ABSTRACT
Uncovering the genetic and molecular basis of barriers to gene flow between populations is key to understanding how new species are born. Intrinsic postzygotic reproductive barriers such as hybrid sterility and hybrid inviability are caused by deleterious genetic interactions known as hybrid incompatibilities. The difficulty in identifying these hybrid incompatibility genes remains a rate-limiting step in our understanding of the molecular basis of speciation. We recently described how whole genome sequencing can be applied to identify hybrid incompatibility genes, even from genetically terminal hybrids. Using this approach, we discovered a new hybrid incompatibility gene, gfzf, between Drosophila melanogaster and Drosophila simulans, and found that it plays an essential role in cell cycle regulation. Here, we discuss the history of the hunt for incompatibility genes between these species, discuss the molecular roles of gfzf in cell cycle regulation, and explore how intragenomic conflict drives the evolution of fundamental cellular mechanisms that lead to the developmental arrest of hybrids.
KEYWORDS: cell cycle, Drosophila, genomic conflict, hybrid incompatibility, speciation
Incomplete incompatibilities
A key step in the origins of new species is the evolution of barriers to gene flow between previously interbreeding populations. Intrinsic postzygotic barriers—such as hybrid sterility or hybrid inviability—are caused by deleterious genetic interactions known as hybrid incompatibilities. While we have a strong theoretical framework to describe how such hybrid incompatibilities may arise, many empirical aspects of the molecular and evolutionary basis of hybrid incompatibilities remain poorly understood.1,2 For a comprehensive understanding of how reproductive barriers evolve, we need to identify the genes that cause hybrid incompatibilities, dissect the molecular basis for hybrid dysfunction, and understand the biological forces that drive changes in the cellular machinery that lead components to become incompatible.3 Addressing these aspects of hybrid incompatibilities across many species may not only reveal whether particular genes and pathways repeatedly play a role in speciation, but will also provide a unique view into the evolution of fundamental developmental processes. We still, however, lack a single case where the genetic, molecular and evolutionary causes of hybrid incompatibilities are fully understood. The single most important bottleneck in speciation research responsible for this gap in our understanding is the difficulty in identifying hybrid incompatibility genes. Even in the case of one of the long studied genetic model systems—Drosophila melanogaster—understanding the nature of hybrid incompatibilities has proven to be one of the longest standing and difficult quandaries in evolutionary genetics.4,5
The quest to understand the nature of hybrid incompatibilities using Drosophila started after Quackenbush, a master's student in T.H. Morgan's fly laboratory, reported his surprising observation of unisexual broods in experimental fly crosses.6 A. H. Sturtevant later showed these skewed progeny ratios were a result of hybrid inviability in crosses between D. melanogaster and its closest sister species, D. simulans.7 When D. melanogaster females are crossed to D. simulans males, they produce only sterile hybrid F1 females; hybrid F1 males from this cross die as late larvae and never develop into adults.
Sturtevant's attempts to describe the genetic basis of hybrid F1 male inviability in crosses between D. melanogaster females and D. simulans males were thwarted by the complete sterility or inviability of all hybrids. Schultz and Dobzhansky attempted to dissect the genetic basis of this hybrid male inviability by crossing triploid D. melanogaster females with D. simulans males, but even these crosses produced only sterile or dead hybrid offspring.8 A major leap in our understanding of the genetic architecture of hybrid incompatibilities between D. melanogaster females and D. simulans came from a series of X-ray experiments by H.J. Muller and G. Pontecorvo.9,10 In a seminal experiment, they crossed triploid D. melanogaster females to heavily irradiated D. simulans males to generate “partial-hybrid” progeny. By tracking the marked D. melanogaster chromosomes in these partial-hybrid progeny, they determined that the D. melanogaster X, D. simulans 2nd, and D. simulans 3rd chromosomes were simultaneously required to cause hybrid male lethality.10 The realization that epistatic interactions between genes from both species contribute to the incompatibility in hybrids fulfills a primary theoretical prediction for the evolution of isolating genes.
Despite Pontecorvo's revelation of the genetic architecture of hybrid male inviability between D. melanogaster and D. simulans, the problem of identifying the causal genes still remained out of reach for several decades. The sterility or inviability of all hybrids between these species provided an insurmountable barrier to approaches that rely on recombination or deletion mapping. This stalemate was broken by the discovery of strains that could produce viable hybrid F1 males. Naturally occurring strains in D. simulans were identified that, when crossed with D. melanogaster males, produced viable hybrid F1 males.11 These D. simulans strains were named Lethal hybrid rescue (Lhr) for their hybrid rescue effect. Similarly, D. melanogaster strains that produce viable hybrid F1 males in crosses with D. simulans males were isolated; these were named Hybrid male rescue (Hmr).12 The discovery of these naturally occurring rescue strains opened the door to the application of classic genetics approaches to identify the causal genes. Deletion mapping and transgenic experiments in D. melanogaster proved that a single X-linked gene that encodes a DNA binding protein at the Hmr locus was responsible for male rescue.13 The discovery of Lhr relied on the insight that it was likely to be rapidly evolving, and Lhr was shown to be a member of the heterochromatin protein family present on the D. simulans 2nd chromosome.14 Neither Hmr nor Lhr are essential for viability in pure species, and the rescue effects of each are due to loss of function alleles of Hmr and Lhr. Together, the discovery of these mutations that can single handedly reverse the lethal hybrid incompatibility between substantially diverged species represents some of the biggest breakthroughs in speciation genetics.
While Hmrmel and Lhrsim have been confirmed as the X and 2nd chromosome incompatibility genes, transgenic expression of Lhrsim in D. melanogaster is insufficient to cause male lethality.14 Together with Pontecorvo's findings, these results suggest that Hmrmel and Lhrsim are insufficient to cause hybrid male lethality; at least one more hybrid incompatibility gene may also be required. Studies to understand the normal function of Hmr and Lhr suggest that they are repressors of centromeric or pericentric heterochromatin associated repetitive sequences and transposable elements.15-17 It still, however, remains unclear how their function relates to the developmental arrest in male hybrids. With a gap in our genetic and molecular understanding of the system, we set out to devise a method to find the missing third hybrid incompatibility gene predicted by Pontecorvo's experiments to reside on the D. simulans 3rd chromosome.18
A genomic screen for hybrid rescue
When D. melanogaster females are crossed to D. simulans males carrying a null allele at Lhr, viable hybrid F1 males are produced. Since at least three loci, including Hmrmel and Lhrsim, are required to kill hybrid F1 males, we reasoned that a null allele of the hybrid incompatibility gene on the D. simulans third chromosome would also rescue male hybrids. In the absence of naturally occurring rescue alleles that correspond to this missing hybrid incompatibility gene – strains that made the discovery of Hmr and Lhr possible – the identification of this missing hybrid incompatibility gene proved impervious to existing genetic approaches. To sidestep these traditional barriers, we designed a screen for mutations in D. simulans that would break the hybrid incompatibility and result in viable hybrid F1 males. Because efficient balancer chromosomes are unavailable in D. simulans, it is not possible to maintain a large collection of mutagenized chromosomes. Instead, we fed D. simulans males the mutagen ethyl methane sulfonate (EMS) and crossed these males to D. melanogaster females. If a D. simulans sperm that carries a null mutation at the third incompatibility gene fertilizes a D. melanogaster egg, the resulting hybrid male is predicted to be viable. Though screening for viable hybrid F1 males by this method is straightforward, the resulting hybrid males are still sterile. Generating stable mapping strains using such a male is, therefore, not feasible.
We isolated six independent bona fide rescue hybrid F1 males. To identify all new mutations the D. simulans complement of their hybrid genomes, we obtained the whole genome sequences of each individual rescued hybrid male. These mutations were scattered at random across the genome, and any single hybrid male carried mutations at hundreds of genes. To isolate the causal rescue mutation from the haystack of mutations in each rescued hybrid male, we focused on the common set of genes disrupted in independently rescued hybrid F1 males. We reasoned that viable hybrid F1 males that are the result of a common mechanism of rescue would have disruptions in a common gene or handful of genes.
All six rescued hybrid males had exactly one commonly disrupted gene, GST-containing FLYWCH zinc-finger protein (gfzf) / Suppressor of Killer-of-prune (Su(Kpn)).19,20 Each male carried a unique mutation at the D. simulans allele of gfzf (gfzfsim), including deletion, missense, nonsense and frameshift mutations. Curiously, we did not isolate any males with mutations in Lhr, suggesting that the screen was not carried to saturation. Because gfzf has a much larger coding sequence than Lhr, our failure to isolate a mutation in Lhr may also be explained by the difference in the mutational target sizes. Regardless, our suite of mutations gave us a strong candidate gene in the form of gfzf.
Our results predicted that removing or reducing the expression gfzfsim in hybrid F1 males should result in a rescue their viability. Because gfzf is a viability essential gene, and no D. simulans gfzf mutants are available, we resorted to using RNA interference to test this prediction. We designed RNAi knockdown constructs that only target the gfzfsim allele. We generated transgenic D. melanogaster females that carried these gfzfsim knockdown constructs and crossed them to D. simulans males. Knockdown of gfzfsim consistently produced a robust rescue of the viability of hybrid F1 males, showing that gfzf is indeed the missing hybrid incompatibility gene predicted by Pontecorvo to exist on the D. simulans 3rd chromosome.18
Molecular function of gfzf
While the exact molecular role of gfzf remains unclear, it appears to play an essential role in cell cycle regulation in Drosophila. A genome-wide RNAi screen to identify G2/M checkpoint genes in D. melanogaster S2 cells identified gfzf as a significant player.21 gfzf has also been shown to play a role in blocking cell proliferation by potentiating the dE2F2/ RBF pathway.22 Other evidence points to a role as a positive regulator of cell proliferation. The Ras pathway is regulated transcriptionally by gfzf and gain of function Ras phenotypes are suppressed by loss-of-function alleles at gfzf.23 It is possible that the interaction of gfzf with the cell cycle is dependent on the context of its interactions with other genes not yet identified. Together, these results suggest an important role for gfzf in regulating the cell cycle in Drosophila and provide some insight into the developmental arrest of hybrid F1 males between D. melanogaster and D. simulans.
Could the role of gfzf in cell cycle regulation directly cause hybrid male inviability in D. melanogaster and D. simulans hybrids? A clue is provided by the observation that hybrid F1 males display cell cycle progression defects, degenerated imaginal discs and die at the late larval stage.24 This is interesting because cell cycle checkpoint activation due to genetic or environmental insults such as X-ray irradiation also causes ablation of the imaginal discs and subsequent lethality at the late larval stage.25 Why are imaginal disc cells particularly susceptible to DNA damage? Drosophila larvae mostly consist of polyploid cells with the exception of imaginal discs and the nervous system, which are diploid. The polyploid larval cells grow by increasing cell size rather than through cell division. In contrast, the diploid imaginal disc cells are under tight cell cycle regulation making them particularly sensitive to insults that activate the cell cycle checkpoint.
We reasoned that the dominant effect of the D. simulans homolog of gfzf is to suppress the proliferation of the imaginal discs, and thereby prevent the larvae from reaching critical mass and initiating the first stages of pupation. To test if gfzfsim knockdown rescued cell proliferation, we examined the growth of hybrid larval brains. These tissues have a well characterized wave of proliferating S-phase cells which is lost in the hybrid males.15 Consistent with our prediction, knockdown of gfzf partially restores cell proliferation in the brains of hybrid F1 male larvae. To further test this hypothesis, we drove the knockdown of gfzfsim in imaginal discs alone and assayed for the rescue of male hybrids. Confining knockdown of gfzfsim to imaginal discs was sufficient to rescue the viability of hybrid F1 males, suggesting that cell cycle defects in the imaginal discs of hybrid males explain the molecular developmental basis of hybrid F1 male lethality.
Suppressor of killer of prune
Interestingly, gfzf also plays an essential role in a different within-species dominant lethal incompatibility. Sturtevant discovered that when prune mutant D. melanogaster females are crossed to males from certain wild type D. melanogaster strains called “Killer-of-prune,” the resulting sons are inviable.26 prune is an X-linked eye color gene and encodes a phosphodiesterase.27 Killer of prune (Kpn) is a single non-synonymous change in the gene abnormal wing discs (awd).28 awd is the Drosophila homolog of the metastasis suppressor gene Nm23, and encodes a nucleoside diphosphate (NDP) kinase.29 Although the Killer of prune allele of awd (awdKpn) substantially reduces its NDP kinase stability, individuals homozygous for this allele are viable and have no observable phenotypic consequence.30,31 The dominant lethal activity of Kpn is seen only in combination with the prune mutation, thus behaving as a within species incompatibility. In a comprehensive genetic screen to isolate suppressors of this lethal interaction, 13 mutants were isolated that rescued the viability of prune-Kpn individuals.20 All of these mutations mapped to gfzf, showing that this gene is an essential third component for this dominant lethal incompatibility (Fig. 1). gfzf is, therefore, also known as Suppressor of Killer of prune (Su(Kpn)). The essential role of a single gene in dominant incompatibilities both within species and between species suggests that there may be limited genetic paths for the evolution of dominant lethal interactions.
Figure 1.
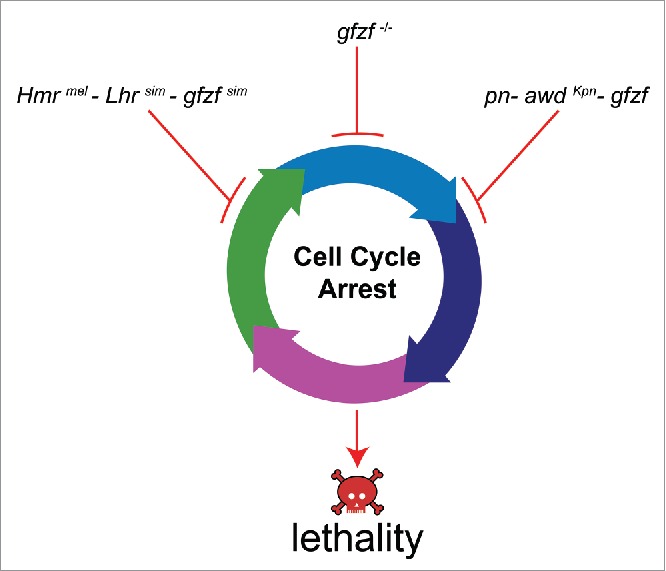
gfzf activity causes an arrest in cell cycle progression. The pn-Kpn-gfzf system and Hmr-Lhr-gfzf hybrid incompatibility both cause an arrest in cell cycle progression similar to gfzf homozygous null mutations. This leads to a developmental arrest and eventual death of larvae. Together, these forms of lethality highlight the surprising role of cell cycle regulation in dominant incompatibilities.
Moving forward
gfzf carries four FLYWCH zinc finger domains that are unrelated by homology to any other gene in the Drosophila genome, and a Glutathione-S-Transferase (GST) domain. The only other protein in Drosophila with FLYWCH domains is the chromatin regulator gene mod(mdg4).32 Though FLYWCH domains are rare in Drosophila, they appear to be present in all metazoans. This could be due to genomic deposition via several classes of transposable elements that contain DNA binding domains with the FLYWCH motif.33,34 An interesting possibility is that these transposable elements were co-opted by Drosophila for gene regulation, similar to the situation in C. elegans, where they regulate gene expression via repression of miRNAs.35 Combined with the evidence that gfzf regulates the abundance of mek transcripts, it is also possible that gfzf is a transcriptional regulator of the cell cycle via miRNA repression.23 gfzf, thus, plays an essential role in cell cycle regulation, but its precise molecular function remains unclear. Our active line of inquiry is to determine how gfzf interacts with the cell cycle and to understand how this function is related to the arrest in proliferation of imaginal discs in hybrids F1 male larvae. Interestingly, only mutations in the D. simulans allele of gfzf rescue hybrid F1 male viability; mutations in the D. melanogaster allele do not produce any hybrid rescue. These results suggest functional differences in the properties of gfzfsim and gfzfmel in hybrids. Our ongoing experiments with chimeric constructs will help identify the causal genetic changes between gfzfmel and gfzfsim that lead to the lethal incompatibility.
With the identities of three hybrid incompatibility genes in hand, we can now begin to formulate hypotheses about how they interact together to cause hybrid male inviability. Proteomic studies for genes that interact with Hmr and Lhr indicate that while they physically interact with each other, there is little evidence to suggest a direct physical interaction with gfzf.14,16 Loss of function mutations in either Hmr and Lhr do not directly affect the cell cycle in Drosophila,13,14 but knockdown of either of these genes using RNAi in S2 cells slows cell progression due to lagging chromosomes during mitosis.16 Other evidence suggests that Hmr and Lhr are repressors of transposable element activity.16,36 In contrast to Hmr and Lhr, all evidence regarding gfzf points to a much more direct role in the cell cycle.21-23 Given the lack of physical interaction between Hmr/Lhr and gfzf, it is possible that an incompatible interaction between Hmr and Lhr causes a perturbation in hybrids that subsequently affects the lethal activity of gfzf.
From a genetic perspective, it is unclear whether Hmrmel, Lhrsim and gfzfsim fully describe the hybrid incompatibility that kills hybrid F1 males. A genetic screen using autosomal deficiencies did not identify any large effect hybrid lethality factors in D. melanogaster.37 Our genetic screen in D. simulans did not isolate mutations in Lhrsim, suggesting that this screen was not saturated. We, therefore, cannot formally rule out the contribution of more hybrid incompatibility genes by D. simulans. If the transgenic introduction of Lhrsim and gfzfsim in D. melanogaster flies produces a lethal phenotype, this would show that the complete set of D. simulans hybrid incompatibility genes responsible for hybrid F1 male lethality has now been identified. Alternatively, if these three genes prove insufficient to reconstitute the hybrid incompatibility, this would suggest the existence of more genes that are essential in the dominant epistatic interaction. In this case, our screen may be modified and carried to saturation to identify these missing partners. Yet another possibility is that a sensitized hybrid genetic background may be required for Hmr, Lhr, and gfzf to cause hybrid lethality.14 For example, scattered heterochromatin interactions17 or de-repression of transposable elements may also be essential for hybrid lethality.16,36 Under this scenario, the hybrid incompatibility genes are still required to cause hybrid lethality, but these genes may be responding to many other changes in the hybrid background.
Understanding the particular biological forces that drive the evolution of hybrid incompatibilities is one of the most critical functions of evolutionary genetics. An increasing amount of evidence points toward the role of intragenomic conflict involving selfish genetic elements as a driving force in the evolution of hybrid incompatibilities (Fig. 2).38,39 In the case of D. melanogaster and D. simulans, the divergence of Hmr, Lhr and gfzf may have been driven by an arms race with selfish genetic elements such as transposable elements or satellite sequences, resulting in the evolution of hybrid male lethality between these species. The discovery of more hybrid incompatibility genes and a deeper understanding of the molecular and evolutionary aspects are necessary for a clearer view of the process of speciation. Our genomics approach may be readily modified to identify hybrid incompatibility genes in other model and non-model species where traditional approaches fall short. An acceleration in the identification of hybrid incompatibility genes may not only provide unique insights into the evolution of fundamental cellular processes such as cell cycle regulation, but may also illuminate how new species are born; a process that Darwin famously called “the mystery of mysteries.”
Figure 2.
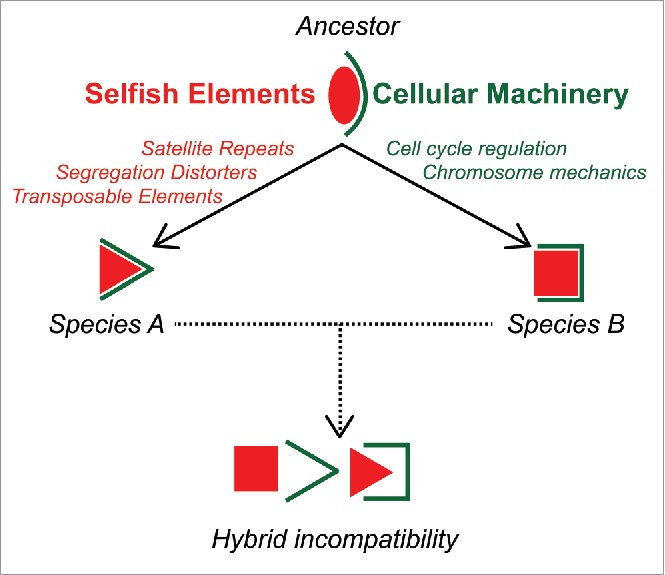
Intragenomic arms races between the selfish genetic elements and the cellular machinery drive the evolution of hybrid incompatibilities. Interactions between selfish elements and host genomes co-evolve as selection favors selfish elements that can evade host defenses. This in turn triggers an evolutionary response favoring host variants that can defend themselves from selfish elements. The genes that are at the interface of these conflicts are predicted to diverge rapidly under selection, and can lead them to become incompatible between species.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Chris Leonard, Harmit S. Malik, Maulik Patel and two anonymous reviewers for helpful comments with this manuscript.
Funding
We are supported by an NIH Developmental Biology Training Grant 5T32 HD0741 (JCC), NIH- R01 GM115914 (NP) and a Mario Capecchi endowed assistant professorship (NP).
References
- [1].Dobzhansky T. Genetics and the Origin of Species. New York: Columbia University Press; 1937. [Google Scholar]
- [2].Muller HJ. Isolating mechanisms, evolution and temperature. Biol Symp 1942; 6:71-125. [Google Scholar]
- [3].Coyne J, Orr HA. Speciation. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- [4].Provine WB. Alfred Henry Sturtevant and crosses between Drosophila melanogaster and Drosophila simulans. Genetics 1991; 129:1-5; PMID:1936952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Barbash DA. Ninety Years of Drosophila melanogaster Hybrids. Genetics 2010; 186:1-8; PMID:20855573; http://dx.doi.org/ 10.1534/genetics.110.121459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Quackenbush LS. Unisexual Broods of Drosophila. Science 1910; 32:183-5; PMID:17839276; http://dx.doi.org/ 10.1126/science.32.814.183 [DOI] [PubMed] [Google Scholar]
- [7].Sturtevant AH. A New Species Closely Resembling Drosophila Melanogaster. Psyche J Entomol 1919; 26:153-5; http://dx.doi.org/ 10.1155/1919/97402 [DOI] [Google Scholar]
- [8].Schultz J, Dobzhansky T. Triploid hybrids between Drosophila melanogaster and Drosophila simulans. J Exp Zool 1933; 65:73-82; http://dx.doi.org/ 10.1002/jez.1400650105 [DOI] [Google Scholar]
- [9].Muller HJ, Pontecorvo G. Recombinants between Drosophila Species the F1 Hybrids of which are Sterile [Internet]. 1940. [cited 2016 Mar 10]; Available from: http://www.nature.com/nature/journal/v146/n3693/abs/146199b0.html [Google Scholar]
- [10].Pontecorvo G. Viability interactions between chromosomes ofDrosophila melanogaster and Drosophila simulans. J Genet 1943; 45:51-66; http://dx.doi.org/ 10.1007/BF02982774 [DOI] [Google Scholar]
- [11].Watanabe TK. A gene that rescues the lethal hybrids between. Drosophila melanogaster and D. simulans. 遺伝學雑誌 1979; 54:325-31.228849 [Google Scholar]
- [12].Hutter P, Ashburner M. Genetic rescue of inviable hybrids between Drosophila melanogaster and its sibling species. Nature 1987; 327:331-3; PMID:3108667; http://dx.doi.org/ 10.1038/327331a0 [DOI] [PubMed] [Google Scholar]
- [13].Barbash DA, Siino DF, Tarone AM, Roote J. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc Natl Acad Sci 2003; 100:5302-7; PMID:12695567; http://dx.doi.org/ 10.1073/pnas.0836927100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA. Two Dobzhansky-Muller genes interact to cause hybrid lethality in drosophila. Science 2006; 314:1292-5; PMID:17124320; http://dx.doi.org/ 10.1126/science.1133953 [DOI] [PubMed] [Google Scholar]
- [15].Bolkan BJ, Booker R, Goldberg ML, Barbash DA. Developmental and cell cycle progression defects in drosophila hybrid males. Genetics 2007; 177:2233-41; PMID:17947412; http://dx.doi.org/ 10.1534/genetics.107.079939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Thomae AW, Schade GOM, Padeken J, Borath M, Vetter I, Kremmer E, Heun P, Imhof A. A pair of centromeric proteins mediates reproductive isolation in drosophila species. Dev Cell 2013; 27:412-24; PMID:24239514; http://dx.doi.org/ 10.1016/j.devcel.2013.10.001 [DOI] [PubMed] [Google Scholar]
- [17].Ferree PM, Gomez K, Rominger P, Howard D, Kornfeld H, Barbash DA. Heterochromatin position effects on circularized sex chromosomes cause filicidal embryonic lethality in Drosophila melanogaster. Genetics 2014; 196:1001-5; PMID:24478337; http://dx.doi.org/ 10.1534/genetics.113.161075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Phadnis N, Baker EP, Cooper JC, Frizzell KA, Hsieh E, Cruz AFA de la, Shendure J, Kitzman JO, Malik HS. An essential cell cycle regulation gene causes hybrid inviability in Drosophila. Science 2015; 350:1552-5; PMID:26680200; http://dx.doi.org/ 10.1126/science.aac7504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dai MS, Sun XX, Qin J, Smolik SM, Lu H. Identification and characterization of a novel Drosophila melanogaster glutathione S-transferase-containing FLYWCH zinc finger protein. Gene 2004; 342:49-56; PMID:15527965; http://dx.doi.org/ 10.1016/j.gene.2004.07.043 [DOI] [PubMed] [Google Scholar]
- [20].Provost E, Hersperger G, Timmons L, Ho WQ, Hersperger E, Alcazar R, Shearn A. Loss-of-Function Mutations in a Glutathione S-Transferase Suppress the prune-Killer of prune Lethal Interaction. Genetics 2006; 172:207-19; PMID:16143620; http://dx.doi.org/ 10.1534/genetics.105.044669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kondo S, Perrimon N. A Genome-Wide RNAi Screen Identifies Core Components of the G2-M DNA Damage Checkpoint. Sci Signal 2011; 4:rs1-rs1; PMID:21205937; http://dx.doi.org/ 10.1126/scisignal.2001350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ambrus AM, Rasheva VI, Nicolay BN, Frolov MV. Mosaic Genetic Screen for Suppressors of the de2f1 Mutant Phenotype in Drosophila. Genetics 2009; 183:79-92; PMID:19546319; http://dx.doi.org/ 10.1534/genetics.109.104661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ashton-Beaucage D, Udell CM, Gendron P, Sahmi M, Lefrançois M, Baril C, Guenier AS, Duchaine J, Lamarre D, Lemieux S, et al.. A functional screen reveals an extensive layer of transcriptional and splicing control underlying RAS/MAPK signaling in drosophila. PLoS Biol 2014; 12:e1001809; PMID:24643257; http://dx.doi.org/ 10.1371/journal.pbio.1001809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sánchez L, Dübendorfer A. Development of imaginal discs from lethal hybrids betweenDrosophila melanogaster andDrosophila mauritiana. Wilhelm Rouxs Arch Dev Biol 1983; 192:48-50; http://dx.doi.org/ 10.1007/BF00848770 [DOI] [PubMed] [Google Scholar]
- [25].Gatti M, Baker BS. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev 1989; 3:438-53; PMID:2498166; http://dx.doi.org/ 10.1101/gad.3.4.438 [DOI] [PubMed] [Google Scholar]
- [26].Sturtevant AH. A Highly Specific Complementary Lethal System in Drosophila Melanogaster. Genetics 1956; 41:118-23; PMID:17247604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Teng DHF, Engele CM, Venkatesh TR. A product of the prune locus of Drosophila is similar to mammalian GTPase-activating protein. Nature 1991; 353:437-40; PMID:1654526; http://dx.doi.org/ 10.1038/353437a0 [DOI] [PubMed] [Google Scholar]
- [28].Biggs J, Tripoulas N, Hersperger E, Dearolf C, Shearn A. Analysis of the lethal interaction between the prune and Killer of prune mutations of Drosophila. Genes Dev 1988; 2:1333-43; PMID:2849580; http://dx.doi.org/ 10.1101/gad.2.10.1333 [DOI] [PubMed] [Google Scholar]
- [29].Biggs J, Hersperger E, Steeg PS, Liotta LA, Shearn A. A Drosophila gene that is homologous to a mammalian gene associated with tumor metastasis codes for a nucleoside diphosphate kinase. Cell 1990; 63:933-40; PMID:2175255; http://dx.doi.org/ 10.1016/0092-8674(90)90496-2 [DOI] [PubMed] [Google Scholar]
- [30].Dearolf CR, Hersperger E, Shearn A. Developmental consequences of awdb3, a cell-autonomous lethal mutation of Drosophila induced by hybrid dysgenesis. Dev Biol 1988; 129:159-68; PMID:3137111; http://dx.doi.org/ 10.1016/0012-1606(88)90170-4 [DOI] [PubMed] [Google Scholar]
- [31].Lascu I, Chaffotte A, Limbourg-Bouchon B, Véron M. A Pro/Ser substitution in nucleoside diphosphate kinase of Drosophila melanogaster (mutation killer of prune) affects stability but not catalytic efficiency of the enzyme. J Biol Chem 1992; 267:12775-81; PMID:1320004 [PubMed] [Google Scholar]
- [32].Thomas SE, Soltani-Bejnood M, Roth P, Dorn R, Logsdon JM Jr, McKee BD. Identification of two proteins required for conjunction and regular segregation of achiasmate homologs in Drosophila male meiosis. Cell 2005; 123:555-68; PMID:16286005; http://dx.doi.org/ 10.1016/j.cell.2005.08.043 [DOI] [PubMed] [Google Scholar]
- [33].Babu MM, Iyer LM, Balaji S, Aravind L. The natural history of the WRKY–GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res 2006; 34:6505-20; PMID:17130173; http://dx.doi.org/ 10.1093/nar/gkl888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Marquez CP, Pritham EJ. Phantom, a new subclass of Mutator DNA transposons found in insect viruses and widely distributed in animals. Genetics 2010; 185:1507-17; PMID:20457878; http://dx.doi.org/ 10.1534/genetics.110.116673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ow MC, Martinez NJ, Olsen PH, Silverman HS, Barrasa MI, Conradt B, Walhout AJM, Ambros V. The FLYWCH transcription factors FLH-1, FLH-2, and FLH-3 repress embryonic expression of microRNA genes in C. elegans. Genes Dev 2008; 22:2520-34; PMID:18794349; http://dx.doi.org/ 10.1101/gad.1678808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Satyaki PRV, Cuykendall TN, Wei KHC, Brideau NJ, Kwak H, Aruna S, Ferree PM, Ji S, Barbash DA. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet 2014; 10:e1004240; PMID:24651406; http://dx.doi.org/ 10.1371/journal.pgen.1004240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cuykendall TN, Satyaki P, Ji S, Clay DM, Edelman NB, Kimchy A, Li LH, Nuzzo EA, Parekh N, Park S, et al.. A Screen for F1 Hybrid male rescue reveals no major-effect hybrid lethality Loci in the Drosophila melanogaster autosomal genome. G3 Genes Genomes Genetics 2014; 4:2451-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tao Y, Hartl DL, Laurie CC. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc Natl Acad Sci 2001; 98:13183-8; PMID:11687638; http://dx.doi.org/ 10.1073/pnas.231478798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Phadnis N, Orr HA. A Single Gene Causes Both Male Sterility and Segregation Distortion in Drosophila Hybrids. Science 2009; 323:376-9; PMID:19074311; http://dx.doi.org/ 10.1126/science.1163934 [DOI] [PMC free article] [PubMed] [Google Scholar]


