Abstract
Background
Consistent localization of cerebellar cortex in a standard coordinate system is important for functional studies and detection of anatomical alterations in studies of morphometry. To date, no pediatric cerebellar atlas is available.
New method
The probabilistic Cape Town Pediatric Cerebellar Atlas (CAPCA18) was constructed in the age-appropriate National Institute of Health Pediatric Database asymmetric template space using manual tracings of 16 cerebellar compartments in 18 healthy children (9–13 years) from Cape Town, South Africa. The individual atlases of the training subjects were also used to implement multi atlas label fusion using multi atlas majority voting (MAMV) and multi atlas generative model (MAGM) approaches. Segmentation accuracy in 14 test subjects was compared for each method to ‘gold standard’ manual tracings.
Results
Spatial overlap between manual tracings and CAPCA18 automated segmentation was 73% or higher for all lobules in both hemispheres, except VIIb and X. Automated segmentation using MAGM yielded the best segmentation accuracy over all lobules (mean Dice Similarity Coefficient 0.76; range 0.55–0.91).
Comparison with existing methods
In all lobules, spatial overlap of CAPCA18 segmentations with manual tracings was similar or higher than those obtained with SUIT (spatially unbiased infra-tentorial template), providing additional evidence of the benefits of an age appropriate atlas. MAGM segmentation accuracy was comparable to values reported recently by Park et al. (2014) in adults (across all lobules mean DSC = 0.73, range 0.40–0.89).
Conclusions
CAPCA18 and the associated multi atlases of the training subjects yield improved segmentation of cerebellar structures in children.
Keywords: Pediatric Probabilistic atlas, cerebellum, multi atlas, CAPCA18
1. Introduction
The human brain is a complex structure and mapping its functional organization presents an ongoing challenge. Recent findings suggest that the cerebellum is functionally heterogeneous, with different topological regions subserving sensory, motor, cognitive, and affective processing (Stoodley and Schmahmann, 2009; Schlerf et al., 2010; Strick et al., 2009; Schmahmann and Sherman, 1998; Makris et al, 2005). As such, it has become increasingly important to identify precisely which lobule is activated in functional imaging studies. Efforts to map cerebellar function have, however, been limited by the fact that available cerebellar atlases are generally limited to gross morphologic relationships (Crosby et al., 1962; Carpenter et al., 1976; DeArmond et al., 1976; Waddington et al., 1984; Roberts et al., 1987; Kretschmann and Weinrich, 1992), that the individual cerebellar lobules are generally not labeled, and only limited sections are depicted in either one or two of the cardinal planes with large gaps between these. Furthermore, the terminology used to identify the fissures and lobules in these atlases is not uniform and is often contradictory.
Schmahmann et al. (2000) presented a human cerebellar atlas with sections at 2 mm intervals in three cardinal planes based on high-resolution T1-weighted Magnetic Resonance (MR) images of a single human cerebellum that was coregistered to the Montreal Neurological Institute (MNI) template (Evans et al., 1993) and annotated using a revised and simplified nomenclature. Using the above MR image atlas of the human cerebellum as a basis for identification of landmarks and fissures, Makris et al. (2005) developed a manual method aided by a set of computer-assisted algorithms to facilitate the parcellation of the cerebellar cortex into 32 parcellation units (PUs) per hemicerebellum in a manageable period of time. In their implementation, the fissures divide the cortex into lobules, while longitudinal divisions separate the vermis from the hemispheres, and subdivide the hemispheres into medial and lateral zones. The large lateral hemispheric region of Crus I and II is divided into a further two zones. The authors found that intraclass correlation coefficients (McGraw and Wong, 1996; Shrout and Fleiss, 1979) for both intra- and inter-rater reliability were significantly improved by clustering PUs according to either lobar divisions, anatomical connectivity, or functional connectivity. Lobar clusters are widely used (Pierson et al., 2002) and divide the cerebellum into anterior, posterior and flocculonodular lobes that are separated by the primary and the posterolateral fissures, respectively. In all these studies only data acquired from adults were used.
Subsequently, Diedrichsen (2006) developed the high-resolution spatially unbiased infra-tentorial template (SUIT) of the cerebellum by normalizing individual cerebella of 20 healthy adults non-linearly to each other before averaging, which improved specificity when labelling regions in functional MRI data.
Although atlases are widely used to assign anatomical labels to locations, there is a high risk for error due to high spatial variability of individual cerebellar anatomy, and even more so between different populations. Probabilistic atlases enable the assignment of labels to specific regions while also providing a quantitative measure of the uncertainty of such assignments. Currently, whole brain probabilistic atlases typically treat the cerebellum as a single structure without any lobular divisions (Hammers et al., 2003; Shattuck et al., 2008). In 2009, the first probabilistic cerebellar SUIT atlas was created (Diedrichsen et al., 2009) based on manual tracings of lobules on T1-weighted MRI scans (1 mm isotropic resolution) of 20 healthy adult participants (10 male, 10 female, age range 19–27 years). The SUIT atlas defines twenty-eight compartments: lobules I–IV and V divided into left and right hemispheres; lobules VI, Crus I, Crus II, VIIb, VIIIa, VIIIb, IX, and X divided into vermal sections in addition to left and right hemispheres. This atlas aims specifically to improve inter-subject co-registration of cerebella to yield improved specificity of cerebellar activations and valid assignments of functional activations to specific cerebellar lobules.
We were interested in examining cerebellar anatomy in children (age 9–13 years) from the Cape Coloured (mixed ancestry) community in Cape Town, South Africa. Since manual tracing is both time intensive and subjective, we wanted to perform automatic cerebellar segmentation. It has been noted previously, however, that a specialized atlas should be created for research in children (Diedrichsen, 2006) as the shape and ratio of gray matter to white matter in the cerebella of children differ significantly from that of adults (Fonov et al., 2011). To our knowledge, no pediatric cerebellar atlas is currently available.
In developing an atlas, an important decision relates to the registration target (template). A template closer to the study population reduces morphometric bias and the amount of nonlinear deformation required to establish spatial alignment (Yoon et al., 2009) between the template and subject. Wilke et al. (2008) developed the ‘Template O Matic’ toolbox for SPM that creates an age specific whole brain template by initially using linear co-registration of subjects and regressing for age and gender in pediatric populations. The resulting template, however, appears smoothed and lacks anatomical detail in the regions of greatest variability.
The unbiased nonlinear National Institutes of Health Pediatric Database (NIHPD) template created by Fonov et al. (2011) provides better spatial resolution and improved contrast compared to the classical International Consortium for Brain Mapping (ICBM152) template. NIHPD templates are available for different age ranges for normal brain development; the pediatric population was grouped into five categories between the ages of 4.5 and 18.5 years. For each category, two templates were constructed, one that preserves asymmetry and another with symmetric hemispheres. Deformation studies using these five different templates have shown that the average magnitude of deformation increases with increasing difference in age between the template used and the subject being studied (Fonov et al., 2011). In the present study we used the NIHPD unbiased nonlinear template that preserves asymmetry closest to the age range of our subject population (7.5 – 13.5 years).
Although recent advances in image segmentation have demonstrated that multi atlas segmentation improves accuracy over standard atlas based approaches, these have rarely been applied to cerebellar segmentation, possibly due to the need for a large number of manually segmented atlases, which is both time intensive to construct and requires extensive expertise. Pipitone et al. (2014) recently developed the Multiple Automatically Generated Templates (MAGeT-Brain) algorithm, which minimizes the number of atlases needed by propagating the atlas segmentations to a template library constructed from a subset of the target images. Using this approach and manually segmented atlases of only 5 adult cerebella, Park et al. (2014) demonstrated good accuracy using voxel wise majority voting compared to “gold standard” manual segmentations in the identification of all lobules (mean Dice Similarity Coefficient [DSC] = 0.73; range 0.40–0.89) and the entire cerebellum (mean DSC = 0.93; range 0.90–0.94) in 20 adults (10 healthy controls; 10 patients with schizophrenia). Bogovic et al. (2013) demonstrated superior performance compared to SUIT atlas based and multi atlas fusion approaches using multiple object geometric deformable models. To the author’s knowledge, no multi atlas cerebellar segmentation pipeline is available that has been tailored to pediatric datasets and is based on training data from children. Further, the parametric generative model (Iglesias et al., 2012; Iglesias et al., 2013) has not been applied to cerebellar segmentation.
In this work, we present the probabilistic Cape Town Pediatric Cerebellar Atlas (CAPCA18) for improved labelling of cerebellar structures in children. The atlas was constructed in the already established age-appropriate NIHPD template space from manual tracings of 16 cerebellar compartments in 18 healthy children (age range 9–13 years, 6 male) according to the nomenclature introduced in the MRI atlas of the human cerebellum (Schmahmann et al., 2000). The probabilistic presentations of each compartment provide a quantitative measure of the spatial variability in that region. In addition, manually traced cerebella of the training subjects were used to implement multi atlas label fusion using both voxel wise multi atlas majority voting (MAMV) and multi atlas generative model based label propagation (MAGM) for comparison to atlas based segmentation. Segmentation accuracy compared to manual tracings was evaluated for the CAPCA18 atlas and each of the two multi atlas methods in an independent sample of 14 healthy children (age range 8.9–11.8 years, 10 male).
2. Materials and Methods
High resolution T1-weighted structural images were acquired on a 3T Allegra (Siemens, Erlangen, Germany) MRI scanner using a magnetization prepared rapid gradient echo (MPRAGE) sequence (TR 2300ms, TE 3.93ms, TI1100ms, 160 slices, flip angle 12 degrees, 1.3×1.0×1.0mm3, 6.03 minutes) in 18 healthy children (mean age 11.8±1.2 years) from the Cape Coloured community in Cape Town, South Africa who were recruited as typically developing controls for ongoing studies of Fetal Alcohol Spectrum Disorder (Jacobson et al., 2008). A further 14 healthy children (mean age 10.5 ± 0.8 years) from the same community were scanned using a volumetric navigated (Tisdall et al., 2012) multiecho (ME) MPRAGE sequence (van der Kouwe et al., 2008) (128 sagittal slices, TR 2530 ms, TE 1.53/3.21/4.89/6.57 ms, TI 1100 ms, flip angle 7 degrees, 1.3×1.0×1.3 mm3). All children were scanned according to protocols that had been approved by the Faculty of Health Sciences Human Research Ethics Committee at the University of Cape Town; parents of all children provided written informed consent and children provided oral assent.
Image Preprocessing
The images were reoriented with the horizontal line defined by the anterior posterior commisure (ACPC orientation) and the sagittal planes parallel to the midline. The images were resampled to isotropic 1mm3 voxels using windowed sinc interpolation in Brain Voyager (Goebel et al., 2006). The resulting images were cropped to a fixed bounding box by removing the empty slices from the volume using a customised script written in MATLAB (www.mathworks.com). This process ensured removal of the neck and generated similarly oriented whole brain images.
Manual Tracing
The cerebella of the 18 training subjects were manually traced in native space according to the revised nomenclature defined by Schmahmann et al. (2000) using Multitracer (Woods et al., 2003) software on a tablet PC by an expert neuroanatomist (CW) who was blind to the age and sex of the children. Although tracings were largely performed in the sagittal view, all three planes were used to ensure three dimensional (3D) continuity. Tracings performed in the coronal and axial planes appeared as dots in the sagittal view and were used to define lateral boundaries of structures. Tracings were performed at four times magnification. The following lobules of the cerebellum were traced on both the left and right hemispheres: lobules I–V, VI, Crus I, Crus II, VIIb, VIII, IX and X.
The right hemispheres of 10 randomly selected subjects were re-traced at a later time by CW in order to compute intra-rater reliabilities. Cerebella of 14 test subjects were traced by a different neuroanatomist (NB) using the same protocol. Inter-rater reliabilities for lobules of eight cerebellar hemispheres traced by both neuroanatomists were compared using intraclass correlation coefficients (ICC).
The midline slice was defined as the sagittal slice in which the cerebral aqueduct was most clearly visible. Left and right hemispheric subregions were traced separately. The anterior lobe comprises lobules I–V; lobule VI, Crus I and II, and lobule VIIb form the superior-posterior lobe; and lobules VIII, IX and X comprise the inferior posterior lobe. For each lobule, the regions demarcated by the drawn contours as belonging to that lobule were masked on all relevant slices and combined to construct a 3D volume of each lobule. The resulting volumes and their surfaces were inspected visually to identify and correct tracing errors (examples shown in Fig.1) in an iterative way. The resulting masks define 16 cerebellar compartments, 8 in each hemisphere, each of which was labeled with a unique integer value. Tracings for one hemisphere of one brain are shown in the sagittal plane in the right panel of Fig. 2, with the corresponding masked regions and their color representations shown in the middle and left panels, respectively. For each subject we also generated a cerebellar mask comprising the sum of the gray matter parcellations and the total cerebellar white matter.
Fig. 1.
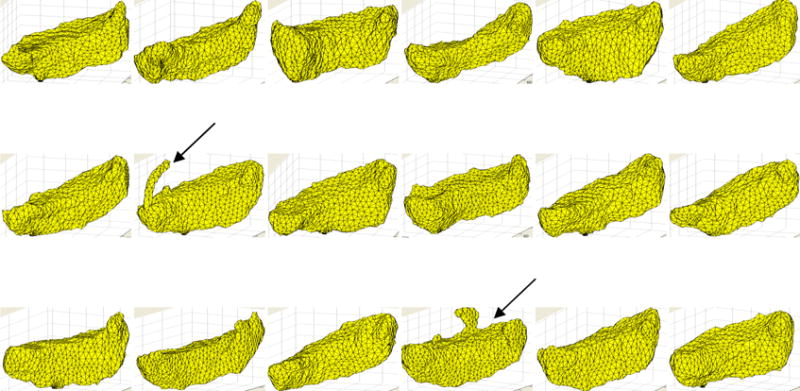
Illustration of CrusII volumes for the 18 training subjects with tracing errors in two (indicated by the black arrows), which were manually corrected by the expert neuroanatomist.
Fig. 2.
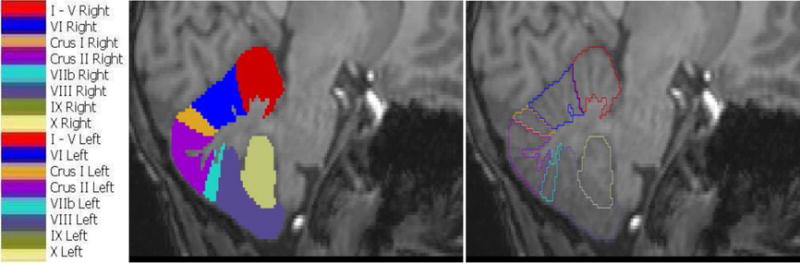
Cerebellar lobules and their color representations. Left and right hemispheric structures are represented using the same colors and unique labels. The image on the right shows manually traced contours in the sagittal view for one subject, while the middle panel shows the corresponding masked regions and their color representations on the left.
Construction of the CAPCA18 Probabilistic Atlas
The T1-weighted images of the 18 training subjects were spatially normalized to the age-appropriate NIHPD asymmetric template (Fonov et al., 2011) using discrete cosine non-linear deformation. We used the SPM5 unified segmentation (Ashburner and Friston, 2005) method that uses a mixture of Gaussian models to simultaneously perform spatial normalization, bias correction, and tissue classification. The tissue probability maps (gray/white/cerebrospinal fluid (CSF)) of the template were used to classify tissues as gray matter, white matter and CSF. Further, all SPM’s default settings were used. Following alignment of the T1 images to the template, individual lobular masks and cerebellar masks of each subject were resampled into the template space using nearest neighbour interpolation.
Co-registered cerebellar masks were averaged in the NIHPD template space to generate an average cerebellar mask. In this work, we chose to set probabilities of voxels with values of 0.3 or less equal to zero, as these voxels were part of the cerebellum in only 30% or fewer of the subjects, and values greater than 0.3 equal to unity. Using a threshold helps to reduce the effects of outliers and individual subject variance. The average of the whole brain images of the training subjects normalised to the NIHPD template space was multiplied by the resulting cerebellar mask to construct an average cerebellar image (Fig. 3A).
Fig. 3.
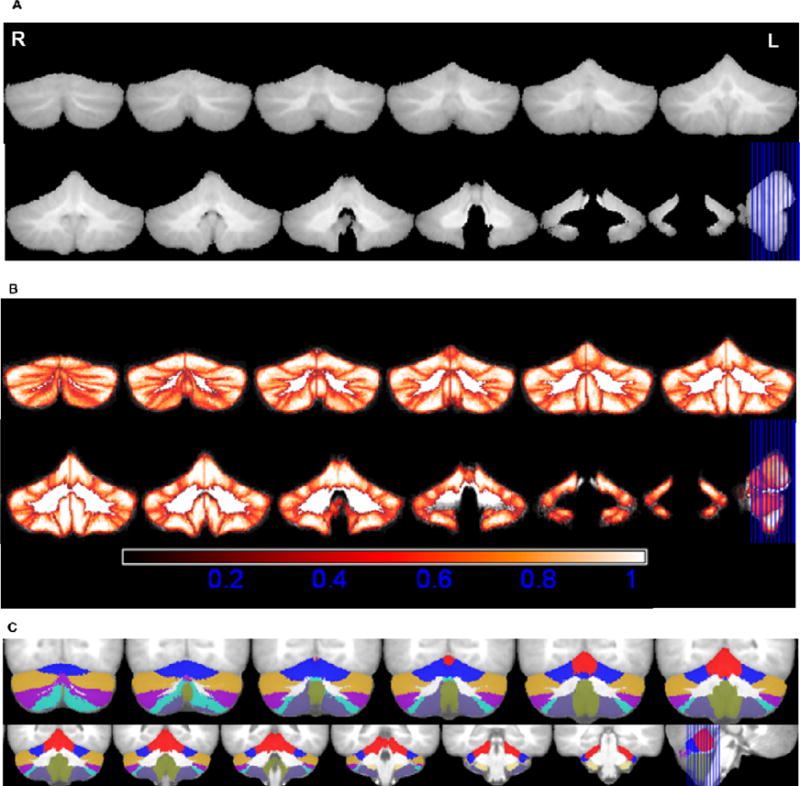
A. Average cerebellar image in normalised space, R-Right, L-Left; B. Maximum Probability maps of individual structures; bright colors (white) indicate voxels with high probability and darker (red) colors indicate voxels with lower probability. C.Maximum likelihood labellings superimposed on the average image generated after spatial normalization with the NIHPD (7.5–13.5 years) asymmetric template. The colors represent the lobules of the cerebellum as per the color look up table in Figure 2. The coronal slices range from MNI coordinates −75 to −33.
In a similar way, resampled lobular masks in normalised space were averaged across subjects to generate probability maps for each structure, where the value in each voxel location denotes the probability of a voxel belonging to said structure. Typically, there is spatial variability of individual cerebellar structures between subjects even after alignment to a reference template. The probability maps indicate the proportion of subjects in whom a specific lobule occupies a location in the reference space and as such also quantifies the spatial variability in different regions. This enables one to visualize both the spatial extent of individual sub-regions and their spatial variability. Fig. 3B shows the maximum probability map generated by combining the averaged lobular masks in normalised space. Each voxel is assigned a value equal to the maximum probability for that location. Further, voxels for which the maximum probability is 0.3 or less are set to zero as this indicates that these voxels belonged to a cerebellar lobule in 30% or fewer of the subjects in the initial dataset.
From the maximum probability map, we constructed an atlas of maximum likelihood labellings, which assigns to every voxel a label indicating the individual cerebellar structure located at that voxel most often. At the boundaries between structures, where a voxel may have equal probability of belonging to two different lobules, the label that occurs most often in a 3×3×3 mm3 region surrounding the voxel was assigned to the voxel (Fig. 3C).
CAPCA18 Atlas Based Segmentation
The images of the 14 test subjects were preprocessed using the same pipeline as the training subjects. The unified normalisation algorithm in SPM5 was applied to simultaneously perform tissue classification and normalisation of each subject’s whole-brain images to the NIHPD asymmetric template. Labels from our probabilistic CAPCA18 atlas were then resampled to subject space using the inverse of the deformation warps obtained during normalisation to obtain CAPCA18 based cerebellar segmentations for each test subject.
We computed for each test subject both individual lobular volumes and total cerebellar gray matter volume, and compared the percentage of total gray matter volume that each lobule occupies in children from our study with values previously reported in adults (Makris et al., 2005).
SUIT Atlas Based Segmentation
For comparison, automated atlas based segmentation was also performed using the SUIT cerebellar atlas available in SPM5 that was developed using adult data. Briefly, each subject’s images were cropped to the region of the cerebellum after co-registration to the ICBM152 template. Individual cerebella of the test subjects were normalised to the SUIT template to obtain deformation maps for each image. Manual editing was required around the anterior borders between the cerebellum and the temporal and occipital lobes for 8 of the test subjects. After editing the cerebellar cortex achieved 78% average overlap with the SUIT template, which is comparable with the 75% previously reported (Diedrichsen et al., 2009). Normalisation was repeated after manual editing and the labels from the SUIT atlas were resampled to the cropped test subject space using the inverse of the deformation warps obtained during normalisation. Finally, labels were resampled to native test subject space using the suit_reslice command.
Comparison of CAPCA18 and SUIT Atlas Based Segmentation
Using Dice’s Similarity Coefficient (DSC; Dice et al., 1945) to quantify of spatial overlap with manual tracings, we compared cerebellar segmentation accuracy using our CAPCA18 atlas and the SUIT atlas. SUIT labelling differs from the conventions followed in this paper, in that the vermis is defined as a single medial entity in SUIT, while it was not traced separately in our work and as such was included in our hemispheric regions. In order to avoid errors due to omission or inclusion of the vermal regions when comparing data from SUIT to manual tracings, the left and right SUIT hemispheric regions and the SUIT vermal region were combined for each lobule to define whole lobules and compared to whole lobules (left plus right) from manual tracings. To facilitate comparison of the two atlases, spatial overlap of CAPCA18 segmentations and manual tracings were also computed in 8 whole lobules. In addition, we compared in each of the 14 test subjects the spatial overlap of the total cerebellar gray matter from manual tracing with that obtained using automated SUIT and CAPCA18 segmentations, respectively.
Multi Atlas Segmentation
The images of each of the training subjects and their respective label maps generated from the manual tracings are collectively referred to as the training atlases.
The whole brain images of the training and test subjects were processed using Freesurfer software (http://surfer.nmr.mgh.harvard.edu/). The processing steps included skull stripping (Ségonne et al., 2004), intensity normalisation (Sled et al., 1998), cortical and subcortical structure segmentation, surface generation and cortical thickness calculation (Dale and Sereno, 1993; Dale et al., 1999; Fischl et al., 1999). The surfaces were superimposed on the T1 weighted images and visually assessed for correctness of gray-white and gray-CSF boundaries. White matter boundaries were edited manually to ensure correct surface generation.
After processing with Freesurfer, the surfaces from the training subjects were used as the moving sources and the surfaces of the test subjects as the targets for pair-wise combined volume- and surface-based (CVS) registration of each training image to each target image. The CVS registration tool (Postelnicu-Zöllei et al., 2009) achieves good correspondence between both cortical folding patterns and subcortical structures. We performed the complex registration on a high performance computer cluster facility which has 127 batch nodes, two quad-core Xeon 5472 3.0 GHz CPUs and 32 GB RAM. All nodes were running the 64-bit version CentOS 6. The pair-wise registration between the training and test surfaces yielded 252 (18×14) registered outputs and their associated deformation warps.
To obtain the final segmentation for each test subject, we combine all the registered atlases obtained by applying to each training atlas the deformation from the CVS registration of the corresponding training image to the relevant test image. Two different label propagation techniques were employed: multi atlas majority voting (MAMV) (Heckemann et al., 2006) and multi atlas generative model based label propagation (MAGM; Iglesias et al., 2012). Figure 4 shows the processing pipeline for our multi atlas based segmentation.
Fig. 4.
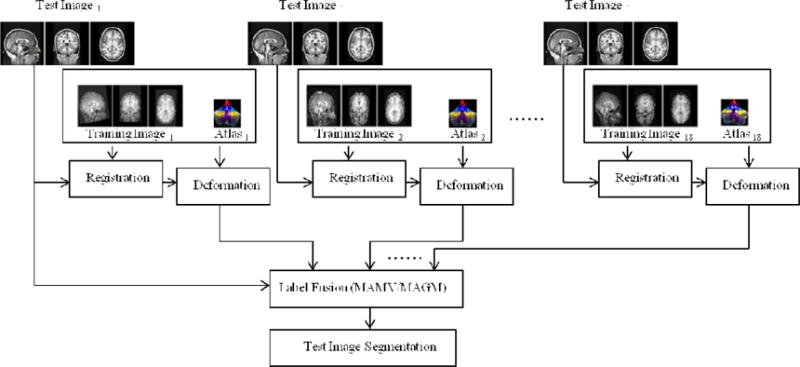
Flow chart showing the processing pipeline for multi atlas based segmentation. Every image in the training set is registered to the individual test subject’s image, whereafter the resulting deformation is applied to each training atlas. Two different label fusion strategies are used to propagate labels from the registered training atlases to the image of the test subject.
Majority voting assigns a label to each voxel of the test image by determining the label that occurs most frequently in the corresponding voxel on all the registered training atlases. All training images are weighted equally and it is assumed that each training atlas represents an accurate segmentation. The effects of independent noise associated with a particular label are reduced using this approach.
The generative model approach uses probabilistic modelling of segmentations from a set of segmented images to generate a second set of final segmentations. The model is independent of the intensity of the training images. The intensity of the test image and the respective deformed outputs are used to model the final segmentation of the test image. The mathematical formulation of the model was derived from the implementations by Sabuncu et al. (2010) and Iglesias et al. (2012). The model includes a single model parameter β, where β = 0 treats each voxel independently, β = ∞ corresponds to a single label map, and β = finite value yields the combined output. In our work we used β=0 to obtain the fused output from MAMV and β=0.3 for MAGM.
Comparison of Atlas Based and Multi Atlas Methods
Segmentation accuracy in the 14 test subjects was compared for CAPCA18, MAMV, and MAGM segmentation using both volumetrics and DSC scores as a measure of spatial overlap with manual tracings.
3. Results
Intra-rater reliabilities for 10 right hemispheres traced on two separate occasions by CW yielded ICCs ranging from 0.72 to 0.96 for different structures (Table 1). Only in the small lobules IX and X were ICCs below 0.85. Inter-rater reliabilities for lobules of 8 cerebellar hemispheres traced by two different neuroanatomists (CW, NB) show ICCs greater than 0.8 in five of eight lobules traced. ICCs in VIIb and X were unacceptably low, indicating discrepancy between the raters, suggesting that these regions are difficult to trace reliably.
Table 1.
Intra-rater reliability for tracings repeated in 10 right hemispheres by an expert neuroanatomist, and inter-rater reliability for lobules traced in 8 hemispheres by two different neuroanatomists.
| Cerebellar Lobules | ICC Intra-rater | ICC Inter-rater |
|---|---|---|
| I–V | 0.91 | 0.92 |
| VI | 0.87 | 0.83 |
| Crus I | 0.85 | 0.81 |
| Crus II | 0.89 | 0.69 |
| VIIb | 0.85 | 0.43 |
| VIII | 0.96 | 0.88 |
| IX | 0.83 | 0.87 |
| X | 0.72 | 0.46 |
Table 2 gives the average lobular volumes as a percentage of total cerebellar gray matter volume for the 14 test subjects after automatic segmentation using the CAPCA18 atlas. The average gray matter volume for our test subjects is 113.2 ± 11.5cm3, which compares favourably with gray matter volumes of 112.9 ± 18.9 cm3 reported by Makris et al. (2005) in adults. Crus I, Crus II and lobule VIIb together occupy 49.4% of the total cerebellar gray matter. For comparison, percentage lobular volumes reported by Makris et al. (2005) for adults are included in Table 2. Proportionally, lobules VI, VIIb and X comprise smaller fractions of the total cerebellar gray matter in children, while lobule IX is proportionately about 50% larger than in adults.
Table 2.
Mean and standard deviation (SD) of percentage lobular volumes (as a percentage of total cerebellar gray matter volume) for 14 test subjects using CAPCA18 atlas based segmentation.
| Lobule | Right Hemispheric Volume (% of total cerebellar gray matter volume) | Left Hemispheric Volume (% of total cerebellar gray matter volume) | Total Hemispheric Volume (% of total cerebellar gray matter volume) | Total Hemisphere Volume (Makris et al., 2005) (% of total cerebellar gray matter volume) | % Difference between relative lobular sizes in pediatric and adult cerebella | ||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | Mean | ||
| I–V | 5.9 | 0.3 | 6.7 | 0.4 | 12.6 | 12.1 | 4.1 |
| VI | 6.4 | 0.4 | 7.0 | 0.5 | 13.4 | 16.3 | −17.8 |
| Crus I | 12.5 | 1.4 | 12.2 | 1.5 | 24.7 | 23.5 | 5.1 |
| Crus II | 9.3 | 1.2 | 8.5 | 1.2 | 17.8 | 16.4 | 8.5 |
| VIIb | 3.3 | 0.4 | 3.7 | 0.5 | 6.9 | 9.0 | −23.3 |
| VIII | 7.2 | 1.0 | 6.6 | 0.8 | 13.8 | 14.9 | −7.4 |
| IX | 4.7 | 0.5 | 4.9 | 0.5 | 9.6 | 6.4 | 50.0 |
| X | 0.6 | 0.1 | 0.6 | 0.0 | 1.2 | 1.4 | −14.3 |
| Total | 49.9 | 4.8 | 50.2 | 5.0 | 100.0 | 100.0 | |
Figure 5 compares for the 14 test subjects DSC scores that quantify spatial overlap in 8 whole lobules between manual tracing and automated segmentation using either the SUIT (red) or CAPCA18 (blue) atlases. CAPCA18 achieves significantly better (student’s T-test) DSC scores with respect to manual segmentations than SUIT in lobules I–V, VI, CrusI, VIII and IX, and overall exhibits lower variability in DSC scores. DSC scores drop below 0.6 in most subjects in lobules VIIb and X using both atlases.
Fig. 5.
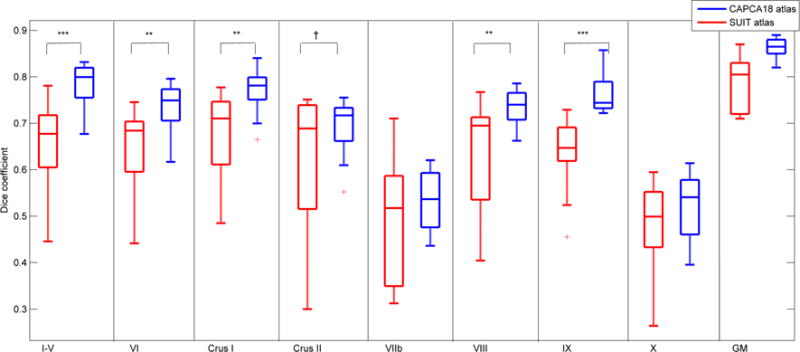
Box-and-whisker plots of Dice coefficients that quantify spatial overlap in 14 test subjects of manual tracing with automatic segmentation using either the SUIT (red) or CAPCA18 (blue) atlases for eight whole lobules and total cerebellar gray matter (GM). †p<0.1; *p<0.05; **p<0.01; ***p<0.001
In all but one of the test subjects, total cerebellar gray matter from CAPCA18 segmentation showed greater spatial overlap with manually traced total cerebellar gray matter than SUIT. On average, gray matter spatial overlap using SUIT before and after manual editing were 75% and 78%, respectively, while it was 86% using the fully automated CAPCA18 atlas based segmentation.
Table 3 presents average lobular volumes for the 14 test subjects using each of the automated segmentation methods compared to “gold-standard” manual segmentation. Volumes of lobules VIIb and X differed from manually traced volumes using all three methods. Further, left Crus I and IX were larger and left Crus II smaller using CAPCA18 atlas based segmentation compared to manual tracing. In all other regions, volumes from automated segmentation were similar to those from manual tracing for all three methods.
Table 3.
Average lobular volumes from automated segmentation using the CAPCA18 atlas, multi atlas majority voting (MAMV), and multi atlas generative model (MAGM), respectively, compared to manual segmentation volumes (Paired Student’s t-test).
| Lobule Name | Manual Tracing volume (cm3) | CAPCA18 segmentation volume (cm3) | t | MAMV segmentation volume (cm3) | t | MAGM segmentation volume (cm3) | t |
|---|---|---|---|---|---|---|---|
| Right Hemisphere | |||||||
| I–V | 6.3 (1.1) | 6.1 (0.3) | 0.9 | 6.2 (1.0) | 0.9 | 6.4 (0.9) | −0.04 |
| VI | 6.8 (1.1) | 6.5 (0.1) | 0.8 | 7.0 (0.8) | −1.8 | 6.9 (0.5) | −0.04 |
| Crus I | 12.0 (2.1) | 12.5 (0.2) | −1.3 | 12.2 (1.4) | −0.7 | 12.1 (1.2) | −0.6 |
| Crus II | 9.5 (1.9) | 9.2 (0.2) | 0.9 | 9.4 (1.6) | 1.3 | 9.4 (1.2) | 0.5 |
| VIIb | 3.6 (0.4) | 3.3 (0.0) | 2.3† | 4.3 (1.1) | −2.8** | 4.4 (1.1) | −2.9** |
| VIII | 7.5 (1.2) | 7.1 (0.2) | 1.4 | 7.4 (0.8) | 0.5 | 7.4 (0.5) | 0.5 |
| IX | 4.2 (0.5) | 4.6 (0.1) | −2.1† | 4.4 (0.5) | −1.2 | 4.3 (0.4) | −0.5 |
| X | 0.4 (0.0) | 0.6 (0.0) | −9.4*** | 0.5 (0.1) | −7.0*** | 0.5 (0.1) | −12.0*** |
| Left Hemisphere | |||||||
| I–V | 6.5 (1.2) | 6.7 (0.3) | −0.8 | 6.6 (0.5) | −0.6 | 6.4 (0.5) | 0.5 |
| VI | 6.7 (1.2) | 7.0 (0.1) | −1.2 | 7.0 (0.8) | −0.8 | 6.9 (0.1) | −0.6 |
| Crus I | 11.0 (1.2) | 12.1 (0.2) | −4.1* | 11.8 (1.3) | −0.8 | 10.9 (1.0) | 0.2 |
| Crus II | 10.4 (2.9) | 8.4 (0.2) | 3.9* | 10.5 (1.2) | −0.2 | 10.3 (1.0) | 0.3 |
| VIIb | 3.0 (0.4) | 3.7 (0.1) | −4.3* | 3.8 (0.5) | −3.1** | 3.6 (0.7) | −2.9** |
| VIII | 7.2 (2.0) | 6.6 (0.0) | 1.5 | 7.4 (0.5) | −0.7 | 7.3 (0.5) | −0.5 |
| IX | 4.3 (0.3) | 4.9 (0.1) | −3.7* | 4.8 (0.4) | −1.5 | 4.7 (1.0) | −1.4 |
| X | 0.4 (0.0) | 0.6 (0.0) | −12.9*** | 0.5 (0.1) | −7.0*** | 0.5 (0.1) | −7.0*** |
Values are Means (standard deviations)
p<0.1;
p<0.05;
p<0.001;
p<0.0001
For comparison purposes, we present in Table 4 a comparison of mean whole lobular volumes (left and right hemispheres and vermis combined) in the 14 test subjects from segmentation using the SUIT atlas with volumes obtained from manual tracings. Hemispheric volumes from SUIT segmentation were significantly different to those from manual tracing in four of eight lobules (paired Student’s T-test), and tended to be larger, although below conventional levels of significance, in a further three lobules. Volumes were equivalent only in lobule VIII. Notably, one of the regions that SUIT overestimates is lobule VI, which is proportionately smaller in children than in adults, while lobule IX, which is underestimated with SUIT, is proportionately larger in children.
Table 4.
Comparison of mean and standard deviation (SD) of volumes of cerebellar lobules obtained from manual tracing and SUIT segmentation in 14 test subjects.
| Cerebellar Lobules | Cerebellar hemispheric volumes | ||
|---|---|---|---|
| Volume from Manual Tracing (cm3) | Volume from SUIT Segmentation (cm3) | t | |
| I–V | 12.8 (2.1) | 14.3 (2.5) | −1.6† |
| VI | 13.5 (2.0) | 18.3 (3.0) | −5.9*** |
| Crus I | 23.0 (1.8) | 20.0 (4.4) | 2.5* |
| Crus II | 19.9 (1.7) | 15.0 (4.2) | 3.8** |
| VIIb | 6.7 (0.9) | 7.5 (1.9) | −1.5† |
| VIII | 14.7 (2.2) | 14.0 (3.2) | 0.8 |
| IX | 8.6 (1.2) | 5.8 (1.2) | 5.3*** |
| X | 0.8 (0.1) | 0.9 (0.2) | −1.6† |
Values are Mean (SD);
p<0.1;
p<0.05;
p<0.001;
p<0.0001
In Figure 6 we compare manual segmentation with automated segmentation for each of the three methods using spatial overlap, expressed as DSC scores. On average, spatial overlap between manual and automated segmentations are 70% or higher in all regions for all three automated segmentation methods, except lobules VIIb and X where it is between 50% and 70%. Notably, these are the two regions where inter-rater reliabilities for manual tracings were unacceptably low. In right VI, VIIb, and X, and left I–V, VI, CrusI, CrusII, and X MAGM produced better spatial overlap with manual tracings than CAPCA18 segmentation, while MAMV produced better spatial overlap than CAPCA18 bilaterally in lobules VIIb and X. The mean DSC score across all lobules for MAGM segmentation was 0.76 (range 0.55 – 0.91), compared to 0.67 (range 0.40 – 0.84) and 0.73 (range 0.52 – 0.89) for CAPCA18 and MAMV segmentation, respectively. The mean DSC of the entire cerebellum was 0.86, 0.88 and 0.90 for CAPCA18, MAMV and MAGM segmentation, respectively.
Fig. 6.
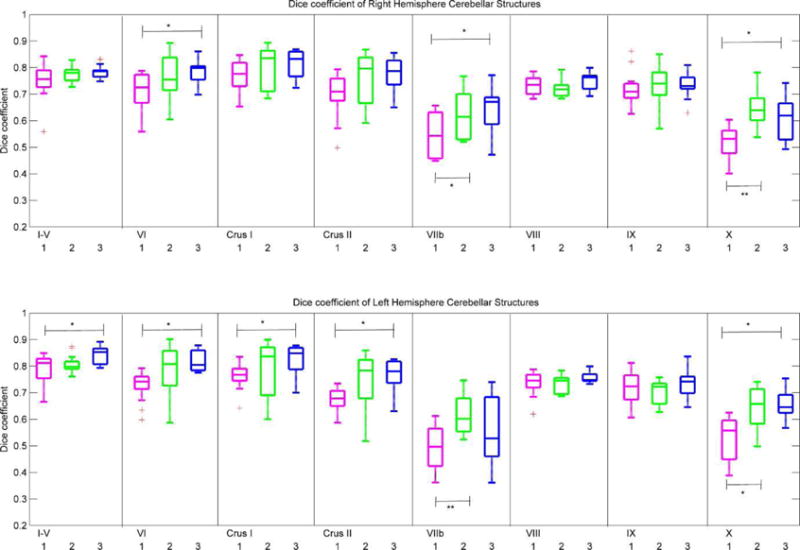
Box-and-whisker plots of Dice coefficients in the right hemisphere (top row) and left hemisphere (bottom row) that quantify spatial overlap in 14 test subjects of manual tracing with each of three automated segmentation strategies, (1): CAPCA18 probabilistic atlas based segmentation (magenta); (2): multi atlas majority voting (MAMV; green); (3): multi atlas generative model (MAGM; blue). †p<0.1; *p<0.05; **p<0.01; ***p<0.001.
4. Discussion
We have presented the CAPCA18 probabilistic three dimensional cerebellar atlas for children with parcellations into 16 macroanatomic structures, as well as a pipeline for multi atlas segmentation that utilises the individual atlases of the 18 training children with either MAMV or MAGM based label fusion. We evaluated in 14 test children the segmentation accuracy of CAPCA18, SUIT, MAMV and MAGM compared to manual tracing. Overall MAGM segmentation yielded the best segmentation accuracy compared to manual tracing across all lobules.
Although manual delineation is extremely labour intensive, it is still considered the gold standard for volumetric assessment. Observer bias may, however, affect the results whenever a manual method is used. Prior studies of the cerebellum have emphasized the challenges associated with manual delineation of Crus II, lobules VIIb and VIII (Diedrichsen et al., 2009; Bogovic et al., 2013) and that raters should consider different image features like size, depth of fissures, location of the fissure, starting and ending points of the fissure, and lobule boundaries when deciding where to draw boundaries. Our expert neuroanatomists completed manual tracing of individual lobules of a complete cerebellum in roughly 30–60 minutes. Intra-class correlation coefficients (ICCs) of individual lobules traced at two different times by the same tracer were greater than 0.8 in all lobules, except lobule X where it was 0.72, confirming the reliability and robustness of the tracing protocol used in this study. Inter-rater reliabilities below 0.5 in lobules VIIb and X indicate that these regions are difficult to trace reliably and may be prone to observer bias. Since the training data that were used in atlas construction were traced by one tracer, and the test data used for validation by the other, we expected poor performance of our automatic segmentation algorithm in these lobules.
Fissures in an average child brain are not as clearly distinguishable as in an average adult brain and the shape and ratio of gray matter (GM) to white matter (WM) also differs, with differences ranging from 4 to 23% (Fonov et al., 2011). As a result, improved co-registration and segmentation results are expected using a cerebellar atlas for children. The CAPCA18 atlas provides both a measure of inter-subject variability in different cerebellar structures and assigns to each voxel a unique label according to the label most often found in that location in the training data. To our knowledge, this is the first pediatric cerebellar atlas that can serve as a better spatial prior to analyze cerebellar morphometry in children.
The average percentage gray matter volume occupied by each lobule in the cerebellar hemispheres was compared with values previously reported by Makris et al. (2005) to examine potential relative size differences in children compared to adults. It was found that lobule IX is proportionately larger in children, while lobules VI, VIIb and X are proportionately smaller. The increased volume in lobule IX in children may be due to increased gray matter around the posterior lobe of the cerebellum. It has been shown previously when deforming pediatric brains to the adult ICBM152 nonlinear template that gray matter around the posterior lobe is increased in children (Fonov et al., 2011).
Spatial overlap of CAPCA18 segmentations with manual tracings was greater than 73% bilaterally in lobules I–V, VI, crus I, crus II, VIII and IX. These values are comparable with DSC scores that have been reported in other studies (Bogovic et al., 2013; Park et al., 2014). Notably, lobules VIIb and X, which yielded low DSC scores (50–70%), are the smallest cerebellar lobules, comprising only 1.2% (lobule X) and 6.9% (lobule VIIb) of the total cerebellar gray matter, so that even small differences will result in low Dice coefficients. These are also the regions that are difficult to trace reliably as evidenced by their poor inter-rater reliabilities. The accuracy of structure segmentation depends on its spatial location and the tissues surrounding the structure. The inferior part of lobule VIIb comprises a very thin layer of gray matter and the inclusion of additional white matter during manual tracing of this structure in the training subjects may explain the low accuracies of automatic segmentation in this region.
Bogovic et al. (2013) who used an automatic classification of cerebellar lobules algorithm that employs an implicit multi-boundary evolution (ACCLAIM) approach, obtained greater spatial overlap with manual tracings than we did with CAPCA18 only in lobules VI (ACCLAIM 0.72–0.83; CAPCA18 0.69 – 0.77) and IX (ACCLAIM 0.78 – 0.88; CAPCA18 0.72 – 0.86). The ACCLAIM method, however, relies on good contrast between CSF and GM and high spatial resolution (0.828 × 0.828 × 1.1mm3) and as such may perform less well in our pediatric data.
Average spatial overlap of the whole cerebellar cortex in the test images with manual gray matter segmentations was 86% with CAPCA18, compared to 78% after normalisation to the SUIT template. Further, CAPCA18 segmentation yielded higher or similar DSC scores than SUIT in all lobules when compared to manual tracing. Our finding that SUIT underestimates the volume of lobule IX is consistent with those of another recent study (Park et al., 2014) and may, in part, be due to the fact that lobule IX is proportionately larger in children than in adults. In contrast, CAPCA18 obtains good spatial overlap with manual tracing in lobule IX and volumes are more similar (albeit bigger on the left) to those from manual segmentation. These findings suggest that our pediatric cerebellar atlas helps to reduce bias and segmentation errors that may result from using an atlas constructed from adult data.
Consistent with previous studies, multi atlas segmentation consistently performed better than CAPCA18 atlas based segmentation. There were no regions where CAPCA18 yielded better DSC scores than either MAMV or MAGM. Using MAGM segmentation, we obtained a mean DSC score across all lobules of 0.76 (range 0.55 – 0.91) and 0.90 (range 0.86–0.93) for the entire cerebellum. These values are in excellent agreement with those reported recently by Park et al. (2014) in adults (across all lobules mean DSC = 0.73, range 0.40– 0.89; entire cerebellum mean DSC = 0.93, range 0.90–0.94).
In the present work our training set comprised 18 cerebella that had been manually traced by an expert neuroanatomist as part of interlinking studies. The cost and time required to perform manual tracings in more children exceeded the resources that were available for the current project. Further, this number is comparable to the number of subjects that have been used in similar works – SUIT used 20 subjects. Park et al. (2014) demonstrated that only 5 atlases could provide accurate segmentation when combined with their MAG-eT Brain algorithm to generate more templates. Since our 18 training subjects yielded, in children of similar age and from the same population, segmentation accuracies that were comparable to those reported in other studies, we deemed the current number to be sufficient. In future studies we will need to evaluate whether this holds true when our algorithms are applied to different populations and different ages. Aljabar et al. (2009) demonstrated that using a subset of atlases, selected from a database of 275 based on image similarity or age, markedly improved spatial overlap with manual tracings compared to using a random subset of atlases. Further, the authors reported that simply using larger and larger numbers of atlases (after selection by image similarity or age) leads to lower accuracy in the resulting segmentation. Approximately 20 atlases produced near maximal accuracy across all structures examined in their study. While the availability of an independent sample of manually traced cerebella were important in the present study to validate the performance of our automated methods, we could increase our multiple atlas database to 32 by combining data from the 18 training and 14 test subjects in future studies.
It has been noted previously that direct comparison of quantitative segmentation results across publications are difficult and not always fair due to inconsistencies and discrepancies in the manual segmentation protocol, the quality of the imaging data, and differences in the patient populations (Wang et al., 2012). Despite these limitations, the present work demonstrates that cerebellar segmentation using the CAPCA18 atlas combined with multi atlas based methods consistently achieved greater segmentation accuracy in the present pediatric data set than the SUIT atlas derived from adult data.
We used publicly available SPM software to construct the CAPCA18 atlas, which ensures that our atlas can be easily used for both segmentation and localisation in morphometric and functional imaging studies, respectively.
For the test subject data used in the current study no manual editing was required when performing CAPCA18 segmentation. It is, however, important to check the images generated after initial preprocessing as well as after each intermediate step to ensure that no errors are introduced in the pipeline when computing the deformations between the atlas and test subjects.
One limitation of the present study is that the number of boys and girls were not equal in either the training or test data sets. In the training data set there were twice as many girls as boys, while 71% of the test subjects were boys. This could have introduced bias into the atlas, which may have resulted in poorer segmentation performance in the test subjects. We examined gender differences by comparing lobular volumes from manual tracings in boys and girls. Although lobular volumes in both hemispheres were equivalent between boys and girls (all p’s > 0.1) in this small sample, we expect that the atlas may be improved if the training data set had equal numbers of boys and girls.
Further, segmentation performance was evaluated in this work using volumetrics and DSC scores. DSC scores are, however, only comparable if they pertain to structures with similar surface-to-volume ratios (Rohlfing et al. 2004). While DSC scores are therefore adequate for comparing segmentation performance within a structure, they are suboptimal for identifying structures that are segmented less accurately than others and a surface distance measure, such as the symmetric Hausdorff distance, could provide additional insights.
5. Conclusion
This paper presents the probabilistic CAPCA18 atlas, and multi atlases constructed from the training data, that can be used to automatically segment the cerebella of children into 16 lobules. CAPCA18 segmentation, MAMV and MAGM label fusion were validated in 14 test subjects from the same age and ethnic group as the children whose data were used in the construction of the atlas. Although CAPCA18 segmentation consistently performed better than SUIT segmentation, MAGM yielded the greatest spatial overlap with manual tracings. CAPCA18 is the first pediatric cerebellar atlas and will be a useful tool in structural and functional imaging studies in children. Future work will focus on improving the accuracy of individual parcellations and validating the atlases in a larger number of subjects, different populations, and different age groups. Combining patch based segmentation with multi atlas label fusion methods can potentially improve the segmentation accuracy.
Highlights.
The probabilistic Cape Town Pediatric Cerebellar Atlas (CAPCA18) provides accurate assignment of 16 hemispheric regions.
In a pediatric dataset (age 9–12 years), automated CAPCA18 atlas based segmentation performs better than SUIT segmentation.
Multi atlas based label fusion using the 18 training atlases improves spatial overlap with manual tracings, compared to CAPCA18 segmentation.
Acknowledgments
Cerebellar neuroimaging data were collected with funding from NIH Fogarty International Research Collaboration (FIRCA; R03 TW007030) and National Institute on Alcohol Abuse and Alcoholism (NIAAA; R01AA016781). Recruitment of the Cape Town longitudinal cohort was funded by two administrative supplements from NIAAA to R01AA09524 and the NIH Office of Research on Minority Health; recruitment of the Cape Town cross-sectional cohort, by NIH FIRCA (R03 TW007030). This research was supported by funding from NIH/NIAAA (R21AA017410), a National Research Foundation of South Africa Focus Area grant (FA2005040800024), South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation of South Africa, Medical Research Council of South Africa, Children’s Bridge grant from the Office of the President of Wayne State University, a seed money grant from the University of Cape Town; and grants from the Joseph Young, Sr., Fund from the State of Michigan. LZ was supported for this research in part by NICHD grants 1K99HD061485-01A1 and R00 HD061485-03. We thank M. September, J. Croxford, and A.S. Marais for their work on recruitment and retention of the cohorts; the staff at the Cape Universities Brain Imaging Centre and our University of Cape Town research staff for collection of the neuroimaging data. We also express our gratitude to the mothers and children who have participated in the Cape Town Longitudinal Cohort Studies.
References
- Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D. Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. Neuroimage. 2009;46(3):726–738. doi: 10.1016/j.neuroimage.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Friston K. Unified segmentation. Neuroimage. 2005;26(3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Bogovic JA, Bazin P-L, Ying SH, Prince JL. (Lecture notes in computer science Volume 7917).Automated segmentation of the cerebellar lobules using boundary specific classification and evolution. 2013:62–73. doi: 10.1007/978-3-642-38868-2_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter MB. Human Neuroanatomy. Williams & Wilkins, Baltimore; Maryland: 1976. [Google Scholar]
- Crosby EC, Humphrey T, Lauer EW. Correlative Anatomy of the Nervous System. Macmillan; NewYork: 1962. pp. 188–192. [Google Scholar]
- Dale AM, Sereno MI. Improved localization of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction: a linear approach. Journal of Cognitive Neuroscience. 1993;5:162–176. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis I : Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- DeArmond SJ, Fusco MM, Dewey MM. A Photographic Atlas: Structure of the Human Brain. Oxford University Press; New York: 1989. [Google Scholar]
- Diedrichsen J. A spatially unbiased atlas template of the human cerebellum. Neuroimage. 2006;33(1):127–138. doi: 10.1016/j.neuroimage.2006.05.056. [DOI] [PubMed] [Google Scholar]
- Diedrichsen J, Balsters JH, Flavell J, Cussans E, Ramnani N. A probabilistic MR atlas of the human cerebellum. Neuroimage. 2009;46(1):39–46. doi: 10.1016/j.neuroimage.2009.01.045. [DOI] [PubMed] [Google Scholar]
- Evans AC, Collins DL, Mills S, Brown E, Kelly R, Peters TM. 3D statistical neuroanatomical models from 305 MRI volumes. Nuclear Science Symposium and Medical Imaging Conference, IEEE Conference Record. 1993:1813–1817. [Google Scholar]
- Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- Fonov V, Evans AC, Botteron K, Almli CR, McKinstry RC, Collins DL. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage. 2011;54(1):313–327. doi: 10.1016/j.neuroimage.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel R, Esposito F, Formisano E. Analysis of functional image analysis contest (FIAC) data with brainvoyager QX: From singlesubject to cortically aligned group general linear model analysis and selforganizing group independent component analysis. Hum Brain Mapp. 2006;27(5):392–401. doi: 10.1002/hbm.20249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammers A, Allom R, Koepp MJ, Free SL, Myers R, Lemieux L, Mitchell TN, Brooks DJ, Duncan JS. Three-dimentsional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp. 2003;19(4):224–247. doi: 10.1002/hbm.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage. 2006;33:115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- Iglesias JE, Sabuncu MR, Van Leemput K. A generative model for multi-atlas segmentation across modalities. 9th IEEE International Symposium on Biomedical Imaging (ISBI) 2012:888–891. doi: 10.1109/ISBI.2012.6235691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias JE, Sabuncu MR, Van Leemput K. A unified framework for cross-modality multi-atlas segmentation of brain MRI. Medical image analysis. 2013;17:1181–1191. doi: 10.1016/j.media.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson SW, Stanton ME, Molteno CD, Burden MJ, Fuller DS, Hoyme HE, Robinson LK, Khaole N, Jacobson JL. Impaired eyeblink conditioning in children with fetal alcohol syndrome. Alcoholism: Clin and Exp Res. 2008;32(2):365–372. doi: 10.1111/j.1530-0277.2007.00585.x. [DOI] [PubMed] [Google Scholar]
- Jacobson SW, Stanton ME, Dodge NC, Pienaar M, Fuller DS, Molteno CD, Meintjes EM, Hoyme HE, Robinson LK, Khaole N, Jacobson JL. Impaired delay and trace eyeblink conditioning in school-age children with fetal alcohol syndrome. Alcoholism: Clin and Exp Res. 2011;35(2):250–264. doi: 10.1111/j.1530-0277.2010.01341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmann HJ, Weinrich W. Cranial neuroimaging and clinical neuroanatomy. Thieme; New York: 1992. [Google Scholar]
- Makris N, Schlerf JE, Hodge SM, Haselgrove C, Albaugh MD, Seidman LJ, Rauch SL, Harris G, Biederman J, Caviness VS., Jr MRI-based surface-assisted parcellation of human cerebellar cortex: an anatomically specified method with estimate of reliability. Neuroimage. 2005;25(4):1146–1160. doi: 10.1016/j.neuroimage.2004.12.056. [DOI] [PubMed] [Google Scholar]
- McGraw KO, Wong S. Forming inferences about some intraclass correlation coefficients. Psychological methods. 1996;1(1):30–46. [Google Scholar]
- Park MT, Pipitone J, Baer HL, Winterburen JL, Yashvi S, Sofia C, Schira MM, Lobaugh JN, Lerch PJ, Voineskos AN, Chakravarthy MM. Derivation of high-resolution MRI atlases of the human cerebellum at 3T and segmentation using multiple automatically generated templates. Neuroimage. 2014;95(1):217–231. doi: 10.1016/j.neuroimage.2014.03.037. [DOI] [PubMed] [Google Scholar]
- Pierson R, Corson PW, Sears LL, Alicata D, Magnotta V, O’Leary D, Andreasen NC. Manual and semiautomated measurement of cerebellar subregions on MR images. Neuroimage. 2002;17(1):61–76. doi: 10.1006/nimg.2002.1207. [DOI] [PubMed] [Google Scholar]
- Pipitone J, Park MT, Winterburn J, Lett TA, Lerch JP, Pruessner JC, Lepage M, Voineskos AN, Chakravarty MM, Alzheimer’s Disease Neuroimaging Initiative Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage. 2014;101:494–512. doi: 10.1016/j.neuroimage.2014.04.054. [DOI] [PubMed] [Google Scholar]
- Postelnicu G, Zöllei L, Fischl B. Combined volumetric and surface registration. IEEE Transaction on Medical Imaging. 2009;28:508–522. doi: 10.1109/TMI.2008.2004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MP, Hanaway J, Morest DK. Atlas of the Human Brain in Sections. Lea & Febiger; Philadelphia: 1987. [Google Scholar]
- Rohlfing T, Brandt R, Menzel R, Maurer RC., Jr Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. Neuroimage. 2004;21:1428–1442. doi: 10.1016/j.neuroimage.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Sabuncu MR, Yeo B, Van Leemput K, Fischl B, Golland P. A generative model for image segmentation based on label fusion. IEEE Transactions on Medical Imaging. 2010;29:1714–1729. doi: 10.1109/TMI.2010.2050897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlerf JE, Verstynen TD, Ivry RB, Spencer RM. Evidence of a novel somatotopic map in the human neocerebellum during complex actions. Journal of neurophysiology. 2010;103:3330–3336. doi: 10.1152/jn.01117.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahmann JD, Doyon J, McDonald D, Holmes C, Lavoie K, Hurwitz AS, Kabani N, Toga A, Evans A, Petrides M. Three-dimensional MRI atlas of the human cerebellum in proportional stereotaxic space. Neuroimage. 1999;10(3):233–260. doi: 10.1006/nimg.1999.0459. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Sherman JC. The cerebellar cognitive affective syndrome. Brain. 1998;121:561–579. doi: 10.1093/brain/121.4.561. [DOI] [PubMed] [Google Scholar]
- Segonne F, Dale A, Busa E, Glessner M, Salat D, Hahn H, et al. A hybrid approach to the skull stripping problem in MRI. Neuroimage. 2004;22:1060–1075. doi: 10.1016/j.neuroimage.2004.03.032. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage. 2008;39(3):1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychological bulletin. 1979;86(2):420. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Transactions on Medical Imaging. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Stoodley CJ, Schmahmann JD. Functional topography in the human cerebellum: a meta-analysis of neuroimaging studies. Neuroimage. 2009;44(2):489–501. doi: 10.1016/j.neuroimage.2008.08.039. [DOI] [PubMed] [Google Scholar]
- Strick PL, Dum RP, Fiez JA. Cerebellum and nonmotor function. Annual review of neuroscience. 2009;32:413–434. doi: 10.1146/annurev.neuro.31.060407.125606. [DOI] [PubMed] [Google Scholar]
- Tisdall MD, Hess AT, Reuter M, Meintjes EM, Fischl B, Van der Kouwe AJ. Volumetric navigators for prospective motion correction and selective reacquisition in neuroanatomical MRI. Magnetic Resonance in Medicine. 2012;68(2):389–399. doi: 10.1002/mrm.23228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Kouwe AJ, Benner T, Salat DH, Fischl B. Brain morphometry with multiecho MPRAGE. Neuroimage. 2008;40(2):559–569. doi: 10.1016/j.neuroimage.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington Margaret M. Atlas of Human Intracranial Anatomy. Rutland, Vt: Academic Books; 1984. [Google Scholar]
- Wang H, Suh JW, Das SR, Pluta JB, Craige C, Yushkevic PA. Multi-atlas segmentation with joint label fusion. Pattern Analysis and Machine Intelligence, IEEE Transactions. 2013;35(3):611–623. doi: 10.1109/TPAMI.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke M, Holland SK, Altaye M, Gaser C. Template-O-Matic: a toolbox for creating customized pediatric templates. Neuroimage. 2008;41(3):903–913. doi: 10.1016/j.neuroimage.2008.02.056. [DOI] [PubMed] [Google Scholar]
- Woods RP. Multitracer: a Java-based tool for anatomic delineation of grayscale volumetric images. Neuroimage. 2003;19(4):1829–1834. doi: 10.1016/s1053-8119(03)00243-x. [DOI] [PubMed] [Google Scholar]
- Yoon U, Fonov VS, Perusse D, Evans AC. The effect of template choice on morphometric analysis of pediatric brain data. Neuroimage. 2009;45(3):769–777. doi: 10.1016/j.neuroimage.2008.12.046. [DOI] [PubMed] [Google Scholar]


