Abstract
Precision medicine is about matching the right drugs to the right patients. Although this approach is technology agnostic, in cancer there is a tendency to make precision medicine synonymous with genomics. However, genome-based cancer therapeutic matching is limited by incomplete biological understanding of the relationship between phenotype and cancer genotype. This limitation can be addressed by functional testing of live patient tumour cells exposed to potential therapies. Recently, several ‘next-generation’ functional diagnostic technologies have been reported, including novel methods for tumour manipulation, molecularly precise assays of tumour responses and device-based in situ approaches; these address the limitations of the older generation of chemosensitivity tests. The promise of these new technologies suggests a future diagnostic strategy that integrates functional testing with next-generation sequencing and immunoprofiling to precisely match combination therapies to individual cancer patients.
An increasing number of safe and effective novel cancer therapies target specific signalling and subcellular machinery; thus, the pressure to match these therapies to individual patients is growing. Most current efforts to match patients to therapies depend on molecular, often genomic, technologies, based on the idea that cancers are just like rare genetic disorders, despite the presence of widespread epigenetic changes, lineage-specific drivers and non-oncogene-driven vulnerabilities (BOX 1). Remarkable advances in next-generation sequencing (NGS) technology have enabled cancer biologists to identify tens of thousands of mutations in patient tumours1,2, which has revolutionized our understanding of the origins of cancer. Now that thousands of cancer genomes have been sequenced, we are reaching the ‘long tail’ of mutations that occur in only a minor subset of patient tumours1, suggesting that the majority of ‘low-hanging-fruit’ driver mutations that affect populations of cancer patients large enough to justify drug discovery efforts have probably been discovered. Even considering the known frequently mutated genes, clinical results of therapeutic matching have yet to replicate the >90% response rate and long-term control achieved with imatinib in patients with chronic-phase BCR–ABL-mutated chronic myeloid leukaemia (CML)3,4 that launched the genomic cancer era. Furthermore, only a small fraction (<10%) of patients have clinically validated and US Food and Drug Administration (FDA)-approved therapies matched to mutations5. We expect that over time some of these newly discovered mutations will be functionally annotated and provide biomarker strategies to guide therapy. Nevertheless, we have little understanding of the functional consequences of the thousands of mutations in tumours, leaving most patients in need of new, complementary strategies to match their tumours to appropriate therapies.
Box 1. Promise and limitations of genetic markers for patient stratification.
The common goal of cancer researchers and clinicians is to better match patients with therapies. Next-generation sequencing (NGS)-based matching has been the most advanced technology applied to this problem up to now. Cancers accumulate genetic alterations, but we may be approaching an asymptotic limit with regard to uncovering these driver oncogenes1. Even the best successes demonstrate that identifying well-characterized mutations in an individual patient may yield transient, if any, benefit from matched single-agent targeted therapy88. Although the dramatic effects of imatinib in chronic myeloid leukaemia (CML) established the modern cancer genomics paradigm, it is remarkable and unfortunate that thus far no other targeted therapy has produced this type of sustained response and survival benefit in populations of cancer patients. More recent examples of successful targeted therapies, such as vemurafenib in BRAFV600E-mutant melanoma89,90, gefitinib in epidermal growth factor receptor (EGFR)-mutant non-small-cell lung cancer (NSCLC)91 and crizotinib in anaplastic lymphoma kinase (ALK)-mutant NSCLC92, have yet to replicate the impressive and durable effects of imatinib in CML, and give often short-lived and partial responses93. Only about half of leukaemia patients with Fms-like tyrosine kinase 3 (FLT3) mutations respond, transiently, to potent and specific FLT3 inhibitors94. In other cases, the response to a driver oncogene seems to be lineage and context specific, such as the lack of strong responses to BRAF inhibitors in BRAFV600E-mutant colorectal cancers95.
On the basis of tumour genomic profiling, these clinically validated mutations are present in <10% of patients, even if >80% of patient tumours contain mutations that suggest novel clinical hypotheses5,96. Most such hypotheses are untested in the clinic or untestable without years of additional drug discovery. Such cross-indication hypotheses are the basis of ‘basket’ or ‘umbrella’ trials such as SIGNATURE, MATCH and Lung-MAP97, but similar trials have faced the challenges of recruitment of patients to arms with rare mutations and have rarely yielded knowledge beyond the well-known responses to inhibition of, for example, EGFR, ALK and BRAF98. Given the measureable but brief responses to single-agent therapy, it is expected that the field will move to combinations of targeted therapies. However, combinations present the even greater, nonlinear challenge of matching to genetic features alone; indeed, in multiplexed gene panels, <5% of patients had two bona fide actionable co-occurring mutations in NSCLC99.
Furthermore, only a small proportion of the thousands of mutations identified from whole-genome sequencing have functional data suggesting that they should be targeted with drugs. Even synonymous mutations can have effects on cellular fitness100. The scale of fitness effects is not known; indeed, it is possible that it is the accumulation of a large number of relatively minor fitness effects that lead to cancer, and targeting any one, or ten, directly, may yield little in terms of cellular effects. Many targeted drugs are successful even though they are matched not to somatic genotype or driver mutation, but rather to lineage: for example, antibodies against CD20, inhibitors of Bruton’s tyrosine kinase (BTK)101 and inhibitors of PI3Kδ in chronic lymphocytic leukaemia (CLL)102. These so-called non-oncogene vulnerabilities103, in addition to widespread epigenetic alterations, exist widely but would not be revealed by genome sequencing. The overwhelming complexity of the cancer genome and epigenome suggests that we are in the earliest phases of interpreting such results and translating that data into knowledge that is useful to clinicians. Thus, new, orthogonal technologies are needed together with NGS to hasten the era of precision medicine in cancer.
Although the rare genetic disease paradigm provides an appealing narrative to apply to cancer owing to successes in other disease areas6, cancer and immunological diseases categorically differ from rare diseases in that these are microevolutionary processes driven by replication. The deployment of one microevolutionary system (the immune system) against another (the tumour) is probably why immuno-oncology approaches sometimes yield durable, long-term remissions in a minority of patients7,8. However, genomic and most traditional pathological techniques are descriptive and static — they represent the sum of the history of the tumour’s development as determined from dead cancer cells. Thus, despite the predictive power of NGS in certain subsets of patients, we argue that additional diagnostic approaches will be necessary to complement NGS and thus guide therapy choice in a greater proportion of patients.
One attractive approach that is dynamic and that evaluates the current vulnerabilities of the tumour cells to intervention is functional testing. Here, we widen the definition of ‘functional’ screening to include any test that monitors live tumour states and is used to guide patient therapy, such as measuring tumour cell death from biopsies after ex vivo drug exposure. This type of testing is routinely used in the infectious disease setting as ‘precision medicine’ to select antibiotics tailored to a patient’s infection9. In cancer, however, although these assays have been used in research for more than half a century, their widespread adoption has been challenged by the lack of adequate proof of clinical utility. Although there are several explanations for the failure of older assay technologies, the idea of ex vivo screening of patient samples remains compelling to oncologists and patients alike10, but is fundamentally untested. We describe recent technological advances in specific areas of functional testing, including tumour manipulation and culture, and assays for measuring antitumour drug responses. These ‘next-generation’ functional tests (FIG. 1) warrant further preclinical and clinical testing to provide predictive, actionable information for oncologists.
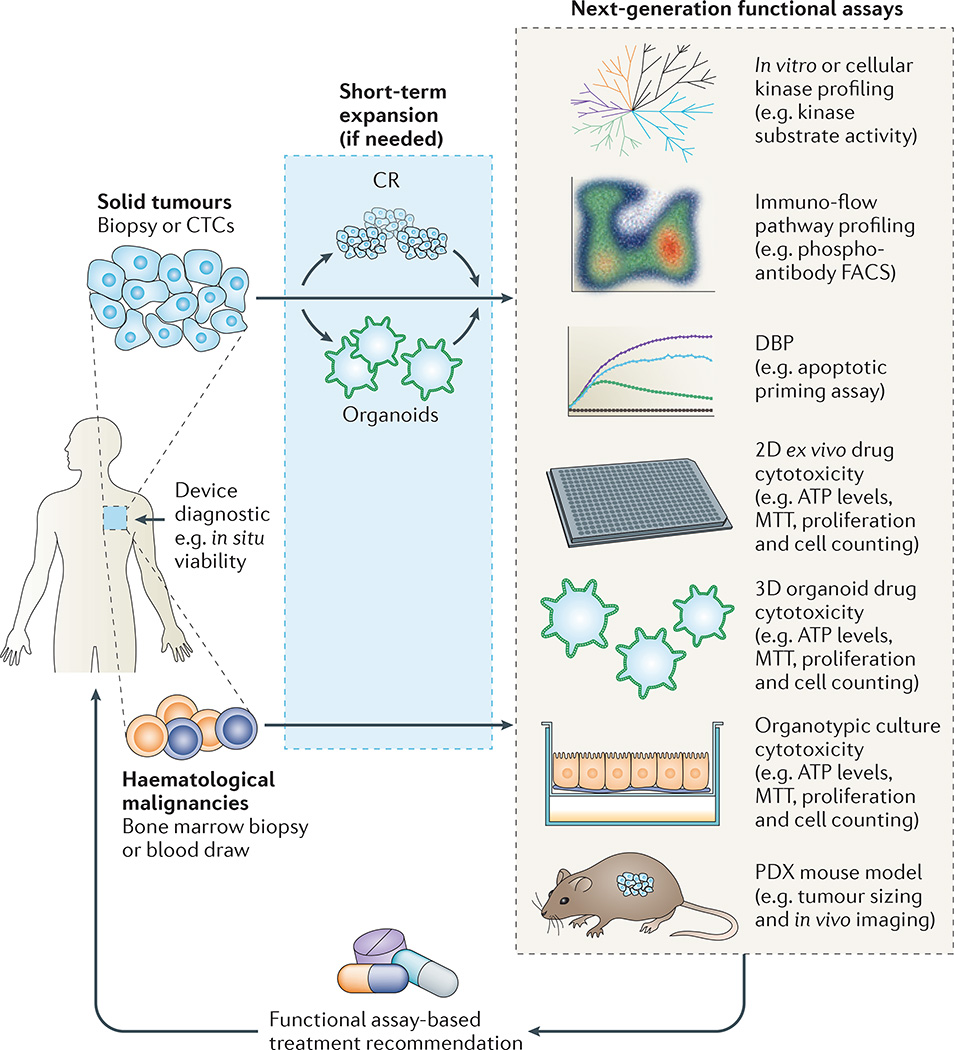
Figure 1. Next-generation approaches for cancer precision medicine.
As discussed in the text, a wide range of new technologies have been developed for the ex vivo determination of live tumour cell responses to drug therapies. These include (arranged from top to bottom of figure) target- and pathway-based methods, direct cytotoxicity (reduction in tumour cell numbers) and in vivo models. All methods end in a recommendation for (or against) a specific therapy for a patient with advanced cancer on an individualized basis. For small tumour samples and depending on the method, some ex vivo expansion may be necessary before a functional measurement can be made. Combinations of these functional approaches may also be needed. Combined with gene sequencing and immunoprofiling, functional diagnostic methods are part of a comprehensive approach to precise matching of novel therapies to patients. CR, conditional reprogramming; CTCs, circulating tumour cells; FACS, fluorescence-activated cell sorting; MTT, metabolic tetrazolium dye; PDX, patient-derived xenograft. The dynamic BCL-2 homology domain 3 (BH3) profiling (DBP) graph is adapted with permission from REF. 81, Elsevier.
First-generation functional diagnostics
To properly predict the behaviour of a complex system such as cancer, one needs both the initial conditions (genes, transcripts, proteins, metabolites and so on) and all of the interactions of the components of that system (for example, gene–RNA, RNA–protein and protein–protein interactions) in all cellular states11,12. However, baseline genomic information provides only the initial conditions of a diverse cancer ecosystem — an ‘archaeological’ exploration of all the insults to the genome of that cell over the decades of cancer development (BOX 1). Although progress is being made on understanding the interactions between components of the cancer cell — for example, protein–protein interactions13 and transcriptional profiles in response to perturbation14 — we still have only rudimentary knowledge of this system to be able to predict novel outcomes. Perturbations of a complex system, however, yield far more information about the organization and behaviour of such networks, thus arguing for the use of functional testing. Newer efforts to obtain genomic information before and after drug treatment by using repeat biopsies or by monitoring changes in circulating tumour DNA15–17 have yielded insight into the mechanisms of resistance to therapies, but do not necessarily lead to a direct, actionable result for the individual patient. For example, identification of a new hotspot mutation in a target after targeted therapy may still require years of drug discovery to develop a new therapy against that mutated target. By contrast, direct live-cell perturbation can directly reveal whether the given sample is sensitive to agents that could be immediately administered clinically.
Intuitively, functional testing is exposure of a patient’s tumour biopsy to drugs. Tumour responses, often measured by cell death, are then used to help to direct therapy choices. ‘First-generation’ versions of these tests, which have been developed and intensively studied over the past 30–40 years, can be divided into a number of categories; a comprehensive review of these older tests is beyond the scope of this article and is well summarized in recent commentaries18–20. The earliest of these include the human tumour clonogenic assay (HTCA)21, differential staining cytotoxicity (DiSC) assay22, extreme drug resistance assay (EDRA)23, ChemoFx assay (Helomics, Pittsburgh, Pennsylvania, USA)24 and subrenal capsule assay (SRCA)25. Several of these protocols, such as HTCA and ChemoFx, grow the patient’s tumour cells until homogeneous tumour colonies or lines are generated; the viability of these cells is then tested after exposure to chemotherapeutic agents using assays such as cell counting, metabolic tetrazolium dye (MTT) or whole-cell ATP-content-based readouts of cell numbers. Other protocols directly test the effects of chemotherapeutic agents on heterogeneous mixtures of tumour cells and stromal cells in patient biopsies after 3–7 days of drug exposure. Most studies have been restricted to traditional cytotoxic chemotherapy agents, and very few of these older tests have been assessed in prospective, controlled, randomized trials that are adequately powered to address the central question of whether the assays improve patient outcomes. One such trial in ovarian cancer used an ATP-based assay after 6 days of tissue drug exposure to guide patients to a range of cytotoxic chemotherapies and showed no statistically significant improvement in progression-free survival26. Not surprisingly, the most recent comprehensive reviews of the clinical data supporting these chemotherapy sensitivity and response assays determined that there were no studies sufficiently conclusive to recommend the routine use of these assays18–20. However, despite the heterogeneity of assays, drugs and study designs, it is notable that most of these studies, over several decades, show a trend towards prediction of resistance to chemotherapies. Nevertheless, none of these tests has become standard of care, as the field has shifted its attention to the genetic causes of tumorigenesis as a path towards therapeutic selection.
New methods for tumour manipulation
Perturbation of patient samples requires patient-derived live tumour cells, but ex vivo cultivation and manipulation of solid tumours can be challenging. The generation of stable cell lines for functional testing from individual patients using traditional 2D cultivation on cell culture dishes in typical media is laborious and generally unsuccessful. Such homogeneous cell lines typically have properties that are distinct from the parental tumour, at both the genotypic level (for example, mutations and expression profiles)27,28 and phenotypic level (much more rapid growth and increased sensitivity to chemotherapies); in particular, these lines have lost the functional and genotypic heterogeneity of patient tumours as well as the tumour–stroma interactions that support tumour growth. These effects are further complicated by the small quantity of live tissue obtained from patients owing to, for example, small core biopsies. Therefore, obtaining sufficient cell numbers for functional testing has been a major roadblock in previous approaches.
Conditional reprogramming to produce patient-derived tumour material
One method recently described for rapidly growing larger quantities of patient tissue is conditional reprogramming (CR). This process uses an irradiated fibroblast feeder layer, growth-factor-enriched media and a RHO-associated protein kinase (ROCK) inhibitor to rapidly expand patient tissues, both benign and malignant29. CR seems to induce a rapid and reversible epigenetic state similar to that seen in adult stem cells, characterized by increased levels of telomerase reverse transcriptase (TERT), integrins, p63 isoforms, CD44 and nuclear β-catenin, and decreased Notch signalling, but without inducing features of induced pluripotent stem cells30. Furthermore, recent data suggest that CR retains intratumour genetic heterogeneity and can expand multiple clones from the original biopsy15. In one case report, a patient with human papillomavirus (HPV)-induced respiratory papillomatosis was given vorinostat based on the ex vivo response of their CR-derived tumour sample to this drug, and the patient showed a durable response to treatment31.
More recently, Crystal et al.32 described the use of CR technology to develop a large number of ex vivo-derived models of tumours from patients with non-small-cell lung cancer (NSCLC) who showed clinical resistance to targeted therapies. Rather than relying on resistance observed or induced in existing cell lines, the method allowed the generation of lines reflecting the ‘real-world’ resistance arising in patients. Importantly, the authors observed the continued presence of the original driver mutation in these ex vivo-derived models and the maintenance of resistance to the single agent for which resistance was originally shown15. These lines were used to identify a novel combination of targeted therapies against MEK and anaplastic lymphoma kinase (ALK) to combat resistance to single-agent ALK inhibition in ALK-mutant NSCLC. Remarkably, sequencing of the patient’s tumour revealed multiple potentially ‘actionable’ mutations; although a MEK mutation was identified that might explain sensitivity to MEK inhibition, other mutations such as a Janus kinase (JAK) mutation were also observed without a corresponding combinatorial effect from a JAK inhibitor. Thus, the authors proposed that drug screening could be used to triage potential vulnerabilities suggested by genetics. Several improvements in CR technology would be necessary before it could be used for routine chemosensitivity testing. Because this method rapidly expands all epithelial components present in the tissues, separation of tumour tissue from non-tumour tissue requires additional steps such as differential trypsinization29, extensive subculturing or single-cell cloning. In addition, extensive growth in 2D monolayers may be required to produce large numbers of homogeneous cells for testing; during this growth period, the cells may drift from their original phenotype, and the delays incurred by using this method may limit the clinical utility of subsequent findings.
Circulating tumour cells
Methods of isolation and downstream manipulation of circulating tumour cells (CTCs) from the blood of patients with cancer have dramatically improved over the past few years. Previous methods required immediate transplantation of CTCs into animals for propagation33,34 or had low success rates35. Recently, CTCs have been isolated from patients with breast cancer using an improved CTC microfluidic chip that was shown to enrich tumour cells for further manipulation36. Specific culture conditions were discovered that support growth of these isolated cells; interestingly, CTC growth in culture was only successful when the cells were grown as a suspension, not as adherent cultures, which is consistent with their circulating phenotype. In addition to making broader and deeper sequencing of CTCs possible through increased tumour material, the long-term stable cultures were used for screening studies. These drug screens showed that the CTC-derived cells retained functional responses to chemotherapies consistent with the patient’s clinical response and suggested novel drug combinations. Given the ease of isolation of CTCs from a simple blood draw, they represent a compelling source of patient tumour cells for eventual downstream testing. Potentially, such screening could even be conducted ‘online’ in the CTC microfluidic chambers in suspension (rather than in traditional screening plates), as has been demonstrated using an optically labelled drug library37; scaling these novel devices, and automating their use to perform routine testing of patient cells by exposure to drugs, requires additional research. As with other CTC methods, however, months of growth are required from the typically <100 cells isolated from patients (although sometimes no cells can be isolated) to obtain enough cells for routine testing, during which time the primary tumour populations may no longer be represented by the expanded CTC population.
Patient-derived organoids
Another source of expanded tumour material is patient-derived organoids38. Organoids are developed by explanting dissociated patient-derived cells into a semi-solid extracellular matrix and expanding these cells in growth-factor-enriched medium39,40. Organoids have the distinct advantage of growing in three dimensions, and they often recreate the endogenous architecture of the tissue from which they were derived, theoretically recapitulating the in vivo tumour environment more closely than 2D cultures on plastic. Recently, organoids have been developed from patients with pancreatic41, prostate42 and colon40 cancer. Organoids maintained the same driver mutations that were identified in the primary tumour41. As with CR cultures, organoids often contain normal epithelial cells and can require weeks to generate sufficient cells for drug testing, and their establishment success rates can be specific to tissue of origin. Further advancements and clinical validation are needed before they can be used for routine testing, but early results are promising: several organoids from patients with colorectal cancer were recently generated prospectively, together with paired normal samples43. The tumour samples displayed heterogeneous morphologies and proliferation rates but retained most of the known cancer-causing mutations, despite half of the samples showing clonal selection during organoid derivation from the original biopsy. Several of these organoids were then subjected to a screen of cancer-relevant drugs, revealing patient-specific vulnerabilities. Although some of the drug sensitivity profiles could be correlated with patient-specific mutations, several drug response differences did not seem to be linked to genotype, emphasizing the importance of functional information in suggesting patient therapies.
Organotypic cultures
One major limitation of methods such as those described above, which may produce homogeneous cells from patient tumours, is that they fail to maintain the 3D environment of the native tumour or to preserve the heterogeneity of the original tumour admixed with stromal cells. Intrapatient tumour cell heterogeneity may be a prognostic and predictive feature that is important to preserve as a biomarker44, and the tumour cell environment can dramatically influence therapeutic responses45. To address these issues, investigators have developed artificial organotypic cultures with multiple patient-derived cell types that maintain gene expression patterns more similar to those of tumours than to those of 2D cultures46; such cultures using cell lines have been shown to be predictive of in vivo results47. In other studies, live thin slices of the original patient solid tumour maintained in commercial culture plate inserts have been treated with drugs; under these conditions, the tumour cells showed appropriate on-pathway responses to inhibitors48. Alternatively, others have used tumour fragments in larger clusters as ‘microspheroids’, rather than single-cell suspensions, for viability testing after exposure to chemotherapeutic agents49.
Recently, investigators have improved on previous organotypic approaches48 to more rigorously recreate the endogenous tumour microenvironment during ex vivo exposure to drugs50. The authors incubated thin slices of patient tumours with combinations of extracellular proteins that they had previously identified as being present in patient tumours and patient-derived (autologous) serum to recreate the heterogeneous tumour environment ex vivo in microtitre plates50. Using this method, they demonstrated more faithful preservation of tumour architecture, proliferation, ATP utilization and signalling pathway activation with the patient-derived environment than with standard, defined reagents. Finally, by interrogating multiple tumour responses (for example, viability, proliferation and apoptosis) and deriving an algorithm combining these readouts, clinical outcomes were correlated with the ex vivo responses of head and neck squamous cell carcinoma and colorectal cancer samples to standard chemotherapy cocktails. Overall, they found 87% accuracy for their ex vivo-response-derived algorithm in predicting patient responses, which provides compelling evidence that such a method may help to direct therapy choice prospectively. Inclusion of immune cells in these 3D cultures may provide more accurate modelling of tumour–immune system interactions51, as newer immunomodulatory agents are developed and predictive markers of response are sought.
In situ functional diagnostics
As no ex vivo approach can exactly match the human tumour context, an alternative approach is to directly test drug effects with micro-dosing in solid tumours within the patient using novel devices. Two reports have recently been published that describe distinct technologies for micro-dosing52,53. In one study, xenograft tumours were removed from animals for histochemical analysis, including measurement of toxicity and signalling pathway activity, 24–72 hours after injection using a microneedle injector device (CIVO; Presage Biosciences, Seattle, Washington, USA) that delivers small drug doses directly into the tumour. Cytotoxicity responses in the injected tumours mirrored those from systemic drug delivery. The device was further used in studies in both humans and dogs with accessible lymphomas, in which the procedure was well tolerated52. In the second study, a small device containing slow-releasing reservoirs of up to 16 different drugs53 was implanted using a biopsy needle, and the device was left in situ in tumours for 24 hours, after which the tumour was retrieved and drug responses were measured. Using mouse xenografts, the authors observed a strong correlation between the apoptotic index of cells surrounding each reservoir and systemic tumour responses to a range of cytotoxic chemotherapies.
There are several potential challenges associated with the development of such devices for clinical care. Notably, their use is restricted to accessible solid tumours of a certain size and requires an additional, potentially hazardous, procedure to retrieve the tumour from patients following drug exposure. Furthermore, controlling, standardizing and analysing drug diffusion from the device and correlating the absolute and relative responses with the pharmacokinetics of known drugs may be challenging, particularly given the interpatient heterogeneity in intratumour hydrodynamic pressure and tumour–stroma admixture. Nevertheless, both devices show the value of drug susceptibility testing in predicting responses to systemic therapy. Even before clinical validation of their diagnostic power, their most immediate use could be in preclinical drug discovery studies to validate multiple novel therapies in vivo simultaneously, followed by early clinical pharmacodynamic proof-of-concept studies of experimental therapeutics.
Mouse models for functional testing
Another technology that may more accurately mimic the endogenous tumour environment is patient-derived xenograft (PDX) mouse models as ‘avatars’ for drug testing of patient tumours. PDX models, in which patient biopsy material is implanted subcutaneously or orthotopically and expanded in vivo, have the theoretical advantage of retaining some of the histology, gene expression and somatic genetics of the patient tumour54. Although PDX models are becoming standard in the drug discovery pharmacology toolbox for testing efficacy, they have also been suggested as avenues for selecting patient therapies55. In these PDX approaches, actionable therapeutics are tested in mice with in vivo-expanded patient tumours. Drugs causing tumour regression or decreased growth are then recommended as therapies for the patients. Although data so far have been limited to anecdotal case studies or small nonrandomized trials, initial data from some PDX platforms are promising, with response rates of 81–88% after PDX-directed therapies56,57.
“Functional screening will become part of an emerging comprehensive diagnostic approach that includes traditional pathology techniques together with genetic sequencing.”
Several practical challenges are associated with xenograft mouse models of patient tumours. The first challenge is the time required to generate sufficient tumour material in enough mice to test one or more regimens; this can often take as long as 6–8 months or more56–58. Second, animal colonies bearing an individual patient’s tumour are practically limited to testing a few agents. Third, the additional time required to generate a sufficient number of cells to test regimens in groups of mice has clear consequences for the representation of various tumour clones in the patient model. For example, in one large study of breast cancer PDX mouse models, all models showed selection during tumour growth, from moderate drifts to dramatic clonal selection, even within the first mouse passage59. Another more limited study similarly showed a large number of differences between mutations found from whole-exome sequencing in patient tumours and those from matched second-passage PDX models50. Finally, PDX models are established in immunodeficient mice, reducing the similarity of the model to the original tumour environment, which includes human stromal components and immune cell interaction. Although data on such PDX models are promising, larger, prospective randomized trials will be needed to obtain more robust evidence for PDX models as a therapeutic selection strategy for patients more broadly.
Functional assays for leukaemia
Most of the novel methods described above attempt to address particularly vexing challenges of the survival, growth and measurement of solid tumours. Leukaemias and other haematological malignancies, conversely, represent the ‘lowest hanging fruit’ for clinical translation of functional screening, given the ease of obtaining large quantities of viable malignant cells in single-cell suspension from simple blood draws or bone marrow biopsies. Over the past few years, several groups have begun testing advanced high-throughput approaches for the measurement of such leukaemic responses ex vivo and the correlation of these responses with patient outcomes.
For example, Tyner et al.60 have developed a high-throughput screening approach for patient samples. Originally designed for RNA interference-based target identification in primary tumour samples, the investigators adapted this system to screen a large panel of therapies, including clinically relevant targeted therapies, against more than 150 primary patient-derived leukaemia samples61. The clinical validity of the inhibitor activities identified by this high-throughput technique in patient samples was confirmed by overlaying the inhibitor target profiles to triangulate on common targets for samples in which multiple drugs led to increased cell killing. Thus, cells from patients with mutant Fms-like tyrosine kinase 3 internal tandem duplication (FLT3-ITD)-positive acute myeloid leukaemia (AML) were killed by drugs that are all known to target FLT3, and cells from patients with BCR–ABL-positive CML were killed by drugs that had overlapping targeting of ABL. In one case study, a patient received the kinase inhibitor sorafenib based on the ex vivo response profile. Strikingly, the ex vivo response of the patient’s follow-up samples to sorafenib mirrored the initial sensitivity and then resistance to sorafenib seen in the clinic, whereas the ex vivo response to another kinase inhibitor, sunitinib, was unchanged, reflecting a sorafenib-specific resistance effect. Of note, the most specific and potent drug varied for each patient, and in many cases a novel off-label indication of an FDA-approved therapy was identified. This platform has been further validated in case studies of paediatric leukaemia62, and in solid tumours for which sufficient cells were available from larger biopsies, including case studies in renal cell carcinoma63 and in a canine osteosarcoma64. The clinical utility of this system is currently being tested in a prospective clinical trial in relapsed AML (ClinicalTrials.gov identifier: NCT01620216).
Using a similar screening assay, Wennerberg and colleagues65 have developed a high-throughput ex vivo assay to test targeted therapies, both singly and in combination, in patient biopsies. For one cohort, after obtaining 28 samples of bone marrow biopsies from patients with AML, the authors developed a drug sensitivity and resistance testing (DSRT) platform that examined the effects of multiple doses of 187 drugs on viability in patient cells after 72 hours of exposure65. To specifically score drug responses, an algorithm (drug sensitivity scoring (DSS)) was developed that integrates the difference in dose–response profiles between the individual patient and a collection of control samples66. Notably, the study identified multiple drugs that are not approved for use in AML but that were effective in killing patient AML cells. Although the samples could be grouped by hierarchical clustering into major susceptibility groups, the most effective drug varied from patient to patient, emphasizing the need to personalize drug selection. The drug responses could also be correlated with known mutation groups, such as the sensitivity of FLT3-mutant AMLs to FLT3 inhibitors, validating the finding that ex vivo responses match known tumour biology. Most recently, this approach was used to identify a novel off-target effect of the vascular endothelial growth factor receptor (VEGFR) inhibitor axitinib: specifically, it inhibited the T315I gatekeeper mutant BCR–ABL in patient samples, and the study found a patient response to this off-label application of the drug67.
One challenge when working with samples of haematological malignancies, particularly AML, is the fragility of the blast population, in that ex vivo manipulation can rapidly cause cell death independently of drug effects. Furthermore, many patients, particularly those who have had repeated cytotoxic therapies, may have hypocellular bone marrow, which limits the number of cells available for testing. Conditions for survival and pre-testing expansion of AML cells have recently been identified and developed to explore the effects of low-dose decitabine on primary AML cells68. Similar to CR of solid tumours, these techniques take advantage of a feeder layer and growth-factor-enriched medium to provide suitable conditions for short-term expansion.
Although ex vivo testing of patient samples has now become routine in investigations of basic biological mechanisms in haematological malignancies and in drug discovery (for example, recently, in 30 patients with chronic lymphocytic leukaemia (CLL), for whom the combination of carfilzomib and ibrutinib was discovered to be effective69), its diagnostic potential has yet to be fully realized. These recent studies demonstrate that such direct patient testing might lead to patient benefit without knowledge of the underlying biology or mutations, which complements clinical scenarios in which treatments have been found to yield benefit when matched to specific mutations.
New assays of ex vivo tumour responses
All functional assays eventually must end with some method to detect drug effects. Most of the above studies use measurement of a cellular end point, either automated or manual, such as intracellular ATP concentration, cell counting or specific cell markers of proliferation or apoptosis. These end points are compelling, as they test the ultimate goal of ex vivo chemosensitivity assays: measurement of cell killing that might be translated into treatment of patients. However, most of the measurement techniques require incubation of the tumour samples with drugs for days to observe such phenotypic effects, during which time the tumour cells are in the cell culture environment, which has altered stromal and multidimensional interactions, oxygen tension, temperature, metabolites and other parameters. Recently, several alternative assays have been described that measure more specific molecular events ex vivo.
Measuring target engagement
Measurement of target engagement by targeted agents can be correlated with patient response in molecularly defined subpopulations. Thus, investigators have developed techniques to measure target engagement using live patient cells or lysates from live cells; this enables prediction of patient responses rather than carrying out static measurement of pathway engagement after drugs have been given. These assays have the advantage of using well-defined molecular reagents and often can be quite rapid. For example, it is well known that patients with BRAF-mutant melanoma have increased MAPK activation and that pathway modulation by mutant-specific or pan-BRAF or MEK inhibitors correlates with response to therapy. One technology uses kinase substrate peptide microarrays over which the lysed sample from the patient is dispensed, followed by detection of phosphorylation of the substrate70. Using tissue samples from patients with melanoma, baseline kinase activities, as measured by substrate phosphorylation, were indistinguishable between major genotypes of melanoma (namely, genotypes with mutations in BRAF, NRAS, cyclin-dependent kinase inhibitor 2A (CDKN2A) or TP53); only during ex vivo exposure of the lysate to the BRAF inhibitor vemurafenib were kinase profiles distinguishable between these genotypes, pointing to the importance of functional data in the classification of patients71. Although such a test can thus be used to determine the response to a specific agent, this assay could also be used to recommend novel therapies among a panel of potential targeted therapies by identifying targets with high activity in patient samples. A different technique that used an ex vivo platform to measure phosphorylated (activated) ERK responses of live melanoma samples to BRAF inhibitors identified and predicted a patient with a BRAF mutation who showed no response to a BRAF inhibitor72.
Measuring pathway activation
Measurement of multiple pathways simultaneously may enable better prediction than measurement of a single pathway. Such profiling of signalling pathways in live patient samples has been performed broadly in leukaemias, in which cell populations are more homogeneous and accessible. Early studies using multiparameter fluorescence-activated cell sorting (FACS) monitored multiple signalling pathways in samples from patients with AML after exposure to several signalling ligands and revealed distinct subpopulations within patient samples and heterogeneity among patients73. Expanding on this work, the same group monitored multiple pathways in the setting of drug screening in leukaemia cell lines and primary mouse splenocytes74. Moving this platform into the clinical setting, others were able to identify a pattern of pathway modulation by both cytokines and cytotoxic chemotherapies that correlated with complete responses in adult and paediatric patients with AML75,76, with high reproducibility (Pearson coefficient >0.8) and an area under the curve of the receiver operating characteristic (AUC of the ROC; a benchmark of the test’s false and true positive rate) of 0.66 to 0.7 (with a maximum value of 1.0), rising to as high as 0.88 in the intermediate-risk AML clinical category, for which new predictive biomarkers are particularly helpful and needed.
BH3 profiling
Most if not all targeted therapies seem to be effective through a final mitochondrial death pathway77. Using this common mechanism of action as a starting point, we have recently described a more rapid approach for detection of ex vivo drug responses within a few hours that may obviate the need for long-term ex vivo culture, which is a major stumbling block for first-generation chemosensitivity assays. Previously, we developed a system for measurement of the ‘death threshold’ of patient tumours ex vivo using a process called BCL-2 homology domain 3 (BH3) profiling78. By exposing permeabilized tumour cells to synthetic BH3 domain-containing peptides and measuring mitochondrial permeability, we observed that the cells of some patients were much more likely to lose mitochondrial potential; these patients, who had multiple types of leukaemia and solid tumours, were more sensitive to cytotoxic chemotherapies79,80. We recently described an improvement on this ‘static BH3 profiling’ called ‘dynamic BH3 profiling’ (DBP)81: in DBP, patient cells are first exposed ex vivo to drugs for 16–24 hours, after which BH3 profiling is performed (FIG. 2). Mitochondrial priming predicted the eventual drug responses using traditional end point assays of viability, which were carried out days later across a range of solid and haematological tumour cell lines, with an AUC of the ROC of 0.89. More importantly, however, DBP outcomes predict patient responses to drugs: the mitochondrial priming induced by imatinib in samples from patients with CML correlated with the clinical response to imatinib, and patients with ovarian cancer whose samples showed increased mitochondrial priming in response to cisplatin demonstrated significantly improved progression-free survival. Interestingly, the results were consistent across a range of molecularly targeted signal transduction inhibitors and cytotoxic chemotherapies. Thus, DBP is probably applicable to most anticancer agents, whether conventional or targeted, that are currently used. The full range of drug categories that can be predicted by DBP remains to be fully explored in larger cohorts of patient samples, and the utility of DBP in clinical decision making will require clinical testing.
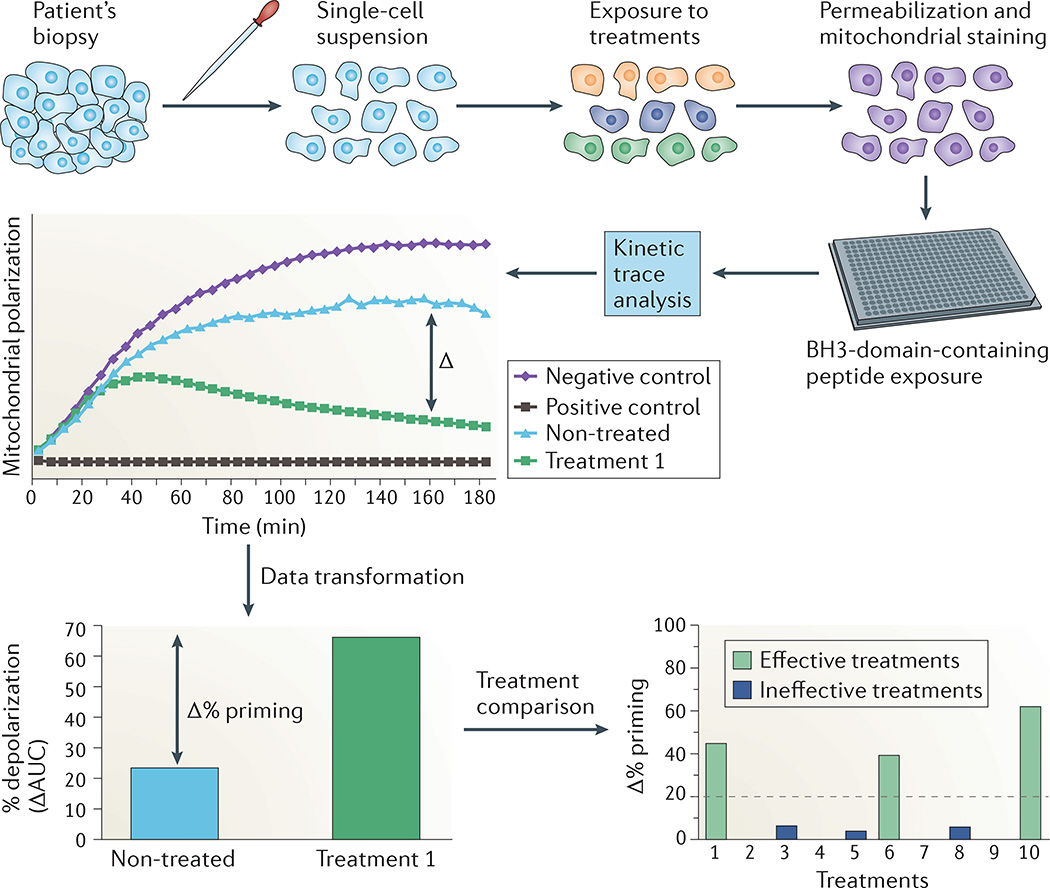
Figure 2. Dynamic BH3 profiling can predict patient responses to cancer therapies.
Dynamic BCL-2 homology domain 3 (BH3) profiling is an example of a newer, more molecularly precise assay that can be used for ex vivo functional screening. Biopsy material from the patient is dispersed (for solid tumours) and briefly (for 16–24 hours) exposed to potential drug treatments. After incubation, cells are permeabilized and exposed to BH3-domain-containing peptides. Mitochondrial outer membrane permeabilization is measured, generating a kinetic trace of mitochondrial polarization (central graph). In this analysis, drug treatments that shift the apoptotic threshold generate a large increase in the difference between the kinetic trace area under the curve (AUC) for a negative control-treated sample and the drug-treated sample. This difference creates the ‘Δ% priming’ metric of apoptotic threshold (lower left graph). By comparing this metric across control and drug-treated samples, one can select drug treatments that preferentially lead to apoptotic priming (lower right graph). The dotted line represents the threshold of effectiveness. Adapted with permission from REF. 81, Elsevier.
These technologies are compelling in that they reduce the complex observations of live cells to well-defined and robust molecular interactions. Correlation with patient outcomes will be needed to confirm that these overactive targets and pathways in patient biopsies are real vulnerabilities or ‘drivers’. Many other novel assays that provide functional readouts from live patient biopsies have been described in the past few years, including impedance spectroscopy to measure ex vivo responses of fragments of live melanoma tumour specimens to chemotherapy82, and metabolic outputs such as oxygen consumption83. Similarly to measurements of proliferation, viability and apoptosis, these assays will require rigorous evaluation of their relevance to prediction of patient outcomes and comparison between them to determine the advantages and disadvantages of each assay beyond technical differences in reagents, timing and analysis.
Functional assay clinical validation
Next-generation functional testing methods are intended to help to guide oncologists in their choice of the best treatment for their patients in practice and as a stratification test in clinical trials of experimental therapeutics. Like all predictive biomarkers, including single-gene and multi-gene panels using NGS, such tests will require rigorous evaluation of their analytical validity, clinical validity and clinical utility to demonstrate their value to patients and to be adopted in the clinic84,85. These test parameters have been extensively considered for molecular technologies, but many of the same standards can be applied to next-generation functional diagnostics, including rigorous quality control and quality assurance measures, significant and meaningful correlation of test outcomes with clinical responses and, finally randomized, controlled trials of clinical utility (BOX 2). Such standards will be crucial in bringing clarity to the regulatory and reimbursement landscape for these functional assays, most of which are likely to be performed as a laboratory-developed test (LDT) in a centralized testing laboratory. In the United States, LDTs have been historically regulated by the Centers for Medicare & Medicaid Services (CMS) under the Clinical Laboratory Improvement Amendments (CLIA), as the FDA has exercised its enforcement discretion in not requiring formal approval before clinical use. However, the agency has recently announced a suggested framework in which formal approval would be required for certain LDTs, particularly those classified as high risk86, a position affirmed by the American Association for Cancer Research85. Given that functional assays directly recommend treatment regimens for patients by design, it is likely that they will be regulated as such if the guidelines are adopted. Thus, well-designed clinical trials will be required to demonstrate the clinical validity and utility of functional testing before it becomes available to the broader market.
Box 2. Validating new diagnostic technologies for clinical cancer care.
Analytical validation
Analytical validation is the documentation showing that a test measures what it is intended to measure with defined precision, accuracy, specificity and sensitivity84. Unlike next-generation sequencing (NGS), in which traditional sequencing methods can be used as a gold standard to benchmark newer methods to the objective presence or absence of a mutation, functional tests are often measured relative to internal controls of cellular activity without clear standards. Furthermore, attention has recently been drawn to noise in cell-screening assays, especially when correlated with genomic features104; such noise can be addressed through rigorous assay standardization105. A functional assay should demonstrate accurate and precise quantification of the phenotype it is purporting to measure by comparing it with an orthogonal, if lower-throughput method of the same cellular end point, and should have well-defined performance characteristics such as reproducibility, concordance and stability across a range of conditions and throughputs. Some analytical parameters of the work flow can be validated using standard cancer cell lines as supportive data.
Clinical validity
Clinical validity, or correlation of the biomarker with clinical response, can be obtained from retrospective studies of patient responses and assay results. For functional screening, such evaluations are more difficult to perform in retrospect because very few patient samples are routinely archived to preserve cell viability. However, given the wealth and strength of data linking a limited number of gene mutations to drug response, an intermediate surrogate of clinical validity for functional tests could be obtained from the correlation of ex vivo responses to such targeted therapies in samples with these mutations. More definitive clinical validity can be obtained from ‘retrospective–prospective’ trials in which samples are collected and tested in parallel with clinical care, and the ex vivo test output is later compared with clinical outcome.
Clinical utility
Given the costs and risks of increasingly complicated clinical biomarkers, there is growing awareness of the need for clinical biomarkers to demonstrate evidence of clinical utility or improvement in patient outcomes. All diagnostic methods, whether genomic or functional, face the challenge of sampling error generated by intrapatient tumour heterogeneity as one cause of test variability, but only through rigorous clinical validity and utility testing will we understand whether this source of error precludes meaningful patient benefit from the tests. The gold standard for such evaluation is the prospective trial randomizing patients to assay-guided therapy versus an appropriately selected control group (for example, non-assay guided but identical drug menu or oncologist’s choice)106. However, given the size and cost required for such trials even for moderate expectations of clinical benefit, functional predictive tests could be examined in smaller, nonrandomized trials measuring overall response rates in advanced cancer populations for which the historical response rate to novel therapies is expected to be low84. Building this approach into clinical practice would facilitate effective triage of patients to standard-of-care therapy versus clinical trials that test novel combination strategies. Of course, for the vast majority of cancer patients for whom molecularly targeted therapies are not available, assay-guided therapy could be immediately evaluated in prospective trials that seek to show that durable responses can be reliably predicted. Just as NGS methods are being evaluated across tumour types or across multiple drugs in ‘basket’ and ‘umbrella’ type trials, so functional assays could be validated with panels of specific drugs for a specific tumour type (umbrella) or a single drug across multiple tumour types (basket), depending on assay flexibility107.
Conclusions
Functional testing complements genetic sequencing by providing the response of patient cells without a priori knowledge of the mechanism of drug activity. Given the intuitive and compelling rationale for functional testing in oncology, it is not surprising that there has been renewed interest in next-generation functional diagnostics in recent years. As with any new clinical biomarker, these assays will require clinical validation that such tests can provide lasting benefit to patients, leading to adoption in routine oncology care. We imagine that such trials over the next 3–5 years will ultimately result in identification of the specific clinical scenarios in which next-generation functional testing can help to guide oncologists in choosing the right therapy for their patients. It follows that such tests can also prevent the unnecessary use of therapeutics to which patients may be resistant, reducing toxicities as well as costs.
Currently, molecular approaches such as NGS are being used based on the hope that somatic cancer mutations can suggest therapies. We believe that for most cancer patients this information alone is ultimately limited. Functional screening will become part of an emerging comprehensive diagnostic approach that includes traditional pathology techniques together with genetic sequencing. Complementing these two approaches, investigating the phenotype and functional responses of patient tumour-infiltrating lymphocytes, as well as tumour antigen prediction and overall mutational load, will be used to help to make recommendations for immuno-oncology therapy choices87. In this paradigm, multiple diagnostic technologies will be used on a single patient biopsy before therapy to enable the best ‘personalized’ choice from an armamentarium of large numbers of single and combination drug regimens. Only through such a comprehensive picture will the emerging cohort of hundreds of targeted therapies and immunotherapies be precisely matched to individual patients with cancer.
Acknowledgments
The authors apologize to investigators whose research they were unable to include owing to space considerations.
Footnotes
Competing interests statement
The authors declare competing interests: see Web version for details.
Contributor Information
Adam A. Friedman, Massachusetts General Hospital Cancer Center, Harvard Medical School, 55 Fruit Street, Boston, Massachusetts 02114, USA
Anthony Letai, Dana-Farber Cancer Institute, Harvard Medical School, 440 Brookline Avenue, Mayer 430, Boston, Massachusetts 02215, USA.
David E. Fisher, Massachusetts General Hospital Cancer Center, Harvard Medical School, 55 Fruit Street, Boston, Massachusetts 02114, USA Dermatology and Cutaneous Biology Research Center, Massachusetts General Hospital, 149 East 13th Street, Charlestown, Massachusetts 02129, USA.
Keith T. Flaherty, Massachusetts General Hospital Cancer Center, Harvard Medical School, 55 Fruit Street, Boston, Massachusetts 02114, USA
References
- 1.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sawyers CL, et al. Imatinib induces hematologic and cytogenetic responses in patients with chronic myelogenous leukemia in myeloid blast crisis: results of a Phase II study. Blood. 2002;99:3530–3539. doi: 10.1182/blood.v99.10.3530. [DOI] [PubMed] [Google Scholar]
- 4.Talpaz M, et al. Imatinib induces durable hematologic and cytogenetic responses in patients with accelerated phase chronic myeloid leukemia: results of a Phase 2 study. Blood. 2002;99:1928–1937. doi: 10.1182/blood.v99.6.1928. [DOI] [PubMed] [Google Scholar]
- 5.Dienstmann R, Jang IS, Bot B, Friend S, Guinney J. Database of genomic biomarkers for cancer drugs and clinical targetability in solid tumors. Cancer Discov. 2015;5:118–123. doi: 10.1158/2159-8290.CD-14-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 2013;12:581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 7.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 8.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jorgensen JH, Ferraro MJ. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis. 2009;49:1749–1755. doi: 10.1086/647952. [DOI] [PubMed] [Google Scholar]
- 10.Dishing out cancer treatment [Editorial] Nat. Biotechnol. 2013;31:85. doi: 10.1038/nbt.2516. [DOI] [PubMed] [Google Scholar]
- 11.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 13.Rolland T, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 15.Piotrowska Z, et al. Heterogeneity underlies the emergence of EGFRT790 wild-type clones following treatment of T790M-positive cancers with a third generation EGFR inhibitor. Cancer Discov. 2015;5:713–722. doi: 10.1158/2159-8290.CD-15-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Allen EM, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johannessen CM, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–142. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burstein HJ, et al. American Society of Clinical Oncology clinical practice guideline update on the use of chemotherapy sensitivity and resistance assays. J. Clin. Oncol. 2011;29:3328–3330. doi: 10.1200/JCO.2011.36.0354. [DOI] [PubMed] [Google Scholar]
- 19.Samson DJ, Seidenfeld J, Ziegler K, Aronson N. Chemotherapy sensitivity and resistance assays: a systematic review. J. Clin. Oncol. 2004;22:3618–3630. doi: 10.1200/JCO.2004.04.077. [DOI] [PubMed] [Google Scholar]
- 20.Schrag D, et al. American Society of Clinical Oncology Technology Assessment: chemotherapy sensitivity and resistance assays. J. Clin. Oncol. 2004;22:3631–3638. doi: 10.1200/JCO.2004.05.065. [DOI] [PubMed] [Google Scholar]
- 21.Von Hoff DD, et al. A Southwest Oncology Group study on the use of a human tumor cloning assay for predicting response in patients with ovarian cancer. Cancer. 1991;67:20–27. doi: 10.1002/1097-0142(19910101)67:1<20::aid-cncr2820670105>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 22.Nagourney RA, Evans SS, Messenger JC, Su YZ, Weisenthal LM. 2 chlorodeoxyadenosine activity and cross resistance patterns in primary cultures of human hematologic neoplasms. Br. J. Cancer. 1993;67:10–14. doi: 10.1038/bjc.1993.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kern DH, Weisenthal LM. Highly specific prediction of antineoplastic drug resistance with an in vitro assay using suprapharmacologic drug exposures. J. Natl Cancer Inst. 1990;82:582–588. doi: 10.1093/jnci/82.7.582. [DOI] [PubMed] [Google Scholar]
- 24.Grendys EC, Jr, et al. Overview of a chemoresponse assay in ovarian cancer. Clin. Transl Oncol. 2014;16:761–769. doi: 10.1007/s12094-014-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maenpaa JU, et al. The subrenal capsule assay in selecting chemotherapy for ovarian cancer: a prospective randomized trial. Gynecol. Oncol. 1995;57:294–298. doi: 10.1006/gyno.1995.1145. [DOI] [PubMed] [Google Scholar]
- 26.Cree IA, et al. A prospective randomized controlled trial of tumour chemosensitivity assay directed chemotherapy versus physician’s choice in patients with recurrent platinum-resistant ovarian cancer. Anticancer Drugs. 2007;18:1093–1101. doi: 10.1097/CAD.0b013e3281de727e. [DOI] [PubMed] [Google Scholar]
- 27.Sandberg R, Ernberg I. The molecular portrait of in vitro growth by meta-analysis of gene-expression profiles. Genome Biol. 2005;6:R65. doi: 10.1186/gb-2005-6-8-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dairkee SH, et al. A molecular ‘signature’ of primary breast cancer cultures; patterns resembling tumor tissue. BMC Genomics. 2004;5:47. doi: 10.1186/1471-2164-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am. J. Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suprynowicz FA, et al. Conditionally reprogrammed cells represent a stem-like state of adult epithelial cells. Proc. Natl Acad. Sci. USA. 2012;109:20035–20040. doi: 10.1073/pnas.1213241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan H, et al. Use of reprogrammed cells to identify therapy for respiratory papillomatosis. N. Engl. J. Med. 2012;367:1220–1227. doi: 10.1056/NEJMoa1203055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crystal AS, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–1486. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baccelli I, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 2013;31:539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 34.Hodgkinson CL, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat. Med. 2014;20:897–903. doi: 10.1038/nm.3600. [DOI] [PubMed] [Google Scholar]
- 35.Zhang L, et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl Med. 2013;5:180ra48. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu M, et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brouzes E, et al. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl Acad. Sci. USA. 2009;106:14195–14200. doi: 10.1073/pnas.0903542106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sachs N, Clevers H. Organoid cultures for the analysis of cancer phenotypes. Curr. Opin. Genet. Dev. 2014;24:68–73. doi: 10.1016/j.gde.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 39.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 40.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 41.Boj SF, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160:324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao D, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159:176–187. doi: 10.1016/j.cell.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van de Wetering M, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mengelbier LH, et al. Intratumoral genome diversity parallels progression and predicts outcome in pediatric cancer. Nat. Commun. 2015;6:6125. doi: 10.1038/ncomms7125. [DOI] [PubMed] [Google Scholar]
- 45.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–354. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 46.Ridky TW, Chow JM, Wong DJ, Khavari PA. Invasive three-dimensional organotypic neoplasia from multiple normal human epithelia. Nat. Med. 2010;16:1450–1455. doi: 10.1038/nm.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kenny HA, et al. Quantitative high throughput screening using a primary human three-dimensional organotypic culture predicts in vivo efficacy. Nat. Commun. 2015;6:6220. doi: 10.1038/ncomms7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaira V, et al. Preclinical model of organotypic culture for pharmacodynamic profiling of human tumors. Proc. Natl Acad. Sci. USA. 2010;107:8352–8356. doi: 10.1073/pnas.0907676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagourney RA, et al. Functional profiling to select chemotherapy in untreated, advanced or metastatic non-small cell lung cancer. Anticancer Res. 2012;32:4453–4460. [PubMed] [Google Scholar]
- 50.Majumder B, et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat. Commun. 2015;6:6169. doi: 10.1038/ncomms7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirt C, et al. ‘In vitro’ 3D models of tumor-immune system interaction. Adv. Drug Deliv. Rev. 2014;79–80:145–154. doi: 10.1016/j.addr.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 52.Klinghoffer RA, et al. A technology platform to assess multiple cancer agents simultaneously within a patient’s tumor. Sci. Transl Med. 2015;7:284ra58. doi: 10.1126/scitranslmed.aaa7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonas O, et al. An implantable microdevice to perform high-throughput in vivo drug sensitivity testing in tumors. Sci. Transl Med. 2015;7:284ra57. doi: 10.1126/scitranslmed.3010564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siolas D, Hannon GJ. Patient-derived tumor xenografts: transforming clinical samples into mouse models. Cancer Res. 2013;73:5315–5319. doi: 10.1158/0008-5472.CAN-13-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aparicio S, Hidalgo M, Kung AL. Examining the utility of patient-derived xenograft mouse models. Nat. Rev. Cancer. 2015;15:311–316. doi: 10.1038/nrc3944. [DOI] [PubMed] [Google Scholar]
- 56.Hidalgo M, et al. A pilot clinical study of treatment guided by personalized tumorgrafts in patients with advanced cancer. Mol. Cancer Ther. 2011;10:1311–1316. doi: 10.1158/1535-7163.MCT-11-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stebbing J, et al. Patient-derived xenografts for individualized care in advanced sarcoma. Cancer. 2014;120:2006–2015. doi: 10.1002/cncr.28696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rubio-Viqueira B, et al. An in vivo platform for translational drug development in pancreatic cancer. Clin. Cancer Res. 2006;12:4652–4661. doi: 10.1158/1078-0432.CCR-06-0113. [DOI] [PubMed] [Google Scholar]
- 59.Eirew P, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2015;518:422–426. doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tyner JW, et al. RNAi screen for rapid therapeutic target identification in leukemia patients. Proc. Natl Acad. Sci. USA. 2009;106:8695–8700. doi: 10.1073/pnas.0903233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tyner JW, et al. Kinase pathway dependence in primary human leukemias determined by rapid inhibitor screening. Cancer Res. 2013;73:285–296. doi: 10.1158/0008-5472.CAN-12-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Glover JM, Loriaux M, Tyner JW, Druker BJ, Chang BH. In vitro sensitivity to dasatinib in lymphoblasts from a patient with t(17;19)(q22;p13) gene rearrangement pre-B acute lymphoblastic leukemia. Pediatr. Blood Cancer. 2012;59:576–579. doi: 10.1002/pbc.23383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kulesz-Martin MF, et al. A molecular case report: functional assay of tyrosine kinase inhibitors in cells from a patient’s primary renal cell carcinoma. Cancer Biol. Ther. 2013;14:95–99. doi: 10.4161/cbt.22960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davis LE, et al. A case study of personalized therapy for osteosarcoma. Pediatr. Blood Cancer. 2013;60:1313–1319. doi: 10.1002/pbc.24512. [DOI] [PubMed] [Google Scholar]
- 65.Pemovska T, et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov. 2013;3:1416–1429. doi: 10.1158/2159-8290.CD-13-0350. [DOI] [PubMed] [Google Scholar]
- 66.Yadav B, et al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci. Rep. 2014;4:5193. doi: 10.1038/srep05193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pemovska T, et al. Axitinib effectively inhibits BCR-ABL1(T315I) with a distinct binding conformation. Nature. 2015;519:102–105. doi: 10.1038/nature14119. [DOI] [PubMed] [Google Scholar]
- 68.Klco JM, et al. Genomic impact of transient low-dose decitabine treatment on primary AML cells. Blood. 2013;121:1633–1643. doi: 10.1182/blood-2012-09-459313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lamothe B, et al. Proteasome inhibitor carfilzomib complements ibrutinib’s action in chronic lymphocytic leukemia. Blood. 2015;125:407–410. doi: 10.1182/blood-2014-07-585364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hilhorst R, et al. Peptide microarrays for profiling of serine/threonine kinase activity of recombinant kinases and lysates of cells and tissue samples. Methods Mol. Biol. 2013;977:259–271. doi: 10.1007/978-1-62703-284-1_21. [DOI] [PubMed] [Google Scholar]
- 71.Tahiri A, et al. Differential inhibition of ex-vivo tumor kinase activity by vemurafenib in BRAF(V600E) and BRAF wild-type metastatic malignant melanoma. PLoS ONE. 2013;8:e72692. doi: 10.1371/journal.pone.0072692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schayowitz A, et al. Functional profiling of live melanoma samples using a novel automated platform. PLoS ONE. 2012;7:e52760. doi: 10.1371/journal.pone.0052760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Irish JM, et al. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 74.Krutzik PO, Crane JM, Clutter MR, Nolan GP. High-content single-cell drug screening with phosphospecific flow cytometry. Nat. Chem. Biol. 2008;4:132–142. doi: 10.1038/nchembio.2007.59. [DOI] [PubMed] [Google Scholar]
- 75.Kornblau SM, et al. Dynamic single-cell network profiles in acute myelogenous leukemia are associated with patient response to standard induction therapy. Clin. Cancer Res. 2010;16:3721–3733. doi: 10.1158/1078-0432.CCR-10-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lacayo NJ, et al. Development and validation of a single-cell network profiling assay-based classifier to predict response to induction therapy in paediatric patients with de novo acute myeloid leukaemia: a report from the Children’s Oncology Group. Br. J. Haematol. 2013;162:250–262. doi: 10.1111/bjh.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hata AN, Engelman JA, Faber AC. The BCL2 family: key mediators of the apoptotic response to targeted anticancer therapeutics. Cancer Discov. 2015;5:475–487. doi: 10.1158/2159-8290.CD-15-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Del Gaizo Moore V, Letai A. BH3 profiling — measuring integrated function of the mitochondrial apoptotic pathway to predict cell fate decisions. Cancer Lett. 2013;332:202–205. doi: 10.1016/j.canlet.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ni Chonghaile T, et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science. 2011;334:1129–1133. doi: 10.1126/science.1206727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vo TT, et al. Relative mitochondrial priming of myeloblasts and normal HSCs determines chemotherapeutic success in AML. Cell. 2012;151:344–355. doi: 10.1016/j.cell.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Montero J, et al. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell. 2015;160:977–989. doi: 10.1016/j.cell.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jahnke HG, et al. Direct chemosensitivity monitoring ex vivo on undissociated melanoma tumor tissue by impedance spectroscopy. Cancer Res. 2014;74:6408–6418. doi: 10.1158/0008-5472.CAN-14-0813. [DOI] [PubMed] [Google Scholar]
- 83.Kleinhans R, et al. Sensor-based cell and tissue screening for personalized cancer chemotherapy. Med. Biol. Eng. Comput. 2012;50:117–126. doi: 10.1007/s11517-011-0855-7. [DOI] [PubMed] [Google Scholar]
- 84.Simon R, Roychowdhury S. Implementing personalized cancer genomics in clinical trials. Nat. Rev. Drug Discov. 2013;12:358–369. doi: 10.1038/nrd3979. [DOI] [PubMed] [Google Scholar]
- 85.Sawyers CL, van ‘t Veer LJ. Reliable and effective diagnostics are keys to accelerating personalized cancer medicine and transforming cancer care: a policy statement from the American Association for Cancer Research. Clin. Cancer Res. 2014;20:4978–4981. doi: 10.1158/1078-0432.CCR-14-2295. [DOI] [PubMed] [Google Scholar]
- 86.U.S. Department of Health and Human Services. Anticipated Details of the Draft Guidance to Industry, Food and Drug Administration Staff and Clinical Laboratories: Framework for Regulatory Oversight of Laboratory Developed Tests (LDTs) FDA [online] 2014 http://www.fda.gov/downloads/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/UCM407409.pdf.
- 87.Kreiter S, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–696. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wheler JJ, et al. Prospective study comparing outcomes in patients with advanced malignancies on molecular alteration-matched versus non-matched therapy. J. Clin. Oncol. 2015;33:S11019. [Google Scholar]
- 89.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Flaherty KT, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mok TS, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 92.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 94.Wander SA, Levis MJ, Fathi AT. The evolving role of FLT3 inhibitors in acute myeloid leukemia: quizartinib and beyond. Ther. Adv. Hematol. 2014;5:65–77. doi: 10.1177/2040620714532123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kopetz S, et al. PLX4032 in metastatic colorectal cancer patients with mutant BRAF tumors. J. Clin. Oncol. 2010;28:15s. [Google Scholar]
- 96.Rubio-Perez C, et al. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell. 2015;27:382–396. doi: 10.1016/j.ccell.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 97.Herbst RS, et al. Lung Master Protocol (Lung-MAP) — a biomarker-driven protocol for accelerating development of therapies for squamous cell lung cancer: SWOG S1400. Clin. Cancer Res. 2015;21:1514–1524. doi: 10.1158/1078-0432.CCR-13-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lopez-Chavez A, et al. Molecular profiling and targeted therapy for advanced thoracic malignancies: a biomarker-derived, multiarm, multihistology Phase II basket trial. J. Clin. Oncol. 2015;33:1000–1007. doi: 10.1200/JCO.2014.58.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kris MG, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lind PA, Berg OG, Andersson DI. Mutational robustness of ribosomal protein genes. Science. 2010;330:825–827. doi: 10.1126/science.1194617. [DOI] [PubMed] [Google Scholar]
- 101.Cameron F, Sanford M. Ibrutinib: first global approval. Drugs. 2014;74:263–271. doi: 10.1007/s40265-014-0178-8. [DOI] [PubMed] [Google Scholar]
- 102.Markham A. Idelalisib: first global approval. Drugs. 2014;74:1701–1707. doi: 10.1007/s40265-014-0285-6. [DOI] [PubMed] [Google Scholar]
- 103.Luo J, Solimini NL, Elledge SJ. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell. 2009;136:823–837. doi: 10.1016/j.cell.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Haibe-Kains B, et al. Inconsistency in large pharmacogenomic studies. Nature. 2013;504:389–393. doi: 10.1038/nature12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hatzis C, et al. Enhancing reproducibility in cancer drug screening: how do we move forward? Cancer Res. 2014;74:4016–4023. doi: 10.1158/0008-5472.CAN-14-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J. Natl Cancer Inst. 2009;101:1446–1452. doi: 10.1093/jnci/djp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tsourounis M, Stuart J, Pignato W, Toscani M, Barone J. Current trends in personalized medicine and companion diagnostics: a summary from the DIA meeting on personalized medicine and companion diagnostics. Ther. Innov. Regul. Sci. 2015;49:530–543. doi: 10.1177/2168479015570330. [DOI] [PubMed] [Google Scholar]