cold PBS and lysed in lysis buffer (20 mM Hepes, pH 7.2, 1% NP-40 substitute, and 150 mM NaCl supplemented with the protease inhibitors benzamidine, PMSF, aprotinin, and leupeptin) on ice. Lysates were clarified by centrifugation and protein concentration was determined by BCA assay (Thermo Fisher Scientific). Lysates were diluted to 1 to 2 mg/ml and incubated with the indicated primary antibody overnight at 4°C. The next day, samples were incubated with protein G–Sepharose (GE Healthcare) for 4 h. Immunoprecipitates were washed five times in lysis buffer and eluted by incubating at 90°C. Samples were separated by SDS-PAGE and transferred to nitrocellulose membrane. Membranes were blocked in 5% milk in PBS plus 0.1% Tween-20 (PBST), and primary antibodies were diluted in blocking buffer or 3% BSA/PBST. Primary antibody incubations were performed overnight at 4°C. The following day, membranes were washed with PBST, blocked again as before, and incubated with HRP-conjugated secondary antibodies, diluted in 5% milk/PBST, for one hour at room temperature. Membranes were then washed with PBST before protein detection with Western Lightning ECL and Plus-ECL kits (PerkinElmer) according to the manufacturer’s instructions.
Isolation of mitochondria and reconstitution of MDV formation in vitro
Mitochondrial isolation from mouse liver was adapted from a protocol by Frezza et al. (2007). Tissue and fractions were kept on ice at all times, and centrifugation was done at 4°C using precooled rotors. Six to eight week-old mice were euthanized with CO2, and the liver was excised and cut into small pieces before homogenization (four passes at 1,600 rpm) in cold isolation buffer (20 mM Hepes, pH 7.4, 220 mM mannitol, 68 mM sucrose, 76 mM KCl, 4 mM KOAc, and 2 mM MgCl2, supplemented with protease inhibitors benzamidine, PMSF, aprotinin, and leupeptin). The postnuclear supernatant was collected by centrifuging the homogenate at 600 g for 10 min in a centrifuge (J-25I; Avanti) using a fixed-angle rotor (SLA-25.50; Beckman Coulter). This supernatant was collected and centrifuged again at 7,000 g for 10 min. The pellet (HM fraction) was resuspended in a large volume of isolation buffer and centrifuged again, and then stored in isolation buffer on ice. The 7,000-g supernatant was then centrifuged at 200,000 g for 90 min in an ultracentrifuge (L-90K; Optima) using a 70-Ti rotor. The resulting supernatant (S cytosolic fraction) was stored on ice, and the pellet (LM fraction) was resuspended in isolation buffer with 2% Triton X-100. Protein concentrations were determined by BCA assay. MDV formation was reconstituted according to Soubannier et al. (2012b). The HM fraction was first washed twice in isolation buffer without protease inhibitors (10,000 rpm for 10 min in a microfuge at 4°C). The S fraction was added to the HM at a 1:4 ratio (by protein weight) in the presence of an ATP regenerating mixture (1 mM ATP, 5 mM succinate, 80 µM ADP, and 2 mM K2HPO4, pH 7.4) and 50 µM antimycin A, topped up with isolation buffer. For small-scale reactions, the total reaction volume was 250 µl (HM, 6 mg/ml; S, 1.5 mg/ml). These were incubated at 37°C for 2 h (vortexing every 30 to 45 min) and then centrifuged at 10,000 rpm for 10 min in a microfuge at 4°C. The pellet was resuspended in SDS-PAGE sample buffer. 0.5 mg/ml trypsin was added to the supernatants, and this was incubated on ice for 10 min, after which 0.5 mg/ml soybean trypsin inhibitor was added to quench the proteolysis, followed by SDS-PAGE sample buffer. In the case of larger-scale reactions (1-ml total volume, with the same fraction concentrations as small-scale reactions), the reaction was centrifuged as above. 900 µl of the supernatant was equilibrated to ∼50% sucrose by mixing with 1.35 ml of 80% sucrose dissolved in isolation buffer. 2 ml of this was loaded at the bottom of a 14 × 89-mm Ultra-Clear ultracentrifuge tube (Beckman Coulter). Subsequent 2-ml steps of 40%, 30%, 20%, and 10% sucrose (dissolved in isolation buffer), followed by buffer alone, were added slowly to establish a discontinuous sucrose gradient. Gradients were centrifuged overnight (minimum of 12 h) at 35,000 rpm in an ultracentrifuge using a swinging-bucket rotor (SW41 Ti). The next day, 500-µl fractions were collected in SDS-PAGE sample buffer.
Transmission EM and immunogold labeling
Cells expressing YFP-Stx17 or GFP-parkin were prepared for immuno-EM by fixation in 5% formaldehyde and 0.1% glutaraldehyde in PBS for 15 min at 37°C. Cells were washed in PBS, quenched with 50 mM glycine in PBS for 15 min, and washed again. Cells were semi-permeabilized in 0.1% saponin and 5% BSA in PBS for 30 min, followed by three 5-min washes in 5% BSA in PBS. Cells were then incubated with anti-GFP antibody (Invitrogen) diluted 1:2,000 in 1% BSA for 1 h, washed in 1% BSA/PBS three times for 5 min each, then incubated with nanogold-conjugated (1.4 nm colloidal gold) goat anti–rabbit IgG (Nanoprobes) for another hour (diluted 1:200 as before). Cells were then washed three times in PBS and postfixed with 1.6% glutaraldehyde for 10 min at room temperature, then rinsed extensively with water. Gold particles were enhanced for 5 min using the HQ Silver Enhancement kit (Nanoprobes) according to the manufacturer’s instructions, and cells were washed extensively in water afterward and stored in 1.6% glutaraldehyde at 4°C until EM processing. For immunogold labeling using 10 nm gold, cells were prepared for immuno-EM as above but incubated with 10 nm gold-conjugated goat anti–rabbit IgG (Abcam) overnight at 4°C (diluted 1:20) and were not subsequently silver enhanced. For preparation of samples for morphological analysis, cells were fixed in 2.5% glutaraldehyde for 2 h at room temperature and then stored at 4°C before processing. Thin sections on grids were observed in a Tecnai 12 BioTwin transmission electron microscope (FEI) at 120 keV, and images were acquired with a charge coupled device camera (XR80C; AMT).
Statistical analyses, graphing, and protein structure/alignment graphic generation
Unless otherwise indicated in the figure legends, all quantitative experiments were performed two to four times. The numbers of cells quantified per experiment are indicated in the figure legends. No statistical method was used to predetermine the experimental sample size. Statistical tests and representations of the data were generated using Prism (GraphPad Software). For histograms, data are given as the mean ± SEM, whereas the mean is indicated on dot plots. Statistical significance was determined by the appropriate statistical test; one- (Fig. 4 C) and two-tailed (Fig. 3 G; Fig. 4 D; Fig. 5, F and G; and Fig. S1 D) t tests, and one- (Fig. 5, G, H, and J; Fig. 7, E, F, and H; and Fig. 9 F) and two-way (Fig. 2 E; Figs. 3, D and H; Fig. 4 D; Fig. 5 C; Fig. 8, B and D; and Fig. S5 B) analyses of variance followed by Bonferroni post hoc tests. Differences were considered significant if P < 0.05. Note that for Fig. 8 (B and D) DMSO-treated samples, of which all but one were left out of the histogram (as they were identical to the DMSO-treated control siRNA sample), were accounted for in the statistical analyses. The MDV half-lives in Fig. 4 D were estimated from fitting a decay plot to the graph with Microsoft Excel, using the antimycin A–treated and untreated data as maximum and baseline MDV levels, respectively (r2 = 0.98 for control siRNA; r2 = 0.91 for siStx17). Diagrams of the structure of the SNARE tetrad and the R-SNARE sequence alignment were created with PyMOL and eBioX, respectively.
Online supplemental material
Fig. S1 examines the effect of Stx17 depletion on the targeting of other MDV populations to other cellular destinations. Fig. S2 demonstrates that VAMP7 and VAMP8 localize to late endosomes and lysosomes. Fig. S3 shows that the targeting of MDVs to LAMP1-positive compartments is rescued in siStx17/siVAMP7 double-knockdown cells by the reintroduction of Stx17 and VAMP7 (see also Fig. 5, I and J). Fig. S4 demonstrates that a 3Q:1R zero-layer ratio is required in the Stx17–VAMP7-containing SNARE complex in order for proper MDV targeting (see also Fig. 7, G and H). Fig. S5 identifies a role for Vps39 and VAMP8 (but not Stx17) in TOM20 turnover by mitophagy (see also Fig. 8, A and B). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201603105/DC1.
Supplementary Material
Acknowledgments
The authors would like to thank Jeannie Mui and the Facility for Electron Microscopy Research (McGill University) for transmission EM sample processing and technical advice.
This work was supported by operating grants from the Canadian Institutes for Health Research to H.M. McBride and E.A. Fon. G.-L. McLelland is supported by a Canada Graduate Scholarship from the Canadian Institutes for Health Research.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- DRP1
- dynamin-related protein 1
- HM
- heavy membrane fraction
- HOPS
- homotypic fusion and vacuole protein sorting
- LAMP
- lysosome-associated membrane protein
- LM
- light membrane fraction
- MDV
- mitochondrial-derived vesicle
- OMM
- outer mitochondrial membrane
- PD
- Parkinson’s disease
- PDH
- pyruvate dehydrogenase complex
- PINK1
- PTEN-induced kinase 1
- S
- soluble fraction
- SD-OSR
- spinning disk Olympus superresolution
- STX
- syntaxin
- TOM20
- translocase of the outer membrane 20 kD
- VAMP
- vesicle-associated membrane protein
- VPS
- vacuolar protein sorting
- WT
- wild type
References
- Arasaki K., Shimizu H., Mogari H., Nishida N., Hirota N., Furuno A., Kudo Y., Baba M., Baba N., Cheng J., et al. . 2015. A role for the ancient SNARE syntaxin 17 in regulating mitochondrial division. Dev. Cell. 32:304–317. 10.1016/j.devcel.2014.12.011 [DOI] [PubMed] [Google Scholar]
- Baker R.W., Jeffrey P.D., Zick M., Phillips B.P., Wickner W.T., and Hughson F.M.. 2015. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 349:1111–1114. 10.1126/science.aac7906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balderhaar H.J., and Ungermann C.. 2013. CORVET and HOPS tethering complexes - coordinators of endosome and lysosome fusion. J. Cell Sci. 126:1307–1316. 10.1242/jcs.107805 [DOI] [PubMed] [Google Scholar]
- Braschi E., Goyon V., Zunino R., Mohanty A., Xu L., and McBride H.M.. 2010. Vps35 mediates vesicle transport between the mitochondria and peroxisomes. Curr. Biol. 20:1310–1315. 10.1016/j.cub.2010.05.066 [DOI] [PubMed] [Google Scholar]
- Byrnes L.J., Singh A., Szeto K., Benvin N.M., O’Donnell J.P., Zipfel W.R., and Sondermann H.. 2013. Structural basis for conformational switching and GTP loading of the large G protein atlastin. EMBO J. 32:369–384. 10.1038/emboj.2012.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X.T., Zhou B., Lin M.Y., Cai Q., and Sheng Z.H.. 2015. Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J. Cell Biol. 209:377–386. 10.1083/jcb.201412046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daumke O., and Praefcke G.J.. 2011. Structural insights into membrane fusion at the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 108:2175–2176. 10.1073/pnas.1019194108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage B.L., and Cookson B.T.. 2012. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect. Immun. 80:1948–1957. 10.1128/IAI.06014-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J., Liu R., Rong Y., Zhao M., Zhang J., Lai Y., Zhou Q., Wilz L.M., Li J., Vivona S., et al. . 2015. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 520:563–566. 10.1038/nature14147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durcan T.M., Tang M.Y., Pérusse J.R., Dashti E.A., Aguileta M.A., McLelland G.L., Gros P., Shaler T.A., Faubert D., Coulombe B., and Fon E.A.. 2014. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 33:2473–2491. 10.15252/embj.201489729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D., Sutton R.B., Brunger A.T., and Jahn R.. 1998. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA. 95:15781–15786. 10.1073/pnas.95.26.15781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratti R.A., Collins K.M., Hickey C.M., and Wickner W.. 2007. Stringent 3Q.1R composition of the SNARE 0-layer can be bypassed for fusion by compensatory SNARE mutation or by lipid bilayer modification. J. Biol. Chem. 282:14861–14867. 10.1074/jbc.M700971200 [DOI] [PubMed] [Google Scholar]
- Frezza C., Cipolat S., and Scorrano L.. 2007. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat. Protoc. 2:287–295. 10.1038/nprot.2006.478 [DOI] [PubMed] [Google Scholar]
- Gao Y., Zorman S., Gundersen G., Xi Z., Ma L., Sirinakis G., Rothman J.E., and Zhang Y.. 2012. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 337:1340–1343. 10.1126/science.1224492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A., Gutiérrez L.M., Carrasco-Serrano C., Alonso M.T., Viniegra S., and Criado M.. 2002. Modifications in the C terminus of the synaptosome-associated protein of 25 kDa (SNAP-25) and in the complementary region of synaptobrevin affect the final steps of exocytosis. J. Biol. Chem. 277:9904–9910. 10.1074/jbc.M110182200 [DOI] [PubMed] [Google Scholar]
- Graham M.E., Washbourne P., Wilson M.C., and Burgoyne R.D.. 2001. SNAP-25 with mutations in the zero layer supports normal membrane fusion kinetics. J. Cell Sci. 114:4397–4405. [DOI] [PubMed] [Google Scholar]
- Guo B., Liang Q., Li L., Hu Z., Wu F., Zhang P., Ma Y., Zhao B., Kovács A.L., Zhang Z., et al. . 2014. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat. Cell Biol. 16:1215–1226. 10.1038/ncb3066 [DOI] [PubMed] [Google Scholar]
- Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. . 2013. Autophagosomes form at ER-mitochondria contact sites. Nature. 495:389–393. 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- Hanson P.I., Roth R., Morisaki H., Jahn R., and Heuser J.E.. 1997. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 90:523–535. 10.1016/S0092-8674(00)80512-7 [DOI] [PubMed] [Google Scholar]
- Hu J., Shibata Y., Zhu P.P., Voss C., Rismanchi N., Prinz W.A., Rapoport T.A., and Blackstone C.. 2009. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 138:549–561. 10.1016/j.cell.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V., Zou P., Rhee H.W., Udeshi N.D., Cracan V., Svinkina T., Carr S.A., Mootha V.K., and Ting A.Y.. 2014. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell. 55:332–341. 10.1016/j.molcel.2014.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., and Mizushima N.. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Jiang P., Nishimura T., Sakamaki Y., Itakura E., Hatta T., Natsume T., and Mizushima N.. 2014. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol. Biol. Cell. 25:1327–1337. 10.1091/mbc.E13-08-0447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane L.A., Lazarou M., Fogel A.I., Li Y., Yamano K., Sarraf S.A., Banerjee S., and Youle R.J.. 2014. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205:143–153. 10.1083/jcb.201402104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L., and Brennwald P.. 2000. Testing the 3Q:1R “rule”: mutational analysis of the ionic “zero” layer in the yeast exocytic SNARE complex reveals no requirement for arginine. Mol. Biol. Cell. 11:3849–3858. 10.1091/mbc.11.11.3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D.G., Ritorto M.S., Hofmann K., Alessi D.R., Knebel A., Trost M., and Muqit M.M.. 2014. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460:127–139. 10.1042/BJ20140334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi-Itakura C., Koyama-Honda I., Itakura E., and Mizushima N.. 2014. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cell Sci. 127:4089–4102. 10.1242/jcs.156034 [DOI] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., and Shimizu N.. 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 392:605–608. 10.1038/33416 [DOI] [PubMed] [Google Scholar]
- Kloepper T.H., Kienle C.N., and Fasshauer D.. 2008. SNAREing the basis of multicellularity: consequences of protein family expansion during evolution. Mol. Biol. Evol. 25:2055–2068. 10.1093/molbev/msn151 [DOI] [PubMed] [Google Scholar]
- Koumandou V.L., Wickstead B., Ginger M.L., van der Giezen M., Dacks J.B., and Field M.C.. 2013. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit. Rev. Biochem. Mol. Biol. 48:373–396. 10.3109/10409238.2013.821444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., et al. . 2014. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 510:162–166. 10.1038/nature13392 [DOI] [PubMed] [Google Scholar]
- Lauer J.M., Dalal S., Marz K.E., Nonet M.L., and Hanson P.I.. 2006. SNARE complex zero layer residues are not critical for N-ethylmaleimide-sensitive factor-mediated disassembly. J. Biol. Chem. 281:14823–14832. 10.1074/jbc.M512706200 [DOI] [PubMed] [Google Scholar]
- Lawrence G.W., and Dolly J.O.. 2002a Ca2+-induced changes in SNAREs and synaptotagmin I correlate with triggered exocytosis from chromaffin cells: insights gleaned into the signal transduction using trypsin and botulinum toxins. J. Cell Sci. 115:2791–2800. [DOI] [PubMed] [Google Scholar]
- Lawrence G.W., and Dolly J.O.. 2002b Multiple forms of SNARE complexes in exocytosis from chromaffin cells: effects of Ca(2+), MgATP and botulinum toxin type A. J. Cell Sci. 115:667–673. [DOI] [PubMed] [Google Scholar]
- Lee S., Zhang C., and Liu X.. 2015. Role of glucose metabolism and ATP in maintaining PINK1 levels during Parkin-mediated mitochondrial damage responses. J. Biol. Chem. 290:904–917. 10.1074/jbc.M114.606798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Kümmel D., Coleman J., Reinisch K.M., Rothman J.E., and Pincet F.. 2014. A half-zippered SNARE complex represents a functional intermediate in membrane fusion. J. Am. Chem. Soc. 136:3456–3464. 10.1021/ja410690m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik B.R., Godena V.K., and Whitworth A.J.. 2015. VPS35 pathogenic mutations confer no dominant toxicity but partial loss of function in Drosophila and genetically interact with parkin. Hum. Mol. Genet. 24:6106–6117. 10.1093/hmg/ddv322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy M.K., Kaganovich A., Rudenko I.N., Ding J., and Cookson M.R.. 2014. Hexokinase activity is required for recruitment of parkin to depolarized mitochondria. Hum. Mol. Genet. 23:145–156. 10.1093/hmg/ddt407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan D.G., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D., Coxon F.P., Miranda de Stegmann D., Bhogaraju S., Maddi K., et al. . 2015. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell. 57:39–54. 10.1016/j.molcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- McLelland G.L., Soubannier V., Chen C.X., McBride H.M., and Fon E.A.. 2014. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 33:282–295. 10.1002/embj.201385902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P., and Chan D.C.. 2016. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 212:379–387. 10.1083/jcb.201511036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuspiel M., Schauss A.C., Braschi E., Zunino R., Rippstein P., Rachubinski R.A., Andrade-Navarro M.A., and McBride H.M.. 2008. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr. Biol. 18:102–108. 10.1016/j.cub.2007.12.038 [DOI] [PubMed] [Google Scholar]
- Ossig R., Schmitt H.D., de Groot B., Riedel D., Keränen S., Ronne H., Grubmüller H., and Jahn R.. 2000. Exocytosis requires asymmetry in the central layer of the SNARE complex. EMBO J. 19:6000–6010. 10.1093/emboj/19.22.6000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier M.A., Hao J.C., Malkus P.N., Chan C., Moore M.F., King D.S., and Bennett M.K.. 1998. Protease resistance of syntaxin.SNAP-25.VAMP complexes. Implications for assembly and structure. J. Biol. Chem. 273:11370–11377. 10.1074/jbc.273.18.11370 [DOI] [PubMed] [Google Scholar]
- Rosenbloom A.B., Lee S.H., To M., Lee A., Shin J.Y., and Bustamante C.. 2014. Optimized two-color super resolution imaging of Drp1 during mitochondrial fission with a slow-switching Dronpa variant. Proc. Natl. Acad. Sci. USA. 111:13093–13098. 10.1073/pnas.1320044111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J.E. 2014. The principle of membrane fusion in the cell (Nobel lecture). Angew. Chem. Int. Ed. Engl. 53:12676–12694. 10.1002/anie.201402380 [DOI] [PubMed] [Google Scholar]
- Ryan B.J., Hoek S., Fon E.A., and Wade-Martins R.. 2015. Mitochondrial dysfunction and mitophagy in Parkinson’s: from familial to sporadic disease. Trends Biochem. Sci. 40:200–210. 10.1016/j.tibs.2015.02.003 [DOI] [PubMed] [Google Scholar]
- Scorrano L. 2013. Keeping mitochondria in shape: a matter of life and death. Eur. J. Clin. Invest. 43:886–893. 10.1111/eci.12135 [DOI] [PubMed] [Google Scholar]
- Sherer N.M., Lehmann M.J., Jimenez-Soto L.F., Ingmundson A., Horner S.M., Cicchetti G., Allen P.G., Pypaert M., Cunningham J.M., and Mothes W.. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic. 4:785–801. 10.1034/j.1600-0854.2003.00135.x [DOI] [PubMed] [Google Scholar]
- Söllner T., Whiteheart S.W., Brunner M., Erdjument-Bromage H., Geromanos S., Tempst P., and Rothman J.E.. 1993. SNAP receptors implicated in vesicle targeting and fusion. Nature. 362:318–324. 10.1038/362318a0 [DOI] [PubMed] [Google Scholar]
- Soubannier V., McLelland G.L., Zunino R., Braschi E., Rippstein P., Fon E.A., and McBride H.M.. 2012a A vesicular transport pathway shuttles cargo from mitochondria to lysosomes. Curr. Biol. 22:135–141. 10.1016/j.cub.2011.11.057 [DOI] [PubMed] [Google Scholar]
- Soubannier V., Rippstein P., Kaufman B.A., Shoubridge E.A., and McBride H.M.. 2012b Reconstitution of mitochondria derived vesicle formation demonstrates selective enrichment of oxidized cargo. PLoS One. 7:e52830 10.1371/journal.pone.0052830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai V.J., Hickey C.M., and Wickner W.. 2008. HOPS proofreads the trans-SNARE complex for yeast vacuole fusion. Mol. Biol. Cell. 19:2500–2508. 10.1091/mbc.E08-01-0077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura A., McLelland G.L., Fon E.A., and McBride H.M.. 2014. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 33:2142–2156. 10.15252/embj.201488104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton R.B., Fasshauer D., Jahn R., and Brunger A.T.. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 395:347–353. 10.1038/26412 [DOI] [PubMed] [Google Scholar]
- Swaney D.L., Rodríguez-Mias R.A., and Villén J.. 2015. Phosphorylation of ubiquitin at Ser65 affects its polymerization, targets, and proteome-wide turnover. EMBO Rep. 16:1131–1144. 10.15252/embr.201540298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Nagy P., Varga Á., Pircs K., Kárpáti M., Varga K., Kovács A.L., Hegedűs K., and Juhász G.. 2013. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201:531–539. 10.1083/jcb.201211160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takáts S., Pircs K., Nagy P., Varga Á., Kárpáti M., Hegedűs K., Kramer H., Kovács A.L., Sass M., and Juhász G.. 2014. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol. Biol. Cell. 25:1338–1354. 10.1091/mbc.E13-08-0449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. . 2004. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 304:1158–1160. 10.1126/science.1096284 [DOI] [PubMed] [Google Scholar]
- Van Laar V.S., Arnold B., Cassady S.J., Chu C.T., Burton E.A., and Berman S.B.. 2011. Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Hum. Mol. Genet. 20:927–940. 10.1093/hmg/ddq531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincow E.S., Merrihew G., Thomas R.E., Shulman N.J., Beyer R.P., MacCoss M.J., and Pallanck L.J.. 2013. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc. Natl. Acad. Sci. USA. 110:6400–6405. 10.1073/pnas.1221132110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Wang X., Fujioka H., Hoppel C., Whone A.L., Caldwell M.A., Cullen P.J., Liu J., and Zhu X.. 2016. Parkinson’s disease-associated mutant VPS35 causes mitochondrial dysfunction by recycling DLP1 complexes. Nat. Med. 22:54–63. 10.1038/nm.3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Katayama N., Takeuchi K., Togano T., Itoh R., Sato M., Yamazaki M., Abe M., Sato T., Oda K., et al. . 2013. Point mutation in syntaxin-1A causes abnormal vesicle recycling, behaviors, and short term plasticity. J. Biol. Chem. 288:34906–34919. 10.1074/jbc.M113.504050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Matsuda N., and Tanaka K.. 2016. The ubiquitin signal and autophagy: an orchestrated dance leading to mitochondrial degradation. EMBO Rep. 17:300–316. 10.15252/embr.201541486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii S.R., and Mizushima N.. 2015. Autophagy machinery in the context of mammalian mitophagy. Biochim. Biophys. Acta. 1853(10, 10 Pt B):2797–2801. 10.1016/j.bbamcr.2015.01.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.