Abstract
Background and Aims Poa subgenus Poa supersect. Homalopoa has diversified extensively in the Americas. Over half of the species in the supersection are diclinous; most of these are from the New World, while a few are from South-East Asia. Diclinism in Homalopoa can be divided into three main types: gynomonoecism, gynodioecism and dioecism. Here the sampling of species of New World Homalopoa is expanded to date its origin and diversification in North and South America and examine the evolution and origin of the breeding system diversity.
Methods A total of 124 specimens were included in the matrix, of which 89 are species of Poa supersect. Homalopoa sections Acutifoliae, Anthochloa, Brizoides, Dasypoa, Dioicopoa, Dissanthelium, Homalopoa sensu lato (s.l.), Madropoa and Tovarochloa, and the informal Punapoa group. Bayesian and parsimony analyses were conducted on the data sets based on four markers: the nuclear ribosomal internal tanscribed spacer (ITS) and external transcribed spacer (ETS), and plastid trnT-L and trnL-F. Dating analyses were performed on a reduced Poa matrix and enlarged Poaceae outgroup to utilize fossils as calibration points. A relaxed Bayesian molecular clock method was used.
Key Results Hermaphroditism appears to be pleisiomorphic in the monophyletic Poa supersect. Homalopoa, which is suggested to have originated in Eurasia 8·4–4·2 million years ago (Mya). The ancestor of Poa supersect. Homalopoa radiated throughout the New World in the Late Miocene–Early Pliocene, with major lineages originating during the Pliocene to Pleistocene (5–2 Mya). Breeding systems are linked to geographic areas, showing an evolutionary pattern associated with different habitats. At least three major pathways from hermaphroditism to diclinism are inferred in New World Homalopoa: two leading to dioecism, one via gynodioecism in South America and another directly from hermaphroditism in North America, a result that needs to be checked with a broader sampling of diclinous species in North America. A third pathway leads from hermaphroditism to gynomonoecism in Andean species of South America, with strictly pistillate species evolving in the highest altitudes.
Conclusions Divergence dating provides a temporal context to the evolution of breeding systems in New World Poa supersect. Homalopoa. The results are consistent with the infrageneric classification in part; monophyletic sections are confirmed, it is proposed to reclassify species of sect. Acutifoliae, Dasypoa and Homalopoa s.l. and it is acknowledged that revision of the infrageneric taxonomy of the gynomonoecious species is needed.
Keywords: Breeding system, diclinism, dioecism, DNA, gynomonoecism, gynodioecism, molecular dating, molecular phylogeny, Poa, Homalopoa, South America, North America
INTRODUCTION
Poa L. (Poaceae, Pooideae, Poeae) is one of the largest genera of grasses, with an estimated 530 species distributed worldwide (Soreng et al., 2007, 2015b; Gillespie et al., 2008), mostly in high altitudes and/or latitudes of both hemispheres (Hartley, 1961). Poa is generally a morphologically well-marked and monophyletic genus (Soreng et al., 1990, 2015a; Gillespie and Soreng, 2005; Gillespie et al., 2007, 2008). Molecular studies have also supported uniting 11 genera with Poa, including six whose species are now considered members of supersect. Homalopoa: Anthochloa Nees & Meyen, Dasypoa Pilg., Dissanthelium Trin. and Tovarochloa T.D. Macfarl. & But from South America; and Austrofestuca (Tzvelev) E. B. Alexeev sensu stricto (s.s.) and Neuropoa W. D. Clayton from Australia (Gillespie and Soreng, 2005; Gillespie et al., 2007, 2008, 2009; Jacobs et al., 2008; Soreng et al., 2009; Refulio-Rodriguez et al., 2012). The South American genus Aphanelytrum (Hack.) Hack. has also been confirmed as part of the Homalopoa clade but has not yet been synonymized under Poa (Gillespie et al., 2008; Refulio-Rodriguez et al., 2012; P. M. Peterson and R. J. Soreng, unpubl. res.).
Recent molecular phylogenetic studies have substantially influenced the classification of Poa. Poa is currently divided into five subgenera: Sylvestres, Ochlopoa, Pseudopoa, Stenopoa and Poa, corresponding to the five major plastid clades in the genus (Gillespie et al., 2007; updated in Gillespie et al., 2008; Soreng et al., 2010). The largest subgenus, Poa, is further divided into two clades corresponding to supersections Poa and Homalopoa. The latter was first recognized as a clade based on plastid restriction site data (Soreng, 1990; Gillespie and Soreng, 2005), and then confirmed with plastid sequence data and formally named (Gillespie et al., 2007); however, when analysing nuclear ribosomal DNA (nrDNA) sequences, species of the two supersections were intermingled in a Poa–Homalopoa clade or resolved in an independent clade (Nosov and Rodionov, 2008; Gillespie et al., 2009; Nosov et al., 2015) for which no corresponding plastid type is known (‘X-clade’ of Gillespie et al., 2009).
Poa supersect. Homalopoa (hereafter excluding species with X-clade nrDNA) is a large and diversified clade in terms of both species and sections, and includes about half of the species in Poa; it is currently divided into ten sections worldwide along with the Punapoa informal assemblage (Fig. 1; Table 1). The New World represents a major centre of diversity with 144 endemic Homalopoa species. With many species and morphological diversity and adaptations to many niches, P. supersect. Homalopoa is difficult to characterize morphologically. Species are ephemeral annuals to long-lived perennials, ranging in size from a few centimetres to 1·5 m tall (Gillespie and Soreng, 2005), upper culm sheaths are generally fused at the base for more than a quarter of the length, the lower sheaths are commonly distinctly to strongly compressed, and lemmas are generally strongly five-veined. They occupy a wide diversity of habitats, including moist temperate deciduous forests, coniferous forests, dry steppe, arid sub-tropical deserts, pampas, paramo, puna, wet saline meadows, dunes, low-arctic meadows and acidic to basic and sometimes ultramafic substrates, among others.
Fig. 1.
Morphological variation among species of Poa supersect. Homalopoa of the New World. (A) Poa cusickii subsp. purpurascens (clade C); (B) P. unispiculata (clade E); (C) P. bergii (clade D), photo: Daniel Testoni; (D) P. cuspidata; (E) P. aequigluma (clade E); (F) Robert Soreng holding a specimen of P. horridula (clade F); (G) P. lanigera (clade D); clades are given following the results as presented in Supplementary Data Fig. S1.
Table 1.
Classification of Poa subg. Poa supersect. Homalopoa (Giussani 2000; Negritto and Anton, 2000; Gillespie et al., 2007; updated in Soreng et al., 2009; Refulio-Rodriguez et al., 2012)
| Sections | New World/BS | Old World/BS | Worldwide | Geographical distribution | ||
|---|---|---|---|---|---|---|
| Acutifoliae | 2 | h, gd | 0 | 2 | S Andes | |
| Anthochloa | 1 | gm | 0 | 1 | Andes | |
| Brizoides | 0 | 48 | h | 48 | Australasia | |
| Dasypoa | 3 | h, gm | 0 | 3 | Andes (1 sp. also in Mexico and Guatemala) | |
| Dioicopoa | 31 | di (gd) | 0 | 31 | S South America (1 sp. S USA) | |
| Dissanthelium s.s. | 7 | gm | 0 | 7 | Andes (1 subsp. in Mexico) | |
| Homalopoa s.l. (incl. Plicatae) | 59 | h, gm, sgm (gd, di) | 28 | h, gm, sgm (gd, di) | 87 | America, Eurasia |
| Madropoa | 22 | di (p, gd, sgm) | 0 | 22 | North America (1 sp. in Chile P. pfisteri) | |
| Monandropoa | 1 | h | 0 | 1 | S South America | |
| Punapoa informal group | 12 | p, gd, gm | 0 | 12 | Andes (2 spp. in Mexico) | |
| Tovarochloa | 1 | h | 0 | 1 | Peru | |
| Unplaced | 3 | h (gm) | 3 | America | ||
| Genus Aphanelytrum (not transferred yet) | 2 | h | 0 | 2 | N Andes | |
| Total | 144 | 76 | 220 | |||
Numbers of species in the Americas (New World), the Old World (including Australasia) and worldwide are given for each section, the Punapoa informal species group and Aphanelytrum (confirmed member of the supersect. Homalopoa clade, but not yet synonymized under Poa) (taxonomic placement data derived from Tzvelev, 1983; Probatova, 2003; Soreng et al., 2003; Zhu et al., 2006; Soreng, 2007; Giussani et al., 2012; Refulio-Rodriguez et al., 2012; Soreng and Peterson, 2012).
Most frequent breeding system (BS) in sections of supersect. Homalopoa (infrequent to rare types in parentheses): h, hermaphrodite; gm, simple gynomonoecy; sgm, sequentially adjusted gynomonoecy; gd, gynodioecy; p, strictly pistillate; di, dioecy.
Section Homalopoa sensu latu (s.l.) is the largest, most widespread and most heterogeneous section in supersect. Homalopoa, and is found in both the Americas and Eurasia (Soreng, 2007). In the strict sense, sect. Homalopoa comprises approximately five Eurasian species [and possibly the North American diploid P. occidentalis (Vasey) Vasey], but remains poorly defined and may also be heterogeneous. In the Old World, only one section is endemic, sect. Brizoides of Australasia. Following Giussani (2000, for Dioicopoa), Negritto and Anton (2000, for gynomonoecious Poa, and sect. Monandropoa Parodi), Soreng et al. (2003), Gillespie et al. (2007, for sect. Anthochloa), Zuloaga et al. (2008) and Refulio-Rodriguez et al. (2012, for sect. Dissanthelium and Tovarochloa), eight sections are restricted to the Americas (Table 1): Anthochloa and Tovarochloa are endemic to the southern Andes; Dasypoa and Dissanthelium are primarily Andean, each with one disjunct species between the Andes and Mesoamerica [P. scaberula Hook. f. and P. calycina (J. Presl) Kunth, respectively]; Dioicopoa is primarily southern South American, with one species in the southern USA (P. arachnifera Torr.); Monandropoa includes a single species close to P. scaberula, endemic to Catamarca and Tucumán provinces of Argentina, above 3000 m; Acutifolae is endemic to the central Chilean–Argentinean Andes; and Madropoa is North American (with one Chilean species, P. pfisteri Soreng, tentatively placed here by Soreng and Peterson, 2008). In addition, Punapoa is recognized as an informal species group of the Andes, with two species disjunct between the Andes and Mexico (P. chamaeclinos Pilg. and P. gymnantha Pilg.) (Soreng et al., 2003; Gillespie et al., 2007; Giussani et al., 2012; Soreng and Peterson, 2012). Aphanelytrum (Hack.) Hack., when synonymized under Poa (P. M. Peterson and R. J. Soreng, unpubl. res.), may represent a new section from the northern Andes.
Species of Poa show an exceptional diversity in breeding systems represented by the occurrence of hermaphroditism to dioecism (Parodi, 1936; Marsh, 1952; Nicora, 1978; Connor, 1979; Anton and Connor, 1995; Giussani, 2000; Negritto and Anton, 2000; Soreng, 1991; Soreng and Keil, 2003). Although hermaphroditism is the most common reproductive system among Poa species, dicliny was estimated to occur in about 30 % of the species (R. J. Soreng, unpubl data; based on approx. 166 diclinous species out of a total of approx. 530 Poa species). Most of the species diversity and variation in dicliny in Poa is found in sections and informal groups of Poa subg. Poa supersect. Homalopoa (Table 1).
While some species spread vegetatively by rhizomes forming patches or spreading, apomixis is also a common mode of asexual propagation in high polyploid species of Poa, partially or fully supplanting sexual reproduction to ensure the production of seed (Kelley et al., 2009) or bulbils. While well known in Poa of the northern hemisphere (Kellogg, 1989; Kelley et al., 2009), apomictic production of seed was assumed or strongly suspected in pistillate populations of Andean species of Poa (Anton and Connor, 1995), and first confirmed anatomically in P. gymnantha in South America (Negritto et al., 2008). Pseudovivipary is another method of vegetative or apomictic reproduction, producing vegetative propagules (bulbils) in the spikelets instead of normal florets; in the Americas, it is mostly associated with species from higher latitude regions of extreme cold weather or areas of high rainfall in the Circumboreal biome and the Patagonian floristic regions (Moore and Doggett, 1976; Pierce et al., 2003), and with differences in the onset of the rainy season (Ofir and Kigel, 2014).
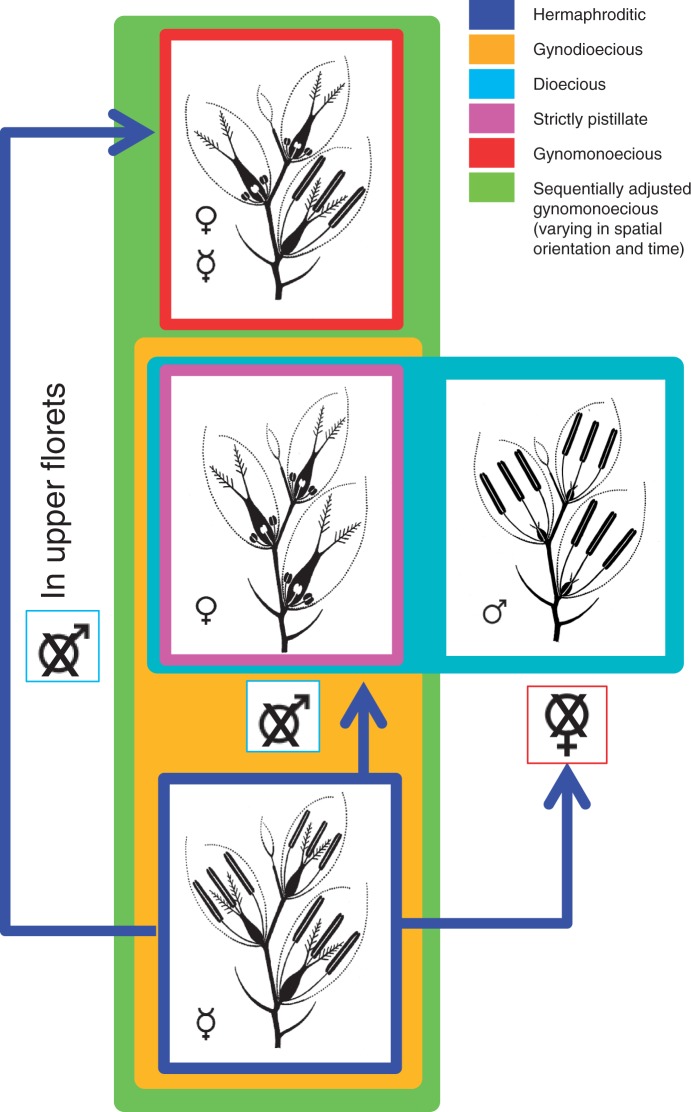
Diclinism, in the broad sense, includes any occurrence or arrangement of unisexual flowers (with or without co-occurrence of hermaphroditic flowers) within or among individuals of a species, hence including monoecy and all its sub-types (gynomonoecy, andromonoecy), androdioecy, gynodioecy, dioecy and trioecy. Figure 2 represents all types of floral arrangements found within Poa; dicliny is well represented in the Americas, and is also found in Asia, New Zealand and the Sub-antarctic islands (Anton and Connor, 1995; Soreng, 2007; Edgar and Connor, 2010; Giussani et al., 2012). Gynomonoecy, in its simple form, where spikelets bear pistillate upper florets and perfect basal florets, is well represented among supersect. Homalopoa species of the informal group Punapoa, sect. Anthochloa, sect. Dissanthelium, sect. Dasypoa and sect. Homalopoa s.l. in South America (Anton, 1978; Negritto and Anton, 2000, 2006; Soreng et al., 2003; Negritto et al., 2008), while simple gynomonoecy is only present in non-Homalopoa species in North America [P. abbreviata R. Br. and P. suksdorfii (Beal) Vasey ex Piper of P. subg. Stenopoa sect. Abbreviatae, and non-native P. annua L., P. infirma Kunth and P. supina Schrad. of subg. Ochlopoa sect. Micrantherae]. As a sub-type of gynomonoecy, sequentially adjusted gynomonoecy (Soreng and Keil, 2003) is found mainly in North American sect. Madropoa and refers to temporal variation in the production of pistillate flowers within spikelets to whole inflorescences, the percentage of pistillate flowers produced in later inflorescences increasing through the growing season in some plants, while inflorescences remain perfect in other plants. Gynodioecious and dioecious species have sexes separated in different individuals within a population. Species having mixtures of individuals with all or mainly hermaphrodite flowers and individuals with only pistillate flowers are known as gynodioecious. Gynodioecism is relatively infrequent in Poa, known in only 12 species (including nine species of supersect. Homalopoa, all from the New World). Dioecism is the most extreme expression of sexual differentiation, with staminate flowers and pistillate flowers separated in different individuals, usually in a ratio of 1:1. In the genus Poa, dioecy is restricted to the Americas within supersect. Homalopoa, and to New Zealand (where it is confined to five species with X-clade nrDNA); sexual dimorphism developed only in species of the South American sect. Dioicopoa. The occurrence of dioecism implies the suppression of both maleness and femaleness in respective individuals, while all other reproductive systems in New World Homalopoa result from different timing and zone of action of maleness suppression (Fig. 2).
Fig. 2.
Breeding systems in Poa supersect. Homalopoa; diagram showing sexes of flowers within spikelets for each breeding system. Blue: hermaphroditic; the diagram shows flowers with well-developed pistils and stamens in all flowers of the spikelet. Yellow: gynodioecious; species having a mixture of individuals with hermaphroditic flowers and individuals with only pistillate flowers. Light blue: dioecious; represents staminate flowers and pistillate flowers separated in different individuals. Pink: represents individuals with strictly pistillate flowers in all spikelets. Red: gynomonoecious; individuals with spikelets with basal flowers hermaphroditic and upper flowers pistillate. Green: sequentially adjusted gynomonoecious; sex of flowers varying in spatial orientation and time: individuals with pistillate flowers within spikelets to whole inflorescences, or pistillate flowers produced in later inflorescences increasing through the growing season in some plants, while inflorescences remain perfect in other plants. Crosses over sex diagrams denote suppression of maleness or femaleness.
Dioecious species are present in disjunct areas in the Americas, and taxonomically grouped in two sections within supersect. Homalopoa: Dioicopoa and Madropoa. Morphologically, sect. Dioicopoa is well diversified and marked by sexual dimorphism. Florets of pistillate plants are usually abundantly pubescent, while those of staminate plants are glabrous or sparsely pubescent. Moreover, pistillate florets are generally larger and fewer per spikelet than staminate florets (Giussani, 2000; Giussani et al., 2000, 2008, 2012). In addition, habitat preference favouring the establishment of pistillate individuals (Giussani et al., 1996; Bertiller et al., 2000, 2002) or different responses of sexes to competition with neighbours under grazing pressure (Giussani and Collantes, 1997; Graff et al., 2013) have been reported. In contrast, dimorphism between sexes in the North American dioecious sect. Madropoa has not been found or is negligible (Soreng, 1991). Although dioecy has presumably originated at least three times within Poa (based on the tree in Gillespie et al., 2009), the monophyly and origins of sect. Dioicopoa and sect. Madropoa are still in need of clarification. The relationship of dioecious species to gynodioecious and gynomonoecious species could reveal possible ancestral pathways to dioecy (Anton and Connor, 1995).
Cytogenetically, Poa is characterized in having medium to large sized chromosomes with a base number of x = 7 and a high frequency of polyploid species. Only about 9 % of the reported species are diploids, with an additional 4–6 % having both diploid and polyploid populations, mostly from Europe and few from Asia (Moore, 1982; Rodionov et al., 2010; Soreng et al., 2010). Only three diploid species (2n = 14), P. lettermanii Vasey, P. pseudoabbreviata Roshev. (sect. Abbreviatae) and P. occidentalis (sect. Homalopoa), are native to North America, and none has been reported from South America (apart from the introduced diploid species P. infirma, P. supina and P. trivialis L.). In the southern hemisphere, only tetraploid and octoploid to high polyploid species have been reported (Hair and Beuzenberg, 1961; Hunziker, 1978; Murray et al., 2005); in South America, dioecious species were found to have 2n = 28 or 56 (Saura, 1943, 1948; Bowden and Senn, 1962; Hunziker, 1978; Guillin et al., 1995), while an exclusively pistillate species, P. gymnantha, was counted as 2n = 70 (Negritto et al., 2008).
Taking into account reproductive systems of species, and the distribution of high frequency of diploids, the origin of the genus was suggested to have been in Eurasia, with hermaphrodite species arriving in North America via Beringia (Soreng, 1990; Anton and Connor, 1995), and then diversifying into different gynomonoecious, gynodioecious and dioecious lineages in North and South America. The most relevant hypotheses on the evolution of sexes within Poa have been presented by Anton and Connor (1995), who postulated that after migrating to North America, part of the migrating hermaphrodite species of Poa derived to dioecism, probably via gynodioecism, and to a much lesser extent from hermaphrodite to gynomonoecism. In South America, they also postulated that part of the migrating hermaphrodite species of Poa differentiated into gynomonoecism, with apomictic pistillate populations evolving in some alpine species, while another part of migrating hermaphrodite species of Poa independently differentiated into gynodioecism, from which dioecism evolved in the same area. Few hypotheses have been offered to relate any geological or climatological events to the origin and diversification of Poa. In Australasia, Poa is suggested to have diversified within the last 4·3 million years, correlating with the appearance of grasslands in the mid-Pliocene (Birch et al., 2014), and a recent rapid radiation was reported for the Australian alpine Poa species (Griffin and Hoffmann, 2014). However, no time for the arrival and diversification of the breeding system has been postulated for species in the Americas.
While the phylogeny of the genus Poa has been investigated in previous studies and the taxonomy has been generally sorted out, we here increase sampling to understand the origin and diversification of Poa supersect. Homalopoa and its breeding systems in North and South America. Dating of major phyletic nodes in Poa is hampered by the absence of fossils; hence, to date divergence time within supersect. Homalopoa, we selected stratigraphically well-dated and taxonomically identifiable micro- and macrofossils of the Poaceae family for calibration points. Molecular phylogenetic tools offer an independent source to test evolutionary hypotheses and to estimate dates on the origin of major lineages. Our aims are to analyse relationships among species of supersect. Homalopoa present in the Americas and the Southern Hemisphere: sections Acutifoliae, Anthochloa, Brizoides, Dasypoa, Dioicopoa, Dissanthelium, Homalopoa, Madropoa and Tovarochloa and the Punapoa informal group; and to date principal nodes within Poa supersect. Homalopoa. Hypotheses concerning the patterns of evolution of diclinous breeding systems in Poa are evaluated based on our phylogenetic results.
MATERIALS AND METHODS
DNA isolation, amplification and sequencing
Plants were field collected and dried in silica gel, and DNA was extracted using the modified CTAB (cetyltrimethylammonium bromide) protocol from Doyle and Doyle (1987). DNeasy Plant Mini Kits (Qiagen) were used to extract DNA from herbarium specimens when fresh material was not available. Alternatively, the silica-based column method of Alexander et al. (2007) was used for both silica gel dried and herbarium material.
A preliminary analysis on the variability of different markers was evaluated. A total of six markers: ITS and ETS (nuclear), and trnT-L, trnL-F, rpoA and rpl16 (plastid) were sequenced and analysed using parsimony. Only the most informative regions were then selected. Subsequently, two nuclear ribosomal markers, ETS and ITS, and two plastid regions, trnT-L and trnL-F, were amplified and sequenced for every sample. The ETS fragment (comprising the 3′ region of the external transcribed spacer of 18S–26S rDNA) was amplified using the forward primer RETS4 designed by Gillespie et al. (2010) and the reverse primer 18S-IGS (Baldwin and Markos, 1998) or 18S-R (Starr et al., 2003). The ITS region, including the internal transcribed spacers ITS 1 and ITS 2 and the 5·8S rDNA gene, was amplified with primers designed by White et al. (1990) using ITS5 as the forward primer and ITS4 as the reverse primer, or using the primer combination KRC (Torrecilla and Catalán, 2002) and AB102 (Douzery et al., 1999). Internal primers ITS2 and ITS3 were also used when sequences were difficult to amplify.
The trnT-L and trnL-F regions were amplified using the primer pairs denoted by a-b, c-d, and e-f as described by Taberlet et al. (1991). These primers amplify the spacer region between the trnT(UGU) and trnL(UAA) 5′ exon (i.e. the trnT-L spacer), the intron of the trnL(UAA) 5′ exon and the trnL(UAA) 3′ exon (i.e. the trnL intron), and the spacer region between the trnL(UAA) 3′ exon and the trnF(GAA) exon (i.e. the trnL-F spacer), respectively.
Polymerase chain reactions were performed in a 25 μL final volume with 50–100 ng of template DNA, 0·2 μm of each primer, 25 μm of DNTPs, 5 mm MgCl2, 1× buffer and 1·5 U of Taq polymerase (Invitrogen). The reaction conditions were: a first period of denaturation at 94 ºC for 5 min, followed by 35 cycles of denaturation at 94 ºC for 30 s, annealing at 48 ºC (52 ºC for ITS) for 1 min and extension at 72 ºC for 90 s. Final extension at 72 ºC for 6 min terminated the reactions. PCR products were run out on a 1 % TBE agarose gel, stained with SYBR safe™ DNA gel stain (Invitrogen) and visualized in a blue light transilluminator. Automated sequencing was performed by Macrogen, Inc. Alternatively, PCR and sequencing followed the methods of Gillespie et al. (2008, 2009). Both strands were sequenced for each fragment.
Phylogenetic sampling
A total of 124 specimens were included in the matrix; 116 represent the genus Poa, of which 105 are specimens of Poa supersect. Homalopoa belonging to a total of 89 species. We here included species of sections Acutifoliae, Anthochloa, Brizoides, Dasypoa, Dioicopoa, Dissanthelium, Homalopoa, Madropoa, Tovarochloa and the Punapoa group (Supplementary Data Appendix S1). Aphanelytrum was represented by two species. Outgroups were selected to include representatives of all other subgenera of Poa known from the Americas: subg. Poa sects Macropoa and Poa, subg. Ochlopoa sects Alpinae and Parodiochloa, subg. Stenopoa sects Pandemos, Secundae and Stenopoa, and subg. Sylvestres. To anchor Poa, we added species of the following genera: Alopecurus (sub-tribe Alopecurinae), Arctagrostis, Arctophila, Nicoraepoa (sub-tribe Poinae s.l.) and Phleum (sub-tribe Phleinae) (following Soreng and Gillespie, 2007; Gillespie et al., 2008, 2010). For classification purposes, we followed the most current taxonomy for each taxon (Table 1; Appendix S1).
Phylogenetic analyses
Sequence editing and assembly were performed using Chromas Pro ver 1·7·6 (Technelysium Pty Ltd, South Brisbane, Queensland, Australia) and BioEdit version 5·0·9 (Hall, 1999) or Geneious vers. 6·1·5 (Biomatters Ltd., http://www.geneious.com). The whole data set was aligned with MAFFT ver. 7 (Katoh and Standley, 2013) and the alignment was then manually checked. When amplification failed for a fragment, the respective positions were coded as missing data in separate or combined matrices. Percentages of missing data (gaps not included) were calculated for each matrix (Table 2).
Table 2.
Information on sequences and matrices derived from the ETS and ITS ribosomal nuclear regions, and trnT-L-F plastid markers, and the combined (ETS + ITS + trnT-L-F) data set
| ETS | ITS | trnT-L-F | Combined | |
|---|---|---|---|---|
| No. of taxa | 124 | 122 without P. pfisteri/P. schizantha | 124 | 124 |
| Length of the alignment | 560 | 685 | 2204 | 3449 |
| Poa shortest sequence | 527 bp = P. apiculata | 665 bp = P. interior | 1751 bp (=P. holciformis + P. trivialis) | – |
| Poa longest sequence | 544 bp = P. trivialis | 670 bp = P. cucullata | 1883 bp (P. alopecurus subsp. alopecurus) | – |
| Missing data (% ) | 1·9 | 4·7 | 2·8 | 3 |
| No. of informative characters | 157 | 135 | 125 | 417 |
| Length of the shortest trees* | 430 | 380 | 223 | 1144 |
| Parsimony indexes | Ci = 0·52, Ri = 0·82 | Ci = 0·51, Ri = 0·74 | Ci = 0·67, Ri = 0·87 | Ci = 0·49, Ri = 0·76 |
| No. of nodes in the strict consensus tree | 32 | 30 | 31 | 61 |
| No. of nodes with bootstrap support >50 % | 36 | 26 | 33 | 54 |
| Unambiguous informative indels within Poa |
|
|
|
Parsimony analyses were conducted with the four regions analysed separately and combined using TNT (Goloboff et al., 2008) under equal weights. Characters were considered unordered, and uninformative characters were excluded from the analyses. The search strategy consisted of heuristic searches performed using 1000 series of random addition sequences followed by TBR (tree bisection and reconnection) branch rearrangements and retaining ten trees per series. Trees recovered were saved in memory and additionally TBR swapped, retaining a maximum of 100 000 trees. Branches with ambiguous length of 0 or 1 were collapsed, according to collapsing rule 1. A 50 % majority rule consensus tree and a strict consensus tree were generated from the most parsimonious trees. Branch support was assessed using bootstrap analyses (Felsenstein, 1985) with 1000 replicates and heuristic searches of ten series using random taxon entry followed by TBR branch swapping; values given as BS in the text. Incongruence among individual data sets was visually checked by comparing strict consensus trees from individual analyses. When analysing the topology obtained by each partition, all partitions retrieved a similar number of nodes, and most clades were equivalent in species composition. Species that were inconsistent among consensus trees of each individual partition will be reported when presenting results for the combined data set.
Bayesian analysis was conducted for the combined data matrix using MrBayes version 3.1.2 (Ronquist and Huelsenbeck, 2003). jModelTest 2.1.4 (Darriba et al., 2012) was employed to determine the sequence evolution model that best fits the data; ITS and ETS markers were set to TIM2 + G, whereas trnT-L and trnL-F were set to GTR + G. Models for each partition were selected by the Akaike information criterion (AIC; Akaike, 1974). We carried out two independent runs of 10 000 000 generations using the default priors and four Markov chains (one cold and three heated chains), sampling one tree every 1000 generations. The program Tracer v.1.6 (Rambaut et al., 2014) was used to examine the output parameters from Bayesian analyses to determine stationarity. Trees prior to reaching stationarity were discarded as burn-in, and the remaining trees were used to compute a 50 % majority rule consensus tree and posterior probabilities (shown as BI).
Data on breeding systems in Poa were obtained from the literature and personal observations (Table 3; Appendix S1). Species that presented variation in the breeding system (P. aequigluma, P. iridifolia, P. matris-occidentalis, P. palmeri and P. plicata) were coded as polymorphic (Appendix S1). Although breeding system was not included in the analyses, it was optimized on the majority rule consensus tree of most parsimonious trees using the command ‘common mapping’ by TNT, where the common optimization is represented in the consensus diagram to determine whether breeding system character states were synapomorphies or homoplasies in the clade of interest. To analyse ambiguous optimization on clades of interest, the reconstruction of this character was also evaluated on one of the most parsimonious trees using the command ‘character reconstruction’ by TNT, where all possible optimizations for ambiguous nodes are determined.
Table 3.
Breeding systems in Poa subg. Poa supersect. Homalopoa in the Americas, with current sectional classification and DNA clade in our study
| New World current classification for supersect. Homalopoa (+P. yaganica) | Species | Clades in our study | Breeding system |
|---|---|---|---|
| Acutifoliae | P. acinaciphylla | D + | Hermaphroditic |
| *P. planifolia | Gynodioecious | ||
| Dioicopoa | P. alopecurus (viviparous sometimes), P. arachnifera, P. bergii, P. bonariensis, P. calchaquiensis, P. cumingii, P. denudata, P. dolichophylla, P. durifolia, P. holciformis, P. hubbardiana, P. huecu, P. lanigera, P. lanuginosa,P. ligularis, P. nubensis,P. obvallata(viviparous sometimes),P. paposana, P. schizantha, P. spiciformis,P. stuckertii | D | Dioecious |
| P. reitzii | C (D) | ||
| P. arechavaletae Parodi, P. gayana E. Desv., P. megalantha (Parodi) Herter, P. pedersenii Nicora, P. pilcomayensis Hack., P. sellowii Nees, *P. umbrosa Trin., P. uruguayensis Parodi | |||
| *P. iridifolia (viviparous rarely) | D + | Gynodiecious/Dioecious | |
| Unplaced | P. spicigera | D | Pistillate |
| Madropoa | P. pfisteri | D + | Dioecious |
| P. chambersii (gd in eastern population), P. cusickii (often p, rarely gd), P. douglasii, P. fendleriana (rarely h, often p), P. macrantha, P. piperi, P. porsildii | C | ||
| P. atropurpurea Scribn., P. pringlei Scribn. (sometimes p), P. rhizomata Hitchc. (subdioecious), P. sierrae J.T. Howell | |||
| P. nervosa, P. wheeleri (p, rarely with stamens) | C | Sequentially Adjusted Gynomonecious | |
| P. cuspidata | Unplaced | ||
| P. arnowiae Soreng, P. diaboli, P. tracyi Vasey | |||
| P. confinis Vasey, P. leibergii Scribn., P. stebbinsii Soreng (trioecious?) | Gynodioecious | ||
| Anthochloa | *P. lepidula (sgm?) | E | Gynomonoecious |
| Unplaced | P. ramifera | E+, G | |
| Punapoa group | P. humillima, P. marshallii | ||
| P. anae Tovar, P. brevis Hitchc., P. denticulata Hack. (?), P. dentigluma Tovar, P. dissanthelioides | |||
| P. aequigluma, P. chamaeclinos, P. gymnantha, P. perligulata | E | Pistillate | |
| P. unispiculata (d?) | Gynodioecious | ||
| Dissanthelium s.s. | P. calycina (h in North America), P. parvifolia, P. serpana | E + Dissanthelium | Gynomonoecious |
| P. arcuata Refulio, P. congesta Refulio, P. rauhii Refulio, P. swallenii Refulio | |||
| *P. macusaniensis (E.H.L. Krause) Refulio (gm?, see Sulekic,1999) | Hermaphroditic | ||
| ex Dissanthelium (unplaced) | P. linearifolia | Sister to E | Gynomonoecious |
| P. thomasii Refulio, P. amplivaginata (Tovar) Refulio | Hermaphroditic | ||
| *P. aequalis (Swallen & Tovar) Refulio | Gynodioecious | ||
| P. deminuta Refulio (?), P. gigantea (Tovar) Refulio, | Gynomonoecious | ||
| P. trollii (Pilg.) Refulio (?) | Dioecious | ||
| P. boliviana Refulio | Unknown | ||
| Tovarochloa | P. apiculata | E + Tovarochloa | Hermaphroditic |
| Genus Aphanelytrum | A. peruvianum, A. procumbens | E + | |
| Dasypoa | P. laetevirens, P. subspicata | Gynomonoecious | |
| P. parviceps Hack. | |||
| P. scaberula | Unplaced | Hermaphroditic | |
| Homalopoa s.l. (incl. sects.Plicatae) | P. atropidiformis, P. matris-occidentalis (gm?), P. mulleri, P. occidentalis, P. plicata (h?) | ||
| *P. aequatoriensis, *P. cucullata, P. hieronymi, *P. huancavelicae, *P. pauciflora | E + | ||
| P. bigelovii Vasey & Scribn., P. bolanderi Vasey, P. bradei Pilg., P. howellii Vasey & Scribn., P. jujuyensis, P. orizabensis Hitchc., P. parviceps, P. reflexa Vasey & Scribn., P. ruprechtii Peyr. (gm?), P. seleri Pilg., P. strictiramea Hitchc. (?, stamens often aborted late in development, possibly due to apomixis), P. tacanae Swallen, P. talamancae R.W. Pohl, P. wendtii Soreng & P.M. Peterson (?) | |||
| P. bajaensis | Sister to F | ||
| *P. candamoana, *P. gilgiana, *P. glaberrima, P. kurtzii, *P. pearsonii | F | Gynomonoecious | |
| P. horridula | Sequentially Adjusted Gynomonecious | ||
| P. fibrifera | E+, G | ||
| P. androgyna Hack. | |||
| P. superata | Unplaced | ||
| *P. ayacuchensis Tovar (sgm?), *P. chirripoensis R.W. Pohl (sgm or gd?), *P. grisebachii R.E. Fr., *P. leioclada Hack., *P. mucuchachensis Luces, *P. mulalensis Kunth, P. myriantha Hack., *P. orthophylla Pilg., P. oscariana Negritto & Anton, *P. petrosa Swallen, P. pilgeri Negritto & Anton, *P. ragonesei Nicora, *P. scabrivaginata Tovar, P. soderstromii Negritto & Anton, *P. trachyphylla Pilg. | Gynomonoecious | ||
| P. linearifolia | Sister to E | ||
| P. lilloi | Unplaced | Gynodioecious | |
| P. cabreriana Anton & Ariza Esp. | |||
| P. palmeri (trioecious?) | D + | Dioecious Gynodioecious | |
| Sect. Poa | *P. yaganica (sgm?) | ||
| Sect. Monandropoa | P. tucumana Parodi (1 anther) | Hermaphroditic |
Variation and uncertainty in breeding system is given in parentheses: h, hermaphroditic; gm, gynomonoecious (pistillate flowers above perfect flowers in spikelets); sgm, sequentially adjusted gynomonoecious; gd, gynodioecious; d, dioecious; p, pistillate; ?, uncertain.
Breeding system type is from Anton (1978), Anton and Connor (1995), Davidse et al. (2010), Giussani (2000), Giussani et al. (2012), Negrito and Anton (2000, 2006), Nicora (1978), Parodi (1936), Peterson et al. (2006), Refulio-Rodriguez et al. (2012), Soreng (1991, 1998, 2007), Soreng and Keil (2003) and Soreng and Peterson (2008).
*New observations (or those differing from the literature).
Species present in the molecular analysis are indicated in bold; for authorship of these species see Appendix S1.
Molecular dating analyses
To estimate dates, we constructed a matrix based on both nuclear and plastid data following recommendations by Christin et al. (2014), who highlighted the importance of using markers from different genomes, considering that nuclear markers are useful complements to plastid markers. We used ITS sequence data as the nuclear partition (ETS was unavailable for most of the outgroup taxa; therefore, it was not considered for this analysis), and trnT-L plus trnL-F as the plastid regions (hereafter trnT-L-F).
We assembled a matrix consisting of 145 terminal taxa of which 96 are species of Poa. The species selection represents all lineages of Poa supersect. Homalopoa; double entries used for phylogenetic analyses were here removed if their positions in the cladogram were confirmed to be close to each other. At least one representative of all New World subgenera of Poa was included (Appendix S1).
In the absence of Poa fossils to date divergence time among major groups within supersect. Homalopoa, we selected appropriate species of the Poaceae family to utilize fossils as calibration points. Outgroup selection comprised representatives of both major Poaceae clades: BOP clade (Bambusoideae, Oryzoideae and Pooideae) and PACMAD (Panicoideae, Aristidoideae, Chloridoideae, Micrairoideae, Arundinoideae and Danthonioideae); taxonomy following Grass Phylogeny Working Group II (2012) and Soreng et al. (2015b). A particular emphasis was placed on including genera of different tribes of subfamily Pooideae that are closely related to Poa.
The topology of the initial tree was constrained to represent relationships among Poaceae as previously reported by Bouchenak-Khelladi et al. (2010), and by the Grass Phylogeny Working Group II (2012). Also, phylogenetic relationships within Poa supersect. Homalopoa were constrained following results of the parsimony/Bayesian analyses (see below).
Fossil evidence, such as micro- and macrofossils, has been used previously to date grasses; this evidence also reflects conflicts among different fossils used as calibration points (Prasad et al., 2005; Strömberg, 2005; Vicentini et al., 2008; Bouchenak-Khelladi et al., 2010; Christin et al., 2014). To take into account different dates proposed for a particular clade, we used a log-normal prior distribution on nodes constraining dates to a minimum age represented by a well-known fossil for the group (offset value) and setting the upper bound (95 %) on a secondary calibration point based on the earliest fossil proposed for the same clade; standard deviation was set to 1.
We calibrated six external nodes for this data set. (1) We used a multiflowered-spikelet fossil which shares characters of both the BOP and PACMAD clades to calibrate the most external node of the Poaceae at 55 million years ago (Mya) (Crepet and Feldman, 1991); also, the date accords with a fossil leaf impression with bambusoid affinities from the Eocene (Frenguelli and Parodi, 1941; Brea and Zucol, 2007). Hence, the offset was set at 55 Mya ranging to 70 Mya (upper bound, 95 %) based on the grass-like pollen fossil described in Linder (1986). (2) Then, divergence time of the Pooideae clade from the PACMAD clade was set at a minimum age of 42 Mya estimated from phytolith grass fossils described by Zucol et al. (2010). Phytoliths were found in sediments of the Gran Barranca member of the Sarmiento Formation in Argentina representing festucoid and pooid morphotypes; similarly, phytoliths were also found in fossil dung beetle brood balls (Coprinisphaera) from the Middle Eocene–Early Miocene Sarmiento Formation by Sánchez et al. (2010). The upper bound (95 %) was set at 67 Mya based on phytolith fossils of dinosaur coprolites from India as stated in Prasad (2005). (3) For the PACMAD clade, a minimum age was estimated at 31 Mya based on phytoliths from the Gran Barranca (Zucol et al., 2010), while the upper bound was set at 52 Mya. (4) Based on phytolith evidence, the offset for the Chloridoids was 14 Mya (Strömberg, 2005) and the upper bound was set at 19 Mya based on Dugas and Retallack (1993). (5) Based on a well conserved stipoid fossil, the silicified reproductive bracts of Berriochloa (Thomasson, 2005), we set the minimum age at 7·6 Mya for the Stipeae node, ranging to 34 Mya (95 %) based on another fossil with stipoid similarities (Prasad et al., 2011). (6) To calibrate internal branches within Panicoids, we used a fossil related to Setaria to set a minimum age of 7 Mya (Elias, 1942) for Panicum as both genera are closely related (Giussani et al., 2001).
We used a relaxed Bayesian molecular clock method to consider variation in evolutionary rates across the phylogeny (Drummond et al., 2006). The analysis was set with prior on the distribution of node ages approximated by a Yule speciation process and evolutionary rates among branches followed a log-normal distribution and considered uncorrelated as implemented in BEAST v 1·8 (Drummond et al., 2002–2013). The substitution models used and the Bayesian analysis were implemented as reported in ‘Phylogenetic analyses’.
Geographic data
Data from herbarium databases of the Darwinion Institute (Documenta Florae Australis: http://www.darwin.edu.ar/iris/), Smithsonian Institution, Canadian Museum of Nature and Negritto’s database were used to score 2786 specimens of the American species of Poa supersect. Homalopoa for geographical distribution (Supplementary Data Appendix S2). Geographic co-ordinates were obtained from specimen labels or georeferenced localities; the package ‘raster’ (Hijmans and van Etten, 2015) available in the R statistical package 3·2·2 (R Development Core Team, 2010) was employed to plot the specimens. Altitude as represented in the map has been obtained from WorldClim–Global Climate Data (http://www.worldclim.org/).
RESULTS
Phylogenetic analyses
A total of 124 specimens were considered for the phylogenetic analyses; only two species (P. pfisteri and P. schizantha) were difficult to amplify and discarded for the ITS data set. Characteristics of individual partitions (trnT-F, trnL-F, ETS and ITS) are presented in Table 2; informative indels are reported and discussed if they have phylogenetic significance. The aligned ETS partition yielded a total of 560 bp; the ITS was 685 bp long, while the plastid trnT-L-F region consisted of 2204 bp.
The combined matrix of nuclear and plastid markers is 3449 bp long and 417 characters are phylogenetically informative (Table 2). Trees obtained from parsimony and Bayesian inference analyses are presented in Supplementary Data Figs S1 and S2, respectively. Similar clades were obtained in both analyses.
Subgenus Poa resolved as monophyletic (BS = 100; BI = 1). Both varieties of P. hachadoensis and P. mendocina were united in a clade (X-clade) that is sister to a group with both supersections of the subgenus Poa: supersect. Poa (BS = 100; BI = 1) and supersect. Homalopoa. However, some discrepancies were found among partitions: the ETS partition showed P. pratensis subsp. alpigena and P. sibirica included within the Homalopoa clade, while in the trnT-L-F and ITS consensus trees Homalopoa was not recovered.
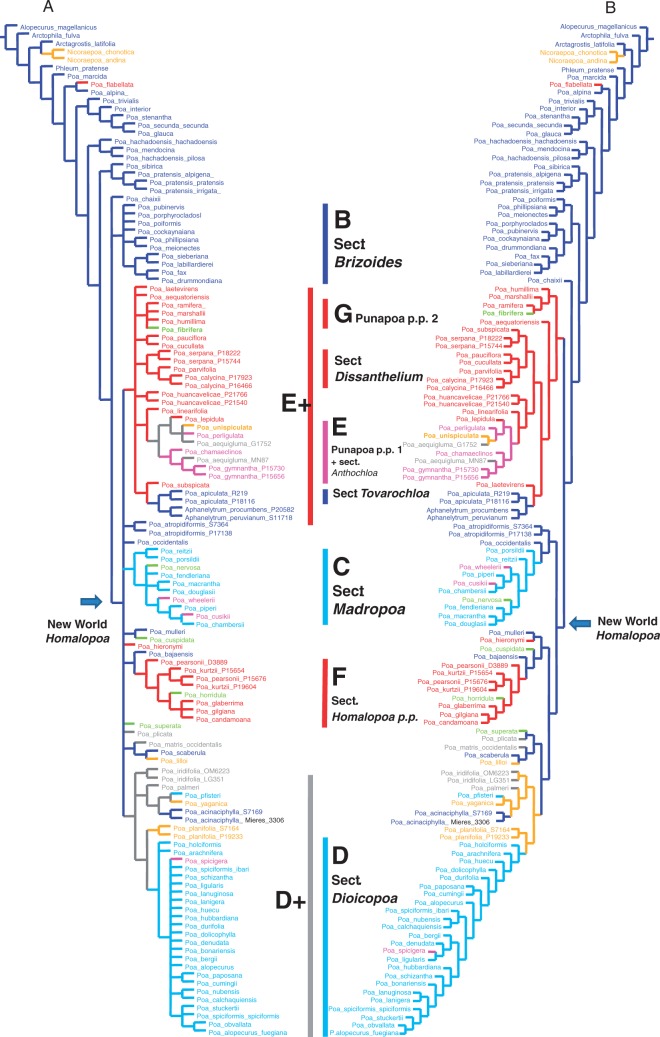
Supersection Homalopoa is monophyletic and supported when the data sets are combined (BS = 98; BI = 0·8). Within this group, major clades and species are resolved in a basal trichotomy: Poa chaixii Vill.; the New World Homalopoa species, clade A (BS = 89; BI = 0·94); and all species of the Australasian sect. Brizoides, clade B (BS = 98; BI = 1). Relationships among major clades within the New World Homalopoa clade collapse in a polytomy in the strict consensus parsimony tree (Fig. S1) and in the Bayesian tree (Fig. S2).
Most of the dioecious species of North America are united in the monophyletic sect. Madropoa, clade C (BS = 83; BI = 0·99), with the Brazilian P. reitzii Swallen, previously classified in sect. Dioicopoa, in a basal position within clade C. The sect. Dioicopoa is monophyletic (clade D, sect. Dioicopoa s.s.: BS = 99; BI = 1) if including all South American dioecious species, except for P. iridifolia Hauman and P. pfisteri Soreng (ITS data lacking), plus the North American dioecious species P. arachnifera. Clade D also includes P. spicigera, a species previously unplaced. The broader moderately supported D+ clade (BS = 73; BI = 1) includes section Dioicopoa s.s. plus other gynodioecious, dioecious and hermaphrodite species (Dioicopoa s.l.). Members of this clade share a 23 bp insertion of the trnT-L region (TATATATGAAAGATATAATAAAG) that is a duplication of the previous segment (Table 2); it is absent only in P. bonariensis, P. holciformis and P. pfisteri, and present in P. reitzii and P. hachadoensis). Poa planifolia Kuntze, a gynodioecious species classified in sect. Acutifoliae from South America (Appendix S1), was resolved as sister taxon to sect. Dioicopoa s.s. Other taxa resolved as members of D+ are: P. iridifolia (classified in sect. Dioicopoa; dioecious and gynodioecious, pers. obs.) of the Argentine Pampas hills; P. palmeri Soreng & P.M. Peterson from Mexico (sect. Homalopoa s.l.; partially dioecious, because of the presence of hermaphrodite, pistillate and staminate individuals); P. yaganica Speg., a southern Patagonian species (subg. Poa supersect. Poa; gynodioecious); and P. acinaciphylla E. Desv. (sect. Acutifoliae; hermaphroditic) from the central Andes of Argentina and Chile. The position of P. reitzii in clade C needs to be corroborated; although resolved with sect. Madropoa in the combined analysis and in both individual phylogenies from ETS and trnT-L-F, it was included within clade D, sect. Dioicopoa s.s., in the ITS parsimony majority rule tree, but was unresolved in the strict consensus tree.
The following groups were supported (all from South America unless noted). The Punapoa group was not monophyletic, but we detected one core group of dwarf species that inhabit the Andean Altiplano and form a highly supported sub-clade E (BS = 100; BI = 1; Punapoa p.p. 1). An extended clade E+ includes species of sect. Dissanthelium (BI = 0·78), both species of Aphanelytrum together with P. apiculata Refulio of sect. Tovarochloa (BS = 100; BI = 1), several species of Homalopoa s.l. and two species of sect. Dasypoa. A group of four dissimilar species are included in subclade G (BS = 62; BI = 0·85): P. humillima Pilg. and P. marshallii Tovar (Punapoa p.p. 2), and P. fibrifera Pilg. and P. ramifera Soreng & P.M. Peterson. A separate group of South American Homalopoa s.l. species (P. candamoana Pilg., P. gilgiana Pilg., P. glaberrima Tovar, P. horridula Pilg., P. kurtzii R.E. Fr. and P. pearsonii Reeder) are included in a highly supported clade F (BS = 99; BI = 1), with P. bajaensis Soreng (from Mexico) resolved as sister. Poa gymnantha Pilg., classified with the core Punapoa group (above), was included unambiguously in subclade E by the ETS, ITS and the combined data set analyses.
Evolution of breeding systems
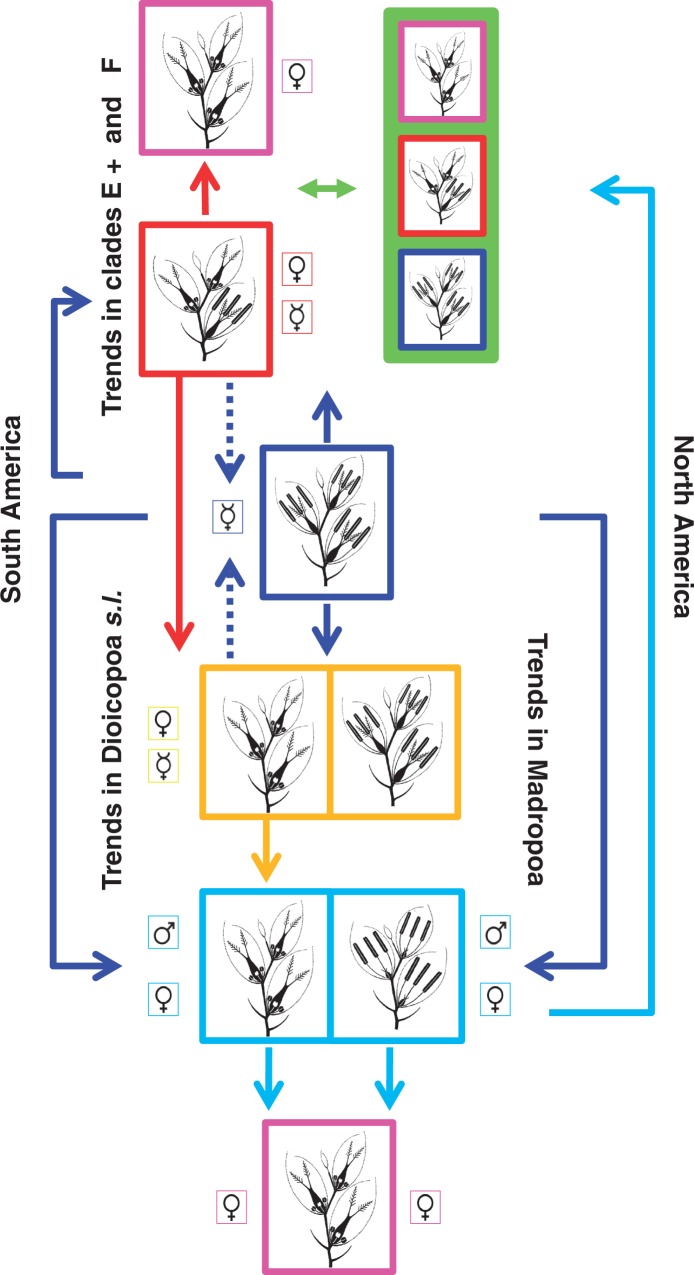
When optimizing the reproductive system onto the most parsimonious trees, hermaphroditism appears as the ancestral state for Poa. Since basal relationships within the New World Homalopoa clade were unresolved, different solutions to the evolution of the reproductive system are possible (Fig. 3). However, ancestral states for each major clade were recovered unambiguously for one reproductive system as seen when optimizing in the majority rule consensus tree (Fig. 3A). Based on possible optimizations of the basal node of the New World Homalopoa clade (there are 1504 possible resolutions for the polytomy), dioecy appeared at least once, if Madropoa (M) and D+ are closely related (an equally parsimonious relationship, although not supported in parsimony analyses), or twice if those clades had independent origins within the New World supersect. Homalopoa. Gynomonoecy, although present in most species of E+ and F clades, is also found in a single species (P. hieronymi Hack.) as a derived state from hermaphrodite ancestors; and a reversal to hermaphroditism appears to have occurred in section Tovarochloa (where there is only one flower per spikelet) plus Aphanelytrum. At least two different reconstructions were possible for the Dioicopoa s.l. (clade D+), with dioecy or gynodioecy being equally parsimonious ancestral states. For sub-clade E, both pistillate and gynomonoecy are possible states for the ancestor of this monophyletic group. Gynodioecy appeared three to four times in Homalopoa clade A: as an ancestral state of Dioicopoa s.l., with a reversal to hermaphroditsm in P. acinaciphylla, or twice within Dioicopoa s.l.; in P. unispiculata (sub-clade E); and in P. lilloi from the Altiplano. A diagram with possible evolutionary pathways of the breeding systems in the New World Homalopoa is presented in Fig. 4.
Fig. 3.
Optimization of the breeding system. (A) Majority rule consensus tree from the most parsimonious trees optimized by ‘common mapping’ in TNT; (B) character reconstruction in one or the most parsimonious trees. Bars and letters indicate principal clades within Poa supersect. Homalopoa as indicated by our results. Colours represent variation of the breeding system among species and follow references as presented in Fig. 2; grey colour represents ambiguous optimization on branches.
Fig. 4.
Evolutionary pathways of the breeding systems in the New World Homalopoa. Colours represent variation of the breeding system among species and follow references as presented in Fig. 2. Principal trends among American clades are shown, with arrows as explained in the text.
Geographic distribution
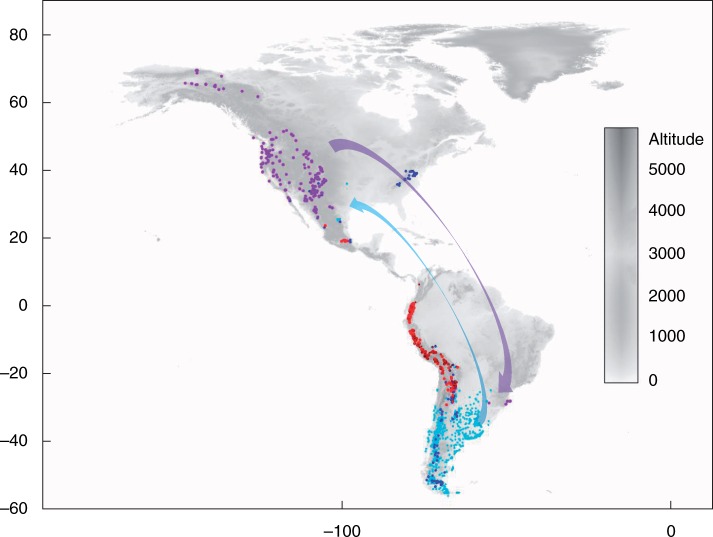
A geographic distribution map was obtained for the American species of Poa supersect. Homalopoa using georeferrenced specimen data (Fig. 5). Major clades were visualized onto the map and showed a particular distribution associated with restricted areas. Dioecy according to phylogenetic results appeared twice in clades C and D both in North and South America, respectively; only P. reitzii and P. arachnifera have a different distribution according to their phylogenetic affiliations. Gynomonoecious species (clades E+ and F) are clearly distributed along the Andes in northern Argentina and Chile to Colombia in South America and in a few localities in Mexico.
Fig. 5.
Current distribution of New World species of Poa supersect. Homalopoa. Colours indicate species as grouped by the phylogenetic results. Light blue, clade D + (sect. Dioicopoa s.l.); violet, clade C (sect. Madropoa); red, clade E+; red circle with black border, clade F; blue, species not grouped. Arrows represent long-distance dispersal events which occurred in the past: the light blue arrow shows two long-distance dispersal events from the South American D+ clade to North America (P. arachnifera and P. palmeri); the violet arrow shows a long-distance dispersal event from the North American clade C to South America (P. reitzii). Specimen coordinates are presented in Appendix S2.
Molecular dating: time of divergence in Poa
The estimated ages for crown and stem nodes are shown in Fig. 6. Divergence dates for both major clades BOP and PACMAD were estimated at 61–55 Mya and yielded similar results to those previously reported (Vicentini et al., 2008; Bouchenak-Khelladi et al., 2010; Wu and Ge, 2012). The crown age of PACMAD was estimated at 43–33 Mya, while the age of the BOP was estimated at 57–43 Mya, agreeing with previous estimations of a radiation in the Middle Eocene to Early Oligocene (Zachos et al., 2001; Strömberg 2005; Inda et al., 2008; Bouchenak-Khelladi et al., 2010; Edwards et al., 2010; Wu and Ge, 2012).
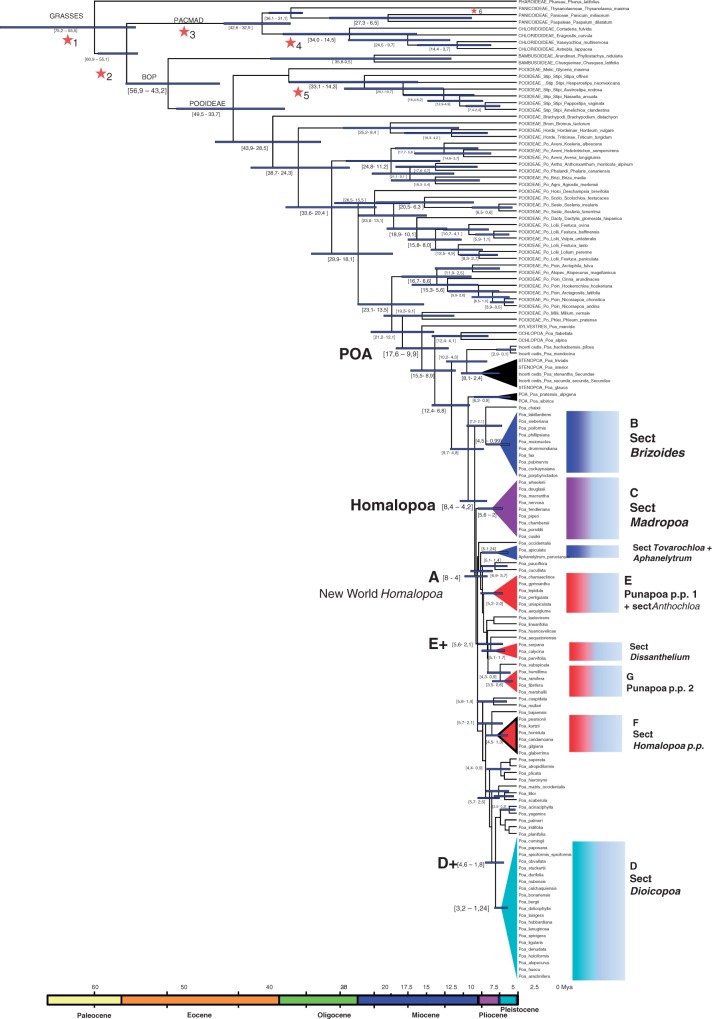
Fig. 6.
Poa chronogram based on a relaxed Bayesian clock. Node bars represent 95 % confidence intervals of divergence times. Stars and numbers indicate dated nodes based on fossil evidence as described in the Materials and Methods. Letters indicate principal clades within Poa supersect. Homalopoa as shown by our results; traditional sectional treatments are also presented as introduced in the text and Table 1. Colours indicate major groupings: light blue, dioecious species in South America + P. arachnifera (sect. Dioicopoa) + P. spicigera; violet, dioecious species in North America (sect. Madropoa) + P. reitzii; red, gynomonoecious species of clade E+; red triangle with black border, gynomonoecious species of clade F; blue, hermaphroditic species of sect. Brizoides and sect. Tovarochloa and Aphanelytrum.
Within the subfamily Pooideae, the crown radiated 50–34 Mya ago and, within this clade, Stipeae diverged between 33 and 14 Mya. Because our major concern is with Poa, results for other groups are presented in Fig. 6 and were checked for constancy with previous dating.
The estimated age for the crown node of the genus Poa is about 18–10 Mya, in the Middle Miocene, while the supersection Homalopoa, although few representatives were included from the Old World, is estimated at about 8·4–4·2 Mya. Radiation of species into the Americas is estimated at almost the same age, in the Late Miocene–Early Pliocene (8–4 Mya). Within the New World Homalopoa clade, a polytomy is shown for basal lineages; major clades diverged at similar ages or periods, mostly in the Pliocene to Early Pleistocene, about 5–2 Mya (Fig. 6). Section Dioicopoa s.s. (clade D) evolved during the Late Pliocene to Early Pleistocene in southern South America, and is one of the most recently evolved lineages within Homalopoa (3·2–1·24 Mya). Section Madropoa in North America is estimated to be older (5·6–2 Mya) than Dioicopoa s.s. Results for other lineages within Homalopoa are presented in Fig. 6.
DISCUSSION
Monophyly of Poa has been extensively discussed, and resolution of phylogenetic relationships among closely related genera has helped to elucidate the identity of Poa (Soreng et al., 1990, 2015a; Gillespie and Soreng, 2005; Gillespie et al., 2007, 2008, 2009, 2010; Refulio-Rodriguez et al., 2012). In our analyses, Poa was resolved as monophyletic, and representatives of the major subgenera were included to anchor relationships among sections of supersect. Homalopoa. Despite high morphological and taxonomic diversity (Fig. 1; Table 1), molecular studies have had little success in resolving relationships within supersect. Homalopoa (Gillespie and Soreng, 2005; Gillespie et al., 2007, 2009; Refulio-Rodriguez et al., 2012). In our work, all American species were recovered in clade A, named the New World Homalopoa clade (Figs S1 and S2). Clade A, the Eurasian Poa chaixii and the Australasian sect. Brizoides together comprise the Poa supersect. Homalopoa clade. Section Brizoides was monophyletic, in agreement with Gillespie et al. (2009) and Refulio-Rodriguez et al. (2012). In their study of Australasian Poa, Birch et al. (2014) greatly increased sampling in sect. Brizoides and resolved three sub-clades, but did not include Homalopoa species from other sections or from outside the region. Our study is the first to provide significant and meaningful structure among New World Homalopoa. Although basal relationships were poorly resolved, seven clades (described in detail below) were resolved that correspond well to the sectional classification and/or to morphological characteristics. Four of these clades (sects. Dioicopoa s.s., Dissanthelium s.s., Madropoa and Tovarochloa + Aphanelytrum) were also detected in previous studies, but mostly with less support or fewer species sampled (Soreng, 1990; Gillespie et al., 2008, 2009; Refulio-Rodriguez et al., 2012).
The evolutionary history of Poa in the Americas is linked to the diversification of breeding systems in the region. While hermaphroditism was revealed as plesiomorphic for the genus Poa and for supersect. Homalopoa, variation in reproductive systems occurred principally within the New World species of supersect. Homalopoa (Fig. 3; Table 3). Reversals to hermaphroditism occur in the Tovarochloa + Aphanelytrum clade, which is embedded within the expanded gynomonoecious clade E+. Reversion to monoecy has also been reported in several groups (Schaefer and Renner, 2010; Ming et al., 2011), and it is also possible in Poa. Other hermaphroditic species such as Poa atropidiformis Hack., P. mulleri Swallen, P. occidentalis and P. scaberula are part of a basal polytomy and have retained the plesiomorphic state. Poa bajaensis, sister to the gynomonoecious clade F, also retains the plesiomorphic state. The hermaphrodite P. acinaciphylla may represent a link to the ancestor of the D+ clade or a reversal to hermaphroditism within the D+ clade. Here we review the breeding systems and groups detected in our study.
Dioecy–gynodioecy
Dioecy appeared three times in Poa. Two origins of dioecy occurred within supersect. Homalopoa, evolving independently in the monophyletic sections Dioicopoa and Madropoa; however, when resolving the major polytomy within the New World Homalopoa, a common ancestor between D+ and Madropoa cannot be ruled out. Another event involves three unclassified dioecious New Zealand species [P. subvestita (Hack.) Edgar, P. sudicola Edgar and P. foliosa (Hook. f.) Hook. f.; not sampled here] which, along with 13 New Zealand and eight New Guinea hermaphroditic species (the latter to which the name Poa sect. Pauciflorae Pilg. ex Potztal applies), align in the nrDNA X-clade and appear to have a reticulate origin (Gillespie et al., 2009; Birch et al., 2014).
In South America, P. acinaciphylla, a hermaphroditic species of Chile and Argentina, appears to be closely related to a group of gynodioecious and dioecious species that together constitute the Dioicopoa s.l. clade (D+): P. iridifolia, P. palmeri, P. planifolia, P. pfisteri, P. yaganica and the Dioicopoa s.s. clade (Table 3; Figs S1 and S2). Poa planifolia is a gynodioecious species that occurs in the same general area as P. acinaciphylla in the Andean region between latitude 32 ° and 34 °S in Argentina and Chile (both placed in sect. Acutifoliae). Soreng and Peterson (2008) suggested a hybrid origin for P. acinaciphylla involving P. planifolia and P. holciformis of sect. Dioicopoa s.s. Our data do not support this hypothesis, although these species are related by ancestral relationships: P. planifolia is sister to the D clade (where P. holciformis is included), and P. acinaciphylla and P. planifolia are included in the D+ clade by plastid data (not resolved by the nrDNA data), showing a close relationship to Dioicopoa s.s. Although Soreng and Peterson (2008) suggested that P. pfisteri might be related to P. diaboli of sect. Madropoa, and P. yaganica to P. pratensis of supersect. Poa sect. Poa, we here confirm their relationship to Dioicopoa s.l. Poa palmeri, an endemic species from the Sierra Madre Oriental in Mexico, is here linked to Dioicopoa s.l.; specimens of this species are most frequently hermaphroditic (although anthers are sometimes aborted late in development), but some plants are completely staminate or pistillate (tentatively identified as trioecious by Soreng and Peterson, 2012), suggestive of a possible step toward dioecy. Poa palmeri and P. arachnifera are the only two taxa included in the D+ clade that are found in North America (Fig. 5). Relationships of both taxa to the South American dioecious clade are probably the result of separate long-distance dispersal events in the past taking place from South to North America. Although the two could be relicts from earlier diversification in North America, P. arachnifera is fully dioecious, with sexual dimorphism and panicles fully developed in the same way as most South American sect. Dioicopoa species, thus supporting more recent long-distance dispersal northward from South America.
Dioicopoa s.s.
Section Dioicopoa as previously circumscribed included 30 South American species and P. arachnifera from North America (Parodi, 1936; Rosengurtt et al., 1970; Anton, 1978; Nicora, 1978; Smith et al., 1981; Longhi-Wagner, 1987; Giussani, 2000; Soreng et al., 2003; Giussani et al., 2012), 23 (80 %) of which were included in the analyses (Table 3). The majority of these sampled species resolved in clade D (21 species), a group which we define here as sect. Dioicopoa s.s. All species of clade D are dioecious except for a Peruvian unplaced species P. spicigera for which only pistillate individuals are known (Tovar, 1993; Giussani, 2000; Giussani et al., 2012). The latter species would be the only strictly pistillate species of the group, having changed to apomictic seed reproduction in Dioicopoa s.s. However, its morphology suggests a relationship to P. perligulata of sub-clade E, hinting at a possible deeper reticulate origin, a hypothesis that needs to be tested.
Dioicopoa s.s. species are adapted to some of the driest and coldest areas in South America, and many of its species occur in Patagonia of Argentina and Chile; some species also inhabit temperate regions in the Pampas in Argentina, Brazil, Paraguay and Uruguay, and the equivalent habitat in the southern Great Plains of North America (P. arachnifera), or high elevations of the ‘Altiplano’ in Argentina (Giussani et al., 2008), Bolivia, Chile and Peru (Fig. 5), or coastal fog belts and sand dunes in arid central Chile.
Surprisingly, P. iridifolia, an endemic species from the Ventania and Tandilia mountain ranges in southern Buenos Aires province of Argentina (Torres, 1970) and considered a member of the P. dolichophylla complex of sect. Dioicopoa (Giussani, 2000), is apart from Dioicopoa s.s. This result, together with new observations by Villamil and Giussani that confirm that dioecy is not completely established in its populations (since this species presents hermaphroditic and pistillate individuals, or, similarly to P. palmeri, hermaphroditic, staminate and pistillate phases), suggests that it does not belong to Dioicopoa s.s. but rather to a group of diclinous species that resolved at the base of the D+ clade (described above).
A synapomorphic state for Dioicopoa s.s. (excluding P. spicigera) is the highly specialized sexual dimorphism. Characters linked to sexes are: the copious hairiness in the pistillate floret (hairs along the nerves and/or internerves of lemmas, and usually woolly or plicate hairs in three well-developed tufts on the callus), with the exceptions of glabrous individuals of P. bergii, and three glabrous species: P. holciformis, P. huecu and P. nubensis; plant size (pistillate plants taller than staminate plants); and quantitative traits of the spikelet (pistillate larger than staminate, and often fewer flowered) (Giussani, 2000). Dimorphism is also related to spatial segregation of sexes adapted to differences in microsite quality and resource availability (Bertiller et al., 2000, 2002), or to grazing pressure (Graff et al., 2013), revealing a high degree of specialization for dispersal and habitat segregation within the section. As a result, Dioicopoa s.s. is remarkably variable in morphology, distribution and habitats. In addition to sexual dimorphism, species are characterized by contracted and linear or elliptic (less often open-pyramidal) panicles, spikelets with three or more florets, and staminate flowers with long fertile anthers [1·5–3·5 (–4·5) mm long]. Plants mainly grow below 3500 m elevation (only P. calchaquiensis and P. nubensis grow above this altitude; Giussani et al., 2008). It is evident that speciation resulted in morphological discontinuities more rapidly than fixation of molecular variation in the DNA markers studied; hence few monophyletic groups are detected within this section in the molecular phylogeny. Several far southern species exhibit pseudoviviparous reproduction in addition to sexual reproduction (Moore and Doggett, 1976).
Madropoa
This section evidently originated in North America and has a centre of diversity in the mountains of the western USA, with the highest species diversity and numerous endemics in California, Oregon and Washington (Fig. 5). Madropoa s.s. (Soreng, 1991) originally included 11 species and one nothospecies of western North America, six of which are dioecious, three are gynodioecious, one trioecious, one with gynodioecious and dioecious populations, and the nothospecies is predominantly pistillate–apomictic. Most of the taxa have folded involute-margined leaf blades that adaxially are usually scabrous or coarsely puberulent on and between the veins, contracted panicles, and occur in open habitats. Eight species of the P. nervosa complex were added (Soreng, 2007) to Madropoa along with a new species, P. diaboli Soreng & D.J. Keil (Soreng and Keil, 2003): five are sequentially gynomonoecious (Soreng and Keil, 2003), one has gynodioecious and dioecious populations, one is sub-dioecious, one is dioecious and one is pistillate–apomictic. These mostly have flat or folded leaf blades that adaxially are smooth and glabrous between the veins (except the pistillate–apomict), most have open panicles, and they are generally confined to forested habitats. Callus hairs in Madropoa may be absent, arise in a single dorsal tuft or be a bit diffuse or arranged as a crown around the callus, but none occurs in three isolated tufts as in Dioicopoa. All 21 Madropoa species are diclinous (Soreng, 2007), but none exhibits simple gynomonoecy. Of these, five have numerous pistillate–apomictic plants, and apomixis occurs in over half or all of their geographic ranges (Table 3). The complexity of breeding systems in Madropoa is well illustrated by the four subspecies of P. cusickii Vasey, two of which are sexually reproducing, gynodioecious and dioecious, or strictly dioecious (subspp. cusickii and pallida Soreng, respectively), both with frequent populations that are strictly pistillate–apomictic in more arid and colder habitats, and two subspecies which are strictly pistillate–apomictic occurring in sub-alpine and low alpine habitats [subspp. epilis (Scribn.) W.A. Weber and purpurascens (Vasey) Soreng] (Soreng, 1991). Sequentially adjusted gynomonoecy is less frequent than dioecy among species of sect. Madropoa (Table 3), occurring mostly in the P. nervosa complex; only P. nervosa is present in our analyses and included in clade C. Poa cuspidata, although sequentially gynomonoecious and included in the P. nervosa complex of sect. Madropoa by Soreng (1991, 2007), is not resolved within this clade, but instead is linked to P. mulleri (hermaphroditic), both in a basal polytomy within the New World Homalopoa clade. The phylogenetic position of other species of sect. Madropoa exhibiting sequentially adjusted gynomonoecy, such as P. arnowiae Soreng, P. diaboli and P. tracyi (Soreng, 2007), still needs to be determined (Table 3). Poa wheeleri Vasey, a high polyploid apomictic species that is pistillate with vestigial (or very rarely developed) anthers, also belongs in clade C; it was suggested to be a hybrid between P. cusickii and a member of the P. nervosa complex (Soreng, 2007) not in our study.
Surprisingly, a Brazilian endemic, P. reitzii, is included in the North American Madropoa clade with high support in the ETS and trnT-L-F trees, and with moderate support in the combined trees. If this relationship is later confirmed, it would imply that an early long-distance dispersal event of an ancestral species took place between North and South America, followed by isolation and speciation, with the southern descendant, P. reitzii, now restricted to high elevations of the Serra Geral in southern Brazil (Fig. 5). However, more samples of this species are needed to confirm this relationship. Its breeding system is thought to be fully dioecious (Smith et al., 1981). Morphologically it seems close to P. iridifolia, with its strongly compressed and wing-keeled sheaths, broad, flat, somewhat firm blades, large loose panicles, presence of three tufts of callus hairs on each pistillate floret, and glabrous staminate florets (a dimorphism characteristic of Dioicopoa s.s.). In contrast to Dioicopoa, sexes of sect. Madropoa are not dimorphic or are only obscurely so, as described for P. fendleriana (Steud.) Vasey by Soreng (1985).
Gynomonoecy and exclusively pistillate species
Simple gynomonoecy is a widespread breeding system among species of Poa, where it is readily diagnosed by spikelets with the upper one (or two) flower pistillate and the lower one (or two) perfect in all individuals. It is well expressed and fixed in at least 40 Andean species of South America (Negritto, 1998; R. J. Soreng, pers. obs.), but only tentatively diagnosed in three species endemic to North America (Anton and Connor, 1995; Soreng and Peterson, 2012); Fig. 5. The frequency of simple gynomonoecism has been found to be six times greater in South America than in the Old World (Anton and Connor, 1995); in the Old World it occurs in about 26 species from Eurasia, Africa and New Zealand, of which only four are confirmed to belong in supersect. Homalopoa (Anton and Connor, 1995; Edgar and Connor, 2010; R. J. Soreng, pers., obs.). It was first identified in P. annua (Hackel, 1904), and later in its mainly Eurasian parents, P. infirma Kunth and P. supina Schrad. (Soreng et al., 2010). Simple gynomonoecy appears in clades E+ and F (Fig. 3); within E+ there appears to be a reversal to hermaphroditism in sect. Tovarochloa + Aphanelytrum.
Sequentially adjusted gynomonoecy diagnoses require studying populations through time (Soreng and Keil, 2003; earlier identified as ‘partial gynodioecy’ by Soreng and Hatch (1983) and Soreng (1985), and its nature confused by Anton and Connor, 1995). This breeding system is known or estimated for five or six species from North America (best studied in the P. nervosa complex) and 5–7 species of South America (where it has been tentatively diagnosed both by R. J. Soreng, pers. obs., and M. A. Negritto, pers. obs.). Sequentially adjusted gynomonoecious species are scattered in our trees, appearing to have evolved multiple times within New World Homalopoa: P. nervosa (Clade C, sect. Madropoa), P. fibrifera (sub-clade G), P. horridula (clade F), P. cuspidata and P. superata (both occur in unresolved clades) and P. palmeri (possibly trioecious, clade D+); P. horridula and P. fibrifera appear to be derived from simple gynomonoecious species. The diverse placements of putative sequentially adjusted gynomonoecious species suggest that this breeding system may represent a transitional state in derived species. It is remarkable that sequentially adjusted gynomonoecy is also expressed in seven Asian species (Zhu et al., 2006; R. J. Soreng, pers. obs.).
In our study, two major clades (clades E+ and F, Figs S1 and S2) of simple gynomonoecious species of the Altoandina and Puneña phytogeographical provinces [including the highest elevations all along the Andes, the former province above 4200 m a.s.l. and the latter from 3200 to 4400 m (Cabrera and Willink, 1980)] are recovered in the phylogeny. The number of times simple gynomonoecy originated depends on the resolution of basal clades, varying from two to four depending upon optimizations (Fig. 3).
Clade F
This is a group of gynomonoecious species from the Andes, ranging from Colombia (P. horridula) to northern Argentina and Chile (Fig. 5). All these species inhabit prairies on slopes at high elevations (3000–4800 m). This group is morphologically coherent, characterized by medium to large plants (30–150 cm), with panicles ovate to pyramidal, open or loosely contracted, lax, with spikelets mostly concentrated at the medial or apical portion of branches. Two groups of species are evident in Figs S1 and S2. Poa kurtzii and P. pearsonii are both densely cespitose, xeromorphic species of southern distribution frequent at medium to high elevations in arid Argentina and Bolivia. Poa candamoana, P. gilgiana, P. glaberrima and P. horridula are mesomorphic, weakly rhizomatous plants (only P. candamoana is cespitose), mostly distributed from Bolivia to Ecuador; P. horridula is the most widely distributed species (Argentina to Colombia) with the largest and most robust plants (up to 150 cm high). All species have a simple gynomonoecious breeding system, except for P. horridula, which is sequentially adjusted gynomonoecious (R. J. Soreng, pers. obs.), or possibly gynodioecious (Negritto, 1998). A possible connection to a hermaphroditic ancestor is based on the sister relationship of the Andean clade F with the hermaphrodite P. bajaensis (although with low probability), a Mexican endemic from Sierra San Pedro Mártir in Baja California. Poa bajaensis grows at medium to high elevations and also presents similar open, lax panicles, with basally naked branches.
Clade E+
This clade unites central and northern Andean species of South America, mostly concentrated at higher elevations of northernmost Argentina, Bolivia, Ecuador and Peru; only P. chamaeclinos and P. gymnantha (both pistillate–apomicts), and a hermaphroditic form of P. calycina (normally simple gynomonoecious) reach Mexico in North America (Fig. 5). Four sub-clades are well supported within E+: sect. Dissanthelium, sect. Tovarochloa + Aphanelytrum, sub-clade E and sub-clade G (Figs S1 and S2). The 12 species placed in the informal group Punapoa (by Soreng et al., 2003), plus P. unispiculata (Davidse et al., 2010), are divided between sub-clades G and E. We consider Punapoa to be an unnatural group in both its initial (Soreng, 1990) and later sense (Soreng et al., 2003; Davidse et al., 2010). Section Dissanthelium s.s., recently recircumscribed by Refulio-Rodriguez et al. (2012), includes eight simple gynomonoecious species and one apparently hermaphroditic species [P. macusaniensis (E.H.L. Krause) Refulio], distributed from Mexico to Argentina; species are cespitose and dwarf, with two-flowered spikelets (upper floret pistillate), glumes equal to or longer than the florets, and anthers 0·2–1·1 mm long. The second sub-clade comprises the monotypic section Tovarochloa (with the single species P. apiculata) and Aphanelytrum, which was also supported in the analyses of Gillespie et al. (2008) and Refulio-Rodriguez et al. (2012). All members are hermaphroditic; if simple gynomonoecism was fixed in the ancestor of clade E+, as appears likely, then this sub-clade represents a reversal to hermaphroditism. Poa apiculata is a rare dwarf annual species with one flower per spikelet that inhabits shallow ephemeral pools at high altitudes (Refulio-Rodriguez et al., 2012). Thus, a reversal to hermaphroditism, at least for sect. Tovarochloa, could be explained by the loss or suppression of the terminal pistillate floret of a simple gynomonoecious ancestor. Aphanelytrum is a genus currently with two species (Sánchez Vega et al., 2007; Refulio-Rodriguez et al., 2012; P. M. Peterson and R. J. Soreng, unpubl. res.) from humid to montane forests in the Andes (from Bolivia to Colombia); its unusual two- to three-flowered spikelets have minute or small glumes, mucronate lemmas with two lobes beside the mucro, and florets separated on long flexuous rachilla internodes. Sub-clade G includes a morphologically and ecologically diverse group of four species, ranging from dwarf (P. humillima), to slight and medium-sized (P. marshallii), to robust plants up to 1 m tall (P. fibrifera and P. ramifera). All of these grow in Peru (with P. humillima also in Argentina and Bolivia), although they are distributed at different altitudes and occupy different environments. Poa fibrifera is probably sequentially adjusted gynomonoecious; the other three species are simple gynomonoecious.
Sub-clade E includes a set of dwarf high Andean species that are mostly exclusively pistillate species. These exclusively pistillate species have stamens reduced to staminodes in all plants over large geographic ranges, except in P. aequigluma and P. gymnantha, where plants in a few localities have well-developed, fertile-appearing, long anthers. Sub-clade E is characterized by plants 2–8 cm tall (sometimes taller in P. gymnantha), with mostly (one) two florets per spikelet, inhabiting humid prairies, vegas and dry steppe at high elevations [(3000–) 4000–5000 m a.s.l.] in the Altoandean province. The simple gynomonoecious species P. lepidula (Nees & Meyen) Soreng & L.J. Gillespie (sect. Anthochloa), a dwarf of the highest sparsely vegetated elevations in the Andes (with 2–7 flabellate florets per spikelet) and the gynodioecious P. unispiculata are firmly nested within sub-clade E, a relationship previously detected by Refulio-Rodriguez et al. (2012). The gynomonoecious species, P. linearifolia Refulio (previously as Dissanthelium longifolium Tovar) from high elevations in Peru, is sister to sub-clade E. Optimization of the breeding system shows a probable simple gynomonoecious origin for sub-clade E.
Breeding system evolution
Hermaphroditism appears as the most primitive state, and diclinism is derived (Fig. 3A, B). Dicliny is rare in sub-tribe Poinae and surrounding sub-tribes, and only Poa exhibits gynomonoecy within the subfamily (Connor, 1981).
The phylogeny helps to infer at least three major pathways from hermaphroditic ancestors to diclinism (Figs 3 and 4). Two major evolutionary pathways indicate a direction to dioecy, one in South America (probably via gynodioecy) and one in North America; a third pathway leads from hermaphroditism to simple gynomonoecism in Andean species of South America. Two derived states evolved independently from hermaphroditic, gynomonoecious or dioecious species. The step leading to strictly pistillate flowering probably included total supression of maleness in flowers and gain of apomixis, allowing species to perpetuate themselves in extreme habitats and produce seed without the need for pollen to stimulate endosperm development. The occurrence of sequentially adjusted gynomonoecy is possibly due to the suppression of maleness in space (partial or whole inflorescence) and in time (developmentally and through the growing season); Figs 2 and 4.
Dioecism appears to have originated twice in the Americas, independently in both hemispheres, representing dissimilar pathways (Figs 3–6). In one case, gynodioecy appeared as an intermediate state to dioecy in the South American Dioicopoa s.l. In the second case, dioecy appears suddenly in the North American Madropoa with no obvious intermediate states (Fig. 3). Soreng (1991, 2000) and Soreng and Keil (2003) suggested that dioecy in Madropoa evolved via intermediate pathways, proceeding from sequentially adjusted gynomonoecism [found in the P. nervosa complex of Madropoa (sensu Soreng, 2007)] to gynodioecy, or to sub-dioecy. A review of phylogenetic studies showed that dioecy can be gained and lost several times once evolved, and that there are more genera with hermaphroditic and dioecious species than genera with intermediate breeding systems (Renner 2014), suggesting possible evidence for this evolutionary pathway. We would emphasize that diagnosing intermediate stages is not a trivial matter, and these may have been overlooked in broad surveys, especially those dependent mainly on herbarium material. However, considering that less than half of section Madropoa was sampled in this study (Table 3), it will be critical to include a full set of Madropoa species to confirm this hypothesis.
Simple gynomonoecism could have originated twice from hermaphroditic ancestors in South American and Mesoamerican Homalopoa. This breeding system also represents an intermediate step towards the evolution of strictly pistillate apomictic populations in extreme habitats of the High Andes. Only the presumed gynodioecious species P. unispiculata (Davidse et al., 2010) resolved within the pistillate sub-clade E reveals a pathway from gynomonoecy to gynodioecy. An alternative hypothesis of gynodioecy as a precursor condition leading to the development of pistillate apomictic populations (Fig 3A) needs further exploration of the reproductive biology and ecological adaptations in this clade. In Poa, phenotypic plasticity (Frenot et al., 1999; Couso and Fernández, 2012) and polyploidy are frequent phenomena that, in conjunction with apomixis, favour the adaptability of pistillate populations, thus fixing genetic variation established via earlier sexual reproduction to maintain successful genotypes in extreme climates with short flowering seasons (Asker and Jerling, 1992). Advanced dicliny may also favour the establishment of pistillate apomictic reproduction when population sizes are low and the chance of pollen meeting stigma is critically reduced, as hypothesized for sect. Madropoa species (Soreng, 2000).
Direct relationships among dioecious and gynodioecious species and exclusively pistillate species are rare (except in sect. Madropoa). Species with only pistillate plants appear independently several times in the phylogeny. In addition to pistillate species of sub-clade E, the Peruvian–Bolivian pistillate species resolved in sect. Dioicopoa s.s. is the only pistillate taxon in this dioecious clade and is geographically isolated from the other taxa in the section. Within sect. Madropoa (clade C), P. cusickii has two alpine subspecies (subspp. epilis and purpurascens) that are exclusively pistillate, and P. wheeleri is almost always pistillate. The other two subspecies of P. cusickii have both sexually reproducing (gynodioecious and dioecious) and pistillate–apomictic populations, as do the three subspecies of P. fendleriana (dioecious), and populations of P. pringlei Scribn. (dioecious in the Oregon and California Coast Ranges only, and pistillate elsewhere) (Soreng and Van Devender, 1989; Soreng, 1991, 2000).
As shown in Fig. 3A and B, reversal to hermaphroditism is only represented in the Tovarochloa + Aphanelytrum clade and in P. acinaciphylla; the first derived from gynomonoecy while P. acinaciphylla is resolved among gynodioecious ancestors. In our phylogeny, reversal to hermaphroditism could be explained through different mechanisms: by the loss of the terminal pistillate floret in sect. Tovarochloa, by a reversal to the expression of maleness in the upper floret of Aphanelytrum spp. and by the loss of male suppression in pistillate plants of P. acinaciphylla.
Dating Poa and the evolution of the breeding system in New World Homalopoa
According to our results, Poa is estimated to have originated around 17·6–9·9 Mya during the Middle Miocene (Fig. 6). Eurasia is considered to be the centre of diversification of the genus (Hartley, 1961; Soreng et al., 1990; Anton and Connor, 1995), particularly the western part where most of the sections and diploids occur (Soreng et al., 2010). Although subg. Sylvestres, which is endemic to North America today, is sister to the rest of the genus (excluding Arctopoa), Soreng (1990) suggested that Sylvetres might have originated from an early dispersal from the European sub-continent when closer to North America. However, there is evidence from reticulate origins of Aniselytron and Arctopoa with ancestral Sylvestres in East Asia (Gillespie et al., 2010), which would be consistent with an early dispersal to North America via Beringia for Sylvestres. The origin of Poa supersect. Homalopoa, a diversified group occurring today in Africa, Australasia, Eurasia, North America and South America, is suggested to have been in Eurasia 8·4–4·2 Mya ago. The expansion of Homalopoa to the Americas also occurred through Beringia, with colonizing opportunities favouring expansion through North and South America (Soreng et al., 2010). Because our results show a single New World Homalopoa clade, there was presumably a single dispersal event for this group from Eurasia to North America; this ancestor of the New World Homalopoa then rapidly diversified and radiated in the Late Miocene–Early Pliocene (8–4 Mya). Analyses including additional species of Homalopoa from Eurasia are needed to test this hypothesis.
Several major geological and environmental events facilitated dispersal to South America and promoted the rapid radiation of Homalopoa in both South and North America. During the Middle Miocene, a widespread area of South America previously flooded by the ‘Paranean Sea’ and the ‘Tethys Waterspout’ was succeeded by plains that extended north from northern Patagonia to central and northern Argentina, Uruguay, along the eastern slopes of the rising Andes of northern Bolivia, southern Peru and Venezuela, and also in the upper Amazon basin (Pascual et al., 1996). During this period known as ‘the Age of the Southern Plains’ (Pascual and Bondesio, 1982; Ortiz-Jaureguizar and Cladera, 2006), the climate was cooler, seasonality was marked, and more varied environmental sub-division occurred (Pascual et al., 1996); consequently, new opportunities were established for colonization of grasses in South America (Simpson, 1983). Although the Isthmus of Panama connected North and South America about 4 Mya ago, partly due to sea level drop from glaciation cycles, Poa presumably crossed between the continents on tropical mountain tops or longer distance hitchhiking via birds. The tangled hairs on the lemma nerves and long tufts of hairs on the callus of Poa are quite effective for seed dispersal by animals (Davidse, 1987), but transporting cool temperate species across the tropical lowlands via mammals seems improbable, leaving birds and winds as the most likely vectors for migrations between North and South America. The uplift of the Andes in South America occurred during the Middle Miocene through Early Pliocene until reaching modern elevations by around 2·7 Mya (Gregory-Wodzicki, 2000). Similarly in North America, regional uplift of major mountains, such as the Sierra Nevada in western North America, and local doming that deformed the peneplain, such as the San Juan Formation (4 Mya), occurred in the transition from Tertiary to Quaternary time (Wallace and Kirtley, 1932; Ruddiman and Kutzbach, 1991). In both hemispheres, mountain uplifts brought severe consequences to landscapes and vegetation, exposing species to drastic climatic and geological changes in a relatively short time. During the Pleistocene (about 2·5 Mya), glacial cycles similarly affected landscapes and life. New habitats, fragmentation of habitats, aridization and colder climates probably favoured speciation in the New World Homalopoa. All principal lineages described in this study, such as sections Dioicopoa, Madropoa, Dissanthelium, Tovarochloa, clade F and the dwarf sub-clade E, diversified in parallel at about the same time during the Pliocene to early Pleistocene (5–2 Mya) in North and South America. At the same time, speciation of major lineages also took place in Australasia, where the ancestor of sect. Brizoides diversified (4·5–0·99 Mya) in agreement with a suggested rapid diversification in the Pleistocene for this Australian group (Gillespie et al., 2009; Hoffmann et al., 2013; Griffin and Hoffmann, 2014), and coincident with the appearance of prominent grasslands or shrubland/grassland mosaic vegetation dating from the mid-Pliocene (Birch et al., 2014).
The evolution of major New World lineages within Homalopoa may have been facilitated by the diversification of the breeding systems that helped to perpetuate successful genotypes in extreme conditions, particularly at higher altitudes of both hemispheres. Possibly dicliny evolved under pressure to circumvent inbreeding depression once the hermaphroditic and self-compatible ancestor(s) established in the New World. Gynomonoecious lineages appear to have given rise to the Altoandinean–Puneña clades (E, F); in sub-clade E, apomictic, strictly pistillate species favoured the establishment of adaptive characteristics to extreme habitats. Most probably, dioecy appeared independently in North and South America with the diversification of Madropoa in mountains of the western USA (5·60–2 Mya) earlier than Diocopoa (3·2–1·24 Mya) in Sub-antarctic and Patagonian regions in southern South America. Gynodioecious species represent intermediate pathways to dioecy (clade D+), and also originated independently as derived states several times in the evolution of New World Homalopoa (such as P. lilloi and P. unispiculata).
Few long-distance dispersal events are the best explanation for the position of some species in the phylogeny. Poa reitzii would have dispersed from North to South America, and P. palmeri in the opposite direction during the Early Pliocene; later, during the Pleistocene, another long-distance dispersal event took the ancestor of P. arachnifera from South to North America. These long-distance dispersal events in dioecious species of Poa were most probably related to the migration of birds, or facilitated by wind dispersal of hairy seeds.
Conclusions
Based on a comprehensive examination of many taxa of Poa, Anton and Connor (1995) proposed evolutionary hypotheses for the diversification of the breeding system that our phylogeny, for the most part, supports. However, the timing and selective pressures favouring the different sexual pathways in Poa, from hermaphroditism to gynomonoecy and to dioecy, are suggested here for the first time based on a robust dated phylogeny. Diversification of the breeding system in the New World Homalopoa occurred at the end of the Miocene and Early Pliocene. As a consequence of the formation of the Panama Isthmus and mountain uplifts in tropical latitudes, connection between continents and the formation of new habitats favoured the expansion of the ancestral species of New World Homalopoa. During the Pliocene, diversification of breeding systems helped to perpetuate genotypes in extreme climatological and environmental conditions. Later, long-distance dispersal events are the best explanation for species to migrate between North and South America, explaining the presence of species from North and South America in the same clades. Speciation accompanied cycles of glaciations during the Pleistocene, giving rise to the extensive variation in morphology and breeding system among species of major lineages found in the New World Homalopoa.
Finally, because the evolutionary history of Poa is linked to the diversification of the breeding system in the Americas, infrageneric taxonomic categories can be accommodated to the phylogenetic results in correlation with the type of reproductive system. A new sectional treatment is needed to classify the gynomonoecious species of clade E+, the dwarf and pistillate species of the South American Punapoa group (sub-clade E) and the group of Andean species of clade F (Table 3), and to reclassify species of sects Acutifoliae, Dasypoa and Homalopoa s.l. In addition, some species need to be realigned in well-supported monophyletic sections, while further work is needed to place other species included in polytomies.
Our study is an important contribution to the understanding of the evolutionary patterns in Poa associated with the sexuality of species, current distribution and historical biogeography; the future challenge is to incorporate all this information into taxonomic categories that reflect evolution within the New World Poas.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: majority rule consensus tree of Poa obtained from the 100 000 most parsimonious trees based on four molecular markers: ITS, ETS, trnT-L and trnL-F. Figure S2: Bayesian 50 % majority rule consensus tree of the combined data matrix using ITS, ETS, trnT-L and trnL-F sequences from species of Poa and related taxa. Appendix S1: voucher information for species used in the phylogenetic and dating analyses. Appendix S2: geographic coordinates of the specimens used to represent species in their geographic range.
ACKNOWLEDGEMENTS
This article is dedicated to the memory of Osvaldo Morrone, with whom I, L.M.G., had fun working and who put trust in me and encouraged me to move forward. We are grateful to Alejandro F. Zucol for suggesting fossil evidences and dates on Poaceae from South America, Francisco Rojas who collaborated with the illustrations on flower diagrams, and Diego L. Salariato for his help on dating analyses. We thank Agostina Sassone who assisted in the organization of the map. Sequences of Dissanthelium species were provided by Nancy Refulio-Rodriguez. We also thank Paul Peterson for providing material of Poa collected in South America, the Smithsonian Institution for logistics and collecting support to R.J.S., staff of the Instituto Darwinion for support with lab work, library and herbarium assistance, staff of the herbaria: BAA, CONC, CORD, K, LP, LPB, SI, US, USM, W, for the loan of and access to the studied materials, and two anonymous reviewers and the Chief Editor for comments on the manuscript. This research was supported by the ‘Consejo Nacional de Investigaciones Científicas y Técnicas-Argentina’ (CONICET-PIP 2010-0233) and the ‘Agencia Nacional de Promoción Científica y Tecnológica- Argentina’ (Préstamo BID-PICT-2013-0298) to L.M.G.; and the Canadian Museum of Nature research grants to L.J.G. L.M.G. thanks Hugo Cota Sánchez who allowed her to work in his laboratory to obtain DNA extraction and PCRs for several species of Poa through a Mini-PEET Awards to Enhance Transfer of Taxonomic Knowledge from the Society of Systematic Biology (SSB) to L.M.G. We appreciate Henry Connor’s work on breeding systems in grasses that was of much help in elucidating the evolution of the breeding system in Poa. We are also indebted to Elisa Nicora for her work and observations on the reproductive systems of Poa species in Patagonia, Argentina.
LITERATURE CITED
- Akaike H. 1974. A new look at the statistical model identification. IEEE Transactions on Automatic Control 19: 716–723. [Google Scholar]
- Alexander PJ, Rajanikanth G, Bacon CD, Bailey CD. 2007. Recovery of plant DNA using a reciprocating saw and silica-based columns. Molecular Ecology Notes 7: 5–9. [Google Scholar]
- Anton AM. 1978. Notas críticas sobre Gramíneas de Argentina III. Contribución al conocimiento de la sexualidad en Poa. Boletin de la Academia Nacional de Ciencias (Córdoba) 52: 267–275. [Google Scholar]
- Anton AM, Connor H. 1995. Floral biology and reproduction in Poa (Poeae: Gramineae). Australian Journal of Botany 43: 577–599. [Google Scholar]
- Asker SE, Jerling L. 1992. Apomixis in plants. Boca Raton, FL: CRC Press. [Google Scholar]
- Baldwin BG, Markos S. 1998. Phylogenetic utility of the external transcribed spacers (ETS) of 18S–26S rDNA: congruence of ETS and ITS trees of Calycadenia (Compositae). Molecular Phylogenetics and Evolution 10: 449–463. [DOI] [PubMed] [Google Scholar]
- Bertiller MB, Ares J, Graff P, Baldi H. 2000. Sex-related spatial patterns of Poa ligularis in relation to shrub patch occurrence in northern Patagonia. Journal of Vegetation Science 11: 9–14. [Google Scholar]
- Bertiller MB, Sain CL, Bisigato AJ, Coronato FR, Ares JO, Graff P. 2002. Spatial sex segregation in the dioecious grass Poa ligularis in northern Patagonia: the role of environmental patchiness. Biodiversity and Conservation 11: 69–84 [Google Scholar]
- Birch JL, Cantrill DJ, Walsh NG, Murphy DJ. 2014. Phylogenetic investigation and divergence dating of Poa (Poaceae, tribe Poeae) in the Australasian region. Botanical Journal of the Linnean Society 175: 523–552. [Google Scholar]
- Bouchenak-Khelladi Y, Verboom GA, Savolainen V, Hodkinson TR. 2010. Biogeography of the grasses (Poaceae): a phylogenetic approach to reveal evolutionary history in geographical space and geological time. Botanical Journal of the Linnean Society 162: 543–557. [Google Scholar]
- Bowden WM, Senn HA. 1962. Chromosome numbers in 28 grass genera from South America. Canadian Journal of Botany 40: 1115–1124. [Google Scholar]
- Brea M, Zucol AF. 2007. Guadua zuloagae sp. nov., the first petrified bamboo culm record from the Ituzaingó Formation (Pliocene), Paraná Basin, Argentina. Annals of Botany 100: 711–723, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera AL, Willink W. 1980. Biogeografía de América Latina. Colección de Monografías Científicas de la Secretaría General de la Organización de los Estados Americanos, Programa Regional de Desarrollo Científico y Tecnológico. 2nd edn. Washington, DC.
- Christin PA, Spriggs E, Osborne CP, Stromberg CA, Salamin N, Edwards EJ. 2014. Molecular dating, evolutionary rates, and the age of the grasses. Systematic Biologist 63: 153–165. [DOI] [PubMed] [Google Scholar]
- Connor HE. 1979. Breeding systems in the grasses: a survey. New Zealand Journal of Botany 17: 547–574. [Google Scholar]
- Connor HE. 1981. Evolution of reproductive systems in the Gramineae. Annals of the Missouri Botanical Garden 68: 48–74. [Google Scholar]
- Couso LL, Fernández RJ. 2012. Phenotypic plasticity as an index of drought tolerance in three Patagonian steppe grasses. Annals of Botany 110: 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepet WL, Feldman GD. 1991. The earliest remains of grasses in the fossil record. American Journal of Botany 78: 1010–1014. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidse 1987. Fruit dispersal in the Poaceae In: Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME, eds. Grass systematics and evolution. Washington, DC: Smithsonian Institution Press, 143–155. [Google Scholar]
- Davidse G, Soreng RJ, Peterson PM. 2010. Poa unispiculata, a new gynodioecious species of cushion grass from Peru with a single spikelet per inflorescence (Poaceae: Pooideae: Poeae: Poinae). Journal of the Botanical Research Institute of Texas 4: 37–44. [Google Scholar]
- Douzery EJP, Pridgeon AM, Kores P, Linder HP, Kurzweil H, Chase MW. 1999. Molecular phylogenetics of Diseae (Orchidaceae): a contribution from nuclear ribosomal ITS sequences. American Journal of Botany 86: 887–899. [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Drummond AJ, Rambaut A, Suchard MA. 2002. –2013. BEAST v1·8·0. Bayesian Evolutionary Analysis Sampling Trees. http://beast.bio.ed.ac.uk/. [DOI] [PMC free article] [PubMed]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. 2006. Relaxed phylogenies and dating with confidence. PLoS Biology 4: e88. doi:10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas DP, Retallack GJ. 1993. Middle Miocene fossil grasses from Fort Ternan, Kenya. Journal of Paleontology, 67: 113–128. [Google Scholar]
- Edgar E, Connor HE. 2010. Flora of New Zealand, Vol. 5, 2nd edn Lincoln, NZ: Mannaaki Whenua Press. [Google Scholar]
- Edwards EJ, Osborne CP, Strömberg CAE, et al. 2010. The origins of C4 grasslands: integrating evolutionary and e cosystem science. Science 328: 587–591. [DOI] [PubMed] [Google Scholar]
- Elias MK. 1942. Tertiary prairie grasses and other herbs from the High Plains. Geological Society of America Special Paper (Regular Studies) 41: 1–176. [Google Scholar]
- Felsentein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Frengüelli J, Parodi LR. 1941. Una Chusquea fósil de El Mirador (Chubut). Notas del Museo de La Plata 6: 235–238. [Google Scholar]
- Frenot Y, Aubry M, Misset MT, Gloaguen JC, Gourret JP, Lebouvier M. 1999. Phenotypic plasticity and genetic diversity in Poa annua L. (Poaceae) at Crozet and Kerguelen Islands (subantarctic). Polar Biology 22: 302–310. [Google Scholar]
- Gillespie LJ, Soreng RJ. 2005. A phylogenetic analysis of the bluegrass genus Poa L. (Poaceae) based on cpDNA restriction site data. Systematic Botany 30: 84–105. [Google Scholar]
- Gillespie LJ, Archambault A, Soreng RJ. 2007. Phylogeny of Poa (Poaceae) based on trnT-trnF sequence data: major clades and basal relationships. Aliso 23: 420–434. [Google Scholar]
- Gillespie LJ, Soreng RJ, Jacobs SWL. 2009. Phylogenetic relationships of Australian Poa (Poaceae: Poinae) including molecular evidence for two new genera, Saxipoa and Sylvipoa. Australian Systematic Botany 22: 413–436. [Google Scholar]
- Gillespie LJ, Soreng RJ, Paradis M, Bull RD. 2010. Phylogeny and reticulation in subtribe Poinae and related subtribes (Poaceae) based on nrITS, ETS, and trnTLF data In: Seberg O, Petersen G, Barfod AS, Davis JI, eds. Diversity, phylogeny, and evolution. In: Proceedings of the Fourth International Conference on the Comparative Biology of the Monocotyledons and the Fifth International Symposium on Grass Systematics and Evolution. Aarhus: Aarhus University Press, 589–617. [Google Scholar]
- Gillespie LJ, Soreng RJ, Bull RD, Jacobs SWL, Refulio-Rodriguez NF. 2008. Phylogenetic relationships in subtribe Poinae (Poaceae, Poeae) based on nuclear ITS and plastid trnT-trnL-trnF sequences. Botany 86: 938–967. [Google Scholar]
- Giussani LM. 2000. Phenetic Similarity patterns of dioecious species of Poa from Argentina and neighboring countries. Annals of the Missouri Botanical Garden 87: 203–233. [Google Scholar]
- Giussani LM, Collantes MB. 1997. Variación fenotípica en el complejo Poa rigidifolia asociada al efecto del pastoreo ovino y al ambiente en Tierra del Fuego, Argentina. Consecuencia taxonómica. Revista Chilena de Historia Natural 70: 421–434. [Google Scholar]
- Giussani LM, Martínez AJ, Collantes MB. 1996. Morphological variation associated with the environment in four dioecious Patagonian Poa species. The Poa rigidifolia complex. Canadian Journal of Botany 74: 762–772. [Google Scholar]
- Giussani LM, Nicora EG, Roig FA. 2000. Poa durifolia y su relación con el patrón fenético de Poa sección Dioicopoa (Poaceae). Darwiniana 38: 47–57. [Google Scholar]
- Giussani LM, Cota-Sanchez JH, Zuloaga FO, Kellogg EA. 2001. A molecular phylogeny of the grass subfamily Panicoideae (Poaceae) shows multiple origins of C4 photosynthesis. American Journal of Botany 88: 1993–2012. [PubMed] [Google Scholar]
- Giussani LM, Fernandez-Pepi MG, Morrone O. 2008. Phenetic patterns in Poa section Dioicopoa: a new species from the Puneña and Altoandina phytogeographical provinces of Argentina. Botanical Journal of the Linnean Society 157: 239–248. [Google Scholar]
- Giussani LM, Negritto MA, Romanutti A, Anton AM, Soreng RJ. 2012. Poa In: Zuloaga FO, Rúgolo de Agrasar ZE, Anton AM, eds. Flora Argentina. Flora vascular de la República Argentina, Monocotyledoneae. Poaceae. Pooideae 3(2) Córdoba: Gráficamente Ediciones, 284–339. [Google Scholar]
- Goloboff PA, Farris JS, Nixon K. 2008. TNT, a free program for phylogenetic analysis. Cladistics 24: 774–786. [Google Scholar]
- Graff BP, Rositano F, Aguiar MR. 2013. Changes in sex ratios of a dioecious grass with grazing intensity: the interplay between gender traits, neighbour interactions and spatial patterns. Journal of Ecology 101: 1146–1157. [Google Scholar]
- Grass Phylogeny Working Group II. 2012. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytologist 193: 304–312. [DOI] [PubMed] [Google Scholar]
- Gregory-Wodzicki KM. 2000. Uplift history of the Central and Northern Andes: a review. Geological Society of America Bulletin 112: 1091–1105. [Google Scholar]
- Griffin PC, Hoffmann AA. 2014. Limited genetic divergence among Australian alpine Poa tussock grasses coupled with regional structuring points to ongoing gene flow and taxonomic challenges. Annals of Botany 113: 953–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillin EA, Giussani LM, Oliva G. 1995. Estudios morfológicos y citológicos en especies dioicas del género Poa XXVI Congreso Argentino de Genética y I Jornada Argentino–Chilena de Genética. XXVIII Sociedad Argentina de Genetica San Carlos de Bariloche, Río Negro, Argentina.
- Hackel E. 1904. Zur biologie der Poa annua. Österreichische Botanische Zeitschrift 54: 273–278. [Google Scholar]
- Hair JB, Beuzenberg EJ. 1961. High polyploidy in a New Zealand Poa. Nature 189: 160.13714992 [Google Scholar]
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hartley W. 1961. Studies on the origin, evolution, and distribution of the Gramineae. IV. The genus Poa L. Australian Journal of Botany 9: 152–161. [Google Scholar]
- Hijmans RJ, van Etten J. 2015. Raster. Geographic analysis and modeling with raster data: R package version 2·4-20. Available from: http://cran.r-project.org/package=raster.
- Hoffmann M, Schneider J, Hase P, Röser M. 2013. Rapid and recent worldwide diversification of bluegrasses (Poa, Poaceae) and related genera. PLoS One 8: e60061. doi:10.1371/journal.pone.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunziker JH. 1978. Cytogenetics and evolution of some species of the genus Poa (Gramineae) In: Drets Brum-Zorrilla ME, Folle GA, eds. Actas III Congreso Latinoamericano de Genética. Montevideo: Asociación Latinoamericana de Genética, 144–149. [Google Scholar]
- Inda L, Segerra-Moragues J, Müller J, Peterson P, Catalán P. 2008. Dated historical biogeography of the temperate Loliinae (Poaceae, Pooideae) grasses in the northern and southern hemispheres. Molecular Phylogenetics and Evolution 46: 932–957. [DOI] [PubMed] [Google Scholar]
- Jacobs SWL, Gillespie LJ, Soreng RJ. 2008. New combinations in Hookerochloa and Poa (Gramineae). Telopea 12: 273–278. [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley AM, Johnson PG, Waldron BL, Peel MD. 2009. A survey of apomixis and ploidy levels among Poa L. (Poaceae) using flow cytometry. Crop Science 49: 1395–1402. [Google Scholar]
- Kellogg EA. 1989. Apomixis in the Poa secunda Complex. American Journal of Botany 74: 1431–1437. [Google Scholar]
- Linder HP. 1986. The evolutionary history of the Poales/Restionales – a hypothesis. Kew Bulletin 42: 297–318. [Google Scholar]
- Longhi-Wagner HM. 1987. Flora Ilustrada do Rio Grande do Sul, Fasc. 17. Gramineae, Tribo Poeae. Boletim Instituto Central de Biociências Universidade Federal do Rio Grande do Sul 41: 1–191. [Google Scholar]
- Marsh VL. 1952. A taxonomic revision of the genus Poa of United States and southern Canada. American Midland Naturalist 47: 202–256. [Google Scholar]
- Ming R, Bendahmane A, Renner SS. 2011. Sex chromosomes in land plants. Annual Review of Plant Biology 62: 485–514. [DOI] [PubMed] [Google Scholar]
- Moore DM. 1982. Flora Europaea check-list and chromosome index. Cambridge: Cambridge University Press. [Google Scholar]
- Moore DM, Doggett MD, 1976. Pseudo-vivipary in Fuegian and Falkland Island grasses. British Antarctic Survey Bulletin 43: 103–110. [Google Scholar]
- Murray BG, de Lange PJ, Ferguson AR. 2005. Nuclear DNA variation, chromosome numbers and polyploidy in the endemic and indigenous grass flora of New Zealand. Annals of Botany 96: 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negritto MA. 1998. Las especies ginomonoicas del género Poa (Gramineae: Poeae) en Sudamérica. PhD thesis, Universidad Nacional de Córdoba, Facultad de Ciencias Exactas, Físicas y Naturales, Argentina.
- Negritto MA, Anton AM. 2000. Revisión de las especies de Poa (Poaceae) del noroeste argentino. Kurtziana 28: 95–136. [Google Scholar]
- Negritto MA, Anton AM. 2006. Three new species of Poa from the Andes of Colombia and Peru. Systematic Botany 31: 83–88. [Google Scholar]
- Negritto MA, Romanutti AA, Acosta MC, Moscone EA, Cocucci AE, Anton AM. 2008. Morphology, reproduction and karyology in the rare Andean Poa gymnantha. Taxon 57: 171–178. [Google Scholar]
- Nicora EG. 1978. Poa In: Correa MN, ed. Gramineae, Flora Patagónica. Colección Científica del INTA 8: 138–207. [Google Scholar]
- Nosov NN, Rodionov AV. 2008. Molecular phylogenetic study of relationships between members of the genus Poa. Botanicheskii Zhurnal 93: 1919–1935. [Google Scholar]
- Nosov NN, Punina EO, Machs EM, Rodionov AV. 2015. Interspecies hybridization in the origin of plant species: cases in the genus Poa sensu lato. Biology Bulletin Reviews 5: 366–382. [Google Scholar]
- Ofir M, Kigel J. 2014. Temporal and intraclonal variation of flowering and pseudovivipary in Poa bulbosa. Annals of Botany 113: 1249–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Jaureguizar E, Cladera GA. 2006. Paleoenvironmental evolution of southern South America during the Cenozoic. Journal of Arid Environments 66: 498–532. [Google Scholar]
- Parodi LR. 1936. Contribución al conocimiento de las especies del género Poa de la flora uruguaya. Revista Argentina de Agronomía 3: 133–152. [Google Scholar]
- Pascual R, Bondesio P. 1982. Un roedor Cardiatheriinae (Hydrochoeridae) de la Edad Huayqueriense (Mioceno tardío) de La Pampa. Sumario de los ambientes terrestres en la Argentina durante el Mioceno. Ameghiniana 19: 19–36. [Google Scholar]
- Pascual R, Ortiz-Jaureguizar E, Prado JL. 1996. Land mammals: paradigm of Cenozoic South American geobiotic evolution. Müncher Geowissenschaftliche Abhandlungen (A) 30: 265–319. [Google Scholar]
- Peterson PM, Soreng RJ, Herrera Arrieta Y. 2006. Poa matri-occidentalis (Poaceae: Pooideae: Poeae: Poinae), a new species from Mexico. Sida 22: 905–914. [Google Scholar]
- Pierce S, Stirling CM, Baxter R. 2003. Pseudoviviparous reproduction of Poa alpina var. vivipara L. (Poaceae) during long-term exposure to elevated atmospheric CO2. Annals of Botany 91: 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad V, Stömberg CAE, Alimohammadian H, Sahni A. 2005. Dinosaur coprolites and the early evolution of grasses and grazers. Science 310: 1177–1180. [DOI] [PubMed] [Google Scholar]
- Prasad V, Strömberg CAE, Leaché AD, et al. 2011. Late Cretaceous origin of the rice tribe provides evidence for early diversification in Poaceae. Nature Communications 2: 480. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1·6 Available from http://tree.bio.ed.ac.uk/software/tracer/.
- R Development Core Team. 2010. R: a language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing. Available from https://cran.r-project.org/bin/windows/base/.
- Refulio-Rodriguez N, Columbus JT, Gillespie LJ, Peterson PM, Soreng RJ. 2012. Molecular phylogeny of Dissanthelium (Poaceae: Pooideae) and its taxonomic implications. Systematic Botany 37: 122–133. [Google Scholar]
- Renner SS. 2014. The relative and absolute frequencies of angiosperm sexual systems: dioecy, monoecy, gynodioecy, and an updated online database. American Journal of Botany 101: 1588–1596. [DOI] [PubMed] [Google Scholar]
- Rodionov AV, Nosov NN, Kima ES, Machsa EM, Puninaa EO, Probatova NS. 2010. The origin of polyploid genomes of bluegrasses Poa L. and gene flow between northern pacific and subantarctic islands. Russian Journal of Genetics 46: 1407–1416. [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rosengurtt B, Arrillaga de Maffei BR, Izaguirre de Artucio P. 1970. Gramíneas uruguayas. Universidad de la República, Departamento de Publicaciones, Montevideo, Uruguay. [Google Scholar]
- Ruddiman WF, Kutzbach JE. 1991. Plateau uplift and climatic change. Scientific American 264: 66–75.1711238 [Google Scholar]
- Sánchez MV, González MG, Genise JF. 2010. Phytolith analysis of Coprinisphaera, unlocking dung beetle behaviour, herbivore diets and palaeoenvironments along the Middle Eocene–Early Miocene of Patagonia. Palaeogeography, Palaeoclimatology, Palaeoecology 285: 224–236. [Google Scholar]
- Sánchez Vega I, Peterson PM, Soreng RJ, Laegaard S. 2007. Aphynelytrum peruvianum (Poaceae: Poinae): a new species from Peru. Journal of the Botanical Research Institute of Texas 1: 841–845. [Google Scholar]
- Saura F. 1943. Cariología de Gramíneas. Géneros Paspalum, Stipa, Poa, Andropogon y Phalaris. Revista de la Facultad de Agronomía y Veterinaria 10: 344–353. [Google Scholar]
- Saura F. 1948. Cariología de Gramíneas en Argentina. Revista de la Facultad de Agronomía y Veterinaria 12: 51–67. [Google Scholar]
- Schaefer H, Renner SS. 2010. A three-genome phylogeny of Momordica (Cucurbitaceae) suggests seven returns from dioecy to monoecy and recent long-distance dispersal to Asia. Molecular Phylogenetics and Evolution 54: 554–560. [DOI] [PubMed] [Google Scholar]
- Simpson BB. 1983. An historical phytogeography of the high Andean flora. Revista Chilena de Historia Natural 56: 109–122. [Google Scholar]
- Smith LB, Wasshausen DC, Klein RM. 1981. Gramíneas. Gêneros: 1. Bambusa até 44. Chloris. In Reitz PR, ed. Flora Ilustrada Catarinense Conselho Nacional de Desenvolvimento Científico e Tecnológico, Instituto Brasileiro de Desenvolvimento Florestal, Secretaria de Agricultura e Abastecimento, US National Science Foundation, Herbário ‘Barbosa Rodrigues’. Itajaí, Santa Catarina, Brasil, 1–436.
- Soreng RJ. 1985. Poa L. in New Mexico, with a key to middle and southern Rocky Mountain species (Poaceae). Great Basin Naturalist 45: 395–422. [Google Scholar]
- Soreng RJ. 1990. Chloroplast-DNA phylogenetics and biogeography in a reticulating group: study in Poa (Poaceae). American Journal of Botany 77: 1383–1400. [Google Scholar]
- Soreng RJ. 1991. Systematics of the ‘Epiles’ group of Poa (Poaceae). Systematic Botany 16: 507–528. [Google Scholar]
- Soreng RJ. 2000. Apomixis and amphimixis comparative biogeography: a study in Poa (Poaceae) In: Jacobs SWL, Everett J, eds. Grasses systematics and evolution. Melbourne, Australia: CSIRO, 294–306. [Google Scholar]
- Soreng RJ. 2007. Poa L In: Barkworth ME, Capels KM, Long SL, Anderton LK, Piep MB, eds. Magnoliophyta: Commelinideae (in part); Poaceae, part 1, Flora of North America north of Mexico, 24 New York: Oxford University Press, 486−601. [Google Scholar]
- Soreng RJ, Hatch SL. 1983. A comparison of Poa tracyi and Poa occidentalis (Poaceae: Poeae). Sida 10: 123–141. [Google Scholar]
- Soreng RJ, Van Devender DR. 1989. Late Quarternary fossils of Poa fendeleriana (muttongrass): Holocene expansions of apomicts. Southwestern Naturalist 34: 35–45. [Google Scholar]
- Soreng RJ, Keil DJ. 2003. Sequentially adjusted sex-ratios in gynomonoecism, and Poa diaboli (Poaceae), a new species from California. Madroño 50: 300–306. [Google Scholar]
- Soreng RJ, Gillespie LJ. 2007. Nicoraepoa (Poaceae, Poeae), a new South American genus based on Poa subgenus Andinae, and emendation of Poa section Parodiochloa of the sub-Antarctic islands. Annals of the Missouri Botanical Garden 94: 821–849. [Google Scholar]
- Soreng RJ, Peterson PM. 2008. New records of Poa (Poaceae) and Poa pfisteri: a new species endemic to Chile. Journal of the Botanical Research Institute of Texas 2: 847–859. [Google Scholar]
- Soreng RJ, Peterson PM. 2012. Revision of Poa L. (Poaceae, Pooideae, Poeae, Poinae) in Mexico: new records, re-evaluation of P. ruprechtii, and two new species, P. palmeri and P. wendtii. Phytokeys 15: 1–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreng RJ, Davis JI, Doyle JJ. 1990. A phylogenetic analysis of chloroplast DNA restriction site variation in Poaceae subfamily Pooideae. Plant Systematics and Evolution 172: 83–97. [Google Scholar]
- Soreng RJ, Davidse G, Peterson PM, et al. 2003. Catalogue of New World grasses (Poaceae): IV. Subfamily Pooideae. Contributions from the United States National Herbarium 48: 1–730. http://mobot.mobot.org/W3T/Search/nwgclass.html. [Google Scholar]
- Soreng RJ, Davis JI, Voionmaa MA. 2007. A phylogenetic analysis of Poaceae tribe Poeae s.l. based on morphological characters and sequence data from three chloroplast-encoded genes: evidence for reticulation, and a new classification for the tribe. Kew Bulletin 62: 425–454. [Google Scholar]
- Soreng RJ, Gillespie LJ, Jacobs SWL. 2009. Saxipoa and Sylvipoa – two new genera and a new classification for Australian Poa (Poaceae: Poinae). Australian Systematic Botany 22: 401–412. [Google Scholar]
- Soreng RJ, Bull RD, Gillespie LJ. 2010. Phylogeny and reticulation in Poa based on plastid trnTLF and nrITS sequences with attention to diploids In: Seberg O, Peterson G, Barfod A., Davis JI, eds. Diversity, phylogeny, and evolution in the monocotyledons. Aarhus: Aarhus University Press, 619–643. [Google Scholar]
- Soreng RJ, Gillespie LJ, Koba H, Boudko K, Bull RD. 2015a. Molecular and morphological evidence for a new grass genus, Dupontiopsis (Poaceae tribe Poeae subtribe Poinae s.l.), endemic to alpine Japan, and implications for the origin of Dupontia and Arctophila within Poinae s.l. Journal of Systematics and Evolution 53: 138–162. [Google Scholar]
- Soreng RJ, Peterson PM, Romaschenko K, et al. 2015b. A worldwide phylogenetic classification of the Poaceae (Gramineae). Journal of Systematics and Evolution 53: 117–137. [Google Scholar]
- Starr JR, Harris SA, Simpson DA. 2003. Potential of the 5′ and 3′ ends of the intergenic spacer (IGS) of rDNA in the Cyperaceae: new sequences for lower-level phyogenies in sedges with example from Uncinia Pers. International Journal of Plant Sciences 164: 213–227. [Google Scholar]
- Strömberg CAE. 2005. Decoupled taxonomic radiation and ecological expansion of open-habitat grasses in the Cenozoic of North America. In: Proceedings of the National Academy of Sciences, USA 102: 11980–11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet PL, Gielly L, Pautou G, Bouvet J. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17: 1105–1109. [DOI] [PubMed] [Google Scholar]
- Thomasson JR. 2005. Berriochloa gabeli and Berriochloa huletti (Gramineae: Stipeae), two new grass species from the late Miocene ash hollow formation of Nebraska and Kansas. Journal of Paleontology 79: 185–199. [Google Scholar]
- Torrecilla P, Catalán P. 2002. Phylogeny of broad-leaved and fine-leaved Festuca lineages (Poaceae) based on nuclear ITS sequences. Systematic Botany 27: 241–251. [Google Scholar]
- Torres MA. 1970. Poa. Flora de la Provincia de Buenos Aires. Colección Científica del INTA 4: 102–125. [Google Scholar]
- Tovar O. 1993. Las Gramíneas del Perú. RUIZIA. Monografías del Real Jardín Botánico. Consejo Superior de Investigaciones Científicas.
- Vicentini A, Barber JC, Aliscioni SS, Giussani LM, Kellogg EA. 2008. The age of the grasses and clusters of origins of C4 photosynthesis. Global Change Biology 14: 2963–2977. [Google Scholar]
- Wallace WA, Kirtley FM. 1932. Physiology and Quaternary geology of the San Juan Mountains, Colorado. US Geological Survey, Professional Paper, 166. Department of the Interior, Washington.
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis M, Gelfand D, Sninsky J, White T, eds. PCR protocols: a guide to methods and applications. San Diego: Academic Press, 315–322. [Google Scholar]
- Wu ZQ, Ge S. 2012. The phylogeny of the BEP clade in grasses revisited: evidence from the whole-genome sequences of chloroplasts. Molecular Phylogenetics and Evolution 62: 573–578. [DOI] [PubMed] [Google Scholar]
- Zachos J, Pagani M, Sloan L, Thomas E, Billups K. 2001. Trends, rhythms, and aberrations in global climate 60 Mya to present. Science 292: 686–693. [DOI] [PubMed] [Google Scholar]
- Zhu GH, Lui L, Soreng RJ, Olonova MV. 2006. Poa L In: Flora of China Editorial Committee, eds. Flora of China. Vol. 22, Poaceae. Saint Louis, MO: Missouri Botanical Garden Press, 257–309. [Google Scholar]
- Zucol AF, Brea M, Bellosi ES. 2010. Phytolith studies in Gran Barranca (central Patagonia, Argentina): the middle-late Eocene In: Madden RH, Carlini AA, Vucetich MG, Kay RF, eds. The paleontology of Gran Barranca. Evolution and environmental change through the Middle Cenozoic of Patagonia. Cambridge University Press, New York. [Google Scholar]
- Zuloaga FO, Morrone O, Belgrano MJ. 2008. Catálogo de las Plantas Vasculares Del Cono Sur: (Argentina, Sur de Brasil, Chile, Paraguay y Uruguay). Monographs in Systematic Botany from the Missouri Botanical Garden, 107. Missouri Botanical Garden Press.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.