Abstract
Aim
To identify and characterize cancer stem cells (CSC) in moderately differentiated buccal mucosa squamous cell carcinoma (MDBMSCC).
Methods
Four micrometer-thick, formalin-fixed, paraffin-embedded MDBMSCC samples from six patients underwent 3,3-diaminobenzidine (DAB) immunohistochemical (IHC) staining for the embryonic stem cell (ESC) markers, NANOG, OCT4, SALL4, SOX2, and pSTAT3; cancer stem cell marker, CD44; squamous cell carcinoma (SCC) marker, EMA; and endothelial marker, CD34. The transcriptional activities of the genes encoding NANOG, OCT4, SOX2, SALL4, STAT3, and CD44 were studied using NanoString gene expression analysis and colorimetric in situ hybridization (CISH) for NANOG, OCT4, SOX2, SALL4, and STAT3.
Results
Diaminobenzidine and immunofluorescent (IF) IHC staining demonstrated the presence of (1) an EMA+/CD44+/SOX2+/SALL4+/OCT4+/pSTAT3+/NANOG+ CSC subpopulation within the tumor nests; (2) an EMA−/CD44−/CD34−/SOX2+/OCT4+/pSTAT3+/NANOG+ subpopulation within the stroma between the tumor nests; and (3) an EMA−/CD44−/CD34+/SOX2+/SALL4+/OCT4+/pSTAT3+/NANOG+ subpopulation on the endothelium of the microvessels within the stroma. The expression of CD44, SOX2, SALL4, OCT4, pSTAT3, and NANOG was confirmed by the presence of mRNA transcripts, using NanoString analysis and NANOG, OCT4, SOX2, SALL4, and STAT3 by CISH staining.
Conclusion
This study demonstrated a novel finding of three separate CSC subpopulations within MDBMSCC: (1) within the tumor nests expressing EMA, CD44, SOX2, SALL4, OCT4, pSTAT3, and NANOG; (2) within the stroma expressing SOX2, SALL4, OCT4, pSTAT3, and NANOG; and (3) on the endothelium of the microvessels within the stroma expressing CD34, SOX2, SALL4, OCT4, pSTAT3, and NANOG.
Keywords: buccal, mucosal, squamous cell carcinoma, cancer, stem cells, oral cavity
Introduction
Oral cavity cancer is the sixth most common cancer worldwide (1), with over 90% being squamous cell carcinoma (SCC) (1). Oral cavity squamous cell carcinoma (OCSCC) may arise from sites of leukoplakia or erythroplakia involving the oral squamous epithelium (2, 3), characterized by its dysplastic morphology. Late-stage disease often presents with nodal metastases and less commonly, metastasis to the lung, brain, bone, and liver (2, 3).
Oral cavity squamous cell carcinoma affects the oral tongue, floor of the mouth, buccal mucosa, hard palate, mandibular and maxillary alveolus, vestibule of mouth, and retromolar trigone (4, 5). While buccal mucosal squamous cell carcinoma (BMSCC) accounts for <10% of all OCSCC in the Western world (4, 6), it is the most prevalent OCSCC in South East Asia and South Asia (6, 7). This high prevalence is primarily due to the practice of betel nut chewing (6, 7), and/or alcohol consumption (8) and/or tobacco use (5), although recent studies suggest other non-lifestyle contributing factors (5). Due to the particularly aggressive nature of this cancer (9), patients with BMSCC have a high locoregional recurrence rate of up to 57% with associated poor survival (10). Despite improvements in diagnosis and treatments, 5-year survival rates have remained around 50% since the early 1980s (4–7). Current treatment of BMSCC involves a multimodal approach (11), usually requiring surgical resection with postoperative adjuvant radiotherapy (RT) and occasionally chemotherapy (ChT), especially in cases with adverse features, such as positive surgical margins and/or extracapsular spread of the nodal metastases (11).
It has been proposed that the development and spread of many cancers (12–14), including OCSCC (15), are driven by a subpopulation of cancer cells known as cancer stem cells (CSC) (12). Recent studies have identified these CSC in OCSCC by their expression of the embryonic stem cell (ESC) markers, NANOG (15), SOX2 (16), SALL4 (17), phosphorylated (activated) form of signal transducer and activator of transcription 3 (pSTAT3) (18), and OCT4 (15, 16). NANOG is a transcription factor that controls cell proliferation, migration, and invasion (19). SOX2 is critical for the self-renewal properties of ESC (16). OCT4 is a POU domain transcription factor associated with cell self-renewal, proliferation, and pluripotency (16) and works synergistically with SOX2 to regulate pluripotency of ESC (19). This pluripotency network also includes additional factors, such as SALL4, a transcriptional activator of POU5f1 that maintains embryonic pluripotency by modulating the expression of OCT4 (20), and pSTAT3, which is required for tumor formation and growth, and suppression of apoptosis (21). The cell surface marker CD44 is a multistructural and multifunctional CSC marker associated with angiogenesis, cell proliferation, migration, and differentiation (22), as well as some progenitor cell properties (23). Unlike progenitor cells and differentiated cells, CSCs possess the ability for self-renewal and multilineage differentiation through either asymmetric or symmetric cell division (12). It has been reported that the presence of CSC is associated with both a greater capacity for tumor growth and a worsening prognosis (13, 15). This is supported by the idea that CSC have unique abilities to resist cell damage and may represent critical mediators for both RT and ChT resistance (24).
Despite recent literature showing the presence of CSC within many cancers, including breast (25), brain (26), and pancreatic (27) cancer, there have been no reports describing the presence of CSC within BMSCC.
This study investigated the expression, within moderately differentiated buccal mucosa squamous cell carcinoma (MDBMSCC), of the ESC markers NANOG, SOX2, SALL4, pSTAT3, OCT4, and CD44, at both the transcriptional and translational level, to identify and characterize the putative CSC population.
Materials and Methods
Tissue Samples
Previously untreated MDBMSCCs from five male and one female patients, aged 38–80 (mean, 59) years, were used in this study, which was approved by the Central Health and Disabilities Ethics Committee (ref. no. 12/CEN/74).
Histochemical Staining and Immunohistochemical Staining
Hematoxylin and eosin staining was performed on 4 μm-thick, formalin-fixed, paraffin-embedded blocks of six MDBMSCC samples, to confirm the appropriate histological grading and the presence of BMSCC in the sections. 3,3-Diaminobenzidine (DAB) immunohistochemical (IHC) staining of the sections was then performed using the Leica Bond Rx auto-stainer (Leica, Nussloch, Germany), as previously described (28). Staining for NANOG (1:2000; cat# NBP1-04320, Novus Biologicals LLC, Littleton, CO, USA), SOX2 (1:500; cat# PA-094, Thermo Fisher Scientific, Rockford, IL, USA), SALL4 (1:30; cat# CM385M-16, Cell Marque, Rocklin, CA, USA), pSTAT3 (1:100; cat# 9145, Cell Signaling Technology, Danvers, MA, USA), and OCT4 (1:200; cat# NBP1-47923, Novus Biologicals LLC, Littleton, CO, USA), CD34 (ready-to-use; cat# PA0212, Leica), CD44 (1:1500; cat# MRQ-13, Cell Marque), and epithelial membrane antigen (EMA, ready-to-use; cat# PA0212, Leica), diluted with Bond™ primary antibody diluent (Leica AR9352), was done for all tissue samples.
To confirm co-expression of two proteins, representative slides of MDBMSCC (n = 2) underwent immunofluorescent (IF) IHC staining using a combination of Vectafluor Excel anti-rabbit 594 (ready-to-use; cat# VEDK-1594, Vector Laboratories, Burlingame, CA, USA) and Alexa Fluor anti-mouse 488 (1:500; cat# A21202, Life Technologies) so as to detect combinations that included NANOG, SOX2, and pSTAT3, and Vectafluor Excel anti-mouse (ready-to-use; cat# VEDK2488, Vector Laboratories) and Alexa Fluor anti-rabbit 594 (1:500; cat# A21207, Life Technologies) to detect combinations that included CD44, EMA, OCT4, or SALL4.
Positive control tissues used for the primary antibodies were human infantile hemangioma (IH) for NANOG (29), and SALL4 (30), myometrium for OCT4 (31), skin for SOX2 (32), tonsil for pSTAT3 (33) and CD44 (34). A negative control for the primary antibody was performed on a MDBMSCC sample from the cohort of patients used for IHC staining.
Nanostring Gene Expression Analysis
RNA was extracted from five snap-frozen MDBMSCC samples from the same cohort of patients used for DAB IHC staining and was used for NanoString nCounter™ Gene Expression Assay (Nanostring Technologies, Seattle, WA, USA). Total RNA was extracted using the MagJET RNA kit (Thermo Fisher Scientific) with the protocol adapted for tissue and run on a KingFisher Duo machine (Thermo Fisher Scientific). RNA samples were then quantitated on a Qubit® 2.0 fluorometer (Invitrogen, Life Technologies) and were subject to RNA integrity analysis via the 2100 Bioanalyzer Instrument (Agilent Technologies). The samples then underwent NanoString nCounter gene expression assay performed by New Zealand Genomics (Dunedin, New Zealand) according to the manufacturer’s protocol. Probes for the genes encoding NANOG (NM_024865.2), SALL4 (NM_020436.3) SOX2 (NM_003106.2), OCT4 (NM_002701.4), CD44 (NM_001001392.1), and STAT3 (NM_139276.2) and the housekeeping gene GUSB (NM_000181.1) were designed and manufactured by NanoString Technologies. Raw data were analyzed using nSolver™ software (NanoString Technologies) using standard settings and normalized against the housekeeping gene.
Colorimetric In Situ Hybridization
Representative 4 μm-thick, formalin-fixed, paraffin-embedded sections of three MDBMSCC samples from the original cohort of six patients used for DAB IHC staining were used for mRNA colorimetric in situ hybridization (CISH). Staining was done on the Leica Bond Rx auto-stainer and detected using the ViewRNA red stain kit (Affymetrix, Santa Clara, CA, USA), as previously described (35). The probes used for NANOG (NM_024865), SOX2 (NM_003106), SALL4 (NM_020436), STAT3 (NM_003150), and OCT4 (NM_002701) were obtained from Affymetrix. Positive controls used were human IH for SALL4 and human seminoma for NANOG, SOX2, OCT4, and STAT3. A negative control for the primary antibody was done on a sample of MDBMSCC from the CISH cohort by omitting the probe.
Image Analysis
Diaminobenzidine IHC- and CISH-stained slides were viewed and imaged using an Olympus BX53 light microscope (Tokyo, Japan). IF IHC-stained images were captured using an Olympus FV1200 biological confocal laser-scanning microscope and processed with the cellSens Dimension 1.11 software using 2D deconvolution algorithm (Olympus).
Results
Histochemical and Immunohistochemical Staining
Hematoxylin and eosin staining confirmed the appropriate histological grading and the presence of MDBMSCC on the slides used for DAB IHC staining (data not shown).
Diaminobenzidine IHC staining showed that cells within the tumor nests stained positively with the SCC marker EMA (Figure 1A, brown). Both SOX2 (Figure 1B, brown) and SALL4 (Figure 1C, brown) showed positive nuclear staining with increased expression especially toward the periphery of the tumor nests. Nuclear staining for OCT4 (Figure 1D, brown) and pSTAT3 (Figure 1E, brown) of the cells within the tumor nests was noted, while primarily cytoplasmic staining with some nuclear positivity for NANOG (Figure 1F, brown) was observed. There was also membranous expression of the CSC marker CD44 (Figure 1G, brown) that was exclusively localized to cells within the tumor nests. Clusters of cells within the stroma also showed nuclear expression of SOX2 (Figure 1B, brown, arrowheads), OCT4 (Figure 1D, brown, arrowheads), and pSTAT3 (Figure 1E, brown, arrowheads), while cytoplasmic and nuclear expression of NANOG (Figure 1F, brown, arrowheads) was observed in these cells. The endothelium of the microvessels within the stroma also stained positively for SOX2 (Figure 1B, brown, thin arrows), SALL4 (Figure 1C, brown, thin arrows), OCT4 (Figure 1D, brown, thin arrows), pSTAT3 (Figure 1E, brown, thin arrows), and NANOG (Figure 1F, brown, thin arrows).
Figure 1.

Representative DAB IHC-stained sections of MDBMSCC demonstrating nuclear expression of EMA of cells within the tumor nests [(A), brown]. Expression of SOX2 was seen in cells within the tumor nests [(B), brown, thick arrows] and the stroma [(B), brown, arrowheads], and on the endothelium of the microvessels within the stroma [(B), brown, thin arrows]. Expression of SALL4 was limited to cells within the tumor nests [(C), brown, thick arrows] and the endothelium of the microvessels [(C), brown, thin arrows]. OCT4 was also expressed in cells within tumor nests [(D), brown, thick arrows] and the stroma [(D), brown, arrowheads], and the endothelium of the microvessels within the stroma [(D), brown, thin arrows]. Expression of pSTAT3 was detected on cells within the tumor nests [(E), brown, thick arrows] and the stroma [(E), brown, arrowheads], and the endothelium of the microvessels within the stroma [(E), brown, thin arrows]. NANOG was also seen in cells within the tumor nests [(F), brown, thick arrows] and the stroma [(F), brown, arrowheads], and the endothelium of the microvessels within the stroma [(F), brown, thin arrows]. CD44 expression was seen as membranous staining of the tumor nest cells [(G), brown]. Original magnification: 400×.
Expected staining patterns for the NANOG (Figure S1A in Supplementary Material, brown), SALL4 (Figure S1B in Supplementary Material, brown), OCT4 (Figure S1C in Supplementary Material, brown), SOX2 (Figure S1D in Supplementary Material, brown), pSTAT3 (Figure S1E in Supplementary Material, brown), and CD44 (Figure S1F in Supplementary Material, brown) were demonstrated in the respective positive controls. The omission of the primary antibody in MDBMSCC samples provided an appropriate negative control (Figure S1G in Supplementary Material).
To demonstrate co-expression of the ESC markers, IF IHC staining was performed on two representative MDBMSCC samples used for DAB IHC staining. pSTAT3 staining (Figures 2A,B, red) was localized to cells within the tumor nests that displayed membranous staining for EMA (Figure 2A, green), the endothelium of the microvessels that stained positively for CD34 (Figure 2B, green) and cells within the stroma. NANOG was expressed by cells within the tumor nests (Figure 2C, red) and the stroma (Figure 2C, red), as well as the endothelium of the microvessels that stained positively for CD34 (Figure 2C, green). A similar staining pattern was also seen with SOX2 for cells within the tumor nests (Figure 2D, red) and the stroma (Figure 2D, red), and the endothelium marked by CD34 (Figure 2D, green). SOX2 (Figure 2E, red) and SALL4 (Figure 2E, green) were co-expressed (appearing as orange) by cells within the tumor nests and the stroma (Figure 2E), and the endothelium of the microvessels (Figure 2E, red). SOX2 (Figure 2F, red) and OCT4 (Figure 2F, green) were also co-localized (appearing as orange) to cells within the tumor nests and the stroma (Figure 2F), and the endothelium of the microvessels (Figure 2F). pSTAT3 (Figure 2G, red) and membranous staining of CD44 (Figure 2G, green) were co-expressed by cells within the tumor nests. Images of the individual stains are presented in Figure S2 in Supplementary Material.
Figure 2.

Representative IF IHC-stained sections of MDBMSCC demonstrating the expression of pSTAT3 [(A), red] and EMA [(A), green] by cells within the tumor nests. There was a CSC subpopulation remonstrating nuclear co-expression of STAT3 [(B), red] and CD34 [(B), green], appearing as orange, on the endothelium of the microvessels within the stroma; and another subpopulation staining only positively for pSTAT3 within the stroma [(B), red]. Nuclear expression of NANOG [(C), red] was demonstrated on the endothelium of the microvessels which expressed CD34 [(C), green] within stroma. The NANOG+ cells [(C), red] that do not express CD34 were seen within the tumor nests and the stroma. SOX2 [(D), red] was also expressed by cells within tumor nests and the stroma, and the endothelium of the microvessels expressing CD34 [(D), green]. Nuclear expression of both SOX2 [(E), red] and SALL4 [(E), green], appearing as orange, was seen on the cells within the tumor nests and the stroma. Expression of both SOX2 [(F), red] and OCT4 [(F), green], appearing as orange, was seen on cells within the tumor nests and the stroma, and the endothelium of the microvessels within the stroma. pSTAT3 [(G), red] and membranous staining CD44 [(G), green] were co-expressed by cells within the tumor nests. Scale bars: 20 μm.
NanoString Gene Analysis
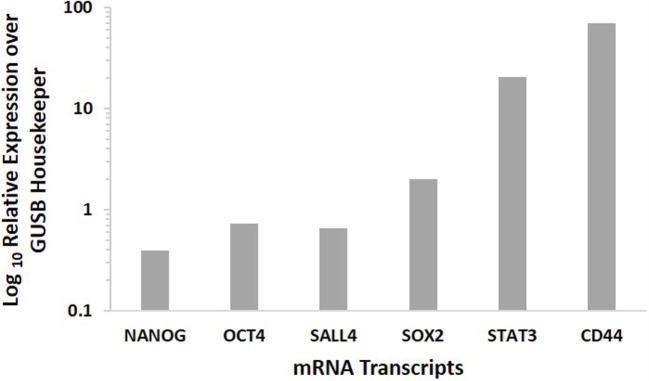
NanoString analysis confirmed the presence of the mRNA transcripts for NANOG, OCT4, SALL4, SOX2, STAT3, and CD44, in MDBMSCC samples of all five patients, while SALL4 was detected in four of the five samples (Figure 3).
Figure 3.
Expression of CSC-related mRNA transcripts of NANOG, OCT4, SALL4, SOX2 STAT3, and CD44 in MDBMSCC samples from five patients. Their expression was normalized over GUSB housekeeper.
Colorimetric In Situ Hybridization
Colorimetric in situ hybridization confirmed the presence of mRNA for SOX2 (Figure 4A, pink, arrows), SALL4 (Figure 4B, pink, arrows), OCT4 (Figure 4C, pink, arrows), STAT3 (Figure 4D, pink, arrows), and NANOG (Figure 4E, pink, arrows) in the cells within all three MDBMSCC samples.
Figure 4.
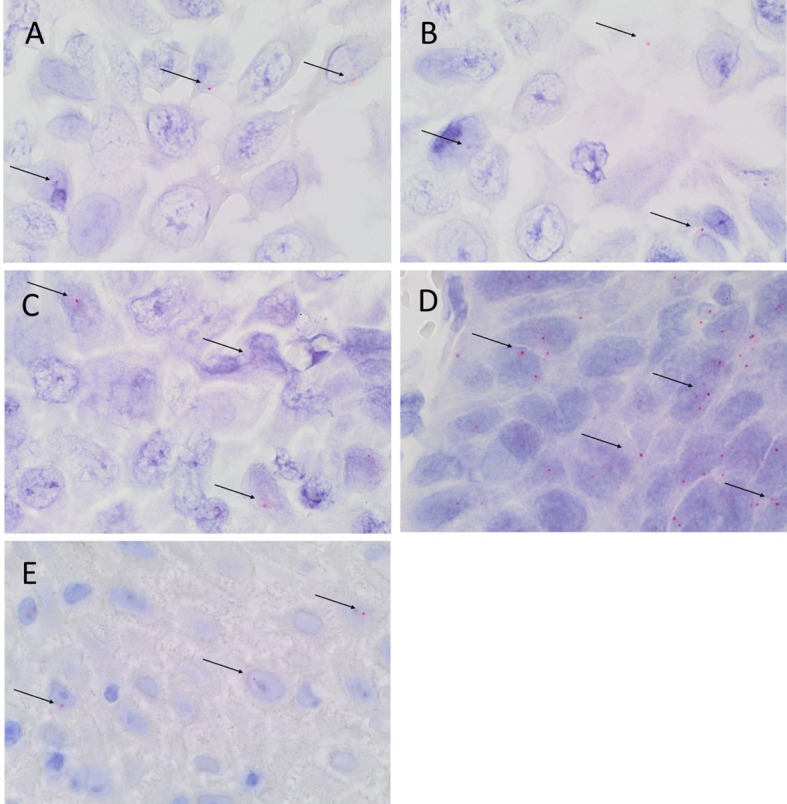
Representative CISH-stained sections of MDBMSCC demonstrating mRNA expression of SOX2 [(A), pink, arrows], SALL4 [(B), pink, arrows], OCT4 [(C), pink, arrows], STAT3 [(D), pink, arrows], and NANOG [(E), pink, arrows]. Original magnification: 1000×.
Discussion
The recent literature supporting the concept of CSC in carcinogenesis has reported on the presence of these cells in multiple cancer types (12–14). The findings of this study further support previous reports on the presence of CSC in OCSCC (15, 36). The discovery of more than one subpopulation of putative CSC within MDBMSCC expressing ESC markers is novel. The detection of a EMA+/CD44+/SOX2+/OCT4+/SALL4+/pSTAT3+/NANOG+ subpopulation within the tumor nests aligns with recent literature reporting localization of CSC in OCSCC (15, 36), particularly at the “tumor front” (37). To the best of our knowledge, this is the first report demonstrating a EMA−/CD34+/CD44−/SOX2+/OCT4+/SALL4+/pSTAT3+/NANOG+ subpopulation on the endothelium of the microvessels, and a separate EMA−/CD44−/CD34−/SOX2+/OCT4+/SALL4+/pSTAT3+/NANOG+ subpopulation within the stroma of MDBMSCC. It is exciting to speculate that the abundant expression of these ESC markers, and therefore CSC, which are known to be associated with cell proliferation, invasion, and inhibition of cell death (19), is a reflection of the particularly aggressive nature of this cancer.
Interestingly, the ESC markers SOX2, OCT4, SALL4, pSTAT3, and NANOG, showed both nuclear and cytoplasmic expression. This novel finding in MDBMSCC is consistent with previous studies reporting on cytoplasmic expression of NANOG in cervical cancer (38) and SALL4 in breast cancer (39) cells, inferring cytoplasmic expression as a predictor of poor prognosis (39). The reasons for this are the topic of further investigation.
This novel demonstration of the co-expression of ESC markers within the endothelium of the microvessels and the cells within the stroma may be a result of an epithelial-to-mesenchymal transition (EMT) and vascular mimicry, which have been previously demonstrated within breast (40) and head and neck (41) cancers. It is exciting to speculate that the CSC in MDBMSCC undergo EMT and preferentially express OCT4, pSTAT3, SOX2, and NANOG and potentially lose the expression of EMA and CD44, as none of these cells outside of the tumor nests express EMA or CD44. A reason for this is the potential loss of the relatively more downstream markers EMA and CD44, as the cells adopt a more stem-like phenotype with the ability to undergo EMT (42). This would be consistent with a previous report demonstrating the increased expression of the EMT-associated genes, twist and snail1, in the stromal population of pharyngeal SCC (43). Alternatively, these three CSC subpopulations may be distinct from one another, each playing a vital role in carcinogenesis. However, this is beyond the scope of this study.
The expression of ESC markers by the endothelium of the microvessels demonstrated by IHC and CISH staining is fascinating and merits further investigation. Curiously, while all five ESC markers were demonstrated using IHC staining, NanoString analysis confirmed the presence of the mRNA transcripts CD44, NANOG, OCT4, SOX2, and STAT3 in all the MDBMSCC samples from all five patients, while SALL4 was detected in only four of the five samples. A possible reason for this is the heterogeneous nature of cancer tissues. The abundance of cells within MDBMSCC expressing stem cell markers demonstrated by us is consistent with similar reports on hypopharyngeal SCC (44). Our findings of OCT4 expression in the tumor nests is similar to that reported by Ge et al. (44) showing OCT4 staining within hypopharyngeal SCC. However, we have also demonstrated the presence of an independent OCT4+ subpopulation within the stroma, consistent with the findings of Huang et al. (16).
Our demonstration of the presence of three putative CSC subpopulations within MDBMSCC adds further insight into the biology of this disease, highlighting a potential vital role for CSC in tumor growth and development, locoregional and distant metastatic spread, and resistance to RT and ChT. A greater understanding of the CSC, by characterizing the subpopulations in well and poorly differentiated BMSCC lesions using these same markers, as well as further investigations into their microenvironment and their regulatory pathways, may reveal novel therapeutic targets for this aggressive cancer.
Take Home Messages
(1) This study demonstrates the novel finding of three CSC subpopulations within MDBMSCC, with; (2) one CSC subpopulation within the tumor nests, expressing EMA, CD44, SOX2, SALL4, OCT4, pSTAT3, and NANOG; (3) a second CSC subpopulation is within the stroma between the tumor nests, expressing SOX2, SALL4, OCT4, pSTAT3, and NANOG; and (4) a third CSC subpopulation expressing CD34, SOX2, SALL4, OCT4, pSTAT3, and NANOG localized to the endothelium of the microvessels within the stroma.
Ethics Approval
This study was approved by Central Regional Health and Disability Ethics Committee (ref. no: 12/CEN/74).
Author Contributions
TI and STT formulated the study hypothesis. TI and STT designed the study. HHY, TF, HDB, and TI interpreted the IHC data. HHY, TF, HDB, and TI interpreted the CISH data. AMC processed the tissues for NanoString analysis and interpreted the data. HHY, TI, PFD, and STT drafted the manuscript. All authors commented on and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. TI, PFD, and STT are inventors of the PCT patent application (No. PCT/NZ2015/050108) Cancer Diagnosis and Therapy.
Acknowledgments
The authors thank Ms. Liz Jones of the Gillies McIndoe Research Institute for their assistance in IHC and CISH staining. HHY and TF were supported by summer scholarships from the Deane Endowment Trust. TI was supported, in part, by the Pacific Emerging Researcher First Grant, Health Research Council of New Zealand (grant no. HRC 16/434).
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fsurg.2016.00046
DAB IHC-stained sections of human infantile hemangioma for NANOG [(A), brown] and SALL4 [(B), brown], myometrium for OCT4 [(C), brown], skin for SOX2 [(D), brown], tonsil for pSTAT3 [(E), brown] and CD44 [(F), brown], and the omission of the primary antibody in a moderately differentiated MDBMSCC sample provided an appropriate negative control [(G), brown]. Cellular nuclei were counterstained with hematoxylin [(A–F), blue]. Original magnification: 400×.
Representative IF IHC-stained sections of MDBMSCC demonstrating nuclear expression of pSTAT3 [(A,C,M), red], EMA [(B), green], NANOG [(E), red], CD34 [(D,F,H), green], SOX2 [(G,I,K), red], SALL4 [(J), green], OCT4 [(L), green], and CD44 [(N), green]. Separated images of the individual stains shown in (A–N). Scale bars: 20 μm.
References
- 1.Arya S, Rane P, Deshmukh A. Oral cavity squamous cell carcinoma: role of pretreatment imaging and its influence on management. Clin Radiol (2014) 69:916–30. 10.1016/j.crad.2014.04.013 [DOI] [PubMed] [Google Scholar]
- 2.Scully C, Bagan J. Oral squamous cell carcinoma overview. Oral Oncol (2009) 45:301–8. 10.1016/j.oraloncology.2009.01.004 [DOI] [PubMed] [Google Scholar]
- 3.Osterkamp RW, Whitten JB. The etiology and pathogenesis of oral cancer. CA Cancer J Clin (1973) 23:28–32. 10.3322/canjclin.23.1.28 [DOI] [PubMed] [Google Scholar]
- 4.Funk GF, Karnell LH, Robinson RA, Zhen WK, Trask DK, Hoffman HT. Presentation, treatment, and outcome of oral cavity cancer: a National Cancer Data Base report. Head Neck (2002) 24:165–80. 10.1002/hed.10004 [DOI] [PubMed] [Google Scholar]
- 5.Zini A, Czerninski R, Sgan-Cohen HD. Oral cancer over four decades: epidemiology, trends, histology, and survival by anatomical sites. J Oral Pathol Med (2010) 39:299–305. 10.1111/j.1600-0714.2009.00845.x [DOI] [PubMed] [Google Scholar]
- 6.Lee K, Veness M, Pearl-Larson T, Morgan G. Role of combined modality treatment of buccal mucosa squamous cell carcinoma. Aust Dent J (2005) 50:108–13. 10.1111/j.1834-7819.2005.tb00349.x [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Huang HC, Lin LM, Lin CC. Primary oral squamous cell carcinoma: an analysis of 703 cases in southern Taiwan. Oral Oncol (1999) 35:173–9. 10.1016/S1368-8375(98)00101-8 [DOI] [PubMed] [Google Scholar]
- 8.Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, et al. The global burden of cancer 2013. JAMA Oncol (2015) 1:505–27. 10.1001/jamaoncol.2015.0735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang Q-G, Shi S, Li Z-N, Zhang X, Liua F-Y, Xu Z-F, et al. Squamous cell carcinoma of the buccal mucosa: analysis of clinical presentation, outcome and prognostic factors. Mol Clin Oncol (2013) 1:531–4. 10.3892/mco.2013.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin C-S, Jen Y-M, Cheng M-F, Lin Y-S, Su W-F, Hwang J-M, et al. Squamous cell carcinoma of the buccal mucosa: an aggressive cancer requiring multimodality treatment. Head Neck (2006) 28:150–7. 10.1002/hed.20303 [DOI] [PubMed] [Google Scholar]
- 11.NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®). Head and Neck Cancers – Head-and-Neck.pdf. (2015). Available from: http://oralcancerfoundation.org/treatment/pdf/head-and-neck.pdf
- 12.O’Connor ML, Xiang D, Shigdar S, Macdonald J, Li Y, Wang T, et al. Cancer stem cells: a contentious hypothesis now moving forward. Cancer Lett (2014) 344:180–7. 10.1016/j.canlet.2013.11.012 [DOI] [PubMed] [Google Scholar]
- 13.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell (2014) 14:275–91. 10.1016/j.stem.2014.02.006 [DOI] [PubMed] [Google Scholar]
- 14.Borah A, Raveendran S, Rochani A, Maekawa T, Kumar DS. Targeting self-renewal pathways in cancer stem cells: clinical implications for cancer therapy. Oncogenesis (2015) 4:e177. 10.1038/oncsis.2015.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiou S-H, Yu C-C, Huang C-Y, Lin S-C, Liu C-J, Tsai T-H, et al. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res (2008) 14:4085–95. 10.1158/1078-0432.CCR-07-4404 [DOI] [PubMed] [Google Scholar]
- 16.Huang C-F, Xu X-R, Wu T-F, Sun Z-J, Zhang W-F. Correlation of ALDH1, CD44, OCT4 and SOX2 in tongue squamous cell carcinoma and their association with disease progression and prognosis. J Oral Pathol Med (2014) 43:492–8. 10.1111/jop.12159 [DOI] [PubMed] [Google Scholar]
- 17.Miettinen M, Wang Z, McCue PA, Sarlomo-Rikala M, Rys J, Biernat W, et al. SALL4 expression in germ cell and non-germ cell tumors: a systematic immunohistochemical study of 3215 cases. Am J Surg Pathol (2014) 38:410–20. 10.1097/PAS.0000000000000116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fasanaro E, Staffieri C, Cappellesso R, Marino F, Ottaviano G, Val M, et al. Prognostic significance of serine-phosphorylated STAT3 expression in pT1-T2 oral tongue carcinoma. Clin Exp Otorhinolaryngol (2015) 8:275–80. 10.3342/ceo.2015.8.3.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hadjimichael C, Chanoumidou K, Papadopoulou N, Arampatzi P, Papamatheakis J, Kretsovali A. Common stemness regulators of embryonic and cancer stem cells. World J Stem Cells (2015) 7:1150–84. 10.4252/wjsc.v7.i9.1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang J, Tam W-L, Tong GQ, Wu Q, Chan H-Y, Soh B-S, et al. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol (2006) 8:1114–23. 10.1038/ncb1481 [DOI] [PubMed] [Google Scholar]
- 21.Suiqing C, Min Z, Lirong C. Overexpression of phosphorylated-STAT3 correlated with the invasion and metastasis of cutaneous squamous cell carcinoma. J Dermatol (2005) 32:354–60. 10.1111/j.1346-8138.2005.tb00906.x [DOI] [PubMed] [Google Scholar]
- 22.Sawant S, Gokulan R, Dongre H, Vaidya M, Chaukar D, Prabhash K, et al. Prognostic role of Oct4, CD44 and c-Myc in radio-chemo-resistant oral cancer patients and their tumourigenic potential in immunodeficient mice. Clin Oral Investig (2016) 20(1):43–56. 10.1007/s00784-015-1476-6 [DOI] [PubMed] [Google Scholar]
- 23.Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene (2006) 25:1696–708. 10.1038/sj.onc.1209327 [DOI] [PubMed] [Google Scholar]
- 24.Chinn SB, Darr OA, Peters RD, Prince ME. The role of head and neck squamous cell carcinoma cancer stem cells in tumorigenesis, metastasis, and treatment failure. Front Endocrinol (2012) 3:90. 10.3389/fendo.2012.00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A (2003) 100:3983–8. 10.1073/pnas.0530291100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res (2003) 63:5821–8. [PubMed] [Google Scholar]
- 27.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res (2007) 67:1030–7. 10.1158/0008-5472.CAN-06-2030 [DOI] [PubMed] [Google Scholar]
- 28.Tan EMS, Chudakova DA, Davis PF, Brasch HD, Itinteang T, Tan ST. Characterisation of subpopulations of myeloid cells in infantile haemangioma. J Clin Pathol (2015) 68:571–4. 10.1136/jclinpath-2014-202846 [DOI] [PubMed] [Google Scholar]
- 29.Itinteang T, Tan ST, Brasch HD, Steel R, Best HA, Vishvanath A, et al. Infantile haemangioma expresses embryonic stem cell markers. J Clin Pathol (2012) 65:394–8. 10.1136/jclinpath-2011-200462 [DOI] [PubMed] [Google Scholar]
- 30.Xu D, O TM, Shartava A, Fowles TC, Yang J, Fink LM, et al. Isolation, characterization, and in vitro propagation of infantile hemangioma stem cells and an in vivo mouse model. J Hematol Oncol (2011) 4:54. 10.1186/1756-8722-4-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ono M, Kajitani T, Uchida H, Arase T, Oda H, Nishikawa-Uchida S, et al. OCT4 expression in human uterine myometrial stem/progenitor cells. Hum Reprod (2010) 25:2059–67. 10.1093/humrep/deq163 [DOI] [PubMed] [Google Scholar]
- 32.Laga AC, Lai C-Y, Zhan Q, Huang SJ, Velazquez EF, Yang Q, et al. Expression of the embryonic stem cell transcription factor SOX2 in human skin: relevance to melanocyte and merkel cell biology. Am J Pathol (2010) 176:903–13. 10.2353/ajpath.2010.090495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skinnider BF, Elia AJ, Gascoyne RD, Patterson B, Trumper L, Kapp U, et al. Signal transducer and activator of transcription 6 is frequently activated in Hodgkin and Reed-Sternberg cells of Hodgkin lymphoma. Blood (2002) 99:618–26. 10.1182/blood.V99.2.618 [DOI] [PubMed] [Google Scholar]
- 34.Lee YS, Lee J-E, Park H-Y, Lim Y-S, Lee J-C, Wang S-G, et al. Isolation of mesenchymal stromal cells (MSCs) from human adenoid tissue. Cell Physiol Biochem (2013) 31:513–24. 10.1159/000350072 [DOI] [PubMed] [Google Scholar]
- 35.Tan EMS, Itinteang T, Chudakova DA, Dunne JC, Marsh R, Brasch HD, et al. Characterisation of lymphocyte subpopulations in infantile haemangioma. J Clin Pathol (2015) 68:812–8. 10.1136/jclinpath-2015-203073 [DOI] [PubMed] [Google Scholar]
- 36.Baillie R, Itinteang T, Yu HH, Brasch HD, Davis PF, Tan ST. Cancer stem cells in moderately differentiated oral tongue squamous cell carcinoma. J Clin Pathol (2016) 69:742–4. 10.1136/jclinpath-2015-203599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jouppila-Mättö A. Epithelial-mesenchymal transition and expression of its transcription factors in pharyngeal squamous cell carcinoma. Publication of the University of Eastern Finland Dissertations in Health Sciences. Koupio: University of Eastern Finland; (2014). Available from: http://epublications.uef.fi/pub/urn_isbn_978-952-61-1624-2/urn_isbn_978-952-61-1624-2.pdf [Google Scholar]
- 38.Gu T-T, Liu S-Y, Zheng P-S. Cytoplasmic NANOG-positive stromal cells promote human cervical cancer progression. Am J Pathol (2012) 181:652–61. 10.1016/j.ajpath.2012.04.008 [DOI] [PubMed] [Google Scholar]
- 39.Yue X, Xiao L, Yang Y, Liu W, Zhang K, Shi G, et al. High cytoplasmic expression of SALL4 predicts a malignant phenotype and poor prognosis of breast invasive ductal carcinoma. Neoplasma (2015) 62:980–8. 10.4149/neo_2015_119 [DOI] [PubMed] [Google Scholar]
- 40.Kotiyal S, Bhattacharya S. Epithelial mesenchymal transition and vascular mimicry in breast cancer stem cells. Crit Rev Eukaryot Gene Expr (2015) 25:269–80. 10.1615/CritRevEukaryotGeneExpr.2015014042 [DOI] [PubMed] [Google Scholar]
- 41.Fan Y-L, Zheng M, Tang Y-L, Liang X-H. A new perspective of vasculogenic mimicry: EMT and cancer stem cells (review). Oncol Lett (2013) 6:1174–80. 10.3892/ol.2013.1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X, Fan D. The epithelial-mesenchymal transition and cancer stem cells: functional and mechanistic links. Curr Pharm Des (2015) 21:1279–91. 10.2174/1381612821666141211115611 [DOI] [PubMed] [Google Scholar]
- 43.Jouppila-Mättö A, Närkiö-Mäkelä M, Soini Y, Pukkila M, Sironen R, Tuhkanen H, et al. Twist and snai1 expression in pharyngeal squamous cell carcinoma stroma is related to cancer progression. BMC Cancer (2011) 11:350. 10.1186/1471-2407-11-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge N, Lin H-X, Xiao X-S, Guo L, Xu H-M, Wang X, et al. Prognostic significance of Oct4 and Sox2 expression in hypopharyngeal squamous cell carcinoma. J Transl Med (2010) 8:94. 10.1186/1479-5876-8-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DAB IHC-stained sections of human infantile hemangioma for NANOG [(A), brown] and SALL4 [(B), brown], myometrium for OCT4 [(C), brown], skin for SOX2 [(D), brown], tonsil for pSTAT3 [(E), brown] and CD44 [(F), brown], and the omission of the primary antibody in a moderately differentiated MDBMSCC sample provided an appropriate negative control [(G), brown]. Cellular nuclei were counterstained with hematoxylin [(A–F), blue]. Original magnification: 400×.
Representative IF IHC-stained sections of MDBMSCC demonstrating nuclear expression of pSTAT3 [(A,C,M), red], EMA [(B), green], NANOG [(E), red], CD34 [(D,F,H), green], SOX2 [(G,I,K), red], SALL4 [(J), green], OCT4 [(L), green], and CD44 [(N), green]. Separated images of the individual stains shown in (A–N). Scale bars: 20 μm.