Abstract
Abstract Abundances of virus-like particles (VLPs, mostly bacteriophages) are high in aquatic environments; therefore, techniques for precise enumeration are essential in ecological monitoring. VLPs were determined after staining with SYBR Gold by conventional epifluorescence microscopy and compared to enumerations performed by confocal laser scanning microscopy (CLSM). In order to assess the potential of CLSM for viral direct counts (VDCs), we processed samples from different freshwater and marine systems. Optical sectioning by CLSM and production of an overlay picture of multiple scans enables the often uneven whole investigated filter area to be brought to the plane of focus. This allows for subsequent image analysis of digitally created high-quality images. Another advantage using the CLSM was that the short spot excitation of the stain via laser beam minimized fading of the stain. The VDC results show that there is no significant difference between the two methods. Regarding the known difficulties of viral abundance estimates on particulate material, CLSM was further applied to enumerate VLPs on a small set of marine transparent exopolymeric particles sampled from the Atlantic Ocean. Our data suggest that CLSM is a useful tool to count viruses in water samples as well as attached to certain types of aquatic aggregates.
Keywords: Viruses, Confocal laser scanning microscopy, Epifluorescence microscopy, SYBR Gold, Microaggregates
Introduction
Following the first reports on the high numbers of virus-like particles (VLPs) in the aquatic environment, much research was done to elucidate and quantify their role in ecological as well as biogeochemical processes in marine and freshwater systems. Viruses are a dynamic member of the microbial community by influencing host abundance, cycling nutrients, and carbon (“the viral shunt”; Wilhelm and Suttle 1999) and affecting the diversity of their host populations (Weinbauer 2004; Peduzzi and Luef 2009). Thus, the development of accurate methods for detecting and enumerating viruses is an important issue in aquatic microbial ecology: precise determination of virus abundance is essential to evaluate their distribution and role in various systems. Many methods, for example assessing viral production and the impact of viruses on bacterial mortality, depend on reliably determining total viral numbers (Luef et al. 2009a).
While transmission electron microscopy (TEM) remains important in estimating virus production parameters such as burst size and frequency of visibly infected cells (Weinbauer et al. 2002; Weinbauer 2004; Peduzzi and Luef 2009), epifluorescence microscopy (EFM) is preferred for virus enumeration. Another important approach is using flow cytometry (FCM), which also makes staining with fluorochromes necessary (Marie et al. 1999; Brussard et al. 2000; Duhamel and Jacquet 2006). FCM requires expensive equipment and expertise in handling the instrument, although high volumes/numbers of samples can be processed without any concentration procedure (Peduzzi and Luef 2009). Several studies have investigated the applicability of nucleic acid stains such as DAPI, Yo-Pro-1, SYBR Green I and II, and SYBR Gold (Proctor and Fuhrman 1992; Hennes and Suttle 1995; Weinbauer and Suttle 1997; Noble and Fuhrman 1998; Weinbauer et al. 1998; Bettarel et al. 2000). They showed that EFM reliably estimates viral abundances. Compared to the TEM method, the EFM approach yielded similar counts at viral abundances <106 ml−1 and even higher counts at numbers >106 ml−1 (Hennes and Suttle 1995; Hara et al. 1996; Weinbauer and Suttle 1997; Noble and Fuhrman 1998; Bettarel et al. 2000). Furthermore, optical microscopy such as EFM is less elaborate and cheaper than TEM. At high virus concentrations, however, the relatively quick fading of the fluorescent dyes might hamper precise enumeration (Bettarel et al. 2000; Chen et al. 2001).
Although the fluorescent yield of the new class of fluorescent dyes (e.g., Yo-Pro-1, SYBR Green I and II, and SYBR Gold) upon binding to nucleic acids is very high and fading can be reduced by appropriate mounting solutions, most of the stains are not tapped to their full potential concerning excitation and emission characteristics. During the last years, more sophisticated microscopic systems such as confocal laser scanning microscopy (CLSM) entered the field of microbial ecology. This provided more exact excitation of dyes via laser beams along with continuously adjustable bandwidths for a more precise detection by the photomultiplier. Moreover, due to digital imaging and the possibility of scanning samples in the z-direction, this technology allows for investigating three-dimensional objects, e.g., biofilms and aggregates in aquatic environments (Holloway and Cowen 1997; Lawrence and Neu 1999; Neu 2000; Battin et al. 2003; Luef et al. 2009b). For example, the spatial and temporal distribution of microbial consortia can now be studied in biofilms and aggregates in more detail (Luef et al. 2009c). Organic aggregates and biofilms in aquatic systems constitute an important component in the turnover and decomposition of organic substances, thus contributing substantially to energy and nutrient cycling (Simon et al. 2002; Battin et al. 2003; Weinbauer et al. 2009).
Using CLSM for routine detection and monitoring of natural viral assemblages is not yet well-established. Viral presence on aggregates was demonstrated by Peduzzi and Weinbauer (1993) and Simon et al. (2002) using TEM and EFM, respectively, as well as by Luef et al. (2009c) applying CLSM. The TEM approach has the disadvantage that it is tedious and that aggregates often shrink during preparation, whereas the EFM approach faces the problem that it is difficult to identify viruses within or on the opposite side of an organic matrix. This study was designed to: (1) assess the potential of CLSM for routinely counting viruses and (2) evaluate this technique for assessing natural virus abundance on aquatic transparent exopolymeric particles.
Material and methods
Study sites and sample collection
Water samples were taken from surface marine and freshwater environments, which differed strongly in productivity and general ecological conditions (Table 1). Furthermore, one sample was collected by DSV Alvin from a Riftia field within the deep-sea waters of the Pacific Ocean in about 2,500 m depth (Alvin# 3730, Riftia Field coordinates 9° N, 50.705, 104° W 17.59; cruise name “Holidays at Sea”; chief scientist C.R. Fisher, The Pennsylvania State University, USA). Samples were preserved in formaldehyde (4 % final concentration) immediately after collection and stored at 4 °C in the dark. They were processed as soon as possible (between 24 and 96 h) and always by both methods at the same time for one particular sample (i.e., a potential storage effect was always the same for both approaches). Marine aggregate samples were taken from surface waters of the Atlantic Ocean near Tenerife (Spain).
Table 1.
Viral direct counts (×107) obtained by conventional epifluorescence microscopy (EFM) and confocal laser scanning microscopy (CLSM)
| Sampling sites | EFM |
CLSM |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| BDC (106 ml−1) | VDC (107 ml−1) | SD (107) | CoVar | VBR | VDC (107 ml−1) | SD×107 | CoVar | EFM/CLSM | |
| Freshwater environments | |||||||||
| Dorfteich (Riegersburg, Austria) | 1.78 | 9.53 | 0.71 | 7.4 | 53.54 | 9.70 | 1.10 | 11.3 | 0.98 |
| Regelsbrunn b (Danube, Austria) | 2.10 | 4.16 | 0.42 | 10.2 | 19.81 | 3.45 | 0.17 | 4.9 | 1.21 |
| Nile (Cairo, Egypt) | 5.21 | 7.42 | 0.24 | 3.2 | 14.24 | 7.75 | 0.52 | 6.7 | 0.96 |
| Lake Victoria (Kenya, Africa) | 13.72 | 9.68 | 1.58 | 16.4 | 7.06 | 6.95 | 0.91 | 13.1 | 1.39 |
| Lake Hakone (Tokyo, Japan) | 2.24 | 3.15 | 0.06 | 1.9 | 14.06 | 3.20 | 0.37 | 11.7 | 0.99 |
| Wadi Hatta | 1.12 | 0.74 | 0.07 | 8.8 | 6.61 | 0.89 | 0.02 | 2.7 | 0.83 |
| Marine environments | |||||||||
| Dubai sea (Arabian Sea) | 1.22 | 2.36 | 0.16 | 6.8 | 19.34 | 2.74 | 0.16 | 5.8 | 0.86 |
| Gran Canaria (Atlantic Ocean) | 0.32 | 0.46 | 0.02 | 4.8 | 14.38 | 0.43 | 0.02 | 4.7 | 1.06 |
| Tokyo Bay (Western Pacific Ocean) | 5.40 | 9.53 | 0.70 | 7.4 | 17.65 | 11.50 | 3.50 | 30.4 | 0.83 |
| Deep sea, Riftia field (Eastern Pacific Ocean) | 2.91 | 1.43 | 0.21 | 14.9 | 4.91 | 1.60 | 0.30 | 18.9 | 0.89 |
| Waddenzee (The Netherlands, North Sea) | 2.27 | 3.59 | 0.10 | 2.7 | 15.81 | 3.32 | 0.16 | 4.9 | 1.08 |
| mean | 7.7 | 10.5 | 1.01 | ||||||
BDC bacterial direct counts, VDC viral direct counts, SD standard deviation, CoVar coefficient of variation, VBR virus-to-bacteria ratio
Enumeration of viruses and bacteria
Bacteria and viruses were stained by slightly modifying the protocol of Noble and Fuhrman (1998) after collecting viruses and bacteria on Anodisc filters (Whatman, 0.02 μm pore size) by using SYBR Gold (10,000× in dimethyl sulfoxide) at a final concentration of 3.3×. This stain is reported to have higher fluorescence yield compared to SYBR Green I or II (Chen et al. 2001). As mounting solution, we used Citifluor AF1 (Citifluor, catalog number R1320) because of its easy application and excellent anti-fading properties. Three subsamples were withdrawn from each sample. Volumes of filtered subsamples varied between 200 and 1,000 μl to obtain a sufficient amount of viruses for counting depending on the investigated sampling sites. For counting, digital images were created using the confocal laser scanning inverted microscope DMIRE 2 with the confocal system TCS SP2 (Leica Microsystems, Germany). Due to the short exposure time to the scanning laser beam, samples were subject to minimal fading compared to the prolonged UV illumination during counting with conventional EFM. The CLSM was equipped with an argon and helium–neon laser with selectable excitation wavelengths of 488, 543, 568, and 633 nm. Virus samples were visualized using a PL APO 100.0×1.40 oil UV lens with the following CLSM settings: beam splitter RSP 500 and a pinhole of 1 Airy. For the excitation of SYBR Gold, we used the argon laser at 488 nm wavelength, a detection range from 500 to 670 nm (SYBR Gold: excitation 495 nm, emission maximum at 537 nm), and a pixel density of 1,024×1,024. The inspected image size comprised 150×150 μm.
Immediately after processing the samples with the CLSM, the same slides were counted with a conventional epifluorescence microscope (Nikon Eclipse 800) equipped with a PL APO 100.0×1.40 oil lens. As site-specific additional information, bacterial abundances were determined, but only with EFM and only for one subsample per sampling site.
Image processing and analysis
A common problem of microphotography of filters for counting microorganisms is that due to unevenness of the filter surface, only a small area is in the plane of focus. Optical sections (z-scans) were therefore performed to correct for unevenness of the filter disk. Furthermore, each single scan per optical section was run two times and averaged to improve the signal-to-noise ratio while imaging. The interval between two sections was 0.2 μm and the consecutive scans were then used to produce an overlay picture, which did not miss any fluorescent signal. These z-scans were further processed with the image analysis software package Optimas 6.5.172 (Media Cybernetics, Bethesda, MD, USA). The obtained images were 150×150 μm in size resulting in an area of 22,500 μm2. For each sample slide, ten images were processed. To identify VLPs, we used an area morphometry set provided by the program, in which the parameters area, perimeter, and circularity were included to distinguish the size, maximum length, and shape. The data were then exported to Excel worksheets. After evaluating the correct values for the parameter set, an auto-filter was applied and subsequently all images of one replicate were processed semi-automatically. The values for the filter set sometimes had to be adjusted between replicates or samples to correct for variability in staining and hence signal strength.
Statistics
The results for viral abundances from both methods were compared by performing a regression analysis and a two-way ANOVA.
Results and discussion
This work was designed to evaluate the applicability of CLSM for enumerating viruses in water samples filtered onto 0.02 μm pore size membranes as well as for estimating virus numbers associated with aquatic particles. Bacterial numbers (Table 1) ranged from 3.2×105 to 1.4×107 ml−1 but were in the order of 106 for most of the sampling sites including the deep-sea sample. Bacterial abundance in the water column of deep-sea waters is often in the range of 104 cells ml−1, and thus it is considerably lower than in our samples. This, however, reinforces the idea that hot vents fuel the surrounding waters and increase the standing stock of bacterial biomass.
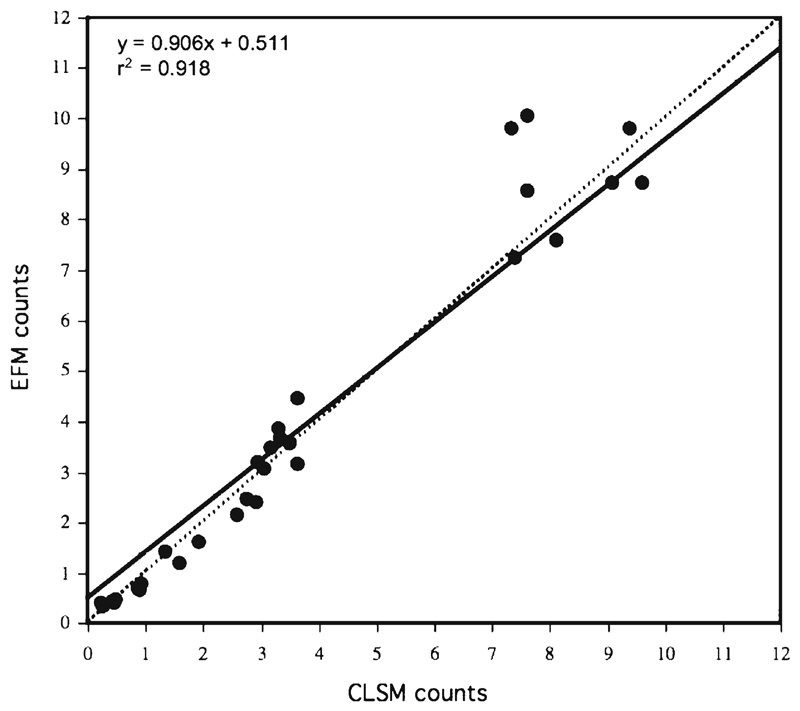
Viral direct counts ranged from 4.3×106 to 1.2×108 ml−1, but at almost all investigated sampling sites, they were in the order of 107 (Table 1). The virus-to-bacterium ratio (VBR) ranged from ca. 5 for the deep-sea sample from the Pacific Ocean to ca. 50 for the sample from Dorfteich (Riegersburg, Austria), a eutrophic pond. The average VBR from all samples was about 18. These values are close to the average values of 10 and 25, respectively, reported for marine and freshwater systems (Peduzzi and Luef 2009; Jacquet et al. 2010). Our numbers are similar to those found in other marine and freshwater environments (Wommack and Colwell 2000; Weinbauer 2004; Peduzzi and Luef 2009). Storage at 4 °C for days can significantly reduce viral abundance (Danovaro et al. 2001; Wen et al. 2004; Parvathi et al. 2011). Thus, viral abundance and VBR were likely underestimated in some of our samples. This, however, does not affect our comparison of viral counts between conventional EFM and the CLSM method because the samples were processed simultaneously. A two-way ANOVA performed on viral abundance from all data revealed no statistically significant difference (p>0.9; n=33) between the two methods. The average coefficient of variation for the EFM counts was 7.7 % (n=11) compared to 11.0 % (two more samples, n=13) for the enumerations performed by CLSM. This suggests similar precision of the two methods and further supports the idea that CLSM is a suitable tool for counting viruses (Table 1). Viral abundances obtained from both methods were highly correlated (r2=0.918; n=33) and the slope was close to one (Fig. 1). Samples from most freshwater environments and from the eutrophic Tokyo Bay are clustering in the upper right corner of this regression analysis. In 64% of the comparisons, the CLSM counts were slightly higher than the EFM numbers (Table 1). These small differences may reflect inaccuracies while counting with EFM due to the fading of the fluorescent signals. While counting viruses by EFM can be biased by the time needed for counting and therefore stain fading, this can be avoided with CLSM. The production of digital images via laser scans strongly reduces the exposure time of the specimen to the light source. At high viral abundance, some counts were higher with conventional EFM (see Fig. 1). This may be due to the higher variance between individually counted smaller subfields in EFM compared to CLSM.
Fig. 1.
Comparison of VDCs from freshwater and marine samples performed by conventional epifluorescence microscopy (EFM) and confocal laser scanning microscopy (CLSM). The numbers indicate VLPs ×107 ml−1. Dashed line: relationship of 1:1. For explanation see text
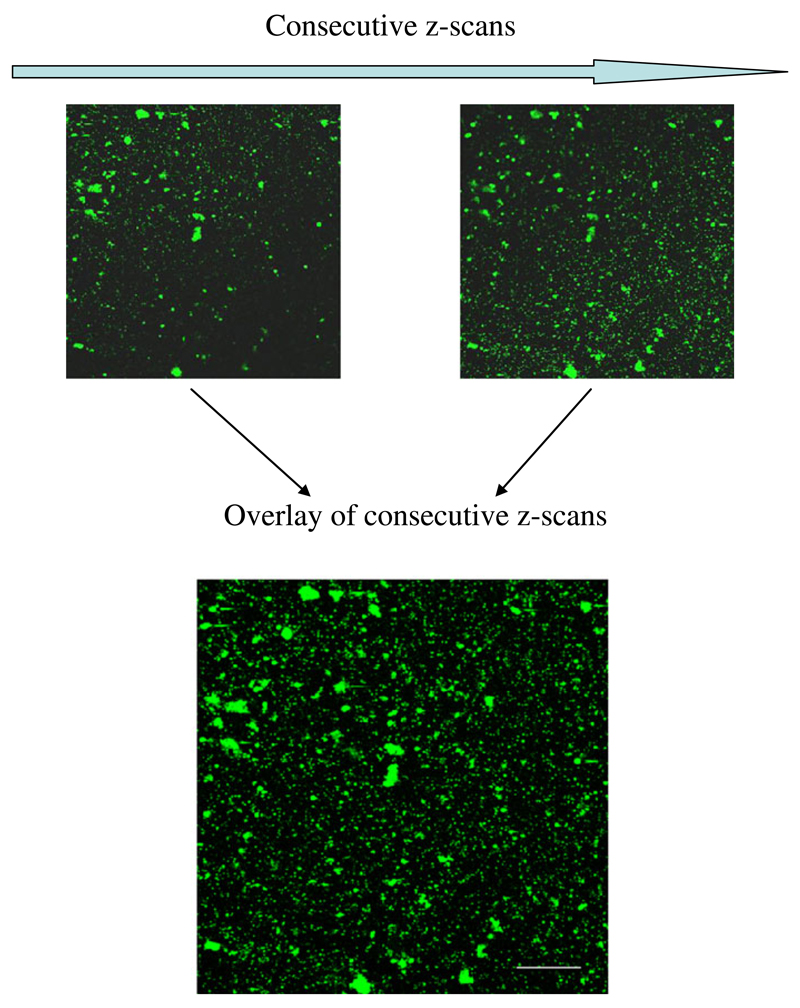
The use of CLSM provides several advantages compared to conventional epifluorescence microscopy. The used fluorescent stain can be excited near or at its maximum excitation wavelength. To receive a good signal, the photomultiplier (detector) can be set to a wide “detection window” around the maximum emission peak of the dye. One advantage of using CLSM is that only light from the plane of focus is detected. This helps to eliminate any other disturbing fluorescence signals from above or underneath (which of course depends on pinhole size) and thus provides a stronger signal at higher fluorescence intensity. This is especially important for detecting viruses on transparent exopolymeric particles (TEPs). Furthermore, in examining a sample by EFM, the plane of focus has to be adjusted repeatedly. While this does not hamper direct counting, it complicates the generation of microphotographs taken at high magnification. When making images with the CLSM for subsequent image analysis, we used the z-sectioning software of the CLSM to perform z-scans of our filter samples to correct for the unevenness of the filter surface. Stacking and overlaying these consecutive z-scans allowed aligning viruses in one plane of focus and thus yielding accurate images from the whole investigated sector (Fig. 2). These high-resolution images can be taken immediately after sampling and then be analyzed right away or stored for later use. Storage of high-quality digital images also provides the opportunity to analyze the samples at any time without the drawback of storing samples in a freezer. Another advantage is the easy acquisition of a larger total counting area; this is advantageous in samples with inhomogeneous distribution of microorganisms (e.g., aggregation) on the filter. In conventional EFM, this would necessitate a substantially higher number of replicated counting fields.
Fig. 2.
Example of digital images of a VDC filter sample created by CLSM after SYBR Gold staining of virus-like particles and bacteria (small and larger dots, respectively). Consecutive z-scans were used to construct an overlay picture (maximum intensity projection) of an uneven filter membrane to ensure that each single fluorescent signal lies in the plane of focus. Scale bar 10 μm
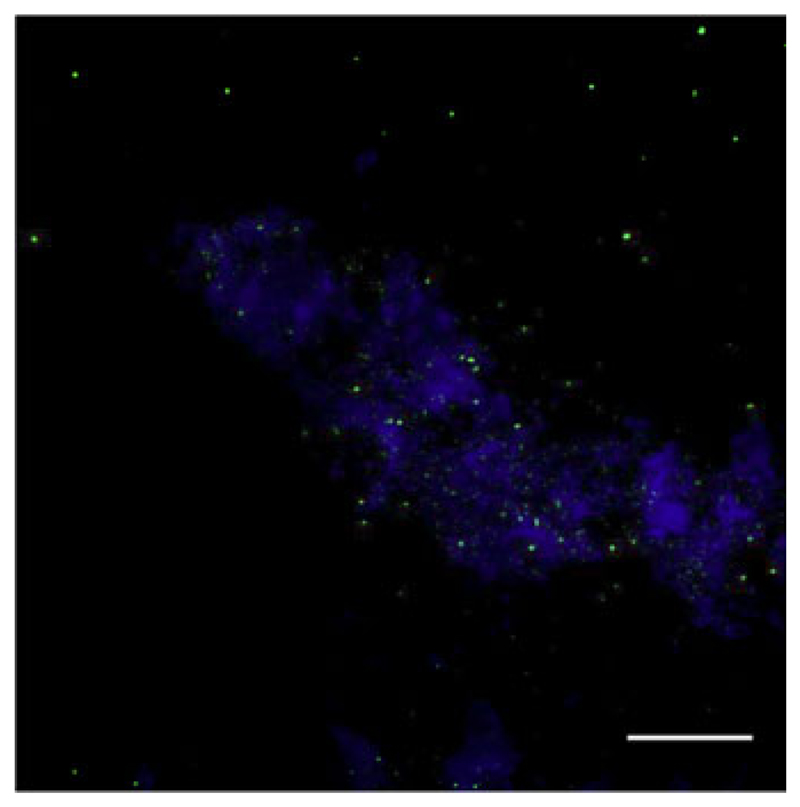
We show that CLSM is a reliable method for counting viruses in ambient water, which is a prerequisite for enumerating viruses on aggregates with this method. The z-scanning function of the CLSM also allows analyzing particulate material (seston and aggregates) and estimating bacterial and virus abundance on fully hydrated material. Investigations of biofilms and aggregates from rivers and lakes and also on marine snow have already been performed (Holloway and Cowen 1997; Lawrence and Neu 1999; Neu 2000; Battin et al. 2003; Luef et al. 2009b), focusing on the compartments and composition of the matrix. Only Luef et al. (2009c) quantitatively analyzed viruses on suspended river particles in detail using CLSM. We therefore applied this technique to marine particles obtained from the Atlantic Ocean. The aggregates consisted mainly of transparent organic material and did not produce high background fluorescence, which allowed precise resolution of single signals produced by viruses and bacteria (Fig. 3). These microaggregates (size <500 μm) constitute a frequent type of particles and are an important factor in the formation and decomposition of macro-aggregates (Simon et al. 2002). In these samples, virus abundance associated with aggregates was 4.6×1010 ml−1 compared to 4.4×106 ml−1 in the ambient water. Bacterial abundance on aggregates was 3.13×109 and 2.16×105 ml−1 in ambient water. This estimate of particle-associated virus abundance is well within the range reported by Peduzzi and Weinbauer (1993), Luef et al. (2009c), and Weinbauer et al. (2009). Moreover, the VBR was only slightly lower in our particles than in the ambient water, supporting the interpretation that particles are a habitable environment for viruses. Counting bacteria attached to aggregates has frequently been done using EFM (Simon et al. 2002), but when single particles are investigated, the precision is low due to fluorescence interference of the stained organic matrix, and bacteria lying on top of each other might not be distinguishable. The need for precise determination of microbial abundances on particulate material is obvious: their activity on aggregates is known to affect the turnover of organic substances in aquatic systems (Simon et al. 2002). Aquatic aggregates are considered to be hot spots of biogeochemical transformation and prokaryotic activity (Weinbauer et al. 2009). Furthermore, viral infection of bacterioplankton attached to biological surfaces, e.g., aggregates and biofilms may be of ecological importance. Viruses are also a major cause of bacterial mortality. Accordingly, they themselves may also affect the formation and decomposition of organic particulates (Peduzzi and Weinbauer 1993).
Fig. 3.
Digital overlay image (maximum intensity projection) of consecutive z-scans of a microaggregate from the Atlantic Ocean. Virus-like particles and bacteria (small and larger green dots, respectively) were stained with SYBR Gold, and particle matrix (blue) was counterstained with a TRITC-labeled lectin from Triticum vulgaris (Neu 2000); scale bar 25 μm
Applying CLSM microscopy will no doubt increase the reliability of abundance estimates also of attached bacteria. Moreover, the distribution pattern of viruses and bacteria on a filter (Fig. 3) can be used to identify aggregates without further staining. This may be of help to quantify microbes on TEP by avoiding possible alterations of the matrix due to the staining procedure, for example with Alcian blue. TEPs are an important class of particles in terms of organic carbon content and interfaces for biogeochemically relevant microbial activity (Smith et al. 1992).
Thus, reliable counts of bacteria and viruses contribute to our understanding of their ecological and biogeochemical roles. Exploiting the advantages of CLSM, including more accurate excitation and detection of the used stain and minimized photo-bleaching of the specimen, combined with z-sectioning, yields high-quality digital images (with all fluorescent signals in the plane of focus) and facilitates subsequent image analysis. The increasing amount of data will help better assess the distribution of free-living versus attached viruses and bacteria and the role they play in these systems. Our study supports the view that CLSM is a powerful technique, particularly when TEPs are abundant and very accurate numbers are required for studying the role of aquatic microorganisms. For routine monitoring of aquatic samples from environments with low abundance of suspended particles, FCM (without any necessary concentration procedure) or EFM might be easier and less time consuming.
Acknowledgments
We are thankful to R. Agis, G. Winkler, and M.G. Weinbauer for providing surface samples from Egypt, Kenya, and The Netherlands and C.R. Fisher, M. Bright, and A. Nussbaumer for the deep-sea samples from the Pacific Ocean. Special thanks are due to M.G. Weinbauer for helpful discussions and comments on the manuscript. This work was supported by a grant from the Austrian Science Foundation FWF to P.P. (FWF P 17798-B03).
References
- Battin TJ, Kaplan LA, Newbold JD, Hansen CM. Contributions of microbial biofilms to ecosystem processes in stream mesocosms. Nature. 2003;426:439–442. doi: 10.1038/nature02152. [DOI] [PubMed] [Google Scholar]
- Bettarel Y, Sime-Ngando T, Amblard C, Laveran H. A comparison of methods for counting viruses in aquatic systems. Applied and Environmental Microbiology. 2000;66:2283–2289. doi: 10.1128/aem.66.6.2283-2289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussard C, Marie D, Bratbak G. Flow cytometric detection of viruses. Journal of Virological Methods. 2000;85:175–182. doi: 10.1016/s0166-0934(99)00167-6. [DOI] [PubMed] [Google Scholar]
- Chen F, Lu JR, Binder BJ, Liu YC, Hodson RE. Application of digital image analysis and flow cytometry to enumerate marine viruses stained with SYBR Gold. Applied and Environmental Microbiology. 2001;67:539–545. doi: 10.1128/AEM.67.2.539-545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danovaro R, Dell’Anno A, Trucco A, Serresi M, Vanucci S. Determination of virus abundance in marine sediments. Applied and Environmental Microbiology. 2001;67:1384–1387. doi: 10.1128/AEM.67.3.1384-1387.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duhamel S, Jacquet S. Flow cytometric analysis of bacteria- and virus-like particles in lake sediments. Journal of Microbiological Methods. 2006;64:316–332. doi: 10.1016/j.mimet.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Hara S, Koike I, Terauchi K, Kamiya H, Tanoue E. Abundance of viruses in deep oceanic waters. Marine Ecology Progress Series. 1996;145:269–277. [Google Scholar]
- Hennes KP, Suttle CA. Direct counts of viruses in natural waters and laboratory cultures by epifluorescence microscopy. Limnology and Oceanography. 1995;40:1050–1055. [Google Scholar]
- Holloway CF, Cowen JP. Development of a scanning confocal laser microscopic technique to examine the structure and composition of marine snow. Limnology and Oceanography. 1997;42:1340–1352. [Google Scholar]
- Jacquet S, Miki T, Noble R, Peduzzi P, Wilhelm S. Viruses in aquatic ecosystems: important advancements of the last 20 years and promising prospects in the field of microbial oceanography and limnology. Advances in Oceanography and Limnology. 2010;1:71–101. [Google Scholar]
- Lawrence JR, Neu TR. Confocal laser scanning microscopy for analysis of microbial biofilms. Methods in Enzymology. 1999;310:131–144. doi: 10.1016/s0076-6879(99)10011-9. [DOI] [PubMed] [Google Scholar]
- Luef B, Luef F, Peduzzi P. Online program “VIPCAL” for calculating lytic viral production and lysogenic cells based on a viral reduction approach. Environmental Microbiology Reports. 2009a;1:78–85. doi: 10.1111/j.1758-2229.2008.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luef B, Neu TR, Zweimüller I, Peduzzi P. Structure and composition of aggregates in two large European rivers, based on confocal laser scanning microscopy and image and statistical analyses. Applied and Environmental Microbiology. 2009b;75:5952–5962. doi: 10.1128/AEM.00186-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luef B, Neu TR, Peduzzi P. Imaging and quantifying virus fluorescence signals on aquatic aggregates: a new method and its implication for aquatic microbial ecology. FEMS Microbiology and Ecology. 2009c;68:372–380. doi: 10.1111/j.1574-6941.2009.00675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie D, Brussard CPD, Thyrhaug R, Bratbak G, Vaulot D. Enumeration or marine viruses in culture and natural samples by flow cytometry. Applied and Environmental Microbiology. 1999;65:45–52. doi: 10.1128/aem.65.1.45-52.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu TR. In situ cell and glycoconjugate distribution in river snow studied by confocal laser scanning microscopy. Aquatic Microbial Ecology. 2000;21:85–95. [Google Scholar]
- Noble RT, Fuhrman JA. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquatic Microbial Ecology. 1998;14:113–118. [Google Scholar]
- Parvathi A, Radhakrishnan S, Sajila MP, Jacob B. Study of changes in bacterial and viral abundance in formaldehyde-fixed water samples by epifluorescence microscopy. Environmental Monitoring and Assessment. 2011;177:227–231. doi: 10.1007/s10661-010-1629-7. [DOI] [PubMed] [Google Scholar]
- Peduzzi P, Luef B. Viruses. In: Likens GE, editor. Encyclopedia of inland waters. Vol. 3. Oxford: Elsevier; 2009. pp. 279–294. [Google Scholar]
- Peduzzi P, Weinbauer MG. Effect of concentrating the virus-rich 2-200-nm size fraction of seawater on the formation of algal flocs (marine snow) Limnology and Oceanography. 1993;38:1562–1565. [Google Scholar]
- Proctor LM, Fuhrman JA. Mortality of marine bacteria in response to enrichments of the virus size fraction from seawater. Marine Ecology Progress Series. 1992;87:283–293. [Google Scholar]
- Simon M, Grossart H-P, Schweitzer B, Ploug H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquatic Microbial Ecology. 2002;28:175–211. [Google Scholar]
- Smith DC, Simon M, Alldredge AL, Azam F. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature. 1992;359:139–142. [Google Scholar]
- Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiology Reviews. 2004;28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Weinbauer MG, Suttle CA. Comparison of epifluorescence and transmission electron microscopy for counting viruses in natural marine waters. Aquatic Microbial Ecology. 1997;13:225–232. [Google Scholar]
- Weinbauer MG, Beckmann C, Höfle MG. Utility of green fluorescent nucleic acid dyes and aluminum oxide membrane filters for rapid epifluorescence enumeration of soil and sediment bacteria. Applied and Environmental Microbiology. 1998;64:5000–5003. doi: 10.1128/aem.64.12.5000-5003.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer MG, Winter C, Höfle MG. Reconsidering transmission electron microscopy based estimates of viral infection of bacterioplankton using conversion factors derived from natural communities. Aquatic Microbial Ecology. 2002;27:103–110. [Google Scholar]
- Weinbauer MG, Bettarel Y, Cattaneo R, Luef B, Maier C, Motegi C, Peduzzi P, Mari X. Viral ecology of organic and inorganic particles in aquatic systems: avenues for further research. Aquatic Microbial Ecology. 2009;57:321–341. doi: 10.3354/ame01363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen K, Ortmann AC, Suttle CA. Accurate estimation of viral abundance by epifluorescence microscopy. Applied and Environmental Microbiology. 2004;70:3862–3867. doi: 10.1128/AEM.70.7.3862-3867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm SW, Suttle CA. Viruses and nutrient cycles in the sea. BioScience. 1999;49:781–788. [Google Scholar]
- Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiology and Molecular Biology Review. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]