Ferguson et al. demonstrate that clathrin coat growth rates can be utilized as quantitative reporters of clathrin-mediated endocytic dynamics in cellular contexts where errors associated with single-particle tracking are significant. They validate the tool within tissues of Drosophila embryos.
Abstract
Current understanding of clathrin-mediated endocytosis (CME) dynamics is based on detection and tracking of fluorescently tagged clathrin coat components within cultured cells. Because of technical limitations inherent to detection and tracking of single fluorescent particles, CME dynamics is not characterized in vivo, so the effects of mechanical cues generated during development of multicellular organisms on formation and dissolution of clathrin-coated structures (CCSs) have not been directly observed. Here, we use growth rates of fluorescence signals obtained from short CCS intensity trace fragments to assess CME dynamics. This methodology does not rely on determining the complete lifespan of individual endocytic assemblies. Therefore, it allows for real-time monitoring of spatiotemporal changes in CME dynamics and is less prone to errors associated with particle detection and tracking. We validate the applicability of this approach to in vivo systems by demonstrating the reduction of CME dynamics during dorsal closure of Drosophila melanogaster embryos.
Introduction
Clathrin-mediated endocytosis (CME) is the major pathway responsible for internalization of lipids and receptor-bound macromolecules from the plasma membrane of eukaryotic cells (Conner and Schmid, 2003). During internalization of a cargo molecule, clathrin triskelions assemble into submicron-sized polyhedral structures upon their recruitment to the plasma membrane by the endocytic clathrin adaptor protein AP2 (Ehrlich et al., 2004; Saffarian and Kirchhausen, 2008; Boucrot et al., 2010; Cocucci et al., 2012; Hong et al., 2015). Live-cell imaging studies designed for tracking of fluorescently tagged clathrin coat components have revealed the dynamics of formation, internalization, and dissolution of distinct classes of clathrin-coated structures (CCSs; Gaidarov et al., 1999; Merrifield et al., 2002; Ehrlich et al., 2004; Loerke et al., 2009; Mettlen et al., 2010; Taylor et al., 2011; Kural and Kirchhausen, 2012; Aguet et al., 2013). The best-characterized structures are highly curved, cage-like assemblies that deform the plasma membrane into pits and vesicles. In conventional fluorescence time-lapse acquisitions, clathrin-coated pits appear as diffraction-limited spots with mean lifetimes of ∼1 min (Ehrlich et al., 2004; Saffarian et al., 2009; Kural et al., 2012; Aguet et al., 2013). CCSs disappearing within the first ∼20 s of their initiation are abortive structures that fail to construct bona fide endocytic carriers (Hong et al., 2015). Flat arrays of clathrin, also known as plaques, are larger than coated pits and slower in their internalization dynamics (Saffarian et al., 2009; Grove et al., 2014). Physiological relevance of clathrin-coated plaques has been equivocal, because they only appear at the substrate contact sites of cultured cells and, because of their long lifetimes, they are not effective endocytic carriers.
Dynamics of endocytic pathways are inversely related to plasma membrane tension, because membrane internalization machinery are required to do work against the two major constituents of tension (i.e., in-plane tension and membrane-cytoskeleton adhesion) to create invaginations (Dai et al., 1997; Raucher and Sheetz, 1999; Sheetz, 2001; Apodaca, 2002; Gauthier et al., 2012; Diz-Muñoz et al., 2013). Tension regulates formation and curvature of clathrin coats reconstructed on giant unilamellar vesicles (Saleem et al., 2015). Studies in yeast and in polarized and mitotic mammalian cells show that CME is inhibited unless plasma membrane tension is counteracted by actin dynamics (Aghamohammadzadeh and Ayscough, 2009; Boulant et al., 2011; Kaur et al., 2014). Regulation of endocytic rates by mechanical cues has important roles in development; during the early stages of Drosophila melanogaster embryogenesis, increased tension inhibits Fog receptor endocytosis, which is required for completion of ventral furrow formation (Pouille et al., 2009).
Our current understanding of CME dynamics is based on in vitro imaging studies that are limited in their potential to mimic physical properties of tissue microenvironments. In a majority of these studies, dynamics of CCSs were monitored at the plasma membrane–coverglass interface, which has no physiological correspondence. Plating conditions, membrane–substrate interactions, and cell spreading area can regulate clathrin dynamics in in vitro experiments (Batchelder and Yarar, 2010; Tan et al., 2015). A holistic understanding of CME requires elucidating clathrin coat dynamics in cells residing within tissues of multicellular organisms.
Determining lifetime distributions of CCSs is the prevalent technique for monitoring CME dynamics. This approach necessitates identifying complete traces of individual CCSs (from initiation to dissolution), which is error prone within high-density particle fields and regimes with low signal to noise (Aguet et al., 2013; Mettlen and Danuser, 2014). CME dynamics have not been reported for any in vivo systems, because determining lifetimes of individual CCSs is more challenging within complex, 3D geometries of living tissues. In this study, we show that the rate of incorporation or dispersion of clathrin coat components during formation of endocytic vesicles can be used as reporters for clathrin dynamics. Because hundreds of clathrin-coated endocytic carriers can be detected within a cell at a given instant, distributions spanning the entire range of formation and disassembly rates can be obtained within temporal windows shorter than the lifetime of clathrin coats. This advantage makes growth rate distributions a superior alternative to clathrin lifetime analyses, especially within cellular contexts where the fidelity of fluorescent particle tracking is low. Using this approach, we provide the first experimental evidence of CME mechanoregulation in tissues of live Drosophila embryos.
Results
CCS growth rates are robust reporters of CME dynamics
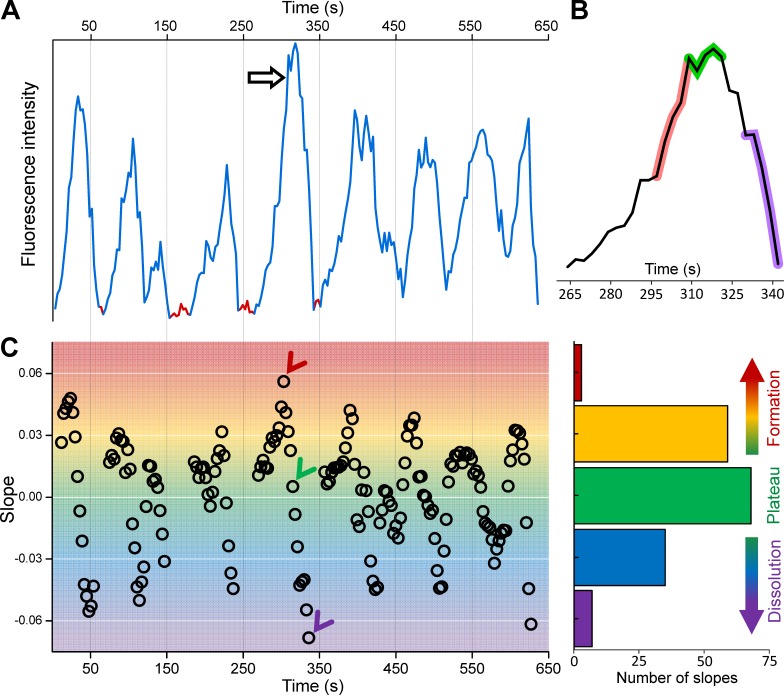
In fluorescence imaging assays, endocytic CCS formation is marked by appearance of a diffraction-limited spot that steadily increases in intensity because of accumulation of fluorescently tagged coat components. The internalization of the clathrin-coated vesicle is followed by a relatively fast dimming of the fluorescence caused by dissolution of the coat (Kural and Kirchhausen, 2012; Fig. 1, A and B). Lifetime (i.e., the time required for formation and internalization of CCSs from the plasma membrane) is an extensively used metric for characterizing CME dynamics. Factors that elongate CCS lifetimes reduce the efficacy of ligand endocytosis (Cureton et al., 2010; Boulant et al., 2011). Any factor that affects the rate of formation and/or dissolution of clathrin coats is a potential regulator of CCS lifetimes and, hence, CME dynamics. To establish whether we can use growth rates of CCS signal as metrics for endocytic dynamics, we quantified formation and dissolution (negative growth) rates of individual CCSs by determining the slope of the normalized fluorescence intensity within 12-s temporal windows (Fig. 1 C and online supplemental software). Because hundreds of CCS traces can be detected within a single cell at a given instant, we were able to assemble distributions of growth rates for each frame of time-lapse acquisitions (Video 1). When applied to cells that produce predominantly clathrin-coated pits, the majority of the growth rates obtained in this way were positive (i.e., corresponding to increasing fluorescence signal; Fig. 2 B). This is consistent with the characteristic intensity profile of pits, which has a steady formation phase followed by a relatively abrupt dissolution (Ehrlich et al., 2004; Massol et al., 2006; Fig. 1 B).
Figure 1.
Determining CCS growth rate distributions. (A) Fluorescence intensity trace of a clathrin hotspot imaged at the ventral surface of a BSC-1 cell expressing AP2-GFP. Traces segments that are not used in growth rate calculation are shown in red, as they were considered the background signal. (B) Zoomed version of the clathrin-coated pit trace marked by the arrow in A. Shaded regions show 12-s-long fragments dwelling at formation (red), plateau (green), and dissolution (purple) phases of the pit. (C, left) For the intensity trace in A, slope values representing the growth rates are determined from 12-s-long fragments centered on each time point. Red, green, and purple arrowheads mark the slopes corresponding to the growth, plateau, and dissolution fragments highlighted in B, respectively. (C, right) Bar plot shows the distribution of the growth rates shown in the left panel. Positive and negative values correspond to formation and dissolution phases, respectively.
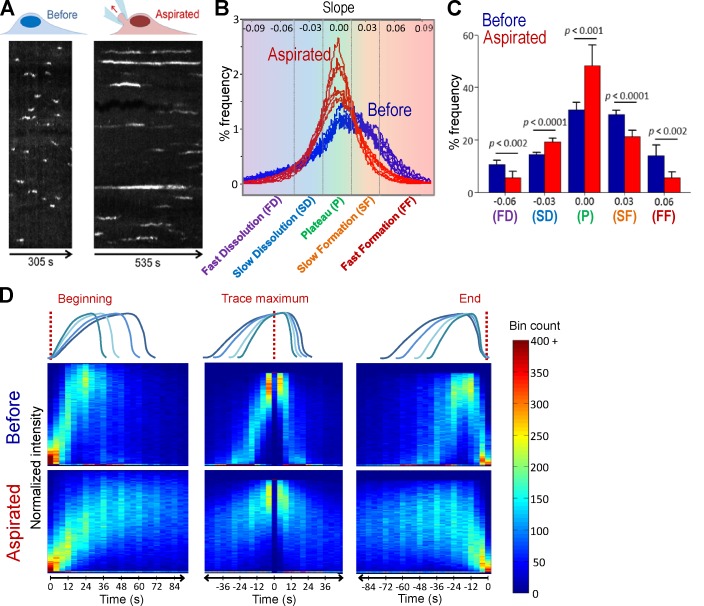
Figure 2.
Using growth rate histograms as reporters of clathrin dynamics. (A) Kymographs are generated from the same BSC-1 cell before and during microaspiration, respectively. Elongated AP2 traces demonstrate longer CCS lifetimes under increased membrane tension. (B) Growth rate distributions are shown for seven different cells before and during aspiration. Change in CCS dynamics induced by micropipette aspiration can be observed in growth rate distributions. In the control experiments (cells before aspiration), CCSs spend more time in the formation phase (i.e., the distribution is inclined toward positive slopes). The asymmetry was abolished upon aspiration and plateau phases got relatively longer (Ncells = 7 and Ntraces = 38136). (C) Growth rate distributions in B are assembled in five bins to better delineate different phases of clathrin-coated vesicle formation (FD, fast dissolution; FF, fast formation; P, plateau; SD, slow dissolution; SF, slow formation). The bars show mean + standard deviation to illustrate dispersion between cells. P-values were obtained using the two-tailed t test. (D) 2D histograms of normalized intensity traces aligned at different time points (beginning, trace maximum, and end) and superposed as represented by the cartoons. In each alignment, the aspirated cells show a significantly widened distribution, demonstrating a preference for slopes lower in magnitude. Bins corresponding to multiples of 12 s are more populated, as they contain trace data from both 3- and 4-s frame rate acquisitions.
As a means to alter CME efficiency, we used micropipette aspiration to increase in-plane membrane tension in cells (Houk et al., 2012). We detected a significant increase in CCS lifetimes when micropipette aspiration was applied (45.5 ± 27.1 s versus 80.1 ± 86.6 s, Ncells = 7, Ntraces= 38,136; Fig. 2 A). Asymmetry of the growth rate distributions is abolished upon aspiration, and a greater fraction of intensity traces were associated with steady levels of fluorescence signal (i.e., the plateau phase). High-magnitude slopes, which represent fast formation and dissolution phases, are diminished with the increasing membrane tension (Fig. 2, B and C). These transformations in the growth rate distributions suggest that obstruction of both coat formation and dissolution is the prevalent factor behind elongation of the mean CCS lifetime under increased membrane tension. We used two independent visualization tools to validate these results. First, we created 2D histograms assembled by superposition of CCS intensity profiles that are synchronized at the beginning, middle, or end of traces (Fig. 2 D). Even though CCSs have a wide distribution of lifetimes, 2D histograms assembled from traces obtained before aspiration displayed the characteristic intensity profile of coated pits (Fig. 2 D, top row, middle column). However, the histograms assembled using aspirated cell traces were wider and displayed significant elongation in formation, plateau, and dissolution phases (Fig. 2 D, bottom row). In the second approach, we used a hierarchical clustering algorithm to create groups of CCS traces that have similar intensity profiles. As expected, clusters obtained from aspirated cells displayed longer plateau phases and slower formation and dissolution rates (Fig. S1).
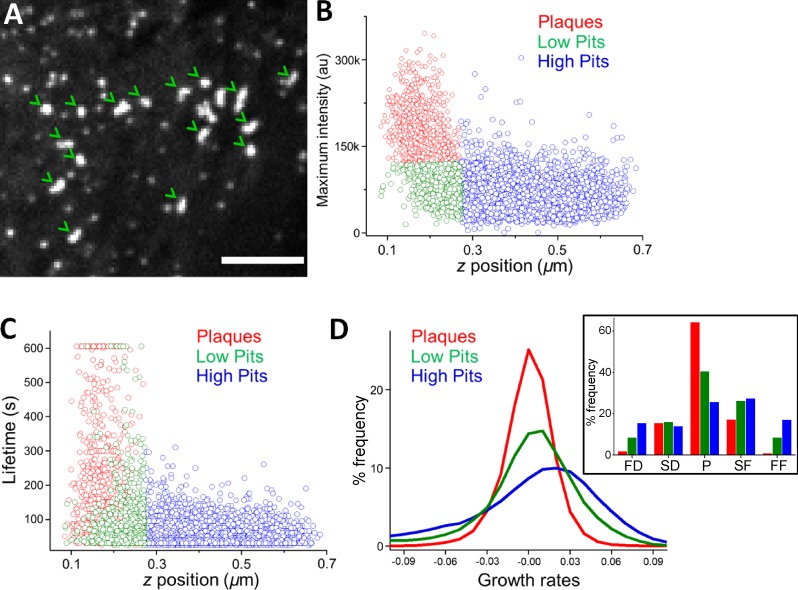
To elucidate the effects of membrane–substrate interactions on CCS growth rates, we used a 3D particle tracking algorithm to determine the relative axial positions and fluorescence intensities of CCSs at the ventral surface of cultured cells (Kural et al., 2012, 2015). Along with pits, we observed formation of clathrin-coated plaques at the ventral surfaces of BSC-1 cells that are plated for >72 h (Fig. 3 A). Based on the intensity and position information, we categorized the CCSs as plaques (bright and close to substrate), “low” pits (dim and close to substrate), and “high” pits (dim and far from the substrate; Fig. 3, B and C). In good agreement with the previous studies, we found that CCSs positioned closer to the substrate contact sites are significantly longer lived (plaques, 174.5 ± 160.5 s; low pits, 75.2 ± 78.3 s; high pits, 48.3 ± 27.8 s, Ncells = 6, Ntraces= 11,482; Fig. 3 C; Batchelder and Yarar, 2010). Growth rate analyses establish that impairment of clathrin dynamics in the proximity of adhesion regions is associated with extension of the plateau and diminishing of the high slope phases, consistent with the measurements performed in aspirated cells (Fig. 3 D). Our combined results demonstrate significant alterations in CCS growth rates caused by physical factors that hinder endocytic dynamics.
Figure 3.
CCSs in proximity of substrate adhesion sites have reduced dynamics. (A) Arrowheads mark clathrin-coated plaques at the ventral surface of a BSC-1 cell stably expressing AP2-GFP. Bar, 5 µm. (B) Scatterplot shows maximum intensity versus the axial positions of CCSs detected at the ventral surface. The positions are relative to the lower bound of the confocal volume used for 3D tracking (Kural et al., 2012). CCS traces are divided into plaque (red) and pit (green and blue) populations based on their maximum intensities. Clathrin pits positioned closer to the substrate are in green (Ncells = 6 and Ntraces = 11,482). (C) Scatterplot shows the lifetime versus axial positions of the CCSs shown in B. (D) Growth rate distributions of the pit and plaque populations. The inset shows the distributions as bar plots.
Accuracy of growth rate distributions does not depend on determining complete traces of CCSs
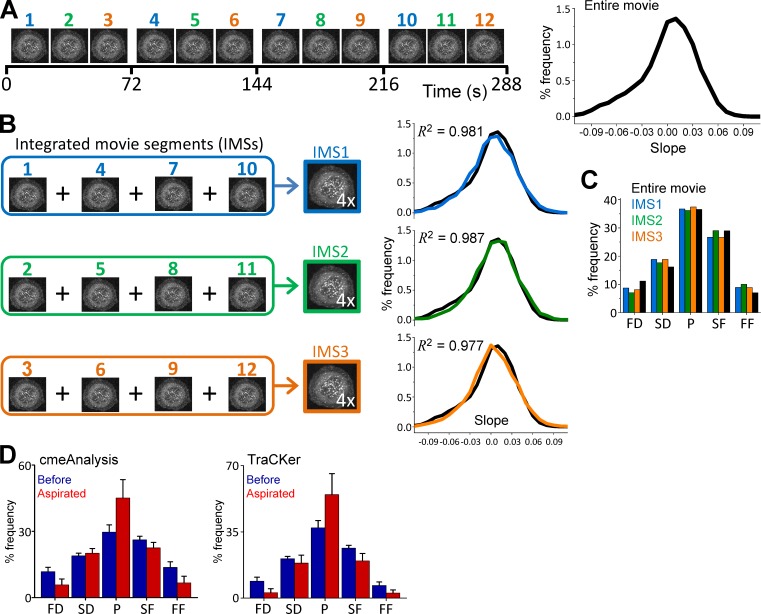
Errors associated with particle detection and tracking result in significant underestimation of CCS lifetimes (Aguet et al., 2013; Mettlen and Danuser, 2014). Slope values populating the growth rate histograms, however, are calculated using trace information within 12-s-long windows and thus do not rely on determining the complete CCS traces. We tested the reproducibility of growth rate distributions on 24-s-long integrated movie segments (IMSs) that are the quadrature sum of four different temporal sections of a movie (Fig. 4, A–C). Faithful assessment of CCS lifetimes from IMSs is impractical because (1) density of CCSs is on average four times greater than the original acquisition, (2) background noise level is increased due to error propagation, and (3) IMSs are significantly shorter than the mean coated pit lifetime (24 s versus 1 min). Despite these impediments, we found that CCS growth rate distributions could be reproduced very accurately from the IMSs (Fig. 4, B and C). We also found that distortion of the growth rate distributions caused by increased tension could be observed in the IMSs of microaspirated cells regardless of the algorithm used for tracking CCSs (Fig. 4 D).
Figure 4.
Growth rate analysis does not depend on determining the complete traces of CCSs. (A, left) Thumbnails represent 12 temporal sections (24 s each) of a 288-s-long fluorescence movie of a BSC-1 cell stably expressing AP2-GFP. (right) CCS growth rate distribution of the entire movie. (B, left) Integrated movie segments (IMSs) are the quadrature sum of four 24-s-long segments in which density of CCSs is approximately four times larger than the original movie. Temporal separation is maximized between the segments to minimize the number of self-overlapping clathrin spots. (right) Growth rate distributions obtained from each IMS is plotted in comparison with the distribution obtained from the entire movie (black). (C) Growth rate distributions of the IMSs and the entire movie are displayed as bar plots. (D) We tested the reproducibility of growth rate distributions using the CCS traces produced by cmeAnalysis (Aguet et al., 2013) and TraCKer (homemade particle tracking program; see online supplemental software). Changes in slope distributions upon micropipette aspiration can be observed in both datasets. Numbers of the analyzed IMSs are 25 and 64 for before and during aspiration conditions, respectively (Ncells = 7). Error bars show standard deviations. FD, fast dissolution; FF, fast formation; P, plateau; SD, slow dissolution; SF, slow formation.
Spatiotemporal variations in CME dynamics can be resolved in real time using CCS growth rates
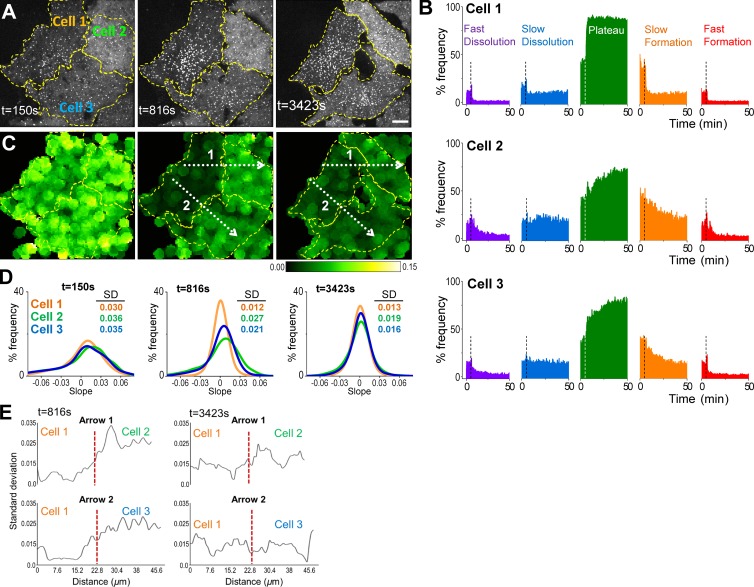
When CCS lifetime distributions are used as metrics for CME efficiency, the mean lifetime of clathrin-coated pits (∼1 min) sets the limit of the temporal resolution for distinguishing the changes in clathrin coat dynamics. Consequently, the factors that influence endocytic processes at shorter time scales become indiscernible. Growth rate distributions, however, are constructed before the completion of traces and thus enable real-time monitoring of CCS dynamics in cells. We used growth rate analysis to monitor gradual changes in CCS dynamics upon acute cholesterol depletion. Structural studies show that cholesterol depletion reduces CME efficiency by replacing clathrin-coated pits with flat clathrin arrays (plaques) at the cell membrane (Subtil et al., 1999; Fig. S2 A). When cells are treated with methyl-β-cyclodextrin (MβCD), in line with our micropipette aspiration experiments, we found that impairment of CCS dynamics is coupled with disappearance of high-magnitude slopes in CCS growth rate distributions (Fig. 5, A and B; and Video 2). This result was anticipated, as cholesterol depletion increases the effective membrane tension through escalation of membrane–cytoskeleton adhesion energy (Khatibzadeh et al., 2012; Fig. S2 B). We used standard deviation of regional CCS growth rates to generate a visualization tool for monitoring spatiotemporal changes in clathrin dynamics in real time (Fig. 5 C). In regions with impeded CCS dynamics, standard deviations of the growth rates were reduced because of the disappearance of the high-magnitude slopes (Fig. 5 D). Using this approach, we found that MβCD treatment can affect CME dynamics at different rates in neighboring cells (Fig. 5 D, E). Cell-to-cell variation may stem from cells’ mitotic state or position with respect to cell islet edges, as described earlier (Snijder et al., 2009).
Figure 5.
Real-time monitoring of CCS dynamics in cholesterol-depleted cells. (A) Fluorescence images show snapshots of three BSC-1 cells stably expressing AP2-GFP. 10 mM MβCD was applied at the sixth minute (t = 360 s) to initiate cholesterol depletion (Video 2). Dashed lines demarcate the cell boundaries. Bar, 10 µm. (B) For the three cells shown in A, percentage frequency of the five growth phases are plotted as a function of time. The dashed lines denote the time point of MβCD addition. The fastest change in CCS dynamics is observed in cell 1. (C) Figures show standard deviation (of local clathrin growth rates for the snapshots shown in A. Each pixel in the image is given the value of standard deviation of the growth rates detected from CCSs found in a radius of 4.8 µm. This representation is used for illustrating temporal and spatial variations in CCS dynamics. Standard deviation is lower in regions where the growth rate distribution is narrow, a signature of impeded CCS dynamics. Cell 1 responds to cholesterol depletion the earliest, as observed at t = 816 s. (D) Growth rate histograms are shown for the three cells at the time points of the snapshots in A and C. Spatial and temporal heterogeneity of CCS dynamics can be detected using the standard deviation of growth rate histograms, which change at different rates for the three cells. (E) Plots show spatial variation in standard deviation of local CCS growth rates over the distances shown by the dashed arrows in C. Positions of the cell boundaries are marked by the dashed red lines.
Reduction in CCS formation rates (positive slopes) upon cholesterol depletion could be attributed to depletion of free clathrin coat components in the cytosol caused by increased CCS lifetime, as they are contained within existing coats. In that case, we would expect dissolution rates to disappear the earliest. However, we found that both positive and negative slopes (i.e., fast formation and dissolution phases) diminish simultaneously (Fig. 5 B), which indicates that flat clathrin arrays have slower formation rates than clathrin-coated pits (Fig. S2 A). Therefore, at variance with earlier interpretations (Subtil et al., 1999), our results indicate that flat clathrin arrays found in cholesterol-depleted cells cannot be considered as direct precursors of clathrin-coated pits, as they have distinct formation dynamics.
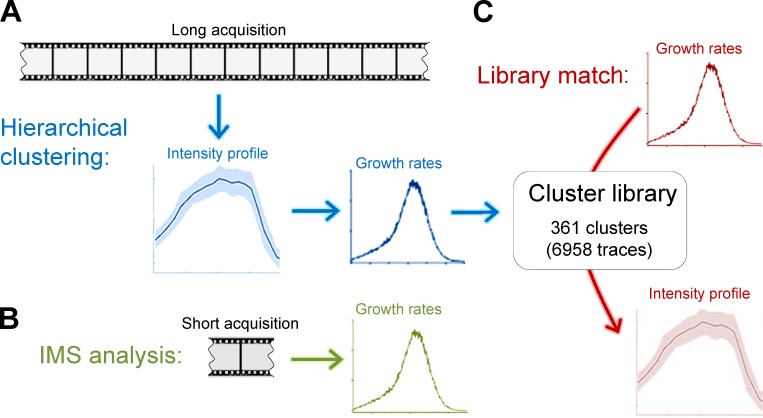
CCS intensity profiles can be reproduced from growth rate distributions
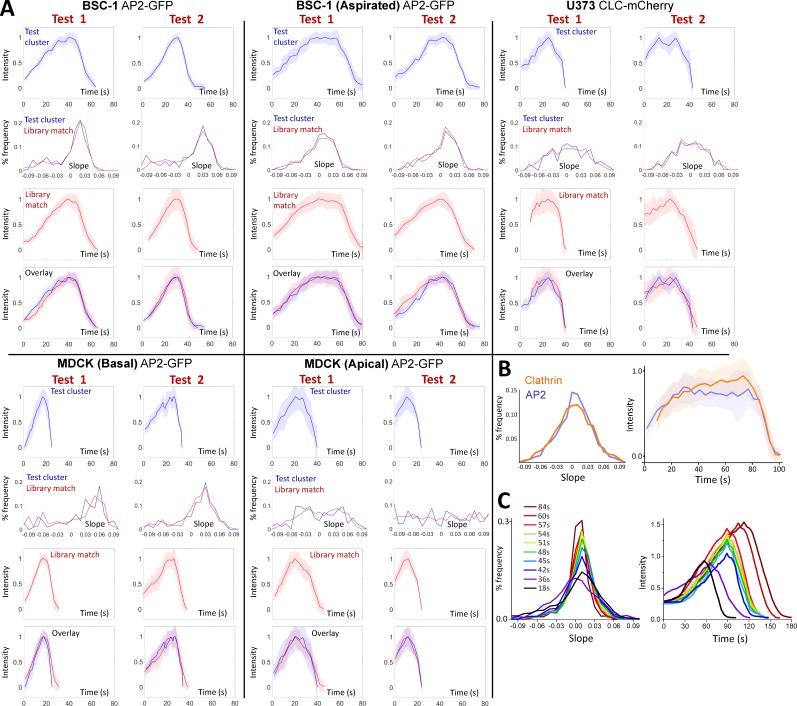
Hierarchical clustering of CCS traces allows us to determine growth rate distributions corresponding to different intensity profiles (Fig. 6 A). An interesting question that arises is whether we can reverse the flow of information in this process (i.e., reproduce the mean intensity profiles using growth rate distributions). Our IMS analyses show that growth rate distributions can be accurately assembled even in the absence of complete CCS intensity profiles (Fig. 6 B). Therefore, whatever the design, such a methodology would be immensely useful for interpreting the CCS growth rates that are obtained from acquisitions in which monitoring complete CCS traces is infeasible. We used a strategy based on determining the analogous growth rate distributions within a library of CCS clusters (Fig. 6 C). We assembled a library of growth rate histograms that are obtained from 361 clusters containing a total count of 6,958 CCS traces. When we used test clusters obtained from various cellular contexts, we were able to make accurate predictions of their intensity profiles solely by determining the most analogous growth rate distribution within the cluster library (Fig. 7 A and online supplemental software). When applied to cells coexpressing fluorescently tagged AP2 and clathrin, we found that the AP2 growth rate distribution has higher frequencies at the low-magnitude slope region, and its predicted intensity profile has a longer plateau before dissolution (Fig. 7 B). This result is in accord with the previous studies, which show that AP2 fluorescence growth halts earlier than the clathrin signal during the completion of the coat (Saffarian and Kirchhausen, 2008). When cells are treated with MβCD, incremental changes in mean CCS intensity profiles taking place within short intervals could be resolved in real time (Fig. 7 C and Video 2). Upon cholesterol depletion, intensity profiles predicted using growth rate distributions became longer lived and reached higher maximum intensities in time. This was expected, as clathrin-coated pits are gradually replaced by larger and less dynamic CCSs in these cells (Fig. S2 A). Although the extent of the cluster library is a critical determinant of the fidelity of predicted intensity profiles, the presented strategy is a powerful tool for analyzing the changes in CCS growth rates obtained from in vivo datasets.
Figure 6.
A novel analytical toolbox for CME dynamics. (A) CCS traces with similar intensity profiles can be grouped using a hierarchical clustering algorithm. This is applicable to acquisitions longer than the mean CCS lifetime. Growth rate distributions obtained from different clusters are used to develop a cluster library. (B) As validated by IMS analysis, growth rate distributions can be assembled using short fragments of CCS traces. (C) For a given growth rate distribution, an accurate estimation of the corresponding intensity profile is possible by determining analogous growth rate distributions in the cluster library.
Figure 7.
Determining CCS intensity profiles using growth rate distributions. (A) Trace clusters from test cells (excluded from the library) are matched with clusters from the CCS library by their growth rate histograms. Successful reproductions of the cluster traces occurred in multiple cell types and conditions. Two clusters and library matches are shown per condition. Intensity profiles of test clusters and corresponding growth rate distributions are shown in blue, and library matches are in red. Shaded regions in intensity profiles mark the standard deviation of the cluster. (B) In a U373 cell coexpressing fluorescently tagged AP2 and clathrin, the growth rate of AP2 is more peaked at low slope magnitudes (left). Corresponding library matches show continuing increase in clathrin signal after AP2 signal plateaued (right). (C) MβCD is added to BSC1 cells at t = 0. Growth rate distributions determined at different time points of the treatment are shown on the left. (right) Predicted intensity profiles for the given growth rate distributions. Gradual changes in growth rates result in increasing trace lifetime and peak intensity.
CME dynamics slow down during dorsal closure of Drosophila embryos
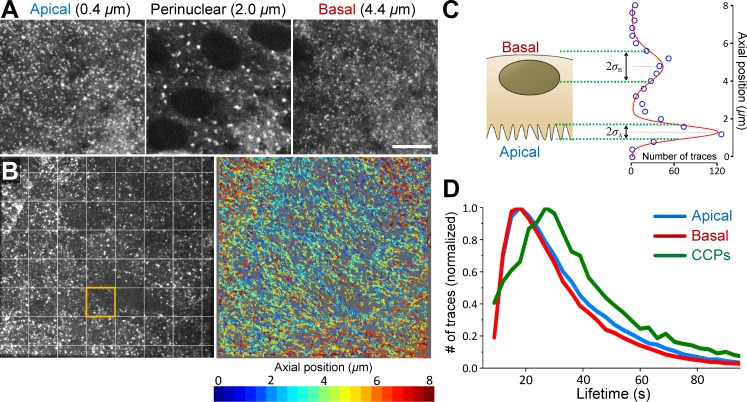
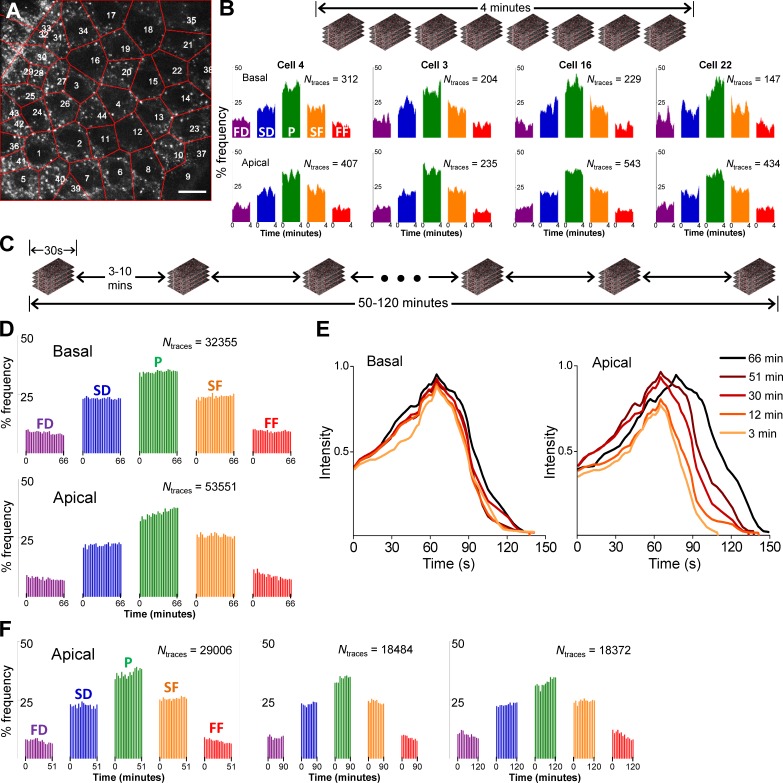
Taking advantage of the new set of analytical tools at hand, we investigated the dynamics of CME in amnioserosa tissue of developing Drosophila embryos. Amnioserosa consists of a single layer of polarized cells that covers the dorsal surface of the embryo after germ band retraction. The tissue is entirely curtained by lateral epidermal cells in ∼4 h by a process called dorsal closure (Jacinto et al., 2002). CCSs originating at the apical and basal surfaces of the amnioserosa can be detected in embryos expressing clathrin light chain (CLC) fused with GFP, using spinning disk confocal fluorescence imaging (Fig. 8 A). CCSs forming on intracellular organelles appear as bright blobs at the perinuclear regions. To determine individual CCS traces at the aminoserosa, we used a robust 2D particle tracking software (Aguet et al., 2013) and processed the output data using a trace combination algorithm to correct for particle disappearances caused by movements in the axial dimension (Fig. 8 B and Video 3). We used local distributions of the number of CCS traces to determine the axial positions of apical and basal surfaces (Fig. 8 C). We found that lifetime distributions for both of the surfaces are dominated by short CCS traces in comparison with the distributions obtained for clathrin-coated pits originating in cultured cells (Fig. 8 D). We believe the anomaly in lifetime distributions obtained from the amnioserosa has no biological basis but is a consequence of single-particle tracking errors caused by high CCS motility in this tissue and increased ratio of incomplete trace fragments (Fig. S3). When we used CCS growth rates as the alternative approach, we noted rapid changes in the distributions obtained from individual amnioserosa cells. However, we found no correlation between the temporal evolution of the distributions at the apical and basal surfaces (Fig. 9, A and B). We extended the duration of the imaging assays by acquiring CCS growth rates from 30-s-long acquisitions that are separated by temporal gaps of 3 or 10 min (Fig. 9 C). In this experimental scheme, monitoring CCS dynamics of individual cells throughout the entire assay was not achievable because the cellular organization of amnioserosa is not preserved over long durations, as the tissue is gradually replaced by lateral epidermis. When the growth rates are calculated for the entire tissue, we found that the intensity traces predicted using CCS growth rates were significantly longer lived than typical clathrin-coated pits (Fig. 9 E), in striking contrast with the results of the lifetime distributions (Fig. 8 D). In all of the four embryos we have analyzed in this way, we recorded significant change in growth rate distributions at the apical surface, which correspond to slower CCS dynamics and elongated lifetimes over time (Fig. 9, D–F).
Figure 8.
3D tracking of CCSs in apical and basal surfaces of Drosophila amnioserosa. (A) Three sections of a confocal z-stack show apical, perinuclear, and basal regions of Drosophila amnioserosa cells expressing clathrin light chain fused with GFP (CLC-GFP). CCSs originating on organelles are observed mostly at the perinuclear regions as nondiffraction-limited bright fluorescent puncta (blobs). Bar, 10 µm. (B, left) Maximum projection image of a z-stack acquired at the amnioserosa tissue of a live Drosophila embryo. The image is divided into windows of 10 × 10 µm for determination of basal and apical surfaces (expanded in C). (right) Snapshot of CCS traces that are detected within the left panel, color-coded according to their axial positions. The projection of the entire 3D time-lapse acquisition and the corresponding CCS traces are shown in Video 3. (C) In each frame of the 3D time-lapse acquisition, axial positions of the apical and basal surfaces are determined separately for the 10 × 10 µm windows. Cartoon represents an amnioserosa cell oriented in a way that its apical surface faces the detection optics. The adjacent histogram shows the distribution of traces detected within the orange square shown in B with respect to their axial positions. The distribution is bimodal because of increased CCS density at the apical and basal surfaces and can be fit with a sum of two Gaussians. CCSs falling within one standard deviation of the respective means (± σ) are considered apical or basal. (D) A comparison of CCS lifetime distributions obtained from apical and basal amnioserosa with clathrin-coated pits (CCPs) detected in cultured BSC-1 cells stably expressing AP2-GFP (Amnioserosa: Nembryos = 2, Ncells = 75, and Ntraces = 124,013; CCPs: Ncells = 3 and Ntraces = 12,002).
Figure 9.
CME dynamics in Drosophila amnioserosa. (A) Amnioserosa tissue of a late Drosophila embryo is imaged for 4 min using confocal z-stacks acquired every 3 s. Representative frame is an image section at the middle of a stack. Red lines represent the boundaries between amnioserosa cell centers, which are marked by numbers. Cell boundaries determined in each frame of a 3D time-lapse acquisition are shown in Video 4. Bar, 10 µm. (B) Histograms show evolution of the growth rates corresponding to different cells selected from the amnioserosa tissue in A. Frequencies of the five phases are plotted as a function of time for the apical and basal surfaces. (C) Thumbnails represent 30-s-long 3D time-lapse acquisitions separated by intermissions. (D) Transformation of CCS growth rates at the basal and apical surfaces of the amnioserosa during dorsal closure of a Drosophila embryo. Each bar in the histograms represents the frequency of growth rates obtained from CCS traces detected in individual 30-s-long acquisitions. A significant change in the growth rates is observed at the apical surface. (E) CCS intensity profiles predicted using the growth rates in (D). The change in the apical CCS dynamics is observed as gradually elongated lifetime (right). No major change is observed in the basal surface dynamics (left). (F) Histograms show the CCS growth rates at the apical amnioserosa of three embryos. Increasing frequency of the plateau phase is a hallmark of reduced CCS dynamics. FD, fast dissolution; FF, fast formation; P, plateau; SD, slow dissolution; SF, slow formation.
Discussion
We show that clathrin coat growth rates can be used as quantitative reporters of CME dynamics in cellular contexts where errors associated with single-particle tracking are significant. Growth rate distributions obtained from short fragments of CCS traces offer the ability to assess clathrin dynamics from acquisitions shorter than the mean CCS lifetime. Gradual changes in endocytic dynamics can be detected in real time. Mean intensity profiles of the CCS traces that are used to generate growth rate distributions can be reconstructed by finding analogous growth rates in a library of trace clusters. These advantages make growth rate analysis a strong alternative to existing methodologies, which rely on quantifying lifetimes of individual CCSs.
Growth rate analyses reveal that physical factors increasing the energy cost of membrane deformation (e.g., in-plane tension, membrane–cytoskeleton adhesion, and membrane–substrate interactions) slow down formation and dissolution of CCSs. It has been shown that the same energy cost also reduces the curvature of CCSs (Saleem et al., 2015). In-plane tension is assumed to be homogenous in a cell, but membrane–cytoskeleton adhesion and membrane–substrate interactions are not uniform and, therefore, may induce spatial heterogeneities in dynamics and geometries of CCSs. This heterogeneity may account for the coexistence of clathrin-coated pits and plaques at the ventral surface of certain cell types (Saffarian et al., 2009). Growth rate analyses suggest that flat clathrin arrays and clathrin-coated pits have distinct formation dynamics. Even though flat lattices may acquire curvature over time (Avinoam et al., 2015), our data suggest that this mechanism cannot account for the formation of clathrin-coated pits. Transition of flat clathrin lattices to curved pits requires fourfold reduction in the projected area of clathrin coats on the x–y plane (Fig. S4). Such abrupt shape transformations are not observed in recent superresolved fluorescence acquisitions displaying the formation of clathrin-coated pits (Li et al., 2015), which suggests that curvature of the clathrin coat is constant during pit formation.
Strategies described here lay the groundwork for other quantitative assays that aim to elucidate endocytic processes in various multicellular contexts. Experimental and analytical deficiencies that distort CCS lifetime distributions are less detrimental to growth rates, as their quantification does not rely on tracing the complete lifetime of CCSs. Our study shows that CME dynamics can be assessed within tissues of developing organisms through quantification of the fluorescence growth rates of individual endocytic assemblies. Lifetime distributions obtained from Drosophila amnioserosa are overpopulated by traces that last <20 s, which would be regarded as abortive structures in in vitro assays. The intensity traces reproduced using growth rate distributions extended to durations longer than the mean clathrin-coated pit lifetime. Longer CCS lifetimes may be a result of the intrinsic tension of the amnioserosa tissue (Kiehart et al., 2000). Likewise, gradual increase in CCS lifetimes during dorsal closure may be induced by the changes in tension levels. This assumption is in accord with a recent study showing that reduction in volume of amnioserosa cells leads to increasing tension during dorsal closure (Saias et al., 2015). Drosophila amnioserosa cells undergo periodic but asynchronous shape changes at shorter time scales (Solon et al., 2009). Further research is needed to elucidate if rapid fluctuations of CCS growth rates observed in individual cells are correlated with temporal changes in amnioserosa mechanics. Such studies may also elucidate whether dynamics of CCSs can be used as reporters for mechanical states of cells and tissues.
Materials and methods
Fluorescence imaging
The imaging system was an Eclipse TI-E microscope (Nikon) equipped with a temperature controlled chamber, a CSU-W1 spinning disk confocal unit (Yokogawa Electric Corporation), a 100× objective lens (Nikon CFI Plan-Apochromat Lambda, NA 1.45) and an EMCCD camera (iXon DU897 Ultra; Andor Technology). 2D and 3D time series were obtained using NIS Elements image acquisition software.
S. Boulant (University of Heidelberg, Heidelberg, Germany) provided BSC-1, MDCK, and U373 cells stably expressing σ2-EGFP. Imaging of cultured cells was performed 8–24 h after plating on glass bottom dishes at 37°C ambient temperature (Greiner Bio-One), unless stated otherwise. Cells were imaged at a rate of 0.25–0.5 Hz and laser exposure of 50–300 ms per frame. Imaging medium was phenol red–free L15 (Thermo Fisher Scientific) supplemented with 10% FBS. Serum-free L15 was used for cholesterol depletion experiments. Final concentration of MβCD (Sigma-Aldrich) was 10 mM in Fig. 5, Fig. 7 C, and Video 2 and 4 mM in Fig. S2 B. Latrunculin B (Sigma-Aldrich) was used at the final concentration of 2 µM.
Drosophila embryos were harvested from a cross between male flies homozygous for GAL4-Arm and female flies homozygous for CLC-GFP (Bloomington Drosophila Stock Center). Eggs laid by this cross were collected and allowed to develop for ∼11 h at 25°C (stage 14 of development). The embryos were then mounted on a slide with glue (ventral side up), immersed in halocarbon oil (to prevent drying out), and covered with a coverslip. Amnioserosa tissues were imaged at 22°C using 3D time-lapse acquisitions containing ∼20 planes (400 nm apart) exposed for 50–100 ms. Temporal gap between adjacent z-stacks was 3 s.
Micropipette aspiration
Glass micropipettes (BF100-58-10) were pulled using a micropipette puller (P-97; Sutter Instrument). A custom stage was built to mount a Sutter Instrument BRM/E micromanipulator to the live-cell imaging platform. Suction pressure was controlled with a Sutter Instrument BRE110/E microinjection system. Cell aspiration was performed with a 5- to 10-µm microneedle at the dorsal surface of cells, and the clathrin activity at the ventral surface was recorded with spinning-disk fluorescence imaging. P. Selvin (University of Illinois at Urbana-Champaign, Urbana, IL) provided the micromanipulator and injection system.
Single-particle tracking
2D particle tracking was performed using the cmeAnalysis software (obtained from http://lccb.hms.harvard.edu/software.html) unless stated otherwise. TraCKer software is a less sophisticated but faster tracking algorithm that lacks the advanced forward-backward-forward rechecking and intelligent thresholding of cmeAnalysis. It also lacks the 2D Gaussian fitting of point-spread functions and uses integrated pixel intensity as the CCS signal. TraCKer uses a simple threshold determined over a Mexican hat filtered image for detection of fluorescent spots. CCS positions are located using the intensity-weighted center of the point-spread function. In this work, TraCKer was used for the sole purpose of testing growth rate analysis on IMSs.
cmeAnalysis software occasionally detects objects that last a single frame or persist consistently in the background without following an intensity path that could be considered as a CCS. We implemented a sorting scheme for these traces, requiring that they be at least three frames long and at some point meet statistical criteria for demonstrating a linear increase or decrease in intensity (corresponding to CCS growth and dissolution, respectively). We went over every three or four consecutive intensity points (three for traces that last for <10 frames and four for longer traces) and performed a least-squares fit. Traces that had no fits with r2 value >0.75 were rejected. Rejected traces were not used in calculation of growth rate distributions. Traces with at least two separate high r2 values of both positive slope (growth) and negative slope (dissolution) were valued most highly. Fig. S5 shows the classification of CCS traces obtained from amnioserosa tissues.
3D tracking of CCSs within amnioserosa tissues
Each z-plane of a 3D time-lapse movie was analyzed using cmeAnalysis software. Detected 2D traces were then run through our trace-rejection scheme. The resulting data were analyzed to combine traces that occur at the same lateral position in two adjacent z-planes. Coincident traces had to be within one pixel (160 nm) x–y distance for at least three consecutive frames. For each time point in the movie, the resulting trace contains the maximum intensity value of all traces considered for combination. Axial positions were assigned by calculating the intensity-weighted mean z-position of all traces considered. The algorithm for trace combination ran from the outermost z-planes to the innermost (alternating between the top and the bottom) to ensure that there was no directional bias and all possible trace combinations were considered. The resulting data structure will have the duplicate traces deleted and contain subplane z-position data where possible.
Classification of apical and basal CCSs and blobs
Because of the curvature of the dorsal surface, cells in the amnioserosa tissue are not coplanar. Therefore, the field of view was divided into 64 equal square regions, and apical and basal surfaces were determined for each region independently (Fig. 8 B). Axial positions of the traces detected inside each square region were put together within in a two-frame temporal radius. These values were binned into discrete z-plane positions, and the resulting count-value graph was fit with two Gaussian functions. The fit for the apical surface has a low standard deviation. The basal surface was found at a higher axial position and the corresponding Gaussian fit had a larger standard deviation in general. Traces found at the mean or within one standard deviation of their respective Gaussian fits were classified as apical or basal for each of the 64 squares. The brightest 2% of all traces were classified as blobs and were excluded from the apical and basal populations.
Determining amnioserosa cell boundaries
To determine the positions of amnioserosa nuclei, the middle imaging plane of each z-stack was filtered with a Gaussian filter. Each frame was then inverted and local maxima were determined using the FastPeakFind function (A. Natan, PULSE Institute, Stanford, CA). After multiple nuclei positions were determined for each frame, they were tracked using the TraCKer algorithm. Once the centers were known and connected for each frame, the boundaries were determined by creating a Voronoi diagram using the cell centers in each frame.
Growth rate distributions
Slopes were extracted from CCS traces that pass the trace rejection scheme. We found that CCS lifetime distributions of obtained from accepted (nonrejected) traces were very similar to those reproduced by others who used different parameters to distinguish genuine endocytic transporters (Fig. S5; Aguet et al., 2013). Each trace was normalized by subtracting a global background and dividing by the new maximum of the trace. From this normalized trace, every 12-s interval was used in a least-squares fit to determine the slope of the trace at all frames (Fig. 1 C). A trace had to be at least 12 s long to be considered for slope extraction. 12 s was chosen because it evenly divides 1-, 2-, 3-, and 4-s frame rates, which are the most common frame rates used. An arbitrary bin width (0.03) was assigned for the histograms displaying the five distinct growth phases.
Hierarchical clustering of intensity profiles and library lookup
Representative traces were created by combining similar intensity traces within a single cell using a standard hierarchical clustering algorithm. The metric used was the mean squared Euclidean distance, with the condition that the lifetimes of the traces were within 10 s of each other. To compare clusters, we used complete linkage clustering; that is, we used the distance between the farthest two component traces. After clustering, clusters with a minimum of eight traces were added to our trace library. For such clusters, the mean of the component traces was calculated. These mean traces were normalized to the mean maximum trace intensity of the cell to compensate for cells’ differing expression levels and imaging conditions. Additionally, for each of these clusters, a single growth rate histogram was generated from the combined data of the constituent traces. The library was assembled from clusters obtained from the following cell types and conditions: BSC-1 cells expressing σ2-GFP (before and during aspiration), polarized MDCK cells expressing σ2-GFP (apical and basal surfaces), and U373 cells expressing LCa-mCherry.
Growth rate histograms of test clusters were compared with the histograms of library clusters using the squared Euclidean metric. If the input histogram was generated from trace data that were confined spatially (e.g., single cell in a culture or tissue) and/or temporally (e.g., half a minute long acquisition of the whole tissue), predicted traces are the weighted averages of the 10 closest cluster matches from the CCS library (Fig. 7 C, Video 2, and Fig. 9 E). For this procedure, the library clusters were sorted from closest to farthest by calculating the squared Euclidean distance between their growth slope histograms, and each cluster’s contribution to the mean is weighted by the factor (10 − i), where i is the cluster’s index in the sorted list and the first index is i = 0.
Online supplemental material
Fig. S1 provides examples of hierarchical clustering in cells before and during micropipette aspiration. Fig. S2 describes the difference between traces found in control cells and in those treated with MβCD and how latrunculin B affects that response. Fig. S3 compares the root mean squared displacement of traces detected in the amnioserosa tissue versus those found in U373 cells. Fig. S4 demonstrates the differences between a flat clathrin array and a curved clathrin pit. Fig. S5 shows the intensity and lifetime distributions of CCS traces detected in amnioserosa cells based on their classification. Video 1 gives a frame-by-frame depiction of CCS growth rates throughout a cell and its corresponding growth rate histogram. Video 2 gives a frame-by-frame depiction of growth rate histograms in three separate cells treated simultaneously with MβCD and displays corresponding intensity traces derived from our hierarchical clustering library. Video 3 shows a 3D maximum projection image of amnioserosa tissue with an overlay marking CCSs classified as blobs. It also shows traces colored to demonstrate the axial position. Video 4 shows the boundaries of individual cells in the amnioserosa over the course of the movie. Finally, the MATLAB-based software used for our analysis is provided to be used or modified as fits the user’s needs. The software package includes a README file, clearly commented code, and example data files. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201604128/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Steve Boulant for BSC-1, MDCK, and U373 cells stably expressing σ2-EGFP; Dr. Paul Selvin for the micromanipulator and injection system; Vannimul Hem (Ohio State University) for the help with the micromanipulation experiments; and Dr. Michael Sheetz (Columbia University) and members of our laboratory for helpful discussions.
This work was supported by National Institutes of Health grant R01 AI121124.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CCS
- clathrin-coated structure
- CLC
- clathrin light chain
- CME
- clathrin-mediated endocytosis
- IMS
- integrated movie segment
- MβCD
- methyl-β-cyclodextrin
References
- Aghamohammadzadeh S., and Ayscough K.R.. 2009. Differential requirements for actin during yeast and mammalian endocytosis. Nat. Cell Biol. 11:1039–1042. 10.1038/ncb1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguet F., Antonescu C.N., Mettlen M., Schmid S.L., and Danuser G.. 2013. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell. 26:279–291. 10.1016/j.devcel.2013.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apodaca G. 2002. Modulation of membrane traffic by mechanical stimuli. Am. J. Physiol. Renal Physiol. 282:F179–F190. [DOI] [PubMed] [Google Scholar]
- Avinoam O., Schorb M., Beese C.J., Briggs J.A.G., and Kaksonen M.. 2015. ENDOCYTOSIS. Endocytic sites mature by continuous bending and remodeling of the clathrin coat. Science. 348:1369–1372. 10.1126/science.aaa9555 [DOI] [PubMed] [Google Scholar]
- Batchelder E.M., and Yarar D.. 2010. Differential requirements for clathrin-dependent endocytosis at sites of cell-substrate adhesion. Mol. Biol. Cell. 21:3070–3079. 10.1091/mbc.E09-12-1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E., Saffarian S., Zhang R., and Kirchhausen T.. 2010. Roles of AP-2 in clathrin-mediated endocytosis. PLoS One. 5:e10597 10.1371/journal.pone.0010597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulant S., Kural C., Zeeh J.-C., Ubelmann F., and Kirchhausen T.. 2011. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat. Cell Biol. 13:1124–1131. 10.1038/ncb2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocucci E., Aguet F., Boulant S., and Kirchhausen T.. 2012. The first five seconds in the life of a clathrin-coated pit. Cell. 150:495–507. 10.1016/j.cell.2012.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner S.D., and Schmid S.L.. 2003. Regulated portals of entry into the cell. Nature. 422:37–44. 10.1038/nature01451 [DOI] [PubMed] [Google Scholar]
- Cureton D.K., Massol R.H., Whelan S.P.J., and Kirchhausen T.. 2010. The length of vesicular stomatitis virus particles dictates a need for actin assembly during clathrin-dependent endocytosis. PLoS Pathog. 6:e1001127 10.1371/journal.ppat.1001127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Ting-Beall H.P., and Sheetz M.P.. 1997. The secretion-coupled endocytosis correlates with membrane tension changes in RBL 2H3 cells. J. Gen. Physiol. 110:1–10. 10.1085/jgp.110.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diz-Muñoz A., Fletcher D.A., and Weiner O.D.. 2013. Use the force: membrane tension as an organizer of cell shape and motility. Trends Cell Biol. 23:47–53. 10.1016/j.tcb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M., Boll W., Van Oijen A., Hariharan R., Chandran K., Nibert M.L., and Kirchhausen T.. 2004. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 118:591–605. 10.1016/j.cell.2004.08.017 [DOI] [PubMed] [Google Scholar]
- Gaidarov I., Santini F., Warren R.A., and Keen J.H.. 1999. Spatial control of coated-pit dynamics in living cells. Nat. Cell Biol. 1:1–7. http://dx.doi.org/10.1038/8971 [DOI] [PubMed] [Google Scholar]
- Gauthier N.C., Masters T.A., and Sheetz M.P.. 2012. Mechanical feedback between membrane tension and dynamics. Trends Cell Biol. 22:527–535. 10.1016/j.tcb.2012.07.005 [DOI] [PubMed] [Google Scholar]
- Grove J., Metcalf D.J., Knight A.E., Wavre-Shapton S.T., Sun T., Protonotarios E.D., Griffin L.D., Lippincott-Schwartz J., and Marsh M.. 2014. Flat clathrin lattices: stable features of the plasma membrane. Mol. Biol. Cell. 25:3581–3594. 10.1091/mbc.E14-06-1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.H., Cortesio C.L., and Drubin D.G.. 2015. Machine-learning-based analysis in genome-edited cells reveals the efficiency of clathrin-mediated endocytosis. Cell Reports. 12:2121–2130. 10.1016/j.celrep.2015.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houk A.R., Jilkine A., Mejean C.O., Boltyanskiy R., Dufresne E.R., Angenent S.B., Altschuler S.J., Wu L.F., and Weiner O.D.. 2012. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell. 148:175–188. 10.1016/j.cell.2011.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto A., Woolner S., and Martin P.. 2002. Dynamic analysis of dorsal closure in Drosophila: from genetics to cell biology. Dev. Cell. 3:9–19. 10.1016/S1534-5807(02)00208-3 [DOI] [PubMed] [Google Scholar]
- Kaur S., Fielding A.B., Gassner G., Carter N.J., and Royle S.J.. 2014. An unmet actin requirement explains the mitotic inhibition of clathrin-mediated endocytosis. eLife. 3:e00829 10.7554/eLife.00829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatibzadeh N., Gupta S., Farrell B., Brownell W.E., and Anvari B.. 2012. Effects of cholesterol on nano-mechanical properties of the living cell plasma membrane. Soft Matter. 8:8350–8360. 10.1039/c2sm25263e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehart D.P., Galbraith C.G., Edwards K.A., Rickoll W.L., and Montague R.A.. 2000. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. J. Cell Biol. 149:471–490. 10.1083/jcb.149.2.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kural C., and Kirchhausen T.. 2012. Live-cell imaging of clathrin coats. Methods Enzymol. 505:59–80. 10.1016/B978-0-12-388448-0.00012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kural C., Tacheva-Grigorova S.K., Boulant S., Cocucci E., Baust T., Duarte D., and Kirchhausen T.. 2012. Dynamics of intracellular clathrin/AP1- and clathrin/AP3-containing carriers. Cell Reports. 2:1111–1119. 10.1016/j.celrep.2012.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kural C., Akatay A.A., Gaudin R., Chen B.-C., Legant W.R., Betzig E., and Kirchhausen T.. 2015. Asymmetric formation of coated pits on dorsal and ventral surfaces at the leading edges of motile cells and on protrusions of immobile cells. Mol. Biol. Cell. 26:2044–2053. 10.1091/mbc.E15-01-0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Shao L., Chen B.-C., Zhang X., Zhang M., Moses B., Milkie D.E., Beach J.R., Hammer J.A. III, Pasham M., et al. . 2015. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 349:aab3500 10.1126/science.aab3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerke D., Mettlen M., Yarar D., Jaqaman K., Jaqaman H., Danuser G., and Schmid S.L.. 2009. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 7:e57 10.1371/journal.pbio.1000057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massol R.H., Boll W., Griffin A.M., and Kirchhausen T.. 2006. A burst of auxilin recruitment determines the onset of clathrin-coated vesicle uncoating. Proc. Natl. Acad. Sci. USA. 103:10265–10270. 10.1073/pnas.0603369103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield C.J., Feldman M.E., Wan L., and Almers W.. 2002. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat. Cell Biol. 4:691–698. 10.1038/ncb837 [DOI] [PubMed] [Google Scholar]
- Mettlen M., and Danuser G.. 2014. Imaging and modeling the dynamics of clathrin-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 6:a017038 10.1101/cshperspect.a017038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen M., Loerke D., Yarar D., Danuser G., and Schmid S.L.. 2010. Cargo- and adaptor-specific mechanisms regulate clathrin-mediated endocytosis. J. Cell Biol. 188:919–933. 10.1083/jcb.200908078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouille P.-A., Ahmadi P., Brunet A.-C., and Farge E.. 2009. Mechanical signals trigger Myosin II redistribution and mesoderm invagination in Drosophila embryos. Sci. Signal. 2:ra16 10.1126/scisignal.2000098 [DOI] [PubMed] [Google Scholar]
- Raucher D., and Sheetz M.P.. 1999. Membrane expansion increases endocytosis rate during mitosis. J. Cell Biol. 144:497–506. 10.1083/jcb.144.3.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffarian S., and Kirchhausen T.. 2008. Differential evanescence nanometry: live-cell fluorescence measurements with 10-nm axial resolution on the plasma membrane. Biophys. J. 94:2333–2342. 10.1529/biophysj.107.117234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffarian S., Cocucci E., and Kirchhausen T.. 2009. Distinct dynamics of endocytic clathrin-coated pits and coated plaques. PLoS Biol. 7:e1000191 10.1371/journal.pbio.1000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saias L., Swoger J., D’Angelo A., Hayes P., Colombelli J., Sharpe J., Salbreux G., and Solon J.. 2015. Decrease in cell volume generates contractile forces driving dorsal closure. Dev. Cell. 33:611–621. 10.1016/j.devcel.2015.03.016 [DOI] [PubMed] [Google Scholar]
- Saleem M., Morlot S., Hohendahl A., Manzi J., Lenz M., and Roux A.. 2015. A balance between membrane elasticity and polymerization energy sets the shape of spherical clathrin coats. Nat. Commun. 6:6249 10.1038/ncomms7249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz M.P. 2001. Cell control by membrane-cytoskeleton adhesion. Nat. Rev. Mol. Cell Biol. 2:392–396. 10.1038/35073095 [DOI] [PubMed] [Google Scholar]
- Snijder B., Sacher R., Rämö P., Damm E.-M., Liberali P., and Pelkmans L.. 2009. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 461:520–523. 10.1038/nature08282 [DOI] [PubMed] [Google Scholar]
- Solon J., Kaya-Copur A., Colombelli J., and Brunner D.. 2009. Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell. 137:1331–1342. 10.1016/j.cell.2009.03.050 [DOI] [PubMed] [Google Scholar]
- Subtil A., Gaidarov I., Kobylarz K., Lampson M.A., Keen J.H., and McGraw T.E.. 1999. Acute cholesterol depletion inhibits clathrin-coated pit budding. Proc. Natl. Acad. Sci. USA. 96:6775–6780. 10.1073/pnas.96.12.6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X., Heureaux J., and Liu A.P.. 2015. Cell spreading area regulates clathrin-coated pit dynamics on micropatterned substrate. Integr Biol (Camb). 7:1033–1043. 10.1039/C5IB00111K [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.J., Perrais D., and Merrifield C.J.. 2011. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 9:e1000604 10.1371/journal.pbio.1000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.