Abstract
It has been suggested that the alkaline form of cytochrome c (cyt c) regulates function of this protein as an electron carrier in oxidative phosphorylation and as a peroxidase that reacts with cardiolipin (CL) during apoptosis. In this form, Met80, the native ligand to the heme iron, is replaced by a Lys. While it has become clear that the structure of cyt c changes, the extent and sequence of conformational rearrangements associated with this ligand replacement remain a subject of debate. Herein we report a high-resolution crystal structure of a Lys73-ligated cyt c conformation that reveals intricate change in the heme environment upon this switch in the heme iron ligation. The structure is surprisingly compact, and the heme coordination loop refolds into a β-hairpin with a turn formed by the highly conserved residues Pro76 and Gly77. Repositioning of residue 78 modifies the intraprotein hydrogen-bonding network and, together with adjustments of residues 52 and 74, increases the volume of the heme pocket to allow for insertion of one of the CL acyl moieties next to Asn52. Derivatization of Cys78 with maleimide creates a solution mimic of the Lys-ligated cyt c that has enhanced peroxidase activity, adding support for a role of the Lys-ligated cyt c in the apoptotic mechanism. Experiments with the heme peptide microperoxidase-8 and engineered model proteins provide a thermodynamic rationale for the switch to Lys ligation upon perturbations in the protein scaffold.
Graphical abstract
Introduction
Conformational switching between distinct protein structures is an important mechanism for biological information transfer,1,2 control of enzymatic activity,3,4 and pathological aggregation.5,6 Furthermore, switchable protein systems are attractive design models for molecular electronics and artificial photosynthesis. However, the structural details of alternatively folded forms are often elusive, as are the mechanisms of the switching process.
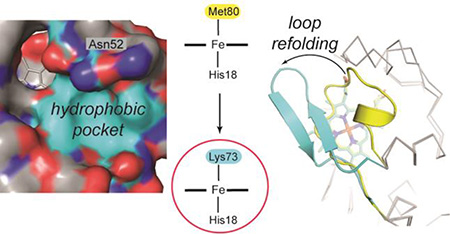
The small heme protein cytochrome c (cyt c) has been a powerful model system for examining factors that control protein conformational dynamics.7 Several conformational forms of cyt c, some with functional implications in oxidative phosphorylation and apoptosis, have been identified by spectroscopic studies.8–10 Among them, the alkaline state of ferric cyt c, which is typically populated at high solution pH, has been a subject of considerable attention.11–17 In this state, the native heme iron ligand Met80 is replaced by either Lys73 or Lys79 within the heme coordination loop (residues 70–85).15 The change in ligation lowers the heme reduction potential, affecting rates of electron transfer (ET) and possibly controlling unidirectionality of physiological ET between cyt c and cyt c oxidase.15,18,19 A Lys-ligated species has also been suggested to form upon cyt c interactions with cardiolipin (CL) membranes.20 These interactions enable cyt c to act as a peroxidase in early stages of apoptosis.21 Experimental as well as theoretical studies have illustrated similarities of the alkaline form to late folding intermediates, suggesting that this species may also model key features of the protein folding energy landscape.9,22
The heme moiety is essential for the structural integrity of cyt c.23,24 The extensive hydrogen-bonding network involving the heme group connects regions of different folding stability.25,26 This network also mediates cross-talk between distant sites in cyt c and contributes to cooperative stabilization of the protein.27,28 Changes in this network may provide the trigger mechanism for the alkaline transition. Indeed, a recent structure of a hydroxide-ligated cyt c has revealed an alteration in this network when Met80 ligation is lost.29 However, despite previous efforts to characterize alkaline cyt c structurally,30 key details of the heme environment in Lys-ligated cyt c have been lacking.
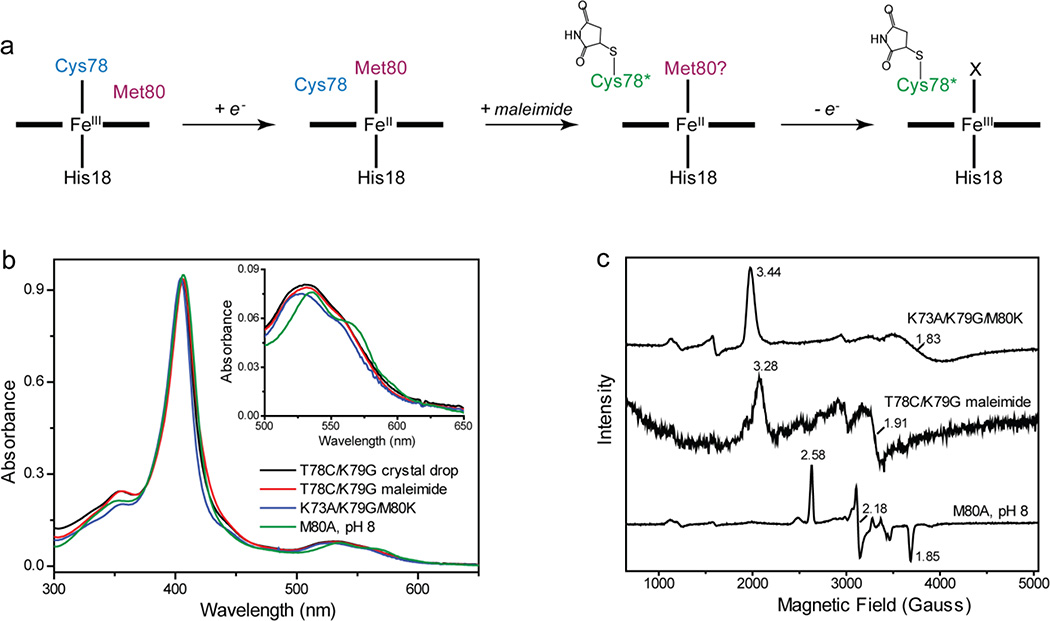
The highly-conserved residue Thr788,31 is a critical part of the intraprotein hydrogen-bonding network in cyt c. We recently showed that in T78C/K79G/M80X (X=L, I, and F) mutants of yeast iso-1 cyt c, the Cys78 thiolate coordinates to the ferric heme iron.32 In order to explore the ability of the protein to sample conformations with distinct ligands to the heme iron, we have now created a new T78C/K79G mutant, which allows for Cys78 coordination while retaining Met80, the ligand in the wild-type protein. Our studies reveal that T78C/K79G acts as a redox-controlled switch: while the ferrous heme iron is Met80-ligated, the ferric heme iron is ligated by Cys78 (major form) or by Lys73 in a Cys78-linked dimer (minor form).
In this report, we describe a high-resolution crystal structure of the Lys73-ligated T78C/K79G. The structure is surprisingly compact and reveals refolding of the coordination loop into a tight β-hairpin. Even though the heme is enclosed, the change in ligation affects the volume of the heme pocket and protein peroxidase activity. With a series of model studies we provide the thermodynamic rationale for Lys ligation to the heme iron when potential ligands Cys and Met are also available. β-sheet formation33 and Lys ligation20 have recently been implicated in CL-mediated switching of cyt c into an apoptotic peroxidase;a our structure illuminates these transformations at atomic resolution.
Results
T78C/K79G Characterization in Solution
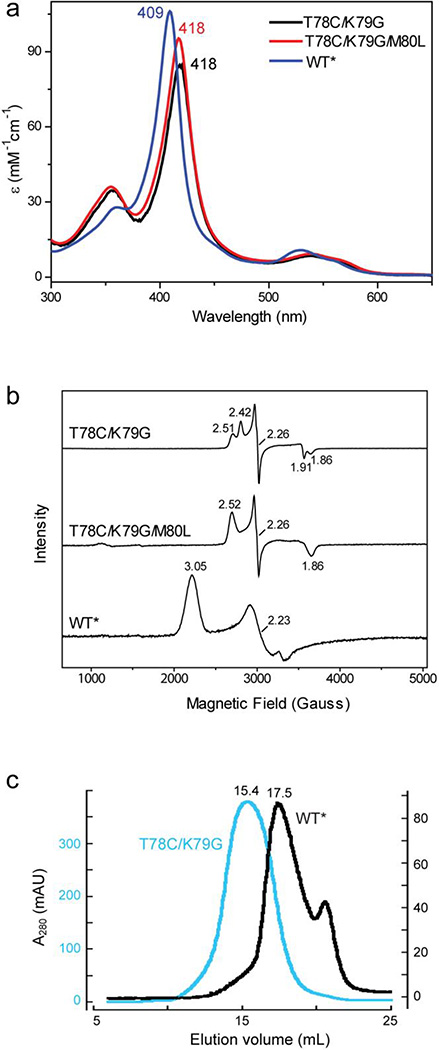
Under conditions that stabilize the ferric state, the electronic absorption (Figures 1a and S1a) and EPR (Figure 1b) spectra of T78C/K79G are similar to those of ferric T78C/K79G/M80X (X=L, I, F).32 The ~10 nm red shift in the Soret and Q-bands compared to those of WT*b in the electronic absorption spectrum as well as the characteristic narrow spread of g values in the EPR spectra support thiolate ligation in the mutant protein. The positions of the heme absorption bands in T78C/K79G are distinct from those of the hydroxide-ligated M80A32,34 arguing in favor of thiolate rather than hydroxide ligation to the heme iron. Furthermore, the two absorption bands at 634 and 738 nm (Figure S1a) are similar to the thiolate-to-Fe(III) charge-transfer bands of the imidazole adduct of P45035 and such bands in other thiolate-ligated variants of cyt c.32,34 As in many other thiolate-ligated heme proteins,36–38 two overlapping EPR signals (gz=2.42 and gz=2.51, Table S1) are observed for T78C/K79G. Thermal melting curves and pH titrations of the ferric protein (Figures S2 and S3 and Tables S2–S4) reveal that Cys78 ligation in the mutant renders the ferric protein more resistant toward heat and acid denaturation than the native Met80 ligation in WT* or K79G. It is noteworthy that the K79G mutation, employed to favor Cys78 coordination and avoid Lys79 coordination to the heme iron in T78C mutant proteins, minimally affects the stability of cyt c (Table S4). While the low-temperature EPR spectra indicate a low-spin state for the ferric heme in T78C/K79G, no hyperfine-shifted heme resonances characteristic of low-spin or high-spin species are observed between 5 and 45 °C by 1H NMR suggesting an intermediate (ms timescale) exchange between distinct heme species in this temperature regime (Figure S4). The downfield-shifted resonance at 22.5 ppm in the T78C/K79G spectrum likely arises from ε1(C-H) His18; if so, the increased line width of this signal compared to that in WT* is also consistent with the proposed dynamics.
Figure 1.
(a) UV-visible and (b) EPR spectra of ferric T78C/K79G, T78C/K79G/M80L and WT* cyt c in a 100 mM sodium phosphate buffer at pH 7.4. (c) Size-exclusion chromatograms of T78C/K79G (cyan) and WT*(black) cyt c in a 10 mM sodium phosphate buffer at pH 7.4.
Both the ferric WT* and K79G exhibit well-defined shifts in the position of their Soret bands upon the transition to the alkaline form with pKa values of 8.8±0.132 and 8.6±0.1, respectively (Table S3). In contrast, the position of the Soret band in ferric T78C/K79G does not readily change at alkaline pH, suggesting that Cys78 thiolate remains bound to the ferric heme iron at pH values as high as 10.8 (Figure S3b). However, the intensity of this band grows yielding a transition with a pKa of 9.7±0.3 (Table S3). This behavior is similar to that of T78C/K79G/M80X (X=L, I, F) variants and reflects a change in protonation of another group (residue) close to the heme.32 As in the case of WT*, interactions of T78C/K79G with CL-containing liposomes modify the heme iron coordination: Cys78 is replaced by another strong-field ligand (Figure S1b).
Under conditions that stabilize the ferrous state, the electronic absorption spectrum of the T78C/K79G is very similar to that of ferrous WT*, with the Soret band at 415 nm and ε415=129 mM−1cm−1 (Figure S3c), which is typical of a low-spin ferrous heme species with a neutral ligand. The hysteresis in the spectroelectrochemicalc results (Table 1 and Figure S1c, black curves) suggests that redox-dependent ligand switching occurs in T78C/K79G. The positive values of the heme-iron midpoint potentials are consistent with the soft-base properties of thioether Met that favor the ferrous heme iron. Results of thermal denaturation and pH titrations of ferrous T78C/K79G and WT* are very similar (Figures S2 and S3 and Tables S2 and S3), further implicating Met80 as the ligand to the heme iron in the ferrous mutant protein. Finally, the 1H NMR spectra of T78C/K79G show upfield-shifted Met80 resonances39 at positions almost the same as those in WT* (Figure S1d). We conclude that for T78C/K79G in solution, the ferric heme iron is predominantly Cys78-ligated, while the ferrous heme iron is Met80-ligated.
Table 1.
Reduction Potentials and Proposed Ligand Exchange Processes for Studied Variants of Yeast iso-1 Cyt c at pH 7.4a,b
| Variant | Em (mV vs SHE) | Proposed Redox Reactions and Ligand Exchange Processes |
|---|---|---|
| T78C/K79G solution | 22 ± 7 (82 ± 3)c |
CysFe(III)→MetFe(III)↔MetFe(II) (MetFe(II)↔MetFe(III)→CysFe(III))c |
| T78C/K79G/M80Ld | −340 | CysFe(III)↔CysFe(II) |
| K73A/K79G/M80K | −94 ± 2 | LysFe(III)↔LysFe(II) |
| K79G | 266 ± 2 | MetFe(III)↔MetFe(II) |
| WT* | 247 ± 4 | MetFe(III)↔MetFe(II) |
| M80A | −80 ± 5 | (OH−)Fe(III)↔Fe(II) |
| T78C/K79G-maleimide | 63 ± 4 (51 ± 5)c |
LysFe(III)→MetFe(III)↔MetFe(II) (MetFe(II)↔MetFe(III)→LysFe(III))c |
| T78C/K79G crystal drope | 83; −160 | CysFe(III)→MetFe(III)↔Met(FeII); (OH−)Fe(III)↔Fe(II) and/or LysFe(III)↔LysFe(II) |
All variants contained two background mutations K72A and C102S, to prevent Lys72 coordination to the heme iron and formation of Cys102-linked dimers, respectively.
The observed potentials are strongly affected when there is coupling to a ligand exchange process.
Reductive (oxidative) direction.
From ref. 32.
A mixture of heme-ligated species, from direct electrochemistry measurements (Figure S11).
T78C/K79G Crystallization and Structure Determination
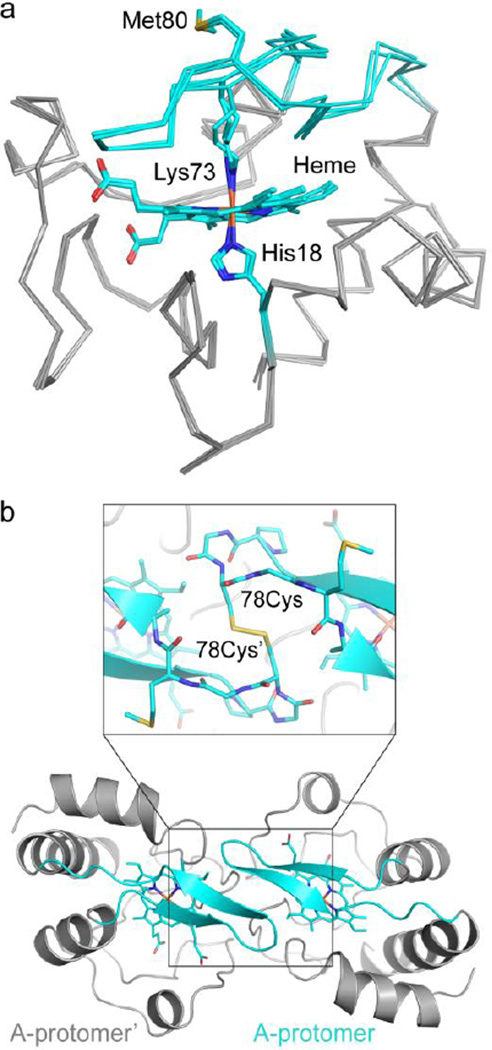
Ferric T78C/K79G crystallized in space group I422 within one week under vapor-diffusion conditions. Although the crystals diffract to high resolution (IσI=4.3 at 1.9 Å; Table S5), structure determination was complicated by pseudo-merohedral twinning with roughly equal twin fractions (Table S5). Nevertheless, following molecular replacement and initial refinement, electron density maps clearly show the position of the heme group, which had not been included in the model (Figure S5a). The refined model contains three highly similar protomers in the asymmetric unit (Figures 2a and S6b), exhibits excellent geometry, and has Rwork and Rfree values of 0.17 and 0.20, respectively (Table S5). Overall, the T78C/K79G structure is similar to other structures of cyt c: the top 100 hits in a distance-alignment search40 are all cyt c proteins.
Figure 2.
The T78C/K79G crystal structure reveals Lys73-coordination to the heme iron. (a) Alignment of the three molecules in the asymmetric unit of T78C/K79G cyt c (gray Cα trace, with cyan coordination loop) by their main chain atoms reveals a similar overall structure, RMSD < 0.3 Å. In all three molecules, the heme iron is coordinated by His18 and Lys73. Met80 is solvent-exposed, as labeled. Heme, His18, Lys73, and Met80 are shown as stick figures, with cyan carbons and non-carbon atoms colored by element: N=blue, O=red, S=yellow, Fe=orange. (b) Each protomer of the asymmetric unit forms a dimer with a molecule related by a two-fold symmetry axis. Here, chain A is shown with respect to its symmetry mate, chain A’ (both in gray cartoon, with coordination loop colored cyan), and the covalent dimer interaction is highlighted. Inset: the Cys78 residue forms a disulfide bond with Cys78’ (proteins in cartoon and stick representation). Figure S5c shows non-covalent interactions at the dimer interface.
While spectroscopic results provide clear evidence for Cys78 ligation to the ferric heme in solution, the heme iron in all three protomers in the crystallographic structure is coordinated by Lys73 (Figure 2a), a ligation also observed in the alkaline form of cyt c. Fe-N(Lys73) bond distances and angles of the Lys/His-coordinated heme iron in T78C/K79G are similar to those found in other Lys-ligated proteins proposed41,42 (Figure S6) or confirmed30 to be in their ferric state. Although photoreduction during synchrotron data collection is less likely for low potential proteins, we nevertheless tested whether it had occurred in this case. We separately processed the first 150 and final 150 oscillation images, and computed difference electron-density maps. No shift in the Fe-N bond length was observed. Independent model fits against each partial data set also yielded identical coordination geometries, strongly suggesting that the original ferric state was preserved. Importantly, the large structural rearrangements associated with Cys78 replacement by Lys73 at the heme iron could not have been caused by heme iron photoreduction in the vitrified crystal, and in the following analysis we focus on the mechanism of this transformation in solution before and during crystallization.
An essential feature of the crystal structure is that Cys78, the ligand to the heme iron of T78C/K79G in solution, instead forms a disulfide bond with its symmetry mate Cys78′ on an adjacent molecule (Figure 2b) in the crystal lattice, stabilizing an interface that also involves several non-covalent interactions between the two monomers (Figure S5c). The K79G substitution, although introduced to avoid alternative Lys coordination, actually appears critical for formation of the observed lattice: a Cβ atom at position 79 would collide with an adjacent dimer in the lattice (Figure S5d). Met80, the ligand to the heme iron of WT*, is solvent exposed.
The observation of Lys-, rather than Cys-ligated protein in the crystal was surprising, but repeated analysis of ferric T78C/K79G samples by nonreducing SDS-PAGE gel electrophoresis uncovered the presence of Cys-linked dimers (<5%) in solution (Figure S7c). Although the electronic absorption and EPR spectra of ferric cyt c differ between the Cys- and Lys-ligated species, simulations with the spectra of the Cys-ligated T78C/K79G/M80L and Lys-ligated model protein K73A/K79G/M80K (see below) suggested that it is difficult to detect this level of heterogeneity using these spectroscopic techniques. However, size-exclusion chromatography (SEC) experiments have uncovered an interesting difference between ferric WT* and T78C/K79G proteins. Each variant shows a dominant elution peak, but despite similar predicted protomer molar masses (~12.5 kDa), the mutant elutes more rapidly than WT* (Figure 1c), emerging before rather than after the RNaseA standard (13.7 kDa, Figures S7a and S7b). These hydrodynamic behaviors are consistent with the presence of alternative conformations and/or oligomeric states of T78C/K79G.
Spectroscopic and Thermodynamic Signatures of Lys Ligation to the Heme Iron
Given the unexpected observation of Lys ligation in the T78C/K79G structure, we investigated the ability of different side-chain ligands to compete as ligands for the heme iron. Studies with the model peptide N-acetyl microperoxidase-8 (AcMP8) permit direct comparison of binding affinities of Cys, Met, and Lys for the heme.32,43 Binding of N-acetyllysine (AcLys) to ferric AcMP8 resulted in clear changes in the heme absorption spectra, most notably a red shift in the Soret band, coupled with a decrease in the intensity of this band (Figure S8). Similar trends have been reported for binding of the terminal amine of the Ala amino acid to AcMP8.43 The titrations shown in Figure S8 yielded an apparent K value (Kapp) of 2.8×102 at pH 9.0. After accounting for the pKa of AcLys (10.2), the association constant Ka for the deprotonated AcLys amine to ferric AcMP8 is 4.7×103, very similar to that of the thiolate AcCys32 and almost three orders of magnitude greater than that of AcMet43 (Table 2). These results suggest that anionic Cys- binding to the heme iron is strongly favored under neutral pH conditions, but that Lys can become a competitive alternative ligand if its deprotonation is assisted by a base.
Table 2.
Binding (Association) Constants (Ka) and Changes in Free Energies (ΔG) for Binding of Amino-Acid Ligands to Ferric AcMP8
The spectroscopic and thermodynamic properties of Lys-heme interaction were also probed with a newly created variant K73A/K79G/M80K. This mutant was designed to replace the WT* ligand Met80 with a Lys and eliminate other potential Lys ligands. The absorption (with the Soret band at 405 nm, Figures 3a and S9) and EPR (distinct from those of the hydroxide-ligated M80A, Table S1) spectra of this variant in the ferric state are in accord with those of cyt c550 M100K and the alkaline form of cyt c,44,45 suggesting that our design succeeded in achieving Lys coordination. The low redox potential of −94±2 mV at pH 7.4 (Table 1) is also consistent with such ligation. The absorption spectrum of K73A/K79G/M80K cyt c changes as a function of pH (Figure S10). The pH dependence of the spectra reveals two transitions at acidic pH and one at alkaline pH, reflecting changes in the heme environment of the ferric protein; their assignments are in Table S3. Results of these pH experiments suggest that the cyt c scaffold allows for Lys coordination to the heme iron within a broad pH range.
Figure 3.
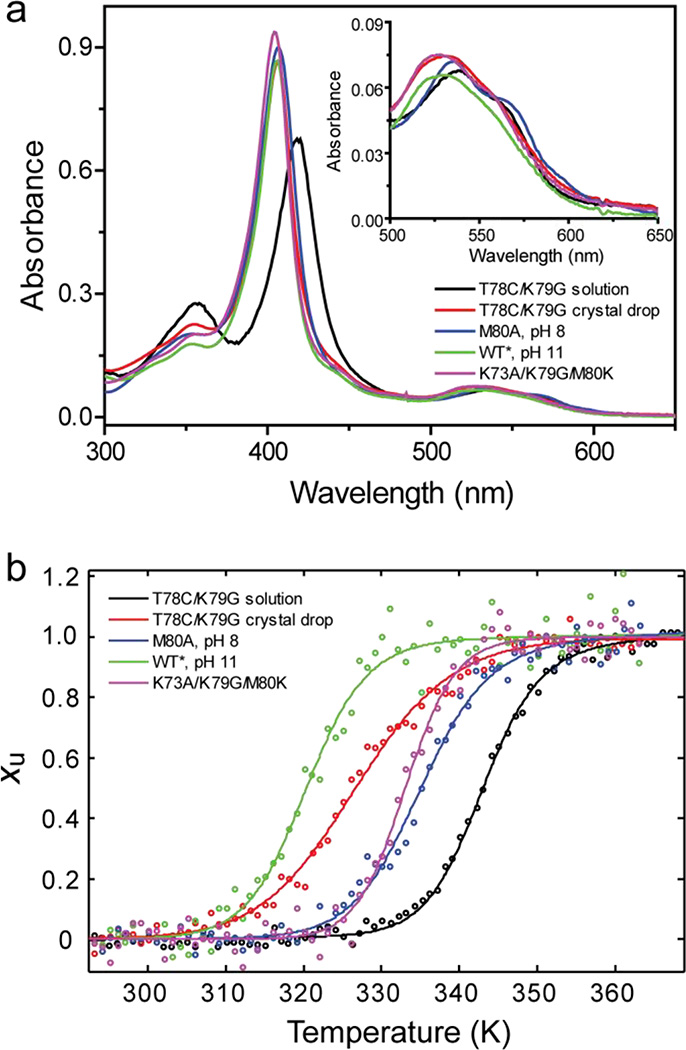
(a) UV-visible spectra and (b) thermal unfolding curves (fraction of unfolded protein Xu versus temperature) from CD measurements are shown for ferric T78C/K79G in solution (black) and recovered from crystal drops (red) as well as K73A/K79G/M80K (magenta) at pH 7.4, M80A at pH 8 (blue), WT* at pH 11 (green). The inset in (a) illustrates different features of Q-bands for the five samples.
Thermodynamic parameters from thermal denaturation experiments with K73A/K79G/M80K (Figure 3b) are listed in Tables S2 and S4. The Tm value of ferric K73A/K79G/M80K is similar to that of K79G. When comparing variants with Cys, Met, and Lys ligation to the ferric heme iron, their ΔGD values are within only 2.4 kcal/mol of each other (Table S4). However, the enthalpic and entropic contributions to stability are different. For the Lys80-ligated K73A/K79G/M80K, as well as the Cys78-ligated T78C/K79G and T78C/K79G/M80L proteins, the enthalpic term ΔHD is more favorable compared to that for WT* (likely owing to the change in intramolecular contacts upon these mutations) but this is compensated by a less favorable entropic (−TΔSD) term. We have previously hypothesized that changes in polypeptide packing and solvation account for the increase in the entropy of the native state in engineered cyt c mutants with nonnative ligands at the heme iron.32 The results of this thermodynamic analysis demonstrate that while Cys-ligation to the heme iron is favored in the ferric protein, the difference in stability of Cys- and Lys-ligated species is modest.
Heme Iron Ligation of the Protein from the Crystal Drop and Effects of Cys78 Displacement
With spectroscopic and thermodynamic signatures of Lys ligation in hand, we sought to characterize the protein in the mother liquor used for crystallization. The absorption spectrum of the protein recovered from crystallization experiments (Figure 3a) no longer shows the features of predominant Cys ligation. Instead, the position of the Soret band at 406 nm is consistent with the main species having a Lys and/or a hydroxide as the heme iron ligand. The broad contour of the Soret band and a δ band at 355 nm suggest that Cys-ligated protein is still present but has become a minor species.
The protein amounts recovered from crystal drops were not sufficient for EPR spectroscopy but direct voltammetry provided additional support for the heterogeneity of protein solution, as well as a loss of the Cys ligation for much of the protein. At slow scan rates, the cyclic voltammetry (CV) of the protein from the crystal drop is dominated by a reductive wave at low potential (Figure S11a). Increasing the scan rate to 10 V/s allows the measurement to outpace the catalytic process responsible for the reductive wave and reveals a reversible couple with an average potential of −180 mV (Figure S11b). The technique of square wave voltammetry (SWV)46,47 has the advantage of subtracting out most of the background electrochemical processes and resolves two peaks: a small one at +83 mV and a large one at −160 mV (Figure S11c). The small +83 mV signal is difficult to assign conclusively. Our best supposition is that it stems from methionine binding combined with structural changes that increase heme accessibility. The large −160 mV (pH 7.4) signal is in between the value of −205 mV (pH 8.45)48 previously reported for alkaline cyt c and −94±2 mV (pH 7.4) for K73A/K79G/M80K (Table 1). This value is thus in accord with Lys ligation. However, the observed voltammetry does not exclude hydroxide as a possible ligand to the heme iron, a ligation that could result from opening of the heme pocket. Adding support to this scenario, the persistence of the catalytic oxygen reduction process even at <10 ppm O2 suggests that the heme is solvent-exposed and readily available for oxygen binding. The observed potential and the catalytic activity of the heme group are consistent with opening of the heme pocket and likely arise from protein destabilization. The greater susceptibility to thermal denaturation of T78C/K79G from crystallization experiments (ΔTm≈ − 16 K; Figure 3 and Table S2) is also consistent with this interpretation.
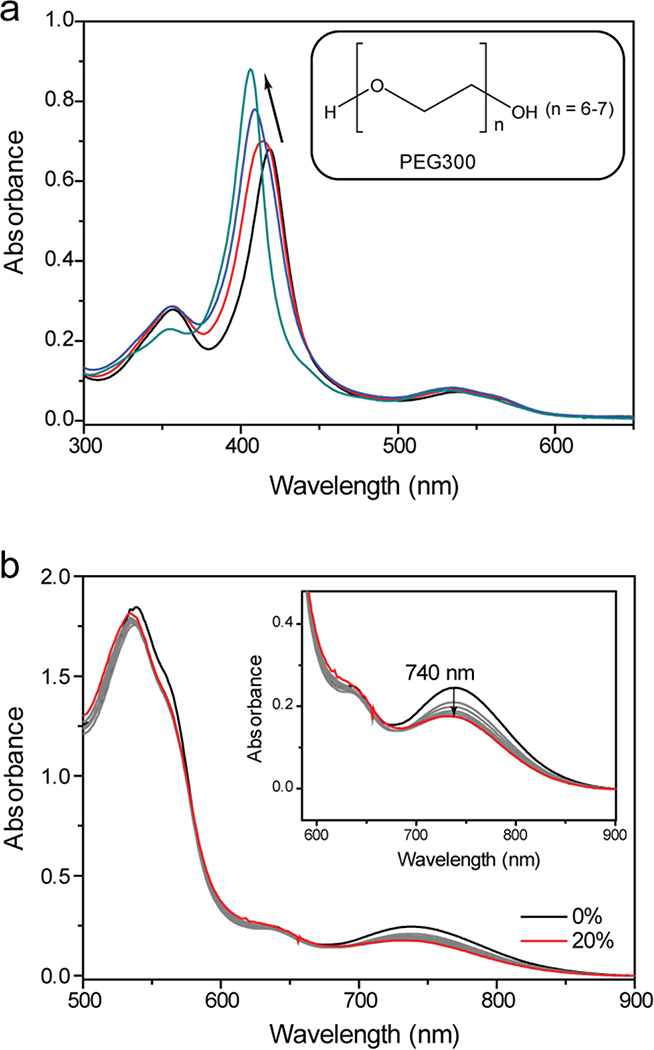
Recent studies have employed polyethylene glycol (PEG) as a precipitant for cyt c crystallization.49–55 Interestingly, several of these previously determined structures revealed the loss of the native Met80 ligand from the heme.49–51 Our findings of the loss of Cys78 ligation in T78C/K79G and hints of protein destabilization prompted us to investigate whether PEG could be the culprit. Addition of PEG300 to T78C/K79G resulted in a blue shift of the Soret band and loss of the charge transfer bands associated with thiolate ligation to the ferric heme iron (Figures 4 and S12). We have also observed similar loss of the 695 nm charge transfer band associated with Met80 ligation with WT* exposed to PEG300. Removal of PEG300 by dialysis only partially restored Cys ligation (Figure S12b), most likely owing to oxidation of Cys78 in the PEG-denatured protein; addition of DTT shifted the Soret band back to 417 nm, a position similar to that in T78C/K79G not treated with PEG. Dissociation of Cys78 from the heme iron upon exposure to PEG could have favored T78C/K79G dimerization during the crystallization experiments, and we find a corresponding increase in the population of the Cys-linked dimers when the protein is incubated with PEG (Figure S7c).
Figure 4.
(a) Changes in the UV-visible spectra of T78C/K79G during crystal growth: shown are spectra (upon dilution, see Materials and Methods) at the start of crystallization (black), the next day (red), after one week (blue), and after two weeks, when crystals became clearly visible (teal). Inset: Structure of polyethylene glycol 300 (PEG300). (b) Visible and near-IR (inset) spectra of T78C/K79G at different concentrations of PEG300 (from 0 to 20% v/v) added to a 100 mM sodium phosphate buffer at pH 7.4.
We wondered whether a simple repositioning of the residue 78 in the heme pocket, e.g., through dimer formation or by other effectors (see below), could trigger Lys coordination to the heme iron. Covalent modification of Cys78 with a small maleimide tag prevented thiolate ligation to the heme iron for T78C/K79G in solution. Instead, spectra of this derivative revealed clear features of Lys ligation, particularly in EPR experiments (Figure 5). The maleimide-modified variant is thermodynamically stable (Table S2), and its positive reduction potential (Table 1), very different from that of the Lys-ligated K73A/K79G/M80K, suggests that the Lys ligand in this derivative is likely replaced by Met upon reduction of the heme iron. The potentials for maleimide-modified T78C/K79G (Lys-bound) and unmodified T78C/K79G in solution (predominantly Cys-bound) were very similar, which likely precluded measurement of a distinct potential for the minor Lys-ligated component in unmodified T78C/K79G. Since the Cys78-modified maleimide derivative and the Cys78-linked dimer both support Lys coordination to the heme iron, they may well have similar conformations of the heme coordination loop.
Figure 5.
(a) Maleimide labeling of T78C/K79G. (b) UV-visible and (c) EPR spectra of ferric T78C/K79G recovered from the crystal drop, maleimide-labeled T78C/K79G, K73A/K79G/M80K at pH 7.4 and M80A at pH 8.0. For T78C/K79G recovered from the crystal drop, the sample amounts were not sufficient to acquire EPR spectra. The inset in (b) illustrates different features of Q-bands for the four samples.
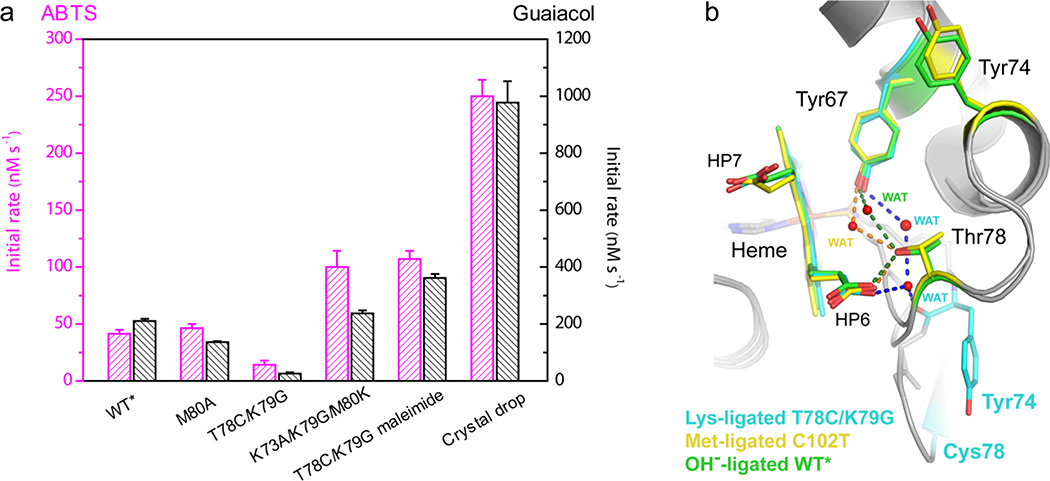
We have also analyzed the intrinsic peroxidase activity of the cyt c variants studied here. Assays with two different substrates have revealed that maleimide-modified T78C/K79G, the solution analogue of our structure, has a higher activity than either the WT* or M80A proteins (Figure 6a). Interestingly, its activity is similar to that of another Lys-ligated variant we prepared, K73A/K79G/M80K. The highest activity we observed is for the protein solution from the crystal drop. This finding is consistent with the above interpretation of the increased exposure of the heme for the major species in this protein mixture owing to PEG-induced protein destabilization and subsequent oxidation of Cys78.
Figure 6.
(a) Rate constants of ABTS (magenta) or guaiacol (black) oxidation by H2O2 catalyzed by different variants of cyt c in a 25 mM HEPES buffer at pH 7.4. The concentrations of protein, guaiacol, and H2O2 were 1 µM, 10 mM, and 5 mM, respectively. The concentrations of protein, ABTS, and H2O2 were 1 µM, 200 µM, and 1 mM, respectively. (b) The hydrogen-bonding networks that involve the heme propionate HP6 in the Lys73-ligated T78C/K79G (cyan carbons), Met80-ligated C102T25 (yellow carbons) and hydroxide-ligated WT*29 (green carbons) structures.
Discussion
Ligand Promiscuity and Protein Fold
Ligation at the heme iron plays an important role in folding and determining the functional state of cyt c. Multiple studies have shown that affinity of Met for the ferric heme iron is low and that the protein scaffold strongly influences Met80 coordination in the protein.32,43,56–58 For example, M80A mutation minimally affects the thermodynamic stability of the ferric protein (Table S2), and small perturbations in the protein fold for WT* are sufficient to yield a structure with Met80 still in the protein sequence, but dissociated from the heme.29 Providing further evidence for to the interplay between Met80-heme interactions and cyt c stability, alterations in the protein hydrogen-bonding network in Y67F increase protein stability, as well as the propensity of Met80 to remain coordinated to the heme with change in solution pH.59,60 Several spectroscopic experiments have demonstrated the switch to a Lys-ligated ‘alkaline’ state at high pH,9,15,30 and our latest mutational study has shown that a T78C mutation yields the Cys78-ligated ferric protein,32 which is more stable than WT*. Higher iron-binding affinities of anionic Cys and neutral Lys (Table 2) provide a thermodynamic rationale for Met replacement upon alteration of intramolecular interactions in the heme coordination loop of cyt c. The pliable nature of this low-stability loop (this region is the second least stable ‘foldon’ in cyt c)61 assists in achieving the nonnative ligation. Dissociation of Met80 in WT* opens up the heme pocket and introduces additional water molecules into the protein interior.29 Ligation by Cys and Lys aids in repatching of the heme pocket, consistent with our findings of increased entropic stabilization of Cys- and Lys-ligated variants.
We originally engineered the T78C/K79G variant to explore switching between Cys78- and Met80-ligated states of the protein. The choice of the ligand depends on the heme-iron oxidation state and solution conditions. In solution, Cys78 and Met80 are the preferred ligands to the heme iron in the ferric and ferrous proteins, respectively. Possibly related to the switchable nature of T78C/K79G, the exchange between distinct heme species is evident in the NMR spectra of the ferric protein, where it abolishes the 1H signals of the heme group. A similar observation of the dynamic equilibrium of spin states responsible for the disappearance of the heme signals in the 1H NMR spectra has recently been made with another heme protein HasAp.62 While the identity of the exchanging species in ferric T78C/K79G is currently unknown, this interesting NMR finding illustrates the dynamic nature of the heme group in the variant. Dissociation of Cys78 from the heme iron and subsequent thiol oxidation during protein crystallization led us to the high-resolution structure of the dimeric Lys73-ligated component in the mixture of ferric T78C/K79G (Figure 2). Thus, the engineered T78C/K79G variant of cyt c is actually able to accommodate three different ligands to the heme iron: Lys73, Cys78, and Met80, all within a compact folded structure.
Structural Rearrangements that Accommodate Lys73 ligation of the Heme Iron
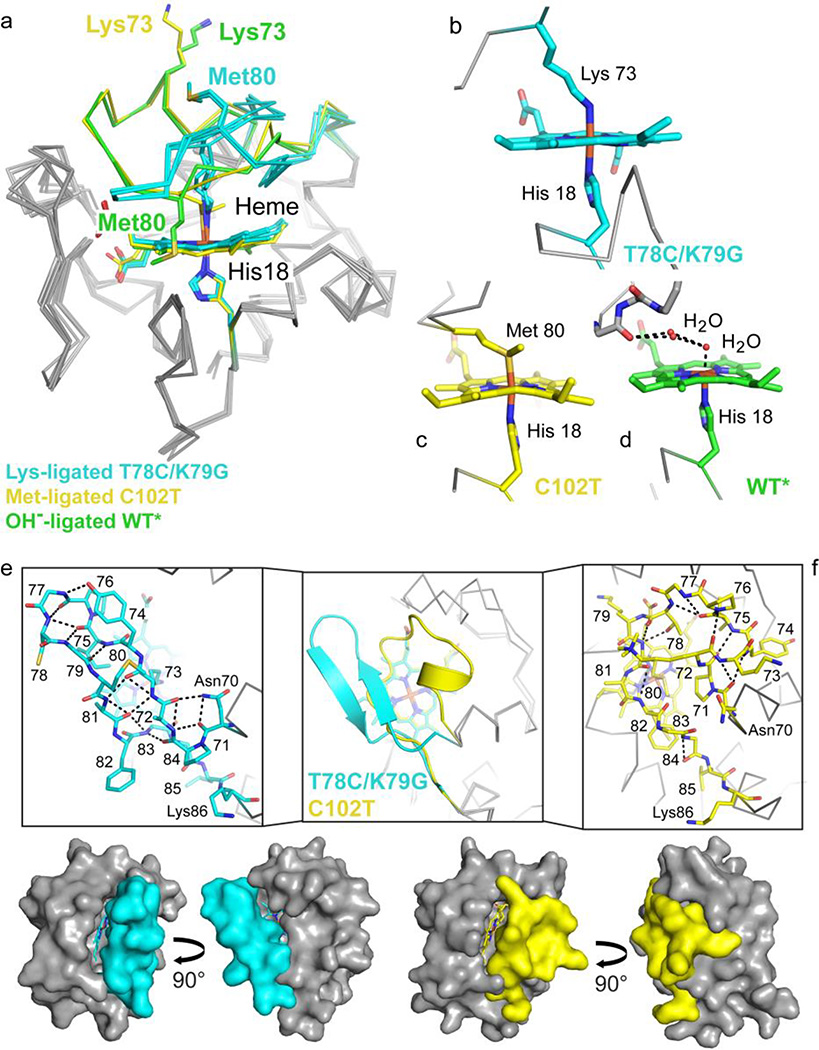
We compared our Lys-ligated structure to several available structures of cyt c with different ligands to the heme iron: the X-ray crystallographic structures of Met80-ligated yeast iso-1 cyt c (PDB ID: 2YCC,25 with the native trimethyllysine Tml72 and background mutation C102T, referred to as C102T), the hydroxide-ligated dimeric horse heart cyt c (PDB ID: 3NBS49) and monomeric yeast WT* (PDB ID: 4MU829), as well as the NMR structure of the Lys73-ligated yeast K72A/K79A/C102T under highly alkaline conditions (PDB ID: 1LMS30).
Our T78C/K79G structure is compact and has a core protein fold very similar to those of Met- and hydroxide-ligated forms of monomeric cyt c (Figure 7a). Large differences are confined to the heme iron ligand (Figures 7b, 7c, and 7d) and to the heme coordination loop. For Met80- and hydroxide-ligated structures, the loop appears to be stabilized by a number of interresidue contacts to positions elsewhere in the protein (Figure 7f). In contrast, the loop in T78C/K79G refolds into a β-hairpin structure with multiple intraloop hydrogen-bonding contacts (Figure 7e). The trigger for this refolding is likely the displacement of the Cys78 side chain from the heme-binding pocket, either through disulfide formation or maleimide labeling.
Figure 7.
Coordination loop differences in cyt c structures with Lys73, hydroxide (PDB ID: 4MU8),29 or Met80 (PDB ID: 2YCC)25 coordinated to the heme iron. Structures are colored by carbon atoms as follows: Lys73-ligated T78C/K79G=cyan, Met80-ligated C102T=yellow, hydroxide-ligated WT*=green. Non-carbon atoms are colored by element: N=blue, O=red, S=yellow, Fe=orange. (a) Alignment of the three structures by main-chain atoms reveal similar core conformations, with RMSD = 0.50 Å and 0.27/0.33 Å (A- and B-protomers), respectively. The Lys73 and Met80 residues are highlighted (side chain stick figures) and labeled for each structure. (b–d) Heme coordination geometry for Lys73-ligated T78C/K79G (b), Met80-ligated C102T (c), and WT* (d), as labeled. (e–f) The heme coordination loops of T78C/K79G (e) and hydroxide-ligated C102T (f) are shown following superposition of the core conformations in cartoon representation (middle box) and as stick figures (left and right boxes). Stick representations of the loops are labeled by residue, and black dashed lines mark hydrogen bonds. Below, the van der Waals surfaces for T78C/K79G (cyan) and C102T (yellow) are shown to highlight the positions of the heme coordination loop.
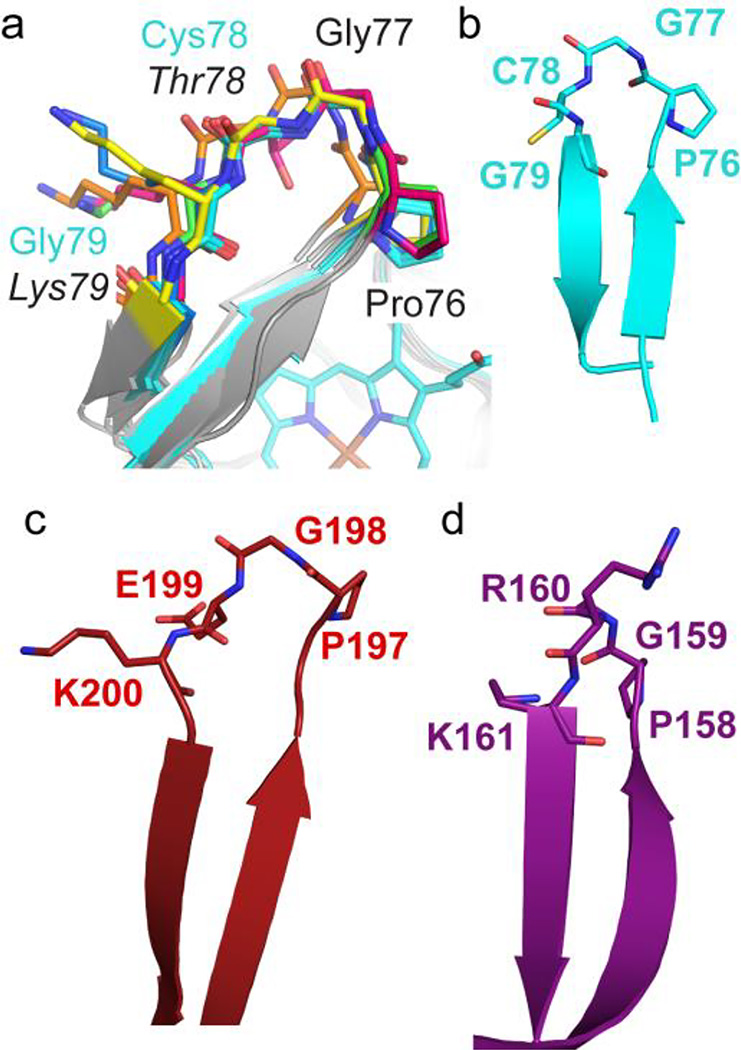
Structural analysis suggests that this conformational switch is accessible to the WT* protein as well as to the T78C/K79G mutant. While the main-chain conformation of the mutant Gly79 residue exhibits glycine-specific torsion angles, modeling with Rosetta Backrub63,64 revealed multiple backbone conformations with minor main-chain adjustments and standard Ramachandran angles at both Thr78 and Lys79 (Figure 8a). Furthermore, the tight turn formed by Pro76 and Gly77, native amino acids that are highly conserved in mitochondrial cyt c proteins,8,31 is also compatible with the native Lys79 side chain. A MaDCaT (Mapping of Distances for the Categorization of Topology)65 search identified two PDB structures in which a similar sequence-specific turn is formed by the sequence P-G-X-K, where X = any amino acid: human protein zinc-α-2-glycoprotein (PDB ID: 3ES6)66, with sequence 197P-G-E-K200, and enterobacteria phage 186 repressor protein CI (PDB ID: 2FKD)67, with sequence 158P-G-R-K161 (Figure 8b–d). The findings of in already existing protein structures as well as our Rosetta Backrub modeling results suggest that the β-hairpin conformation could also be accessible to the protein with the loop sequence of the native cyt c.
Figure 8.
A conserved stretch of the wild-type cyt c sequence, 76PGTK79, is capable of forming a β-hairpin structure. (a) Backbone analysis of T78C/K79G (cyan) using RosettaBackrub,64 with an in silico-reverted wild-type sequence (indicated by italics), revealed that only minor structural changes are required to produce allowed Ramachandran angles for Lys79, according to MolProbity.108 Ribbon diagrams (gray) with 76P-G-T/C-K/G79 (stick representation in different colors) are shown for five of the ten structures with the lowest predicted energies. (b–d) Using the structural motif PDB searching program, MaDCaT,65 we identified two additional β-hairpin structures similar to that in T78C/K79G (b) with the sequence P-G-X-K. They are: (c) human protein zinc-α-2-glycoprotein (PDB ID: 3ES6),66 with sequence 197P-G-E-K200, and (d) enterobacteria phage 186 repressor protein CI (PDB ID: 2FKD),67 with sequence 158P-G-R-K161. All structures are shown in cartoon and stick representation.
The tightly-folded β-hairpin we see with T78C/K79G is distinct from the conformation of the loop in the NMR structure of the Lys73-ligated cyt c at pH 11.1 (PDB ID: 1LMS, Figure S13).30 Under highly alkaline conditions, the loop appears loosely constrained, without many hydrogen bonds either within the loop or with the rest of the protein. At the same time, all NOESY restraints of the NMR structure are consistent with the distances we find in the crystallographic structures of T78C/K79G and C102T, illustrating the preservation of the core in both alkaline and mutant (T78C/K79G) forms of Lys73-ligated cyt c.
The structure of the Cys78-linked dimer of T78C/K79G is also distinct from the previously-reported structure of the dimeric horse heart cyt c (PDB ID: 3NBS, Figure S14).49 The latter structure was obtained after exposure of cyt c to alcohol and revealed swapping of the C-terminal helices of the two monomers. In both of the proteins crystallized as dimers, ours and horse heart cyt c, Met80 no longer coordinates to the heme iron. Instead, it is exposed to the solvent in T78C/K79G, but remains within the heme pocket in the other structure (Figure S14a). These differences may be related to the need for the loop rotation and refolding to accommodate Lys73 coordination to the heme iron (Figure S14b). The contact sites and orientations of the two protomers within these dimeric structures are also distinct (Figure S14c).
The Heme Pocket of the Lys-Ligated T78C/K79G
In native cyt c residue 78 contributes to the critical hydrogen-bonding network that connects heme propionate HP6 and Tyr67 via internal water molecules.25,26 With the expulsion of residue 78 from the heme pocket in the Cys78-linked T78C/K79G dimer, the repositioned main-chain carbonyl of Tyr74 and an additional water molecule complete the network (Figure 6b). This water chain is reminiscent of the one observed in hydroxide-ligated WT*,29 but provides a shorter path to the protein surface. The hydrogen bond from Tyr67 to its neighboring internal water molecule is longest in the Lys-ligated T78C/K79G structure, followed by those in the hydroxide-ligated WT*29 and then by those in Met-ligated C102T25 (Figure S15). Another notable change in T78C/K79G is a shift in the position of the 50s helix in the vicinity of Asn52, which avoids a steric clash with the residues of the refolded coordination loop (Figures S16a and S16b). These structural changes collectively result in opening of the heme pocket in T78C/K79G. Although the solvent accessible surface area of the heme is very similar in both structures (97% of the potential solvent accessible surface area is buried in both structures), the overall volume of the heme pocket is about 25% larger in T78C/K79G (varies between 1020 and 1332 Å3 for the three protomers) than the one in C102T (943 Å3) (Figures 7e and 7f).
The resultant pocket in T78C/K79G is composed of residues Ser40, Ile53, Val57, Trp59, and Tyr67 and has the potential to accommodate a hydrophobic moiety. Indeed, we see positive difference electron density in the vicinity of Asn52 (Figure S16c) for all three protomers in the T78C/K79G structure, although it is most pronounced in protomers A and C. Based on our crystallization conditions, the most likely source of this additional electron density is a PEG molecule. Supporting the non-accidental nature of these interactions, PEG has been shown to bind to cyt c at a hydrophobic patch near the heme.68 However, we are unable to definitively determine the source of this observed electron density and therefore chose not to include PEG, or another molecule, in our final structural model.
Lys-ligated Conformer Trapped at Near Neutral pH Adds Insights to the Mechanism of the Alkaline Transition
Residue Lys73 is strictly conserved among eukaryotic cyt c8 proteins and its coordination to the heme iron has been shown to take place in the alkaline form of the protein at high solution pH.15,30 Simulations of the alkaline transition in horse heart cyt c9 and structural modeling of Lys-ligated iso-1 yeast cyt c69 have suggested that the alkaline transition preserves the protein core, but introduces changes in the region around residues 70–80 (the bulk of the coordination loop). NMR studies of Lys73-ligated K72A/K79A/C102T at pH 11.1 have revealed an increase in the solvent exposure of the heme by about 20% compared to that of the wild-type cyt c, unfolding of the heme coordination loop, as well as changes in the positions of several Ω loops (Figure S13).30 In addition, a reduced number of NOEs and small residual dipolar couplings suggested the increased conformational flexibility of the structure.70 At the high pH used in these experiments some protein denaturation is likely and may explain the large extent of structural perturbations observed. Indeed, we find a sizeable drop in Tm values from thermal denaturation of WT* at pH 11 (Figure 3 and Table S2). As the heme coordination loop unfolds, coordination by Lys, a strong ligand to the ferric heme iron in its deprotonated (neutral) form, becomes possible. When this low-stability loop is destabilized in the presence of urea, another denaturant, a population of the protein with Lys-ligated heme iron can also be detected by NMR.71
In T78C/K79G, the Lys ligation is possible at almost neutral pH, in the presence of the native Met80 ligand and without loss of the protein compact structure. What then could be the process that facilitates Lys deprotonation and coordination to the heme iron? Mechanistic studies of the alkaline transition in cyt c have revealed that deprotonation of a yet unknown ‘trigger’ group precedes ligand replacement.9,22 A buried water molecule, one of the heme propionates, Tyr67, and His18 have all been suggested as possible candidates for such a trigger group. As described earlier, displacement of Cys78 from the heme pocket in T78C/K79G is associated with a cascade of structural changes. For native Thr78 to adopt an orientation similar to that of Cys78 the hydrogen bonds to the nearby water (W166 in 2YCC)25 and to HP6 have to be broken, processes that can be affected by acid-base reactions.
The heme propionate HP6 has previously been suggested to have an unusually high pKa value (≥9) in folded cyt c.72,73 In T78C/K79G and T78C/K79G/M80X(X=L, I, F)32, Cys78 remains coordinated to the heme iron at alkaline pH, but titrations of these variants cause changes in the protonation state of some group in the vicinity of the heme with pKa values between 8 and 10 (Table S3), loosely consistent with the above estimate. If indeed the pKa of HP6 is that high, this group is a strong candidate for a site of deprotonation at alkaline pH. However, it has been puzzling how this group could recover its proton upon Lys coordination to the heme iron, a mechanistic proposal inferred from proton inventory experiments.17 In fact, the rearrangement of the hydrogen-bonding network and capping of the heme pocket by the new residue Tyr74 (Figure S15c) suggests a possible explanation for the subsequent recovery of the HP6 protonation state in the compact protein structure with the Lys-ligated heme iron. Adding support to the importance of the hydrogen bond(s) mediated by Thr78 in the mechanism of the alkaline transition, replacements at this position alter the protein stability and the pKa of the transition.74,75
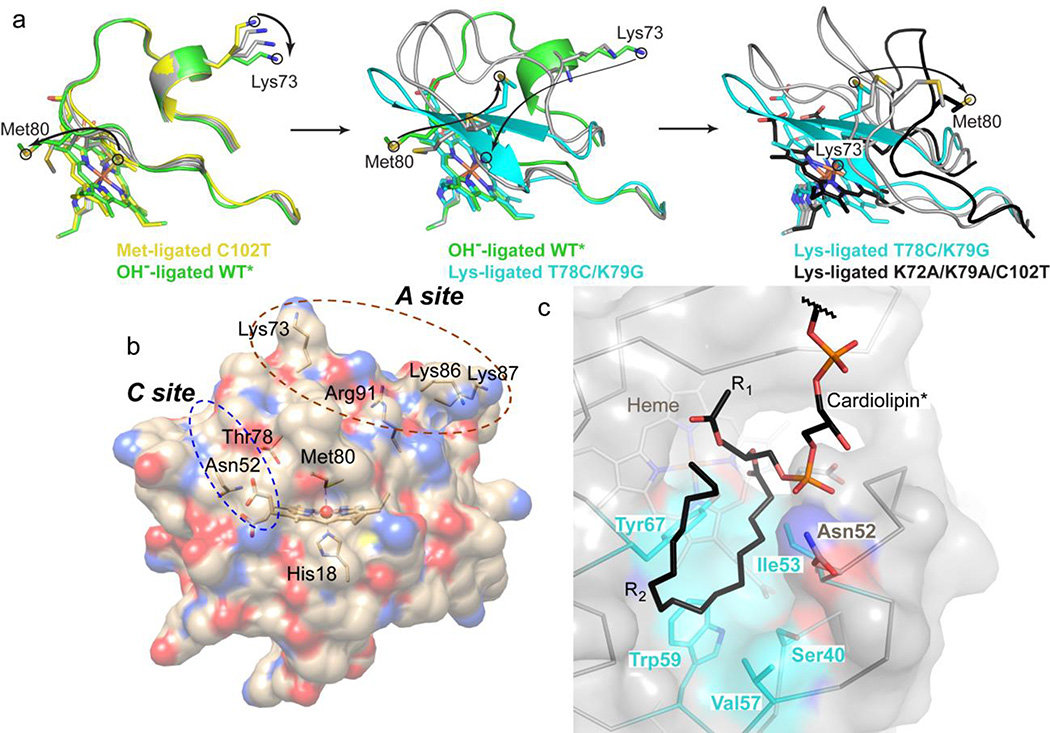
Structural data suggest that Met-to-Lys ligand replacement involves a spectrum of conformational changes. Dissociation of Met80 from the heme iron changes the position of this native ligand within the protein. For hydroxide-ligated dimeric horse heart cyt c49 or monomeric yeast WT*,29 the hydrophobic Met80 continues to reside in the protein interior or near the heme pocket, respectively. With Lys73 coordinated to the heme iron in our structure, Met80 becomes surface-exposed (Figure 2). With Met80 fully expelled from the heme pocket, the polypeptide can form a compact structure around the heme. Interestingly, the NMR structure of the alkaline form with the unfolded coordination loop also has Met80 solvent-exposed and away from the heme.30 Structural snapshots from four different structures illustrate the progression of the alkaline conformational change (Figure 9a). Met80 starts as the heme iron ligand, it crowds the protein interior upon dissociation from the heme iron, then escapes from the heme pocket in the refolded loop, and finally moves even further as the loop denatures at high pH. Similarly, the structures track the position of Lys73 as this residue gets closer to the heme and becomes the iron ligand.
Figure 9.
(a) Model of the Met80-to-Lys73 structural transition in cyt c. To aid in visualization of the transition, “intermediate” states were generated in PyMol using the Met80-ligated C102T25 (yellow), hydroxide-ligated WT*29 (green), Lys73-ligated T78C/K79G (cyan) and Lys73-ligated K72A/K79A/C102T30 (black) structures. Each panel (Met80-to-hydroxide, left; hydroxide-to-Lys73, middle; and further unfolding of the Lys73-ligated conformer, right) contains two “intermediate” states generated by morphing (gray carbons). Arrows indicate movements of Met80 and Lys73 to accommodate the change. (b) Structure of cyt c showing the proposed CL binding sites. At the hydrophobic C site, hydrogen bonding of the CL phospholipid group to Asn52 has been suggested to aid in the acyl insertion into the protein interior.96 (c) Structural model of CL binding into the identified hydrophobic pocket created using Coot105 and based on the positive electron density in the Fo−Fc map (Figure S16c).
β-Sheet Structure in Cyt c and Its Physiological Significance
Many cyt c variants with nonnative heme-iron ligands show increased structural disorder.30,76–78 In contrast, the Lys73-ligated species in this study revealed the coordination loop folded into a tight β-hairpin. The conserved Pro76-Gly77 sequence8,31 is a critical element of this hairpin, but the formation of the hairpin does not require proline isomerization: the imide bond involving Pro76 is trans in the native protein79 as well as in the refolded loop. Nevertheless, the flexibility of this polypeptide fragment is likely important for the rearrangement. Variations in kinetics of His-to-Met ligand exchange in His mutants of yeast cyt c effectively illustrate the critical role of coordination loop dynamics in facilitating ligand replacement.17 Further supporting this argument, the P76G mutation in yeast iso-2 cyt c lowers the apparent pKa of the alkaline transition and increases its rate.79,80
A protein’s ability to refold into an alternative conformation is intriguing on its own, but it bears particular functional significance in the case of cyt c. On the one hand, switching of cyt c between two compact forms offers a fine control of regulation of cyt c function in oxidative phosphorylation, without deleterious effects of proteolysis and/or high reactivity of an exposed heme. The degree of burial of potential surface area is essentially unchanged from that of the Met-ligated C102T (2YCC, Figures 7 and S13). At a low proton gradient across the inner mitochondrial membrane (high pH), conversion of cyt c to the alkaline form may serve as a feedback switch. The Met-to-Lys ligand substitution would lower the heme reduction potential and thus increase the driving force for the reaction of cyt c with cyt c oxidase. Indeed, two conformers of cyt c, one having a low redox potential, have been suggested in the studies of ET between these two physiological partners.19
On the other hand, cyt c refolding into a β-sheet structure may also confer new functions on this protein. Several studies have reported formation of β-sheet structures in denatured cyt c.33,81,82 Among them, the UV-resonance Raman experiments by Spiro and coworkers have suggested that β-strands could extend into the 60s and 70s region of cyt c.33 Consistent with these predictions, the hairpin we observed involves residues Thr69 to Glu88. Since β-sheet structures often mediate intermolecular interactions,83,84 the refolded loop may serve as a possible precursor to cyt c aggregates, including those found in Lewy bodies.85 Within the monomeric form found in mitochondria, such refolding alters the cyt c heme pocket and thus is likely to affect the protein peroxidase activity, a function critical in early stages of apoptosis.21 Indeed, the increase in population of β-sheet structures in thermally denatured cyt c at low pH is correlated with an increase in peroxidase activity.82 Under these experimental conditions, β-sheet conversion also involves the 60s region and there are additional structural changes in the 40s Ω loop and in interloop contacts. Collectively, these structural rearrangements increase the heme exposure.
Cyt c Interactions with CL
Extended (open) cyt c conformers formed upon interaction of the protein with CL-rich membranes provide easy access to the heme for substrates.86 These species exhibit high peroxidase activity, but compact conformers could also add to the reactivity of the ensemble.29 Our earlier work has shown that partitioning between compact and extended conformers depends on a number of experimental variables, including the membrane CL content and available surface area.87 The heme iron ligation is also altered in the compact CL-bound cyt c, where one of the nearby Lys residues has been proposed to take the place of Met80.20 The structure of a Lys-ligated conformer offers a glimpse of how this species may look.
For both the native cyt c and Lys-ligated species, the sixth ligand has to dissociate from the heme to enable coordination of a peroxide. The ligand dissociation is likely rate-limited by opening of the heme pocket.88 Our activity assays suggest that at near-neutral pH, the activity of the Lys-ligated variants is higher than that of WT*. With refolding of the coordination loop in the Cys78-linked dimer, the volume of the pocket increases, and a similar effect is likely to take place in the solution analogue of this species, the maleimide derivative of T78C/K79G. The more open pocket favors dissociation from the heme iron of the Lys73 ligand. Furthermore, the water channel next to the heme propionate HP6 could participate in proton transfer in the peroxidase reaction. The increase in peroxidase activity of our Lys variants contrasts with earlier findings of an activity drop at pH 7.0 for the M100K mutant of cyt c55089; the well-packed heme pocket in this cyt c550 mutant42 as well as possible effects on both Km and kcat could account for the differences with our study. Another relevant finding is the inhibition of peroxidase activity of cyt c under alkaline conditions by Lys coordination to the heme iron.89 However, at near-neutral pH, Lys is protonated and its affinity for the heme diminishes (Figure S8 and Table 2) enabling it to more readily dissociate from the heme iron. Consistent with this proposal, at more acidic pH, the activities of Lys-ligated mutants cyt c550 M100K42 and our K73A/K79G/M80K are greater than those of the corresponding wild-type proteins.
The observed changes in the heme pocket in the Lys-ligated T78C/K79G may also be relevant to interactions of cyt c with CL. The displacement of the conserved residue 78 from the heme pocket is intimately linked to a rearrangement in the intraprotein hydrogen-bonding. The network connects Thr78 to Asn52 and Tyr67, two residues that greatly affect cyt c dynamics and are implicated in the protein’s peroxidase function. Linked directly to the native ligand to the heme iron, Tyr67 forms a hydrogen bond with the sulfur of Met80.25,90–92 Mutations at residue 67 increase peroxidase activity of cyt c.93 Previous work has suggested that Asn52 is close to one of the CL-binding sites in the protein (Figure 9b),94,95 specifically to the location where CL acyl chains could possibly insert into the cyt c interior.96 Consistent with this residue being a gate to the protein interior, replacement of Asn52 with a more hydrophobic and bulky Ile removes the internal water molecule from the hydrogen-bonding network, rigidifies the protein structure, increases the protein stability, and shifts the alkaline transition to higher pH.26,97 Furthermore, in studies of cyt c interactions with CL, this mutation has been shown to greatly decrease the rate of Met80 loss from the heme iron.98 Modeling suggests that the hydrophobic pocket in our structure, which is formed upon repositioning of residues 52, 74, and 78, could easily accommodate the acyl chain of CL (Figure 9c). Given the proximity of Asn52 and Thr78 to the CL binding site and their importance for cyt c structure and dynamics, it is conceivable that CL binding could trigger similar perturbations in the intraprotein hydrogen-bonding network that ultimately change the heme pocket and ligation.
Conclusions
The alkaline form of cyt c with Lys ligation to the heme iron has been linked to critical functions of the protein in both ET and apoptosis. Our structure suggests that this ligation is possible within a compact protein conformation. The much higher binding affinity of the deprotonated Lys for the ferric heme compared to that of Met offers a thermodynamic rationale for Lys ligation when the tertiary contacts of the native cyt c are perturbed. Cycling between well-folded compact conformations may provide better integrity for functional transitions, without deleterious effects of protein aggregation or unintended reactivity of the heme group.
The structure illustrates the refoldable nature of the heme coordination loop and the ability of cyt c to access β-sheet conformations. The tight β-hairpin with a turn formed by conserved Pro76 and Gly77 covers the heme pocket. Even though the Lys-ligated structure is compact, the volume of the heme pocket is larger and the protein peroxidase activity is enhanced. In parallel, repositioning of residue 78 modifies the protein hydrogen-bonding network in direct proximity to the heme. These changes suggest a mechanism by which binding to CL membranes in the vicinity of Asn52 can trigger a switch to Lys ligation to the heme iron and opening of the heme pocket in cyt c. Given the importance of lability of axial ligand for peroxidase function of cyt c, the set of Lys-ligated variants we describe here provides valuable models for examining this process quantitatively and correlating these dynamics to perturbations in protein structure around the heme.
Materials and Methods
Site-Directed Mutagenesis, Protein Expression, and Purification
The plasmid Rbs (WT*) containing genes CYC1 and CYC3 that encode the yeast iso-1 cyt c and yeast cyt c heme lyase, respectively, was a template for all the mutants. This parent pseudo–wild-type cyt c construct WT* contained two additional mutations, K72A and C102S, to prevent Lys72 coordination to the heme iron and formation of Cys102-linked dimers, respectively. The mutants were constructed with a QuikChange Kit (Stratagene), DNA was extracted and purified with the Miniprep Kit (Qiagen), and the correct sequence was verified at the Molecular Biology and Proteomics Core Facility (Dartmouth College, Hanover, NH). All of the mutants were expressed and purified as previously described.32 Protein conversion to ferric and ferrous forms has been accomplished by adding excess of potassium ferricyanide and sodium dithionite, respectively.
Maleimide Labeling of Cys78
The purified ferric Cys78-ligated T78C/K79G protein was converted to the ferrous Met80-ligated protein by adding 5 mM sodium dithionite and letting the reaction proceed for at least 30 min in a glove box under a nitrogen atmosphere (COY Laboratory Products). In addition, 5 mM dithiothreitol (DTT) was included in this mixture to prevent formation of disulfide-linked adducts. Excess reducing agents were removed by passing the protein solution through a PD-10 column (GE Healthcare) pre-equilibrated with a 100 mM sodium phosphate buffer at pH 7.4. Then, an approximately ten-fold molar excess of maleimide was added to the protein solution eluted from the PD-10 column to initiate labeling. The reaction was quenched after 6 h via addition of 5 mM DTT and the mixture solution was dialyzed overnight against a 10 mM sodium phosphate buffer at pH 7.4. The labeled protein was further purified using a HiTrap SP HP column and labeling was confirmed by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry measurements (ABI Voyager-DE Pro MALDI-TOF mass spectrometer, Molecular Biology and Proteomics Core Facility, Dartmouth College).
Preparation of MP8
AcMP8 was prepared by proteolytic degradation of horse heart cyt c (Sigma-Aldrich) and subsequent reaction of the heme-containing eight-residue peptide with acetic anhydride as previously described.32
Spectroscopic Measurements
Absorption spectra were acquired on Agilent 8453 diode-array, Shimadzu UV-1201 (Shimadzu Scientific), or JASCO V-630 scanning spectrophotometers. CD spectra and CD thermal melting curves were recorded on a J815 CD spectropolarimeter equipped with a variable temperature Peltier cell device (JASCO). EPR spectra were recorded at 10 K on a Bruker EMX 300 X-band EPR spectrometer using the following conditions: microwave frequency of 9.49 GHz, microwave power of 3.21 mW, modulation frequency of 100 kHz, modulation amplitude of 1.00 G, and time constant of 20.48 ms.
1H NMR spectra were recorded on a 500 MHz Bruker AVANCE500 spectrometer equipped with a temperature controller. Samples of the ferrous protein (1–1.5 mM) were prepared in a 50 mM sodium phosphate buffer at pH 7.4 containing 10% D2O with the addition of 10 mM sodium dithionite in a nitrogen-filled glove box (COY Laboratory Products), transferred into a sealable anaerobic tube (Wilmad Lab Glass) and then examined by NMR at 25°C. Water suppression was achieved by using excitation sculpting with gradients.99 Samples of the ferric protein (0.4–1 mM) were prepared in a D2O buffer containing 50 mM sodium phosphate (pH 7.0, pH meter reading without correction) or 50 mM sodium borate (pH 9.2) by repeated centrifugal ultrafiltration. The 1D 1H NMR spectra were collected at 5°C, 25°C and 45°C by using a superWEFT pulse sequence100 with a recycle delay of 220 ms for low-spin heme species of cyt c or 110 ms for horse heart myoglobin and potential high-spin species of cyt c. The chemical shifts were calibrated with respect to 3-(trimethylsilyl)-1-propanesulfonic acid (DSS) at each temperature.
Protein extinction coefficients were determined by the pyridine hemochrome method. All experiments with ferrous proteins were performed under anaerobic conditions in a nitrogen-filled glove box (COY Laboratory Products). Thermal denaturation, pH titrations, and studies of AcLys (Acros Organics) binding to AcMP8 were performed according to the published procedures.32 At pH 5.0 and low concentrations of protein (<15 µM), thermal denaturation of all the variants is ≥89% reversible (Figure S17); these favorable conditions were employed to get the detailed thermodynamic characterization of these proteins (Table S4). Higher concentrations of protein and more basic solution pH promote formation of His-misligated and Cys-linked oligomers in the denatured samples.
Crystallization and Structure Determination
Purified ferric T78C/K79G was dialyzed at 4 °C into a 10 mM Tris buffer at pH 7.4 and concentrated to 15 mg mL−1. The crystals used for data collection were obtained by vapor diffusion, with a reservoir buffer containing 200 mM ammonium phosphate, 100 mM Tris and 30% (v/v) polyethylene glycol (PEG) 300 at pH 8.5. 2 µL of the protein solution were added to 2 µL reservoir buffer and equilibrated in a hanging-drop format at 18 °C. Crystals appeared after 3–5 days, and grew for up to 2 months. Prior to data collection crystals were transferred to a cryoprotectant buffer composed of the reservoir buffer including in addition 20% (v/v) PEG400 and frozen with liquid nitrogen.
Diffraction data were collected on the X6A beamline, at the National Synchrotron Light Source (NSLS), Brookhaven National Laboratory. Oscillation data were collected over a total of 180° in Δφ=0.2° slices, at wavelength λ=1.0781 Å and a detector distance of 215 mm. Data were processed using XDS.101 Molecular replacement (MR) was performed using Phenix, with a protomer of yeast iso-2 cyt c (PDB ID: 1YEA)102 as a search model. The heme was omitted from the search model. Electron density for the heme confirmed the MR solution. Model building, refinement, and assessment were performed using Coot, Phenix and REFMAC.103–107 Critically, Phenix.XTRIAGE identified two pseudo-merohedral twin operators. Automated twin amplitude refinement in REFMAC revealed nearly equal twin fractions in the final refined model (Table S4). Structural integrity was assessed using the programs MolProbity, the PDB validation server, and the DALI server.40,108–111
Structural Analysis
All figures were rendered using the program PyMol.112 Alignments were performed in PyMol, using main-chain atoms as indicated. Solvent accessible surface area was calculated using AREAIMOL, in the CCP4 suite of programs. The program RosettaBackrub63,64 was used to assess whether or not the wild-type sequence could adopt the β-hairpin structure seen in the T78C/K79G structure, with minimal conformational shifts. The web interface of the program MaDCaT (Mapping of Distances for the Categorization of Topology) was used to search the PDB65 for β-hairpin structures with the sequence motif, P-G-X-K (X is any amino acid); the 1000 top matches were filtered in the analysis. Heme pocket geometry was assessed using the program CASTp.113
Analysis of the Protein Oligomerization State
T78C/K79G and WT* cyt c solutions were analyzed by analytical size exclusion chromatography (SEC) using a Superdex 75 HR 10/30 column (GE Healthcare Life Sciences) equilibrated with a 10 mM sodium phosphate buffer at pH 7.4. Under these solution conditions the elution peak of standards chymotrypsin (25 kDa) and RNase (13.7 kDa) eluted at 13.9 mL and 16.0 mL, respectively.
SDS-PAGE gels were run with and without DTT to analyze Cys-linked oligomers, followed by Coomassie-staining to visualize the protein bands. The percentage of the dimer species was estimated by the analysis of band intensities in gels of multiple samples using the program ImageJ.
Protein Changes during Crystal Growth
One 24-well crystallization tray was set up under the optimized crystallization condition. Two crystal drops from the tray were taken out on a daily basis to measure absorption spectra and run non-reducing SDS-PAGE experiments for one week, until crystal growth was observed. Before the absorption measurements, 2 µL of the crystal drop was diluted into 200 µL of a 100 mM sodium phosphate buffer at pH 7.4.
Two additional 24-well crystallization trays were set up for EPR, peroxidase activity, thermal denaturation, and electrochemistry measurements. The crystal drops were harvested after growing crystals for approximately two weeks and dissolved into a 100 mM sodium phosphate buffer at pH 7.4. Depending on the subsequent experiments, the protein was then dialyzed into the appropriate buffer.
Cyt c - PEG Interactions
Changes were monitored in the absorption spectra of T78C/K79G upon protein interaction with PEG-containing buffers (0, 3.3, 6.7, 10, 13.3, 16.7, and 20% (v/v) PEG300, dissolved in a 100 mM sodium phosphate buffer at pH 7.4). Before the measurements, the protein (200 µM for the spectra of Q- and charge-transfer bands and 10 µM for the spectra of the Soret band) was incubated with a PEG300-containing buffer for 1 h under anaerobic conditions in a glove box.
Electrochemistry
Spectroelectrochemistry experiments were performed as previously described.32 For the protein recovered from the crystal drop, the sample amounts were not sufficient for such measurements and direct electrochemistry was performed instead.
Voltammetry was performed using a three-electrode cell, in which the saturated calomel reference electrode (SCE) was in a side chamber connected to the main compartment through a Luggin capillary. All potentials were corrected to the standard hydrogen electrode (SHE) by the relationship SHE = SCE + 243 mV at 22 °C.47 The gold working electrode was cleaned through mechanical abrasion using 0.3 µm and 0.05 µm alumina followed by sonication and electrochemical cleaning at high potentials.114 The electrode was then soaked in a 10 mM ethanolic solution of 3-mercaptopropanol overnight and rinsed with ethanol and deionized water before use. A platinum wire was used as the counter electrode. Ultra high purity argon (< 10 ppm O2) was passed through an oxygen-removal catalyst before being used to flush oxygen from the electrochemical cell. In addition, the cell was “submerged” in a small plastic tub that was also flushed with argon. A wire-mesh Faraday cage surrounded the entire apparatus and shared electrical ground with the potentiostat (CH Instruments). The protein was dialyzed into a mixed buffer at pH 7.4 consisting of 85 mM sodium acetate and 5 mM each of MES, MOPS, and TAPS. Before voltammetric measurements, the protein sample was allowed to equilibrate in the cell for at least 30 minutes.
Assays of Peroxidase Activity
Guaiacol and ABTS assays were performed as previously described.94,115 The final concentrations of protein, guaiacol, and H2O2 were 1 µM, 10 mM, and 5 mM, respectively. The final concentration of protein, ABTS, and H2O2 were 1 µM, 200 µM, and 1 mM, respectively. Formation of tetraguaiacol (ε470 = 26.6 mM−1cm−1)116 or oxidized ABTS (ε735 = 14 mM−1cm−1)117 was observed every second for 1000 s by absorbance measurements. The initial rates were determined from the slopes of the linear phase of the reaction progress curves.
Supplementary Material
Acknowledgments
This work was supported by NSF CAREER CHE-0953693 (E.V.P), NIH RO1-GM098502 (E.V.P.) and T32-GM008704 (J.F.A.) grants. Additional support came from a Dartmouth College John Zabriskie ’61 Undergraduate Research Fellowship Award (M.Q.Z.), the Berry College Faculty Development and the Richards Family Funds (K.R.H.). We thank Dr. B. Bowler (University of Montana) for the Rbs(WT*) plasmid, Dr. V. Stojanoff, Dr. J. Jakoncic, and E. Lazo (NSLS) for assistance with data collection, and Dr. J. McLellan (Geisel School of Medicine) and Dr. M. Rivera (University of Kansas) for helpful advice. We dedicate this paper to the memory of Dr. Bernard L. Trumpower.
Footnotes
At this point there is no indication of the role of cyt c in apoptosis in yeast. The yeast iso-1 cyt c, however, is very similar in its sequence and structure to mammalian cyt c proteins and has served as a useful model for studies of cyt c structure, including those related to the mechanism of apoptosis.
A pseudo-wild-type variant of iso-1 yeast cyt c with mutations K72A and C102S, to prevent Lys72 coordination to the heme and formation of Cys102-linked dimers, respectively. All variants in this work contain these two background mutations.
We also attempted to measure the potential of this mutant by direct electrochemistry (as described in Materials and Methods). However, the thiolate-ligated conformer has a rather low reduction potential and we were unable to achieve reduction of T78C/K79G before hitting the effective lower limit set by reductive desorption of 3-mercaptopropanol on gold (approximately – 0.45 V). We continue to investigate other electrode surfaces for this purpose.
Supporting Information Available. Figures showing charge-transfer absorption bands for T78C/K79G, T78C/K79G/M80L, and WT*, effects of CL liposomes on the absorption spectra of T78C/K79G, spectroelectrochemistry curves for T78C/K79G, T78C/K79G/M80L, and WT*, and 1D 1H and 2D 1H NOESY NMR spectra of ferrous T78C/K79G; results of thermal denaturation for ferric and ferrous T78C/K79G, T78C/K79G/M80L, and WT*; pH effects on the T78C/K79G absorption spectra; 1D 1H NMR spectra of ferric WT*, T78C/K79G and myoglobin; electron density for the heme in the crystal structure, arrangement of protomers and dimerization contacts; heme iron coordination geometry in T78C/K79G and other proteins with Lys ligation at the heme iron; analysis of the dimer content in solution; binding of AcLys to AcMP8; spectra of ferric and ferrous K73A/K79G/M80K compared to that of WT*; pH effects on the absorption spectra of ferric K73A/K79G/M80K; voltammograms of the crystal drop sample; changes in the absorption spectra of T78C/K79G after addition of PEG300; comparison of structures with Lys73 coordinated to the heme iron solved by X-ray crystallography and NMR; comparison of the T78C/K79G dimer to the dimeric structure (after alcohol treatment) of horse heart cyt c; environment of the heme propionate HP6; analysis of the shift in the 50s helix; reversibility of thermal denaturation; and tables listing results of thermal denaturation; EPR parameters; and pKa values for the studied variants; data collection and refinement statistics; and comparison of thermodynamic parameters of different variants. This material is available free of charge via the Internet at http://pubs.acs.org.
Coordinates and structure factors were deposited in the Protein Data Bank, with accession code 4Q5P.
References
- 1.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, Dunlap JC, Crane BR. Science. 2007;316:1054. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lätzer J, Shen T, Wolynes PG. Biochemistry. 2008;47:2110. doi: 10.1021/bi701350v. [DOI] [PubMed] [Google Scholar]
- 3.Fraser JS, Clarkson MW, Degnan SC, Erion R, Kern D, Alber T. Nature. 2009;462:669. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giganti D, Albesa-Jové D, Urresti S, Rodrigo-Unzueta A, Martínez MA, Comino N, Barilone N, Bellinzoni M, Chenal A, Guerin ME, Alzari PM. Nat. Chem. Biol. 2015;11:16. doi: 10.1038/nchembio.1694. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Nature. 2004;428:323. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 6.Nerelius C, Sandegren A, Sargsyan H, Raunak R, Leijonmarck H, Chatterjee U, Fisahn A, Imarisio S, Lomas DA, Crowther DC, Strömberg R, Johanssona J. Proc. Natl. Acad. of Sci. USA. 2009;106:9191. doi: 10.1073/pnas.0810364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winkler JR. Curr. Opin. Chem. Biol. 2004;8:169. doi: 10.1016/j.cbpa.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Moore GR, Pettigrew GW. Cytochromes c: Evolutionary, Structural, and Physicochemical Aspects. New York: Springer-Verlag; 1990. [Google Scholar]
- 9.Weinkam P, Zimmermann J, Sagle LB, Matsuda S, Dawson PE, Wolynes PG, Romesberg FE. Biochemistry. 2008;47:13470. doi: 10.1021/bi801223n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Battistuzzi G, Bortolotti CA, Bellei M, Di Rocco G, Salewski J, Hildebrandt P, Sola M. Biochemistry. 2012;51:5967. doi: 10.1021/bi3007302. [DOI] [PubMed] [Google Scholar]
- 11.Theorell H, Åkesson Å. J. Am. Chem. Soc. 1941;63:1804. [Google Scholar]
- 12.Theorell H, Åkesson Å. J. Am. Chem. Soc. 1941;63:1812. [Google Scholar]
- 13.Theorell H, Åkesson Å. J. Am. Chem. Soc. 1941;63:1818. [Google Scholar]
- 14.Theorell H. J. Am. Chem. Soc. 1941;63:1820. [Google Scholar]
- 15.Rosell FI, Ferrer JC, Mauk AG. J. Am. Chem. Soc. 1998;120:11234. [Google Scholar]
- 16.Martinez RE, Bowler BE. J. Am. Chem. Soc. 2004;126:6751. doi: 10.1021/ja0494454. [DOI] [PubMed] [Google Scholar]
- 17.Cherney MM, Bowler BE. Coord. Chem. Rev. 2011;255:664. [Google Scholar]
- 18.Hodges HL, Holwerda RA, Gray HB. J. Am. Chem. Soc. 1975;96:3132. doi: 10.1021/ja00817a019. [DOI] [PubMed] [Google Scholar]
- 19.Döpner S, Hildebrandt P, Rosell FI, Mauk AG, von Walter M, Buse G, Soulimane T. Eur. J. Biochem. 1999;261:379. doi: 10.1046/j.1432-1327.1999.00249.x. [DOI] [PubMed] [Google Scholar]
- 20.Bradley JM, Silkstone G, Wilson MT, Cheesman MR, Butt JN. J. Am. Chem. Soc. 2011;133:19676. doi: 10.1021/ja209144h. [DOI] [PubMed] [Google Scholar]
- 21.Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Nat. Chem. Biol. 2005;1:223. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- 22.Hoang L, Maity H, Krishna MM, Lin Y, Englander SW. J. Mol. Biol. 2003;331:37. doi: 10.1016/s0022-2836(03)00698-3. [DOI] [PubMed] [Google Scholar]
- 23.Stellwagen E, Rysavy R, Babul G. J. Biol. Chem. 1972;247:8074. [PubMed] [Google Scholar]
- 24.Dumont ME, Corin AF, Campbell GA. Biochemistry. 1994;33:7368. doi: 10.1021/bi00189a043. [DOI] [PubMed] [Google Scholar]
- 25.Berghuis AM, Brayer GD. J. Mol. Biol. 1992;223:959. doi: 10.1016/0022-2836(92)90255-i. [DOI] [PubMed] [Google Scholar]
- 26.Berghuis AM, Guillemette JG, McLendon G, Sherman F, Smith M, Brayer GD. J. Mol. Biol. 1994;236:786. doi: 10.1006/jmbi.1994.1189. [DOI] [PubMed] [Google Scholar]
- 27.Redzic JS, Bowler BE. Biochemistry. 2005;44:2900. doi: 10.1021/bi048218b. [DOI] [PubMed] [Google Scholar]
- 28.Gu J, Yang S, Rajic AJ, Kurnikov IV, Prytkova TR, Pletneva EV. Chem. Commun. 2014;50:5355. doi: 10.1039/c3cc47943a. [DOI] [PubMed] [Google Scholar]
- 29.McClelland LJ, Mou TC, Jeakins-Cooley ME, Sprang SR, Bowler BE. Proc. Natl. Acad. of Sci. USA. 2014;111:6648. doi: 10.1073/pnas.1323828111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Assfalg M, Bertini I, Dolfi A, Turano P, Mauk AG, Rosell FI, Gray HB. J. Am. Chem. Soc. 2003;125:2913. doi: 10.1021/ja027180s. [DOI] [PubMed] [Google Scholar]
- 31.Zaidi S, Hassan MI, Islam A, Ahmad F. Cell Mol. Life Sci. 2014;71:229. doi: 10.1007/s00018-013-1341-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong F, Lisi GP, Collins DP, Dawson JH, Pletneva EV. Proc. Natl. Acad. of Sci. USA. 2014;111:E306. doi: 10.1073/pnas.1317173111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balakrishnan G, Hu Y, Spiro TG. J. Am. Chem. Soc. 2012;134:19061. doi: 10.1021/ja307100a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Y, Casimiro DR, Bren KL, Richards JH, Gray HB. Proc. Natl. Acad. Sci. USA. 1993;90:11456. doi: 10.1073/pnas.90.24.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKnight J, Cheesman MR, Thomson AJ, Miles JS, Munro AW. Eur. J. Biochem. 1993 doi: 10.1111/j.1432-1033.1993.tb17808.x. [DOI] [PubMed] [Google Scholar]
- 36.Blanke SR, Martinis SA, Sligar SG, Hager LP, Rux JJ, Dawson JH. Biochemistry. 1996;35:14537. doi: 10.1021/bi961512m. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds MF, Shelver D, Kerby RL, Parks RB, Roberts GP, Burstyn JN. J. Am. Chem. Soc. 1998;120:9080. [Google Scholar]
- 38.Cheesman MR, Little PJ, Berks BC. Biochemistry. 2001;40:10562. doi: 10.1021/bi0100081. [DOI] [PubMed] [Google Scholar]
- 39.Gao Y, Boyd J, Williams RJP, Pielak GJ. Biochemisry. 1990;29:6994. doi: 10.1021/bi00482a007. [DOI] [PubMed] [Google Scholar]
- 40.Holm L, Rosenström P. Nucleic Acids Res. 2010;38:W545. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodrigues ML, Oliveira TF, Pereira IAC, Archer M. EMBO J. 2006;25:5951. doi: 10.1038/sj.emboj.7601439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Worrall JAR, Van Roon A-MM, Ubbink M, Canters GW. FEBS J. 2005;272:2441. doi: 10.1111/j.1742-4658.2005.04664.x. [DOI] [PubMed] [Google Scholar]
- 43.Tezcan FA, Winkler JR, Gray HB. J. Am. Chem. Soc. 1998;120:13383. [Google Scholar]
- 44.Ubbink M, Campos AP, Teixeira M, Hunt NI, Hill HAO, Canters GW. Biochemistry. 1994;33:10051. doi: 10.1021/bi00199a032. [DOI] [PubMed] [Google Scholar]
- 45.Silkstone GG, Cooper CE, Svistunenko D, Wilson MT. J. Am. Chem. Soc. 2005;127:92. doi: 10.1021/ja045719b. [DOI] [PubMed] [Google Scholar]
- 46.Osteryoung J. Acc. Chem. Res. 1993;26:77. [Google Scholar]
- 47.Bard AJ, Faulkner LR. Electrochemical Methods: Fundamentals and Applications. 2nd. New York: John Wiley & Sons; 2001. [Google Scholar]
- 48.Barker PD, Mauk AG. J. Am. Chem. Soc. 1992;114:3619. [Google Scholar]
- 49.Hirota S, Hattori Y, Nagao S, Taketa M, Komori H, Kamikubo H, Wang Z, Takahashi I, Negi S, Sugiura Y, Kataoka M, Higuchi Y. Proc. Natl. Acad. of Sci. USA. 2010;107:12854. doi: 10.1073/pnas.1001839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parui PP, Deshpande MS, Nagao S, Kamikubo H, Komori H, Higuchi Y, Kataoka M, Hirota S. Biochemistry. 2013;52:8732. doi: 10.1021/bi400986g. [DOI] [PubMed] [Google Scholar]
- 51.Deshpande MS, Parui PP, Kamikubo H, Yamanaka M, Nagao S, Komori H, Kataoka M, Higuchi Y, Hirota S. Biochemistry. 2014;53:4696. doi: 10.1021/bi500497s. [DOI] [PubMed] [Google Scholar]
- 52.Mirkin N, Jaconcic J, Stojanoff V, Moreno A. Proteins. 2008;70:83. doi: 10.1002/prot.21452. [DOI] [PubMed] [Google Scholar]
- 53.Liu Z, Lin H, Ye S, Liu QY, Meng Z, Zhang CM, Xia Y, Margoliash E, Rao Z, Liu XJ. Proc. Natl. Acad. of Sci. USA. 2006;103:8965. doi: 10.1073/pnas.0603327103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McGovern RE, Fernandes H, Khan AR, Power NP, Crowley PB. Nat. Chem. 2012;4:527. doi: 10.1038/nchem.1342. [DOI] [PubMed] [Google Scholar]
- 55.Rajagopal BS, Edzuma AN, Hough MA, Blundell KLIM, Kagan VE, Kapralov AA, Fraser LA, Butt JN, Silkstone GG, Wilson MT, Svistunenko DA, Worrall JAR. Biochem. J. 2013;456:441. doi: 10.1042/BJ20130758. [DOI] [PubMed] [Google Scholar]
- 56.Cohen DS, Pielak GJ. J. Am. Chem. Soc. 1995;117:1675. [Google Scholar]
- 57.Pletneva EV, Gray HB, Winkler JR. Proc. Natl. Acad. Sci. USA. 2005;102:18397. doi: 10.1073/pnas.0509076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye T, Kaur R, Senguen FT, Michel LV, Bren KL, Elliott SJ. J. Am. Chem. Soc. 2008;130:6682. doi: 10.1021/ja801071n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luntz TL, Schejter A, Garber EAE, Margoliash E. Proc. Natl. Acad. of Sci. USA. 1989;86:3524. doi: 10.1073/pnas.86.10.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berghuis AM, Guillemette JG, Smith M, Brayer GD. J. Mol. Biol. 1994;235:1326. doi: 10.1006/jmbi.1994.1086. [DOI] [PubMed] [Google Scholar]
- 61.Maity H, Maity M, Englander SW. J. Mol. Biol. 2004;343:223. doi: 10.1016/j.jmb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 62.Kumar R, Matsumura H, Lovell S, Yao H, Rodríguez JC, Battaile KP, Moënne-Loccoz P, Rivera M. Biochemistry. 2014;53:2112. doi: 10.1021/bi500030p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith CA, Kortemme T. J. Mol. Biol. 2008;380:742. doi: 10.1016/j.jmb.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lauck F, Smith CA, Friedland GF, Humphris EL, Kortemme T. Nucl. Acids Res. 2010;38:W569. doi: 10.1093/nar/gkq369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang J, Grigoryan G. Meth. Enzymol. 2013;523:21. doi: 10.1016/B978-0-12-394292-0.00002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hassan MI, Bilgrami S, Kumar V, Singh N, Yadav S, Kaur P, Singh TP. J. Mol. Biol. 2008;384:663. doi: 10.1016/j.jmb.2008.09.072. [DOI] [PubMed] [Google Scholar]
- 67.Pinkett HW, Shearwin KE, Stayrook S, Dodd IB, Burr T, Hochschild A, Egan JB, Lewis M. Mol. Cell. 2006;21:605. doi: 10.1016/j.molcel.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 68.Crowley PB, Brett K, Muldoon J. ChemBioChem. 2008;9:685. doi: 10.1002/cbic.200700603. [DOI] [PubMed] [Google Scholar]
- 69.Ness SR, Lo TP, Mauk AG. Israel Journal of Chemistry. 2000;40:21. [Google Scholar]
- 70.Assfalg M, Bertini I, Turano P, Mauk AG, Winkler JR, Gray HB. Biophysical Journal. 2003;84:3917. doi: 10.1016/S0006-3495(03)75119-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Russell BS, Melenkivitz R, Bren KL. Proc. Natl. Acad. Sci. USA. 2000;97:8312. doi: 10.1073/pnas.150239397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hartshorn RT, Moore GR. Biochem. J. 1989;258:599. doi: 10.1042/bj2580595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tonge P, Moore GR, Wharton CW. Biochem. J. 1989;258:599. doi: 10.1042/bj2580599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wallace CJ, Mascagni P, Chait BT, Collawn JF, Paterson Y, Proudfoot AE, Kent SB. J. Biol. Chem. 1989;264:15199. [PubMed] [Google Scholar]
- 75.Rafferty SP, Guillemette JG, Berghuis AM, Smith M, Brayer GD, Mauk AG. Biochemistry. 1996;35:10784. doi: 10.1021/bi960430v. [DOI] [PubMed] [Google Scholar]
- 76.Feinberg BA, Liu X, Ryan MD, Schejter A, Zhang C, Margoliash E. Biochemistry. 1998:13091–13091. doi: 10.1021/bi981037n. [DOI] [PubMed] [Google Scholar]
- 77.Lan W, Wang Z, Yang Z, Zhu J, Ying T, Jiang X, Zhang X, Wu H, Liu M, Tan X, Cao C, Huang ZX. PLoS One. 2011;6:e27219. doi: 10.1371/journal.pone.0027219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bandi S, Baddam S, Bowler BE. Biochemistry. 2007;46:10643. doi: 10.1021/bi700992y. [DOI] [PubMed] [Google Scholar]
- 79.Wood LC, White TB, Ramdas L, Nall BT. Biochemistry. 1988;27:8562. doi: 10.1021/bi00423a009. [DOI] [PubMed] [Google Scholar]
- 80.Nall BT, Zuniga EH, White TB, Wood LC, Ramdas L. Biochemistry. 1989;28:9834. doi: 10.1021/bi00451a043. [DOI] [PubMed] [Google Scholar]
- 81.Speare JO, Rush TSI. Biopolymers. 2003;72:193. doi: 10.1002/bip.10337. [DOI] [PubMed] [Google Scholar]
- 82.Balakrishnan G, Hu Y, Oyerinde OF, Su J, Groves JT, Spiro TG. J. Am. Chem. Soc. 2007;129:504. doi: 10.1021/ja0678727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bellesia G, Shea JE. J. Chem. Phys. 2009;130:145103. doi: 10.1063/1.3108461. [DOI] [PubMed] [Google Scholar]
- 84.Cheng PN, Liu C, Zhao M, Eisenberg D, Nowick JS. Nat. Chem. 2012;4:927. doi: 10.1038/nchem.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hashimoto M, Takeda A, Hsu LJ, Takenouchi T, Masliah E. J. Biol. Chem. 1999;274:28849. doi: 10.1074/jbc.274.41.28849. [DOI] [PubMed] [Google Scholar]
- 86.Hanske J, Toffey JR, Morenz AM, Bonilla AJ, Schiavoni KH, Pletneva EV. Proc. Natl. Acad. Sci. USA. 2012;109:125. doi: 10.1073/pnas.1112312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hong Y, Muenzner J, Grimm SK, Pletneva EV. J. Am. Chem. Soc. 2012;134:18713. doi: 10.1021/ja307426k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sutin N, Yandell JK. J. Biol. Chem. 1972;247:6932. [PubMed] [Google Scholar]
- 89.Diederix RE, Ubbink M, Canters GW. Biochemistry. 2002;41:13067. doi: 10.1021/bi0260841. [DOI] [PubMed] [Google Scholar]
- 90.Takano T, Dickerson RE. Proc. Natl. Acad. Sci. USA. 1980;77:6371. doi: 10.1073/pnas.77.11.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Takano T, Dickerson RE. J. Mol. Biol. 1981;153:95. doi: 10.1016/0022-2836(81)90529-5. [DOI] [PubMed] [Google Scholar]
- 92.Alvarez-Paggi D, Castro MA, Tórtora V, Castro L, Radi R, Murgida DH. J. Am. Chem. Soc. 2013;135:4389. doi: 10.1021/ja311786b. [DOI] [PubMed] [Google Scholar]
- 93.Ying T, Wang ZH, Lin YW, Xie J, Tan X, Huang ZX. Chem. Commun. 2009:4512. doi: 10.1039/b904347k. [DOI] [PubMed] [Google Scholar]
- 94.Snider EJ, Muenzner J, Toffey JR, Hong Y, Pletneva EV. Biochemistry. 2013;52:993. doi: 10.1021/bi301682c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Muenzner J, Pletneva EV. Chem. Phys. Lip. 2014;179:57. doi: 10.1016/j.chemphyslip.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rytömaa M, Kinnunen PKJ. J. Biol. Chem. 1995;270:3197. doi: 10.1074/jbc.270.7.3197. [DOI] [PubMed] [Google Scholar]
- 97.Hickey DR, Berghuis AM, Lafond G, Jaeger JA, Cardillo TS, McLendon D, Das G, Sherman F, Brayer GD, McLendon G. J. Biol. Chem. 1991;266:11686. [PubMed] [Google Scholar]
- 98.Sinibaldi F, Howes BD, Piro MC, Polticelli F, Bombelli C, Ferri T, Coletta M, Smulevich G, Santucci R. J. Biol. Inorg. Chem. 2010;15:689. doi: 10.1007/s00775-010-0636-z. [DOI] [PubMed] [Google Scholar]
- 99.Hwang TL, Shaka AJ. J. Magn. Reson. A. 1995;112:275. [Google Scholar]
- 100.Inubushi T, Becker ED. J. Magn. Reson. 1983;51:128. [Google Scholar]
- 101.Kabsch W. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Murphy ME, Nall BT, Brayer GD. J. Mol. Biol. 1992;227:160. doi: 10.1016/0022-2836(92)90689-h. [DOI] [PubMed] [Google Scholar]
- 103.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McCoy AJ. Acta Crystallogr. D Biol. Crystallogr. 2007;63:32. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Emsley P, Lohkamp B, Scott WG, Cowtan K. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 108.Chen VB, Arendall WBr, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Feng Z, Westbrook J, Berman HM. NUCheck. NDB-407. New Brunswick, NJ: Rutgers University; 1998. [Google Scholar]
- 110.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J. Appl. Cryst. 1993;26:283. [Google Scholar]
- 111.Vaguine AA, Richelle J, Wodak SJ. Acta Crystallogr. D Biol. Crystallogr. 1999;55:191. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 112.DeLano W. Palo Alto, CA: DeLano Scientific LLC; 2008. [Google Scholar]
- 113.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. Nucleic Acids Res. 2006;34:W116. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jeuken LJC, McEvoy JP, Armstrong FA. J. Phys. Chem. B. 2002;106:2304. [Google Scholar]
- 115.Vincelli AJ, Pottinger DS, Zhong F, Hanske J, Rolland SG, Conradt B, Pletneva EV. Biochemistry. 2013;52:653. doi: 10.1021/bi3014938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Baldwin DA, Helder MM, Pratt JM. J. Inorg. Biochem. 1987;30:203. doi: 10.1016/0162-0134(87)80064-8. [DOI] [PubMed] [Google Scholar]
- 117.Wang Z, Matsuo T, Nagao S, Hirota S. Org. Biomol. Chem. 2011;9:4766. doi: 10.1039/c1ob05552f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.